SUMMARY
Circadian clock-gated cell division cycles are observed from cyanobacteria to mammals via intracellular molecular connections between these two oscillators. Here we demonstrate WNT-mediated intercellular coupling between the cell cycle and circadian clock in 3D murine intestinal organoids (enteroids). The circadian clock gates a population of cells with heterogeneous cell-cycle times that emerge as 12-hr synchronized cell division cycles. Remarkably, we observe reduced-amplitude oscillations of circadian rhythms in intestinal stem cells and progenitor cells, indicating an intercellular signal arising from differentiated cells governing circadian clock-dependent synchronized cell division cycles. Stochastic simulations and experimental validations reveal Paneth cell-secreted WNT as the key intercellular coupling component linking the circadian clock and cell cycle in enteroids.
In Brief
Paneth cell-dependent circadian secretion of WNT is a key mechanism linking circadian rhythms and cell-cycle progression in murine enteroids. This finding implicates Paneth cells as local pacemakers that regulate the timing of cell divisions in intestinal stem cells and progenitor cells in the context of circadian rhythmicity.
INTRODUCTION
The circadian clock and cell cycle are biological oscillators whose coupling is observed across several species (Hong et al., 2014; Yang et al., 2010). At the single-cell level, clock-cell cycle coupling in mammals has been recently described in separate reports using NIH 3T3 cells, transformed mouse embryonic fibroblasts (Bieler et al., 2014; Feillet et al., 2014). Both groups showed a coupling ratio between the clock and cell cycle of ~1:1 in homogeneous cell populations, with single cell-level analyses of the cell cycle and circadian clock. These findings support earlier reports showing that several cell cycle-related genes are under clock control. For example, expression of the cell-cycle checkpoint kinase WEE1 and the cyclin-dependent kinase inhibitor P21 are regulated by the circadian clock transcription factors BMAL1 and REV-ERBα/β in the mouse liver (Gréchez-Cassiau et al., 2008; Matsuo et al., 2003). In addition, the core clock protein PER1 activates check point kinase 2 in human cancer cells (Gery et al., 2006), whereas PER1 and PER2 activate the cyclin-dependent kinase inhibitor P16 in mice (Gery et al., 2006; Kowalska et al., 2013). Together, these molecular connections orchestrate the intracellular coupling of the clock and cell cycle.
Prior work connecting the circadian clock and cell cycle in transformed and primary cell types represents fundamentally important observations; however, the coupling of the clock and cell cycle is likely to be more complex in heterogeneous, multicellular systems and tissues. To that end, intestinal organoids (enteroids) have recently emerged as a powerful platform for understanding adult stem cell dynamics, intestinal epithelial differentiation, and gut pathophysiology (Sato et al., 2009). Mouse enteroids arise from Lgr5-positive intestinal stem cells and self-organize into 3D structures with crypt-, villus- and lumen-like domains (Sato et al., 2009). Importantly, enteroids contain adult intestinal stem cells (ISCs), progenitor cells (PCs), and differentiated cells (DCs), including enterocytes, goblet cells, enteroendocrine cells, and Paneth cells. Paneth cells are secretory cells that reside at the crypt base, where the self-renewal capacity of ISCs and PCs is maintained by Paneth cell-secreted WNT (Clevers et al., 2014).
We have recently reported robust circadian rhythms in mouse enteroids established from PER2::LUCIFERASE (PER2::LUC) mice as well as autonomous synchronization of enteroid PER2 oscillations (Moore et al., 2014). PER2 is a core clock component that plays an essential role in maintaining the circadian clock in mammals (Zhang and Kay, 2010). In this report, we demonstrate self-synchronized, 12-hr cell division cycle progression from a population of enteroids; heterogeneous cell-cycle times (CCTs) with a multimodal distribution; stochastic simulations that explain 12-hr cell division cycles emerging from a heterogeneous population; low-amplitude oscillations of the circadian clock in intestinal stem cells and progenitor cells relative to differentiated cells; and intercellular coupling of the circadian clock and cell cycle via circadian regulation of WNT production and secretion by Paneth cells. Together, these results demonstrate that circadian clock-cell cycle coupling in a dynamic adult stem cell culture system is mediated by an intercellular coupling factor and indicate a novel function for Paneth cells as a peripheral circadian pacemaker for the intestinal epithelium.
RESULTS
Synchronized Oscillations of Cell-Cycle Progression in Mouse Enteroids
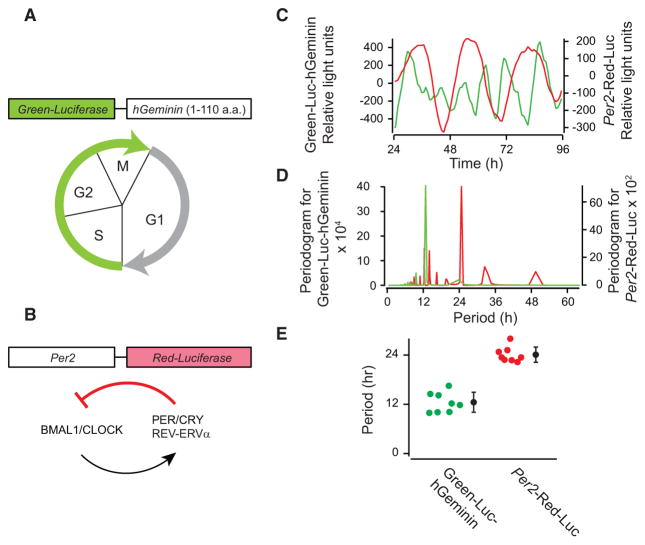
We developed enteroids with dual luciferase reporters to facilitate real-time measurements of cell cycle and circadian clock progression in groups of 20–25 co-cultured mouse enteroids. We genetically modified the fluorescent ubiquitination-based cell-cycle indicator (FUCCI) system (Sakaue-Sawano et al., 2008) to generate a bioluminescent S-G2-M phase reporter by creating a fusion protein of Green-Luciferase (Noguchi et al., 2008) and hGeminin (Sakaue-Sawano et al., 2008; Figure 1A). Circadian oscillations were monitored with Red-Luciferase (Noguchi et al., 2008) under control of the Per2 promoter (Yoo et al., 2004; Figure 1B). As shown in Figure 1C, we observed synchronized circadian clock and cell cycles in a population of enteroids. Interestingly, cell-cycle oscillations displayed two peaks during a single circadian cycle (Figure 1C). Fast Fourier transform (FFT) analysis of the time traces indicated a period of 12.4 ± 2.4 hr and 24.1 ± 1.9 hr for the cell cycle and clock, respectively (mean ± SD; Figures 1D, 1E, and S1A–S1H, available online). These results suggest circadian clock-gated cell division cycles with a 1:2 coupling ratio in populations of mouse enteroids.
Figure 1. Population-Level Analysis of Cell-Cycle and Circadian Clock Progression in Mouse Enteroids.
(A) A schematic representation of a luciferase-based cell-cycle sensor. Green-Luciferase was connected with 1–110 aa of hGeminin, which expresses during S-G2-M phase.
(B) A schematic representation of the luciferase-based circadian sensor.
(C) Representative traces of signal changes of Green-Luciferase-hGeminin (green) and Per2-Red-Luciferase (red) in mouse enteroids.
(D) Representative spectra of the FFT analysis of Green-Luciferase-hGeminin (green) and Per2-Red-Luciferase (red) traces in mouse enteroids.
(E) Distribution of peak periods from FFT data of Green-Luciferase-hGeminin (green dots) and Per2-Red-Luciferase (red dots) traces. Black dots and bars correspond to the average and SEM (n = 8).
See also Figure S1.
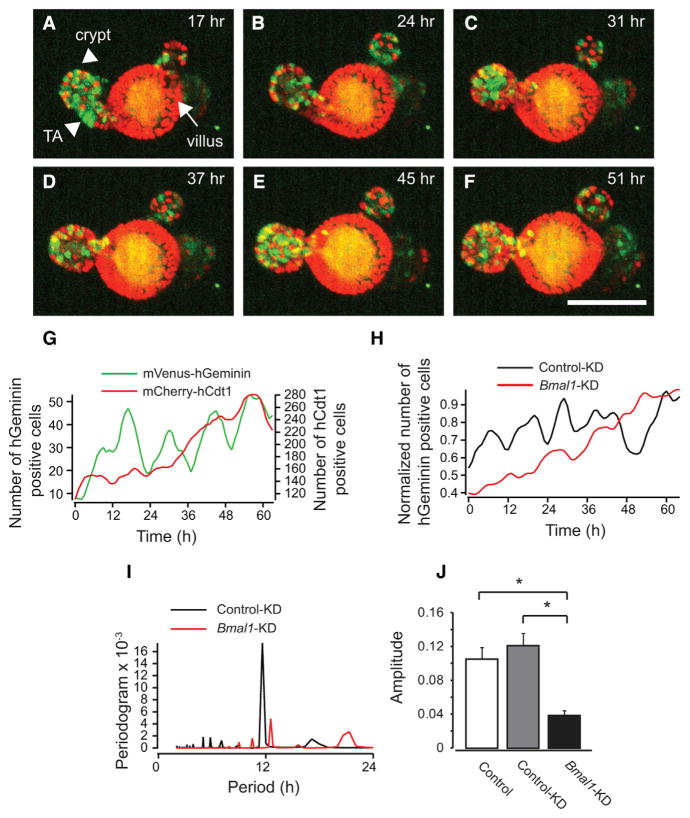
Synchronized oscillations of cell-cycle progression were also observed in single enteroids. To observe cell-cycle progression in single enteroids, we derived enteroids from FUCCI2 transgenic mice (Abe et al., 2013), which express a fluorescent S-G2-M phase reporter, mVenus-hGeminin, and a fluorescent G0/G1 phase reporter, mCherry-hCdt1. Green fluorescence from mVenus-hGeminin was mainly observed at the crypt base and transit-amplifying (TA) domains, where intestinal stem cells and progenitor cells reside, respectively (arrowheads, Figure 2A). In contrast, the red fluorescent mCherry-hCdt1 signal was primarily observed within villus-like domains, where terminally differentiated enterocytes reside (arrow, Figure 2A). Total numbers of both mCherry-hCdt1- and mVenus-hGeminin-expressing cells increased over time because of enteroid growth. The number of mCherry-hCdt1-expressing cells increased more dramatically with accumulation of terminally differentiated cells (G0 phase). We observed that the number of mVenus-hGeminin-positive cells oscillates in single enteroids (Figures 2A–2F; Movie S1) with a period of 12.9 ± 3.8 hr (mean ± SD; Figure 2G), consistent with the population-level analysis obtained with the Green-Luciferase-hGeminin bioluminescent reporter (Figure 1C). We obtained similar results in enteroids derived from H2B-EGFP transgenic mice (Figures S2A–S2D). To determine the extent to which a functional circadian clock is required for synchronized cell division cycles, we knocked down a core circadian clock gene, Bmal1, in PER2::LUC and FUCCI2 enteroids by expressing short hairpin RNA (shRNA) with a lentivirus vector. Bmal1 knockdown (KD) enteroids demonstrated significantly lower amplitude PER2::LUC oscillations (Figures S2E–S2G), indicating impaired circadian transcriptional-translational feedback loop (TTFL) activity. Importantly, Bmal1 KD also showed dramatically lower amplitude oscillations of synchronized cell division cycles compared with control KD (Figures 2H–2J and S2H–S2N). Likewise, circadian arrhythmic enteroids derived from Per1/2 double knockout (Per1/2 DKO) mice also displayed abolished synchronized cell-cycle progression at the population level (Figure S2O). These results indicate that the circadian clock is necessary to maintain synchronized cell-cycle progression.
Figure 2. Single-Enteroid Analyses of Cell-Cycle Progression.
(A–F) Images showing the spatial distribution of mVenus-hGeminin (green, S-G2-M) and mCherry-hCdt1 (red G0/G1) at 17 (A), 24 (B), 31 (C), 37 (D), 45 (E), and 51 hr (F). Arrowheads, crypt and TA domains; arrow, villus domain. Scale bar, 100 μm.
(G) Representative traces of the number of mVenus-hGeminin-positive (green) and mCherry-hCdt1-positive (red) cells in a single FUCCI2 enteroid.
(H) Representative traces of the number of mVenus-hGeminin-positive cells in control KD (black) and Bmal1 KD (red) FUCCI2 enteroids.
(I) Representative periodogram of FFT analysis of mVenus-hGeminin-positive cells from control KD (black) and Bmal1 KD (red) FUCCI2 enteroids.
(J) Average amplitudes of oscillations of mVenus-hGeminin-positive cells in control (non-treatment, n = 13), control KD (n = 7), and Bmal1 KD (n = 12) FUCCI2 enteroids. Error bars correspond to the SEM. *p < 0.05, Tukey-Kramer test.
Circadian Gating of the Cell Cycle in Enteroids
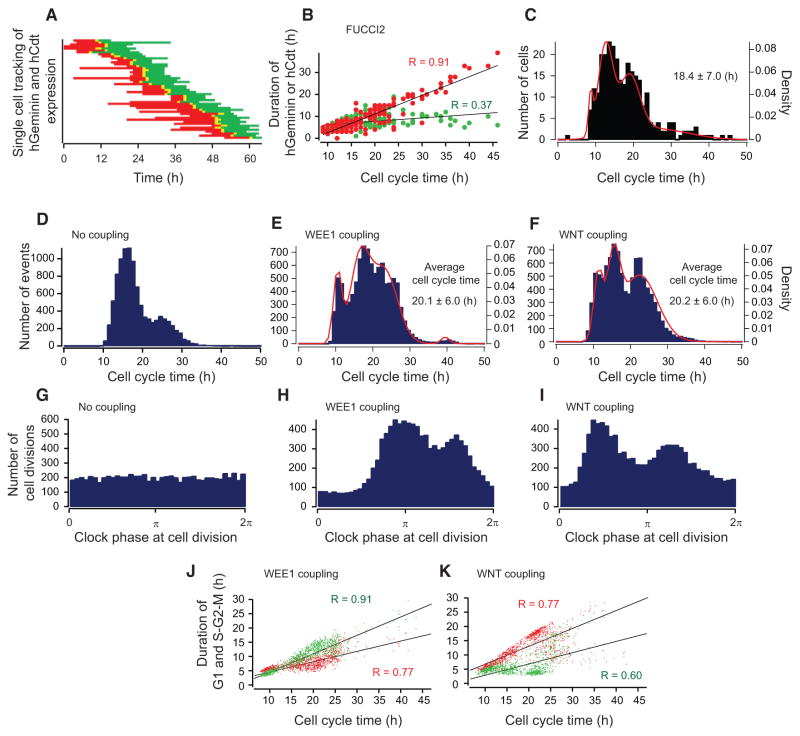
To explore the coupling of the circadian clock and cell cycle in further detail, we tracked cell-cycle progression in individual cells within FUCCI2 enteroids. Cells in FUCCI2 enteroids displayed red, yellow, and green signals during G0/G1, transition from G1 to S, and S-G2-M phases, respectively, and these signals eventually disappeared when cells divided. FUCCI2 enteroids thus enabled us to measure the duration of G1, S/G2/M, and individual CCTs from the beginning of G1 (red) to the end of mitosis (green). Figure 3A shows representative results of single-cell tracking in a single enteroid, with each line displaying color changes (i.e., cell-cycle phases) of a single cell. Of note, we observed a larger variability of G1 duration (red) compared with S/G2/M (green) (G1, 9.6 ± 6.8 hr; S/G2/M, 7.5 ± 2.8 hr (mean ± SD), p < 0.01, F test). We discovered that the duration of CCT strongly correlates with the duration of G1 phase (red), but not with the duration of S/G2/M (green) (Figure 3B). In contrast to ~12-hr oscillations observed at the population level, single-cell analysis revealed a multimodal distribution of cell-cycle times with an average CCT of 18.4 ± 7.0 hr (mean ± SD). This distribution was fitted to a mixture of univariate Gaussian distribution equations, revealing four peaks at 8.7 ± 0.6 hr, 12.6 ± 1.9 hr, 18.9 ± 2.8 hr, and 25.8 ± 8.7 hr (mean ± SD; Figures 3C and S3A).
Figure 3. Multimodal Distribution of CCT in FUCCI2 Enteroids and Stochastic Simulation.
(A) Single-cell tracking of the timing and duration of hCdt1 (red) and hGeminin (green) expression in a single FUCCI2 enteroid. Both hGeminin and hCdt1 express at the transition of G1 to S (yellow).
(B) Relationship between CCT and duration of hGeminin (S-G2-M, green dots) or hCdt1 (G0/G1, red dots). The black lines are the regression lines. Correlation coefficients are shown in the graph (n = 174).
(C) A histogram of the distribution of CCT (total duration from emergence of red to disappearance of green, 283 cells).
(D) Simulated histogram of the distribution of CCT in an uncoupled model (10,000 cells).
(E and F) Simulated histogram of the distribution of CCT in WEE1 (E) and WNT (F) models (10,000 cells). The distributions in E and F were fitted to a mixture of a univariate Gaussian distribution equation.
(G–I) Simulated histograms of the timing of cell divisions relative to clock phase in no coupling (G), WEE1 (H), and WNT (I) models (10,000 cells).
(J and K) Relationship between CCT and duration of S-G2-M (green dots) or G1 (red dots) in WEE1 (J) and WNT (K) models (1,000 cells). The black lines are the regression lines. Correlation coefficients are shown in the graph.
See also Figures S3 and S4.
How can a multimodal distribution of CCTs give rise to synchronized 12-hr cell cycle as a population? We used mathematical modeling to investigate potential mechanisms for the observed phenotypes. Specifically, we modified our previous model of the circadian clock and the cell cycle as coupled oscillators (Zámborszky et al., 2007) (Supplemental Experimental Procedures) to simulate the distributions of the CCTs and the timing of divisions from two distinct molecular coupling mechanisms. First, we simulated the consequences of coupling at the G2-M checkpoint through the circadian regulation of WEE1 (Matsuo et al., 2003). The heterodimeric circadian clock transcription factors CLOCK/BMAL1 regulate the expression of WEE1, which is a cell-cycle kinase that phosphorylates and inactivates the CyclinB/CDK complex to block entry to mitosis (Matsuo et al., 2003). Then, we tested a coupling of the cell cycle and circadian rhythms at the G1-S transition through WNT (Tetsu and McCormick, 1999). WNT is secreted from Paneth cells and controls self-renewal of ISCs and PCs (Clevers et al., 2014), and it has been shown that BMAL1 regulates WNT signaling (Guo et al., 2012; Lin et al., 2013). For these simulations, we assumed that there exist two cell populations with distinct CCTs based on previous literature and our cell-cycle measurements with enteroids from Lgr5-EGFP transgenic mice, indicating fast and slow cycling populations in the mouse small intestine: PCs and ISCs with average CCTs of 16 and 26 hr, respectively (Figures S4A–S4E; Carulli et al., 2014).
In the absence of coupling, we observed a bimodal distribution of CCTs reflecting our assumptions of 16- and 26-hr populations with a larger number of PCs compared with ISCs (Figure 3D). Figures 3E and 3F show simulation results with either WEE1 or WNT as potential coupling mechanisms in the model, respectively. Both coupling mechanisms qualitatively reproduced the observed multimodal distribution of CCTs with expected average CCTs close to the experimental results (WEE1, 20.1 ± 6.0 hr; WNT, 20.2 ± 6.0 hr; mean ± SD; Figures 3E and 3F). Simulation results for separate subpopulations (PCs, ISCs, and combinations thereof) are shown in Figures S3B–S3E. It is important to note that it was necessary to assign appropriate coupling strength to reproduce multimodal distributions (Figures S3B–S3F). Both PC and ISC subpopulations showed multimodal distributions of CCTs when coupled to the circadian clock. Furthermore, we discovered that these multimodal distributions of CCTs appear as bimodal distributions when in silico cells are plotted against the circadian phase (Figures 3H, 3I, and S3G). In other words, the circadian clock influences the timing of the mitosis of cells with heterogeneous CCTs, resulting in two peaks of cell divisions during 24 hr. In contrast, there was no specific timing of cell divisions in the uncoupled case (Figure 3G). These simulations indicate that the cell cycle is gated by the circadian clock in our model, within the described set of parameters.
Intriguingly, we observed a key difference between WEE1 versus WNT coupling scenarios in our simulation results. We discovered that WEE1 coupling results in a bimodal distribution of cell division frequencies, with the time between major peaks much shorter than 12 hr (Figure 3H), which is incompatible with the 12-hr cell divisions we observed (Figures 1C and 2G). In contrast, WNT coupling reproduced a bimodal distribution of cell-cycle events with 12-hr cell division cycles (Figure 3I). These observed differences were due to distinct mechanisms of coupling. The circadian clock-dependent activation of WEE1 results in an impediment of cell division cycles at specific phases of the circadian cycle, with a smaller number of cell divisions when WEE1 is high and a greater number of cell divisions when WEE1 is low. In contrast, WNT coupling facilitates the transition of the G1-S phases via CyclinD1 and does not induce prolonged impediments of cell division cycles. We further tested another scenario of clock-cell cycle coupling by inhibition of G1-S transition via P21 and discovered uneven durations between peaks of cell division in a single clock phase, which did not match our experimental data (Figures S3A–S3G). We then performed simulations to measure the correlation between the duration of G1 or S/G2/M and simulated CCTs from Figures 3E and 3F. We observed that the duration of S/G2/M was highly correlated with WEE1 coupling (Figures 3J and S3H). In contrast, the duration of G1 was highly correlated with WNT coupling (Figures 3K and S3H), which is qualitatively in agreement with our experimental data (Figure 3B). Our simulation results further suggest that canonical WNT signaling-mediated cell-cycle regulation at the G1-S transition is more likely to connect cell-cycle and circadian rhythms versus G2-M transition regulation by WEE1.
Complex Interactions of the Circadian Clock and Cell Cycle in Enteroids
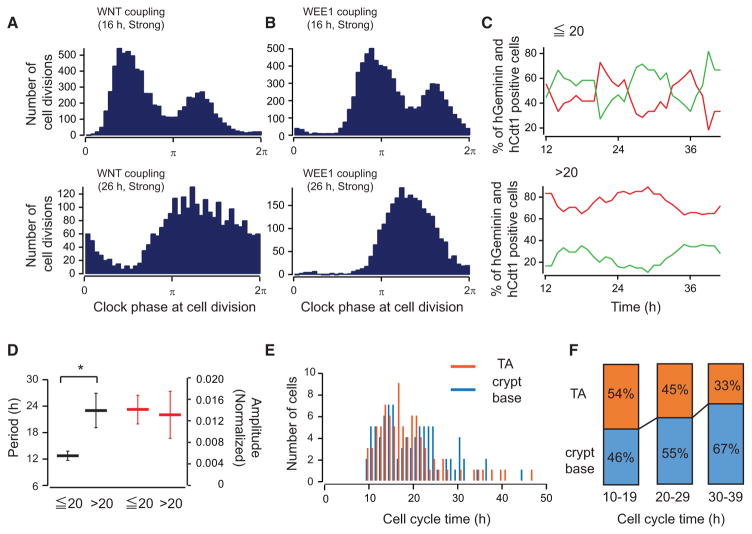
For the stochastic simulations, we assumed that enteroids consist of two populations of proliferating cells that have average CCTs of either 16 or 26 hr in the absence of circadian connections. We explored our simulation results more in depth by segregating these two populations and plotting the number of cell divisions as a function of circadian phase. We observed two peaks of cell division events over one circadian cycle in the population with short CCTs (16 hr), indicating 1:2 coupling (Figures 4A and 4B, top). In contrast, we observed 1:1 coupling for the population with longer CCTs (26 hr), with a single peak of cell division events over one circadian cycle (Figures 4A and 4B, bottom). Furthermore, the simulation results showed that the population with longer CCTs had a cell-cycle distribution ranging from 20–40 hr (Figures S3C–S3E). To compare these in silico results with our experimental data, we segregated the single-cell data from FUCCI2 enteroids into two populations: cells with a CCT of less than 20 hr (Tcc < 20 hr) and cells with a CCT of more than 20 hr (Tcc ≥ 20 hr). The population with shorter CCTs (Tcc < 20 hr) showed 12-hr rhythms (Figure 4C, top), which is 1:2 coupling with respect to the circadian clock (12.6 ± 2.6 hr, n = 3, mean ± SD; Figure 4D). In contrast, the population with longer CCTs (Tcc ≥ 20 hr) demonstrated 1:1 coupling with an average CCT of 24.5 ± 7.8 hr (n = 3, mean ± SD; Figure 4D). This distribution of CCTs in the crypt base versus the TA domain indicates that a greater number of cells with longer CCTs (Tcc ≥ 20 hr) reside in the crypt base (Figures 4E and 4F), consistent with earlier findings on the longer cell-cycle duration of Lgr5-positive intestinal stem cells (Tcc ≥ 20 hr). Enteroids reveal two different coupling ratios between the circadian clock and the cell cycle depending on the intrinsic CCTs. Similar data were also observed in enteroids derived from Lgr5-EGFP-IRES-CreERT2+/−/Rosa-CAG-LSL-tdTomato-WPRE+/− mice (Figures S5A and S5B). Based on these data, we hypothesized that the circadian clock influences a population of cells in 3D adult stem cell cultures, resulting in different coupling ratios for ISCs and PCs.
Figure 4. Complex Interactions of the Circadian Clock and Cell Cycle.
(A and B) Simulated histograms of the timing of cell divisions relative to clock phase in WEE1 (A) and WNT (B) models (10,000 cells). The results from the 16-hr CCT population are shown at the top, and the results from the 26-h CCT population are shown at the bottom.
(C) Representative traces of percentage changes of the number of hCdt1-positive (red) and hGeminin-positive (green) cells in single-cell tracked data from a single FUCCI2 enteroid. Traces of cells whose CCT was shorter than 20 hr (Tcc < 20 hr) are shown at the top, and traces of the cells whose CCT was longer than 20 hr (Tcc > 20 hr) are shown at the bottom.
(D) Average periodicity (black bars) and amplitudes (red bars) detected by FFT. The data of Tcc ≤ 20 hr and Tcc > 20 hr are shown. Error bars correspond to SEM (n = 3).
(E) Histograms of the distribution of CCT of the TA (orange, 86 cells) and crypt base (light blue, 88 cells).
(F) Percentage of single tracked cells from the TA and crypt base. The percentages of CCT of 10–19, 20–29, and 30–39 hr are shown.
See also Figure S5.
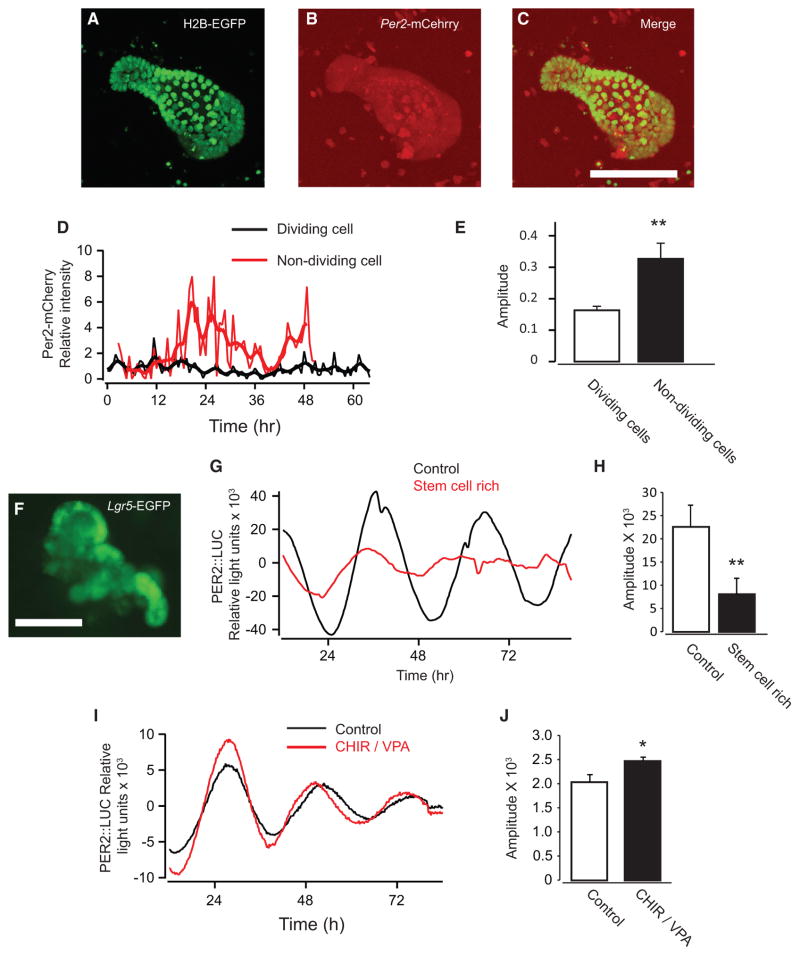
We then asked whether circadian rhythms are robust in intestinal stem cells. To test this, we developed enteroids with dual fluorescent reporters of H2B-EGFP;Per2-mCherry to characterize circadian rhythms from single cells (Figure 5A–5C). Intriguingly, we observed no discernable circadian oscillations in dividing ISCs and PCs in contrast to clear oscillations in non-dividing DCs (Figures 5D and 5E). Furthermore, PER2::LUC enteroids cultured under ISC- and PC-rich conditions (Yin et al., 2014) showed significantly lower amplitude oscillations (Figures 5F and S6A–S6C) compared with normal culture conditions (Figures 5G and 5H). These data suggest weak or absent circadian rhythms in individual ISCs and PCs; hence, intracellular coupling is unlikely to be a major contributor to circadian regulation of the cell cycle in ISCs and PCs. The ISC- and PC-rich culture condition did not reduce the amplitude of PER2::LUC oscillations in fibroblasts, further suggesting that ISC and PC differentiation is required for robust circadian rhythms in enteroids (Figures 5I and 5J).
Figure 5. The Circadian Clock in ISCs and PCs.
(A–C) Fluorescence images of H2B-EGFP (A), Per2-mCherry (B), and the merge (C) in mouse enteroids.
(D) Representative traces of the single-cell Per2-mCherry signals from dividing (black) and non-dividing cells (red). The thick lines are their moving averages.
(E) Average amplitude of dividing (white bar, n = 39) and non-dividing cells (black bar, n = 20).
(F) A fluorescence image of EGFP of the enteroid derived from Lgr5-EGFP transgenic mice (Lgr5-EGFP-IRES-CreERT2+/−/Rosa-CAG-LSL-tdTomato-WPRE+/−) under ISC- and PC-rich culture conditions.
(G) Population level analysis of PER2::LUC oscillations with enteroids derived from PER2::LUC mice. Shown are representative traces of PER2::LUC enteroids cultured under control (n = 3, black) and ISC- and PC-rich conditions (n = 4, red).
(H) Average amplitude of PER2::LUC enteroids cultured under control and ISC- and PC-rich conditions (black).
(I) Representative traces of signal changes of PER2::LUC in mouse fibroblasts cultured with normal medium (black) and stem cell-rich conditioned medium (CHIR99021 [CHIR]/Valproic acid [VPA], red).
(J) Average amplitudes of oscillations of PER2::LUC in mouse fibroblasts cultured with normal medium (white bar, n = 3) and stem cell-rich conditioned medium (black bar, n = 3). *p < 0.05; **p < 0.01, Student’s t test.
Error bars correspond to SEM. Scale bars, 100 μm.
See also Figure S6.
WNT Is a Key Coupling Factor between the Circadian Clock and the Cell Cycle in Mouse Enteroids
ISCs and PCs showed relatively low-amplitude expression of Per2 gene expression. This result suggests that WEE1-mediated intracellular coupling of the circadian clock and the cell cycle may not be a key coupling mechanism in mouse enteroids. Importantly, our simulation results unequivocally demonstrated that WNT-mediated coupling better reproduced existing data, including the correct bimodal distribution of CCTs over the circadian phase and a stronger correlation of the duration of G1 with the observed CCTs (Figures 3H and 3I). Based on these supportive data, we tested whether WNT is a major coupling component connecting the circadian clock to the cell cycle in mouse enteroids.
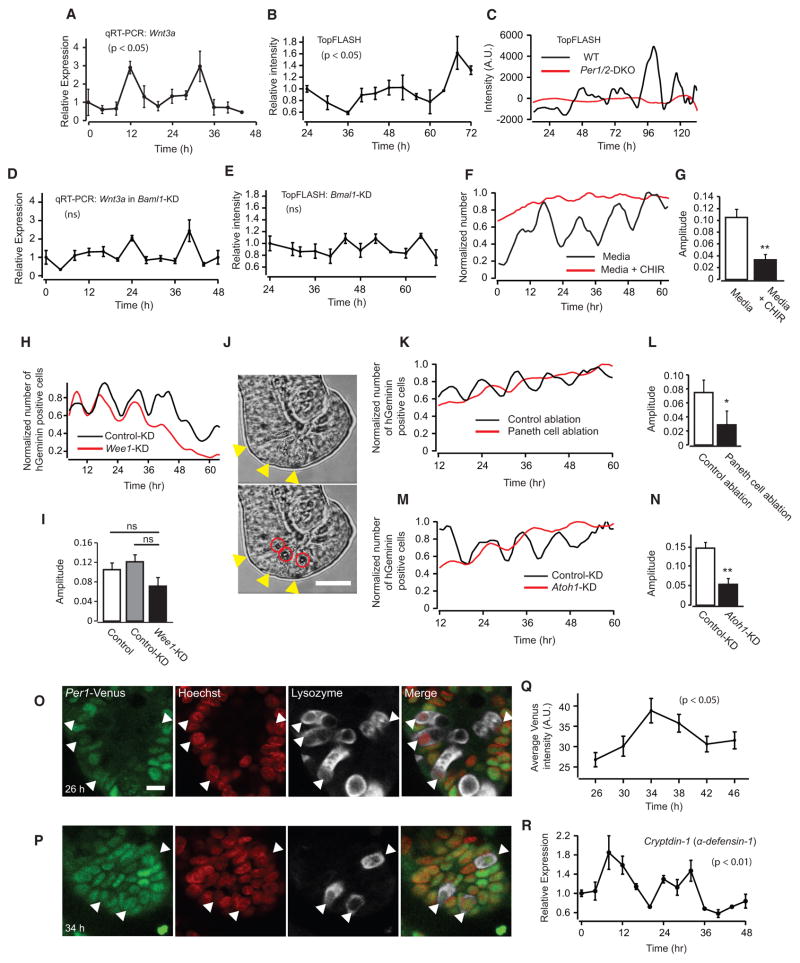
We first discovered that Wnt3a mRNA demonstrates circadian oscillations in mouse enteroids (Figures 6A and S7A). Importantly, we also observed rhythmic WNT activity by TopFlash promoter luciferase assay (Figures 6B and 6C), which reports canonical WNT/β-catenin signaling (Fuerer and Nusse, 2010; Molenaar et al., 1996). These WNT oscillations were abolished in Per1/2 DKO and Bmal1 KD enteroids (Figures 6C–6E). Furthermore, saturated WNT signaling by a WNT agonist, the GSK-3β inhibitor CHIR99021, induced dramatic reductions of synchronized 12-hr cell-cycle oscillations in FUCCI2 enteroids (Figures 6F and 6G), demonstrating the importance of WNT oscillations for coupled oscillations of the clock and cell cycle. Wee1 KD did not affect 12-hr cell division cycles (Figures 6H, 6I, and S7B). These results indicate that WNT-mediated intercellular coupling between the cell cycle and the circadian clock is a key mechanism determining circadian clock-dependent synchronized cell division cycles.
Figure 6. Rhythmic Expression of WNT by Paneth Cells Couples the Circadian Clock and Cell Cycle in Mouse Enteroids.
(A) qRT-PCR data of mRNA expressions of Wnt3a (n = 3).
(B) Secretion of WNT detected by TopFlash luciferase assay. The collected culturing media from enteroids at the indicated times were subjected to TopFlash cells (n = 3).
(C) Secretion of WNT from the enteroids was measured by real-time imaging of luciferase activity from TopFlash cells co-cultured with WT or Per1/2 DKO enteroids.
(D) qRT-PCR data of mRNA expressions of Wnt3a in Bmal1 KD enteroids (n = 3).
(E) Secretion of WNT detected by TopFlash luciferase assay. The collected culturing media from control Bmal1 KD enteroids at the indicated times were subjected to TopFlash cells (n = 3).
(F) Representative traces of the number of mVenus-hGeminin-positive cells in a single FUCCI2 enteroid cultured with normal medium (black) or medium containing CHIR99021 (red).
(G) Average amplitudes of oscillations of mVenus-hGeminin-positive cells in single enteroids cultured with normal medium (n = 13, white bar) and medium containing CHIR99021 (n = 12, black bar).
(H) Representative traces of the number of mVenus-hGeminin-positive cells in a single control KD enteroid (black) and Wee1 KD (red) FUCCI2 enteroids.
(I) Average amplitudes of oscillations of mVenus-hGeminin-positive cells in single WT (n = 13, white), control KD (n = 7, gray), and Wee1 KD (n = 7, black) FUCCI2 enteroids. p > 0.05, Tukey-Kramer and Student’s t test.
(J) Laser irradiation of Paneth cells. Shown are images of a crypt before (top) and after (bottom) laser irradiation. Arrowheads indicate Paneth cells. Circles indicate pores made by laser irradiation.
(K) Representative traces of the number of mVenus-hGeminin-positive cells in a single FUCCI2 enteroid damaged by control (black) and Paneth cell (red) ablation.
(L) Average amplitudes of oscillations of mVenus-hGeminin-positive cells in control-ablated (n = 6, white) and Paneth cell-ablated (n = 5, black) FUCCI2 enteroids.
(M) Representative traces of the number of mVenus-hGeminin-positive cells in a single control KD enteroid (black) and Atoh-1 KD FUCCI2 enteroids.
(N) Average amplitudes of oscillations of mVenus-hGeminin-positive cells in control KD (n = 7, white) and Atoh-1 KD (n = 5, black) FUCCI2 enteroids.
(O and P) Immunostaining of Per1-Venus, a circadian reporter, and Lysozyme, a Paneth cell marker, in enteroids derived from Per1-Venus transgenic mice. The enteroids were serum-shocked to reset the circadian clock and fixed after 26–46 hr. Images taken 26 hr (O) and 34 hr (P) after serum shock are shown in (O) and (P), respectively. Per1-Venus, nuclear (Hoechst), Lysozyme, and the merge of Per1-Venus, Lysozyme, and nuclear images are shown.
(Q) Time course change of intensities of Per1-Venus signal in Paneth cells.
(R) qRT-PCR data of mRNA expressions of Cryptdin-1, a Paneth cell-specific antimicrobial peptide gene (n = 3).
Scale bars, 10 μm. *p < 0.05, **p < 0.01, Student’s t test. Error bars correspond to SEM. See also Figure S7 and Movies S2, S3, S4, and S5.
Paneth cells constitute the Lgr5+ intestinal stem cell niche and are the major secretory source of WNT in intestinal crypts. We confirmed a requirement for WNT in synchronized cell-cycle progression by Paneth cell ablation experiments. We used two distinct methods to ablate Paneth cells: physical ablation by laser irradiation (Figures 6J, S7C, and S7D; Movies S2 and S3) and knockdown of Atoh1, an essential transcription factor for Paneth cell differentiation (Figures S7E–S7M; Durand et al., 2012). As expected, we observed significantly reduced synchronized cell-cycle progression in FUCCI2 enteroids under both experimental conditions (Figures 6K–6N; Movies S4 and S5), suggesting that Paneth cells regulate synchronized cell division cycles via rhythmic production of WNT. Importantly, enteroids derived from Per1-Venus mice demonstrated the existence of circadian rhythms in Paneth cells by immunostaining (Figures 6O–6Q), along with circadian expression of the Paneth cell-specific gene Cryptdin-1 by qRT-PCR (Figure 6R).
Periodic Budding in Enteroids
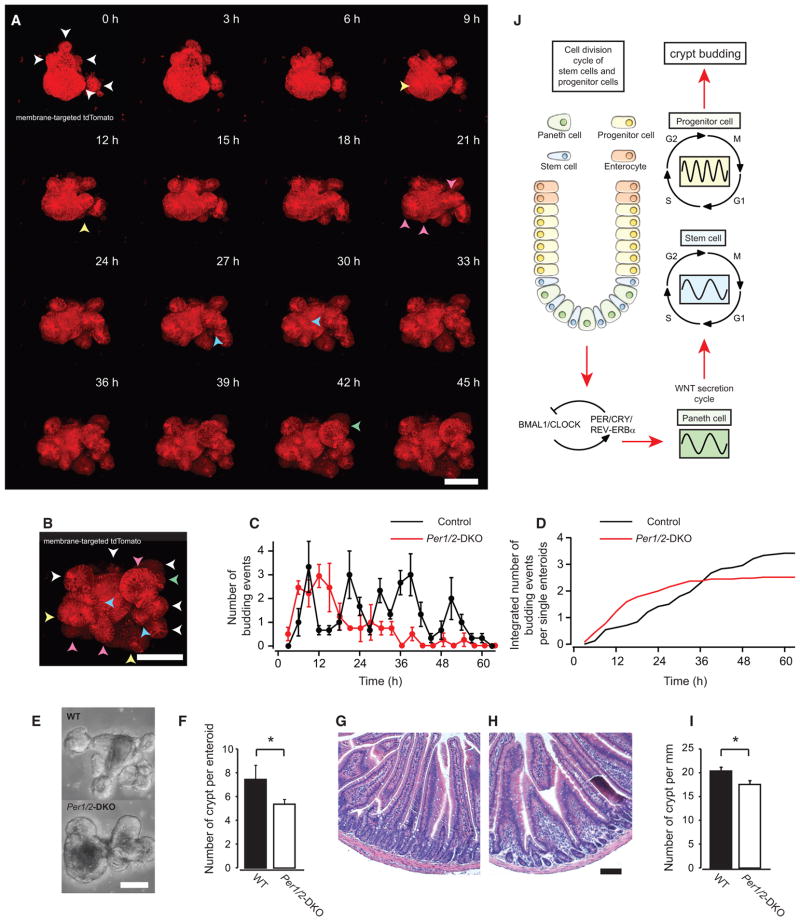
What is the tissue level consequence of the synchronized cell division cycles? We hypothesized that enteroid crypt budding events would occur rhythmically because enteroid crypt domains undergo continuous budding events reminiscent of in vivo crypt fission (Sato et al., 2009). As expected, we observed rhythmic formation of new crypt buds with a period of 12 hr in wild-type (WT), but not in Per1/2 DKO, enteroids (Figures 7A–7C). Moreover, Per1/2 DKO enteroids showed a reduced number of budding events and total crypt domains (Figures 7C–7E) relative to wild-type enteroids. In vivo, we observed a reduced number of crypts in the jejunum of Per1/2 DKO mice (Figures 7F–7H), suggesting that rhythmic cell-cycle progression facilitates crypt budding and formation.
Figure 7. Periodic Budding in Mouse Enteroids.
(A and B) Time-course fluorescence images of membrane-targeted tdTomato in a single mouse enteroid derived from Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo mice (A). White arrows indicate established crypt domains. Yellow, pink, blue, and green arrowheads indicate newly formed crypt domains around 10, 20, 30, and 40 hr, respectively. All arrowheads in A are merged on the enteroid image at 45 hr (B).
(C) Average number of crypt budding events for WT (n = 3) and Per1/2 DKO (n = 4) enteroids. Each experiment included 5–11 enteroids.
(D) Integrated number of crypt budding events in WT (n = 24) and Per1/2 DKO (n = 27) enteroids during confocal imaging.
(E) Phase contrast images of WT (top) and Per1/2 DKO (bottom) enteroids after confocal imaging.
(F) Number of crypts in WT (black bar, n = 24) and Per1/2 DKO (white bar, n = 27) enteroids after confocal imaging.
(G and H) H&E-stained transverse sections of small intestine (jejunum) from WT (G) and Per1/2 DKO (H) mice.
(I) Number of crypts in WT (black bar, n = 3) and Per1/2 DKO (white bar, n = 3) jejunum sections.
(J) A schematic of intercellular coupling of the circadian clock and CCT through oscillatory WNT production and secretion in mouse intestinal epithelium.
*p < 0.05, Student’s t test. Error bars correspond to SEM. Scale bars, 100 μm.
DISCUSSION
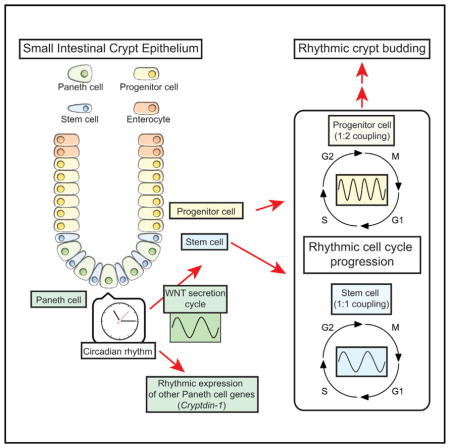
Circadian gating of the cell cycle in mammalian cells remains a controversial subject. Nagoshi et al. (2004) reported the coupling of the circadian clock and the cell cycle in NIH 3T3 mouse fibroblasts. However, two studies reported a lack of clock regulation in the mammalian cell cycle (Pendergast et al., 2010; Yeom et al., 2010). More recently, two reports by Feillet et al. (2014) and Bieler et al. (2014) showed 1:1 coupling of the clock and the cell cycle in NIH 3T3 cells with single-cell resolution imaging and mathematical modeling. Our findings of coupled oscillations of circadian rhythms and cell cycle in a complex 3D adult intestinal stem cell culture system, together with a WNT-mediated mechanism for intercellular coupling, add strong evidence not only for the existence of circadian gating but also for a specific mechanism of coupling, with dynamic consequences of this coupling in different contexts. In this report, we demonstrate that the circadian clock influences a heterogeneous population of cells in a non-transformed, complex 3D adult intestinal stem cell culture system. The circadian clock synchronizes the timing of cell division cycles with 1:1 or 1:2 coupling ratios in different subpopulations of cells, depending on their inherent CCTs (Figure 4C). Intriguingly, these synchronized cell division cycles emerge from heterogeneous CCTs with a multimodal distribution as a result of the coupling (Figure 3; Aihara et al., 2015; Nagoshi et al., 2004). Importantly, we also uncover WNT-mediated intercellular coupling between circadian rhythms and the cell cycle in 3D enteroids.
The multiple types of intestinal epithelial cells in enteroids are organotypic of the complexity observed in vivo in the intestinal epithelium. Within this complexity, we found two populations of cells with a 1:1 and 1:2 coupling ratio of the clock and the cell cycle. Given the length of the CCTs in these populations, it is possible that the 1:1 and 1:2 coupled populations mainly consist of ISCs and PCs, respectively. Similar to other adult stem cells, ISCs have active and quiescent states, and the CCT of PCs may vary depending on the expression level of LGR5, as shown in our imaging data from Lgr5-EGFP enteroids. Taken together, it is possible that there exists a greater abundance of coupling ratios arising from the interactions of the clock and the cell cycle in vivo.
Yagita et al. (2010) previously reported the lack of circadian rhythms in mouse embryonic stem cells, with the circadian clock developing during differentiation. We observed dramatically lower amplitude oscillations of PER2::LUC from enteroids cultured under ISC- and PC-enriched conditions. During mammalian development, pluripotent cells differentiate to give rise to lineage-committing adult stem cells and progenitor cells (Sugawara et al., 2012). Therefore, it is possible that ISCs are “midstream” in development of the circadian TTFL. WEE1 is an important intracellular coupling factor for the circadian clock and cell cycle. However, the expression level of Wee1 in mouse enteroids was one quarter of that in NIH 3T3 cells, immortalized cells commonly used for the study of circadian-gated cell-cycle progression (Nagoshi et al., 2004; Figure S7B). Expression of Wee1 was also high in other immortalized cells (Figure S7B), suggesting compensatory overexpression of Wee1 for suppression of cell-cycle progression in cells with increased rates of proliferation.
Several critical cell cycle components are clock-controlled, and their interactions are thought to establish an intracellular coupling of the clock and cell cycle. ISCs and PCs displayed lower-amplitude oscillations of clock gene expression. Therefore, it is plausible that diffusible factors, including WNT, from secretory cells mediate the intercellular connection between the circadian clock and the cell cycle in ISCs and PCs. Interactions of the circadian clock and WNT signaling have been described previously in other contexts (Guo et al., 2012; Lin et al., 2013), and many genes in the WNT pathway are known to have rhythmic expression (Soták et al., 2014). Hence, not only WNT secretion but also downstream WNT signaling may contribute to clock-dependent synchronized cell-cycle phenotypes in vivo. In contrast to ISCs and PCs, human epidermal stem cells have TTFL of clock genes, with stem cell functions regulated by the circadian clock (Janich et al., 2011, 2013). Interestingly, epidermal stem cells self-renew by autocrine WNT (Lim et al., 2013). WNT signaling is also known to regulate the self-renewal of various adult stem cells in the gut, mammary gland, hematopoietic system, and neuronal system (Ring et al., 2014). These results indicate the possibility of a general role of WNT for coupling of the circadian clock and cell cycle in adult stem cells.
Paneth cells secrete growth factors, including WNT, but also substantial quantities of antimicrobial peptides. Our findings that Paneth cells not only have circadian oscillation of clock genes but also the defensin gene Cryptdin-1 (Figure 6R) strongly supports earlier in vivo evidence of the circadian regulation of Paneth cell immune function (Froy and Chapnik, 2007; Froy et al., 2005). The result suggests that not only cell cycle, crypt budding, and innate immunity but also other physiological events are rhythmically controlled via secretory factors from Paneth cells (Figure 7I). In addition to Paneth cells, three other types of secretory cells exist in the intestinal epithelium: enteroendocrine, goblet, and tuft cells. Gut hormones like glucagon-like peptide 1 and peptide YY are secreted in a circadian manner from the intestinal epithelium (Gil-Lozano et al., 2014; Hill et al., 2011). The intestinal epithelium also secretes glucocorticoid, whose signaling induces circadian phase changes in peripheral tissue cells, including enteroids (Balsalobre et al., 2000; Moore et al., 2014; Noti et al., 2009). Serotonin (5-hydroxytryptamine [5-HT]) is an important regulator of the circadian clock of the suprachiasmatic nucleus and is also secreted from the intestinal epithelium (Balsalobre et al., 2000; Gribble and Reimann, 2016), which may connect the circadian clock to the brain-gut axis.
In conclusion, we uncovered a mechanism of WNT-mediated intercellular coupling between the circadian clock and the cell cycle in enteroids that results in 1:1 and 1:2 coupling ratios between these two oscillators in different intestinal epithelial cell subpopulations. This clock-controlled cell-cycle progression also manifested as rhythmic budding of crypt domains in enteroids. We further determined that WNT is not only an essential niche signaling agent that maintains self-renewal of intestinal stem cells but that it also serves as an intercellular node linking circadian rhythms and the cell cycle. Finally, we identify a key role of Paneth cells as local pacemakers that regulate the timing of cell divisions in intestinal stem cells and progenitor cells in the context of circadian rhythmicity.
EXPERIMENTAL PROCEDURES
Gene Constructions
Green-Luciferase-PEST-hGeminin, Per2-Red-Luciferase-PEST-PGK-puror, and Per2-mCherry were constructed using PCR and yeast recombination. The details of the procedures are given in the Supplemental Experimental Procedures.
Preparation of Enteroids
Using the methods of Sato et al. (2009), we prepared intestinal organoids (enteroids) by isolating fresh mid-jejunal crypts from mice. The details of the procedures are given in the Supplemental Experimental Procedures.
Lentivirus Induction
Green-Luciferase-hGeminin-, Per2-Red-Luciferase-, Per2-mCherry-, control-shRNA-, Bmal1-shRNA, Wee1-shRNA-, and Atoh1-shRNA-expressing enteroids were made by lentivirus induction. The details of the procedures are given in the Supplemental Experimental Procedures.
Bioluminescence Recording
Enteroids or fibroblasts were plated on a 35-mm plastic dish and placed in a Kronos Dio AB-2550 incubating luminometer (ATTO) for real-time periodic quantification of PER2 protein, Green-Luciferase-hGeminin, Per2-Red-Luciferase, or TopFlash reporter (Molenaar et al., 1996) abundance by bioluminescence recording (Malloy et al., 2012). The details of the procedures are given in the Supplemental Experimental Procedures.
Fluorescence Imaging
Enteroids were plated on an 8-well chamber slide (Lab-tech) 2 days before the imaging experiments. Image acquisition was performed with an LSM710 LIVE Duo confocal microscope (Zeiss). Single-cell ablations of enteroid cells were performed through LSM510 two-photon microscope (Zeiss). The details of the procedures are given in the Supplemental Experimental Procedures.
Data Analysis
Data analysis was done with Imaris (Bitplane) software for imaging data, ImageJ software for tissue sections, Kronos (ATTO) software for bioluminescence data, and R for FFT analysis. The details of the procedures are given in the Supplemental Experimental Procedures.
Gaussian Mixture Model
The Gaussian mixture model was estimated by expectation-maximization (EM) algorithm (Day, 1969; Denpster et al., 1977). We implemented the model fitting in R with the package “mixtools.” The histogram indicated small numbers of mixture components for all datasets. Gaussian mixtures with two to six components were compared, and the optimal numbers of components were selected according to the Akaike information criterion (AIC) (Akaike, 1973, 1974).
qPCR
PER2::LUC enteroids and Bmal1 KD B6 enteroids were plated into 12-well plates at the start of the experiment. Organoids for each time point were plated into a separate plate to limit manipulation or exposure to possible resetting cues. End of serum shock is indicated by circadian time 0 for comparison with bioluminescence recordings performed in parallel. The details of the procedures are given in the Supplemental Experimental Procedures.
Histological Analysis of Mouse Jejunum
We sacrificed animals using CO2 inhalation followed by cervical dislocation. The jejunum was dissected from the mouse, flushed with ice-cold PBS, fixed by 10% formalin, and processed for H&E staining for histological evaluation. All animals were maintained and sacrificed according to the oversight, rules, and regulations of the Institutional Animal Care and Use Committee at Cincinnati Children’s Hospital Medical Center.
Statistical Analysis
All values are expressed as mean ± SE. Statistical difference between the considered groups was evaluated by Student’s t test or Tukey-Kramer multiple comparisons test. Time-course qRT-PCR data and TopFlash assay data were analyzed by Kruskal-Wallis test to find differences in the series of values. p < 0.05 was considered significant.
Coupled Mathematical Model of the Circadian Clock and Cell Cycle
For our simulations, we adapted the coupled model of the circadian clock and cell cycle (Zámborszky et al., 2007). The details of the procedures are given in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The circadian clock gates cell-cycle progression in the intestinal epithelium
The circadian clock is less robust in intestinal stem and progenitor cells
WNT secreted from Paneth cells couples the circadian clock and cell cycle
Acknowledgments
We thank Dr. John B. Hogenesch (University of Cincinnati) for discussions and donating the green- and red-luciferase genes. We thank Drs. Atsushi Miyawaki (RIKEN BSI) and Hiroyuki Miyoshi (RIKEN BRC) for donating plasmid vectors to express mVenus-hGeminin. We thank Drs. Hiroshi Kiyonari, Takaya Abe, and Yasuhide Furuta (RIKEN CDB) for donating FUCCI2 (CDB0264K) and H2B-EGFP (CDB0238K) mice. We thank Dr. Jeffery Whitsett (Cincinnati Children’s Hospital Medical Center [CCHMC]) for donating R-spondin-conditioned cells. We thank Drs. Leesa Samson (CCHMC), Roel Nusse (Stanford University), and Ms. Jungin Kwon for donating the Lgr5-EGFP-IRES-CreERT2+/−/Rosa-CAG-LSL-tdTomato-WPRE+/− mice, the 7TFP plasmid (Addgene plasmid 24308), and technical help, respectively. We thank the Integrative Morphology Core of the Digestive Health Center (NIH P30 DK078392) at CCHMC for H&E staining of mouse jejunum sections and for providing support for live microscopy. This work was supported by Department of Interior grant D12AP00005 (to S.R.M. and C.I.H.).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, two tables, and five movies and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.10.015.
AUTHOR CONTRIBUTIONS
T.M., T.Z., S.L., S.R.M., and C.I.H. designed the experiments. A.D. performed the coupled mathematical model of the circadian clock and cell cycle. J.R. performed the population level experiments and analysis of enteroids. T.M. and Y.R. performed the FFT analysis. Y.R. and T.M. performed the mixture model analysis. T.M. and E.A. performed fluorescent imaging, immunostaining of enteroids, and analysis. T.M., H.L., and C.L. performed tissue preparation and analysis of H&E-stained sections. T.M., A.D., J.R., E.A., H.L., Y.R., A.E.R., T.Z., M.H.M., S.L., S.R.M., K.O., and C.I.H. wrote the manuscript. T.M., S.L., S.R.M., and C.I.H. supervised the experiments.
References
- Abe T, Sakaue-Sawano A, Kiyonari H, Shioi G, Inoue K, Horiuchi T, Nakao K, Miyawaki A, Aizawa S, Fujimori T. Visualization of cell cycle in mouse embryos with Fucci2 reporter directed by Rosa26 promoter. Development. 2013;140:237–246. doi: 10.1242/dev.084111. [DOI] [PubMed] [Google Scholar]
- Aihara E, Mahe MM, Schumacher MA, Matthis AL, Feng R, Ren W, Noah TK, Matsu-ura T, Moore SR, Hong CI, et al. Characterization of stem/progenitor cell cycle using murine circumvallate papilla taste bud organoid. Sci Rep. 2015;5:17185. doi: 10.1038/srep17185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike H. Information theory and an extension of the maximum likelihood principle. In: Parzen E, Tanabe K, Kitagawa G, editors. Selected Papers of Hirotugu Akaike. Springer; 1973. pp. 199–213. [Google Scholar]
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schutz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- Bieler J, Cannavo R, Gustafson K, Gobet C, Gatfield D, Naef F. Robust synchronization of coupled circadian and cell cycle oscillators in single mammalian cells. Mol Syst Biol. 2014;10:739. doi: 10.15252/msb.20145218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carulli AJ, Samuelson LC, Schnell S. Unraveling intestinal stem cell behavior with models of crypt dynamics. Integr Biol (Camb) 2014;6:243–257. doi: 10.1039/c3ib40163d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346:1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- Day NE. Estimating the components of a mixture of normal distributions. Biometrika. 1969;56:463–474. [Google Scholar]
- Denpster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J R Stat Soc Series B Stat Methodol. 1977;39:1–38. [Google Scholar]
- Durand A, Donahue B, Peignon G, Letourneur F, Cagnard N, Slomianny C, Perret C, Shroyer NF, Romagnolo B. Functional intestinal stem cells after Paneth cell ablation induced by the loss of transcription factor Math1 (Atoh1) Proc Natl Acad Sci USA. 2012;109:8965–8970. doi: 10.1073/pnas.1201652109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feillet C, Krusche P, Tamanini F, Janssens RC, Downey MJ, Martin P, Teboul M, Saito S, Lévi FA, Bretschneider T, et al. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc Natl Acad Sci USA. 2014;111:9828–9833. doi: 10.1073/pnas.1320474111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froy O, Chapnik N. Circadian oscillation of innate immunity components in mouse small intestine. Mol Immunol. 2007;44:1954–1960. doi: 10.1016/j.molimm.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Froy O, Chapnik N, Miskin R. Mouse intestinal cryptdins exhibit circadian oscillation. FASEB J. 2005;19:1920–1922. doi: 10.1096/fj.05-4216fje. [DOI] [PubMed] [Google Scholar]
- Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS ONE. 2010;5:e9370. doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gery S, Komatsu N, Baldjyan L, Yu A, Koo D, Koeffler HP. The circadian gene per1 plays an important role in cell growth and DNA damage control in human cancer cells. Mol Cell. 2006;22:375–382. doi: 10.1016/j.molcel.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Gil-Lozano M, Mingomataj EL, Wu WK, Ridout SA, Brubaker PL. Circadian secretion of the intestinal hormone GLP-1 by the rodent L cell. Diabetes. 2014;63:3674–3685. doi: 10.2337/db13-1501. [DOI] [PubMed] [Google Scholar]
- Gréchez-Cassiau A, Rayet B, Guillaumond F, Teboul M, Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535–4542. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- Gribble FM, Reimann F. Enteroendocrine cells: chemosensors in the intestinal epithelium. Annu Rev Physiol. 2016;78:277–299. doi: 10.1146/annurev-physiol-021115-105439. [DOI] [PubMed] [Google Scholar]
- Guo B, Chatterjee S, Li L, Kim JM, Lee J, Yechoor VK, Minze LJ, Hsueh W, Ma K. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 2012;26:3453–3463. doi: 10.1096/fj.12-205781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill BR, De Souza MJ, Williams NI. Characterization of the diurnal rhythm of peptide YY and its association with energy balance parameters in normal-weight premenopausal women. Am J Physiol Endocrinol Metab. 2011;301:E409–E415. doi: 10.1152/ajpendo.00171.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong CI, Zámborszky J, Baek M, Labiscsak L, Ju K, Lee H, Larrondo LF, Goity A, Chong HS, Belden WJ, Csikász-Nagy A. Circadian rhythms synchronize mitosis in Neurospora crassa. Proc Natl Acad Sci USA. 2014;111:1397–1402. doi: 10.1073/pnas.1319399111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P, Pascual G, Merlos-Suárez A, Batlle E, Ripperger J, Albrecht U, Cheng HY, Obrietan K, Di Croce L, Benitah SA. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- Janich P, Toufighi K, Solanas G, Luis NM, Minkwitz S, Serrano L, Lehner B, Benitah SA. Human epidermal stem cell function is regulated by circadian oscillations. Cell Stem Cell. 2013;13:745–753. doi: 10.1016/j.stem.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Kowalska E, Ripperger JA, Hoegger DC, Bruegger P, Buch T, Birchler T, Mueller A, Albrecht U, Contaldo C, Brown SA. NONO couples the circadian clock to the cell cycle. Proc Natl Acad Sci USA. 2013;110:1592–1599. doi: 10.1073/pnas.1213317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim X, Tan SH, Koh WL, Chau RM, Yan KS, Kuo CJ, van Amerongen R, Klein AM, Nusse R. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science. 2013;342:1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F, Chen Y, Li X, Zhao Q, Tan Z. Over-expression of circadian clock gene Bmal1 affects proliferation and the canonical Wnt pathway in NIH-3T3 cells. Cell Biochem Funct. 2013;31:166–172. doi: 10.1002/cbf.2871. [DOI] [PubMed] [Google Scholar]
- Malloy JN, Paulose JK, Li Y, Cassone VM. Circadian rhythms of gastrointestinal function are regulated by both central and peripheral oscillators. Am J Physiol Gastrointest Liver Physiol. 2012;303:G461–G473. doi: 10.1152/ajpgi.00369.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science. 2003;302:255–259. doi: 10.1126/science.1086271. [DOI] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Moore SR, Pruszka J, Vallance J, Aihara E, Matsuura T, Montrose MH, Shroyer NF, Hong CI. Robust circadian rhythms in organoid cultures from PERIOD2:LUCIFERASE mouse small intestine. Dis Model Mech. 2014;7:1123–1130. doi: 10.1242/dmm.014399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Ikeda M, Ohmiya Y, Nakajima Y. Simultaneous monitoring of independent gene expression patterns in two types of cocultured fibroblasts with different color-emitting luciferases. BMC Biotechnol. 2008;8:40. doi: 10.1186/1472-6750-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noti M, Sidler D, Brunner T. Extra-adrenal glucocorticoid synthesis in the intestinal epithelium: more than a drop in the ocean? Semin Immunopathol. 2009;31:237–248. doi: 10.1007/s00281-009-0159-2. [DOI] [PubMed] [Google Scholar]
- Pendergast JS, Yeom M, Reyes BA, Ohmiya Y, Yamazaki S. Disconnected circadian and cell cycles in a tumor-driven cell line. Commun Integr Biol. 2010;3:536–539. doi: 10.4161/cib.3.6.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring A, Kim YM, Kahn M. Wnt/catenin signaling in adult stem cell physiology and disease. Stem Cell Rev. 2014;10:512–525. doi: 10.1007/s12015-014-9515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Soták M, Sumová A, Pácha J. Cross-talk between the circadian clock and the cell cycle in cancer. Ann Med. 2014;46:221–232. doi: 10.3109/07853890.2014.892296. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Nishino K, Umezawa A, Akutsu H. Investigating cellular identity and manipulating cell fate using induced pluripotent stem cells. Stem Cell Res Ther. 2012;3:8. doi: 10.1186/scrt99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Yagita K, Horie K, Koinuma S, Nakamura W, Yamanaka I, Urasaki A, Shigeyoshi Y, Kawakami K, Shimada S, Takeda J, Uchiyama Y. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci USA. 2010;107:3846–3851. doi: 10.1073/pnas.0913256107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Pando BF, Dong G, Golden SS, van Oudenaarden A. Circadian gating of the cell cycle revealed in single cyanobacterial cells. Science. 2010;327:1522–1526. doi: 10.1126/science.1181759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom M, Pendergast JS, Ohmiya Y, Yamazaki S. Circadian-independent cell mitosis in immortalized fibroblasts. Proc Natl Acad Sci USA. 2010;107:9665–9670. doi: 10.1073/pnas.0914078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11:106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zámborszky J, Hong CI, Csikász Nagy A. Computational analysis of mammalian cell division gated by a circadian clock: quantized cell cycles and cell size control. J Biol Rhythms. 2007;22:542–553. doi: 10.1177/0748730407307225. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Kay SA. Clocks not winding down: unravelling circadian networks. Nat Rev Mol Cell Biol. 2010;11:764–776. doi: 10.1038/nrm2995. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.