Abstract
Toll-like receptor (TLR) activation contributes to premalignant hematologic conditions, such as myelodysplastic syndromes (MDS). TRAF6, a TLR-effector with ubiquitin (Ub) ligase activity, is overexpressed in MDS hematopoietic stem/progenitor cells (HSPC). Here we show that TRAF6 overexpression in mouse HSPC resulted in impaired hematopoiesis and bone marrow failure. Through the use of a global Ub screen, we identified hnRNPA1, an RNA-binding protein and auxiliary splicing factor, as a substrate of TRAF6. TRAF6 ubiquitination of hnRNPA1 regulated alternative splicing of Arhgap1, which resulted in Cdc42 activation and accounted for hematopoietic defects in TRAF6-expressing HSPC. These results implicate Ub signaling in coordinating RNA processing by TLR pathways during an immune response and in premalignant hematologic diseases, such as MDS.
TLR regulate myelopoiesis and innate immune responses, but chronic stimulation of TLR can lead to HSPC dysfunction1. Most work has described TLR pathways in immune effector cells, and more recently, in coordinating RNA processing during hematopoietic differentiation2,3. TLRs and their coreceptors are expressed on hematopoietic stem cell (HSC) and important for HSPC function4. Chronic TLR stimulation, such as by lipopolysaccharide (LPS) or during sepsis, induces durable changes in bone marrow (BM) physiology, including reduced HSC self-renewal, myeloid-biased differentiation, and neutropenia1. Thus it is not surprising that chronic TLR signaling is implicated in mediating HSC defects and contributing to MDS5–7. MDS are clonal HSC disorders defined by cytopenia, myeloid cell dysplasia, and ineffective hematopoiesis8,9. Multiple genetic mechanisms contribute to TLR pathway activation in MDS5,6,10–13, which converge on the central mediator of innate immune and TNF superfamily receptor signaling, TRAF6. For example, deletion of miR-146a and/or TIFAB, two suppressors of TRAF6 associated with del(5q) MDS result in TRAF6 overexpression5,14. TRAF6 is an E3 ligase that synthesizes lysine (K) 63-linked Ub chains on substrates leading to downstream pathway activation. Despite the association between chronic TLR signaling, diminished HSC fitness, and MDS pathogenesis, the underlying cellular and molecular mechanisms remain unknown. Here we used a hematopoietic-specific TRAF6 overexpression mouse model to describe a mechanism of RNA processing involving ubiquitination of hnRNPA1, an RNA binding protein, by TRAF6, which under conditions of chronic TRAF6 signaling alters RNA splicing and cell-intrinsic HSC defects associated with MDS.
Results
Overexpression of TRAF6 impairs HSC function
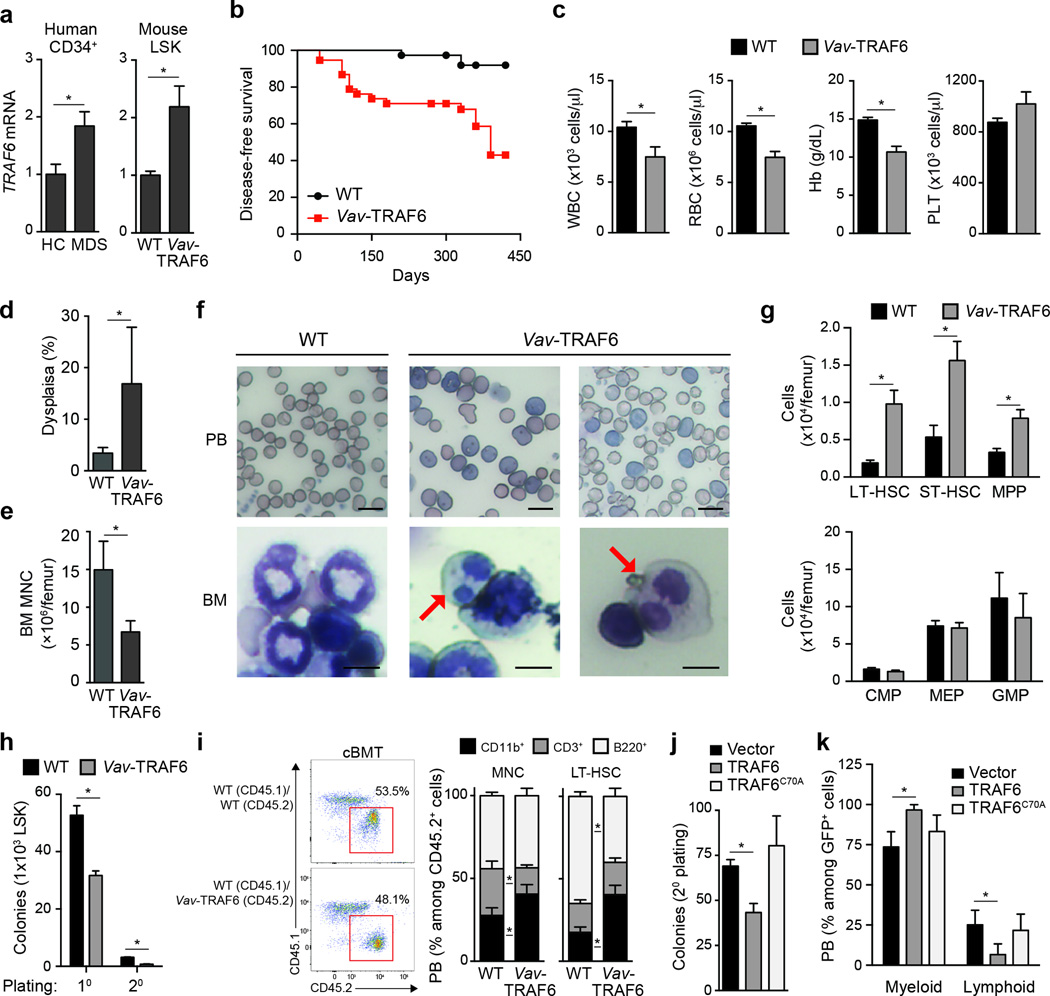
TRAF6 is overexpressed in ~40% of normal karyotype or del(5q) CD34+ cells from MDS patients as compared to CD34+ cells from healthy individuals (Fig. 1a). To model TRAF6 overexpression and genetically-driven chronic TLR activation observed in MDS HSPC, we generated transgenic mice that overexpress TRAF6 under the control of hematopoietic-specific Vav regulatory elements (called Vav-TRAF6 mice hereafter)15,16 (Supplementary Fig. 1a–c). To avoid supraphysiological expression of TRAF6, we selected transgenic mice in which TRAF6 expression was 2-fold higher as compared to endogenous TRAF6 in wild-type HSPC (LSK, Lineage−Sca1+cKit+), and consistent with TRAF6 overexpression in MDS CD34+ cells (Fig. 1a). Beginning at ~5 months, Vav-TRAF6 mice succumbed to a BM failure-like disease, with 50% lethality beyond 15 months (Fig. 1b). Moribund Vav-TRAF6 mice developed leukopenia and anemia, neutrophil dysplasia and reduced BM cellularity (Fig. 1c–f), and exhibited enlarged spleens due to extramedulary erythro- and myelopoiesis (Supplementary Fig. 1d–e). Despite the BM failure, the number of CD34−CD135−LSK (LT-HSC), CD34+CD135−LSK (ST-HSC) and CD34+CD135+LSK (MPP) were elevated in the BM of Vav-TRAF6 mice compared to wild-type mice (Fig. 1g). In methylcellulose assays, Vav-TRAF6 LSK formed fewer colonies compared to wild-type LSK (Fig. 1h). To assess the consequences of TRAF6 overexpression on HSC function in vivo, we competitively transplanted BM mononuclear cells or LT-HSC (CD34−CD135−LSK) from Vav-TRAF6 or wild-type donor mice with equal numbers of wild-type BM cells into lethally-irradiated congenic wild-type recipients. The hematopoietic contribution of Vav-TRAF6 BM mononuclear cells to peripheral blood chimerism in primary recipients was moderately reduced compared to wild-type donor BM mononuclear cells or LT-HSC HSC (Supplementary Fig. 2a,b). Similarly, hematopoietic contribution of Vav-TRAF6 LT-HSC to peripheral blood chimerism following serial transplantation of BM cells in secondary wild-type recipients was reduced compared to wild-type donor LT-HSC (Supplementary Fig. 2a,b). Reduced peripheral blood chimerism of Vav-TRAF6 BM mononuclear cells- or LT-HSC-derived progeny in recipient mice was mainly due to diminished numbers of CD3+ and B220+ lymphoid cells as compared to lymphoid cells in recipient mice reconstituted with wild-type BM mononuclear cells or LT-HSC (Supplementary Fig. 2c). However, BM chimersim of CD34−CD135−LSK (LT-HSC), CD34+CD135−LSK (ST-HSC) and CD34+CD135+LSK (MPP) was similar between Vav-TRAF6 and wild-type donor BM mononuclear cells or LT-HSC (Supplementary Fig. 2d). We next examined mature immune cells in recipient mice competitively transplanted with BM mononuclear cells or LT-HSC (CD34−CD135−LSK) from Vav-TRAF6 or wild-type donor mice and equal number of wild-type BM cells. Peripheral blood CD11b+ myeloid populations were expanded at the expense of CD3+ and B220+ lymphoid populations within CD45.2+ donor-derived Vav-TRAF6 BM mononuclear cells (from 27% to 41%) or LT-HSC (from 18% to 40%) when compared to either wild-type donor-derived BM mononuclear cells or LT-HSC cells, or to CD45.1+ competitor-derived BM mononuclear cells from wild-type mice after 12 weeks (Fig. 1i, Supplementary Fig. 2e). These observations suggest that TRAF6 overexpression results in HSPC defects that are cell-intrinsic and associated with myeloid-biased differentiation.
Figure 1. Overexpression of TRAF6 results in hematopoietic stem/progenitor cell defects.
(a) TRAF6 mRNA in CD34+ BM cells from low-risk MDS patients (n = 36) and age-matched healthy controls (HC; n = 7) (left), and LSK cells from Vav-TRAF6 (n = 6) and WT FVB/NJ (n = 4) mice (primers are designed to measure both endogenous mouse Traf6 and ectopic human TRAF6). P = 0.02. (b) Disease-free survival of Vav-TRAF6 (n = 60) and WT FVB/NJ mice (n = 40) *, P = 0.009. (c) Complete blood counts for WT FVB/NJ (n = 33) and moribund Vav-TRAF6 (n = 18) mice. *. P < 0.01. (d) BM neutrophil dysplasia in age-matched WT (n = 3) and Vav-TRAF6 (n = 5) FVB/NJ mice. At least 100 cells were examined for each mouse. *, P = 0.04. (e) BM mononuclear cell counts from WT (n = 3) and Vav-TRAF6 FVB/NJ mice (n = 4). *, P = 0.004. (f) Representative Wright-Giemsa-stained peripheral blood smears (PB) and bone marrow cytospins (BM) from WT (FVB/NJ) and 2 moribund Vav-TRAF6 mice. The red arrow indicates dysplastic myeloid cells with Pseudo-Pelger Hüet anomaly. Scale bars, 10 µm (top) and 7.5 µm (bottom) (g) Number of long-term hematopoietic stem cells (LT-HSC, P = 0.0056), short-term HSC (ST-HSC, P = 0.0011), multipotent progenitors (MPP, P = 0.014), common myeloid progenitors (CMP), megakaryocyte-erythroid progenitor (MEP), and granulocyte-monocytic progenitor (GMP) in the BM of WT (C57Bl/6) (n = 4) and Vav-TRAF6 (n = 4) ~6 month old mice. (h) Colony numbers were determined after primary and secondary plating LSK BM cells from WT (FVB/NJ; n = 3) and Vav-TRAF6 (n = 3) mice in methylcellulose. *, P < 0.001. (i) FACS analysis of WT (C57Bl/6) or Vav-TRAF6 donor-derived (CD45.2+) and competitor-derived (CD45.1+) mononuclear cells from peripheral blood 12 weeks post competitive transplantation (left panel). Proportion of myeloid (CD11b+) and lymphoid (CD3+ and B220+) cells within the donor-derived compartment (gated on CD45.2+) after competitive transplantation with either BM mononuclear cells (MNC, left) or LT-HSC (right). (n = 8 mice per genotype) (right panel). *, P < 0.05. (j) Colony formation in methylcellulose after serial replating of GFP-sorted Lin− BM cells (C57Bl/6) transduced with retroviral vectors (MSCV-IRES-GFP) encoding WT (n = 3) or an E3 ligase-mutant TRAF6 (TRAF6C70A, n = 3). *, P < 0.01. (k) Proportion of donor-derived (gated on CD45.2+;GFP+) peripheral blood myeloid (Cd11b+) and lymphoid (CD3+ and B220+) cells were examined by FACS for competitively transplanted GFP-sorted Lin− BM cells (C57Bl/6) transduced with retroviral vectors encoding WT (n = 5) or an E3 ligase-mutant TRAF6 (TRAF6C70A, n = 10) co-expressing GFP (MSCV-IRES-GFP). *, P < 0.05. Data are from three experiments (a, c–g; means and s.e.m.) or from two experiments (h–k; means and s.e.m.).
To assess if the Ub ligase activity of TRAF6 is required for the observed hematopoietic phenotype, we retrovirally expressed wild-type (TRAF6-LSK) and E3 ligase-defective TRAF6 (TRAF6-C70A-LSK) in BM LSK, which express low levels of endogenous Traf6, and examined hematopoietic progenitor colony formation in methylcellulose. TRAF6-LSK formed fewer total colonies upon serial replating as compared to vector-transduced LSK (Fig. 1j). In contrast, TRAF6-C70A-LSK cells formed similar number of colonies as vector-transduced LSK cells (Fig. 1j). We next examined the differentiation potential of TRAF6-LSK and TRAF6-C70A-LSK following competitive transplantation with an equal number of wild-type unfractionated BM cells into lethally-irradiated congenic recipients. Peripheral blood CD11b+ myeloid populations were significantly expanded at the expense of CD3+ and B220+ lymphoid populations within GFP+ donor-derived TRAF6-LSK cells compared to donor-derived TRAF6-C70A-LSK or vector-transduced LSK cells after 8 weeks (Fig. 1k). Thus, HSPC expressing the TRAF6 transgene were functionally impaired and showed a myeloid bias in vivo that was dependent on the Ub ligase activity of TRAF6.
RNA binding proteins are TRAF6 substrates
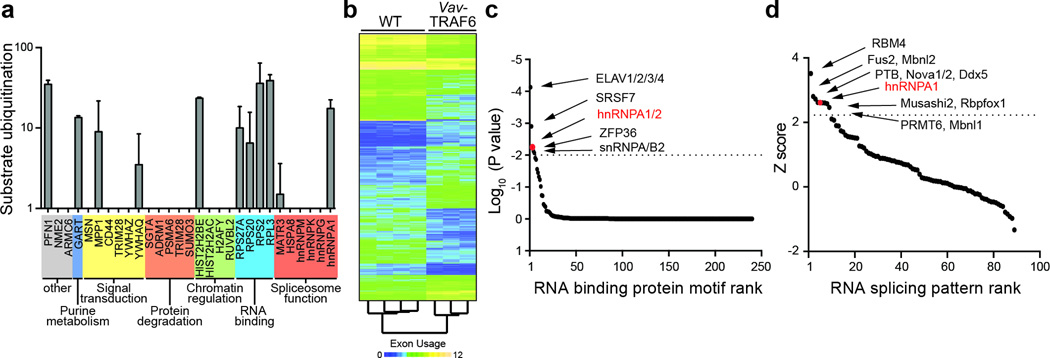
To determine the molecular pathways regulated by chronic TRAF6 overexpression, we performed microarray gene expression analysis in LSK sorted from the BM of 6 month-old Vav-TRAF6 and wild-type mice. 190 genes were differentially expressed in Vav-TRAF6 LSK compared to wild-type LSK (Supplementary Table 1, Supplementary Fig. 3a). Gene set enrichment analysis revealed that Vav-TRAF6 LSK have downregulated genes characteristically expressed by quiescent LT-HSC17, such as Msi2, Esr1, and Bcl2l11, and upregulated expression profiles associated with mature hematopoietic cells and myeloid differentiation (Supplementary Fig. 3b), including genes encoding Myc, Mpo, E2f8 and Ccr2 (Supplementary Fig. 3a). Although NF-κB and immune signaling is associated with normal TRAF6 function, gene expression profiles related to immune and inflammatory responses were not upregulated in Vav-TRAF6 LSK compared to wild-type LSK (Supplementary Fig. 3b). The amount of phosphorylated IKKβ was similar in Vav-TRAF6 and wild-type Lin- BM cells (not shown), cytokines, such as Il1a, Il2, Il3, Ifng, and Il6 were comparable in the peripheral blood of Vav-TRAF6 and wild-type mice (not shown), and inflammation or immune cell infiltration was not detected in moribund Vav-TRAF6 mice (not shown), indicating that the hematopoietic phenotype in Vav-TRAF6 mice was not associated with activation of canonical NF-κB signaling and/or inflammation. As such, we broadened our mechanistic investigation by searching for ubiquitinated substrates of TRAF6. For this, we performed global semi-quantitative Ub capture proteomics in a TRAF6-overexpressing human leukemia cell line (TF1) engineered to express a doxycycline-inducible shRNA targeting TRAF6 (Fig. 2a). Ubiquitinated peptides immunoprecipitated from doxycycline-treated TF1 cells to induce TRAF6 knockdown or from vehicle-treated TF1 cells as controls were analyzed by mass spectrometry (Supplementary Fig. 4a). The proteomic analysis identified 40 unique peptides from 29 proteins that were ubiquitinated in vehicle-treated TF1 cells, but not in doxycycline-treated TRAF6-knockdown TF1 cells (Fig 2a, Supplementary Fig. 4b, Supplementary Table 2). We performed ubiquitiation assays with recombinant ubiquitin activating enzyme E1 (UBE1), E2 ubiquitin conjugating enzyme Ubc13 (UBE2N/UBE2V1), TRAF6 and Ub with the 29 putative protein substrates. Eleven of the 29 candidate proteins, hnRNPA1, MATR3, RPL3, RPS2, RPS20, RPS27A, HIST2H2BE, YWHAQ, MPP1, GART, and PFN1 were ubiquitinated by TRAF6 in at least one replicate assay (Fig. 2a, Supplementary Table 2). Gene ontology analysis indicated that 6 of the 11 TRAF6 substrates, hnRNPA1, MATR3, RPL3, RPS2, RPS20 and RPS27A represented RNA-binding proteins (Fig. 2a, Supplementary Table 2). Thus, we conclude that TRAF6 overexpression resulted in transcriptional changes and ubiquitination of RNA-binding proteins in HSPC.
Figure 2. TRAF6 regulates ubiquitination of RNA binding proteins and RNA splicing.
(a) In vitro Ub reconstitution assays were performed on the identified candidates from the cell-based screen with purified TRAF6 (n = 29). Shown is the substrate ubiquitination in the presence of TRAF6 (F532 CyDye) as compared to without TRAF6 (B532 CyDye) for 2 independent replicates. (b) Exon-level microarray expression analysis of LSK from 6 month WT (FVB/NJ, n = 4) and Vav-TRAF6 mice (n = 3). (c) Motif enrichment in differentially spliced exons from Vav-TRAF6 LSK as compared to WT LSK in the form of position frequency matrix motifs from the CisBP-RNA database. Each point represents one motif. Motifs are sorted left to right in decreasing order of significance. Motifs enriched in Vav-TRAF6 LSK are above the horizontal dotted line. hnRNPA1, P = 9.7x10−5. (d) Enrichment scores for exon usage patterns for Vav-TRAF6 LSK as compared to 46 known splicing factor exon usage patterns. Each point represents the enrichment Z-score for an individual splicing pattern of the splicing factor patterns examined. Exon usage patterns enriched in Vav-TRAF6 LSK are above the horizontal dotted line. hnRNPA1, P = 0.017. Data are from two experiments (a; means and s.d.).
TRAF6 regulates RNA splicing via hnRNPA1
RNA-binding proteins have pleotropic effects on RNA stability, splicing and translation. To explore whether ubiquitination of RNA-binding proteins by TRAF6 results in altered RNA splicing, we performed a microarray exon expression analysis in LSK cells sorted from Vav-TRAF6 and wild-type mice. Two splicing metrics (Splicing Index and FIRMA), which calculate the relative expression of individual exons normalized to all exons within the corresponding gene18,19, were implemented. Three hundred sixteen genes exhibited changes in exon usage in Vav-TRAF6 LSK as compared to wild-type LSK (n = 521 exons using the Splicing Index; n = 1919 exons using the FIRM Index) (Fig. 2b, Supplementary Table 3). A subset of exons with altered usage in Vav-TRAF6 LSK was validated by RT-PCR. Among the 10 exons selected, 6 were alternatively spliced in independent cohorts of Vav-TRAF6 LSK as compared to wild-type LSK: Mllt1 exon 2, Cep164 exon 7, Mtmr14 exon 18, Slmap exon 10, Sema4g exon 13 and Arhgap1 exon 2 (Supplementary Fig. 5). To identify the mechanistic link between the ubiquitinated RNA-binding proteins and altered RNA exon usage in Vav-TRAF6 HSPC, we performed RNA-binding motif- and RNA-splicing pattern-enrichment analyses of alternatively-spliced cassette exons in sorted Vav-TRAF6 LSK. For the motif-enrichment analysis, we examined RNA-binding models in the form of position frequency matrix motifs in the CisBP-RNA database to identify over-representation of RNA-protein binding sites within alternatively-spliced cassette exons in Vav-TRAF6 LSK20. Significant enrichment of binding motifs for ELAV1/2/3/4, SRSF7, hnRNPA1, ZFP36 and snRNPA/B2 were found in the alternatively spliced exon sequences in Vav-TRAF6 LSK as compared to wild-type LSK (Fig. 2c, Supplementary Table 4). These findings were independently confirmed using a hypergeometric test, which measures consensus-based binding site enrichment (not shown). We next performed an RNA splicing signature analysis to compare exon-usage patterns of Vav-TRAF6 LSK with the RNA exon splicing patterns associated with RNA-binding proteins from published data sets. Exon-usage patterns associated with 11 RNA-binding proteins, namely RBM4, Fus2, Mbnl2, PTB, Nova1/2, Ddx5, hnRNPA1, Musashi2, Rbfox1, PRMT6 and Mbnl1 were significantly enriched in Vav-TRAF6 LSK compared to wild-type LSK (Fig. 2d). hnRNPA1 emerged as the only RNA-binding protein that was ubiquitinated by TRAF6 whose binding motifs and global RNA splicing pattern were significantly enriched in LSK from Vav-TRAF6 mice. Thus, TRAF6 overexpression in HSPC results in RNA splicing changes associated with hnRNPA1.
TRAF6 directly ubiquitinates hnRNPA1
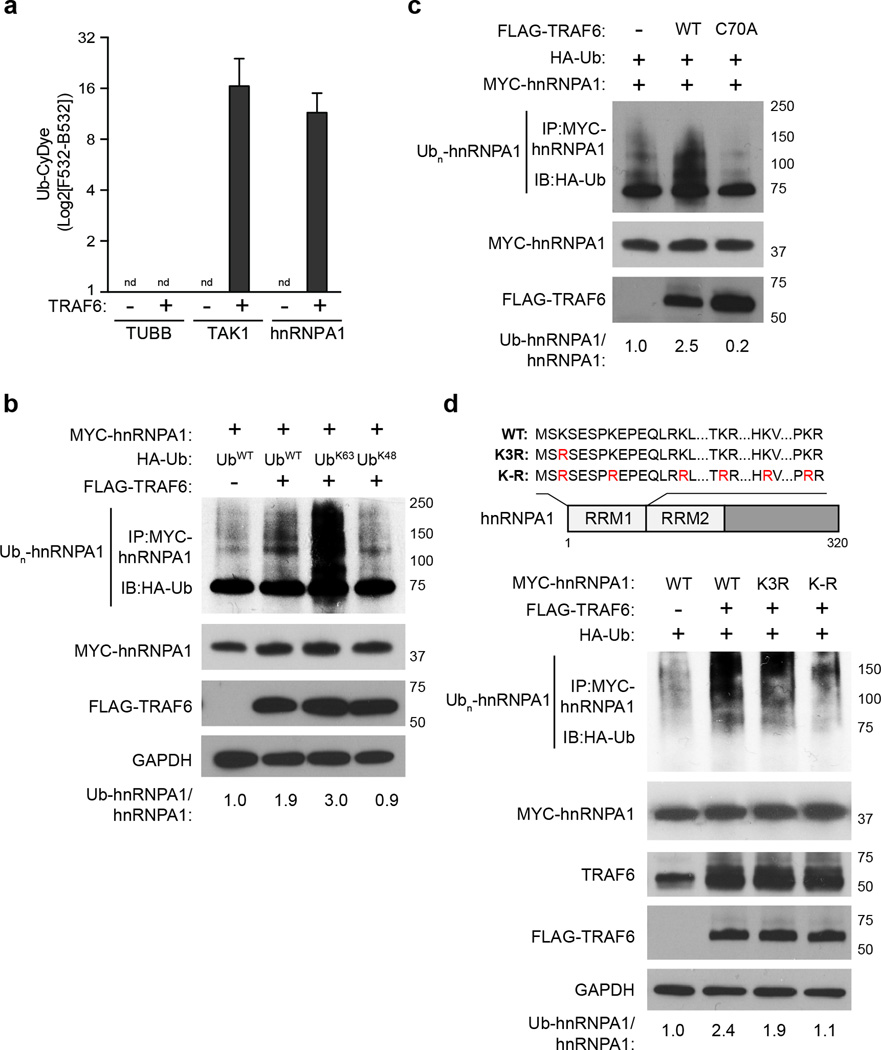
Because hnRNPA1 has been implicated in human myeloid malignancies21, we selected it for further validation. Recombinant protein ubiquitination assays showed that hnRNPA1 was ubiquitinated by TRAF6 to a similar magnitude as TAK1, a known TRAF6 substrate22 (Fig. 3a), indicating that hnRNPA1 is a direct substrate of TRAF6. Doxycycline-treated TRAF6-knockdown TF1 cells showed reduced hnRNPA1 ubiquitination compared to vehicle-treated TF1 cells (Supplementary Fig. 4c). Conversely, transfection of TRAF6 in HEK293 cells resulted in increased hnRNPA1 ubiquitination in the presence of wild-type Ub or Ub that only forms K63-linkages (UbK63) compared to HEK293 cells transfected with empty vector (Fig. 3b, Supplementary Fig. 4d). In contrast, expression of TRAF6 together with Ub that only forms K48-linkages (UbK48) did not result in hnRNPA1 ubiquitination in HEK293 cells (Fig. 3b). Compared to wild-type TRAF6, transfection of TRAF6-C70A in HEK293 cells did not result in ubiquitination of hnRNPA1 (Fig. 3c), indicating that the E3 ligase domain of TRAF6 was required for hnRNPA1 ubiquitination.
Figure 3. TRAF6 ubiquitinates hnRNPA1.
(a) In vitro ubiquitin reconstitution assay performed in duplicate and analyzed by CyeDye fluorescence (F532 subtracted from background, B532) for hnRNPA1, TAK1, and Tubulin (TUBB; a negative control). Individual assays contained recombinant Ub, UBE1, Ubc13/UBE2V1, ATP, and the indicated substrates. nd, not detectable. (b) HEK293 cells transfected with MYC-hnRNPA1, FLAG-TRAF6, and either WT HA-Ub or mutant Ub that form only K63- (UbK63) or K48- (UbK48) linkages were immunoblotted for HA (Ub) on IP MYC-hnRNPA1. (c) HEK293 cells transfected with MYC-hnRNPA1, and either WT or E3 ligase defective (C70A) TRAF6 were IB for IP MYC-hnRNPA1 (anti HA-Ub). (d) Schematic representation of hnRNPA1 protein, and corresponding lysine-to-arginine mutations within the RNA recognition motif (RRM) (above image). HEK293 cells transfected with WT, Lys3-mutant hnRNPA1 (K3R), or hnRNPA1 with mutation of the lysines within the first RRM domain (K–R) were immunoprecipitated (IP) for MYC-hnRNPA1 and then immunoblotted for HA (Ub). Data are from two experiments (a; means and s.d.).
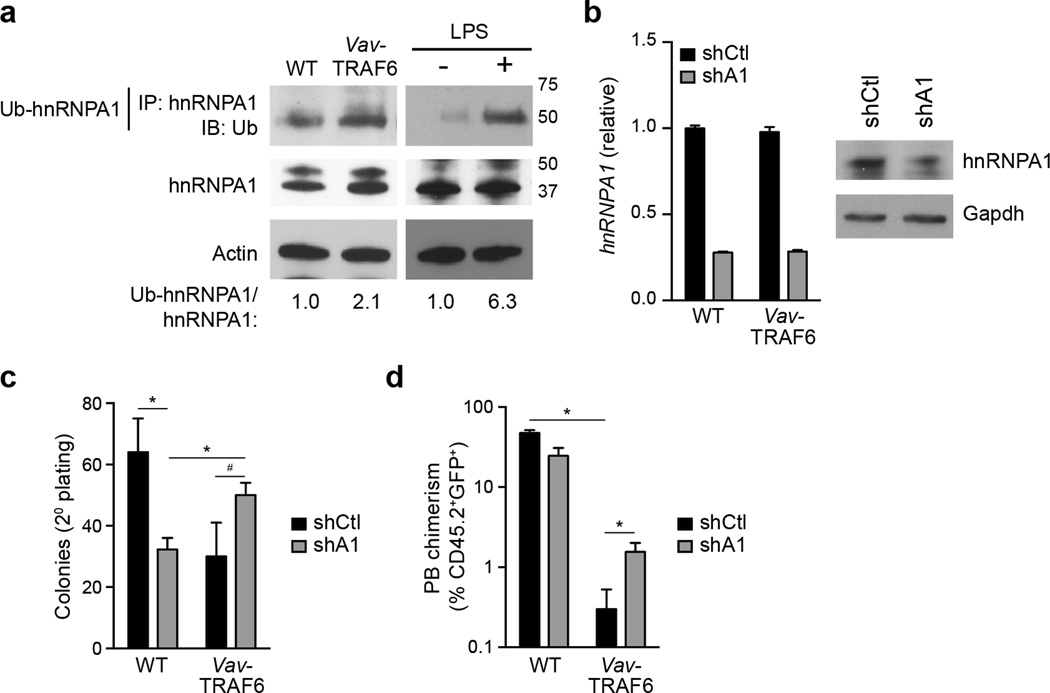
The proteomic analyses indicated that hnRNPA1 was ubiquitinated on Lys3 (K3) (Fig. 3d), which is adjacent to the first RNA-recognition motif (RRM1) of hnRNPA123. To determine whether hnRNPA1 RRM1 is ubiquitinated by TRAF6, we transfected TRAF6 with a mutant hnRNPA1 protein, in which Lys3 was substituted for arginine (hnRNPA1K3R) in HEK293 cells. Compared to wild-type hnRNPA1, ubiquitination of hnRNPA1K3R by TRAF6 was modestly reduced (Fig. 3d), suggesting additional Lys residues in hnRNPA1 can be ubiquitinated by TRAF6. Because ubiquitination of a substrate motif can be promiscuous, resulting in modification of adjacent Lys residues, we additionally mutated the 5 Lys residues within RRM1 adjacent to Lys3 (hnRNPA1K-R hereafter). Transfection of TRAF6 with hnRNPA1K-R in HEK293 cells resulted in further reduction in hnRNPA1 ubiquitination as compared to ubiquitination of wild-type hnRNPA1 or hnRNPA1K3R by TRAF6 (Fig. 3d), indicating that hnRNPA1 ubiquitination by TRAF6 primarily occurs at RRM1. We next examined the ubiquitination of hnRNPA1 by TRAF6 in primary hematopoietic stem and progenitor Lin− BM cells. Immunoprecipitation of endogenous hnRNPA1 indicated that ubiquitination of hnRNPA1 was increased in Vav-TRAF6 Lin− BM cells or in LPS-stimulated Lin− BM cells compared to wild-type Lin− BM cells (Fig. 4a). We next used shRNAs to knock down hnRNPA1 in wild-type and Vav-TRAF6 Lin− BM cells (Fig. 4b). Knockdown of hnRNPA1 in Vav-TRAF6 Lin− BM cells (shA1-Vav-TRAF6) resulted in modestly increased total colony formation after serial replating compared to Vav-TRAF6 Lin− BM cells expressing a non-targeting shRNA (shCtl-Vav-TRAF6)(Fig. 4c). Next, shA1 or shCtl Vav-TRAF6 and wild-type Lin− BM cells were competitively transplanted with wild-type BM mononuclear cells into lethally-irradiated wild-type mice. shA1-Vav-TRAF6 Lin− BM showed improved engraftment of peripheral blood CD45.2+GFP+ cells from 0.3% to 1.6% as compared to shCtl-Vav-TRAF6 Lin− BM cells, while shA1-wild-type Lin- BM cells showed reduced engraftment as compared to shCtl-wild-type Lin− BM cells (Fig. 4d). These findings suggest that hnRNPA1 was ubiquitinated by TRAF6 and this interaction played a role in the hematopoietic phenotype of Vav-TRAF6 mice.
Figure 4. hnRNPA1 contributes to the hematopoietic phenotype in Vav TRAF6 mice.
(a) Immunoprecipitation and immunobloting of ubiquitinated hnRNPA1 from WT (FVB/NJ) and Vav-TRAF6 Lin− BM cells and LPS-treated WT (C57Bl/6) (Traf6+/+) and TRAF6-deficient (Traf6−/−) Lin− BM cells. (b) hnRNPA1 mRNA and protein expression was determined following expression of shRNAs targeting hnRNPA1 (shA1) or non-targeting shRNA (shCtl). (c) Colony formation in methylcellulose after serial replating of GFP-sorted WT (FVB/NJ, n = 3) and Vav-TRAF6 (n = 3) Lin− BM cells transduced with shRNAs targeting hnRNPA1 (shA1) or control shRNA (shCtl) co-expressing GFP. *, P < 0.05; #, P = 0.08. (d) Chimerism of donor-derived (CD45.2+GFP+) peripheral blood cells from lethally-irradiated recipient mice receiving Lin− BM cells transduced with shRNAs targeting hnRNPA1 (shA1) or control shRNA (shCtl) co-expressing GFP competitively transplanted with wild-type (CD45.1+) total BM cells (n = 8 per genotype). *, P < 0.05. Data are from two experiments (b–d; means and s.e.m.).
TRAF6 induces splicing of Arhgap1 RNA
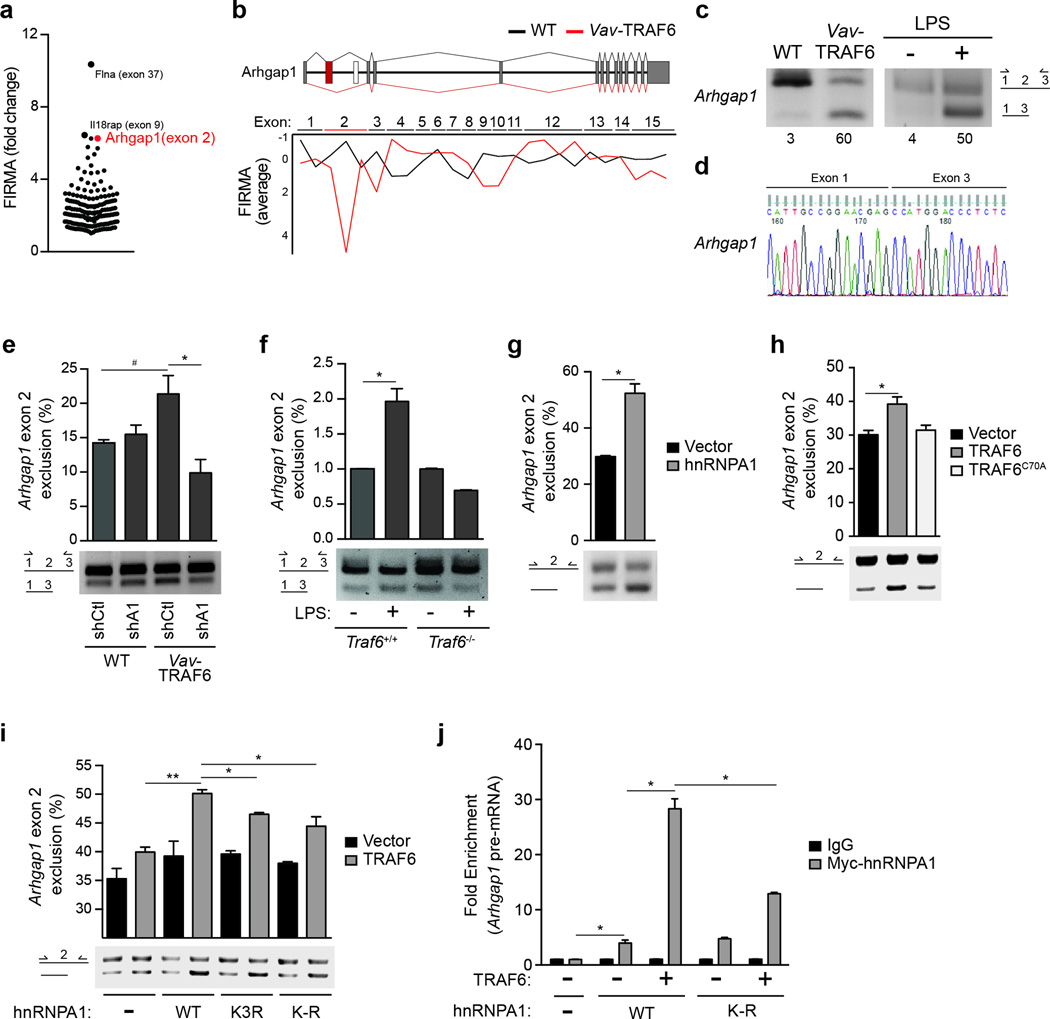
hnRNPA1 regulates RNA splicing by directly binding RNA consensus sites24–26 or indirectly through protein-RNA interactions27. To identify alternatively spliced genes implicated in the hematopoietic phenotype of Vav-TRAF6 mice we focused on the most significantly altered exons in Vav-TRAF6 LSK, as revealed by the exon expression analysis. The top genes with differential exon usage included Flna, Il18rap, Arhgap1, Mcm6, Itpr1, Laptm5, Celf1 and Fbxw2 (Fig. 5a, Supplementary Table 3). Arhgap1 (or Cdc42GAP), an inhibitor of Cdc42 by hydrolysing GTP, was previously implicated in LT-HSC self-renewal and differentiation28–30 and as such emerged as the most relevant candidate. Exon 2 of Arhgap1 was excluded in Vav-TRAF6 LSK, but not wild-type LSK (Fig. 5b). RT-PCR using primers flanking Arhgap1 exon 2 and Sanger sequencing showed exclusion of exon 2 in Vav-TRAF6 Lin− BM cells and in LPS-stimulated wild-type Lin− BM cells (Fig. 5c,d). To establish whether hnRNPA1 is required for TRAF6-induced exclusion of Arhgap1 exon 2, Vav-TRAF6 or wild-type Lin− BM cells were transduced with hnRNPA1-shRNA (shA1) or a non-targeting-shRNA (shCtl). Knockdown of hnRNPA1 in Vav-TRAF6 Lin− BM cells significantly reduced Arhgap1 exon 2 exclusion compared to sh-Ctl-Vav-TRAF6 Lin− BM cells (Fig. 5e). We also investigated Arhgap1 exon 2 exclusion in wild-type and Traf6−/− Lin− BM cells following LPS stimulation. LPS treatment resulted in exclusion of Arhgap1 exon 2 in wild-type Lin− BM cells but not in Traf6−/− Lin− BM cells (Fig. 5f), suggesting that Arghap1 exon 2 exclusion requires TRAF6.
Figure 5. TRAF6 regulates RNA splicing of Arhgap1.
(a) Significantly excluded or included exons as determined by FIRMA index (−4.0 ≥ a ≤ 4.0) in Vav-TRAF6 versus WT (FVB/NJ) LSK (MIDAS P < 0.05). Each point represents a differentially spliced exon. Highlighted are the top 3 differentially spliced exons in Vav-TRAF6 LSK. (b) Schematic representation of the mouse Arhgap1 gene structure with the approximate position of exons and introns. Overview of Arhgap1 exon usage is shown for WT (FVB/NJ) (black lines) and Vav-TRAF6 LSK (red lines). The average of FIRMA index values for individual exons of Arhgap1 in WT (n = 4) and Vav-TRAF6 LSK (n = 3) are shown below. FIRMA index scores indicate exon exclusion (negative values) or exon retention (~0). (c) RT-PCR analysis (using primers flanking exon 2) of the Arhgap1 cassette exon in Vav-TRAF6 and WT (FVB/NJ) LSK (left), and in LPS-treated WT Lin− BM cells (right). Values under the plot represent the short isoform as a percentage of the short and long isoforms. (d) Sequence analysis of the Arhgap1 cassette exon using primers flanking skipped exon 2 isolated from Vav-TRAF6 LSK. (e) RT-PCR analysis (using primers flanking exon 2, as depicted in the schematic) of the Arhgap1 cassette exon in WT (FVB/NJ) or Vav-TRAF6 Lin− BM cells were transduced with non-targeting shRNA (shCtl) or shRNA targeting mouse hnRNPA1 (shA1). Summary of Arhgap1 exon 2 exclusion calculated based on the short isoform as a percentage of the short and long isoforms is shown above. n = 3 per group; #, P = 0.06; *, P = 0.04. (f) RT-PCR analysis (using primers flanking exon 2, as depicted in the schematic) of the Arhgap1 cassette exon in WT (C57Bl/6) (Traf6+/+;Mx1Cre) and TRAF6-deficient Lin− BM cells (Traf6fl/fl;Mx1Cre mice treated with PolyIC) stimulated with 10 ng/mL of LPS for 2 hrs. Summary of Arhgap1 exon 2 exclusion calculated based on the short isoform as a percentage of the short and long isoforms is shown above. n = 3 per group; *, P < 0.05. (g) RT-PCR analysis of a minigene plasmid containing the Arhgap1 cassette exon 2 with 50 bp of flanking intron sequences (using primers flanking exon 2, as depicted in the schematic on the left) in HEK293 cells (HEK293-Arhgap1exon2) transfected with hnRNPA1. Summary of Arhgap1 exon 2 exclusion from the minigene calculated based on the short isoform as a percentage of the short and long isoforms is shown above from 2 independent experiments. *, P = 0.02. (h) RT-PCR analysis of a minigene plasmid containing the Arhgap1 cassette exon 2 (using primers flanking exon 2, as depicted in the schematic on the left) in HEK293-Arhgap1exon2 cells transfected with WT (n = 7) and an E3 ligase-mutant (C70A; n = 5) TRAF6. Summary of Arhgap1 exon 2 exclusion from the minigene was calculated based on the short isoform as a percentage of the short and long isoforms is shown above from 2 independent experiments. *, P = 0.03. (i) RT-PCR analysis of a minigene plasmid containing the Arhgap1 cassette exon 2 (using primers flanking exon 2, as depicted in the schematic on the left) in HEK293-Arhgap1exon2 cells transfected with TRAF6 and hnRNPA1 (WT and lysine mutants). Summary of Arhgap1 exon 2 exclusion from the minigene was calculated based on the short isoform as a percentage of the short and long isoforms is shown above from 2 independent experiments (n = 2 per group). Vec, vector; A1WT, hnRNPA1 WT; A1K3R, hnRNPA1 with K3R mutation; A1K-R, hnRNPA1 with mutation of all lysines within the first RNA recognition motif. *, P < 0.01; **, P = 0.003. (j) qRT-PCR of Arhgap1 cassette exon 2 expressed from the minigene plasmid following RNA immunoprecipitation of Myc-hnRNPA1 in HEK293-Arhgap1exon2 cells transfected with Myc-hnRNPA1 (WT and K-R) and TRAF6. Shown is the average of 2 independent experiments (n = 2 per group). *, P < 0.01. Data are from two experiments (e–j; means and s.e.m.).
To gain further insight into the regulation of Arhgap1 exon 2 exclusion we generated an Arhgap1 exon 2 cassette splicing reporter in HEK293 cells (HEK293-Arhgap1exon2)31. Arhgap1 exon 2 or the flanking introns do not contain a conserved hnRNPA1 RNA binding motif (not shown). However, transfection of hnRNPA1 into HEK293-Arhgap1exon2 cells resulted in exclusion of Arhgap1 exon 2 from the splicing reporter compared to vector-transfected HEK293-Arhgap1exon2 cells (Fig. 5g). In addition, transfection of TRAF6, but not of the E3 ligase-defective TRAF6-C70A, into HEK293-Arhgap1exon2 cells lead to exclusion of Arhgap1 exon 2 from the splicing reporter (Fig. 5h). To investigate whether TRAF6-mediated exon skipping is through an hnRNPA1 ubiquitination-dependent mechanism, we co-expressed TRAF6 and hnRNPA1, hnRNPA1K3R or hnRNPA1K-R in HEK293-Arhgap1exon2 cells. Expression of TRAF6 and hnRNPA1 resulted in increased exclusion of Arhgap1 exon 2 from the splicing reporter as compared to TRAF6 alone (Fig. 5i), while expression of hnRNPA1K-R, and to a lesser extent hnRNPA1K3R, were not as effective as hnRNPA1 to induce Arhgap1 exon 2 exclusion (Fig. 5i). To evaluate whether ubiquitination of hnRNPA1 affected it’s binding to Arhgap1 exon 2, we performed RNA-immunopreciptation for Arhgap1 exon 2 in HEK293-Arhgap1exon2 cells transfected with TRAF6 and hnRNPA1 or hnRNPA1K-R. Transfection of TRAF6 and hnRNPA1 resulted in increased binding of hnRNPA1 to Arhgap1 cassette exon 2 by 4-fold compared to transfection of only hnRNPA1 (Fig. 5j). In contrast, transfection of TRAF6 and hnRNPA1K-R resulted in increased binding of hnRNPA1K-R to Arhgap1 cassette exon 2 by only 2-fold as compared to hnRNPA1 alone (Fig. 5j). These data suggest that ubiquitination of Lys residues within the RRM1 of hnRNPA1 by TRAF6 enhanced the binding to Arhgap1 cassette exon 2, resulting in exon exclusion.
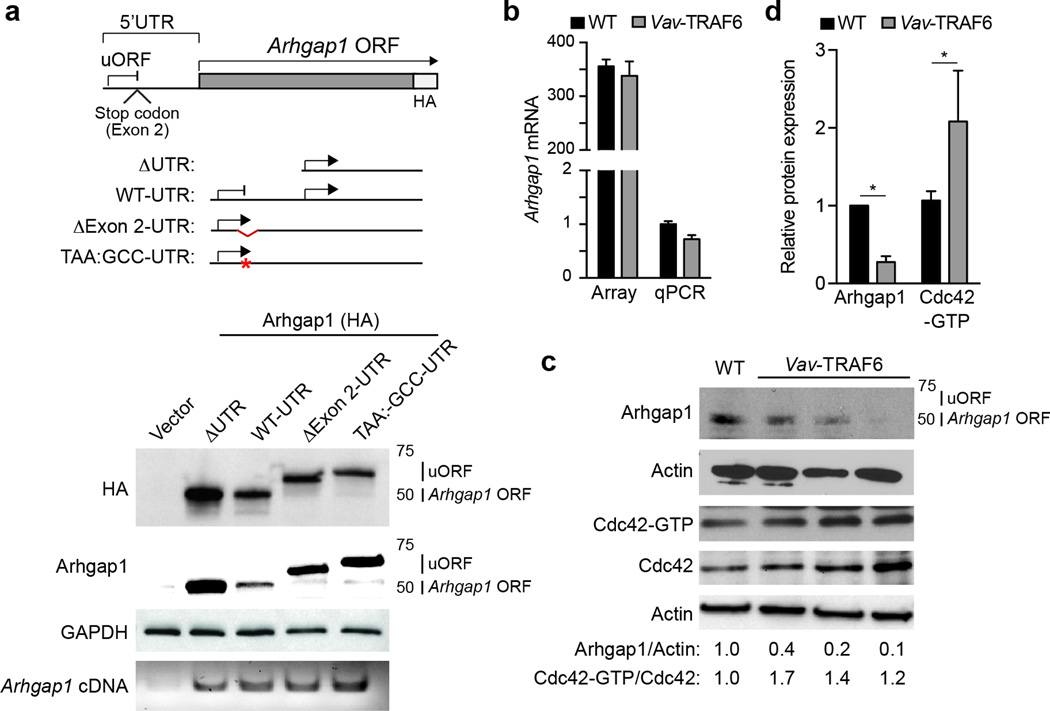
Exclusion of exon 2 results in reduced Arhgap1 protein
Arhgap1 contains a conventional open-reading frame (ORF) initiated within exon 4 and an upstream ORF (uORF) within exon 1. The uORF is terminated by a stop codon encoded within exon 2 (Fig. 6a). To examine the consequences of exon 2 exclusion on Arhgap1 protein, we transfected hemagglutinin (HA)-tagged Arhgap1 cDNAs with the Arhgap1 5’ UTR (WT-UTR-Arhgap1) or without the Arhgap1 5’ UTR (ΔUTR-Arhgap1) in HEK293 cells and evaluated protein expression by immunoblotting with Arhgap1 or HA antibodies. Transfection of ΔUTR-Arhgap1 or WT-UTR-Arhgap1 resulted in translation of Arhgap1 from the conventional ORF (Arhgap1 ORF) as evident by the 50kD Arhgap1 protein (Fig. 6a). In contrast, deletion of exon 2 within the 5’ UTR (ΔExon2-UTR-Arhgap1) resulted in absence of Arhgap1 protein expression from the conventional Arhgap1 ORF (Fig. 6a). Moreover, transfection of ΔExon2-UTR-Arhgap1 resulted in translation from the uORF and generated a chimeric Arghap1 protein with 40 additional N-terminal amino acids (Fig. 6a). Mutation of the stop codon within exon 2 (TAA:GCC-UTR-Arhgap1) resulted in reduced expression of Arhgap1 protein from the conventional ORF (Fig. 6a) and utilization of the uORF to produce a chimeric Arhgap1 protein with 65 additional N-terminal amino acids (Fig. 6a). These findings revealed that exclusion of Arhgap1 exon 2 prevented translation of Arhgap1 from the ORF and induced alternative ORF usage.
Figure 6. Exclusion of exon 2 results in reduced Arhgap1 protein and Cdc42 activation.
(a) Schematic of the Arhgap1 5’-UTR and coding region. uORF, upstream open reading frame; HA, hemagglutinin. The indicated Arhgap1 5’-UTR cDNA fusions were transfected (pCMV-GFP) into HEK293 cells and evaluated by Arhgap1 and HA immunoblotting. Transfected Arhgap1 5’-UTR cDNA were measured by RT-PCR using primers aligned to Arhgap1 coding sequences. (b) Microarray and qRT-PCR analysis of Arhgap1 total RNA from LSK of WT (FVB/NJ) and Vav-TRAF6 mice. (c) Immunoblotting of Arhgap1 in Lin− BM cells from WT (FVB/NJ) and Vav-TRAF6 mice. Cdc42 activity (Cdc42-GTP) in WT (FVB/NJ) and Vav-TRAF6 Lin− BM cells was determined by a pull-down/immunoblot assay. Shown is a representative blot from one WT (FVB/NJ) and three Vav-TRAF6 mice. (d) Densitometric analysis of Arhgap1 protein and Cdc42-GTP (WT, n = 3; Vav-TRAF6, n = 5). *, P < 0.05. Data are from three experiments (b,d; means and s.e.m.).
The consequences of exon 2 exclusion on Arhgap1 RNA and protein expression were next examined in Vav-TRAF6 LSK. As assessed by microarray and qRT-PCR, Arhgap1 mRNA expression was comparable in wild-type and Vav-TRAF6 LSK (Fig. 6b). In wild-type Lin− BM cells, Arhgap1 protein was expressed from the conventional ORF as determined by immunoblotting (Fig. 6c). In contrast, Arhgap1 protein expression from the conventional ORF was reduced in Vav-TRAF6 compared to wild-type Lin− BM cells (Fig. 6c,d), suggesting that exon 2 is required for proper Arhgap1 protein expression. The chimeric Arhgap1 protein derived from the uORF was not detectable in primary Vav-TRAF6 Lin− BM cells (Fig. 6c) for reasons that remain unclear. However, we observed that Cdc42-GTP was significantly higher in Vav-TRAF6 Lin− BM cells compared to wild-type Lin− BM cells (Fig. 6c,d), suggesting increased Cdc42 activation. These findings indicate that TRAF6 overexpression induces exclusion of Arhgap1 exon 2 leading to diminished Arhgap1 protein and Cdc42 activation in HSPC.
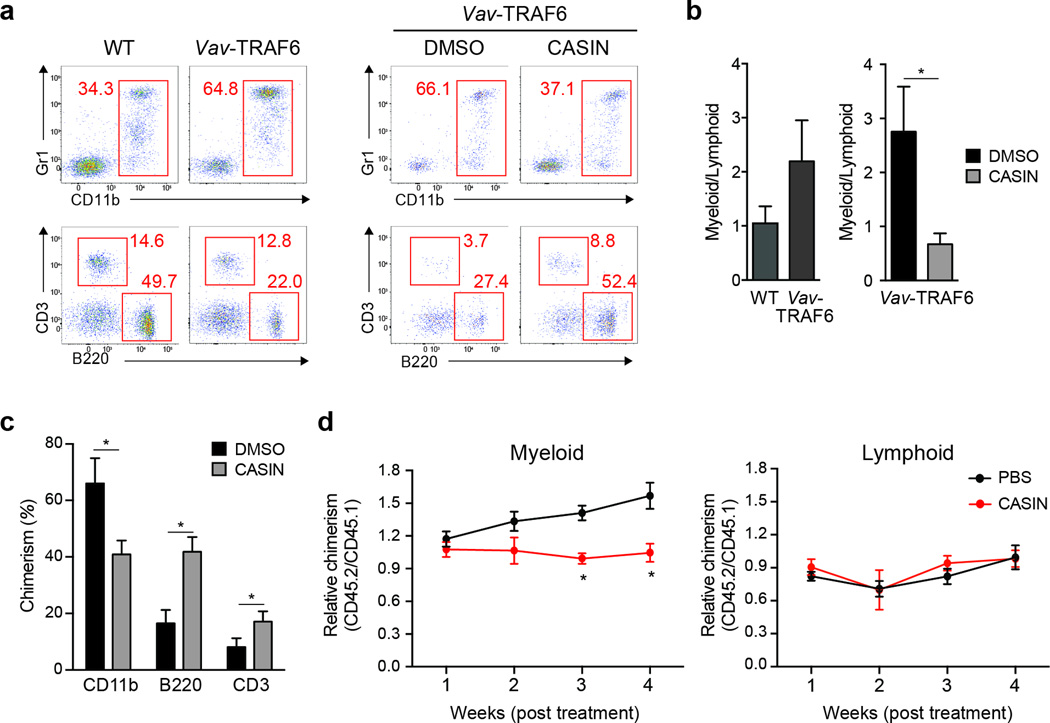
The hematologic phenotype and reduced Arhgap1 protein expression in Vav-TRAF6 mice was reminiscent of the phenotype of Arhgap1-deficient mice, which exhibit increased Cdc42 activity, impaired hematopoiesis and myeloid-biased differentiation28–30. Similar with Vav-TRAF6 mice, Arhgap1+/− neutrophils exhibit a dysplastic phenotype (Supplementary Fig. 6). To investigate whether Cdc42 activation contributes causally to the hematopoietic defects observed in Vav-TRAF6 mice, we used the selective inhibitor CASIN to inhibit Cdc42 in HSC28,32. Two hundred LT-HSC (CD34−CD135−LSK) sorted from CD45.2+ Vav-TRAF6 mice were either directly transplanted with 2x105 wild-type CD45.1+ BM cells into irradiated wild-type congenic mice, or treated with CASIN (or control DMSO) for 24 hr and then transplanted with 2x105 wild-type CD45.1 BM cells into irradiated wild-type mice. Vav-TRAF6 LT-HSC (transplanted either directly or after overnight culture in media containing DMSO) had generated ~65% Gr1+CD11b+ myeloid cells of the CD45.2+ donor-derived peripheral blood compared to ~35% wild-type Gr1+CD11b+ cells 8 weeks post transplantation (Fig. 7a,b). However, CASIN-treated Vav-TRAF6 LT-HSC formed fewer Gr1+CD11b+ myeloid cells among donor (CD45.2+)-derived peripheral blood from 37% to 66% compared to DMSO-treated Vav-TRAF6 LT-HSC after 8 weeks (Fig. 7a,b). Reduced chimerism from CASIN-treatment of Vav-TRAF6 LT-HSC resulted from fewer CD45.2+-donor Gr1+CD11b+ myeloid cells and increased CD3+ and B220+ lymphoid cells relative to wild-type CD45.1+ myeloid and lymphoid cells in peripheral blood (Fig. 7c). In addition, intraperitoneal injection of CASIN or PBS as control for 16 continuous days after adoptive transfer of 200 Vav-TRAF6 LT-HSC and 2x105 wild-type BM competitor cells into lethally-irradiated wild-type mice, resulted in expansion of Vav-TRAF6 CD45.2+CD11b+ myeloid cells by ~50% as compared to wild-type CD45.1+CD11b+ over 4 weeks in PBS-treated chimeric mice, whereas Vav-TRAF6 CD45.2+CD11b+ frequencies did not increase over 4 weeks in CASIN-treated chimeras (Fig. 7d). These findings suggest that Cdc42 activation can cause the myeloid bias observed in the Vav-TRAF6 mice.
Figure 7. Cdc42 contributes to HSPC defects in Vav TRAF6 mice.
(a) FACS analysis of donor-derived (gated on CD45.2+) myeloid (CD11b+Gr1+) and lymphoid (B220+ and CD3+) proportions 8 weeks post transplantation for competitively transplanted WT (C57Bl/6) and Vav-TRAF6 LT-HSC, and for Vav-TRAF6 LT-HSC treated in vitro with CASIN (or DMSO) for 24 hr. *, P < 0.05. (b) Summary of independent experiments of myeloid and lymphoid proportions of donor-derived (gated on CD45.2+) peripheral blood shown as a ratio (CD11b/[B220 + CD3]). (c) Peripheral blood myeloid (CD11b+) and lymphoid (B220+ and CD3+) chimerism (CD45.2+) of donor-derived cells determined 8 weeks after competitive transplantation with Vav-TRAF6 LT-HSC treated with CASIN. (n = 6 per group). *, P < 0.005. (d) Donor-derived LT-HSC cells (CD45.2+) from Vav-TRAF6 mice and competitor BM cells (CD45.1+) were transplanted into lethally-irradiated recipient mice (CD45.1+). Three month post secondary transplantation, the recipients were treated with 30 mg/kg CASIN and vehicle control (PBS) for 16 days. Ratio of donor-derived (gated on CD45.2+) and competitor-derived (gated on CD45.1+) myeloid (CD11b) and lymphoid (CD3 and B220) cells was determined by flow cytometry 4 weeks post-CASIN treatment in vivo. Donor-derived chimerism is normalized (1.0) for each mouse at week 1 (n = 5 per group). *, P = 0.007. Data are from two experiments (b–d; means and s.e.m.).
Cdc42 activation is associated with human MDS
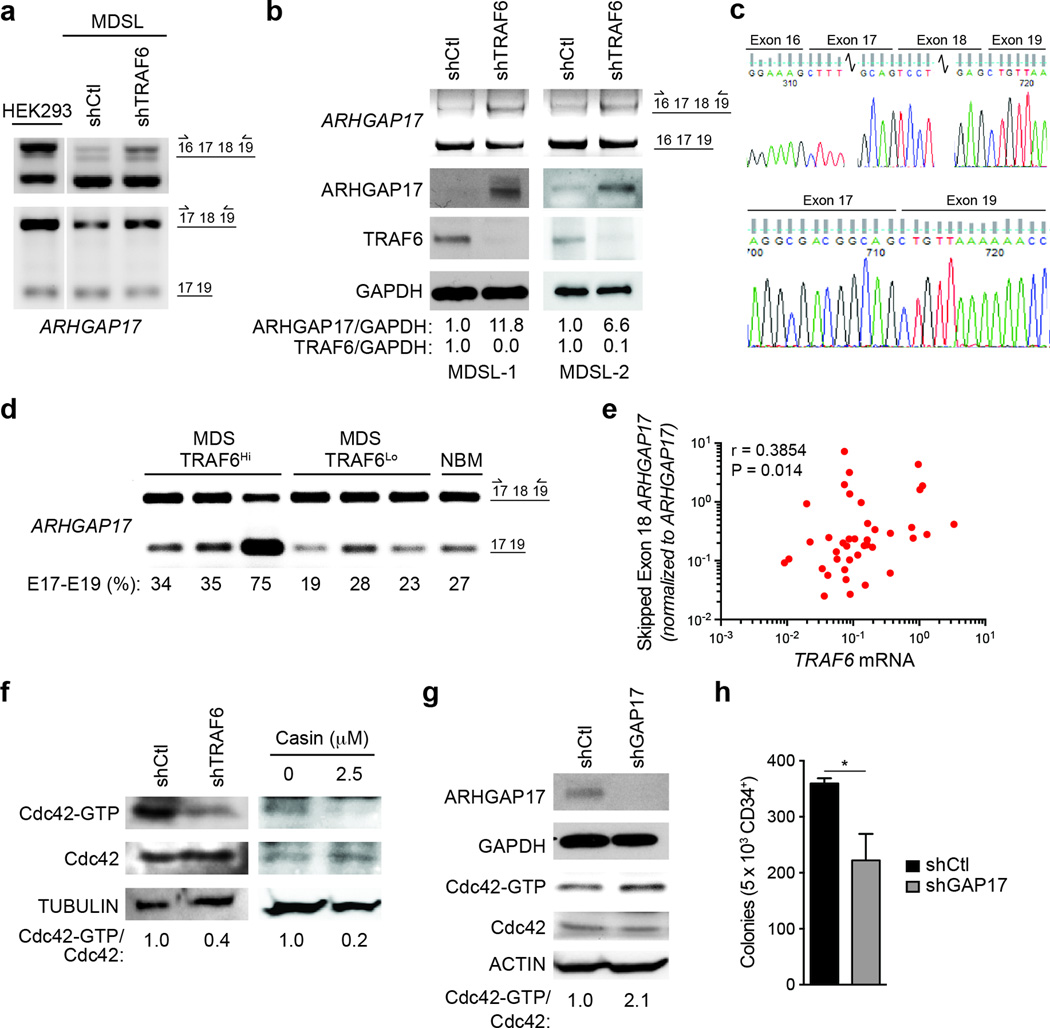
Next, we assessed alternative RNA splicing changes of Arhgap family members in publically available data sets from human MDS-AML samples and cell lines33–35. We identified significant RNA splicing alterations of ARHGAP11b, ARHGAP17, ARHGAP19 and ARHGAP22 in MDS and AML BM CD34+ cells and cell lines. Among these, ARHGAP17 undergoes exclusion of exon 18 in a human monocytic leukemic cells (THP1) following LPS stimulation34 and in an MDS patient-derived human cell line (MDSL) (Fig. 8a, Supplementary Fig. 7a). To establish the contribution of TRAF6 to ARHGAP17 exon 18 exclusion, MDSL cells were transduced with shRNAs targeting TRAF6 (shTRAF6-MDSL) or a non-targeting shRNA (shCtl-MDSL). RT-PCR using primers flanking exons 17 and 18 or exon 18 of ARHGAP17 showed that shTRAF6-MDSL have restored expression of the ARHGAP17 exon 17, 18 and 19 cassette compared to shCtl-MDSL cells (Fig. 8a,b). In human MDS BM cells, exclusion of exon 18 resulted in premature termination of ARHGAP17 protein (Supplementary Fig. 7a), while inclusion of ARHGAP17 exon 18 in shTRAF6-MDSL cells resulted in restored ARHGAP17 protein expression (Fig. 8b), indicating that exon 18 is essential for proper ARHGAP17 protein expression. The splice junctions between exons 16, 17, 18 and 19, and between exons 17 and 19 of ARHGAP17 in MDSL cells were independently confirmed by Sanger sequencing (Fig. 8c). As we observed in shTRAF6-MDSL cells, shRNA-mediated knockdown of hnRNPA1 in MDSL cells (shA1-MDSL) resulted in restoration of full-length ARHGAP17 mRNA, and expression of ARHGAP17 protein (Supplementary Fig. 7b–d).
Figure 8. Cdc42 activation is associated with primary human MDS.
(a) RT-PCR using primers flanking endogenous ARHGAP17 exon 18 or exons 17 and 18 (as depicted in the schematic on the left) in HEK293 or MDSL cells expressing shRNAs targeting TRAF6 (shTRAF6) or a non-targeting control (shCtl). (b) Immunoblotting of ARHGAP17 in independently transduced MDSL cells (MDSL-1-2) expressing a non-targeting shRNA (shCtl) or shRNA targeting TRAF6 (shTRAF6). (c) Sequence analysis of the ARHGAP17 exons 16, 17, 18, and 19 in MDSL cells. (d) RT-PCR using primers flanking endogenous ARHGAP17 exon 18 (as depicted in the schematic on the left) in MDS patient BM mononuclear cells with high (>2-fold TRAF6, n = 3) or low (< 1.0-fold TRAF6, n =3) TRAF6 mRNA, and 1 normal BM sample. (e) qRT-PCR of ARHGAP17 exon 18 using primers to the exon 17–19 junction and normalized to total ARHGAP17 in MDS/AML BM cells (n = 35). Spearman correlation: r = 0.3854, P = 0.014. (f) Cdc42-GTP pulldown assays in MDSL cells expressing non-targeting shRNA (shCtl) or shRNA targeting TRAF6 (shTRAF6), or in MDSL treated with 2.5 µM CASIN. (g) Cdc42-GTP pulldown assays in MDSL expressing non-targeting shRNA (shCtl) or shRNA targeting ARHGAP17 (shGAP17). (h) Colony formation in methylcellulose of human CD34+ cells expressing a non-targeting shRNA (shCtl) or shRNA targeting ARHGAP17 (shGAP17) (n = 3). *, P = 0.047. Data are from three experiments (h; means and s.e.m.).
We next determined whether ARHGAP17 exon 18 exclusion is associated with TRAF6 overexpression in primary human MDS. RT-PCR analysis of BM mononuclear cells showed that increased TRAF6 expression correlated with exclusion of ARHGAP17 exon 18 in MDS patients and normal BM cells (Fig. 8d). In addition, we examined TRAF6 expression and ARHGAP17 exon 18 exclusion by qRT-PCR in larger cohort of primary MDS and AML BM mononuclear cells. Using primers aligning to the splice junction of exons 17–19 of ARHGAP17, we found that increased TRAF6 mRNA positively correlated with exclusion of exon 18 from ARHGAP17 in MDS/AML samples (Fig. 8e), indicating that TRAF6 overexpression is associated with alternative RNA splicing of ARHGAP17. Because ARHGAP17 is a negative regulator of Cdc4236, we evaluated the amount of Cdc42-GTP in MDSL cells following TRAF6 knockdown. Cdc42-GTP was significantly lower in shTRAF6-MDSL cells compared to shCtl-MDSL cells (Fig. 8f), indicating that restored ARHGAP17 protein expression suppresses Cdc42 activity. In contrast, knockdown of ARHGAP17 in MDSL cells with a GAP17-shRNA resulted in increased Cdc42-GTP relative to MDSL cells expressing a non-targeting shRNA (shCtl-MDSL) (Fig. 8g). Moreover, transduction of GAP17-shRNA in primary human CD34+ cells resulted in reduced total colony formation as compared to non-targeting shRNA (shCtl) CD34+ cells (Fig. 8h). As such, lower ARHGAP17 expression, as observed following exclusion of exon 18 in MDS and increased Cdc42 activation impairs human CD34+ HSPC function. Taken together, alternative splicing of Arhgap proteins contributes to increased Cdc42 activation and aberrant hematopoiesis following activation of TRAF6 in MDS (Supplementary Fig. 7e).
Discussion
Our results show that TRAF6 affects HSPC function by regulating exon specification through ubiquitination of the RNA binding protein hnRNPA1, and provide insight into how hematopoietic cells regulate RNA isoform expression during TLR signaling. We identified several hnRNP family members as potential TRAF6 substrates. Thus, it is possible that TRAF6 signaling downstream of TLR, IL-1R, and/or TNF superfamily of receptors coordinate RNA processing via ubiquitination of multiple auxiliary RNA splicing factors. Our mechanistic interrogation revealed that in the presence of TRAF6, hnRNPA1 induces exclusion of Arhgap1 exon 2. The precise mechanism by which ubiquitination of hnRNPA1 controls Arhgap1 exon 2 splicing or globally regulates RNA processing is not conclusive, as hnRNPs can couple RNA splicing with nuclear export, and/or affect RNA splicing directly by binding RNA response elements or in trans through protein interactions with other RNA splicing proteins37. Multiple independent genetic alterations and molecular mechanisms have been reported that suggest chronic immune-related signaling contributes to the pathogenesis of MDS. These MDS-associated alterations converge on activating the TRAF6 signaling complex38. Chronic inflammation mediates pleotropic affects on hematopoiesis by cell autonomous and non-autonomous mechanisms. Moreover, chronic immune signaling has durable and irreversible effects on HSC function1,39,40. Although an inflammatory BM milieu and/or altered immune effector cells may contribute to HSC dysfunction and MDS, our results clearly demonstrate that TRAF6 induces cell-intrinsic HSC defects. TRAF6 mediates RNA splicing of Arhgap1 and activation of the small GTPase, Cdc42, which confers at least part of the hematopoietic phenotype in Vav-TRAF6 mice. Mice with constitutively elevated Cdc42 activity, as a consequence of Arhgap1 deletion, display pre-mature aging phenotypes in multiple tissues, including HSCs41. Innate immune signaling and inflammation are also thought to be implicated in aged HSC1,42, which would provide a mechanistic explanation for Cdc42 activation in aged HSC. In summary, we demonstrate that RNA processing in hematopoietic cells is regulated by TRAF6-mediated ubiquitination of a RNA binding protein, hnRNPA1, and that chronic innate immune signaling via TRAF6 impairs HSPC function and contributes to the pathogenesis of MDS through altered splicing of HSC-requisite genes.
Methods
Mice and primary murine bone marrow cell culture
Generation of the Vav-TRAF6 mice was accomplished by cloning a flag tagged human TRAF6 cDNA downstream of the Vav promoter, followed by pronuclear microinjection of linearized Vav-FLAG-TRAF6 fragment into fertilized eggs. Mouse and human TRAF6 protein have 92% amino acid identity and functional conservation. The eggs were then transferred into pseudopregnant FVB/NJ mice. Vav-TRAF6 mice were also subsequently backcrossed onto C57Bl/6 genetic background. TRAF6fl/fl mice (C57Bl/6) were a kind gift from Dr. Yongwon Choi (University of Pennsylvania)43. Traf6fl/fl mice were crossed with Mx1-Cre and Vav-Cre mice for inducible or conditional deletion of TRAF6 (Traf6fl/fl;Mx1-Cre) and (Traf6fl/fl;Vav-Cre), respectively. To delete TRAF6, Cre transgene expression in Traf6fl/fl;Mx1-Cre mice was induced by five polyinosinic-polycytidylic acid (pIpC) injections. BM cells were obtained by crushing the femur, tibia, and pelvic bone, and maintained in RPMI1640 with 10% fetal bovine serum. The media was supplemented with recombinant mouse stem cell factor (rmSCF, 02931, StemCell Technologies), mouse interleukin 3 (rmIL-3, 02903, StemCell Technologies) and human interleukin 6 (rhIL-6, 02606, StemCell Technologies) at 10 ng/mL. All mice were bred, housed and handled in the Association for Assessment and Accreditation of Laboratory Animal Care-accredited animal facility of Cincinnati Children’s Hospital Medical Center.
BM transplantation
For competitive repopulation, 3 x 106 CD45.2+ MNCs from WT (C57Bl/6) and Vav-TRAF6 (C57Bl/6) mice (2 donor mice, 3 months old) were mixed with 3x106 CD45.1+ B6.SJLPtprca Pep3b/BoyJ MNCs (6–10 weeks of age), and then transplanted into lethally-irradiated recipient mice (CD45.1+ B6.SJLPtprca Pep3b/Boy; 6–10 weeks of age). Alternatively, 200 LT-HSC (Lin−cKit+Sca1+CD34−CD135−) from WT (C57Bl/6) and Vav-TRAF6 (C57Bl/6) mice (2 donor mice, 6–8 moths old) were mixed with 2 x 105 CD45.1+ MNCs (6–10 weeks of age), and then transplanted into lethally-irradiated recipient mice (CD45.1+ B6.SJLPtprca Pep3b/BoyJ; 6–10 weeks of age). For ex vivo CASIN experiments, 200 LT-HSC (Lin−cKit+Sca1+CD34−CD135−) from Vav-TRAF6 mice (6–8 months old) were treated with 5 µM CASIN for 24 hours, and were then mixed with 2 x 105 CD45.1+ B6.SJLPtprca Pep3b/BoyJ MNCs (6–10 weeks of age) prior to transplantation into lethally-irradiated recipient mice (CD45.1+ B6.SJLPtprca Pep3b/BoyJ; 6–10 weeks of age). For in vivo delivery of CASIN, CASIN (30 mg/kg mixed with Dextran) and control vehicle PBS were intraperitoneally injected in recipient mice for 16 consecutive days.
Retroviral expression of TRAF6 in mouse HSPC BM cells
For WT and E3 ligase-defective (C70A) TRAF6 overexpression, HSPC-enriched Lin- BM cells were obtained from C57Bl/6 or Traf6fl/fl;Vav-Cre mice, and then infected with retroviral vectors (co-expressing GFP, see plasmid description below). Cells were flow-sorted for GFP+ cells after 72 hours and then examined for colony formation in methylcellulose or for PB reconstitution in lethally-irradiated mice (CD45.1+ B6.SJLPtprca Pep3b/BoyJ; 6–10 weeks of age).
Hematological and histological analysis
Peripheral blood was collected by cheek piercing or cardiac puncture. Blood counts were measured with a hemacytometer (HEMAVET®). Spleens and tibias were fixed with 4% PFA, sectioned and stained with H&E for histopathological analysis. Cytospin was performed with 2 x 105 BM cells. Blood smear and cytospin were stained with Wright-Giemsa for morphological analysis. Dysplastic cells were evaluated on at least 100 cells per mouse and confirmed by a pathologist. For neutrophils with Pelger-Huet anomalies, hyposegmentation as a bilobed nucleus was present.
Exon array analysis
HSPC-enriched LSK cells were sorted from WT (FVB/NJ) and Vav-TRAF6 mice (FVB/NJ) (6 months of age). Total RNA was extracted from LSK cells and purified with Quick-RNA MiniPrep Kit (Zymogen). RNA quality was tested using the Agilent Bioanalyzer 2100 (Hewelett Packard). Total RNA was reverse transcribed and labeled, and hybridized onto GeneChip® Mouse Exon 1.0 ST Array (Affymetrix). Scanning was performed with the Affymetrix GeneChip Scanner 3000 7G and evaluated with the Genechip Operating Software (Affymetrix). Data mining was performed with GeneSpring software (Agilent). Gene set enrichment analysis was performed on a JAVA-based dataset supported by the Broad Institute. ToppGene analysis (http://toppgene.cchmc.org) was performed to identify enriched pathways.
Exon usage pattern analysis
Detection of alternative splicing events between pairs of biological groups was performed with AltAnalyze (v.2.0.6)44, a python-based program. Briefly, a ‘robust multi-chip analysis’ (RMA) summarization on the selected CEL files was initially generated, and only the probesets that can be aligned to a single ENSEMBL gene were included. This step generated the gene-level expression statistics and annotations for each ENSEMBL gene and each biological comparison group. The probeset-level expression values were further filtered based on DABG P-values cutoff and absolute expression levels for each biological comparison group. Based on these values, the standard alternative exon statistics (i.e. FIRMA, MiDAS P-value, normalized P-value) were computed for each analyzed probeset relative to the determined gene expression levels. For FIRMA analysis19, Affymetrix probe residuals for all probes associated with a gene (metaprobeset) were obtained by performing a second round of RMA summarization (using APT) for all probesets associated with AltAnalyze core probeset, “extended” probeset, or “full” probeset definitions. FIRMA scores for each probeset and sample were further summarized by AltAnalyze along user-defined biological groups and comparisons to obtain a FIRMA fold change and P-value (two-tailed t-test assuming unequal variance). We used the 0.05 as P-value threshold. CORE probesets were used to perform general alternative splicing analysis, but “extended” and “full” probesets were used to visualize the global missplicing patterns using a heatmap. Specifically, probe sets with the core annotation include all Affymetrix core probe sets that specifically overlap with a single ENSEMBL gene, along with any probe set that overlaps with an ENSEMBL or UCSC exon. For exon/intron splicing assay of WT and Vav-TRAF6 LSK cells, the parameters for AltAnalyze program were as follows: dabg (P < 0.05); raw expression threshold (1.0); alt exon fold variable (2.0); gene expression cutoff (3.0); P < 0.05. For FIRMA, the median of the residuals for each probe set for each array sample is compared to the median absolute deviation for all residuals and samples for the gene, as previously described19,44. To study splicing patterns of LPS-treated and untreated THP-1 cells, Gene Expression Omnibus dataset (GSE32141; Affymetrix HuEx1.0 ST exon array) was analyzed with the following parameters in AltAnalyze: dabg (P < 0.05); raw expression threshold (1.0), alt exon fold variable (2.0); gene_expression cutoff (3.0); P < 0.05. Details on the Splicing Index analysis are previously described18.
Splicing pattern enrichment analysis
Identification of alternative exons for 46 previously published splicing factor knockdown, over-expression or mutation datasets (mouse or human) was obtained using AltAnalyze, followed by over-representation analysis in GO-Elite (Supplemental Table 5). Enrichment Z scores for alternative exons from Vav-TRAF6 LSK among regulated exons from 46 evaluated splicing factors (siRNA, knock-out, over-expression, mutation). Only results for splicing factors with at least 4 matching regulated exons in comparison to TRAF6-overexpression patterns are shown. Enrichment was performed using GO-Elite separately for upregulated (increased exon-inclusion) and downregulated (increased exon-exclusion) separately for the input exons and evaluated splicing factor regulated exons. Splicing-factors with at least 10 exons regulated in common with the indicated splicing factor are shown. The splicing factor predicted expression (up or down-regulated) is indicated based on expression of the enriched exons in Vav-TRAF6 LSK.
RNA binding protein site enrichment analysis
Exon and intron sequences were retrieved using R/Bioconductor’s (http://www.bioconductor.org) BSgenome packages. All exons and flanking introns (with explicit start-end coordinates) defined in TXdb (for SpliceTrap) and EnsMart65 (for AltAnalyze) were used as search spaces to compute the total occurrence and location of each predicted binding site (see below). Entire exon blocks (merged Ensembl transcript models from AltAnalyze) were scanned, along with 5’ and 3’ intron region sequences (50nt, 500nt and full intron windows from adjacent splice sites). Exon-inclusion, exon-exclusion and combined differentially regulated probe sets (FIRMA) were scanned separately. Only the exons passing basal expression filters and having explicit start-end coordinates were considered for further analysis (219219 for human RNA-seq data, 261178 for mouse exon array data). To examine these exon and intron sequences for putative binding sites for specific RNA binding proteins (RBPs), we used a public library of 228 human RBP binding models in the form of position frequency matrix motifs obtained from the CisBP-RNA database20, and supplemented them with 12 additional motifs for hnRNP family members. This collection of binding models was used to create a custom motif library in the HOMER software package45. We then used the HOMER algorithm to systematically estimate the significance of the presence of matches to each motif in each input sequence set, scanning only the sense strand in the RNA sequences.
As a complementary approach, we employed a consensus-based binding site enrichment analysis, wherein regular expression pattern searches were determined for a given consensus site on skipped and non-skipped exon sequences. For hnRNPA1 binding motif enrichment analysis, human and mouse consensus binding sites (AGNUAGNN, GUAGUAGU, UAGGGA, UAGGGU)24–26 were utilized. For exons containing binding site matches at multiple locations for a given consensus site, we counted multiple occurrences as “1” for our enrichment tests. Target exons were categorized into 4 groups: with binding site skipped; without binding site skipped; with binding site included; and without binding site included. Binding site enrichment was then performed using the hypergeometric test46.
For human AML, 179 patient (Level3) sample data sets were downloaded from TCGA website. 13 samples with high and 9 samples with low level of TRAF6 (in RPKM) were selected for splicing assay. Corresponding TCGA level-1 data (RNA-seq BAM files) were obtained through dbGaP (ttps://dbgap.ncbi.nlm.nih.gov) and CGHub UCSC (https://cghub.ucsc.edu/datasets/data_sets.html). Each BAM file was converted into two FASTQ files using PICARD’s SamToFastq tool (v1.98) and used as inputs to SpliceTrap47. To quantify exon inclusion ratios based on paired-end reads using SpliceTrap (v0.90.5), each exon was analyzed with respect to adjacent exons flanking the middle spliced exons. The unpaired T-test was used in order to assess the difference of exon usage between these two groups. The exon inclusion ratio was defined as the expression level of the inclusion isoform divided by the total expression level of both isoforms (inclusion and skipped) derived from an exon trio. Inclusion ratio is used to separate constitutive exons (CS, inclusion ratio > 0.9) and cassette/alternative exons (CA, inclusion ratio < 0.9). Since SpliceTrap does not provide per-group analysis, we made arbitrary cutoff (>75% in each group) for exons of interest showing the consistent alternative splicing patterns. As above, hnRNPA1 motif enrichment was determined by the hypergeometric test.
TRAF6 ubiquitin substrate identification by subtractive proteomics
Identification of ubiquitinated peptides was performed in accordance to the manufacturer recommendations (PTMScan Ubiquitin Proteomics System, Cell Signaling Technology). TF1 cells transduced with a doxycycline-inducible TRAF6 shRNA were grown in RPMI1640 supplemented with FBS. Cells were treated with or without doxycycline for 24 hours. After treatment, cells were lysed in denaturing lysis buffer. Lysates were digested with trypsin followed by peptide desalting. Peptides were immunoprecipitated with αnti-diGly coupled to protein A agarose beads for 1 hour at 4°C. Eluted peptides were analyzed by LC-MS/MS on an ABSciex 5600 TripleTOF mass spectrometer coupled to an Eksigent nanoLC.ultra nanoflow HPLC system as described48. MS/MS spectra searches of the nano-LC-MS/MS data (*.wiff file) from the enriched diGly peptides were processed for protein identification using ProteinPilot™ software (version 4.2, revision 1297) supplied by ABSciex. The data were searched against a SwissProt database of Homo sapiens protein sequences with “Biological Modifications” (which included diGly) as a specified defaults in the ProteinPilot software. The resulting *.group file was then used to generate a spreadsheet as a peptide summary report. Peptides identified with a minimum of 99% confidence are reported. Data obtained from TRAF6-expressing (without doxycycline) and in TRAF6-depleted (with doxycycline) TF1 cells was used to generate a non-redundant list of diGly sites.
In vitro ubiquitination assay
Validation of hnRNPA1, TAK1, and Tubulin ubiquitination was performed in accordance with the manufacturer recommendations (E3 Substrate ID, LifeSensors, Inc). In vitro ubiquitin reconstitution assays were performed on two protein arrays (Invitrogen ProtoArray). Recombinant E1 (UBE1), E2 (UBE2N/UBE2V1), wild-type ubiquitin, and ATP were included in each reaction. One array also received a mix containing purified human TRAF6 protein. Arrays were visualized with Tandem Ubiquitin Binding Entities (TUBEs) and FK2 followed by the corresponding Cy-dye labeled secondary detection reagents. To calculate ubiquitination of hnRNPA1, TAK1, and Tubulin, median intensities subtracted from background intensities were utilized from biological replicates (F532 Median - B532 Median = ubiquitin intensity).
RNA immunoprecipitation
RNA immunoprecipitation was performed as previously described49. Briefly, HEK 293 cells were crosslinked with 1% formaldehyde. Crosslinking was stopped by the addition of 200mM glycine. 500ug protein lysate was precleared with protein G Dynabeads (Life Technologies, 10007D), and immunoprecipitation was performed with 10ug of Myc antibody (Santa Cruz, sc-40) with rabbit IgG1 (Cell Signaling, 54153; negative control). RNA bound to the immunoprecipitated proteins was recovered by the treatment with proteinase K and DNase, and then measured with qRT-PCR. Primers to measure pre-mRNA are 5’-ATCTGAGAGAGTGGCTTTTTGC-3’ and 5’-CTGCTTCTGGTGCCTTCTG-3’.
Patient samples
Informed consent was obtained according to protocols approved by the review boards of MD Anderson Cancer Center and Cleveland Clinic. Diagnoses were reviewed and adapted, when required, to WHO 2008 criteria. For qRT-PCR analysis of TRAF6 transcript and ARHGAP17 missplicing, BM aspirates were collected from 36 MDS/AML patients and 7 age-matched healthy controls (Supplemental Table 6). RT-PCR analysis of ARHGAP17 splicing was performed on TRAF6high and TRAF6low MDS/AML patients (as determined by qRT-PCR and selected for > 2-fold or < 1-fold TRAF6 mRNA expression) and healthy controls.
Cell lines and CD34+ cells
TF1 and HEK293 cells were purchased from the American Type Culture Collection. MDSL was provided by Dr. Kaoru Tohyama (Kawasaki Medical School, Okayama, Japan). TF1 and MDSL cells were cultured in RPMI 1640 medium plus 10% FBS and 10ng/mL human recombinant interleukin 3 (rhIL-3, 02603, StemCell Technologies). HEK293 cells were maintained in DMEM media with 10% FBS. Human CD34+ cord blood cells were prestimulated in StemSpan SFEM supplemented with recombinant human stem cell factor (rhSCF; 02830 Stemcell Technologies), recombinant human Flt3 ligand (rhFL; 02840, Stemcell Technologies), recombinant human thrombopoietin (rhTPO; 02522, PeproTech) at 100ng/mL for 48 hours before transduction, and then maintained in rhSCF, rhFL, rhTPO, rhIL-3, and recombinant human IL-6 (rh IL-6; 02606 Stemcell Technologies) at 10ng/mL.
Colony forming assay
Murine LSK or BM mononuclear cells (MNC) were harvested from WT (FVB/NJ) and Vav-TRAF6 mice (FVB/NJ) (~4 months of age), and plated in methylcellulose (3434; Stemcell Technologies). Colonies were counted at 12–14 days after plating. For knockdown of mouse hnRNPA1, Lin− BM cells from WT (FVB/NJ) and Vav-TRAF6 mice were transduced with shRNA-expressing lentivirus, sorted for GFP+ cells by FACS, and plated in methylcellulose (3434; Stemcell Technologies). In addition, Lin− BM cells from WT (C57Bl/6) mice were transduced with retrovirus encoding TRAF6 or ligase inactive TRAF6C70A, and sorted for GFP+ cells by FACS, and plated in methylcellulose (3434; Stemcell Technologies). For knockdown of human ARHGAP17, CD34+ cells were transduced with shRNA-expressing lentivirus, sorted for GFP+ cells by FACS, and plated in methylcellulose (4434, Stemcell Technologies).
Flow cytometry
BM MNC were washed and incubated for 30 minutes with biotin conjugated lineage markers (CD11b, Gr1, Ter119, CD3, B220, Mouse Hematopoietic Lineage Biotin Panel, 88-7774-75, eBioscience), followed by staining with streptavidin APC-Cy™7 (554.63, BD Pharmingen), Sca-1 PE-cy7 (25-5981-82, eBioscience), c-Kit APC (17-1171-81, eBiosciences), CD34 FITC (11-0341-81, eBiosciences), CD135 (12-1351-81, eBiosciences). Long-term HSCs (LT-HSCs) were purified based on surface markers as Lin−Sca−1+c-Kit+CD34−CD135−. For colony forming assay using LSK cells, murine BM MNC were washed and incubated for 30 minutes with biotin conjugated lineage markers (CD11b, Gr1, Ter119, CD3, B220, Mouse Hematopoietic Lineage Biotin Panel, 88-7774-75, eBioscience), followed by staining streptavidin eFluor® 450 (48-4317-82, eBioscience), Sca-1 PE (12-5981-82, eBioscience), c-Kit APC (17-1171-81, eBiosciences). LSK cells were enriched based on surface markers as Lin−Sca−1+c-Kit+.
For immunophenotypic analysis of HSPC, murine BM were washed and incubated for 30 minutes with biotin conjugated lineage markers (CD11b, Gr1, Ter119, CD3, B220, Mouse Hematopoietic Lineage Biotin Panel, 88-7774-75, eBioscience), followed by staining with streptavidin eFluor® 450 (48-4317-82, eBioscience), Sca-1 PE-cy7 (25-5981-82, eBioscience), c-Kit APC-eFluor® 780 (47-1170-80, eBioscience), CD34 PE (551387, BD Biosciences), CD135 APC (17-1351-80, eBioscience), CD16/32 APC (17-0161-81, eBioscience). Short-term HSCs (ST-HSCs) were identified based on surface markers as Lin−Sca−1+c-Kit+CD34+CD135−, multipotential progenitors (MPPs) were with Lin−Sca−1+c-Kit+CD34+CD135+, common myeloid progenitor cells (CMP) were with Lin−Sca−1−c-Kit+CD34+CD16/32−, megakaryocyte-erythroid progenitor cells (MEP) were with Lin−Sca−1−c-Kit+CD34−CD16/32−, granulocyte-macrophage progenitor cells (GMP) were with Lin−Sca−1−c-Kit+CD34+CD16/32+. To study myeloid and lymphoid lineages, PB samples were treated with 1xRBC lysis buffer, washed and incubated with CD11b PE-cy5 (15-0112-82, eBioscience), Gr1 eFluor-450 (48-5931-82, eBioscience), CD3 PE (12-0031-83, eBioscience), B220 APC (17-0452-82, eBioscience). Otherwise, BM cells and splenocytes were stained with CD11b PE-cy5, Gr1 eFluor-450, CD3 PE, B220 APC, Ter119 PE-cy7 (25-5921-81, eBiosciences). Myeloid cells, including granulocytes and monocytes, were based on CD11b+ (Gr1+ or Gr1−), B lymphocytes were with B220+, T lymphocytes were with CD3+, erythroblasts were with Ter119+. After BMT and competitive transplantation, PB were stained with CD45.1 PE-cy7 (25-0453-82, eBioscience), CD45.2 APC-eFluor® 780 (47-0454-82, eBioscience), CD11b PE-cy5, CD3 PE, B220 APC, while BM were with lineage markers plus streptavidin eFluor® 450, CD45.1 PE-cy7, CD45.2 FITC (561874, BD Biosciences), Sca-1 PE (12-5981-81, eBioscience), c-Kit APC-eFluor® 780, CD135 PE-Cy5 (15-1351-81, eBiosciences), CD16/32 PerCP-Cy5.5 (45-0161-80, eBioscience), CD34 eFluor® 660 (50-0341-82, eBioscience) for lineage distribution and chimerism study. Analysis was performed using FACSCanto and/or LSRII flow cytometers with either Diva or FlowJo software.
Reagents and related assays
Lin− BM cells were exposed to 10 ng/mL LPS (TLRL-PEKLPS, InvivoGen) RNA was extracted to study exon skipping.
RT-PCR and qRT-PCR
Total RNA was extracted and purified using Quick-RNA MiniPrep (Zymo research, R1055) and reverse transcription was carried out using SuperScript VILO cDNA Synthesis Kit (Invitrogen). PCR was performed with the following primers: Arhgap1 forward 5’-CAGAGCTGCAAGATGACCTG-3’ and reverse 5’-AAGAGCTTGCTGTGGTCCAG-3’; Sema4g forward 5’-CCTCCAGTTTCCTCTGTCCA-3’ and reverse 5’-CTTGCTCCCATTGAGTAGCC-3’; Mllt1 forward: 5’-CCCGACAGACAATGAGAGC-3’ and reverse 5’-ATAGCCCGACTCCTCCACTT-3’; Cep164 forward 5’-CAACGAGGAGGATGAAGAGG-3’ and reverse 5’-CTCTTCCTTGGGACTTGCTG-3’. Mtmr14 forward 5’-AGTCACTCATCCTCCCCACA-3’ and reverse 5’-TGCTGTATGCAGCCAAGAAG-3’; Slmap forward 5’-ACAAGAGCTCCAGGCAAAAA-3’ and reverse 5’-ATCCATCTGGGCGTCTGTAG-3’; ARHGAP17 forward on exon 16 5’-CTCACCACCCCGAGTTCTAA-3’, on exon 17 5’-GAGCTCTAGGGCTGAAAGCA-3’, reverse on exon 19 5’-GACTGGGTGGATGCTGAGAT-3’. Densitometry was determined with Image J. Quantitative PCR was performed with Taqman Master Mix (Life Technologies) for human TRAF6 (Cat# 4331182, Assay# Hs00371512_g1), human hnRNPA1 (Cat# 4453320, Assay# Hs01656228_g1), mouse hnRNPA1 (Cat# 4448892, Assay# Mm02528230_g1), human GAPDH (Cat# 4331182, Assay# Hs02758991_g1), mouse GAPDH (Cat# 4453320, Assay# Mm99999915_g1) on an Applied Biosystems StepOne Plus Real-Time PCR System. All the probes were purchased from Applied Biosystems. To measure total TRAF6 transcript level in Vav-TRAF6 LSK, primers were designed to amply both endogenous (mouse) and ectopic (human) TRAF6, with forward 5’-CTCAGCGCTGTGCAAACTATATATCCC-3’ and reverse 5’-GGCGTATTGTACCCTGGAAGGG-3’. Mouse GAPDH was used as an internal control with forward primer 5’-ACTCAAGATTGTCAGCAATGCATCCTGC-3’ and reverse primer 5’-ATGAGCCCTTCCACAATGCCAAAGTTG-3’. To quantitatively measure exon skipping, misspliced genes were selected with a smaller exon (< 200bp) and ones that were less likely to undergo non-sense mediated decay. Primers were designed as follows: Cep164 forward 5’-GACAACCAGGAGTCTCCAAG-3’ and reverse 5’- TTGAAGAAGCCTTGGGATCA-3’; ARHGAP17 forward for exon 17 and exon 19 boundary 5’-CAGGCGACGGCAGCTGTTAAAAAAC-3’, and reverse 5’-GACTGGGTGGATGCTGAGAT-3’; Mouse GAPDH and human β-actin were used as internal controls for murine and human samples, respectively. Primers for β-actin are forward 5’-CTCTTCCAGCCTTCCTTCCT-3’ and reverse 5’-AGCACTGTGTTGGCGTACAG-3’.
Plasmids
Mouse Arhgap1 exon2 together with its 61 bp upstream and 52 bp downstream intronic sequence were amplified and inserted into pFlare minigene plasmids31. The control pFlare minigene plasmid containing a modified human β-globin gene cassette has been previously described31. Myc-tagged full-length human hnRNPA1 was synthesized and subcloned into pcDNA3.1. hnRNA1 mutant K3R and RRM K-R were generated with Quick Change Multisite-directed Mutagenesis Kit (200515, Agilent Technologies). FLAG-tagged TRAF6 vectors have been described previously50. Mouse Arhgap1 5-UTR fusions were synthesized by Blue Heron Biotech and cloned into pCMV6-A-GFP.
Transfection
Transfection was performed with HEK293 cells with TransIT®-LT1 Transfection Reagent (MIR2305, Mirus) according to the manufactures’ recommendation.
Immunoblotting and immunoprecipiation
Cells were lysed in cold RIPA buffer in the presence of PMSF, sodium orthovanadate, protease and phosphatase inhibitors. Protein concentration was evaluated by a BCA assay (32106, Pierce). For cytosol and nuclei protein fractionation, cytosol proteins were recovered in 10 mM Hepes pH 7.9, 10 mM KCl, 0,1 mM EDTA, 0,1 mM EGTA, 0,4 % NP40, 1 mM DTT, 0,1 mM PMSF, 10 µg/ml aprotonin, 1 mM OVA. For immunoprecipitation, TRAF6, hnRNPA1 or M2 antibodies (2µg) were added to cell lysates (1–5 mg) for 3 h at 4°C and captured by the addition of Protein A/G Plus beads (sc-2003; Santa Cruz) as described before51. The immune complexes were recovered and detected by immunoblotting. Western blot analysis was performed with the following antibodies: TRAF6 (sc-7221; Santa Cruz)52, ubiquitin (sc-8017; Santa Cruz), Myc-tag (2278; Cell Signaling), M2 (F3105; Sigma), Arhgap1/Cdc42gap (discontinued; BD Laboratories)29,53, Cdc42 (ab64533; Abcam), hnRNPA1 (sc-10030; Santa Cruz), Tubulin (ab6160; Abcam), β-actin (4968; Cell Signaling), GAPDH (5174; Cell Signaling).
Cdc42-GTP pulldown assay
Cells were lysed with 20mM Tris/HCl pH 7.6, 100mM NaCl, 10mM MgCl, 1% Triton X-100, 0.2% SDS with protease and phosphatase inhibitors, incubated with glutathione-agarose beads coated with GST-PAK1 and processed further as described previously28.
Lentivirus production
Lentiviral shRNA expression vector (pLKO.1, OpenBiosystems) were obtained from the Lentiviral core at CCHMC and used to express shCTL (empty or scrambled control) and shRNA for human hnRNPA1 (TRCN0000006583, TRCN0000006586), mouse hnRNPA1 (TRCN0000055265) and human TRAF6 (TRCN0000007348), human ARHGAP17 (TRCN0000047451). For knockdown of hnRNPA1, the majority of experiments were performed with clone TRCN0000006583. Lentivirus was generated as previously described52. An inducible knockdown of human TRAF6 was performed in pTRIPZ system (RHS4696-101315918; OpenBiosystems). After transduction of pTRIPZ, 3µg of puromycin was used for selection. Doxycycline was used to induce knockdown of TRAF6.
Statistical analysis
Unless otherwise specified, results are depicted as the mean ± standard error of the mean. Statistical analyses were performed using Student’s t-test. For Kaplan-Meier analysis, Mantel-Cox test was used. GraphPad Prism (v5, GraphPad) was used for statistical analysis. For correlative analyses, Spearman rank test was used.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank members of the Starczynowski laboratory for discussions. This work was supported by Cincinnati Children’s Hospital Research Foundation, American Society of Hematology (ASH), National Institute of Health (RO1HL111103, RO1DK102759, RO1HL114582), Gabrielle’s Angel Foundation for Cancer Research, and Edward P. Evans Foundation grants to D. Starczynowski. D. Starczynowski. is an Leukemia Lymphoma Society Scholar. Cores are supported through the NIDDK Center’s of Excellence in Experimental Hematology (P30DK090971). We thank J. Bailey and V. Summey for assistance with transplantations (Comprehensive Mouse and Cancer Core). We also thank L. Salati (West Virginia University) for advice on the RIP assays, and H. Singh (CCHMC) for helpful discussion. MDSL cells were kindly provided by K. Tohyama (Kawasaki Medical University). Traf6-floxed mice were kindly provided by Y. Choi (University of Pennsylvania).
Footnotes
Accession numbers
The GEO accession number for the microarray data reported in this paper is GSE64542: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=whibqkqkfrapbgn&acc=GSE64542
Author contributions
J.F., L.B., M-D.F., K.G., H.G., Y.Z. and D.T.S. designed and performed experiments; X.L., S.C., S.A., and R.K. provided technical assistance; K.C. provided bioinformatics support. X.C., N.S. and M.T.W. performed the RNA splicing and exon binding analysis; P.S. provided reagents and advice on RNA splicing. D.W. provided histopathology advice. K.G. performed the mass spectrometry analysis. G.G-M. and J.P.M. provided patient samples. J.F. and D.T.S. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interest.
References
- 1.Esplin BL, et al. Chronic exposure to a TLR ligand injures hematopoietic stem cells. J Immunol. 2011;186:5367–5375. doi: 10.4049/jimmunol.1003438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong JJ, et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013;154:583–595. doi: 10.1016/j.cell.2013.06.052. [DOI] [PubMed] [Google Scholar]
- 3.Shalek AK, et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature. 2013;498:236–240. doi: 10.1038/nature12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagai Y, et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Starczynowski DT, et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 6.Kristinsson SY, et al. Chronic immune stimulation might act as a trigger for the development of acute myeloid leukemia or myelodysplastic syndromes. J Clin Oncol. 2011;29:2897–2903. doi: 10.1200/JCO.2011.34.8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei Y, et al. Toll-like receptor alterations in myelodysplastic syndrome. Leukemia. 2013;27:1832–1840. doi: 10.1038/leu.2013.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caputi M, Mayeda A, Krainer AR, Zahler AM. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nimer SD. Myelodysplastic syndromes. Blood. 2008;111:4841–4851. doi: 10.1182/blood-2007-08-078139. [DOI] [PubMed] [Google Scholar]
- 10.Wei Y, et al. American Society of Hematology. Atlanta, GA: 2012. Deregulation of TLR2-JMJD3 Innate Immunity Signaling, Including a Rare TLR2 SNP As a Potential Somatic Mutation, in Myelodysplastic Syndromes (MDS) [Google Scholar]
- 11.Barreyro L, et al. Overexpression of interleukin 1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 2012 doi: 10.1182/blood-2012-01-404699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dimicoli S, et al. Overexpression of the toll-like receptor (TLR) signaling adaptor MYD88, but lack of genetic mutation, in myelodysplastic syndromes. PLoS One. 2013;8:e71120. doi: 10.1371/journal.pone.0071120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, et al. Induction of myelodysplasia by myeloid-derived suppressor cells. J Clin Invest. 2013;123:4595–4611. doi: 10.1172/JCI67580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varney ME, et al. Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling. J Exp Med. 2015;212:1967–1985. doi: 10.1084/jem.20141898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogilvy S, et al. Promoter elements of vav drive transgene expression in vivo throughout the hematopoietic compartment. Blood. 1999;94:1855–1863. [PubMed] [Google Scholar]
- 16.Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 2005;106:287–295. doi: 10.1182/blood-2004-12-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanova NB, et al. A stem cell molecular signature. Science. 2002;298:601–604. doi: 10.1126/science.1073823. [DOI] [PubMed] [Google Scholar]
- 18.Clark TA, Sugnet CW, Ares M., Jr Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 19.Purdom E, et al. FIRMA: a method for detection of alternative splicing from exon array data. Bioinformatics. 2008;24:1707–1714. doi: 10.1093/bioinformatics/btn284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iervolino A, et al. hnRNP A1 nucleocytoplasmic shuttling activity is required for normal myelopoiesis and BCR/ABL leukemogenesis. Mol Cell Biol. 2002;22:2255–2266. doi: 10.1128/MCB.22.7.2255-2266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan Y, et al. Lysine 63-linked polyubiquitination of TAK1 at lysine 158 is required for tumor necrosis factor alpha- and interleukin-1beta-induced IKK/NF-kappaB and JNK/AP-1 activation. J Biol Chem. 2010;285:5347–5360. doi: 10.1074/jbc.M109.076976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, et al. Crystal Structures and RNA-binding Properties of the RNA Recognition Motifs of Heterogeneous Nuclear Ribonucleoprotein L: INSIGHTS INTO ITS ROLES IN ALTERNATIVE SPLICING REGULATION. J Biol Chem. 2013;288:22636–22649. doi: 10.1074/jbc.M113.463901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han K, Yeo G, An P, Burge CB, Grabowski PJ. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zearfoss NR, Clingman CC, Farley BM, McCoig LM, Ryder SP. Quaking regulates Hnrnpa1 expression through its 3’ UTR in oligodendrocyte precursor cells. PLoS Genet. 2011;7:e1001269. doi: 10.1371/journal.pgen.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huelga SC, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcarcel J. hnRNP A1 proofreads 3’ splice site recognition by U2AF. Mol Cell. 2012;45:314–329. doi: 10.1016/j.molcel.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 28.Florian MC, et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell. 2012;10:520–530. doi: 10.1016/j.stem.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Yang L, Filippi MD, Williams DA, Zheng Y. Genetic deletion of Cdc42GAP reveals a role of Cdc42 in erythropoiesis and hematopoietic stem/progenitor cell survival, adhesion, and engraftment. Blood. 2006;107:98–105. doi: 10.1182/blood-2005-05-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang L, et al. Rho GTPase Cdc42 coordinates hematopoietic stem cell quiescence and niche interaction in the bone marrow. Proc Natl Acad Sci U S A. 2007;104:5091–5096. doi: 10.1073/pnas.0610819104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci U S A. 2008;105:11218–11223. doi: 10.1073/pnas.0801661105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florian MC, et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature. 2013;503:392–396. doi: 10.1038/nature12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolatshad H, et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia. 2015;29:1092–1103. doi: 10.1038/leu.2014.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iglesias MJ, et al. Combined chromatin and expression analysis reveals specific regulatory mechanisms within cytokine genes in the macrophage early immune response. PLoS One. 2012;7:e32306. doi: 10.1371/journal.pone.0032306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research, N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wells CD, et al. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 2006;125:535–548. doi: 10.1016/j.cell.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 37.Jean-Philippe J, Paz S, Caputi M. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci. 2013;14:18999–19024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elias HK, et al. Stem cell origin of myelodysplastic syndromes. Oncogene. 2014;33:5139–5150. doi: 10.1038/onc.2013.520. [DOI] [PubMed] [Google Scholar]
- 39.Baldridge MT, King KY, Boles NC, Weksberg DC, Goodell MA. Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature. 2010;465:793–797. doi: 10.1038/nature09135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Essers MA, et al. IFNalpha activates dormant haematopoietic stem cells in vivo. Nature. 2009;458:904–908. doi: 10.1038/nature07815. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Yang L, Debidda M, Witte D, Zheng Y. Cdc42 GTPase-activating protein deficiency promotes genomic instability and premature aging-like phenotypes. Proc Natl Acad Sci U S A. 2007;104:1248–1253. doi: 10.1073/pnas.0609149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chambers SM, et al. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han D, et al. Dendritic cell expression of the signaling molecule TRAF6 is critical for gut microbiota-dependent immune tolerance. Immunity. 2013;38:1211–1222. doi: 10.1016/j.immuni.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emig D, et al. AltAnalyze and DomainGraph: analyzing and visualizing exon expression data. Nucleic Acids Res. 2010;38:W755–W762. doi: 10.1093/nar/gkq405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLeay RC, Bailey TL. Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics. 2010;11:165. doi: 10.1186/1471-2105-11-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J, et al. SpliceTrap: a method to quantify alternative splicing under single cellular conditions. Bioinformatics. 2011;27:3010–3016. doi: 10.1093/bioinformatics/btr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wijeratne AB, Manning JR, Schultz Jel J, Greis KD. Quantitative phosphoproteomics using acetone-based peptide labeling: method evaluation and application to a cardiac ischemia/reperfusion model. J Proteome Res. 2013;12:4268–4279. doi: 10.1021/pr400835k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cyphert TJ, Suchanek AL, Griffith BN, Salati LM. Starvation actively inhibits splicing of glucose-6-phosphate dehydrogenase mRNA via a bifunctional ESE/ESS element bound by hnRNP K. Biochim Biophys Acta. 2013;1829:905–915. doi: 10.1016/j.bbagrm.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rhyasen GW, et al. Targeting IRAK1 as a Therapeutic Approach for Myelodysplastic Syndrome. Cancer Cell. 2013;24:90–104. doi: 10.1016/j.ccr.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Starczynowski DT, et al. TRAF6 is an amplified oncogene bridging the RAS and NF-kappaB pathways in human lung cancer. J Clin Invest. 2011;121:4095–4105. doi: 10.1172/JCI58818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang J, et al. Cytotoxic effects of bortezomib in myelodysplastic syndrome/acute myeloid leukemia depend on autophagy-mediated lysosomal degradation of TRAF6 and repression of PSMA1. Blood. 2012 doi: 10.1182/blood-2012-02-407999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang L, Yang L, Burns K, Kuan CY, Zheng Y. Cdc42GAP regulates c-Jun N-terminal kinase (JNK)-mediated apoptosis and cell number during mammalian perinatal growth. Proc Natl Acad Sci U S A. 2005;102:13484–13489. doi: 10.1073/pnas.0504420102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.