ABSTRACT
SIRT7, a member of the sirtuin family of NAD+-dependent protein deacetylases, is a key mediator of many cellular activities. SIRT7 expression is linked to cell proliferation and oncogenic activity, connecting SIRT7-dependent regulation of ribosome biogenesis with checkpoints controlling cell cycle progression, metabolic homeostasis, stress resistance, aging and tumorigenesis. Despite this important functional link, the enzymatic activity, the molecular targets and physiological functions of SIRT7 are poorly defined. Here, we review recent progress in SIRT7 research and elaborate the main pathways in which SIRT7 participates.
KEYWORDS: metabolism, SIRT7, stress response, transcription, tumorigenesis
Introduction
Members of the silent information regulator-2 (Sir2) protein family, termed sirtuins, are NAD+-dependent protein deacetylases or ADP ribosyltransferases, which are highly conserved from prokaryotes to eukaryotes. The seven mammalian sirtuins, SIRT1–7 (Fig. 1), share a conserved NAD+-dependent catalytic core domain, localize in different subcellular compartments, target different substrates and govern diverse important biological processes including energy metabolism, stress resistance, maintenance of genomic stability, aging and tumorigenesis (for review, see Refs.1-3). Sirtuins have been implicated in the pathogenesis of several human diseases including cancer, type II diabetes, dyslipidemia and cardiovascular or neurodegenerative disorders.4 While SIRT6 and SIRT7 are nuclear proteins, SIRT3–5 localize mainly in mitochondria, and SIRT1 and SIRT2 are found both in the nucleus and the cytoplasm.5 Although SIRT7 has been shown to promote the invasiveness and metastasis of cancer cells, it is the least understood member of the human sirtuin family. This is to a large extent due to its low enzymatic activity in vitro and the few molecular substrates identified so far. We herein review the recent progress in SIRT7 biology, focusing on transcriptional regulation and the main cellular pathways that are affected by SIRT7.
Figure 1.
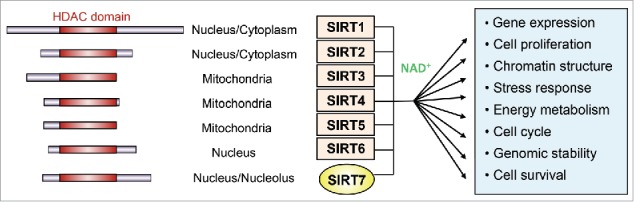
The sirtuin family of NAD+-dependent deacetylases. The seven mammalian sirtuins (SIRT1–7) share a conserved catalytic core domain (HDAC domain) with varying N- and C-terminal sequences. They localize to different cellular compartments, use NAD+ as a co-factor and serve functions in multiple cellular processes.
SIRT7 promotes pre-rRNA transcription and processing
Previous studies have shown that SIRT7 levels positively correlate with ribosome biogenesis and cell proliferation, SIRT7 expression being abundant in metabolically active cells and low or even absent in non-proliferating cells.5,6 SIRT7 is enriched in nucleoli, where it facilitates RNA polymerase I (Pol I)-dependent transcription of rRNA genes (rDNA) by interacting with the transcription factor UBF and the Pol I subunit PAF53 (polymerase-associated factor 53), the mammalian homolog of the yeast Pol I subunit A49.7 Hyperacetylation of PAF53 at lysine 373 by CBP decreases rDNA occupancy of Pol I, whereas deacetylation by SIRT7 leads to increased DNA binding and enhanced pre-rRNA synthesis. The interconnected pathways that link ribosome biogenesis and cell cycle progression provide an intracellular network through which SIRT7 may regulate cell proliferation.
In support of this notion, recent work has shown that non-canonical Wnt signaling suppresses the growth of breast cancer cells by counteracting SIRT7 function.8,9 Wnt5a was found to trigger nucleolar accumulation of Dishevelled 1 (DVL1), a tumor suppressor that binds to rDNA. Increased levels of DVL1 displace SIRT7 from rDNA, hence repressing Pol I transcription.9
In accord with pre-rRNA transcription and maturation being functionally coupled,10,11 SIRT7 is also required for proper processing of pre-rRNA. A recent screening in human cells identified 286 proteins involved in rRNA maturation, among them SIRT7.12 Mechanistically, SIRT7 deacetylates U3–55k, a core component of the U3 snoRNP complex.13-15 Acetylation of U3–55k by the histone acetyltransferase PCAF impairs the association with U3 snoRNA, a prerequisite for proper processing. Deacetylation by SIRT7, on the other hand, facilitates the interaction of U3–55k with U3 snoRNA, thus promoting pre-rRNA cleavage. Importantly, knockdown of U3–55k protein led to the same defects in pre-rRNA processing as those observed upon knockout of SIRT7, underscoring the intimate link between SIRT7 activity and acetylation-dependent U3–55k function.15 Thus, SIRT7 plays a dual function in ribosome biogenesis, coupling rDNA transcription and pre-rRNA processing by deacetylating PAF53 and U3–55k (Fig. 2).
Figure 2.
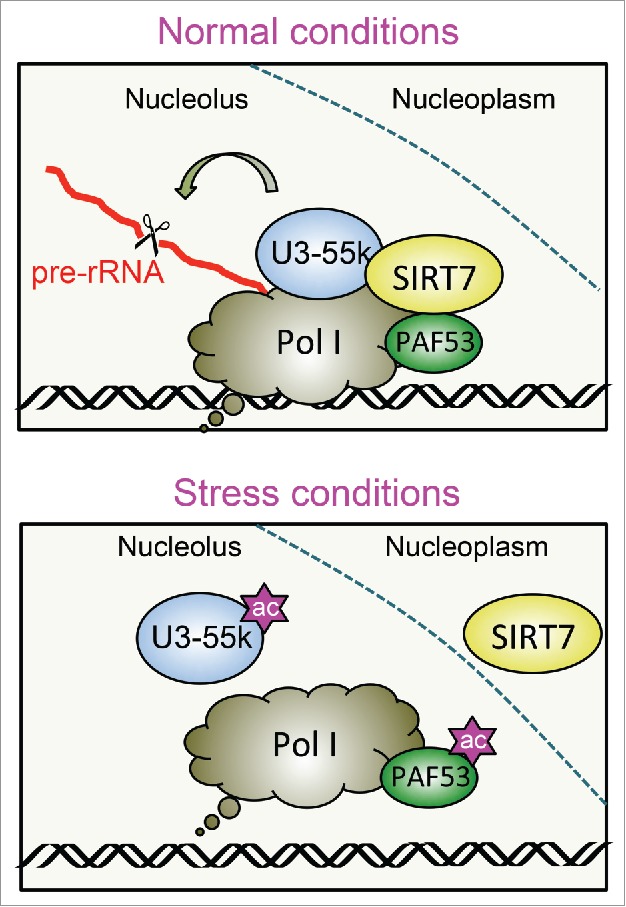
Reversible acetylation of PAF53 and U3–55k regulates pre-rRNA synthesis and processing. In normal growth conditions, SIRT7 keeps the Pol I-associated factor PAF53 hypoacetylated, which is required for rDNA transcription. Deacetylation of the U3–55k protein by SIRT7 facilitates the interaction of U3–55k with U3 snoRNA, thus promoting pre-rRNA cleavage. Nucleolar release of SIRT7 in response to environmental or metabolic stress enhances acetylation of PAF53 and U3–55k, which impairs Pol I transcription and pre-rRNA processing and attenuates ribosome biogenesis.
SIRT7 regulates transcription of all three nuclear RNA polymerases
SIRT7 expression correlates with cell growth, being high in metabolically active cells, and low or even absent in non-proliferating cells.5,6,16,17 High expression of SIRT7 is propelling cells toward tumorigenesis, whereas depletion of SIRT7 leads to decreased cell proliferation, induction of apoptosis and reduced tumor growth.18,19 Given the vital role of SIRT7 in cellular homeostasis, it is not surprising that its function is not restricted to pre-rRNA synthesis and processing. Proteomic approaches have shown that SIRT7 is associated with numerous non-nucleolar target proteins with functions in transcription, ribosome biogenesis and translation.20,21 Consistent with the multifaceted role of SIRT7 in diverse cellular processes, SIRT7 interacts with chromatin remodeling complexes, such as B-WICH, NoRC and SWI/SNF, which are required for the establishment of a specific chromatin structure.20 Furthermore, SIRT7 was found to interact with the Pol III-specific transcription factor TFIIIC2 and to occupy genes that are transcribed by Pol III. In accord with SIRT7 playing a role in Pol III transcription, knockdown of SIRT7 decreased the level of tRNAs in HeLa cells.22 However, whether this effect on Pol III transcription represents a physiological role for SIRT7 remains to be further validated.
In addition to Pol I and Pol III transcription activation, SIRT7 also exerts a positive effect on Pol II transcription. Unpublished results from our lab have revealed that SIRT7 is associated with Pol II and regulates transcription of snoRNAs and other genes transcribed by Pol II. Mechanistically, SIRT7 promotes the release of P-TEFb from the inactive 7SK snRNP complex and deacetylates CDK9, a subunit of the elongation factor P-TEFb, which activates transcription by phosphorylating serine 2 within the C-terminal domain (CTD) of Pol II. SIRT7-mediated activation of P-TEFb promotes phosphorylation of the CTD and stimulates transcription elongation.
Similar to other sirtuins, SIRT7 depletion did not globally change the acetylation levels of nucleolar and nuclear proteins, indicating that it exhibits a weak or substrate-specific deacetylase activity.5 SIRT7 has extremely low basal deacetylase activity on histones and no measurable deacetylase activity of SIRT7 has been detected in vitro. Nevertheless, recent work has identified novel targets of SIRT7, including histone H3, transcription factors and enzymes. SIRT7 has been reported to be recruited by specific transcription factors, e.g., ELK4 and Myc, to deacetylate lysine 18 of histone H3 (H3K18), hypoacetylation of H3K18 compromising transcription of specific target genes.18,23,24 Several studies using purified histones or histone peptides have failed to detect a clear enzymatic activity in in vitro deacetylation assays. On chromatin substrates, however, SIRT7 displayed robust NAD+-dependent H3K18 deacetylase activity, indicating that native chromatin rather than histones is targeted by SIRT7.25
Consistent with SIRT7 mediating many cellular activities, proteins like p53, PAF53, U3–55k, NPM1 and GABPβ1 have been identified as substrates of SIRT7.7,21,26,27 As described above, SIRT7-dependent deacetylation of the nucleolar proteins PAF53 and U3–55k augments pre-rRNA synthesis and processing (Fig. 2). Moreover, SIRT7 has been shown to deacetylate GABPβ1, a key regulator of mitochondrial functions. Hypoacetylation leads to heterotetramer formation with GABPα, which induces transcription of nuclear-encoded mitochondrial genes.27 SIRT7 has also been reported to regulate the level and the function of the tumor suppressor p53.26 The role of SIRT7 in deacetylation of p53, however, remains controversial, as deacetylation of p53 by SIRT7 was only shown in vitro.5,18,26,28 Hence, further studies are needed to decipher the impact of SIRT7 on p53 function.
Interestingly, recent studies suggest that SIRT7 also has NAD+-dependent histone desuccinylase activity, which catalyzes local desuccinylation of H3K122 in response to genomic insults that induce double-strand DNA breaks.29 Both desuccinylation of H3K122 and deacetylation of H3K18 result in chromatin compaction and lead to transcription inhibition.
SIRT7 maintains metabolic homeostasis
With NAD+ serving as a cofactor for SIRT7 activity, there is a direct link between SIRT7 function and the metabolic state of the cell. Therefore, it is not surprising that several studies have uncovered a role of SIRT7 in regulation of metabolic homeostasis. SIRT7 has been shown to suppress endoplasmatic reticulum (ER) stress and prevent the development of fatty liver disease. Mechanistically, SIRT7 is recruited to the promoters of ribosomal protein genes by interaction with the transcription factor Myc to repress ribosomal protein expression and alleviate the ER stress.24 While SIRT7 knockout mice develop hepatic steatosis resembling human fatty liver disease, overexpression of SIRT7 reverted the fatty liver phenotype in obese mice. This study underscores the role of SIRT7 in metabolic regulation and suggests that SIRT7 can be targeted to restore metabolic homeostasis. However, as other studies failed to reach a consensus about the pathways through which SIRT7 exerts its effect on lipid metabolism,27,30 further studies are required to decipher the role of SIRT7 in hepatic homeostasis.
The impact of SIRT7 on cellular energy metabolism is further substantiated by recent work showing that SIRT7 maintains energy homeostasis by deacetylating GABPβ1, a central regulator of mitochondrial function.27 As deacetylation is required for GABPβ1 activity, SIRT7−/− mice displayed pathologies, which resemble phenotypes of mitochondrial diseases.31 In accord with widespread functions of SIRT7 in response to metabolic demands, energy starvation induces ubiquitin-independent degradation of SIRT7, which leads to attenuation of ribosome biogenesis and maintenance of cellular energy homeostasis.32 Furthermore, SIRT7 negatively regulates the levels and activity of the transcription factors HIF1α and HIF2α, which are central players in energy metabolism and the cellular response to varying oxygen levels.33 Together, these studies demonstrate that SIRT7 has widespread functional implications in the metabolic state of the cell.
SIRT7 attenuates cellular stress
One of the most intriguing roles of the nucleolus, long regarded as a mere ribosome-producing factory, is its participation in monitoring cellular stress signals and transmitting them to the RNA polymerase I (Pol I) transcription machinery. Nucleolar proteins are highly dynamic, showing dramatic changes after exposure to stress stimuli, e.g., hypoxia, ER-stress due to unfolded protein response as well as transcriptional, nutritional or genotoxic stress conditions.34-37 Under normal conditions, SIRT7 is associated with elongating Pol I, contacting both PAF53 and nascent pre-rRNA. Downregulation of Pol I transcription upon exposure to osmotic or transcriptional stress is accompanied by release of SIRT7 from nucleoli, which leads to hyperacetylation of PAF53. As a consequence, binding of Pol I to DNA is reduced and the assembly of transcription complexes is precluded.7 These findings connect the spatial distribution of SIRT7 with the acetylation status of PAF53, highlighting a mechanism that links external cues to acetylation of an essential component of the Pol I transcription apparatus.
Given that the activity of Pol I transcription and pre-rRNA processing are intertwined, it is not surprising that stress-dependent release of SIRT7 from the nucleolus leads to defects in rRNA maturation. Upon stress, the U3–55k protein, which is required for all subsequent processing events, remains acetylated and specific processing intermediates accumulate.15 Thus, nucleolar release of SIRT7 in response to environmental or metabolic cues enhances acetylation of PAF53 and U3–55k, which in turn inhibits Pol I transcription and pre-rRNA processing (Fig. 2).
As SIRT7 function is not restricted to Pol I transcription, it might also target specific Pol II and Pol III genes to promote cellular survival and stress resistance. The predicted role of SIRT7 as a pro-survival adaptor molecule in conditions of cellular stress is supported by recent studies showing that SIRT7 can regulate molecules like HIF1α and HIF2α. Knockdown of SIRT7 increased expression levels of HIF proteins upon hypoxic conditions.33 Interestingly, this upregulation of HIF proteins was independent of the catalytic activity of SIRT7, indicating that SIRT7 can regulate downstream effectors by non-enzymatic mechanisms. In support of this notion, SIRT7 was shown to mediate gene silencing by interaction with the positive transcription factor c-Myc. Upon ER stress, SIRT7 associates with c-Myc and throttles transcription of ER stress response genes.17,24 Thus, a growing body of evidence supports that SIRT7 is a critical modulator of the stress response, which adapts cells to environmental challenges and promotes survival in times of adversity.
SIRT7 promotes tumorigenesis
Several studies have unanimously ascribed SIRT7 an oncogenic potential. In epithelial prostate carcinomas and gastric cancer, high SIRT7 levels are associated with aggressive cancer phenotypes, metastatic disease and poor patient prognosis.19,38 Other cancer entities have also been linked to increased levels of SIRT7 including hepatic, ovarian, breast or lung cancer, ascribing SIRT7 an oncogenic function.28,39-41 However, SIRT7 by itself did not cause oncogenic transformation of primary fibroblasts.18 Therefore, the tumor-promoting properties of SIRT7 may be a secondary effect, which may be brought about by its positive effect on ribosome biogenesis. In support of this view, depletion of SIRT7 or overexpression of a catalytically inactive point mutant led to inhibition of cell proliferation, induction of apoptosis, reduction of tumor growth, suppression of metastases and reversion of the transformed state of tumor cells.19,28 This effect on cell proliferation may be due to decreased expression levels of p21 and increased levels of cyclin D1, which lead to impaired G1/S progression.28 Furthermore, SIRT7 may exert its oncogenic properties by deacetylation of H3K18, which attenuates transcription of target genes linked to tumor suppression18 and by upregulation of rRNA synthesis to meet the increased demand for ribosomes in rapidly growing tumor cells.6,7
Regarding the mechanisms underlying the oncogenic potential of SIRT7, analysis of a large cohort of HCC patients revealed that two microRNAs, miR-125a-5p and miR-125b, are transcriptionally repressed in HCC, whereas SIRT7 expression is upregulated. Ectopic expression of miR-125a-5p and miR-125b in liver cancer cells reduced SIRT7 levels and inhibited cell growth.28 Moreover, SIRT7 was shown to promote gastric cancer growth by repressing microRNA miR-34a expression through deacetylation of H3K18 in the promoter region.38 These studies suggest that microRNAs that target SIRT7 may function as tumor suppressors by controlling aberrant expression of SIRT7.
SIRT7 promotes genome stability
Sirtuins are key players in mechanisms that maintain genomic integrity. Consistent with SIRT7 playing an important role in DNA repair, SIRT7 knockout mice displayed signs of replicative stress and an impaired DNA damage response.42 Moreover, recent studies have revealed that SIRT7 also exhibits NAD+-dependent histone desuccinylase activity, which catalyzes local desuccinylation of H3K122 and is implemented in DNA-damage response and cell survival.29 Mechanistically, SIRT7 is recruited to DNA double-strand breaks in a PARP1-dependent manner, where it catalyzes desuccinylation of H3K122 and promotes chromatin condensation and DSB repair. Reciprocally, depletion of SIRT7 sensitizes cells to genotoxic stresses and impairs chromatin compaction during DNA-damage response.29,42,43 Furthermore, PARP1-mediated recruitment of SIRT7 to DNA damage sites modulates the local H3K18Ac levels. H3K18Ac in turn affects recruitment of the damage response factor 53BP1 to DNA DSBs, thereby influencing the efficiency of non-homologous end joining.42 These results reveal a direct role for SIRT7 in DSB repair and establish a functional link between SIRT7-mediated H3K18 deacetylation, H3K122 desuccinylation and the maintenance of genome integrity.
A link between SIRT7 and premature senescence
Various studies have linked sirtuin functions to cellular senescence and lifespan-extension. Although most of these functions have been primarily attributed to SIRT1, recent work has established an emerging role of SIRT7 in age-related processes. SIRT7-knockout mice had shorter lifespan and were suffering from increased embryonic lethality and age-dependent inflammatory cardiomyopathy.26 In addition, SIRT7 deficiency correlates with premature aging, showing symptoms of kyphosis, decreased gonadal fat pad content and reduced IGF-1 plasma levels.42 SIRT7 expression is reduced in aged human stem cells (HSCs), which are characterized by increased apoptosis, loss of quiescence and decreased reconstitution capacity, features resembling those observed in SIRT7 knockout mice. Reintroduction of SIRT7 in aged HSCs improved their regenerative capacity, underscoring the link between SIRT7 and aging.44
In accord with SIRT7 sensing and responding to external stress signals, overexpression of SIRT7 delayed stress-induced premature senescence in cells treated with doxorubicin, a chemotherapeutic drug that induces senescence or apoptosis in a dose-dependent manner. After treatment with low dose of doxorubicin, the levels of the senescence marker p53 and its downstream target p21 were reduced indicating that overexpression of SIRT7 delayed the onset of premature senescence.17 As cellular senescence and genome stability are tightly linked, the number of γH2AX foci, early markers of DNA damage, was decreased upon overexpression of SIRT7.17 This suggests that increased DNA damage may be responsible for premature senescence in SIRT7-deficient cells. Furthermore, the nucleolar protein nucleophosmin (NPM1) was shown to be deacetylated by SIRT7, hyperacetylation of NPM1 in senescent cells correlating with decreased levels of SIRT7.21 However, the acetylation status of NPM1 has not yet been functionally linked to age-related processes, and therefore it needs to be seen which processes are affected by NPM1 acetylation. Although most studies just show a correlation between SIRT7 levels and aging, they demonstrate the close link between SIRT7 and cellular senescence.
Concluding remarks
SIRT7 has emerged as a critical regulator of cellular homeostasis, influencing multiple biological processes, such as transcription, ribosome biogenesis, chromatin structure and cell proliferation. Since the first functional studies showing that SIRT7 activates ribosome biogenesis and promotes cell proliferation,6 the field of SIRT7 research has steadily expanded (Fig. 3). Global proteomic studies have identified several SIRT7 target proteins, and functional studies using SIRT7 knockout mice have characterized cellular processes that are regulated by SIRT7. These studies revealed that SIRT7 is implicated in regulation of nucle(ol)ar RNA polymerases by various mechanisms, most of them depending on the catalytic activity of SIRT7. SIRT7 regulates gene expression by either directly affecting transcription and/or processing at specific target genes, or by deacetylating H3K18, which represses transcription and stabilizes the cancerous phenotype by epigenetic reprogramming. Consistent with its central role in the promotion of cell proliferation and tumorigenesis, SIRT7 was found to be upregulated in all cancer types studied so far. Depletion of SIRT7, on the other hand, has been associated with increased DNA damage, apoptosis and various diseases. As SIRT7 also serves a vital role in attenuating stress conditions, it emerges as a pro-survival molecule that mediates the cellular stress response. Although more studies are required to decipher the mechanisms underlying the physiological and disease-related functions of SIRT7, the implication of SIRT7 in several human pathologies make it a promising target for novel therapeutic approaches for a variety of diseases.
Figure 3.
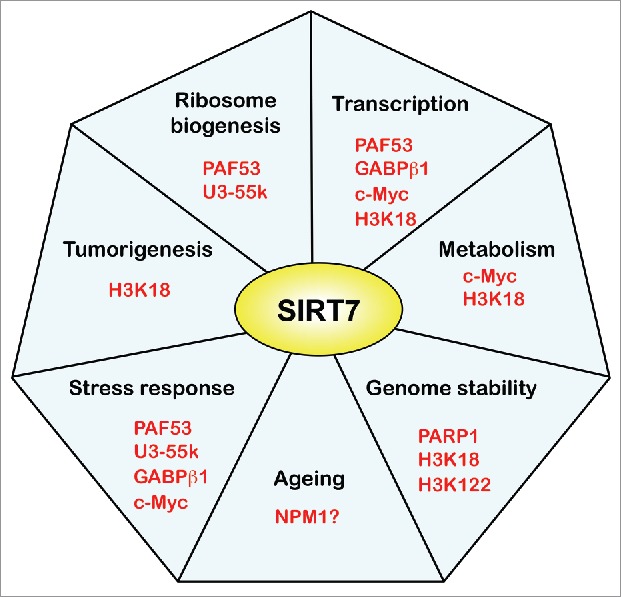
The seven faces of SIRT7. The cartoon depicts the main pathways in which SIRT7 is involved. The proteins that are targeted by SIRT7 or mediate SIRT7 function are highlighted.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
Work from the authors' laboratory was supported by grants from the Deutsche Forschungsgemeinschaft (GR475/22–2; SFB1036) and the CellNetworks Cluster of Excellence (EcTop 5).
References
- [1].Oellerich MF, Potente M. FOXOs and sirtuins in vascular growth, maintenance, and aging. Circ Res 2012; 110:1238-1251; PMID:22539757; http://dx.doi.org/ 10.1161/CIRCRESAHA.111.246488 [DOI] [PubMed] [Google Scholar]
- [2].Yu J, Auwerx J. The role of sirtuins in the control of metabolic homeostasis. Ann N Y Acad Sci 2009; 1173 Suppl 1:E10-E19; PMID:19751409; http://dx.doi.org/ 10.1111/j.1749-6632.2009.04952.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nat Rev Cancer 2015; 15:608-624; PMID:26383140; http://dx.doi.org/ 10.1038/nrc3985 [DOI] [PubMed] [Google Scholar]
- [4].Taylor DM, Maxwell MM, Luthi-Carter R, Kazantsev AG. Biological and potential therapeutic roles of sirtuin deacetylases. Cell Mol Life Sci 2008; 65:4000-4018; PMID:18820996; http://dx.doi.org/ 10.1007/s00018-008-8357-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 2005; 16:4623-4635; PMID:16079181; http://dx.doi.org/ 10.1091/mbc.E05-01-0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 2006; 20:1075-1080; PMID:16618798; http://dx.doi.org/ 10.1101/gad.1399706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen S, Seiler J, Santiago-Reichelt M, Felbel K, Grummt I, Voit R. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol Cell 2013; 52:303-313; PMID:24207024; http://dx.doi.org/ 10.1016/j.molcel.2013.10.010 [DOI] [PubMed] [Google Scholar]
- [8].McDonald SL, Silver A. The opposing roles of Wnt-5a in cancer. Br J Cancer 2009; 101:209-214; PMID:19603030; http://dx.doi.org/ 10.1038/sj.bjc.6605174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dass RA, Sarshad AA, Carson BB, Feenstra JM, Kaur A, Obrdlik A, Parks MM, Prakash V, Love DK, Pietras K. Wnt5a signals through DVL1 to repress ribosomal DNA transcription by RNA polymerase I. PLoS Genet 2016; 12(8):e1006217; PMID:27500936; http://dx.doi.org/ 10.1371/journal.pgen.1006217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Prieto JL, McStay B. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev 2007; 21:2041-2054; PMID:17699751; http://dx.doi.org/ 10.1101/gad.436707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kos M, Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol Cell 2010; 37:809-820; PMID:20347423; http://dx.doi.org/ 10.1016/j.molcel.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tafforeau L, Zorbas C, Langhendries JL, Mullineux ST, Stamatopoulou V, Mullier R, Wacheul L, Lafontaine DL. The complexity of human ribosome biogenesis revealed by systematic nucleolar screening of Pre-rRNA processing factors. Mol Cell 2013; 51:539-551; PMID:23973377; http://dx.doi.org/ 10.1016/j.molcel.2013.08.011 [DOI] [PubMed] [Google Scholar]
- [13].Lübben B, Marshallsay C, Rottmann N, Lührmann R. Isolation of U3 snoRNP from CHO cells: a novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res 1993; 21:5377-5385; PMID:8265352; http://dx.doi.org/ 10.1093/nar/21.23.5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing, and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 2012; 3:397-414; PMID:22065625; http://dx.doi.org/ 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- [15].Chen S, Blank MF, Iyer A, Huang B, Wang L, Grummt I, Voit R. SIRT7-dependent deacetlyation of the U3-55k protein controls pre-rRNA processing. Nat Commun 2016; 7:10734; PMID:26867678; http://dx.doi.org/ 10.1038/ncomms10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Grob A, Roussel P, Wright JE, McStay B, Hernandez-Verdun D, Sirri V. Involvement of SIRT7 in resumption of rDNA transcription at the exit from mitosis. J Cell Sci 2009; 122:489-498; PMID:19174463; http://dx.doi.org/ 10.1242/jcs.042382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kiran S, Chatterjee N, Singh S, Kaul SC, Wadhwa R, Ramakrishna G. Intracellular distribution of human SIRT7 and mapping of nuclear/nucleolar localization signal. FEBS J 2013; 280:3451-3466; PMID:23680022; http://dx.doi.org/ 10.1111/febs.12346 [DOI] [PubMed] [Google Scholar]
- [18].Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K et al.. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012; 487:114-118; PMID:22722849; http://dx.doi.org/ 10.1038/nature11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Malik S, Villanova L, Tanaka S, Aonuma M, Roy N, Berber E, Pollack JR, Michishita-Kioi E, Chua KF. SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci Rep 2015; 5:9841; PMID:25923013; http://dx.doi.org/ 10.1038/srep09841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics 2012; 11:60-76; http://dx.doi.org/ 10.1074/mcp.A111.015156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee N, Kim DK, Kim ES, Park SJ, Kwon JH, Shin J, Park SM, Moon YH, Wang HJ, Gho YS et al.. Comparative interactomes of SIRT6 and SIRT7: Implication of functional links to aging. Proteomics 2014; 14:1610-1622; PMID:24782448; http://dx.doi.org/ 10.1002/pmic.201400001 [DOI] [PubMed] [Google Scholar]
- [22].Tsai YC, Greco TM, Cristea IM. SIRT7 plays a role in ribosome biogenesis and protein synthesis. Mol Cell Proteomics 2014; 13:73-83; PMID:24113281; http://dx.doi.org/ 10.1074/mcp.M113.031377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Seligson DB, Horvath S, McBrian MA, Mah V, Yu H, Tze S, Wang Q, Chia D, Goodglick L, Kurdistani SK. Global levels of histone modifications predict prognosis in different cancers. Am J Pathol 2009; 174:1619-1628; PMID:19349354; http://dx.doi.org/ 10.2353/ajpath.2009.080874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K et al.. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep 2013; 5:654-665; PMID:24210820; http://dx.doi.org/ 10.1016/j.celrep.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Tong Z, Wang Y, Zhang X, Kim DD, Sadhukhan S, Hao Q, Lin H. SIRT7 is activated by DNA and deacetylates histone H3 in the chromatin context. ACS Chem Biol 2016; 11:742-747; PMID:26907567; http://dx.doi.org/ 10.1021/acschembio.5b01084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res 2008; 102:703-710; PMID:18239138; http://dx.doi.org/ 10.1161/CIRCRESAHA.107.164558 [DOI] [PubMed] [Google Scholar]
- [27].Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, Lee JU, Zeviani M, Romand R, Hottiger MO et al.. A SIRT7-dependent acetylation switch of GABPβ1 controls mitochondrial function. Cell Metab 2014; 20:856-869; PMID:25200183; http://dx.doi.org/ 10.1016/j.cmet.2014.08.001 [DOI] [PubMed] [Google Scholar]
- [28].Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Shen Q, Park WS et al.. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology 2013; 57:1055-1067; PMID:23079745; http://dx.doi.org/ 10.1002/hep.26101 [DOI] [PubMed] [Google Scholar]
- [29].Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, He L, Li W, Yi X, Sun L et al.. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun 2016; 7:12235; PMID:27436229; http://dx.doi.org/ 10.1038/ncomms12235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yoshizawa T, Karim MF, Sato Y, Senokuchi T, Miyata K, Fukuda T, Go C, Tasaki M, Uchimura K, Kadomatsu T et al.. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab 2014; 19:712-721; PMID:24703702; http://dx.doi.org/ 10.1016/j.cmet.2014.03.006 [DOI] [PubMed] [Google Scholar]
- [31].Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell 2012; 148:1145-1159; PMID:22424226; http://dx.doi.org/ 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sun L, Fan G, Shan P, Qiu X, Dong S, Liao L, Yu C, Wang T, Gu X, Li Q et al.. Regulation of energy homeostasis by the ubiquitin-independent REGγ proteasome. Nat Commun 2016; 7:12497; PMID:27511885; http://dx.doi.org/ 10.1038/ncomms12497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hubbi ME, Hu H, Kshitiz Gilkes DM, Semenza GL. Sirtuin-7 inhibits the activity of hypoxia-inducible factors. J Biol Chem 2013; 288:20768-20775; PMID:23750001; http://dx.doi.org/ 10.1074/jbc.M113.476903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature 2005; 433:77-83; PMID:15635413; http://dx.doi.org/ 10.1038/nature03207 [DOI] [PubMed] [Google Scholar]
- [35].Boisvert FM, Lamond AI. p53-dependent subcellular proteome localization following DNA damage. Proteomics 2010: 10:4087-4097; PMID:21080495; http://dx.doi.org/ 10.1002/pmic.201000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell 2010; 40:216-227; PMID:20965417; http://dx.doi.org/ 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, Pepperkok R, Lamond AI, Laiho M. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics 2011; 10:M111.009241; PMID:21778410; http://dx.doi.org/ 10.1074/mcp.M111.009241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, Hu Y, Cai T. SIRT7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep 2015; 5:9787; PMID:25860861; http://dx.doi.org/ 10.1038/srep09787 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [39].Aljada A, Saleh AM, Alkathiri M, Shamsa HB, Al-Bawab A, Nasr A. Altered Sirtuin 7 expression is associated with early stage breast cancer. Breast Cancer (Auckl) 2015; 9:3-8; PMID:25922576; http://dx.doi.org/ 10.4137/BCBCR.S23156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang HL, Lu RQ, Xie SH, Zheng H, Wen XM, Gao X, Guo L. SIRT7 exhibits oncogenic potential in human ovarian cancer cells. Asian Pac J Cancer Prev 2015; 16:3573-3577; PMID:25921180; http://dx.doi.org/ 10.7314/APJCP.2015.16.8.3573 [DOI] [PubMed] [Google Scholar]
- [41].Shi H, Zhang D, Liu Y, Fang P. MicroRNA-3666-induced suppression of SIRT7 inhibits the growth of non-small cell lung cancer cells. Oncol Rep 2016; 36:3051-3057; PMID:27599551; http://dx.doi.org/ 10.3892/or.2016.5063 [DOI] [PubMed] [Google Scholar]
- [42].Vazquez BN, Thackray JK, Simonet NG, Kane-Goldsmith N, Martinez-Redondo P, Nguyen T, Bunting S, Vaquero A, Tischfield JA, Serrano L. SIRT7 promotes genomic integrity and modulates non-homologous end joining DNA repair. EMBO J 2016; 35:1488-1503; PMID:27225932; http://dx.doi.org/ 10.15252/embj.201593499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ogiwara H, Ui A, Otsuka A, Satoh H, Yokomi I, Nakajima S, Yasui A, Yokota J, Kohno T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene 2011; 30:2135-2146; PMID:21217779; http://dx.doi.org/ 10.1038/onc.2010.592 [DOI] [PubMed] [Google Scholar]
- [44].Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM. Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 2015; 347:1374-1377; PMID:25792330; http://dx.doi.org/ 10.1126/science.aaa2361 [DOI] [PMC free article] [PubMed] [Google Scholar]


