Abstract
Global expression profiling of neurologic or psychiatric disorders has been confounded by variability among laboratories, animal models, tissues sampled, and experimental platforms, with the result being that few genes demonstrate consistent expression changes. We attempted to minimize these confounds by pooling dentate granule cell transcriptional profiles from 164 rats in seven laboratories, using three status epilepticus (SE) epilepsy models (pilocarpine, kainate, self-sustained SE), plus amygdala kindling. In each epilepsy model, RNA was harvested from laser-captured dentate granule cells from six rats at four time points early in the process of developing epilepsy, and data were collected from two independent laboratories in each rodent model except SSSE. Hierarchical clustering of differentially-expressed transcripts in the three SE models revealed complete separation between controls and SE rats isolated 1 day after SE. However, concordance of gene expression changes in the SE models was only 26–38% between laboratories, and 4.5% among models, validating the consortium approach. Transcripts with unusually highly variable control expression across laboratories provide a ‘red herring’ list for low-powered studies.
Subject terms: Transcriptomics, Epilepsy
Background and Summary
Epileptogenesis is the process that causes networks in a normal brain to become a source of epileptic seizures. Among the numerous triggers of this process is a de novo bout of convulsive status epilepticus (SE)1, which is defined operationally as seizures lasting more than 5 min (typically >30 min), or a cluster of seizures that occur so closely together that full consciousness is not regained between seizures2. Both epidemiological and preclinical observations support the idea that status epilepticus (SE) at any age can result in epilepsy later in life. Between 13–82% of children who experience a febrile convulsive SE eventually develop epilepsy, with a long latent period of up to 10 years3, whereas about 30% of teens or adults who experience SE develop epilepsy within two years1. Uncertainty surrounding the mechanisms underlying epileptogenesis has limited progress towards interventions, although the extended time required in both rodents and man for clinically obvious seizures to develop suggests that gene expression changes might contribute to epileptogenesis. Numerous attempts have been made to employ transcriptional expression profiles in animal models to identify early events in epileptogenesis (e.g., refs 4–10) but, aside from transcripts pointing to an expected inflammatory state, no consensus has developed regarding common transcriptional drivers of epileptogenesis.
Discrepancies among studies likely reflect a combination of low statistical power and technical factors such as differences in the selection of species, variability in model methods, different brain regions selected for study, and the predominant cell type that was investigated. To address these issues, we assembled a group of laboratories into an ‘epilepsy microarray consortium’ to create epileptic rats using a variety of models, with the broad objective of creating a reliable dataset of transcriptional profiles in a single neuron type during the early phase of epileptogenesis. We reasoned that common transcriptional changes among the diverse models would identify changes fundamental to epileptogenesis, irrespective of the model used. We selected Sprague-Dawley rats because they are outbred and they avoid the substrain differences often found with inbred mouse strains. We laser-captured dentate granule cells to avoid the interpretive complications of pooling information from multiple cell types, to avoid changes in cellular makeup due to injury, to minimize the contribution of cell death pathways compared to analysis of dying CA1 or CA3 pyramidal neurons11, and because these neurons control generalization of seizures through hippocampal circuits12–15. Additionally, the number of differentially expressed transcripts in dentate granule cells 1 day after brief kainate-induced seizures was 4–10 times higher than in the CA1 or CA3 cell layers16, so the transcriptional response of these neurons to seizures is high. To minimize model-specific conclusions, four different epilepsy models were studied: SE caused by systemic pilocarpine or kainate, or electrical stimulation (SSSE); a fourth model, amygdala kindling, is not based on SE but instead relies on twice-daily 1 s duration electrical stimulation of the amygdala.
Each model was studied in two independent laboratories to mitigate laboratory-to-laboratory variability. Dentate granule cells were harvested from tissue generated by each laboratory. Rats were produced at each of three time points plus controls, and hemibrains from all rats were examined histologically in a single laboratory to identify outliers that had much more (or less) neurodegeneration than expected. Figure 1 shows the study design. The initial goals of this study were to: a) identify model-independent transcriptional changes in dentate granule cells that might point to novel intervention targets for epileptogenesis, b) characterize the basal transcriptional profile of dentate granule cells, and c) identify genes that have highly variable expression. The data, deposited in NCBI GEO, are the source of several ongoing analyses (e.g., Roopra and Dingledine, in preparation; Srivastava et al., in preparation), and should be a valuable resource for future efforts to understand the process of epileptogenesis based on analyses of transcriptional profiles across animal models and laboratories. Moreover, the data from untreated rats were obtained from 41 animals across seven laboratories, and thus provide a comprehensive picture of the basal transcriptional profile of dentate granule cells.
Figure 1. Workflow scheme.
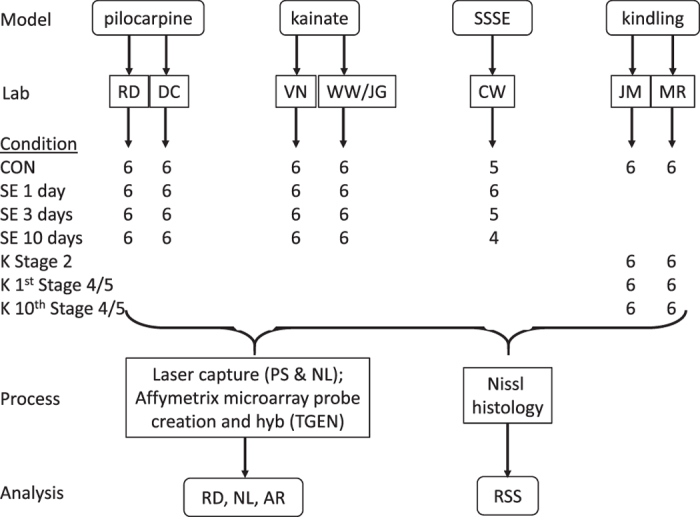
The number of rats from which high quality RNA was obtained is shown for each condition tested. Control in the case of kindling is sham stimulation. Abbreviations: RD, DC etc on the ‘Lab’, ‘Process’ or ‘Analysis’ rows are initials of the responsible authors; CON—control rats that had been injected with vehicle (pilocarpine and kainate models), or implanted but not stimulated (SSSE and kindling models); SE 1 day—rats sacrificed 1 day after experiencing status epilepticus (SE); K stage 2—rats undergoing kindling that were sacrificed 24 h after the first Stage 2 seizure. SSSE—self-sustained status epilepticus, produced by continuous electrical stimulation of the angular bundle.
Methods
Overview of experimental design
We attempted to harmonize methods and, where possible, have procedures performed by a single laboratory or individual to reduce technical variability. Thus, a) each of the two paired laboratories for each animal model agreed upon the procedure to be followed; b) all tissue was sent to a single laboratory (RD), who distributed it for histology and laser capture microdissection (LCM); c) all histology was done in a blinded fashion at a single laboratory (RSS), and LCM at two laboratories (PS and RD); d) the microarray procedures were carried out at a single site (TGEN, Phoenix, AZ).
Animal models and tissue preparation
All animal procedures were performed in accordance with NIH guidelines or EU Directive for the protection of laboratory animals, and were approved by the IACUC of each consortium members’ institution. The animals used in this study were male Sprague-Dawley rats obtained from Charles River. Individually housed animals were handled and allowed to acclimatize for at least 1 week prior to experiments, with food and water available ad libitum. Each laboratory provided tissue from 24 animals, six per group (controls and three groups post-SE or at defined kindling stages). Control animals were collected at each of the three time points.
Pilocarpine model (RD, DC)
Sprague-Dawley rats (180–200 g) were purchased from Charles-River (Raleigh, NC). Rats were given an injection of methylscopolamine (1 mg kg−1, i.p., Sigma) to minimize peripheral cholinergic effects of pilocarpine. After 30 min, animals received pilocarpine hydrochloride (300–350 mg kg−1 sc, Sigma) or saline. Seizures were classified according to Racine17 and Schauwecker and Steward18 with slight modifications11. SE was defined by continuous seizure activity consisting of stage 4 to 6 seizures. SE was terminated with pentobarbital (25 mg kg−1, i.p., McKesson) after 90 min of SE activity. Animals were monitored daily and given dextrose in lactate Ringer solution (~1 ml, i.p.) as needed. Control rats were given methylscopolamine and pentobarbital, but saline instead of pilocarpine.
Kainate model (WW/JG and VN)
Sprague Dawley rats (175–250 g) were purchased from Charles-River, Raleigh (VN) or Harlan, Netherlands (WW/JG). The Netherlands group used an initial intraperitoneal injection of 7.5 mg kg−1 kainic acid, followed by 5 mg kg−1 kainate hourly until SE induction; seizure activity was stopped with pentobarbital 90 min after SE onset. The Duke group injected rats with kainic acid (5 mg kg−1, i.p.) hourly until status epilepticus was achieved and then allowed SE to self-terminate after 6–8 h. All animals received 5–6 injections before SE onset. SE onset was defined by continuous stage 4 and stage 5 seizure activity. Control rats were given saline at the same schedule.
Self-sustained status epilepticus (SSSE) model (AP, CW)
Male Sprague-Dawley rats were purchased from Charles River at 280–300 g. The animals were implanted with a stimulating electrode in the left angular bundle (in reference to lambda AP +0.5 mm, ML +4.5 mm, depth −4.0 mm from surface of brain). Electrodes were positioned under isoflurane anesthesia (4% in O2), and animals were treated for post-operative pain with buprenorphine for three days following surgery. Rats were allowed to recover for 14 days before initiating SE. EEG was recorded from skull screws located over the dorsal hippocampi. The stimulation wave form consisted of 10 s trains of 20 Hz biphasic square waves delivered at 1 train per minute superimposed on continuous 2 Hz monophasic square waves. All square waves were 1 msec in duration. Animals received no treatment following stimulation of the perforant path and were included in the study if they remained in SE 10 min after the end of perforant path stimulation19. Intermittent stage 1–5 kindling-like seizures, and electrographic seizures that were initially nearly continuous and later became separated by periods of low-voltage activity, continued for many hours. The mean duration of SE was 8.2±2.1 h20. Control animals were implanted but not stimulated.
Kindling (JM, MR)
Male Sprague-Dawley rats, 180–200 g at time of receipt, were obtained from Charles River. One week later a bipolar electrode was stereotaxically implanted in the right amygdala under pentobarbital anesthesia (60 mg kg−1, i.p.), and animals allowed to recover for 5–14 days. The electrographic seizure threshold was determined by administering a 1 s train of 1 msec biphasic rectangular pulses at 60 Hz beginning at 60 μA. Additional stimulus trains increasing by 20 μA were administered at one minute intervals until an electrographic seizure lasting at least 5 s was detected on the electroencephalogram (EEG) recorded from the amygdala. Stimulation was subsequently administered at this intensity twice daily. Unstimulated control animals underwent surgical implantation of an electrode in amygdala but were not stimulated. Behavioral seizure class was scored according to Racine’s classification17: Stage 0, no behavioral change; Stage 1, facial clonus; Stage 2, head nodding; Stage 3, unilateral forelimb clonus; Stage 4, rearing with bilateral forelimb clonus; Stage 5, rearing and falling (loss of postural control). Datasets were generated from animals early in the kindling process (24 h after the first Stage 2 seizure), at the completion of kindling (24 h after the first Stage 4 or 5 seizure), and after kindling had been established and was stable over time (24 h after the 10th Stage 4 or 5 seizure).
Tissue handling
Animals that entered status epilepticus (SE) were killed for tissue harvest at 1 day (23–25 h), 3 days (70–74 h), or 10 days after SE onset. Brains were removed from kindled rats 24 h after the first stage 2 seizure, 24 h after the first stage 5 seizure or 24 h after the tenth stage 5 seizure. At each time point, the animal was decapitated following deep ether or isoflurane anesthesia. Brains were removed and longitudinally bisected. The left half of each brain was immersed in 4% (w/v) paraformaldehyde in 0.1 M sodium phosphate buffer, pH 7.4 and stored at 4 °C, while the right half was frozen on dry ice and stored at −80 °C in foil. Each consortium laboratory followed the same procedure for brain tissue preparation. All tissue was sent to one laboratory (RD) and subsequently the fixed brains were sent to RSS for Nissl stain histology and to assess the extent of damage incurred. The frozen half of the brains from the pilocarpine and kainate epilepsy models were sent to PS for laser capture microscopy of dentate granule cells. Laser capture for the brains produced by SSSE was done at Emory University. Our goal was to process tissue from 6 rats at each time point from each laboratory, although as Fig. 1 indicates, some brains had thawed in shipment and were thus unusable.
Laser capture microdissection and RNA isolation
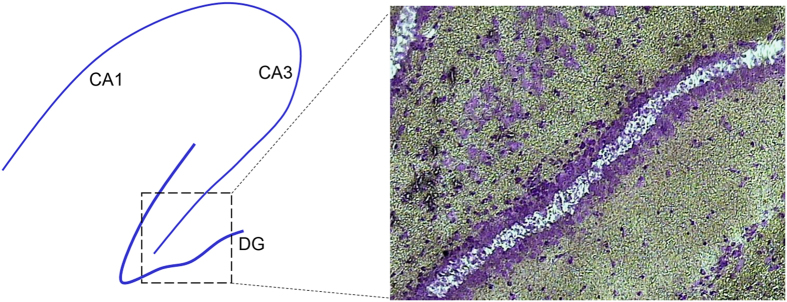
Ten micron coronal sections were taken through the hippocampus and collected onto uncoated microscope slides, refrozen on dry ice, and stored at −80 ° C until staining for LCM. For staining, sections were fixed in ice-cold 70–75% ethanol for 30 s, rinsed in water 10–15 s, dipped in cresyl violet stain 10–15 s, rinsed in H2O for 10 s, then dehydrated through alcohols over 2 min to xylene. Sections were dried in a fume hood and LCM was performed within 1 hour. LCM was performed using a Veritas or Pixcell IIe system with transmission illumination (Arcturus, CA) and the following parameters: spot size=25–30 μm; power=50–65 mW; duration=1,700–3,000 μs per hit. The middle of the dentate gyrus was harvested (Fig. 2) from 10–16 sections per subject in order to avoid the neurogenic zone of the hilar-granule cell border. LCM HS Caps (Arcturus) were used to collect dentate granule cells with 2–5 sections per cap, resulting in 2–8 LCM Caps used per subject and at least 10 ng total RNA for amplification.
Figure 2. Laser capture harvesting of cells in the middle of the dentate granule cell layer in the outer leaf.
Left: diagram of hippocampus with CA1 and CA3 pyramidal cell layers and the dentate gyrus (DG). Laser capture was done from the boxed region and shown in the right panel. Cresyl violet stain of a section from a rat treated with pilocarpine 10 days before. The region of captured dentate granule cells is indicated by the absence of blue stain.
Total RNA was extracted from each LCM cap using the PicoPure Isolation Kit (Arcturus) with DNase digestion (Qiagen Rnase-free DNase Set), and stored at −80 °C until RNA quality verification and quantification using Agilent RNA 6,000 PicoChips run on the Agilent 2,100 Bioanalyzer. The results of the PicoChip determined whether the RNA was of high enough quality to keep for amplification and later hybridization. Total RNA with 28S:18S rRNA ratio >0.8 and RNA Integrity Number (RIN)>6.9 was accepted from each LCM cap in order to obtain one pooled sample from each rat with a minimum of 10 ng total RNA. Once a total RNA pool was complete for each subject, the sample was assessed by the BioAnalyzer, quantity and quality recorded, and samples were frozen at −80 °C until shipment to TGEN for RNA amplification and microarray hybridization.
Microarray hybridization
The NINDS NIMH Microarray Consortium at the Translational Genomics Institute in Phoenix, AZ (TGEN) performed RNA amplification, sample labeling, microarray hybridization, and submission of the data to NCBI GEO. Briefly, TGEN amplified the RNA once, reverse-transcribed the RNA to cDNA and used this to produce biotinylated cRNA with the EnzoBioArray High Yield RNA Transcript Labeling Kit (Affymetrix, CA). Samples (10 μg) were hybridized for 16 h at 45 °C on the GeneChip Rat Genome 230 2.0 Array. GeneChips were washed and stained in the Affymetrix Fluidics Station 400. Probe creation and hybridization was done by two technicians at TGEN; JM, MR, VN and CW samples were processed by one technician, whereas RD, DC and WW samples were processed by a second technician. The RAE230A is a high-density microarray that surveys over 10,000 unique transcripts. GeneChips were scanned using the GeneArray Scanner G2500A. The data were analyzed with Microarray Suite version 5.0 (MAS 5.0) using Affymetrix default analysis settings and global scaling as normalization method. The mean target intensity of each array was set to 150. All image files and data from hybridizations are available at the NCBI GEO website, with access number GSE47752 (Data Citation 1).
Neuropathology
Fixed half-brains from each laboratory were sent to RSS for histological assessment of cell damage and neuronal death in the hippocampus. For each brain, 40-μm-thick coronal sections were cut in 0.1 M Tris (hydroxymethylaminomethane) buffer pH 7.6, with a Vibratome. Regularly spaced sections were mounted on subbed slides for subsequent Nissl (1% cresyl violet) staining. After staining, all slides were dehydrated in graded ethanols and xylene and then coverslipped with Permount21. All tissue from SE-induced animals was assessed against that of controls for cell damage in various areas of the hippocampus, including the CA1, CA3, hilus, dentate gyrus, and piriform cortex. An outlier of SE-brains was defined as either a) one with no discernable neuron death in CA3 or CA1, or b) a brain with an extremely injured hippocampus that includes substantial granule cell death. No outliers were found, so data from all rats were included in the analysis.
Microarray data processing
Affymetrix CEL and Rat230_2.cdf files were used for RMA normalization with background adjust, quantile normalization and median polish. Annotation was performed using netaffx build 35. RMA normalization was performed using all probes regardless of Affymetrix ‘Present’ (P) or ‘Absent’ (A) calls. Subsequently, only probes with the ‘_at’ suffix were retained to ensure that only probes specific to a single gene were carried forward for further analysis. Probe level data were then collapsed to gene symbols with the symbol being designated ‘Present’ in a RNA sample if at least 50% of probes for that symbol were designated as ‘Present’. To designate genes as expressed or not over the entire experiment the following rules were applied: A symbol was designated ‘above background’ in a laboratory for a given status epilepticus time point and epilepsy model (e.g., Day 1 after pilocarpine SE, see Fig. 1) if it was labelled ‘Present’ in at least 3 of the animals at that time point. A gene symbol was designated as ‘above background’ for a specific time point if it was labelled ‘Present’ in at least 4 of the 5 SE laboratories (i.e., no more than 1 lab could call the symbol ‘Absent’). For the control condition, the symbol was considered above background if it was called Present in at least 6 of the 7 laboratories (i.e., no more than 1 laboratory could call the symbol ‘Absent’). Finally, a symbol was designated ‘above background’ over the entire analysis if it was called as Present in any time point. For each gene symbol accepted for the entire analysis, expression values for every animal were included regardless of its Absent or Present call in any individual animal. In summary: A symbol is expressed above background in a laboratory at a given time point if it is labeled as Present in at least 3 animals. A symbol is above background in SE if it is Present in at least 4 laboratories. For controls, the symbol is above background if it is labeled Present in at least 6 laboratories.
To cluster expression values pooled across SE models and laboratories, the expression level of each gene symbol in a laboratory was collapsed to its median log2 value across animals in that laboratory. A total of 9,614 symbols were Present in at least 1 of the 4 conditions (control, 1- day, 3-days, 10-days after SE). Paired t-tests compared expression levels after SE with control values and n=5 labs, then a correction22 was made for multiple comparisons to adjust the false discovery rate (FDR) to 0.05. Altogether there were 2,646 differentially expressed genes after SE at any of the three time points examined in this analysis. Unsupervised hierarchical clustering was performed using median values of the 368 genes that were differentially expressed at least 2-fold between controls and SE rats, with uncentered Pearson correlations and complete clustering. To determine the degree of overlap between differentially expressed genes 1 day after SE among the three models, unpaired t-tests were run separately on data from each of the 5 laboratories (FDR<0.05)22 and a Venn diagram approach used.
Data Records
CEL and CHIP files associated with this analysis are deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/) with the accession number GSE47752 (Data Citation 1). Detailed information about each sample is in Table 1 (available online only).
Table 1. Sample description.
| GEO-ID | organism | tissue | sample description | analysis |
|---|---|---|---|---|
| Data descriptors for all animals. | ||||
| GSM1155788 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-control-rat 1 | microarray |
| GSM1155864 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-control-rat 2 | microarray |
| GSM1155878 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-control-rat 3 | microarray |
| GSM1155804 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-control-rat 4 | microarray |
| GSM1155820 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-control-rat 5 | microarray |
| GSM1155841 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-control-rat 6 | microarray |
| GSM1155858 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 10-rat 1 | microarray |
| GSM1155872 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 10-rat 2 | microarray |
| GSM1155797 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 10-rat 3 | microarray |
| GSM1155814 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 10-rat 4 | microarray |
| GSM1155835 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 10-rat 5 | microarray |
| GSM1155850 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 10-rat 6 | microarray |
| GSM1155832 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 1-rat 1 | microarray |
| GSM1155865 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 1-rat 2 | microarray |
| GSM1155789 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 1-rat 3 | microarray |
| GSM1155806 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 1-rat 4 | microarray |
| GSM1155822 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 1-rat 5 | microarray |
| GSM1155843 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 1-rat 6 | microarray |
| GSM1155793 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 3 -rat 3 | microarray |
| GSM1155854 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 3-rat 1 | microarray |
| GSM1155868 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 3-rat 2 | microarray |
| GSM1155810 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 3-rat 4 | microarray |
| GSM1155826 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 3-rat 5 rehyb | microarray |
| GSM1155846 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 3-rat 6 | microarray |
| GSM1155897 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-control-rat 1 | microarray |
| GSM1155898 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-control-rat 2 | microarray |
| GSM1155899 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-control-rat 3 | microarray |
| GSM1155900 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-control-rat 4 | microarray |
| GSM1155901 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-control-rat 5 | microarray |
| GSM1155902 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-control-rat 6 | microarray |
| GSM1155903 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 10-rat 1 | microarray |
| GSM1155904 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 10-rat 2 | microarray |
| GSM1155905 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 10-rat 3 | microarray |
| GSM1155906 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 10-rat 4 | microarray |
| GSM1155907 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 10-rat 5 | microarray |
| GSM1155908 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 10-rat 6 | microarray |
| GSM1155909 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 1-rat 1 | microarray |
| GSM1155910 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 1-rat 2 | microarray |
| GSM1155911 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 1-rat 3 | microarray |
| GSM1155912 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 1-rat 4 | microarray |
| GSM1155913 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 1-rat 5 | microarray |
| GSM1155914 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 1-rat 6 | microarray |
| GSM1155915 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 3-rat 1 | microarray |
| GSM1155916 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 3-rat 2 | microarray |
| GSM1155917 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 3-rat 3 | microarray |
| GSM1155918 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 3-rat 4 | microarray |
| GSM1155919 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 3-rat 5 | microarray |
| GSM1155920 | Rattus norvegicus albino | dentate granule neurons | kainate-Wadman-day 3-rat 6 | microarray |
| GSM1155786 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-control-rat 1 | microarray |
| GSM1155862 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-control-rat 2 | microarray |
| GSM1155876 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-control-rat 3 | microarray |
| GSM1155802 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-control-rat 4 | microarray |
| GSM1155818 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-control-rat 5 | microarray |
| GSM1155839 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-control-rat 6 | microarray |
| GSM1155833 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-stage 2-rat 1 | microarray |
| GSM1155866 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-stage 2-rat 2 | microarray |
| GSM1155790 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-stage 2-rat 3 | microarray |
| GSM1155807 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-stage 2-rat 4 | microarray |
| GSM1155823 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-stage 2-rat 5 | microarray |
| GSM1155844 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-stage 2-rat 6 | microarray |
| GSM1155855 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-first stage 4/5-rat 1 | microarray |
| GSM1155869 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-first stage 4/5-rat 2 | microarray |
| GSM1155794 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-first stage 4/5-rat 3 | microarray |
| GSM1155811 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-first stage 4/5-rat 4 | microarray |
| GSM1155828 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-first stage 4/5-rat 5 | microarray |
| GSM1155847 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-first stage 4/5-rat 6a | microarray |
| GSM1155859 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-10th stage 4/5-rat 1 | microarray |
| GSM1155873 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-10th stage 4/5-rat 2 | microarray |
| GSM1155798 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-10th stage 4/5-rat 3 | microarray |
| GSM1155815 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-10th stage 4/5-rat 4 | microarray |
| GSM1155836 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-10th stage 4/5-rat 5 | microarray |
| GSM1155851 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-10th stage 4/5-rat 6 | microarray |
| GSM1155787 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-control-rat 1 | microarray |
| GSM1155863 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-control-rat 2 | microarray |
| GSM1155877 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-control-rat 3 | microarray |
| GSM1155803 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-control-rat 4 | microarray |
| GSM1155819 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-control-rat 5 | microarray |
| GSM1155840 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-control-rat 6 | microarray |
| GSM1155834 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-stage 2-rat 1 | microarray |
| GSM1155867 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-stage 2-rat 2 | microarray |
| GSM1155791 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-stage 2-rat 3 | microarray |
| GSM1155808 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-stage 2-rat 4 | microarray |
| GSM1155824 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-stage 2-rat 5 | microarray |
| GSM1155845 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-stage 2-rat 6 | microarray |
| GSM1155856 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski--first stage 4/5-rat 1 | microarray |
| GSM1155870 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski--first stage 4/5-rat 2 | microarray |
| GSM1155795 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski--first stage 4/5-rat 3 | microarray |
| GSM1155812 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski--first stage 4/5-rat 4 | microarray |
| GSM1155829 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski--first stage 4/5-rat 5 | microarray |
| GSM1155849 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski--first stage 4/5-rat 6 | microarray |
| GSM1155860 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-10th stage 4/5-rat 1 | microarray |
| GSM1155874 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-10th stage 4/5-rat 2 | microarray |
| GSM1155799 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-10th stage 4/5-rat 3 | microarray |
| GSM1155816 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-10th stage 4/5-rat 4 | microarray |
| GSM1155837 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-10th stage 4/5-rat 5 | microarray |
| GSM1155852 | Rattus norvegicus albino | dentate granule neurons | kindling-Rogawski-10th stage 4/5-rat 6 | microarray |
| GSM1155880 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-control-rat 1 | microarray |
| GSM1155881 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-control-rat 2 | microarray |
| GSM1155882 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-control-rat 3 | microarray |
| GSM1155883 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-control-rat 4 | microarray |
| GSM1155884 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-control-rat 5 | microarray |
| GSM1155885 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-control-rat 6 | microarray |
| GSM1155921 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 10-rat 1 | microarray |
| GSM1155922 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 10-rat 2 | microarray |
| GSM1155923 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 10-rat 3 | microarray |
| GSM1155924 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 10-rat 4 | microarray |
| GSM1155925 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 10-rat 5 | microarray |
| GSM1155926 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 10-rat 6 | microarray |
| GSM1155927 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 1-rat 1 | microarray |
| GSM1155928 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 1-rat 2 | microarray |
| GSM1155929 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 1-rat 3 | microarray |
| GSM1155930 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 1-rat 4 | microarray |
| GSM1155931 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 1-rat 5 | microarray |
| GSM1155932 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 1-rat 6 | microarray |
| GSM1155933 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 3-rat 1 | microarray |
| GSM1155934 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 3-rat 2 | microarray |
| GSM1155935 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 3-rat 3 | microarray |
| GSM1155936 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 3-rat 4 | microarray |
| GSM1155937 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 3-rat 5 | microarray |
| GSM1155938 | Rattus norvegicus albino | dentate granule neurons | pilo-Coulter-day 3-rat 6 | microarray |
| GSM1155887 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 1a | microarray |
| GSM1155889 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 2a | microarray |
| GSM1155891 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 3a | microarray |
| GSM1155893 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 4a | microarray |
| GSM1155895 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 5a | microarray |
| GSM1155896 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 6 | microarray |
| GSM1155939 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 10-rat 1 | microarray |
| GSM1155940 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 10-rat 2 | microarray |
| GSM1155941 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 10-rat 3 | microarray |
| GSM1155942 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 10-rat 4 | microarray |
| GSM1155943 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 10-rat 5 | microarray |
| GSM1155944 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 10-rat 6 | microarray |
| GSM1155945 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 1-rat 1 | microarray |
| GSM1155946 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 1-rat 2 | microarray |
| GSM1155947 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 1-rat 3 | microarray |
| GSM1155948 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 1-rat 4 | microarray |
| GSM1155949 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 1-rat 5 | microarray |
| GSM1155950 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 1-rat 6 | microarray |
| GSM1155951 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 3-rat 1 | microarray |
| GSM1155952 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 3-rat 2 | microarray |
| GSM1155953 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 3-rat 3 | microarray |
| GSM1155954 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 3-rat 4 | microarray |
| GSM1155955 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 3-rat 5 | microarray |
| GSM1155956 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-day 3-rat 6 | microarray |
| GSM1155785 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-control-rat 1 | microarray |
| GSM1155831 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-control-rat 2 | microarray |
| GSM1155853 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-control-rat 3 | microarray |
| GSM1155857 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-control-rat 4 | microarray |
| GSM1155861 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-control-rat 6 | microarray |
| GSM1155825 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 10-rat 1 | microarray |
| GSM1155830 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 10-rat 2 | microarray |
| GSM1155838 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 10-rat 3 | microarray |
| GSM1155842 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 10-rat 6 | microarray |
| GSM1155871 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 1-rat 1 | microarray |
| GSM1155875 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 1-rat 2 | microarray |
| GSM1155879 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 1-rat 3 | microarray |
| GSM1155792 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 1-rat 4 | microarray |
| GSM1155796 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 1-rat 5 | microarray |
| GSM1155800 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 1-rat 6 rehyb | microarray |
| GSM1155805 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 3-rat 2 | microarray |
| GSM1155809 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 3-rat 3 | microarray |
| GSM1155813 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 3-rat 4 | microarray |
| GSM1155817 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 3-rat 5 | microarray |
| GSM1155821 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 3-rat 6 | microarray |
| Unused samples (preliminary hybridizations or those that failed Affymetrix Q/C) |
||||
| GSM1155827 | Rattus norvegicus albino | dentate granule neurons | kainate-Nadler-day 3-rat 5 | microarray |
| GSM1155848 | Rattus norvegicus albino | dentate granule neurons | kindling-McNamara-stage 4-rat 6 | microarray |
| GSM1155886 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 1 | microarray |
| GSM1155888 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 2 | microarray |
| GSM1155890 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 3 | microarray |
| GSM1155892 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 4 | microarray |
| GSM1155894 | Rattus norvegicus albino | dentate granule neurons | pilo-Dingledine-control-rat 5 | microarray |
| GSM1155801 | Rattus norvegicus albino | dentate granule neurons | SSSE-Wasterlain-day 1-rat 6 | microarray |
Technical Validation
Several aspects of the experiment were designed to improve the quality of the data. First, RNA was harvested from the middle of the dentate granule cell layer (Fig. 2) both to minimize contributions from multiple cell types that would have occurred had we used whole tissue (e.g., hippocampus) instead, and to avoid contamination by cells undergoing neurogenesis at the hilar-granule cell border. However, low to moderate levels of RNA from glial processes found in the cell layer are included; e.g., the microglial transcript that encodes Iba1 is expressed at a level of 104, the oligodendrocyte marker MBP is 612 and the astrocytic marker GFAP is 2,219. Second, we gathered data in parallel from 4 epilepsy models to provide a database from which model-independent conclusions can be drawn. Third, we intended that two independent laboratories study each animal model to reduce lab-to-lab influences; this was successful except for the SSSE model as described above. Fourth, 6 rats (occasionally 4–5) were provided for each treatment group from each laboratory; e.g., the control group was derived from 41 rats in 7 laboratories. Fifth, to minimize technical variation the LCM harvest of RNA from 144 of the 164 samples was done in one laboratory by the same technician, and the remainder (the SSSE samples) in a second laboratory; likewise RNA amplification, hybridization probe generation, microarray hybridization and initial data reduction were done at a single site (TGEN in Phoenix, AZ) by two technicians. Finally, RNA quality was controlled by accepting pooled samples from a rat if the RIN≥6.9, the ratio of 28S to 18S rRNA was ≥0.8, and at least 10 ng of total RNA had been obtained from the granule cells.
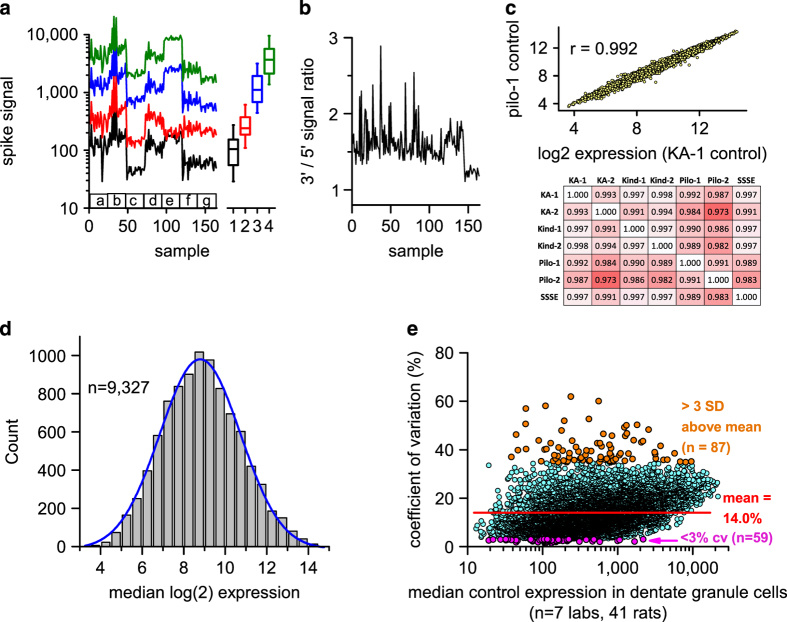
To explore the technical quality and uniformity of the raw microarray data, we carried out two diagnostic tests. Into each RNA sample Affymetrix routinely spikes RNAs from three E. coli genes (bioB, bioC, bioD) and the bacteriophage cre recombinase to assess the overall success of probe synthesis, hybridization and microarray imaging. We plotted the array signals from each of these 4 spikes over the 164 rat samples in Fig. 3a. Success requires that signal intensities from these 4 spikes show increasing values. The associated boxplots of median expression of these spikes demonstrate that the 4 RNA spikes are present at the expected increasing levels. Importantly, the lowest level spike was called ‘present’ in all 164 samples. Second, we plotted the median ratio of signal intensities for probes hybridizing to the 3’ and 5’ regions of 7 spiked RNAs and 3 endogenous RNAs to assess RNA degradation during the process. Figure 3b shows that all 164 samples had a 3’/5’ ratios<3, indicative of acceptable RNA quality.
Figure 3. Quality control assessment for microarrays of the RNA samples from 164 rats.
(a) Signal values from 4 bacterial or viral RNAs that had been spiked into each sample RNA before probe labeling are plotted for each of the 164 RNA samples. Boxplots of median expression across all arrays are shown to the right. Labels a-g denote data from the seven laboratories. Labels 1–4 are different prokaryotic or phage RNAs spiked at increasing levels. (b) Median 3′/5′ intensity ratios from 10 transcripts in each sample are plotted. (c) Excellent correlation between expression in the control condition of the 9,327 transcripts in the first kainate laboratory (KA-1) and the first pilocarpine laboratory (pilo-1). The heatmap below shows Pearson’s correlations between median control expression in all combinations of the 7 laboratories; correlations range from 0.973 to 0.998. (d) Histogram of the median expression level of each expressed gene in the control condition across all 7 laboratories is normal (Kolmogorov-Smirnov test). (e) The coefficient of variation is plotted versus the median expression for each of 9,327 transcripts in the control condition across 7 laboratories, revealing a subset of 59 genes with very tight expression across laboratories (<3% cv) and a subset of 87 genes with very variable expression. (c–e): A transcript was selected for inclusion if at least 6 of the 7 laboratories had no more than 3 absent calls each (mean of 0.26 absent calls/gene/lab).
To further examine the quality of the data and the consequences of our experimental design we carried out several additional tests on data that were filtered as described above. To determine variation of expression levels across laboratories we considered the 41 controls provided by all 7 laboratories. The samples provided by each lab showed high correlation of signal intensities over the 9,327 transcripts studied (Fig. 3c). Pearson correlation coefficients ranged from 0.973 to 0.998 as shown in the heatmap in Fig. 3c. The median expression intensities of each transcript were then calculated across the 7 laboratories, with each laboratory donating a single expression value for each transcript derived from the median of all control rats. A histogram of median log2 values of expression in the control condition across the 7 sites was normally distributed (Fig. 3d), with no ‘excess’ representation of low-expressing noise transcripts. A plot of the coefficient of variation of expression level across laboratories for each of the 9,237 transcripts versus its median expression level (Fig. 3e) showed that the mean CV of expression was 14%. Moreover, this plot revealed the presence of transcripts with highly stable expression across sites (n=59 with CV<3%, see Table 2 (available online only)), and transcripts with highly variable expression (n=87 with CV>3 SDs above the mean, see Table 3 (available online only)). The high variability group of transcripts represent potential ‘red herrings’ in transcriptional profiling studies that have low power.
Table 2. Transcripts in dentate granule cells with coefficient of variation <3% for expression level in the control animals (n=7 independent laboratories, each averaging expression over 5–6 rats).
| Gene symbol | group median | group coef. Var (%) |
|---|---|---|
| Expression levels and SDs of genes in control condition with <3% coefficient of variation. | ||
| Dock6 | 108 | 1.16 |
| Ankrd13b | 131 | 1.35 |
| Lrba | 109 | 1.40 |
| Zcchc8 | 87 | 1.74 |
| Cad | 106 | 1.77 |
| Zfp758 | 45 | 1.81 |
| Timm8a1 | 151 | 1.87 |
| Ints9 /// LOC102549712 | 577 | 1.93 |
| Pikfyve | 310 | 2.01 |
| Golph3l | 138 | 2.03 |
| Rps6kb1 | 1,684 | 2.07 |
| Zzef1 | 121 | 2.10 |
| Khnyn | 74 | 2.12 |
| Lrfn1 | 462 | 2.15 |
| Cryz | 109 | 2.16 |
| Fkbp15 | 126 | 2.19 |
| Setd1b | 107 | 2.19 |
| Pacs1 | 152 | 2.24 |
| Mlxip | 214 | 2.27 |
| Mtmr10 | 115 | 2.29 |
| Strn | 186 | 2.32 |
| Ddx26b | 1,001 | 2.33 |
| Lrrc61 | 167 | 2.43 |
| Chm | 46 | 2.45 |
| Stx3 | 106 | 2.47 |
| Synm | 151 | 2.49 |
| Ddx52 | 123 | 2.49 |
| Ripk1 | 73 | 2.49 |
| LOC102549467 | 19 | 2.51 |
| Nr2c1 | 44 | 2.60 |
| Spin4 | 33 | 2.61 |
| Isoc2b | 92 | 2.63 |
| Klf2 | 106 | 2.64 |
| Ubfd1 | 133 | 2.66 |
| Nol10 | 1,068 | 2.67 |
| RGD1562608 | 109 | 2.67 |
| Creb3l2 | 38 | 2.68 |
| Gpr137 | 216 | 2.70 |
| Blm | 212 | 2.74 |
| RGD1305422 | 162 | 2.75 |
| Arfgef1 | 381 | 2.76 |
| LOC102546678///Prrg3 | 120 | 2.78 |
| Bnip1 | 113 | 2.81 |
| LOC102552625 | 212 | 2.82 |
| LOC100911725///Pfkfb4 | 69 | 2.83 |
| Man1b1 | 229 | 2.86 |
| Lepre1 | 111 | 2.88 |
| Optc | 22 | 2.89 |
| LOC100912473 | 70 | 2.89 |
| Gmds | 485 | 2.89 |
| Adssl1 | 34 | 2.89 |
| Scp2d1 | 28 | 2.93 |
| Sult2b1 | 97 | 2.94 |
| Zbtb12 | 34 | 2.95 |
| Casp9 | 105 | 2.95 |
| Palm3 | 116 | 2.98 |
| Sap30bp | 224 | 2.98 |
| Zfp367 | 32 | 2.99 |
| Med27 | 2,190 | 2.99 |
Table 3. Transcripts in dentate granule cells with coefficient of variation >3 SDs above the mean for expression level in the control condition (n=7 independent laboratories, each averaging expression over 5–6 rats).
| Gene symbol | group median | group coef. Var (%) |
|---|---|---|
| Expression levels and SDs of genes in control condition with cv >3SD’s above mean cv. | ||
| LOC100909504 | 2,944 | 82.9 |
| Mis18a | 238 | 61.9 |
| Ttr | 557 | 60.0 |
| Prkcb | 109 | 58.0 |
| Coch | 59.8 | 56.9 |
| Ptgds | 415 | 52.6 |
| Slc17a6 | 195 | 51.9 |
| Fam111a | 59.6 | 50.6 |
| Atp1a3 | 1,821 | 50.3 |
| Dnajb6 | 1,310 | 50.0 |
| LOC257642 | 1,229 | 49.2 |
| Caly | 2,598 | 48.3 |
| Htr2c | 179 | 47.9 |
| Hba1///Hba2 | 1,331 | 46.6 |
| Cartpt | 649 | 46.5 |
| LOC102546420 | 44.5 | 46.2 |
| Got1 | 842 | 45.7 |
| Adam22 | 1,267 | 45.7 |
| Aplp1 | 1,636 | 43.9 |
| Tbl1xr1 | 89.1 | 43.7 |
| Smarca2 | 249 | 43.7 |
| Ctxn1 | 2,175 | 43.5 |
| Enc1 | 2,074 | 43.0 |
| Psmd14 | 582 | 42.7 |
| LOC100911806 | 128 | 42.6 |
| Myo5b | 538 | 42.6 |
| Cdh13 | 779 | 42.0 |
| LOC100911286///Pcsk1n | 3,750 | 41.5 |
| Slitrk3 | 614 | 41.4 |
| Atxn2 | 280 | 41.0 |
| Arhgap12 | 164 | 40.9 |
| Wasl | 816 | 40.6 |
| Slc35g2 | 112 | 40.3 |
| Sostdc1 | 60.4 | 40.2 |
| Prpf40a | 107 | 40.1 |
| Colec12 | 47.0 | 39.8 |
| Cacnb3 | 384 | 39.6 |
| Pfkl | 186 | 39.6 |
| Tsc22d2 | 446 | 39.4 |
| Pianp | 301 | 39.2 |
| Gria1 | 3,116 | 38.9 |
| Arl8b | 229 | 38.5 |
| Ccnd2 | 1,095 | 38.5 |
| Atp6v0a1 | 790 | 38.5 |
| LOC100364062///Pkm | 5,747 | 38.5 |
| Brinp1 | 1,396 | 38.4 |
| Nsg2 | 3,176 | 38.2 |
| Enpp2 | 375 | 37.8 |
| Slc17a7 | 1,062 | 37.7 |
| Tanc2 | 144 | 37.7 |
| Slitrk1 | 817 | 37.6 |
| LOC679818 | 160 | 37.3 |
| Pkib | 596 | 37.0 |
| F5 | 38.5 | 36.9 |
| Rad17 | 227 | 36.8 |
| Map1b | 327 | 36.7 |
| Mlf2 | 3,229 | 36.4 |
| Cox5a | 318 | 36.3 |
| Cth | 150 | 36.3 |
| Lsamp | 717 | 36.2 |
| Ppp1r1b | 950 | 36.2 |
| Hpcal4 | 596 | 36.2 |
| Myt1l | 1,582 | 36.2 |
| Cnih2 | 4,609 | 35.9 |
| Tspyl2 | 684 | 35.8 |
| Atp5a1 | 4,092 | 35.8 |
| Nmrk1 | 264 | 35.6 |
| Sh3bgrl | 343 | 35.6 |
| Tecpr1 | 428 | 35.6 |
| Sorl1 | 877 | 35.5 |
| Prps1 | 783 | 35.5 |
| Atg12 | 191 | 35.5 |
| Glyr1 | 1,255 | 35.4 |
| RGD1564664 | 216 | 35.3 |
| Mthfd1l | 139 | 35.3 |
| Pfn2 | 196 | 35.3 |
| Dnajc5 | 415 | 35.2 |
| Chsy3///LOC100910780 | 96.1 | 35.2 |
| Slc12a5 | 9,635 | 35.1 |
| Faim2 | 881 | 35.1 |
| Cacna2d1 | 833 | 35.1 |
| Atp6v0c | 6,859 | 35.0 |
| Nucb1 | 784 | 35.0 |
| RGD1565798///Tpt1 | 8,075 | 34.8 |
| Rcan1 | 161 | 34.8 |
| Cck | 576 | 34.8 |
| Apod | 210 | 34.8 |
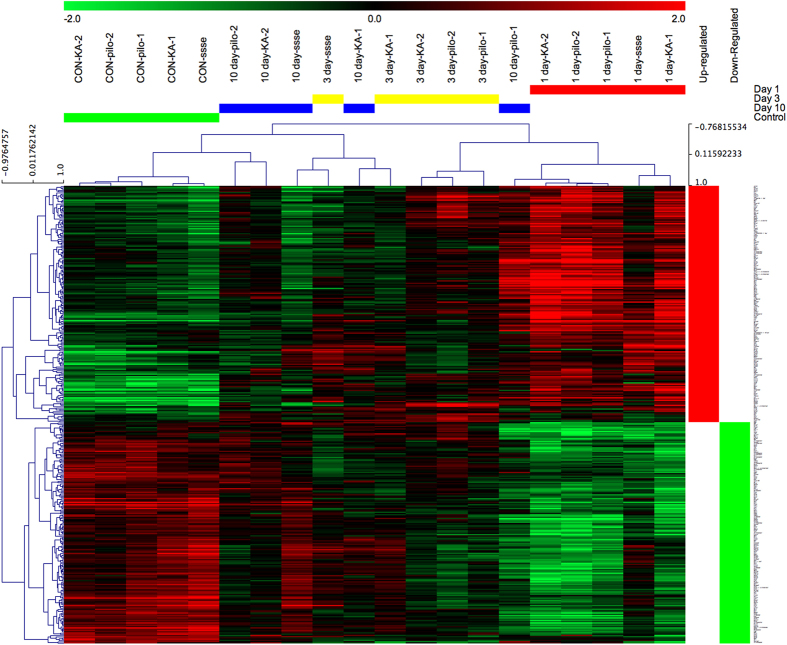
To determine whether our strategy of comparing animal models and of having two independent laboratories study each animal model was worthwhile, we focused on the SE models for which we have the most data. We first performed a hierarchical clustering of expression levels across all four time points and three SE models (control and 1, 3 or 10 days after SE), with data from all rats in each group from each laboratory having been condensed to their median expression level for each transcript. Figure 4 shows complete separation of the 5 control groups and the 5 SE groups 1 day after SE, whereas profiles from the 3 day and 10 day groups were somewhat intermixed. This suggests that the datasets are sufficiently robust at the group level to distinguish SE from control rats reliably in all laboratories, which is a prerequisite for future studies of the whole dataset. Second, to determine whether our experimental design incorporating multiple laboratories to study each animal model was necessary, we used a Venn diagram approach to compare transcript levels 1 day after SE in the pilocarpine, kainate and SSSE models. We found low congruence between paired laboratories and especially among the three models (Fig. 5). Very few transcripts were differentially expressed in any of the kindling groups, and none of those overlapped with the SE groups. These results validate the importance of a consortium strategy for the goal of identifying the relatively small number of transcripts that are differentially expressed in a laboratory- and model-independent fashion. Thus, 1 day after SE only 73 out of 1,638 transcripts differentially expressed in any model in any laboratory (4.5%) were in common across all laboratories and models. These transcripts (Table 4 (available online only)) represent genes whose differential expression in response to SE was independently replicated for each model and laboratory combination. The large group of transcripts that do not overlap in the Venn diagrams may reflect expression changes unique to each treatment model or laboratory. The results suggest that all three SE models converge at the one day time point on a rather small common set of gene expression changes associated with the early development of epilepsy, or potentially with nearby brain injury. Our dataset of transcripts with the most reproducible changes in expression (Table 4 (available online only)) should be useful for numerous analyses of seizure-induced gene expression changes in the rat. None of these transcripts appears in Tables 2 and 3 (available online only).
Figure 4. Hierarchical clustering of differentially expressed genes across laboratories and SE epilepsy models.
Unsupervised hierarchical clustering was performed using median log2 values of the 368 genes that were differentially expressed (FDR<0.05) at least 2-fold between controls and SE-experienced rats, in any model and any lab, with uncentered Pearson correlations and complete clustering.
Figure 5. Low congruence of differentially-expressed genes across laboratories and animal models of epilepsy 1 day after SE.
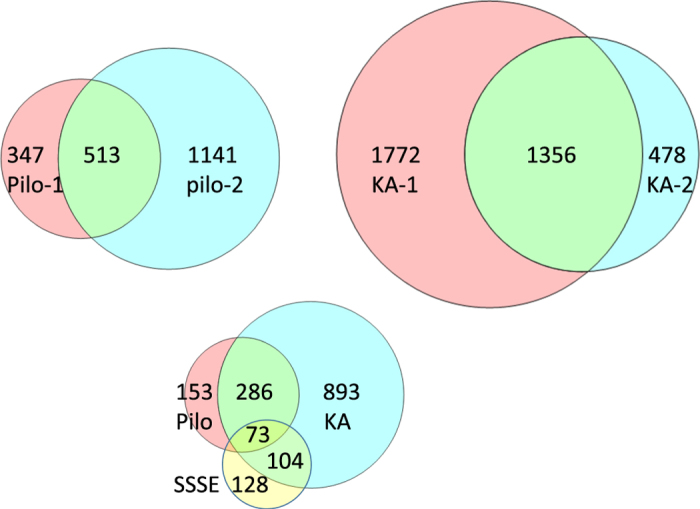
The circles in the top row show overlap between laboratories studying the same epilepsy model, and the bottom compares differentially expressed genes among the three epilepsy models.
Table 4. Differentially expressed genes in common among kainate, pilocarpine and SSSE models in all 5 laboratories 1 day after SE.
| Aard |
| Abcd2 |
| Acvr1c |
| Arsg |
| Boc |
| C1s |
| Ca10 |
| Car11 |
| Cbfb |
| Clmp |
| Col25a1 |
| Crim1 |
| Cyb561 |
| Dclk3 |
| Ddit4l///LOC100363484 |
| Ddr2 |
| Enox2 |
| Extl1 |
| Fam129b |
| Fam69c |
| Fat1 |
| Gabrd |
| Gadl1 |
| Gdf10 |
| Gpc3 |
| Htr5b |
| Ifitm10 |
| Khdrbs3 |
| Kifc3 |
| Klf15 |
| Klhl5 |
| Lingo2 |
| LOC100910632 |
| LOC100910797 |
| LOC100911253 |
| LOC100912459 |
| LOC102552294 |
| LOC102548876 |
| LOC102551251 |
| LOC691995 |
| Lox |
| Lrig1 |
| Mmp9 |
| Mpp6 |
| Nhlh1 |
| Nmb |
| Npdc1 |
| Ntf3 |
| Nubpl |
| Plk5 |
| Podxl2 |
| Prkacb |
| Prss23 |
| Ptpn5 |
| Ptprn |
| Rab26 |
| Rbks |
| Rbp1 |
| Rnasel |
| Rreb1 |
| Rsph10b |
| Scn4b |
| Serinc2 |
| Shb |
| Sim2 |
| Ss18 |
| Ssbp3 |
| Tbc1d2b |
| Trh |
| Tut1 |
| Wls |
| Zfp259 |
| Zmiz1 |
The deposited data on transcriptional responses of dentate granule cells to SE 1, 3 and 10 days later, and throughout the kindling process, provide a reference dataset that can be extended in multiple ways. It would be useful to compare dentate granule cells with other cell types thought to be involved in epileptogenesis, for example the principal neurons in layer II of the medial entorhinal cortex23 or astrocytes24,25. It would also be worthwhile to examine transcriptional responses earlier in the process (e.g., 1–6 h after SE onset), during which neurogenic inflammation is initiated26. Finally, other epilepsy models such as febrile SE27, post-traumatic epilepsy28 or genetic epilepsies that do not involve overt neurodegeneration29, could be considered. The genetic epilepsies might avoid transcriptional changes due to a bystander effect of nearby dying neurons; mice bearing epilepsy genes with late expression onset would lend themselves to the kind of temporal analysis done here but could suffer from effects on development unrelated to epileptogenesis. The cost of RNAseq analyses has now ebbed to the point at which it could be used in future studies and would open the analysis to splicing isoforms of individual transcripts and to non-coding RNAs e.g., miRNAs and lncRNAs. Regardless how this study is extended, it is worth recognizing the value shown here of utilizing multiple models and multiple investigators, and harvesting RNA from single cell types to limit noise in the transcriptome.
Additional Information
How to cite this article: Dingledine, R. et al. Transcriptional profile of hippocampal dentate granule cells in four rat epilepsy models. Sci. Data 4:170061 doi: 10.1038/sdata.2017.61 (2017).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We appreciate the contributions of expert animal modelists Karin Borges, Xiao-Ping He, Yiqun Jiao, Erwin van Vliet, Shareen Nelson, and Alicia White. We thank Dr Randall Stewart (NINDS) for making supplemental funds available to defray much of the cost of the microarray service, and Renee Shaw for technical assistance. This work was supported by NIH grants U01 NS058158 and R01NS097776 (RD), R21 NS03364 (AR & RD), R21 NS095187 (AR), CURE Challenge Award (AR), R01 NS056217 (JM), R01NS‐38108 (JVN), Nationaal Epilepsie Fonds (07-19), R01 NS038572 and R01082046 (DC), the Academy of Finland grants 272249 and 273909 (AP), RO1 NS13515, UO1 NS074926 and VHA Research Service (CW). RD and AR had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
The authors declare no competing financial interests.
Data Citations
- Dingledine R. 2013. NCBI Gene Expression Omnibus. GSE47752
References
- Hesdorffer D. C., Logroscino G., Cascino G., Annegers J. F. & Hauser W. A. Risk of unprovoked seizure after acute symptomatic seizure: effect of status epilepticus. Annals of Neurology 44, 908–912 (1998). [DOI] [PubMed] [Google Scholar]
- Lowenstein D. H., Bleck T. & Macdonald R. L. It's time to revise the definition of status epilepticus. Epilepsia 40, 120–122 (1999). [DOI] [PubMed] [Google Scholar]
- Raspall-Chaure M., Chin R. F., Neville B. G. & Scott R. C. Outcome of paediatric convulsive status epilepticus: a systematic review. The Lancet Neurology 5, 769–779 (2006). [DOI] [PubMed] [Google Scholar]
- Lukasiuk K., Kontula L. & Pitkänen A. cDNA profiling of epileptogenesis in the rat brain. Eur J Neurosci. 17, 271–279 (2003). [DOI] [PubMed] [Google Scholar]
- Elliott R. C., Miles M. F. & Lowenstein D. H. Overlapping microarray profiles of dentate gyrus gene expression during development- and epilepsy-associated neurogenesis and axon outgrowth. J Neurosci. 23, 2218–2227 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S., Chung H., Xia H., Mahadevia A. & Song Y. Environmental enrichment reverses the impaired exploratory behavior and altered gene expression induced by early-life seizures. J Child Neurol. 20, 796–802 (2005). [DOI] [PubMed] [Google Scholar]
- Gorter J. A. et al. Potential new antiepileptogenic targets indicated by microarray analysis in a rat model for temporal lobe epilepsy. J Neurosci. 26, 11083–11110 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winden K. D. et al. A systems level, functional genomics analysis of chronic epilepsy. PLoS ONE 6, e20763 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen K. F., Sakamoto K., Pelz C., Impey S. & Obrietan K. Profiling status epilepticus-induced changes in hippocampal RNA expression using high-throughput RNA sequencing. Sci Rep. 4, 6930 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland S. et al. The transcription factor NRSF contributes to epileptogenesis by selective repression of a subset of target genes. Elife 3, e01267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges K. et al. Neuronal and glial pathological changes during epileptogenesis in the mouse pilocarpine model. Experimental Neurol 182, 21–34 (2003). [DOI] [PubMed] [Google Scholar]
- Heinemann U. et al. The dentate gyrus as a regulated gate for the propagation of epileptiform activity. Epilepsy Res 7(Suppl): 273–280 (1992). [PubMed] [Google Scholar]
- Lothman E. W., Stringer J. L. & Bertram E. H. The dentate gyrus as a control point for seizures in the hippocampus and beyond. Epilepsy Res Suppl. 7, 301–313 (1992). [PubMed] [Google Scholar]
- Dengler C. G. & Coulter D. A. Normal and epilepsy-associated pathologic function of the dentate gyrus. Prog Brain Res. 226, 155–178 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook-Magnuson E. et al. In vivo evaluation of the dentate gate theory in epilepsy. J Physiol. 593, 2379–2388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges K., Shaw R. & Dingledine R. Gene expression changes after seizure preconditioning in the three major hippocampal cell layers. Neurobiol Dis. 26, 66–77 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine R. J. Modification of seizure activity by electrical stimulation. II. Motor seizure. Electroencephalogr. Clin. Neurophysiol. 32, 281–294 (1972). [DOI] [PubMed] [Google Scholar]
- Schauwecker P. E. & Steward O. Genetic determinants of susceptibility to excitotoxic cell death: implications for gene targeting approaches. Proc. Natl. Acad. Sci. USA 94, 4103–4108 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazarati A. M. et al. in: Models of Seizures and Epilepsy (eds Pitkanen A., Moshe S. Jr, Schwartzkroin P.) 449–464 (Elsevier Academic Press, 2005). [Google Scholar]
- Mazarati A. M., Wasterlain C. G., Sankar R. & Shin D. Self-sustaining status epilepticus after brief electrical stimulation of the perforant path. Brain Res. 801, 251–253 (1998). [DOI] [PubMed] [Google Scholar]
- Sloviter R. S., Zappone C. A., Harvey B. D. & Frotscher M. Kainic acid-induced recurrent mossy fiber innervation of dentate gyrus inhibitory interneurons: possible anatomical substrate of granule cell hyper-inhibition in chronically epileptic rats. J Comp Neurol. 494, 944–960 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y. & Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist. Soc B 57, 289–300 (1995). [Google Scholar]
- Armstrong C. et al. Target-selectivity of parvalbumin-positive interneurons in layer II of medial entorhinal cortex in normal and epileptic animals. Hippocampus 26, 779–793 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetherington J., Serrano G. & Dingledine R. Astrocytes in the epileptic brain. Neuron 58, 168–178 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhäuser C., Seifert G. & Bedner P. Astrocyte dysfunction in temporal lobe epilepsy: K+ channels and gap junction coupling. Glia 60, 1192–1202 (2012). [DOI] [PubMed] [Google Scholar]
- Jiang J. et al. Therapeutic window for cyclooxygenase-2 related anti-inflammatory therapy after status epilepticus. Neurobiol Dis. 76, 126–136 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K. P. et al. Rapid, Coordinate Inflammatory Responses after Experimental Febrile Status Epilepticus: Implications for Epileptogenesis. eNeuro 2 ENEURO.0034-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrosio R. et al. Post-traumatic epilepsy following fluid percussion injury in the rat. Brain 127(Pt 2): 304–314 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noebels J. Pathway-driven discovery of epilepsy genes. Nat Neurosci. 18, 344–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Dingledine R. 2013. NCBI Gene Expression Omnibus. GSE47752