ABSTRACT
The functional capacity of cells is defined by the transcriptome. Many recent studies have identified variations in the transcriptome of tumors due to alternative splicing changes, as well as mutations in splicing factors and regulatory signals in most tumor types. Some of these alterations have been linked to tumor progression, metastasis, therapy resistance, and other oncogenic processes. Here, we describe the different mechanisms that drive splicing changes in tumors and their impact in cancer. Motivated by the current evidence, we propose a model whereby a subset of the splicing patterns contributes to the definition of specific tumor phenotypes, and may hold potential for the development of novel clinical biomarkers and therapeutic approaches.
KEYWORDS: alternative splicing, cancer, biomarkers, RNA processing, splicing regulation, therapy
Introduction
Cancer arises from genetic and epigenetic alterations that interfere with essential mechanisms of the normal life cycle of cells such as DNA repair, replication control, and cell death.1,2 For example, DNA mutations occurring at genes and regulatory sites may cause the activation or suppression of crucial functions that lead to uncontrolled proliferation. These alterations also impact the transcriptome, which in turn can induce and sustain multiple mechanisms related to the progression of the tumor. In fact, genetic and epigenetic alterations could impair RNA processing before their effect is even visible at protein level, thereby defining the functional capacity of the cells. Accordingly, identifying the alterations of the transcriptome becomes relevant to advance our understanding of tumor biology.
Among all the steps during gene expression, alternative splicing (AS) provides perhaps the largest potential for molecular diversity and controlled regulation in the cell.3 Genes are transcribed into pre-mRNA molecules that require extensive processing. For most genes, this processing involves the removal of introns through the process of splicing. Multiple molecular complexes, composed of RNA-binding proteins (RBPs), structural RNAs, and other protein factors, bind to the pre-mRNA at various locations (RNA-binding motifs) and mediate the splicing process. On the other hand, different mature RNA molecules can be produced from the same pre-mRNA through the mechanism of AS. AS takes place through the controlled changes in the expression and activity of the complexes acting on the regulatory sequences on the pre-mRNA or as a consequence of the alterations in these complexes and motifs. AS is therefore, a critical mechanism not only in normal physiological processes, but also in multiple pathologies, including cancer.4
Splicing alterations in cancer
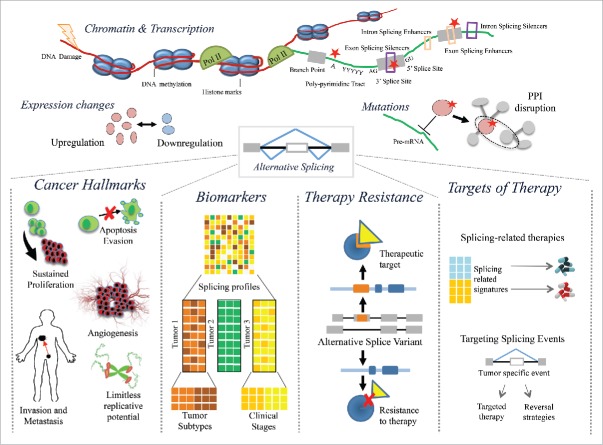
Multiple AS changes have been described that essentially recapitulate cancer-associated phenotypes. A large body of work has been devoted to determine the different alterations that lead to these AS splicing changes observed in cancer. We describe below some of them (Fig. 1).
Figure 1.
Alterations that lead to alternative splicing changes in cancer and their implication for the development of the disease and possible therapeutic strategies. Alterations include expression changes in splicing factors, mutations in splicing factors and splicing regulatory sequences, alterations in the transcription and chromatin state, and DNA damage. These alterations can lead to alternative splicing changes in tumors, which may recapitulate cancer hallmarks, like cell proliferation, disruption of apoptosis, cell motility and invasion, angiogenesis, and limitless replicative potential. Splicing patterns provide predictive signatures for tumor subtypes and clinical properties, and may be indicative of therapy resistance. Finally, some splicing changes are emerging as direct targets of therapy, and the splicing properties of a tumor, as well as the mutational status of splicing factors, can be informative for selection of specific therapies.
Expression changes in splicing regulators
Multiple splicing regulatory factors have been observed to trigger tumorigenic properties in cells when overexpressed or downregulated, and have been characterized as oncogenes or tumor-suppressors, respectively, through the changes they induce in AS.5,6 Some factors recapitulate this role across multiple tumor types, whereas others show a context-dependent expression pattern that may reflect the tissue of origin.7 The expression alteration of splicing regulators may have different origins, like copy number alterations7 or through changes in post-transcriptional modifications that are under the control of cell signaling pathways, which are frequently deregulated in tumors.2 Additionally, multiple splicing factors are transcriptionally controlled by the oncogene MYC, which is frequently overexpressed in tumors and leads to multiple oncogenic splicing changes through the upregulation of splicing factors.6,8 The expression changes in splicing factors is also linked to the metabolic transformations associated to tumors, often triggered by specific cellular microenvironments, which lead to AS changes in genes involved in metabolic processes.9 The link between MYC, splicing, and cancer has been further emphasized recently. Components of the spliceosome appear to be essential for the activity of MYC as oncogene, which underscores the central role of splicing in cancer.10,11
It has been further observed that gene expression alterations in cancer appear to recapitulate partially or extensively physiological pathways. For instance, breast tumors show a pattern in the expression of splicing factors and splicing events that resemble that of undifferentiated cells, including the downregulation of MBNL1 and a splicing change in NUMB.7 Similarly, AS analysis during metastatic colonization12 shows extensive overlap with the changes that occur during epithelial-to-mesenchymal transition.13 However, it is not yet clear whether such cellular programs are fully recapitulated or whether they co-exist with other alterations that appear in tumors, thereby providing tumor cells with a variety of molecular repertoires.
Mutations in splicing regulators
Access to the genome sequence from multiple tumors has uncovered recurrent mutations in core and auxiliary components of the spliceosome in various tumor types. They occur predominantly in hematological malignancies and often involve the factors SF3B1, U2AF1, SRSF2, and ZRSR2 (reviewed in ref.5). Although generally at lower rate, splicing factors also appear mutated in solid tumors, including SF3B1 in breast cancer and melanoma,14,15 U2AF1 and RBM10 in non-small cell lung tumors,16 and HNRNPL in colon tumors.7 An analysis of genes coding for known and putative RBPs has shown that mutations in known and putative regulators of splicing is mostly limited to these cases in solid tumors.7 Additionally, expression changes in splicing factors appear to produce more splicing changes in the events, compared with those related with mutations in splicing factors7 or regulatory regions,17,18 and both types of alterations do not seem to produce the same splicing changes. For instance, modulating the expression of SF3B1 in cells does not recapitulate the changes observed when SF3B1 is mutated.19 The identification of the splicing changes related to mutations in splicing factors is instrumental to understand their relevance for cancer development and therapy and is currently an active area of research.14,19-21
Mutations on splicing regulatory sequences
Somatic mutations that disrupt splicing regulatory motifs can also be a source of splicing changes in cancer. For instance, mutations at the exon–intron boundaries have been associated with intron retention in tumor suppressors such as TP53, ARID1A, PTEN, CHD1, MLL2, and PTCH1.22 Similarly, mutations on synonymous sites on coding exons appear enriched in oncogenes and have been proposed to disrupt the splicing of cancer drivers such as ITK, ALK, IDH1, and BCL6.22 Since splicing regulatory sequences on exons span 4–6 nucleotides, hence possibly covering multiple codons, it is likely that mutations on non-synonymous sites also lead to splicing changes in cancer drivers.23 Intronic mutations also appear to play a crucial role in cancer such as therapy resistance. For instance, a point mutation 51nt upstream of the 3′ splice-site of intron 8 of BRAF promotes a splice variant that confers resistance to Vemurafenib treatment.24 However, in contrast to exonic mutations, not many recurrent intronic mutations have been described so far beyond the exon–intron boundaries, despite the fact that a significant fraction of the splicing regulation is controlled by intronic regulatory sequences, either through the branch-point and poly-pyrimidine tract sequences, or through intronic splicing enhancers and silencers.25 This could be due to the fact that intronic regulatory motifs often present positional variability with respect to the exon–intron boundaries and are, therefore, less straightforward to identify. Although deep intronic mutations may be harder to characterize, they could also affect splicing. For instance, a considerable number of introns harbor distant branch-points located further than 50nt upstream of the 3′ splice-site,26 and the structure of the RNA plays a role in its processing and may bring together distant regions.27 By harnessing the power of characterizing the relevant intronic regulatory regions, we will be able to gain further insights into the disruption of splicing in cancer.
Chromatin and transcription dependent effects
Most of the mechanisms related to gene expression take place in a coordinated way that couples transcription with pre-mRNA processing. Co-transcriptional splicing seems to be quite prevalent and advantageous for the efficiency of splicing.28 There is also plenty of evidence showing that splicing regulation depends on the coupling with the dynamics of RNA polymerase II (RNAPII). This is controlled by, among other elements, the activity of promoters and transcriptional enhancers, the chromatin state, and the recruitment of splicing factors by RNAPII or to the chromatin context.28,29 Accordingly, alterations in cancer that affect transcription or chromatin may also impact splicing. For instance, the Histone methyltransferase SETD2 appears frequently mutated in kidney tumors, which has been related to alterations in RNA processing and splicing.30,31 It is thus conceivable that many of the splicing changes observed in tumors are direct or indirect effects due to global or local somatic alterations of transcription and chromatin.
DNA damage
DNA damage through the exposure to either radiation or toxic chemicals has been shown to induce AS in genes involved in cellular processes such as apoptosis, cell-cycle control, and DNA repair.32,33 The splicing response to DNA damage comes through various modulations such as the post-transcriptional modification of splicing factors that affect protein interactions with other splicing factors or with RNA. For example, dephosphorylation of SRSF10 through DNA damage prevents the interaction with hnRNP F/H, favoring a splicing change in BCL2L1 to enhance the production of its pro-apoptotic isoform.33 On the other hand, stable formation of ribonucleoprotein complexes prevents the appearance of RNA:DNA duplexes, which would otherwise promote mutations and genome instability.32 For instance, the disruption of a complex of BRCA1 with RNA has been found to be related to defective DNA damage repair.34 As the functional control of splicing emerges as essential for DNA damage repair, splicing-related alterations may contribute to a genome instability phenotype and to the accumulation of mutations.
Functional impact of the splicing alterations in cancer
The analyses of transcriptomes from multiple patient tumor samples have highlighted frequent splicing changes during tumor progression and metastasis transformation,12,35 as well as in association to somatic alterations.14,19,20 However, the functional impact of these AS changes and their significance in cancer is only starting to be elucidated.
Alternative splicing recapitulates hallmarks of cancer
Several AS events have been shown to recapitulate cancer-associated phenotypes. For instance, an exon inclusion change in NUMB has been shown to promote cell proliferation.36 Similarly, an exon-skipping event in MST1R has been related to the acquisition of cell motility during cancer cell invasion.37 Moreover, the modulation of these events can recapitulate the tumor phenotype or revert to a normal phenotype.36,38 Therefore, understanding the general functional effects of AS potentially leads to the discovery of novel oncogenic mechanisms and therapeutic targets.
AS changes have been proposed to remodel the network of protein–protein interactions in a tissue-specific manner.39,40 It is, therefore, possible that splicing changes in cancer also impact the network of protein–protein interactions, but in a disruptive, non-regulated way. In this direction, a recent study shows that an AS change in NFE2L2 that occurs in various tumor types leads to the loss of a protein interaction with its negative regulator KEAP1, thereby providing an alternative way to activate the Nrf2 pathway.41 This may in fact be a general mechanism whereby splicing alterations disrupt protein–protein interactions of cancer drivers and related pathways, providing other means to impact cell function that are equivalent to classical somatic mutations in drivers. Additionally, AS may also induce degradation of the transcripts through non-sense mediated decay,42 a mechanism that was associated to somatic mutations on the splice-sites that induce intron-retention in tumor suppressors.18
Alternative splicing as biomarkers
Despite the abundance of splicing changes observed in tumors, only few cases have been characterized for their functional impact. It is possible that the majority of the splicing changes in tumors are passengers, merely reflecting upstream genetic mechanisms and the deregulation of splicing fidelity mechanisms. Yet, they may provide tale-tell signs of specific tumor characteristics. In this context, splicing changes have been shown to separate tumor types and subtypes17 and have been related to tumor stage and patient survival,35,43 so they have the potential to be used as biomarkers for specific clinical conditions. This could be relevant for cases for which a known prognostic marker is either not present in the sample or does not exist, as for pediatric tumors.44
Alternative splicing and therapy resistance
Alterations in AS also appear essential for understanding drug resistance.21 For instance, a considerable proportion of patients that do not respond to targeted treatment against BRAF mutations express a BRAF isoform lacking exons 4–8, which encompass the RAS-binding domain.45 Interestingly, small-molecule modulators of pre-mRNA splicing are capable of restoring the original BRAF splicing and reduce growth of therapy-resistant cells.24 Similarly, AS also impacts immunotherapy in leukemia due to the disrupted activity of the splicing factor SRSF3.46 These results highlight the importance of characterizing the transcriptome for therapy and suggest that specific splicing alterations may provide a selective advantage to tumors.
Alternative splicing as target of therapy
There is a growing interest to search for splicing-related alterations for which specific therapies could be developed. One of the strategies being tested at the moment consists in the synthetic design of antisense oligonucleotides (AONs) that target-specific splicing events. AONs are able to revert AS events to restore normal cellular phenotypes36,38 and have reached already clinical trial stage for some splicing-related disorders.50 Another promising strategy for cancer therapeutics is the use of small molecule compounds that modulate the activity of splicing factors.21,51 These therapies have a wide range of effects depending on the tumor type or the mutational status of the targeted splicing factor. Thus, it becomes essential to know which patients may benefit from splicing-related therapies. One such possible class includes patients with overexpressed MYC in tumors, which are more dependent on the activity of the spliceosome.10,11
AS events are also emerging as direct actionable alterations for targeted therapies. This is the case of the skipping of MET exon 14 observed in some lung cancer patients, resulting in a deletion of the protein region that inhibits its kinase catalytic activity.47 Importantly, the skipping of this exon is sufficient for MET activation and tumors that harbor the event respond to MET-targeted therapies.48,49 Although this splicing change in MET has been explained so far as a result of somatic mutations on exon 14 or on its splice-sites, it is conceivable that the same splicing change could occur due to other mechanisms yet to be discovered. These results raise the interesting possibility that an AS event could be used as direct target of therapy. Thus, either as direct targets or as a means to characterize the tumor, the splicing properties may become fundamental to identify therapeutic vulnerabilities and potential resistance. This may be particularly relevant for tumors lacking somatic mutations in genes with known targeted therapy, as these patients cannot benefit from currently available therapies.
Combinatorial control of RNA splicing and possible implications for cancer
AS changes that characterize and contribute to the pathophysiology of cancer are triggered by alterations in a complex network of different mechanisms. These combinatorial effects have some interesting implications. Different alterations in tumors may in turn impact RNA processing and splicing in similar ways. For instance, mutations in RBM10 or downregulation of QKI lead to the same splicing change in NUMB that promotes cell proliferation.36,52 This suggests that the splicing alterations observed in tumors may be indicative of a phenotypic advantage, and some may even phenocopy somatic mutations in cancer drivers to induce similar functional impacts. Accordingly, a subset of the splicing changes in cancer may play an important role in the neoplastic process independently of or in conjunction with the already characterized genetic alterations.
It is not clear yet whether a single splicing change may be sufficient to induce an oncogenic transformation in a normal tissue context, or even whether splicing events can be considered cancer drivers. It is possible that the splicing-related effects are additive, contributing to, and maintaining specific properties or favoring certain cellular environments that modulate the oncogenic impact of somatic mutations. Consistent with this, there is a relation between specific tumor microenvironments and AS.53 Additionally, somatic mutations in splicing factors are generally heterozygous and appear to require a normal functional splicing machinery to exert their oncogenic function.21,54 For example, the ratio of both mutant and wild-type U2AF1 splicing factor influences the splice-site selection in lung adenocarcinomas, questioning the functional significance of the mutant U2AF1 cells.54 This suggests a context-dependent effect, by which somatic alterations may become relevant in the presence of certain splicing-related signatures. This is further supported by recent findings showing that tumors with overexpressed MYC are highly dependent on the splicing machinery for survival and may be more sensitive to splicing-related therapies.10,11
In conclusion, as selection on the tumor clones is exerted on the phenotype rather than on the genotype, we propose that the splicing patterns may define relevant molecular phenotypes in tumors, despite their genetic heterogeneity. The characterization of tumor transcriptomes – with respect to splicing – thus becomes essential to understand their clinical properties and to select appropriate therapeutic strategies.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank E. Sebestyén, O. Abdel-Wahab, and G. Dujardin for comments on earlier versions of the manuscript, and E. Porta-Pardo, H. Climente-González, J. Yokota, L. Montuenga, and R. Pío for useful discussions.
Funding
This work was supported by the MINECO and FEDER (BIO2014–52566-R) and AGAUR (SGR2014–1121).
References
- [1].Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-674; PMID:21376230; http://dx.doi.org/ 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- [2].Feinberg AP, Koldobskiy MA, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016; 17:284-299; PMID:26972587; http://dx.doi.org/ 10.1038/nrg.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fu X-D, Ares M. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 2014; 15:689-701; PMID:25112293; http://dx.doi.org/ 10.1038/nrg3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chabot B, Shkreta L. Defective control of pre-messenger RNA splicing in human disease. J Cell Biol 2016; 212:13-27; PMID:26728853; http://dx.doi.org/ 10.1083/jcb.201510032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer 2016; 16:413-430; PMID:27282250; http://dx.doi.org/ 10.1038/nrc.2016.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Anczuków O. Krainer AR Splicing-factor alterations in cancers. RNA 2016; 22:1285-1301; PMID:27530828; http://dx.doi.org/22545246 10.1261/rna.057919.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sebestyén E, Singh B, Miñana B, Pagès A, Mateo F, Pujana MA, Valcárcel J, Eyras E. Large-scale analysis of genome and transcriptome alterations in multiple tumors unveils novel cancer-relevant splicing networks. Genome Res 2016; 26:732-744; PMID:27197215; http://dx.doi.org/22545246 10.1101/gr.199935.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Das S, Anczuków O, Akerman M, Krainer AR. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep 2012; 1:110-117; PMID:22545246; http://dx.doi.org/ 10.1016/j.celrep.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010; 463:364-368; PMID:20010808; http://dx.doi.org/ 10.1038/nature08697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hsu TY-T, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S et al.. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 2015; 525:384-388; PMID:26331541; http://dx.doi.org/ 10.1038/nature14985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP, Al-Haddawi M, Tan SY, Osato M, A Sabò et al.. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015; 523:96-100; PMID:25970242; http://dx.doi.org/ 10.1038/nature14351 [DOI] [PubMed] [Google Scholar]
- [12].Lu Z, Huang Q, Park JW, Shen S, Lin L, Tokheim CJ, Henry MD, Xing Y. Transcriptome-wide landscape of pre-mRNA alternative splicing associated with metastatic colonization. Mol Cancer Res 2015; 13:305-318; PMID:25274489; http://dx.doi.org/ 10.1158/1541-7786.MCR-14-0366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. An emt-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 2011; 7:e1002218; PMID:21876675; http://dx.doi.org/ 10.1371/journal.pgen.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, Bailey SL, Bhavsar EB, Chan B et al.. Cancer-associated SF3B1 hotspot mutations induce cryptic 3′ splice site selection through use of a different branch point. Cell Rep 2015; 13:1033-1045; PMID:26565915; http://dx.doi.org/ 10.1016/j.celrep.2015.09.053 [DOI] [PubMed] [Google Scholar]
- [15].Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S et al.. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov 2013; 3:1122-1129; PMID:23861464; http://dx.doi.org/ 10.1158/2159-8290.CD-13-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J et al.. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One 2014; 9:e87361; PMID:24498085; http://dx.doi.org/ 10.1371/journal.pone.0087361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sebestyen E, Zawisza M, Eyras E. Detection of recurrent alternative splicing switches in tumor samples reveals novel signatures of cancer. Nucleic Acids Res 2015; 43:1345-1356; PMID:25578962; http://dx.doi.org/ 10.1093/nar/gku1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, Hong D, Park PJ, Lee E. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet 2015; 47:1242-1248; PMID:26437032; http://dx.doi.org/ 10.1038/ng.3414 [DOI] [PubMed] [Google Scholar]
- [19].Alsafadi S, Houy A, Battistella A, Popova T, Wassef M, Henry E, Tirode F, Constantinou A, Piperno-Neumann S, Roman-Roman S et al.. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat Commun 2016; 7:10615; PMID:26842708; http://dx.doi.org/ 10.1038/ncomms10615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A, Li Y, Chung YR, Micol JB, Murphy ME et al.. SRSF2 mutations contribute to myelodysplasia by mutant-specific effects on exon recognition. Cancer Cell 2015; 27:617-30; PMID:25965569; http://dx.doi.org/ 10.1016/j.ccell.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee SC-W, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med 2016; 22:976-986; PMID:27603132; http://dx.doi.org/ 10.1038/nm.4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell 2014; 156:1324-1335; PMID:24630730; http://dx.doi.org/ 10.1016/j.cell.2014.01.051 [DOI] [PubMed] [Google Scholar]
- [23].Sterne-Weiler T, Sanford JR. Exon identity crisis: disease-causing mutations that disrupt the splicing code. Genome Biol 2014; 15:201; PMID:24456648; http://dx.doi.org/ 10.1186/gb4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Salton M, Kasprzak WK, Voss T, Shapiro BA, Poulikakos PI MT. Inhibition of vemurafenib-resistant melanoma by interference with pre-mRNA splicing. Nat Commun Dis 2015; 6:7103; PMID: 25971842; http://dx.doi.org/26992833 10.1038/ncomms8103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Diederichs S, Bartsch L, Berkmann JC, Fröse K, Heitmann J, Hoppe C, Iggena D, Jazmati D, Karschnia P, Linsenmeier M et al.. The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med 2016; 8:442-57; PMID:26992833; http://dx.doi.org/ 10.15252/emmm.201506055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Corvelo A, Hallegger M, Smith CWJ, Eyras E. Genome-wide association between branch point properties and alternative splicing. PLoS Comput Biol 2010; 6:e1001016; PMID:21124863; http://dx.doi.org/ 10.1371/journal.pcbi.1001016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S et al.. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol 2013; 20:1434-1442; PMID:24213538; http://dx.doi.org/ 10.1038/nsmb.2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Naftelberg S, Schor IE, Ast G, Kornblihtt AR. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu Rev Biochem 2015; 84:165-198; PMID:26034889; http://dx.doi.org/ 10.1146/annurev-biochem-060614-034242 [DOI] [PubMed] [Google Scholar]
- [29].Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell 2011; 144:16-26; PMID:21215366; http://dx.doi.org/ 10.1016/j.cell.2010.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Grosso AR, Leite AP, Carvalho S, Matos MR, Martins FB, Vítor AC, Desterro JM, Carmo-Fonseca M, de Almeida SF. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife 2015; 4:e09214; PMID:26575290; http://dx.doi.org/ 10.7554/eLife.09214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].JM Simon, Hacker KE, Singh D, Brannon AR, Parker JS, Weiser M, Ho TH, Kuan PF, Jonasch E, Furey TS et al.. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res 2014; 24:241-250; PMID:24158655; http://dx.doi.org/ 10.1101/gr.158253.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shkreta L, Chabot B. The RNA splicing response to DNA damage. Biomolecules 2015; 5:2935-2977; PMID:26529031; http://dx.doi.org/ 10.3390/biom5042935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shkreta L, Toutant J, Durand M, Manley JL, Chabot B. SRSF10 connects DNA damage to the alternative splicing of transcripts encoding apoptosis, cell-cycle control, and DNA repair factors. Cell Rep 2016; 17:1990-2003; PMID:27851963; http://dx.doi.org/ 10.1016/j.celrep.2016.10.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Savage KI, Gorski JJ, Barros EM, Irwin GW, Manti L, Powell AJ, Pellagatti A, Lukashchuk N, McCance DJ, McCluggage WG et al.. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol Cell 2014; 54:445-459; PMID:24746700; http://dx.doi.org/ 10.1016/j.molcel.2014.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Trincado JL, Sebestyén E, Pagés A, Eyras E. The prognostic potential of alternative transcript isoforms across human tumors. Genome Med 2016; 8:85; PMID:27535130; http://dx.doi.org/ 10.1186/s13073-016-0339-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bechara EG, Sebestyén E, Bernardis I, Eyras E, Valcárcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell 2013; 52:720-733; PMID:24332178; http://dx.doi.org/ 10.1016/j.molcel.2013.11.010 [DOI] [PubMed] [Google Scholar]
- [37].Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol Cell 2005; 20:881-890; PMID:16364913; http://dx.doi.org/ 10.1016/j.molcel.2005.10.026 [DOI] [PubMed] [Google Scholar]
- [38].Ghigna C, De Toledo M, Bonomi S, Valacca C, Gallo S, Apicella M, Eperon I, Tazi J, Biamonti G. Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed: therapeutic potential of bifunctional oligonucleotides and indole derivatives. RNA Biol 2010; 7:495-503; PMID:20864806; http://dx.doi.org/ 10.4161/rna.7.4.12744 [DOI] [PubMed] [Google Scholar]
- [39].Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks. Mol Cell 2012; 46:871-883; PMID:22749400; http://dx.doi.org/ 10.1016/j.molcel.2012.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ellis JD, Barrios-Rodiles M, Colak R, Irimia M, Kim T, Calarco JA, Wang X, Pan Q, O'Hanlon D, Kim PM et al.. Tissue-specific alternative splicing remodels protein-protein interaction networks. Mol Cell 2012; 46:884-92; PMID:22749401; http://dx.doi.org/ 10.1016/j.molcel.2012.05.037 [DOI] [PubMed] [Google Scholar]
- [41].Goldstein LD, Lee J, Gnad F, Klijn C, Schaub A, Reeder J, Daemen A, Bakalarski CE, Holcomb T, Shames DS et al.. Recurrent loss of NFE2L2 exon 2 is a mechanism for Nrf2 pathway activation in human cancers. Cell Rep 2016; 16(10):2605-2617; PMID:27568559; http://dx.doi.org/ 10.1016/j.celrep.2016.08.010 [DOI] [PubMed] [Google Scholar]
- [42].Green RE, Lewis BP, Hillman RT, Blanchette M, Lareau LF, Garnett AT, Rio DC, Brenner SE. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics 2003; 19 (Suppl 1):i118-121; PMID:12855447; http://dx.doi.org/ 10.1093/bioinformatics/btg1015 [DOI] [PubMed] [Google Scholar]
- [43].Shen S, Wang Y, Wang C, Wu YN, Xing Y. SURVIV for survival analysis of mRNA isoform variation. Nat Commun 2016; 7:11548; PMID:27279334; http://dx.doi.org/ 10.1038/ncomms11548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C et al.. The genetic landscape of the childhood cancer medulloblastoma. Science 2011; 331:435-439; PMID:21163964; http://dx.doi.org/ 10.1126/science.1198056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT et al.. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011; 480:387-390; PMID:22113612; http://dx.doi.org/ 10.1038/nature10662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, Sussman R, Lanauze C, Ruella M, Gazzara MR et al.. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov 2015; 5:1282-1295; PMID:26516065; http://dx.doi.org/ 10.1158/2159-8290.CD-15-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, Holcomb T, Pujara K, Stinson J, Fu L et al.. Somatic mutations lead to an oncogenic deletion of Met in lung cancer. Cancer Res 2006; 66:283-289; PMID:16397241; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-2749 [DOI] [PubMed] [Google Scholar]
- [48].Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, Akimov M, Bufill JA, Lee C, Jentz D et al.. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015; 5:850-860; PMID:25971938; http://dx.doi.org/ 10.1158/2159-8290.CD-15-0285 [DOI] [PubMed] [Google Scholar]
- [49].Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, Ginsberg MS, Borsu L, Schultz N, MF6 Berger, Rudin CM et al.. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015; 5:842-849; PMID:25971939; http://dx.doi.org/ 10.1158/2159-8290.CD-14-1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Havens MA, Hastings ML. Splice-switching antisense oligonucleotides as therapeutic drugs. Nucleic Acids Res 2016; 44 6549-6563; PMID:27288447; http://dx.doi.org/ 10.1093/nar/gkw533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Salton M, Misteli T. Small molecule modulators of pre-mRNA splicing in cancer therapy. Trends Mol Med 2016; 22:28-37; PMID:26700537; http://dx.doi.org/ 10.1016/j.molmed.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H et al.. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014; 10:e1004289; PMID:24722255; http://dx.doi.org/ 10.1371/journal.pgen.1004289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Brosseau J-P, Lucier JF, Nwilati H, Thibault P, Garneau D, Gendron D, Durand M, Couture S, Lapointe E, Prinos P et al.. Tumor microenvironment-associated modifications of alternative splicing. RNA 2014; 20:189-201; PMID:24335142; http://dx.doi.org/ 10.1261/rna.042168.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Fei DL, Motowski H, Chatrikhi R, Prasad S, Yu J, Gao S, Kielkopf CL, Bradley RK, Varmus H et al.. Wild-type U2AF1 antagonizes the splicing program characteristic of U2AF1-mutant tumors and is required for cell survival. PLoS Genet 2016; 12:e1006384; PMID:27776121; http://dx.doi.org/ 10.1371/journal.pgen.1006384 [DOI] [PMC free article] [PubMed] [Google Scholar]