Abstract
National policies designed to control infectious diseases should allocate resources for interventions based on regional estimates of disease burden from surveillance systems. For many infectious diseases, however, there is pronounced seasonal variation in incidence. Policy-makers must routinely manage a public health response to these seasonal fluctuations with limited understanding of their underlying causes. Two complementary and poorly described drivers of seasonal disease incidence are the mobility and aggregation of human populations, which spark outbreaks and sustain transmission, respectively, and may both exhibit distinct seasonal variations. Here we highlight the key challenges that seasonal migration creates when monitoring and controlling infectious diseases. We discuss the potential of new data sources in accounting for seasonal population movements in dynamic risk mapping strategies.
Toward dynamic risk mapping of seasonal infections
Some of the most important infectious diseases of humans, including influenza, cholera, malaria, dengue, and measles, exhibit epidemic dynamics, with incidence often peaking at particular times of the year. Seasonal dynamics are driven both by human factors, such as changing patterns of human aggregation (Figure 1), population-level susceptibility to infection, or importation of pathogens that spark periodic epidemics, and also by biological aspects of transmission such as climatic suitability for disease spread or the availability of vector species. For example, immunizing childhood infections such as measles, rubella, and pertussis, exhibit strong seasonal dynamics, generally reflecting ‘term-time forcing’ where periods of high transmission echo children’s attendance of school [1]. Seasonal migration linked to agriculture, fisheries, and pastoralism has been part of the cultural landscape of many parts of the world for generations (e.g., Niger [2], Ghana [3], India [4]), and has also been implicated in seasonal epidemics of measles, for example [5]. In contemporary populations across both high and low income settings, domestic and international movements linked to work, holidays, and school terms are of significant magnitude, and are likely to shape both the spatial spread and timing of epidemics, as well as modulating the surveillance systems that we use to observe outbreaks (Figure 2). Despite this, the human drivers of infectious disease seasonality are still poorly understood, particularly in low income regions where the burden of infectious diseases is highest.
Figure 1. Seasonal movement and infectious disease dynamics.
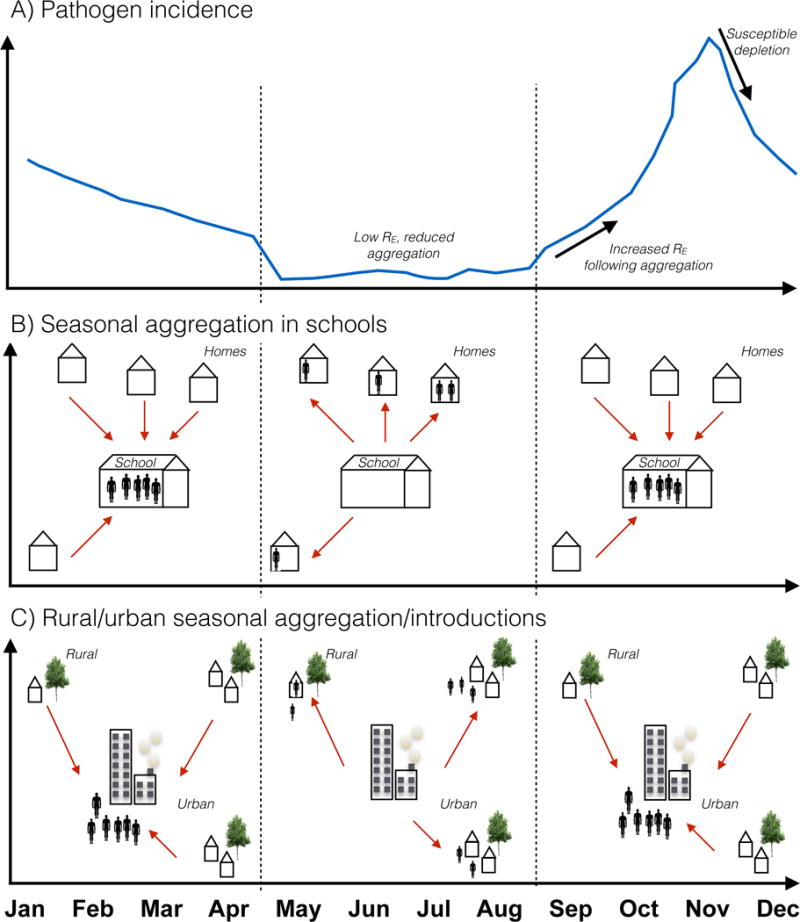
A) Schematic incidence of an infection (blue line) showing cases (y axis) against time of year (x axis). A seasonal peak of incidence follows the summer. The underlying driver of this seasonal cycle is changes in the net reproductive number RE, or the number of new infections per infectious individual, because RE is the product of the basic reproductive number (R0, or the number of new infections in a completely susceptible population), and S, the size of the accessible susceptible population, which varies seasonally following human behavior, but also susceptible depletion by infection. Two possible scenarios are illustrated: B) Aggregation of children in schools after the summer holiday (red arrows) increases effective contact among susceptible individuals, thus altering RE and increasing incidence (reported for many childhood infections [1, 50, 54]); or C) Aggregation in cities after the period of agricultural work where communities have migrated out to rural settings (red arrows) results in an increase in RE (reported for measles in Niger [5])
Figure 2. Seasonal reporting biases in disease incidence.
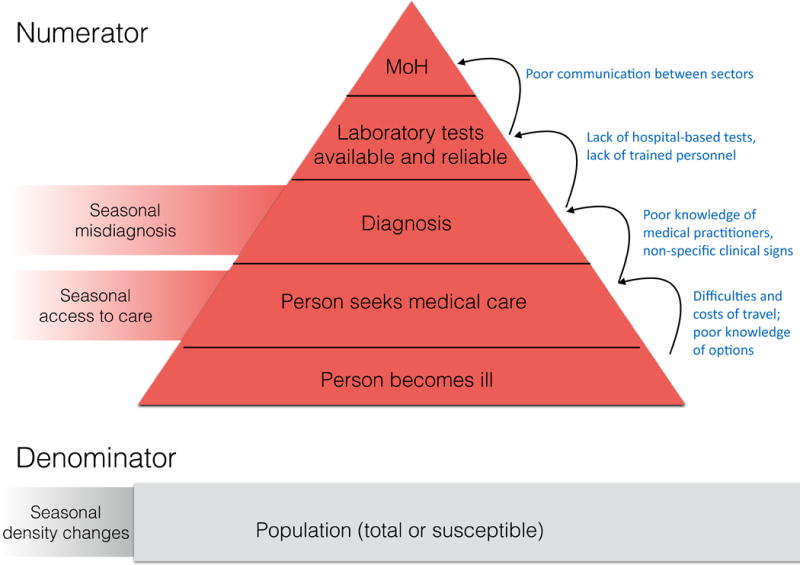
In resource poor settings, the numerator of incidence generally under-estimates cases that have occurred as a result of under-reporting and information loss at a range of scales; here, depicted by a triangle where the narrow point indicates the final data that filters through the surveillance system to the ministry of health (MoH). The filter might also fluctuate seasonally: access to medical health centers where reporting occurs is likely to fluctuate seasonally, for reasons ranging from roads being washed out, to no one being available to accompany sick individuals during busy times such as harvest season. Misdiagnosis might also vary seasonally, for example, where at particular times of year, the expectation is that malaria is the core cause of fever. Finally, the denominator might also vary seasonally as a result of large-scale movements of population, for example linked to agriculture.
For many pathogens, environmental variables are equally important drivers of seasonal variation in disease incidence, and interact with human movements in complex ways. Influenza seasonality, for example, is likely to reflect both term-time forcing [6] and variation in absolute humidity [7]. Vector-borne infections like malaria and dengue fever are strongly influenced by the seasonality of mosquito populations, which are in turn dependent on oscillations in rainfall and temperature [8]. Similarly, in regions with seasonal cholera epidemics, such as Bangladesh, monsoon rains and flooding bring high risk of the water-borne infection each year [9]. Thus, the seasonal cycles of the environment and in human societies interact to produce large oscillations in disease risk. If we can disentangle the key drivers of seasonal epidemics, however, the relative regularity of human behaviors and climatic changes mean that we may be able to predict and prepare for outbreaks. Whereas spatial heterogeneity is increasingly being taken into account to guide resource allocation for disease control [10–12], most risk maps are currently static, providing a single snapshot of incidence. We propose that for many infectious diseases, new approaches for measuring and modeling seasonal population movements may offer robust predictive power for understanding temporal changes in incidence, in addition to spatial heterogeneities.
To establish the impact of seasonal human movements on disease incidence, quantifying human distributions and mobility patterns will be necessary in many cases, but this has proved extremely challenging, particularly in low-income settings and on relevant time scales [13]. For some infections such as measles, the biological basis of transmission is sufficiently well understood that the human dynamics driving seasonal epidemics can be inferred from epidemiological data [14, 15], but this is not the norm. New approaches are being developed to directly observe human mobility using mobile phone data and satellite imagery [16], and these have the potential to transform surveillance tools and public health interventions. However, important analytical and logistical challenges remain, particularly related to integrating diverse data-streams across scales, managing varying resolution in infectious disease surveillance data, and measuring driver variables such as growing seasons for agriculture crops [10, 17]. In addition to these analytical problems, there are multiple political hurdles to overcome in protecting individual privacy and forming partnerships necessary to integrate these approaches into control programs [18]. Here we outline the implications of seasonality – particularly due to human movement – for the surveillance and control of infectious diseases, and argue that we are now in a position to move towards dynamic risk mapping for many infections.
Seasonal population movements may bias estimates of disease burden
Two human factors underlie an epidemic; a sufficient density of susceptible people, and – in places where the disease is not endemic – the introduction of an infectious individual, providing the “spark” to ignite it. In populations that are too small to sustain transmission (i.e., below the Critical Community Size for immunizing infections [19]), or where control has been successfully implemented to achieve elimination, temporal patterns of outbreaks depend on the timing of these introduction events, often from some larger population center. For example, smaller cities in England and Wales lag behind London in the timing of their measles epidemics, as people spread infection outwards from the capital [20].
Introduction events also spark seasonal epidemics of influenza, which occur when novel strains are imported into populations; and evidence from the 2009 H1N1 outbreak indicates a role for both school opening schedules and local, small scale movement [6]. For vector-borne epidemic diseases like malaria and the more recently emerging dengue fever, both novel pathogen strains entering susceptible populations (facilitated by human movement [21]) and seasonally changing environmental suitability for the mosquito vector determine seasonal and epidemic dynamics [9, 12, 22, 23]. Being able to make predictions about the severity, location, and timing of seasonal infectious disease epidemics would not only greatly improve public health preparation and response, and facilitate surveillance and monitoring of population susceptibility, but also allow for more efficient, timed interventions. Improved understanding of the interaction between seasonal population movements and climatic oscillations in driving epidemics will facilitate better predictive frameworks and dynamic risk estimates.
The extent to which seasonal movements will impact infectious disease burden and control policies will depend on the epidemiology of the pathogen and the surveillance strategies used to measure and respond to it. Surveillance forms the basis of infectious disease control policy and the allocation of financial resources from Ministries of Health to smaller administrative units, but it can be biased in many ways. Passive surveillance is the norm for many infections: disease incidence is reported from local clinics and hospitals as the number of cases occurring over some time period relative to the population at-risk, which is assumed to correspond to a census-based, or projected, estimate of the local population size. Data is often then further aggregated, for example into yearly time-steps (e.g., see the World Health Global Health Observatory data repository [24]) or broad age groups; perhaps a legacy of times when data storage and data transmission were more arduous. Such temporal aggregation very obviously impedes estimates of seasonality in burden, but even where data is not aggregated to yearly time-steps, there are many ways in which seasonal fluctuations may bias passive surveillance (Figure 2).
Seasonal movements may cause both the number of cases reported and the at-risk population to be inaccurate in complex ways [25, 26]. First, in determining the frequency of local disease cases (i.e. measuring the numerator of incidence and prevalence estimates) seasonal movements can impact the number of individuals reporting to clinics, since travelers may seek treatment wherever they become sick. For infections with longer latent periods, residents who become infected while traveling, but only suffer symptoms once they come home, can inflate estimates of local transmission. These issues are particularly important for infections with non-specific symptoms that may be treated without confirmation of the causal pathogen, and for infections with a large proportion of asymptomatic cases. For example, malaria, influenza, and dengue fever can all cause acute febrile illness in some, particularly non-immune individuals, and asymptomatic infections in others. Clinicians may either under- or over-estimate the probability that a patient has one or the other infection, depending on their assessment of local transmission. In many tropical regions, dengue fever, Chikungunya, influenza, and malaria, may all be circulating, but with varying frequency in different populations and at different times, due to regional differences in climate or transmission patterns. Without a good understanding of the changing patterns of risk for each of these infections, clinicians may systematically misdiagnose patients [27], resulting in inappropriate treatment and misreporting to the central surveillance system (Figure 2).
In addition to altering the number of reported cases, seasonal population movements affect the size of the at-risk population (i.e. the denominator for prevalence and incidence calculations [13], Figure 2). Population densities may change dramatically in regions where seasonal mobility is pronounced, biasing both the estimated transmission parameters -calculated assuming a particular population density [28]- as well as the requirements for preventative measures or treatment options. For example, in Niger, many people migrate northwards during the rainy season for the agricultural opportunities in the low density northern regions, returning to the cities in the dry season [2]. In this case, for a given number of cases, incidence will be over-estimated in places with high numbers of migrants and under-estimated in areas where many residents are absent. An important consideration for the impact of these shifting population densities is the demographic structure of the mobile populations [22, 29]; in East Africa for example, an analysis of human population movements from census data shows that different demographic groups travel different routes and variable amounts [29–31]. Often, seasonal movements for work means that young men are the most mobile. This causes the profile of disease susceptibility and transmission potential to vary in both the origin and destination populations.
Surveillance may also be active, moving beyond passive recording of clinical cases to a system that directly surveys the population. For immunizing infections where a vaccine is available, serological surveys that characterize the immune status of individuals in different age categories [32] are often used to inform the basis of decision-making for vaccine requirements and delivery. Epidemic-prone infections without an effective vaccine rely on surveillance for allocation of resources for prevention, case management, and transmission reducing measures. This might include distribution of bed-nets and targeted insecticide spraying (prevention) and drug delivery for malaria, for example. Here, biases in both the numerator and the denominator as a result of human movement are possible; and the issue becomes the timing of the survey relative to the ebbs and flows in population movements. Similar concerns arise as with passive surveillance, but the timing of serological surveys can be adjusted to take these fluctuations into account, providing hope that better understanding of population movements could reduce bias substantially. Overall, both active and passive surveillance may be biased by seasonal movement; and Table 1 illustrates the impact of seasonal movements for different surveillance approaches, highlighting the disease-specific aspects to biases.
Table 1.
Surveillance methods for particular pathogens, target information on population, and possible sources of bias introduced by seasonal movement.
| Pathogen | Surveillance method | Targeted information | Challenges introduced by seasonal movement |
|---|---|---|---|
| General across pathogens | Case reports | Local and regional incidence and prevalence estimates | Over or under estimation of incidence/prevalence due to non-accounting for denominator changes |
| Measles / rubella | Serological surveys using IgG (i.e., detecting long term immunity) | -Susceptibility in the population; average age of infection, and related variables (e.g., R0, susceptibility in women of childbearing age for rubella, etc). | -Timing of survey deployment may result in missed seasonally migrant susceptible groups, underestimating size of the susceptible pool and thus outbreak risk. -Timing of survey deployment relative to peak of incidence shaped by seasonal movement can alter the inferred average age or rate of acquisition of infection, modifying estimates of R0, etc. |
| Vaccine preventable diseases generally | Vaccine card monitoring | Vaccination coverage | Timing of survey deployment relative to seasonal pulses in birth as well as seasonally migrant groups, and incidence may bias estimates of coverage. |
| Malaria | Clinic-based reporting of symptomatic cases (sometimes confirmed by microscopy or RDTa, often not) | Bed net and IRS requirements by region, ACT allocation (in Vietnam, e.g., this is based on the previous years’ case reports) | -Seasonal epidemics can lead to ACT stock-outs-possibility of too many bed nets in the wrong places, too few in the right places -Unnecessary coverage of bednets/spraying where little local transmission (all imported) -Failure to effectively target “source” locations-misdiagnosis and incorrect treatment |
| Dengue | Clinic-based reporting of cases (a fraction of which would be lab-confirmed) | Hospital preparedness, vector control | -Hospital overflow issues -Unnecessary or insufficient vector control in the affected region -Misdiagnosis and treatment of dengue cases |
Abbreviations: ACT, artemisinin combination therapy; IRS, indoor residual spraying; RDT, rapid diagnostic test.
Policy implications of biases
Misallocation of resources resulting from biased surveillance estimates may occur in either direction, with the need for both prevention and treatment options in different regions being under- or over-estimated as a result. For countries with constrained financing for public health, over-allocation can be just as problematic as under-allocation, wasting resources and taking money away from interventions in other regions where they are urgently needed. In the case of malaria, for example, frequent imported cases to a particular area may lead to an over-estimate of local vectorial capacity and a misallocation of bed nets and insecticides to populations that are in fact dominated by imported infections, and away from the high transmission settings where cases originate [33]. For infections that cause sporadic outbreaks where a vaccine is available, responding in a targeted, timely manner can be essential [34–36]. Reactive vaccination campaigns for cholera or meningococcal outbreaks, for example – which are generally initiated when local cases reach a particular threshold and take several weeks to roll out – may be triggered ineffectively or in the wrong places if surveillance is biased. Similarly, if particular demographic groups (for example low income families who migrate seasonally, or particular age brackets) are absent at certain times of the year, estimates may not reflect the true distribution of susceptibility and the need for vaccine doses. The same issues may also critically impact estimates from randomized control trials of interventions, where differences in incidence in treatment versus control populations underlie efficacy measures. In addition to these population-level impacts, for many infections that cause general or ambiguous symptoms (e.g. influenza-like illness, which may be caused by any number of pathogens), individual case management may also be affected by incorrect assessments of local infection risk, as described above.
Despite these issues, which are generally acknowledged but poorly measured, spatial and temporal heterogeneities in infection risk are currently not adequately accounted for, even though the drivers – both climatic and human – of seasonal population movements are potentially predictable. For example, although seasonal aggregation is recognized to drive annual and multi-annual dynamics of measles, estimates of the overall burden of measles [37], as well as projection of the burden resulting from disruptions to the health care system post-Ebola [38], have tended to neglect this, and single time point estimates of disease burden or static risk maps are the norm for many distinctly seasonal infections. Significant progress has been made in generating risk maps for malaria that reflect spatial heterogeneities in prevalence (e.g. www.map.ox.ac.uk), for example, as well as incorporating mobility patterns [21, 33], but these generally do not account for the pronounced seasonal peaks in prevalence driven by rainfall, temperature, and resulting mosquito population dynamics (although work is beginning in this direction, see [12, 23, 39–42]).
The need for efficient use of resources calls for temporal variation in both surveillance and control efforts that reflect fluctuating risk for many seasonal infectious diseases. Logistical restrictions emerge as an important consideration here – for example, vaccination campaigns may take years to prepare [43], and thus are hard to deploy reactively. Even when successfully implemented, there is often spatial heterogeneity in coverage achieved, with remote communities remaining underserved [44]. Combined, these two features provide strong motivation to time campaigns to the period during which seasonally mobile populations are most likely to be in areas where vaccination has highest uptake. For other time-varying interventions, such as seasonal malaria chemoprophylaxis, the interaction between human movements and climatic fluctuations that drive vector availability will directly impact the efficacy of the intervention. Thus, although surveillance, prevention and outbreak control strategies may be suboptimal in the face of large seasonal population movements, evidence-based adjustments in both the spatial allocation and timing of prevention and intervention strategies could potentially mitigate these effects.
New approaches to understanding seasonality
Unlike epidemics driven by rare emergence events and/or natural disasters (e.g. the 2014 Ebola epidemic or the 2010 cholera epidemic in Haiti following an earthquake [45]), outbreaks that are broadly repeatable due to annual weather patterns and seasonal movements should be amenable to predictive modeling and targeted interventions. For many infections, this relies on a clear understanding of both the environmental and human drivers of epidemics. Although the relationship between transmission and climate remains poorly understood for many pathogens, the expanding availability of temporally and spatially resolved incidence data, paired with ever more accurate climate measurements has spurred efforts ranging from wavelet analysis [46] to detailed dynamic model investigation of drivers for a range of pathogens [47]; and this is likely to expand in the coming years. At the more local scale, new diagnostic approaches are likely to mitigate the issue of seasonal mis-diagnosis (Figure 2). However, human mobility is likely to be an ongoing challenge to measure and predict, and our focus on emerging approaches for tackling aspects of seasonality and control of infectious disease that are linked to human mobility. We see three distinct aspects to these approaches: first, in quantifying how people are distributed in time and space, second in assessing the directionality of human fluxes, and third in meaningfully combining these two aspects with transmission parameters of interest.
The first challenge that must be met to accurately encompass seasonal human movement in estimates of seasonal disease burden is to adequately characterize the population at risk (the denominator, Figure 2). New methods and Bayesian approaches to integrate multiple sources of data into nuanced mapping of populations are likely to provide part of the solution to defining the denominator [28], particularly as seasonally varying sources of information such as mobile phone data or night lights [48, 49] could be deployed within this framework (Figure 3). A complication for many infectious diseases, and particularly those for which there is some form of immunity, is that past locations of individuals, and specifically, what this past implies for history of infection (or vaccination) will also determine whether they are at risk. Similarly, interpreting seasonal variation in the numerator (Figure 2) may require combining the incidence information with knowledge of the source and destination populations of individuals presenting as cases, both because this will determine where efforts should be targeted for interventions, but also because this may reveal the core processes underlying infection (Figure 1)[33]. A complementary and potentially powerful way to improve the estimation of the denominators for these calculations would be to report cases testing negative for infection during routine surveillance, in addition to positive cases, providing insights into the number and distribution of patients reporting to health facilities around a country. For diseases where elimination planning is underway (e.g. polio or malaria), this information is critical for finding remaining reservoirs of infection, and for evaluating elimination status. Together, these attributes imply a need to explicitly characterize the underlying patterns of human movement shaping the numerator and denominator (in addition to accurately mapping numbers of people in a particular place at a particular time).
Figure 3. New directions for characterizing human seasonal movement, and evaluating its effects on infectious disease dynamics.
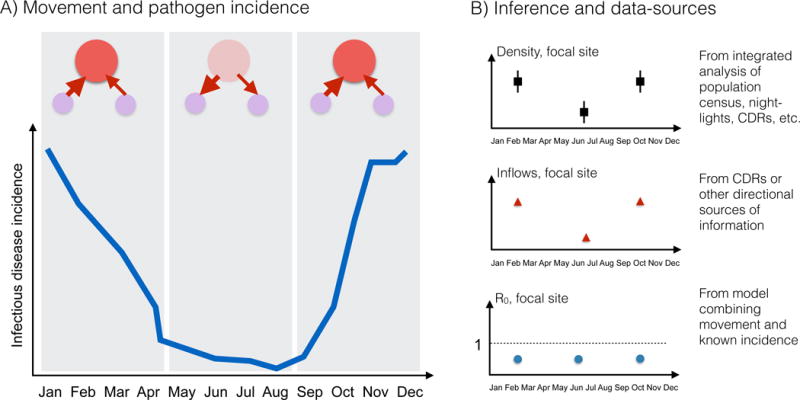
A) A hypothetical population consisting of three patches, a focal site (red), where density varies from low (pale red) to high density (dark red) over the course of the year (x axis) following movement to two smaller patches (red arrows) which are unevenly visited (arrow size); associated pathogen incidence (y axis) over the same time-course (x axis) is shown below (blue line); BB) Potential sources of inference into this system include integrated Bayesian models of population density (top panel) that incorporate seasonally fluctuating sources (extending on [49]); directional information on population flows, such as those available from call data records (CDRs) (middle panel [23, 50, 55]); and methods which combine information on human density and movement with that on infectious disease incidence (bottom panel) using mathematical models to strengthen inference into core parameters such as R0 [33].
Second, we must leverage new methods for quantifying the human dynamics that lead to these spatial distributions. Measuring human travel patterns remains challenging, particularly in low income settings in regions with low population density. However, new approaches to understanding human movements are being developed that utilize the “digital exhaust” produced by mobile phones and other devices, which provide time-stamped locations for millions of individuals. The utility of this approach varies with the density of mobile phone towers, the data available for analysis, phone ownership and usage rates, and the behaviors of subscribers, but is nevertheless promising. Where this data is available, and particularly where it can be combined with data on seasonal disease incidence, there is potential for considerable advances in understanding the role played by human movement in shaping seasonal disease dynamics. Previous work has suggested, for example, that seasonal mobility in Kenya shapes the magnitude of transmission of rubella [50], which has implications in turn for spatial dynamics of the infection, and can determine the burden of Congenital Rubella Syndrome [51]. Likewise, predictive frameworks for dengue outbreaks are more accurate when they incorporate both climatic variations and human movements measured by mobile phones [52], allowing for vector control and hospital preparation in advance of epidemics. With sufficient longitudinal data, it is also possible that the predictability of seasonal movements allows for effective models of disease transmission even in the absence of ongoing access to mobile phone or nightlight data.
Once the location and seasonal dynamics of populations are well described, there are promising possibilities for using modeling approaches to estimate transmission parameters that drive disease spread. In particular, for many important infections, movement patterns shaping transmission may be predictable from year to year, so generalizable models can be used even in the absence of direct data. For example, knowledge of human movement patterns can be a powerful tool for identifying locations where infections are persisting in the absence of re-introductions. Formally, these can be characterized as locations where the basic reproduction number R0<1; and a meta-population model of infectious disease dynamics building on availability of human movement data allows these to be formally identified, enabling targeting for control [33]. So far, such approaches have not considered seasonality, but building on biological knowledge of constraints of vector life-cycles, in the case of malaria or dengue, there is considerable scope for extending this, and refining targeting to be limited in time as well as space.
In addition, combining meta-population frameworks with other sources of information about seasonality can provide validation of mobility models. Travel surveys and other sociological tools will remain a key individual-level source of information about the demographics driving, and motivations for, seasonal population movements [22]. Travel surveys and census data often highlight important gaps in our knowledge about the link between individual- and population-level measures of mobility [29, 53]. Other sources of metadata offer powerful tools for inferring movements indirectly. For example, pathogen genomics provide additional insights into mixing patterns between pathogen populations circulating in different regions, as well as information about where imported infections are coming from. Although integrating seasonal changes in pathogen population sizes is still difficult within population genetic models, the external (inferences from mobile phone data) and internal (pathogen genomics) measures of migration between populations will in many cases provide power to estimate spatial spread of disease due to human movements. Another way to validate and improve seasonal models is by leveraging the differential seasonal peaks from multiple pathogens, it is possible that we can gain further insight into the regional human travel that underlies all of them – providing inference even in the absence of ongoing travel data. Clearly, policy-relevant models will be optimized with as many different, complementary sources of metadata as possible for a given location.
Concluding Remarks
Seasonal travel of one sort or another is a fact of life for nearly every country and socioeconomic group, and its impact on infectious disease outbreaks and control policies will depend on the pathogens and regions in question [30]. Disentangling the variable human travel patterns that contribute to seasonal epidemics will be an important step towards preparing for, and responding to, seasonal infections, and may in some cases provide valuable opportunities to time interventions most effectively (see Outstanding Questions). As climate data and estimates of human mobility improve and we gain access to better diagnostics and rapid sequencing technologies, our ability to measure the drivers of seasonal infections and respond to them efficiently will also improve.
Outstanding Questions.
Who is driving seasonal population movements? Highly mobile groups are often assumed to be some of the most vulnerable populations. Mobile populations are by definition hard to keep track of and access for surveillance and interventions, and poor and stigmatized populations can be more likely to actively avoid national control programs or surveillance. Seasonal mobility is not driven by a single demographic or ethnic group, however, and mobility per se cannot be simply categorized as a risk factor. The first step towards understanding the migratory patterns that underlie seasonal epidemics, therefore, is careful quantitative measurement and nuanced sociological analysis of the dynamics of populations of interest in each region.
How do we incorporate seasonality into mathematical models effectively? Dynamical models of infectious disease transmission, statistical methods for handling spatial data, and population genetic models, all struggle to incorporate temporal fluctuations. These methods primarily deal with cross-sectional data and/or assume fixed population sizes, and new theoretical developments are needed, not only to effectively integrate seasonal fluctuations into analyses, but also to bring together analytical approaches from different fields.
How can promising approaches be integrated into control programs? To date, the use of different types of metadata to augment estimates of disease risk has been largely opportunistic. These diverse data streams have been collected for different purposes, and on different spatial and temporal scales, brought together through academic and political networks rather than as part of an integrated approach to disease control. The biggest challenges, therefore, may be integrating these potentially powerful data streams and analytical approaches with public health programs in real world contexts, where politics and lack of sustained funding and capacity may prove to be important barriers to their implementation.
Trends Box.
Regular, seasonal fluctuations in infectious disease incidence are common. Although human population movements are likely to be important drivers of seasonality, they are poorly quantified and rarely taken into account when planning interventions.
Seasonal population movements may cause systematic biases in surveillance estimates of disease burden, leading to misallocation of resources.
New data sources and analytical approaches offer tools for quantifying and accounting for seasonality in infectious disease risk.
Dynamic population mapping is an important goal for the development of policies to control many infectious disease threats.
Acknowledgments
This work was funded by a Wellcome Trust Sustaining Health Grant, 106866/Z/15/Z (COB, AJT, CJEM), the Bill and Melinda Gates Foundation (CJEM, AJT), and the Models of Infectious Disease Agent Study program (cooperative agreement 1U54GM088558) (COB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bjørnstad ON, et al. Endemic and epidemic dynamics of measles: Estimating epidemiological scaling with a time series SIR model. Ecological Monographs. 2002;72:169–184. [Google Scholar]
- 2.Rain D. Eaters of the dry season: circular labor migration in the West African Sahel. Westview Press; 1999. [Google Scholar]
- 3.Primavera C. Rural-Rural Migration in Ghana: The effects of out-migration on the sustainability of agriculture in the Upper West Region Ghana International School for Humanities and Social Sciences. University of Amsterdam; 2005. [Google Scholar]
- 4.Haberfeld Y, et al. Seasonal migration of rural labor in India. Population Research and Policy Review. 1999;18(5):471–487. [Google Scholar]
- 5.Ferrari MJ, et al. The dynamics of measles in sub-Saharan Africa. Nature. 2008;451:679–684. doi: 10.1038/nature06509. [DOI] [PubMed] [Google Scholar]
- 6.Gog JR, et al. Spatial transmission of 2009 pandemic influenza in the US. 2014 doi: 10.1371/journal.pcbi.1003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaman J, et al. Real-time influenza forecasts during the 2012–2013 season. Nature communications. 2013;4:2837. doi: 10.1038/ncomms3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brady OJ, et al. Global temperature constraints on Aedes aegypti and Ae. albopictus persistence and competence for dengue virus transmission. Parasites and Vectors. 2014;7:338. doi: 10.1186/1756-3305-7-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cash BA, et al. Cholera and shigellosis: different epidemiology but similar responses to climate variability. PloS One. 2014;9(9):e107223. doi: 10.1371/journal.pone.0107223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraemer MU, et al. Progress and Challenges in Infectious Disease Cartography. Trends in parasitology. 2016;32(1):19–29. doi: 10.1016/j.pt.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 11.Hay SI, et al. Global mapping of infectious disease. Philosophical Transactions of the Royal Society of London B: Biological Sciences. 2013;368(1614):20120250. doi: 10.1098/rstb.2012.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen JM, et al. Rapid case-based mapping of seasonal malaria transmission risk for strategic elimination planning in Swaziland. Malar J. 2013;12:61. doi: 10.1186/1475-2875-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatem AJ. Mapping the denominator: spatial demography in the measurement of progress. International health. 2014;6(3):153–155. doi: 10.1093/inthealth/ihu057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grenfell BT, et al. Endemic and epidemic dynamics of measles: Scaling predictability, noise and determinism with the time series SIR model. Ecological Monographs. 2002;72:185–202. [Google Scholar]
- 15.Xia Y, et al. Measles metapopulation dynamics: a gravity model for epidemiological coupling and dynamics. American Naturalist. 2004;164:267–281. doi: 10.1086/422341. [DOI] [PubMed] [Google Scholar]
- 16.Tatem AJ. Mapping population and pathogen movements. International health. 2014;6(1):5–11. doi: 10.1093/inthealth/ihu006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sturrock HJ, et al. Targeting asymptomatic malaria infections: active surveillance in control and elimination. PLoS Med. 2013;10(6):e1001467. doi: 10.1371/journal.pmed.1001467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Omumbo JA, et al. How well are malaria maps used to design and finance malaria control in Africa? PloS one. 2013;8(1):e53198. doi: 10.1371/journal.pone.0053198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartlett MS. The Critical Community Size for Measles in the United States. Journal of the Royal Statistical Society. Series A. 1960;123:37–44. [Google Scholar]
- 20.Grenfell BT, et al. Travelling waves and spatial hierarchies in measles epidemics. Nature. 2001;414:716–723. doi: 10.1038/414716a. [DOI] [PubMed] [Google Scholar]
- 21.Wesolowski A, et al. Quantifying the impact of human mobility on malaria. Science. 2012;338(6104):267–270. doi: 10.1126/science.1223467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall JM, et al. Key traveller groups of relevance to spatial malaria transmission: a survey of movement patterns in four sub-Saharan African countries. Malaria journal. 2016;15:200. doi: 10.1186/s12936-016-1252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatem AJ, et al. Integrating rapid risk mapping and mobile phone call record data for strategic malaria elimination planning. Malaria journal. 2014;13(1):52. doi: 10.1186/1475-2875-13-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.World Health Organization. Global Health Observatory. 2016 http://www.who.int/gho/en/ (accessed June 2016)
- 25.Prothero M. Migration and Health. African Population Studies. 1990;(4) http://dx.doi.org/10.11564/4-0-425.
- 26.Prothero RM. Population mobility and trypanosomiasis in Africa. Bulletin of the World Health Organization. 1963;28(5–6):615. [PMC free article] [PubMed] [Google Scholar]
- 27.Diarra A, et al. Seasonal performance of a malaria rapid diagnosis test at community health clinics in a malaria-hyperendemic region of Burkina Faso. Parasites & vectors. 2012;5(1):1. doi: 10.1186/1756-3305-5-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alegana VA, et al. Fine resolution mapping of population age-structures for health and development applications. Journal of The Royal Society Interface. 2015;12(105):20150073. doi: 10.1098/rsif.2015.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pindolia DK, et al. The demographics of human and malaria movement and migration patterns in East Africa. Malar J. 2013;12(397):10.1186. doi: 10.1186/1475-2875-12-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prothero RM. Population movements and problems of malaria eradication in Africa. Bulletin of the World Health Organization. 1961;24(4–5):405. [PMC free article] [PubMed] [Google Scholar]
- 31.Pindolia DK, et al. Human movement data for malaria control and elimination strategic planning. Malaria journal. 2012;11(1):1. doi: 10.1186/1475-2875-11-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gay N, et al. Interpretation of serological surveillance data for measles using mathematical models: implications for vaccine strategy. Epidemiology and Infection. 1995;115(01):139–156. doi: 10.1017/s0950268800058209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruktanonchai NW, et al. Identifying Malaria Transmission Foci for Elimination Using Human Mobility Data. PLOS Comput Biol. 2016;12(4):e1004846. doi: 10.1371/journal.pcbi.1004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grais RF, et al. Time is of the essence: exploring a measles outbreak response vaccination in Niamey, Niger. Journal of the Royal Society Interface. 2008;6:67–74. doi: 10.1098/rsif.2007.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrari MJ, et al. Time is (still) of the essence: quantifying the impact of emergency meningitis vaccination response in Katsina State, Nigeria. International health. 2014;6(4):282–290. doi: 10.1093/inthealth/ihu062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Azman AS, et al. Urban cholera transmission hotspots and their implications for reactive vaccination: evidence from bissau city, Guinea bissau. PLoS neglected tropical diseases. 2012;6(11):e1901. doi: 10.1371/journal.pntd.0001901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simons E, et al. Assessment of the 2010 global measles mortality reduction goal: results from a model of surveillance data. The Lancet. 2012;379(9832):2173–2178. doi: 10.1016/S0140-6736(12)60522-4. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi S, et al. Reduced vaccination and the risk of measles and other childhood infections post-Ebola. Science. 2015;347:1240–1242. doi: 10.1126/science.aaa3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiss DJ, et al. Air temperature suitability for Plasmodium falciparum malaria transmission in Africa 2000–2012: a high-resolution spatiotemporal prediction. Malaria journal. 2014;13(1):1–11. doi: 10.1186/1475-2875-13-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhatt S, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526(7572):207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alegana VA, et al. Estimation of malaria incidence in northern Namibia in 2009 using Bayesian conditional-autoregressive spatial–temporal models. Spatial and spatio-temporal epidemiology. 2013;7:25–36. doi: 10.1016/j.sste.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alegana VA, et al. Modelling the incidence of Plasmodium vivax and Plasmodium falciparum malaria in Afghanistan 2006–2009. PloS one. 2014;9(7):e102304. doi: 10.1371/journal.pone.0102304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cutts FT, et al. Measles elimination: progress, challenges and implications for rubella control. Expert review of vaccines. 2013;12(8):917–932. doi: 10.1586/14760584.2013.814847. [DOI] [PubMed] [Google Scholar]
- 44.Metcalf CJE, et al. Transport networks and inequities in vaccination: remoteness shapes measles vaccine coverage and prospects for elimination across Africa. Epidemiology and Infection. 2015;143(7):1457–1466. doi: 10.1017/S0950268814001988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bengtsson L, et al. Improved response to disasters and outbreaks by tracking population movements with mobile phone network data: a post-earthquake geospatial study in Haiti. PLoS Med. 2011;8(8):e1001083. doi: 10.1371/journal.pmed.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Panhuis WG, et al. Region-wide synchrony and traveling waves of dengue across eight countries in Southeast Asia. Proceedings of the National Academy of Sciences. 2015;112(42):13069–13074. doi: 10.1073/pnas.1501375112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaman J, Kohn M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proceedings of the National Academy of Sciences. 2009;106(9):3243–3248. doi: 10.1073/pnas.0806852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bharti N, et al. Explaining seasonal fluctuations of measles in Niger using nighttime lights imagery. Science. 2011;334(6061):1424–1427. doi: 10.1126/science.1210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deville P, et al. Dynamic population mapping using mobile phone data. Proceedings of the National Academy of Sciences. 2014;111(45):15888–15893. doi: 10.1073/pnas.1408439111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wesolowski A, et al. Quantifying seasonal population fluxes driving rubella transmission dynamics using mobile phone data. Proceeding of the National Academy of Sciences USA. 2015;112:11114–11119. doi: 10.1073/pnas.1423542112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Metcalf CJE, et al. Rubella meta-population dynamics and importance of spatial coupling to the risk of Congenital Rubella Syndrome in Peru. Journal of the Royal Society Interface. 2011;8:369–376. doi: 10.1098/rsif.2010.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wesolowski A, et al. Impact of human mobility on the emergence of dengue epidemics in Pakistan. Proceedings of the National Academy of Sciences. 2015;112(38):11887–11892. doi: 10.1073/pnas.1504964112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wesolowski A, et al. Quantifying travel behavior for infectious disease research: a comparison of data from surveys and mobile phones. Scientific reports. 2014;4:5678. doi: 10.1038/srep05678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Metcalf CJE, et al. The epidemiology of rubella in Mexico: seasonality, stochasticity and regional variation. Epidemiology and Infection. 2011;139:1029–1038. doi: 10.1017/S0950268810002165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu X, et al. Unveiling hidden migration and mobility patterns in climate stressed regions: A longitudinal study of six million anonymous mobile phone users in Bangladesh. Global Environmental Change. 2016;38:1–7. [Google Scholar]


