SUMMARY
ADP-ribosyltransferases (ARTD1–16) have emerged as major downstream effectors of NAD+ signaling in the cell. Most ARTDs (ARTD7–8, 10–12, 14–17) catalyze the transfer of a single unit of ADP-ribose from NAD+ to target proteins, a process known as mono-ADP-ribosylation (MARylation). Progress in understanding the cellular functions of MARylation has been limited by the inability to identify the direct targets for individual mono-ARTDs. Here we engineered mono-ARTDs to use an NAD+ analogue that is orthogonal to wild-type ARTDs. We profiled the MARylomes of ARTD10 and ARTD11 in vitro, identifying isoform-specific targets and revealing a potential role for ARTD11 in nuclear pore complex biology. We found that ARTD11 targeting is dependent on both its regulatory and catalytic domains, which has important implications for how ARTDs recognize their targets. We anticipate that our chemical genetic strategy will be generalizable to all mono-ARTD family members based on the similarity of the mono-ARTD catalytic domains.
INTRODUCTION
ADP-ribosylation – the transfer of the ADP-ribose (ADPr) moiety from nicotinamide adenine dinucleotide (NAD+) to amino acids in proteins – is a reversible posttranslational modification essential for cellular function in mammals (Hottiger et al., 2010). The enzymes that catalyze ADP-ribosylation, known as ADP-ribosyltransferases (ARTDs, 15 active family members in humans) or poly-ADP-ribose polymerases (PARPs), have been implicated in a number of physiological roles, including gene regulation (Zhang et al., 2012), differentiation (Hu et al., 2013), and signal transduction (Strosznajder et al., 2005); as well as a number of diseases - notably neurodegeneration (Cosi and Marien, 1999) and cancer (Masutani and Fujimori, 2013). As such, the cellular functions of each ARTD family member and their downstream targets have generated significant biological interest; however, the targets of most ARTDs are unknown, which has hampered efforts to delineate their specific roles in cellular processes.
While ARTDs were termed polymerases based on their homology to the catalytic domain of the founding member ARTD1 (a verified polymerase), most ARTD family members (ARTDs 7–8, 10–12, and 14–17) catalyze mono-ADP-ribosylation (MARylation) and not poly-ADP-ribosylation (PARylation) as previously thought (Vyas et al., 2014). The ARTDs that catalyze MARylation, referred to here as mono-ARTDs, are not understood in nearly as much detail as the ARTDs that catalyze PARylation, referred to here as poly-ARTDs. This is due, in part, to the lack of chemical tools to study MARylation in the cell. PARylated and MARylated proteins can be enriched using different protein domains (e.g. macro) (Jungmichel et al., 2013) or the modification of the ADPr adduct with chemical tags (e.g. biotin, boronate resin) (Jiang et al., 2010; Zhang et al., 2013) followed by protein identification by liquid-chromatography and tandem mass spectrometry (LC-MS/MS). But, none of these methods are able to distinguish between MARylation and PARylation and, most importantly, they cannot determine which mono-ARTD is responsible for a given modification. This means that advances in mono-ARTD biology have been painstaking, requiring the identification of targets through traditional molecular biology approaches. Complicating matters further, the mono-ARTD family members are known to form complexes with each other in the cell and could be playing semi-redundant roles in signal transduction (Leung et al., 2011). To push this field forward, new strategies are needed to link a given mono-ARTD to its direct protein targets.
Herein, we report a chemical genetic (“bump-hole”) strategy to label the specific targets of a single engineered mono-ARTD with a clickable NAD+ analogue containing a benzyl substituent at the C-5 position of the nicotinamide ring and an alkyne tag at the N-6 position of the adenosine ring (5-Bn-6-a-NAD+). In this strategy the benzyl substituent acts as a “bump,” preventing 5-Bn-6-a-NAD+ from being used as a substrate for wild-type ARTDs. The engineered mono-ARTD contains a unique pocket, or “hole”, that can accommodate the benzyl substituent and efficiently use 5-Bn-6-a-NAD+ as a substrate. Thus, 6-a-ADPr from 5-Bn-6-a-NAD+ will only be incorporated by an engineered mono-ARTD, facilitating orthogonal - and mono-ARTD specific - target identification. When combined with LC-MS/MS analysis we successfully identified a set of 140 preferred ARTD10-specific targets that are involved in a wide-array of biological processes. We also identified a set of 21 preferred ARTD11-specific targets that are primarily involved in nuclear pore complex biology (Natalizio and Wente, 2013). This identification of cellular ARTD11 targets implicates ARTD11 in a previously uncharacterized biological role. Finally, we implemented our strategy to explore the requirements for target recognition at both the NAD+ active site and the modular n-terminal regulatory domains of ARTDs 10 and 11. We provide evidence that the structurally conserved ARTDcat domains and the non-conserved modular n-terminal regulatory domains in the mono-ARTD family play specific, and necessary, roles in precise target recognition.
RESULTS
Identification of Engineered mono-ARTD – Modified NAD+ Analogue Pairs
We recently adapted a sensitized enzyme-modified substrate (“bump-hole”) method for identifying the direct protein targets of poly-ARTDs (Carter-O’Connell and Cohen, 2015; Carter-O’Connell et al., 2014). This method involved mutating an active site lysine residue (Lys903 in human ARTD1, referred to here as the “ceiling” position) to an alanine to create a unique pocket for accommodating a C-5 ethyl group on the nicotinamide ring of the NAD+ analogue, 5-Et-6-a-NAD+. This NAD+ analogue contains an alkyne at the N-6 position of the adenine ring to aid in target identification using click conjugation to a rhodamine-azide or biotin-azide. We showed that 5-Et-6-a-NAD+ was used as a selective substrate for K903A (KA-), but not wild-type (WT-) ARTD1, and mutation of the ceiling lysine to an alanine in the other poly-ARTDs gave similar results (Carter-O’Connell and Cohen, 2015; Carter-O’Connell et al., 2014).
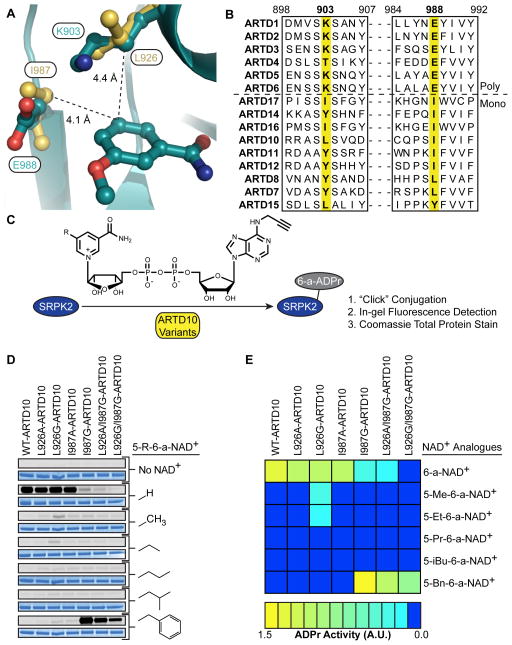
Unlike the poly-ARTDs, the mono-ARTDs do not have a lysine at the ceiling position; rather, they contain a leucine (ARTD10, 15), an isoleucine (ARTD16, 17), or a tyrosine (ARTD7, 8, 11, 12, and 14) as demonstrated by a structure-based sequence alignment (Figure 1A). Overlay of the crystal structures of 3-aminobenzamide-bound ARTD10 (Karlberg et al., unpublished data) and ARTD1 (Ruf et al., 1998) reveals that Leu926 in ARTD10 occupies a similar space as Lys903 in ARTD1 (Figure 1B), suggesting that mutation of the ceiling position in mono-ARTDs to a smaller amino acid (e.g. alanine or glycine) would accommodate 6-a-NAD+ analogues containing a substitution at the C-5 position of the nicotinamide ring.
Figure 1. Identification of Engineered mono-ARTD – Modified NAD+ Analogue Pairs.
(A) Sequence alignment of the nicotinamide binding site of the poly-ARTDs (above dashed line) and the mono-ARTDs (below).
(B) Overlay of the crystal structures of ARTD1cat (dark green) (PDB ID: 3PAX, Ruf et al., 1998) and ARTD10cat (yellow) (PDB ID: 3HKV, Karlberg et al., unpublished data) showing the nicotinamide binding sites. The distance between the key amino acids identified in ARTD10, L926 and I987, and the C-5 position of 3-methoxybenzamide are indicated.
(C) ARTD10 variants were incubated with the ARTD10 target, SRPK2, in the presence of each individual NAD+ analogue. Modified SRPK2 was subjected to “click” conjugation with a fluorogenic probe and total MARylation was observed using in-gel detection.
(D) Results from orthogonal SRPK2 MARylation screen. Engineered ARTD10 variants are listed above the gels. C-5 substitutions on the nicotinamide ring are indicated. For each modified NAD+ analogue tested the same gel was first fluorescently imaged to detect SRPK2 MARylation (top gel, gray) and then stained to detect total SRPK2 (bottom gel, blue).
(E) A heat map depicting the global MARylation efficiency normalized against the total loaded protein for the engineered pairs tested in (D).
See also Figure S1.
We first sought to determine if mutation of Leu926 to an alanine or glycine in ARTD10 would confer sensitivity to C-5 substituted 6-a-NAD+ analogues. In addition to our original 6-a-NAD+ analogue, 5-Et-6-a-NAD+, we synthesized a panel of analogues containing either a methyl, propyl, iso-butyl, or benzyl group at the C-5 position (5-Me-6-a-NAD+, 5-Pr-6-a-NAD+, 5-iBu-6-a-NAD+, and 5-Bn-6-a-NAD+, respectively) to further probe the unique binding pockets in engineered mono-ARTDs (Scheme S1). To test engineered ARTD10 – C-5 substituted 6-a-NAD+ analogue pairs, we monitored ARTD10 catalytic domain (ARTD10cat)-mediated MARylation of the known substrate SRSF protein kinase 2 (SRPK2) (Haikarainen et al., 2013; Morgan and Cohen, 2015) by click conjugation to a rhodamine-azide probe and subsequent in-gel fluorescence detection (Figure 1C). 6-a-NAD+ was used as a substrate to mediate SRPK2 MARylation by WT-ARTD10cat, and to a lesser extent by L926A- and L296G-ARTD10cat (Figure 1D, E). Importantly, none of the C-5 substituted 6-a-NAD+ analogues were used by WT-ARTD10cat (Figure 1D, E). 5-Me-6-a-NAD+ and 5-Et-6-a-NAD+ were used by L296G-ARTD10cat, but were very poor substrates (~5% MARylation activity compared to WT-ARTD10cat with 6-a-NAD+, Figure 1D, E). Based on these results, we sought an alternative position within the nicotinamide binding site of mono-ARTDs that when mutated to a smaller amino acid, might confer sensitivity to our orthogonal NAD+ analogues.
We hypothesized that Ile 987 (human ARTD10 numbering) in the “floor” position of ARTD10 was a good candidate for our enzyme-engineering approach for two reasons: (1) it makes van der Waals contacts with the C-5 position of the benzamidine ring of 3-aminobenzamide (Figure 1B); and (2) it is well-conserved across the mono-ARTD subfamily (Figure 1A). We therefore mutated Ile987 in ARTD10cat to either an alanine or glycine and determined if these engineered ARTD10cat mutants could use C-5 substituted 6-a-NAD+ analogues as substrates. We found that 5-Bn-6-a-NAD+ was used efficiently by I987G-ARTD10cat (~140% MARylation activity compared to WT-ARTD10cat with 6-a-NAD+, Figure 1D, E); by contrast, 6-a-NAD+ was a poor substrate for I987G -ARTD10cat (~5% MARylation activity compared to WT-ARTD10cat with 6-a-NAD+, Figure 1D, E). The apparent KM (KM(app)) for 5-Bn-6-a-NAD+ for IG-ARTD10cat was similar to the KM(app) for 6-a-NAD+ for WT-ARTD10cat (KM(app) = 79.7 versus 69.6 μM, respectively, Figure S1A). Taken together, these results demonstrate that mutation of Ile987 in the floor position of ARTD10 results in an orthogonal switch in substrate specificity from 6-a-NAD+ to 5-Bn-6-a-NAD+.
We next sought to determine if mutating the floor positioning in another mono-ARTD would confer sensitivity to 5-Bn-6-a-NAD+. We expressed WT-ARTD7cat and L659G-ARTD7cat and tested their in vitro MARylation activity with both 6-a-NAD+ and 5-Bn-6-a-NAD+. Similar to I987G-ARTD10cat, L659G-ARTD7cat used 5-Bn-6-a-NAD+ selectively to MARylate SRPK2 (Figure S2). Importantly, WT-ARTD7cat did not use 5-Bn-6-a-NAD+ (Figure S2). As all of the mono-ARTDs contain either a leucine or isoleucine at the I987 position (with the exception of ARTD16), this result suggests that either residue can be mutated to glycine to generate a 5-Bn-6-a-NAD+ sensitive allele throughout the mono-ARTD subclass.
The IG-ARTD10 – 5-Bn-6-a-NAD+ Pair Specifically Label Direct Protein Targets in Multiple Cell Lines
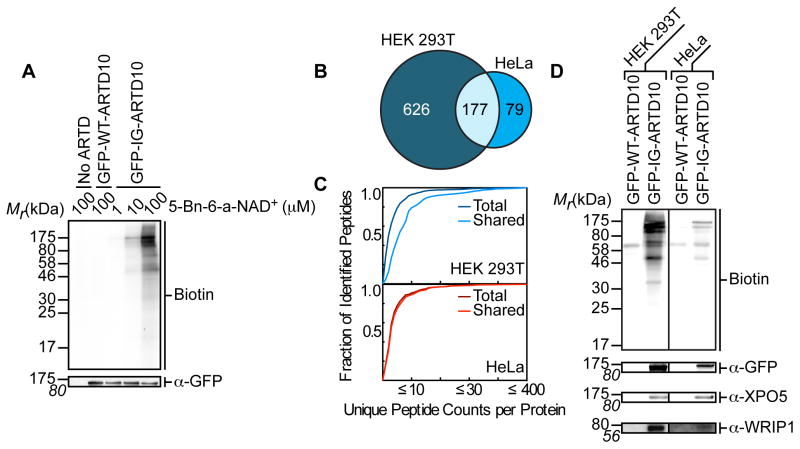
We next determined if I987G-ARTD10 could be used to label direct protein targets in a cellular context. GFP-I987G-ARTD10 (IG-ARTD10) or GFP-WT-ARTD10 (WT-ARTD10) were expressed in human embryonic kidney (HEK) 293T cells and lysates were prepared and incubated with increasing concentrations of 5-Bn-6-a-NAD+ (1–100 μM), followed by click conjugation with biotin-azide. Only at 100 μM 5-Bn-6-a-NAD+, which is near the KM(app) for 5-Bn-6-a-NAD+ for IG-ARTD10 (Figure S1A), did we reliably detect the labeling of several bands (especially lower molecular weight products) with the predominant band corresponding to the size of auto-MARylated IG-ARTD10 (Figure 2A). By contrast, treatment of lysates from WT-ARTD10 transfected cells or non-transfected cells with 100 μM 5-Bn-6-a-NAD+ resulted in a low-level of background labeling – most likely due to endogenously biotinylated proteins (Figure 2A). These results demonstrate that the IG-ARTD10 – 5-Bn-6-a-NAD+ pair can be used to label direct MARylation targets of ARTD10.
Figure 2. IG-ARTD10 Orthogonally Labels Protein Targets in the Presence of 5-Bn-6-a-NAD+.
(A) Lysate labeling by WT-ARTD10 and IG-ARTD10 in the presence of 5-Bn-6-a-NAD+. HEK 293T cells were transfected with either WT-ARTD10 or IG-ARTD10 and the resulting lysate was incubated for 2 hours in the presence of varying amounts of 5-Bn-6-a-NAD+. MARylation of direct protein targets was observed using streptavidin-HRP (Biotin). The faint bands in the WT-ARTD10 lane correspond to endogenous biotinylated proteins. Expression of ARTD10 was confirmed via immunoblot detection of GFP. Shown is a representative image from duplicate measurements.
(B) Venn diagram comparing the IG-ARTD10 targets identified via single LC-MS/MS runs in either HEK 293T or HeLa cells.
(C) Observed distribution functions for the IG-ARTD10 targets identified via single LC-MS/MS runs in either HEK 293T (top) or HeLa (bottom) cells. The distributions for the total protein pool (total) as well as the subset of proteins that were identified in both HEK 293T and HeLa (shared) are indicated. The shared targets identified in HEK 293T cells display significantly elevated peptide counts per identified protein as compared to the total target pool (p < 0.05, non-parametric Mann-Whitney U test). The shared targets identified in HeLa cells also display elevated peptide counts per protein, but the difference compared to the total target pool is not significant.
(D) Immunoblot detection of the LC-MS/MS identified ARTD10 targets (GFP-ARTD10, XPO5, WRIP1) following NeutrAvidin enrichment. MARylation levels were determined using streptavidin-HRP (Biotin). Differences in labeling efficiency between HEK 293T and HeLa lysate required separate immunoblot exposures.
We next sought to use our labeling method to identify the direct MARylation targets of ARTD10 using LC-MS/MS. HEK 293T lysates generated from cells expressing WT-ARTD10 or IG-ARTD10 were treated with 5-Bn-6-a-NAD+ (100 μM). MARylated proteins were conjugated to biotin-azide, enriched using NeutrAvidin agarose, digested with trypsin, and subjected to LC-MS/MS (Figure S3A). We identified 803 ARTD10-specific protein targets (Table S1, thresholds discussed in methods). This represents a much broader target set than that found for either ARTD1 or ARTD2 (42 and 301 proteins, respectively), which we found surprising (Carter-O’Connell et al., 2014). We also find no overlap with the ARTD10 targets identified using protein microarrays (Feijs et al., 2013). This could be due to differences in the context in which the labeling reactions are performed. Nonetheless, we have identified a set of ARTD10 targets in a complex cellular context. As all other PAR and MAR detection methods rely on non-family member specific labeling methods, these findings provide an identification of ARTD10-specific targets in a cellular context.
Given the scarcity of data regarding the physiological role of ARTD10, including its basal activity in different cell types, we thought that perhaps the choice of cell type could be inflating the actual target list of ARTD10 targets. To address this possibility, we repeated the labeling experiment in HeLa cells (Figure S3A). In HeLa cells we identified 256 direct ARTD10 targets (Table S2). The overall lower number of direct targets identified in HeLa versus HEK 293T cells likely reflects the lower expression of GFP-ARTD10 in HeLa compared to HEK 293T cells. Nonetheless, a comparison with the list of ARTD10 targets identified in HEK 293T cells revealed that a majority of the targets found in HeLa cells (69%) were also identified in the HEK 293T samples (Figure 2B).
To identify the most relevant cellular targets of ARTD10, we ranked each of the ARTD10 direct protein targets based on the number of peptide counts per protein identified in the LC-MS/MS analysis. We reasoned that preferred ARTD10 targets would be labeled more efficiently and would thus be enriched to a higher degree. Based on this enrichment, preferred ARTD10 target peptide fragments would also appear more frequently in the LC-MS/MS run. Importantly, the control sample generated from lysates expressing WT-ARTD10 allowed us to remove any proteins that would be enriched for non-enzymatic reasons from this analysis. The bulk of IG-ARTD10 targets identified in HEK 293T cells were identified based on a median of 2 peptides. Selecting for proteins that were also identified in HeLa cells causes a shift in median peptide counts from 2 to 6 peptides per protein. The cumulative distribution frequency of peptide counts per identified protein target for the shared protein pool (HEK 293T and HeLa targets) is elevated significantly above the cumulative distribution generated from the total pool of HEK 293T protein targets (p<0.0001, Mann-Whitney test, Figure 2C). As the majority of HeLa targets are shared with HEK 293T there was not a significant difference in the cumulative distribution frequencies between the shared and total HeLa target pools (Figure 2C). While we acknowledge that some of the targets identified with lower peptide counts in the LC-MS/MS analysis might still represent relevant cellular targets of ARTD10, the ranking of proteins based on peptide frequency counts - as well as their likelihood to be found in multiple cell lines - provides a starting point for linking ARTD10-specific MARylation to cellular processes.
To confirm our LC-MS/MS results, we selected the shared HeLa and HEK 293T targets XPO5 and WRIP1 for identification by Western blot with target-specific antibodies after NeutrAvidin enrichment. We found that XPO5 and WRIP1, as well as auto-MARylated IG-ARTD10, were selectively enriched from lysates generated from either HEK 293T or HeLa cells expressing IG-ARTD10 and treated with the 5-Bn-6-a-NAD+ (Figure 2D and Figure S3B). Taken together, our results demonstrate that our method can be used for the identification of direct MARylation targets of ARTD10 in a complex mixture.
ARTD11 and ARTD10 MARylate Separate Target Pools Involved in Distinct Cellular Processes
We next wanted to demonstrate the generalizability of our method by identifying the direct MARylation targets of another mono-ARTD. We were particularly interested in exploring the target profile of ARTD11 for a number of reasons: (1) ARTD11 is comprised of a fairly simple modular structure as compared to the other mono-ARTDs – with only a WWE domain attached to the catalytic ARTD domain – which would allow us to explore how non-catalytic domains dictate target selection; (2) ARTD11 has an isoleucine (I313, ARTD11 numbering) at the ARTD10-I987 position, but a tyrosine at the ARTD10-L926 position, allowing us to confirm that our method will work with mono-ARTDs with different amino acids at the L926-I987 interface; (3) the comparison of two separate mono-ARTD target profiles would allow us to examine the level of redundant target selection in the mono-ARTD family; and (4) recent work has implicated ARTD11 in nuclear membrane maintenance (Meyer-Ficca et al., 2015) providing us with a potential biological pathway to probe our target list against.
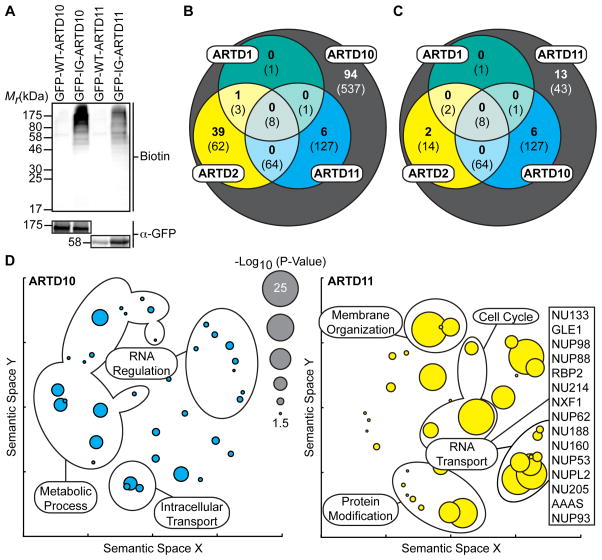
We first compared the MARylation activity of GFP-I313G-ARTD11 (IG-ARTD11) to IG-ARTD10 in HEK 293T lysates. As with WT-ARTD10, the KM(app) of WT-ARTD11 for 6-a-NAD+ was ~80 μM (Figure S1B) so we kept the 5-Bn-6-a-NAD+ concentration in lysates at 100 μM. Treatment of lysates from IG-ARTD11 transfected cells with 5-Bn-6-a-NAD+ resulted in labeling of several bands (Figure 3A). Minimal background labeling was detected in lysates expressing WT-ARTD11, further demonstrating the inability of non-engineered mono-ARTDs to use 5-Bn-6-a-NAD+ as a substrate for MARylation. The banding pattern for IG-ARTD11 is different from that produced by ARTD10, indicating that ARTD11 and ARTD10 are indeed targeting distinct and family-member specific proteins (Figure 3A).
Figure 3. IG-ARTD10 and IG-ARTD11 MARylate Separate, and Family-Member Specific, Protein Targets.
(A) Lysate labeling by IG-ARTD10 or IG-ARTD11 in the presence of 5-Bn-6-a-NAD+. HEK 293T cells were transfected with either WT- or IG-ARTD10 or -ARTD11 and the resulting lysate was incubated for 2 hours in the presence of 5-Bn-6-a-NAD+. MARylation of direct protein targets was observed using streptavidin-HRP (Biotin). The faint bands in the WT-ARTD lanes correspond to endogenous biotinylated proteins. Expression of each ARTD was confirmed via immunoblot detection of GFP. Shown is a representative image from duplicate experiments.
(B) Venn diagram comparing the total IG-ARTD10 target pool with both the current IG-ARTD11 and the previously identified KA-ARTD1 and KA-ARTD2 (Carter-O’Connell et al., 2014) target pools. The protein counts in bold represent the protein targets identified in both LC-MS/MS IG-ARTD10 replicates while the counts in parentheses represent targets identified in at least one replicate. IG-ARTD10 specific targets are shown in the gray circle.
(C) IG-ARTD11 LC-MS/MS targets treated as in (B).
(D) Circle plots depicting enriched GO terms attached to the IG-ARTD10 (left, cyan) or IG-ARTD11 (right, yellow) specific LC-MS/MS identified targets in either replicate. GO term enrichment was performed using the PANTHER toolkit. Significantly enriched GO terms (p < 0.05) were condensed using Revigo and similar terms were plotted based on semantic similarity. Select groups of terms are indicated. Circle radii are scaled proportionally to the −log10(p-value). The IG-ARTD11 specific proteins associated with RNA transport are listed.
Next, we sought to identify the direct targets of ARTD11 using LC-MS/MS. HEK 293T lysates generated from cells expressing WT-ARTD11 or IG-ARTD11 were treated with 5-Bn-6-a-NAD+ (100 μM). MARylated proteins were conjugated to biotin-azide, enriched using NeutrAvidin agarose, digested with trypsin, and subjected to LC-MS/MS (Figure S3A). We identified a total of 260 direct IG-ARTD11 targets (thresholds discussed in methods). Of the 803 and 260 protein targets identified for ARTD10 and ARTD11, respectively, in this study, we identified 140 and 21 proteins in duplicate biological replicates (Table 1). For the subsequent analysis comparing ARTD10 and ARTD11 MARylation, we focused on targets that were present in both replicates. We reasoned that the identification of the same target in multiple independent replicates would increase our confidence that a given protein was a real ARTD target. It is possible that proteins identified in a single replicate still represent cellular ARTD10/ARTD11 targets, so we have compiled full data sets for the combined protein pools (Tables S1, S3). Based on our previous observations with KA-ARTD1 and KA-ARTD2, as well as the current study, it is apparent that the ARTD family displays a spectrum of target specificity. In the case of both ARTD2 and ARTD10, we observe a broad range of cellular targets, while ARTD1 and ARTD11 have a much narrower target profile (Table 1). Taken together, our strategy has yielded a data set capable of distinguishing the PAR and MAR targeting preferences for multiple ARTD family members.
Table 1.
Direct Protein Targets Identified by LC-MS/MS
| KA-ARTD1a | KA-ARTD2a | IG-ARTD10 | IG-ARTD11 | |
|---|---|---|---|---|
| Total Proteins Identified | 123 | 488 | 961 | 479 |
| Proteins with ≥ 2 Unique Peptides | 91 | 428 | 848 | 294 |
| Proteins Enriched Above Backgroundb | 38 | 279 | 803 | 260 |
| Proteins Identified in Duplicate | 16 | N.D.d | 140 | 21 |
| ARTD Family-Member Specific Proteinsc | 13/14 | 117/N.D.d | 537/94 | 43/13 |
KA-ARTD1 and KA-ARTD2 targets were identified as previously described (Carter-O’Connell et al., 2014)
Defined as ≥ 2-fold enrichment in the IG-ARTD sample versus the WT-ARTD sample
Targets that were identified for a single ARTD family-member from the collected datasets from either at least a single replicate (left) or in duplicate (right)
KA-ARTD2 identification was completed for a single replicate and is not included in the duplicate analysis
Our LC-MS/MS analysis allowed us to address a critical outstanding question in the ARTD field: How much redundancy in target selectivity exists between different ARTD family-members? Comparing our collected protein target lists for KA-ARTD1, KA-ARTD2, IG-ARTD10, and IG-ARTD11 we have been able to identify the extent of overlap between these ARTD family-members. Interestingly, the ARTD10 target list overlaps to a greater degree with each of the ARTD family-members than ARTD11 (Figure 3B, C). ARTD10 and ARTD2 in particular share 40 (29% of the total ARTD10 target pool) protein targets (Figure 3B) while ARTD11 only shares 2 targets with ARTD2 and no targets with ARTD1 (Figure 3C). Comparing the combined target pools also allows us to isolate the protein targets that are specific for a given ARTD family-member. The bulk of the identified protein targets are actually unique to either ARTD10 (67%) or ARTD11 (62%).
Using the target datasets for each of the ARTDs detailed above, we next wanted to identify potential cellular roles for ARTD10- and ARTD11-mediated MARylation. Using the set of ARTD10- and ARTD11-specific target proteins, we searched for gene ontology (GO) terms that were significantly enriched (p<0.05, Bonferoni correction) within either the ARTD10 or ARTD11 target list (Table S4). The GO term enrichment profile for ARTD10 displayed a wide spectrum of biological processes, with enriched terms such as cellular metabolism (p=1.73e-12), intracellular protein transport (p=2.26e-10), protein targeting to the ER (p=1.75e-07), and mRNA metabolism (p=6.68e-05) (Figure 3D and Table S4). Interestingly, a number of the ARTD10 targets are involved in ubiquitin transfer and mRNA regulation. Given that ARTD10 contains both ubiquitin-interaction motifs (UIMs) and RNA recognition motifs (RRMs), these GO results seem to align with the proposed functions of ARTD10. For ARTD11, we noticed that the proteins identified in duplicate were primarily nuclear pore proteins or proteins involved in nuclear membrane organization (13 of 21 proteins). The enrichment of nuclear pore proteins led to enrichment of processes related to nuclear envelope organization (p=7.27e-24) and RNA transport (p=2.62e-24) (Figure 3D and Table S4). Taken together, the GO term profiles for each of the ARTD enzymes are distinct, with the ARTD11 profile implicating a specific and novel biological role for ARTD11 MARylation in nuclear pore complex regulation.
Although our method relies on the identification of direct targets of mono-ARTDs in cell lysates, we find that the majority of the protein targets for both ARTD10 and ARTD11 are found in similar cellular compartments as our GFP-ARTD constructs. ARTD10 is localized primarily to the cytoplasm (Figure S4), though it is thought to be able to traffic to the nucleus under the correct stimulus paradigm (Kleine et al., 2012). We find that our IG-ARTD10 construct indeed modifies both cytoplasmic and nuclear targets. GFP-ARTD11 is enriched at the nuclear membrane (Figure S4) and the majority of ARTD11 targets are nuclear pore associated proteins. GFP-ARTD11 is also found in lower levels in the cytoplasm, accounting for the few cytoplasmic proteins we identified. Taken together, our results suggest that the IG-ARTD targets maintain correct targeting in cell lysates.
Both the mono-ARTDcat and the Modular N-Terminal Domains are Necessary for Accurate ARTD11-mediated MARylation
The ARTD family is defined by the presence of a conserved ARTDcat domain (Ame et al., 2004). Each of the mono-ARTDs is then differentiated by the presence of at least one separate modular domain (i.e. WWE, Zn fingers, macro, etc.) found on the n-terminus of the mono-ARTD protein (Schreiber et al., 2006). A major unanswered question in the ARTD field is whether the n-terminal regulatory domain alone, the ARTDcat domain alone, or both together mediate substrate targeting. One of the unique advantages of our engineered mono-ARTD – modified NAD+ analogue pairs is the ability to decouple proximal (i.e. ARTDcat) and distal (i.e. n-terminal domain) elements of mono-ARTD protein targeting and address this question on a proteome-wide scale.
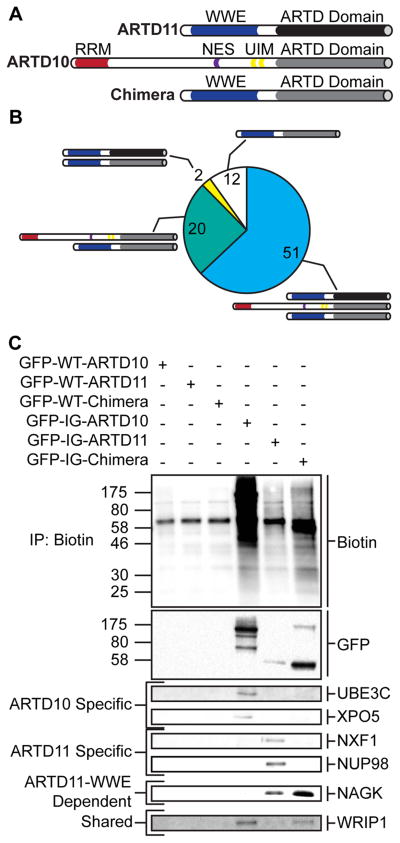
The ARTDcat domain from ARTD10 is attached to an RRM, a nuclear export sequence, and a set of UIMs, whereas the ARTD11cat domain is attached only to a WWE domain (Figure 4A). To address the differential protein target selection requirements for each of these domains, we fused the IG-ARTD10cat domain to the WWE domain from ARTD11 (Figure 4A). The resulting chimeric protein now possesses the distal targeting features of ARTD11 and the proximal targeting features of ARTD10. Interestingly, the chimeric protein is no longer actively exported from the nucleus (like ARTD10) nor localized selectively to the nuclear membrane (like ARTD11) (Figure S4), allowing access to the full range of protein targets in the cell. By comparing the direct protein targets of the IG-chimera with the targets of IG-ARTD10 and IG-ARTD11 we can identify which targets are selected based on proximal and/or distal interactions.
Figure 4. ARTD11cat and ARTD11 WWE Domains are Necessary but Insufficient to Drive ARTD11 Specific MARylation.
(A) Domain architecture of ARTD11, ARTD10, and the chimeric protein (Chimera) created by fusing the ARTD11 n-terminus to the ARTD10cat domain.
(B) Pie chart representing the total MARylated protein targets identified via LC-MS/MS for the chimeric protein. Shared protein targets are indicated by the protein schematics depicted in (A). Shared protein targets were identified based on their presence in at least one of the IG-ARTD10 or IG-ARTD11 LC-MS/MS replicates.
(D) Immunoblot detection of select LC-MS/MS identified ARTD targets (GFP-ARTD, UBE3C, XPO5, NXF1, NUP98, NAGK, WRIP1) following NeutrAvidin enrichment. Overall MARylation levels were determined using streptavidin-HRP (Biotin). ARTD10-specific, ARTD11-specific, ARTD11-WWE dependent, and shared chimera targets are indicated to the left.
We next performed LC-MS/MS analysis on HEK 293T lysate from cells expressing IG-chimera that were treated with the 5-Bn-6-a-NAD+ (Figure S3A). We identified a total of 85 IG-chimera-specific protein targets (Table S5). 60% of the IG-chimera targets are shared with both IG-ARTD10 and IG-ARTD11 (Figure 4B). When the shared targets of ARTD10, ARTD11, and the chimera are compared to the IG-chimera targets that are only shared with ARTD10 (85% of the IG-chimera targets), it is clear that the ARTD10cat domain plays an important role in target selection (Figure 4B). However, it is also apparent that the loss of the ARTD10 n-terminus has drastically reduced the number of proteins that can be targeted by the ARTD10cat domain. We also noted that of the proteins identified in duplicate ARTD11 LC-MS/MS runs, only 2 were shared with the IG-chimera protein (NAGK and WRIP1, Table S3 and S5). All of the nuclear pore proteins require both the n-terminus and the ARTD11cat domain for MARylation. It is possible that the loss of ARTD11cat-dependent enrichment to the nuclear membrane accounts for the loss of nuclear pore protein targeting (Figure S4). Taken together, these results suggest that both proximal and distal substrate interactions are necessary for proper target selection and that the structurally similar ARTDcat domains are playing distinct roles in target selection.
To confirm all of our LC-MS/MS results with IG-ARTD10, IG-ARTD11, and the IG-chimera protein, we selected ARTD10-specific (UBE3C and XPO5), ARTD11-specific (NXF1 and NUP98), ARTD11-WWE-dependent (NAGK), and shared targets (WRIP1) for identification by Western blot with target specific antibodies after NeutrAvidin enrichment. For all of the IG constructs examined, we observe robust enrichment of the auto-MARylated proteins using the GFP antibody (Figure 4C and S3C). We only observe enrichment of UBE3C and XPO5 in the ARTD10 lane and NXF1 and NUP98 in the ARTD11 lane (Figure 4C and S3C). This result confirms that our method is capable of distinguishing between the specific targets of multiple mono-ARTD family members from a complex mixture. The ARTD11-specific target, NAGK, is enriched in both the ARTD11 and chimera lane and is therefore dependent primarily on ARTD11 n-terminal recognition for labeling (Figure 4C and S3C). Finally, the mono-ARTD pan-selective target, WRIP1, is enriched in all three IG variant lanes (Figure 4C and S3C). In each case, none of the selected targets are enriched from HEK 293T lysates expressing the WT constructs, confirming the necessity of the IG mutation for mono-ARTD family-member specific MARylation using 5-Bn-6-a-NAD+ (Figure 4C and S3C). Our method is able to identify direct family-member specific mono-ARTD targets in complex lysates.
MARylation Targets of the Engineered ARTD – 5-Bn-6-a-NAD+ Pairs are Bona Fide ARTD Substrates
We next sought to validate the direct targets of mono-ARTDs identified using our chemical genetic strategy. We focused on the putative ARTD11 target NXF1, an mRNA binding protein that participates in nucleocytoplasmic shuttling (Griffis et al., 2003; Kang and Cullen, 1999). Using immunofluorescence, we found that HA-NXF1 partially co-localized with GFP-WT-ARTD11 at the nuclear membrane in HEK 293T cells (Figure 5A). This result confirms that ARTD11 is found at sites where NXF1 participates in nucleocytoplasmic shuttling (Griffis et al., 2003; Kang and Cullen, 1999). NXF1 was also previously identified as a cellular ARTD target in two separate studies exploring the non-ARTD specific PAR/MARylome (Jungmichel et al., 2013; Zhang et al., 2013). Further, 54% of the specific ARTD10/11 targets identified herein were previously identified as general ARTD targets in at least one study (Table S6). Together, these data suggest that our ARTD10/11 specific targets represent physiologically relevant MARylation targets.
Figure 5. Validation of MARylation Targets Identified via GFP-IG-ARTD – Modified 6-a-NAD+ Pairs.
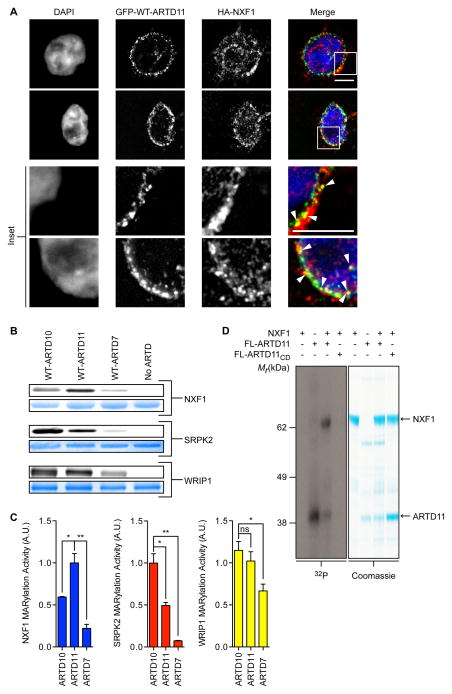
(A) GFP-ARTD11 and HA-NXF1 are partially co-localized at the nuclear membrane. HEK 293T cells co-expressing GFP-ARTD11 and HA-NXF1 were fixed with paraformaldehyde and processed for immunofluorescence. DNA was stained with DAPI. Scale bar = 5 μm. Inset: white arrowheads show co-localization.
(B) In vitro WT-ARTDcat MARylation assays demonstrate that NXF1 is a preferred ARTD11 substrate. WT-ARTD10cat, -ARTD11cat, and -ARTD7cat were screened for MARylation activity using recombinant NXF1, SRPK2, and WRIP1 in the presence of 6-a-NAD+. The same gel was first fluorescently imaged to detect substrate MARylation (top gel, gray) and then stained to detect total substrate (bottom gel, blue).
(C) Quantification of results shown in (B). The bar graphs below depict the MARylation activity for each substrate with each WT-ARTD (mean ± S.E.M., n=3). (*) represents p-value < 0.05 and (**) represents p-value < 0.01, two-tailed student t-test. ns = not significant.
(D) Results from NXF1 in vitro MARylation assay using full-length ARTD11 and 32P-NAD+. Full-length ARTD11 or ARTD11CD was incubated with NXF1 in the presence of 32P-NAD+ and ADPr transfer was visualized using autoradiography (left, gray) and stained to detect total substrate (right, blue). Arrows indicate FL-ARTD11 and NXF1.
See also Figures S4 and S5.
We next determined the in vitro selectivity of the WT-ARTD10cat, ARTD11cat, and ARTD7cat enzymes for recombinant substrates identified using our orthogonal pipeline. MARylation of NXF1 was significantly higher in the presence of ARTD11 as compared to ARTD10 and ARTD7 (Figure 5B, C). By contrast, MARylation of SRPK2, a previously validated ARTD10 target (Haikarainen et al., 2013; Morgan and Cohen, 2015), was significantly higher in the presence of ARTD10 as compared to ARTD11 and ARTD7 (Figure 5B, C). Finally, WRIP1, the dual ARTD10/ARTD11 target discussed above, was MARylated to a similar degree by both ARTD11 and ARTD10 (Figure 5B, C). We confirmed that an active ARTD is required for selective MARylation by incubating NXF1, SRPK2, and WRIP1 with the IG/LG-ARTD variants in the presence of 6-a-NAD+. Without the 5-Bn-6-a-NAD+ analog, the IG/LG-ARTD variants will not transfer ADP-ribose onto target proteins and act as non-enzymatic MARylation controls. We did not observe any labeling with the IG/LG-ARTD variants (Figure S5A). We also confirmed that auto-MARylation of the individual ARTDcat domains was not interfering with trans-MARylation; regardless of substrate we observed no appreciable auto-MARylation (Figure S5B). As neither the IG-ARTD10 nor IG-Chimera label NXF1 at all in lysate, these results confirm our previous observations that the n-terminal domains play an important role in target discrimination. Yet, clearly the ARTDcat domains retain some ability to differentiate between mono-ARTD family-member specific targets.
Finally, we wanted to confirm that NXF1 was a substrate for full-length WT-ARTD11 (FL-ARTD11) using non-modified 32P-NAD+. While FL-ARTD11 undergoes auto-MARylation, consistent with previous studies (Vyas et al., 2014), we found that FL-ARTD11 preferentially MARylates NXF1 (Figure 5D). MARylation of NXF1 was dependent on ARTD11 catalytic activity, as the catalytically inactive H197A mutant (FL-ARTD11CD) did not modify NXF1 (Figure 5D). Together, these results show that NXF1 is a bona fide target of ARTD11.
DISCUSSION
Our chemical genetic strategy to identify the direct targets of mono-ARTDs works with multiple mono-ARTD family members (ARTD10, 11, and 7). We discovered that the mutation that facilitated target identification with poly-ARTDs did not work for the mono-ARTDs (L926 in the ceiling position in ARTD10). This could be due to important differences in the manner in which the ARTDs identify and modify substrate targets. Further, the pocket that we ultimately engineered was more efficient with a glycine rather than an alanine at the key position (the I987G versus I987A mutant). It is possible that the extra bulk of the alanine side-chain prevents binding of the benzyl adduct, or that the glycine confers a greater deal of conformational flexibility to the β-strand near the NAD+-binding site. Future structural studies with the various IG-ARTD variants and modified NAD+ analogues could help discern how orthogonal NAD+ substrate switching occurs in our IG-ARTD mutants. Nonetheless, our strategy provides a key approach for identifying the family-member specific targets for multiple mono-ARTDs.
The target lists generated from ARTD10 and ARTD11 revealed how individual mono-ARTDs exhibit markedly different targeting patterns in cellular lysate. While we focused on identifying the protein targets for ARTD10 and ARTD11, we believe that our strategy will be applicable for the identification of the direct protein targets of the remaining mono-ARTD subclass. Combined with our results identifying the targets of the poly-ARTD subclass, we have now established a clear path towards assigning PARylated and MARylated protein targets to each individual ARTD family-member with a specific ADP-ribose transfer. Moreover, we have generated a database of ARTD10 and ARTD11 MARylation targets that can be used immediately to examine the biological role of these mono-ARTDs in the cell.
The ARTD10 targets we identified share notable overlap with previously reported cellular functions of ARTD10. In particular, we noted the presence of a number of ubiquitin ligases (e.g. UBE3C) in our target dataset. ARTD10 contains two UIMs that were shown to interact with ubiquitinylated tumor-necrosis factor-receptor associated factor (TRAF) to aid ARTD10 in targeting the NF-κB essential modulator (NEMO) for MARylation (Verheugd et al., 2013). An intriguing possibility is that ARTD10 regulates the ubiquitin signal cascade through MARylation of ubiquitin ligases. Additionally, ARTD10 has been implicated in the coordination of cellular trafficking (Kleine et al., 2012) and we indeed find a number of cellular trafficking proteins in our target dataset (Tables S1 and S2).
However, while the target list for ARTD10 most likely contains a number of targets that are involved in specific signaling events mediated by ARTD10, we were still surprised by the broad array of cellular targets that are MARylated by ARTD10 and have no clear functional relationship to each other. We propose that the broad promiscuity evidenced by ARTD10 might actually play a role in the function of ARTD10 in the cell. ARTD10 has been shown to interact with p62, a ubiquitin receptor associated with autophagy (Kleine et al., 2012). In certain conditions, ARTD10 forms cytosolic clusters that bind p62, which implicates ARTD10 in trafficking targets to the autophagosome. It is therefore possible that in this role, ARTD10 is modifying a broad array of targets that are being sent to the autophagosome for degradation. Further exploration of the role for ARTD10 in autophagy will be required to determine if the broad targeting of ARTD10 is important for its function in this pathway.
Compared to ARTD10, relatively little is known regarding the function of ARTD11 in the cell. Recent work has linked ARTD11 to nuclear shaping in spermatids undergoing nuclear condensation and differentiation (Meyer-Ficca et al., 2015), yet the ARTD11-specific targets responsible for this process are unknown. The ARTD11 targets we identified appear to be directly related to the coordination of the nuclear envelope and the organization of nuclear pores (Table S3). One of the ARTD11-specific targets we identified, the nuclear pore complex protein Nup98-Nup96 (NUP98), was previously found to interact with ARTD11 (www.biogrid.org). Our target list for ARTD11 provides a clear point of entry for exploring in molecular detail how ARTD11 MARylation regulates nuclear pore complex biology.
One of the remaining challenges in understanding the relationship between ARTD family-member specific targeting and cellular function is the identification of the specific amino acids targeted by a given mono-ARTD. It appears that while PARylation primarily targets acidic amino acids (i.e. glutamate and aspartate), MARylation may be more promiscuous in its site selection. Indeed, a recent study demonstrated that mono-ARTDs could be auto-MARylated not only on glutamate and aspartate, but also on lysine and cysteine (Vyas et al., 2014). The identity of the MARylated amino acids will be essential for more complete understating of mono-ARTDs functions in cells. We envision that the method described here could be coupled with a recently described method to globally identify PARylated and MARylated sites in protein targets (Daniels et al., 2014). Optimally, we would like to expand our method to include membrane permeant versions of the NAD+ analogues to identify the changes in mono-ARTD targeting at specific cellular locations during various stimuli. Combined with current replacement strategies for swapping the genomic version of a given mono-ARTD with our sensitized allele (e.g. CRISPR-Cas systems), we can use our strategy to connect ARTD targeting to a specific site on a protein substrate and delve deeper into the functional role of MARylation in the cell.
EXPERIMENTAL PROCEDURES
Cell Culture and Immunofluoresence
HEK 293T and HeLa cells were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS, HyClone), penicillin/streptomycin (Invitrogen), and 1X glutamax (Gibco) at 37°C and 5% CO2. Transient transfections of HEK 293T cells with 20 μg of GFP-tagged expression vectors per 10 cm dish (~70% confluency) were performed using the CalPhos system (Clontech) according to manufacturer’s instructions. Transient transfections of HeLa cells were performed with 15 μg GFP-tagged expression vectors and 30 μg lipofection reagent per 10 cm dish using Lipofectamine 2000 (Invitrogen). Cells were lysed in HEPES buffer supplemented with cOmplete EDTA-free protease inhibitor (Roche) and cell debris was cleared by centrifugation at 14,000 g for 5 min at 4°C. Immunofluoresence localization experiments with GFP-ARTD11 and HA-NXF1 were peformed as previously described (Griffis et al., 2002). Images were collected on an ApoTome microscope (Zeiss) and were processed using ImageJ. Z-stacks were compressed as maximal 2D projections and background subtraction was completed using the Rolling Ball Background Subtraction plugin (radius = 8).
Chemical Synthesis
Synthesis of 6-a-NAD+ and 5-Et-6-a-NAD+ was completed as previously described (Carter-O’Connell et al., 2014). See the Supplemental Experimental Procedures for details regarding the synthesis of the remaining C-5 substituted NAD+ analogues (Scheme S1).
SRPK2 MARylation Assay
1 μM of each ARTD10cat variant was incubated with 3 μM SRPK2 and 100 μM of each modified NAD+ analogue for 2 hours at 30°C in a 20 μL reaction volume consisting of 50 mM HEPES, pH 7.5, 100 mM NaCl, 10 mM MgCl2, and 0.5 mM TCEP. Click conjugation was performed with 1.5 mM THPTA, 750 μM CuSO4, 300 μM sulforhodamine B-PEG3-azide, and 7.5 mM sodium ascorbate in 1X PBS for 1 hour at rt. SRPK2 labeling was quantified using Image Lab v5.2 (Bio-Rad).
ARTDcat Selectivity Assay
250 nM of WT-ARTD10cat, -ARTD11cat, or -ARTD7cat was incubated with 3 μM of SRPK2, NXF1, or WRIP1 and 100 μM of 6-a-NAD+ for 2 hours at 30°C in a 12 μL reaction volume consisting of 50 mM HEPES, pH 7.5, 100 mM NaCl, 15 mM MgCl2, and 0.5 mM TCEP. Click conjugation was performed with 100 uM TBTA, 1 mM CuSO4, 100 μM sulforhodamine B-PEG3-azide, 1% SDS, and 1 mM sodium ascorbate in 1X PBS for 30 minutes at rt. Substrate labeling was quantified using Image Lab v5.2 (Bio-Rad) and normalized against the total substrate load as determined using SafeStain (Thermo). The ARTD activity was normalized for each individual substrate.
FL-ARTD11 Activity Assay
Reactions containing either 0.2 μM NXF1, 0.2 μM FL-ARTD11 (recombinantly expressed and purified from e. coli), or both were performed in assay buffer (50 mM Tris-HCL 7.5, 50 mM NaCl, 2 mM MgCl2) at rt using a mixture of cold NAD+ (5 μM) and [32P]-NAD+ (2 μCi per reaction). After 30 min reactions were stopped by adding SDS-PAGE loading buffer and boiled at 80°C for 5 min. All reactions were resolved on 4–12% SDS–PAGE gels. ADP-ribosylated proteins were visualized by autoradiography.
NeutrAvidin Enrichment and LC-MS/MS Analysis
1 mg of total protein from either HEK 293T or HeLa lysate from cells expressing WT- or IG-tagged ARTD variants was incubated with 100 μM 5-Bn-6-a-NAD+ for 2 hours at 30°C, click conjugated to biotin-PEG3-azide, subjected to enrichment using NeutrAvidin agarose (Pierce), and proteolysis as previously described (Carter-O’Connell and Cohen, 2015; Carter-O’Connell et al., 2014). MS experiments were performed using an Orbitrap Fusion (Thermo) equipped with a capillary HPLC system. MS processing and analysis thresholds are discussed in supplemental experimental procedures.
Supplementary Material
Acknowledgments
We thank John Klimek for assistance with the MS analysis at the OHSU proteomics core, Jeffrey Huang for assistance with immunofluorescence microscopy, and P. Chang for the GFP-ARTD10 construct. We thank the members of the Cohen lab for many helpful discussions regarding the manuscript and experimental design. This work was funded by the National Institutes of Health (NIH 1R01NS088629) to M.S.C. and I.C.O. was supported by a Postdoctoral Fellowship (PF-15-008-01-CDD) from the American Cancer Society. The authors declare no competing financial interests.
Footnotes
ACCESSION NUMBERS
Raw and processed MS data files are available at peptideatlas.org (ID: PASS00764).
AUTHOR CONTRIBUTIONS
I.C.O. and M.S.C. designed the experiments and I.C.O. conducted the experiments with the exceptions of the LC-MS/MS setup, which was performed by L.L.D., and the radioactive FL-ARTD11 assay, which was performed by R.K. and I.A. MS analysis was performed by I.C.O. in the OHSU proteomics core. R.K.M. and H.J. synthesized the modified NAD+ analogues. I.C.O. prepared the manuscript with guidance from M.S.C.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. BioEssays : news and reviews in molecular, cellular and developmental biology. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Carter-O’Connell I, Cohen MS. Identifying Direct Protein Targets of Poly-ADP-Ribose Polymerases (PARPs) Using Engineered PARP Variants—Orthogonal Nicotinamide Adenine Dinucleotide (NAD+) Analog Pairs. Current protocols in chemical biology. 2015;7:121–139. doi: 10.1002/9780470559277.ch140259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-O’Connell I, Jin H, Morgan RK, David LL, Cohen MS. Engineering the substrate specificity of ADP-ribosyltransferases for identifying direct protein targets. Journal of the American Chemical Society. 2014;136:5201–5204. doi: 10.1021/ja412897a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosi C, Marien M. Implication of poly (ADP-ribose) polymerase (PARP) in neurodegeneration and brain energy metabolism. Decreases in mouse brain NAD+ and ATP caused by MPTP are prevented by the PARP inhibitor benzamide. Annals of the New York Academy of Sciences. 1999;890:227–239. doi: 10.1111/j.1749-6632.1999.tb07998.x. [DOI] [PubMed] [Google Scholar]
- Daniels CM, Ong SE, Leung AK. Phosphoproteomic approach to characterize protein mono- and poly(ADP-ribosyl)ation sites from cells. Journal of proteome research. 2014;13:3510–3522. doi: 10.1021/pr401032q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feijs KL, Kleine H, Braczynski A, Forst AH, Herzog N, Verheugd P, Linzen U, Kremmer E, Luscher B. ARTD10 substrate identification on protein microarrays: regulation of GSK3beta by mono-ADP-ribosylation. Cell communication and signaling : CCS. 2013;11:5. doi: 10.1186/1478-811X-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Molecular biology of the cell. 2002;13:1282–1297. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Xu S, Powers MA. Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Molecular biology of the cell. 2003;14:600–610. doi: 10.1091/mbc.E02-09-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haikarainen T, Venkannagari H, Narwal M, Obaji E, Lee HW, Nkizinkiko Y, Lehtio L. Structural basis and selectivity of tankyrase inhibition by a Wnt signaling inhibitor WIKI4. PloS one. 2013;8:e65404. doi: 10.1371/journal.pone.0065404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends in biochemical sciences. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Hu B, Wu Z, Hergert P, Henke CA, Bitterman PB, Phan SH. Regulation of myofibroblast differentiation by poly(ADP-ribose) polymerase 1. The American journal of pathology. 2013;182:71–83. doi: 10.1016/j.ajpath.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Kim JH, Frizzell KM, Kraus WL, Lin H. Clickable NAD analogues for labeling substrate proteins of poly(ADP-ribose) polymerases. Journal of the American Chemical Society. 2010;132:9363–9372. doi: 10.1021/ja101588r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmichel S, Rosenthal F, Altmeyer M, Lukas J, Hottiger MO, Nielsen ML. Proteome-wide identification of poly(ADP-Ribosyl)ation targets in different genotoxic stress responses. Molecular cell. 2013;52:272–285. doi: 10.1016/j.molcel.2013.08.026. [DOI] [PubMed] [Google Scholar]
- Kang Y, Cullen BR. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes & development. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H, Herrmann A, Lamark T, Forst AH, Verheugd P, Luscher-Firzlaff J, Lippok B, Feijs KL, Herzog N, Kremmer E, et al. Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell communication and signaling : CCS. 2012;10:28. doi: 10.1186/1478-811X-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani M, Fujimori H. Poly(ADP-ribosyl)ation in carcinogenesis. Molecular aspects of medicine. 2013;34:1202–1216. doi: 10.1016/j.mam.2013.05.003. [DOI] [PubMed] [Google Scholar]
- Meyer-Ficca ML, Ihara M, Bader JJ, Leu NA, Beneke S, Meyer RG. Spermatid head elongation with normal nuclear shaping requires ADP-ribosyltransferase PARP11 (ARTD11) in mice. Biology of reproduction. 2015;92:80. doi: 10.1095/biolreprod.114.123661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RK, Cohen MS. A Clickable Aminooxy Probe for Monitoring Cellular ADP-Ribosylation. ACS chemical biology. 2015;10:1778–1784. doi: 10.1021/acschembio.5b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natalizio BJ, Wente SR. Postage for the messenger: designating routes for nuclear mRNA export. Trends in cell biology. 2013;23:365–373. doi: 10.1016/j.tcb.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf A, de Murcia G, Schulz GE. Inhibitor and NAD+ binding to poly(ADP-ribose) polymerase as derived from crystal structures and homology modeling. Biochemistry. 1998;37:3893–3900. doi: 10.1021/bi972383s. [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nature reviews Molecular cell biology. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- Strosznajder RP, Jesko H, Zambrzycka A. Poly(ADP-ribose) polymerase: the nuclear target in signal transduction and its role in brain ischemia-reperfusion injury. Molecular neurobiology. 2005;31:149–167. doi: 10.1385/MN:31:1-3:149. [DOI] [PubMed] [Google Scholar]
- Verheugd P, Forst AH, Milke L, Herzog N, Feijs KL, Kremmer E, Kleine H, Luscher B. Regulation of NF-kappaB signalling by the mono-ADP-ribosyltransferase ARTD10. Nature communications. 2013;4:1683. doi: 10.1038/ncomms2672. [DOI] [PubMed] [Google Scholar]
- Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, Chang P. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nature communications. 2014;5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Yao J, DuMond ME, Krishnakumar R, Ruhl DD, Ryu KW, Gamble MJ, Kraus WL. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. The Journal of biological chemistry. 2012;287:12405–12416. doi: 10.1074/jbc.M111.304469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nature methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.