Two cases of metastatic colorectal cancer with a POLE mutation, both of which were ultramutated and microsatellite stable, are presented and discussed from the standpoint of the basic biochemical mechanisms leading to a unique phenotype in POLE deficiency, the challenges faced with interpreting the genomic profiling of tumors in this important subset of patients, and the potential clinical implications.
Abstract
Deficiencies in DNA repair due to mutations in the exonuclease domain of DNA polymerase ɛ have recently been described in a subset of cancers characterized by an ultramutated and microsatellite stable (MSS) phenotype. This alteration in DNA repair is distinct from the better‐known mismatch repair deficiencies which lead to microsatellite instability (MSI) and an increased tumor mutation burden. Instead, mutations in POLE lead to impaired proofreading intrinsic to Pol ɛ during DNA replication resulting in a dramatically increased mutation rate. Somatic mutations of Pol ɛ have been found most frequently in endometrial and colorectal cancers (CRC) and can lead to a unique familial syndrome in the case of germline mutations. While other key genomic abnormalities, such as MSI, have known prognostic and treatment implications, in this case it is less clear. As molecular genotyping of tumors becomes routine in the care of cancer patients, less common, but potentially actionable findings such as these POLE mutations could be overlooked unless appropriate algorithms are in place. We present two cases of metastatic CRC with a POLE mutation, both of which are ultramutated and MSS. The basic biochemical mechanisms leading to a unique phenotype in POLE deficiency as well as challenges faced with interpreting the genomic profiling of tumors in this important subset of patients and the potential clinical implications will be discussed here. The Oncologist 2017;22:497–502
Key Points.
Clinicians should recognize that tumors with high tumor mutation burden and that are microsatellite stable may harbor a POLE mutation, which is associated with an ultramutated phenotype.
Work‐up for POLE deficiency should indeed become part of the routine molecular testing paradigm for patients with colorectal cancer.
This subset of patients may benefit from clinical trials where the higher number of mutation‐associated neoantigens and defect in DNA repair may be exploited therapeutically.
Patient #1
The first case is a 49‐year‐old man with recurrent metastatic colon cancer. At age 45, the patient presented with abdominal pain and was found to have a mass at the hepatic flexure on colonoscopy. Staging computed tomography (CT) showed no evidence of metastatic disease. The patient underwent a right hemicolectomy and biopsy of a peritoneal nodule. Pathology from both the colon resection and peritoneal nodule demonstrated signet ring cell adenocarcinoma (Fig. 1). Additional initial testing showed that the patient's tumor had no mutations at codons 12 and 13 in exon 2 of the KRAS gene and was microsatellite stable (MSS) by polymerase chain reaction. He received FOLFOX and bevacizumab for 12 cycles. The patient had no evidence of disease until almost 2 years later when he developed urinary retention. Workup including cystoscopy revealed a prostate mass. Pathology from subsequent bladder biopsy and transurethral resection of the prostate revealed poorly differentiated adenocarcinoma with mucinous and signet ring cell features compatible with the patient's previous colonic primary. He resumed FOLFOX and bevacizumab with stabilization of his disease. At the time of progression, he was enrolled in a clinical trial with FOLFIRI and ziv‐aflibercept. Genomic profiling was performed on the primary colon tumor to assess the patient's eligibility for clinical trials and identified mutations in KRAS (Q61), BRCA2, FGFR2, MSH2, NF1, CDK12, NTRK3, POLE, APC, ATR, CDC73, CHD4, CTNNA1, GRIN2A, KDM6A, KLHL6, LRP1B, MAGI2, PIK3R1, SLIT2, SMAD4, and 71 additional alterations of uncertain significance. The specimen was deemed MSS but revealed an extremely high tumor mutation burden (TMB), with ∼116 mutations per mega base. A course of pembrolizumab was started given the presence of the MSH2 mutation and high TMB, two factors that have been associated with responses to checkpoint inhibitors. Repeat imaging after 12 weeks of therapy, however, showed rapid and clear‐cut progression of disease, and the patient was initiated on regorafenib. The patient continued to experience disease progression and was ultimately transitioned to hospice care.
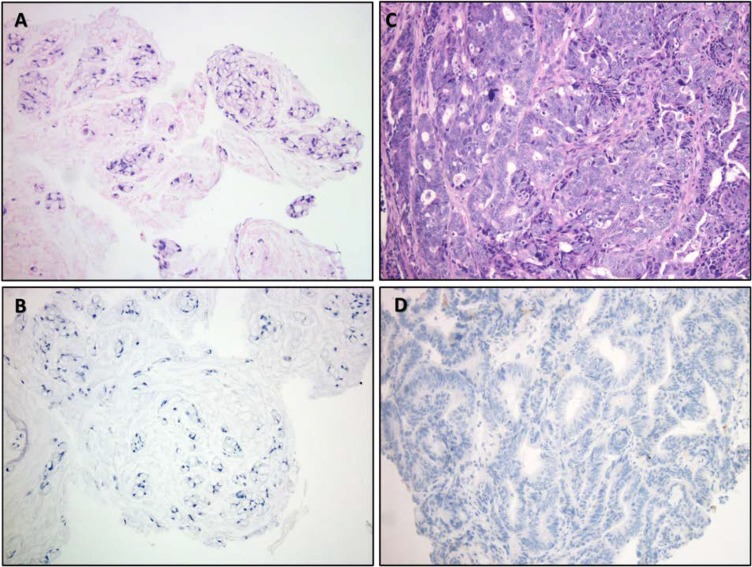
Figure 1.
Histomorphology images from patients 1 and 2. (A): Histologic appearance of hematoxylin and eosin stained tumor from patient #1 demonstrating a discohesive mucinous tumor with signet ring morphology without true glandular formation. (B): No discernable proliferation of tumor‐infiltrating lymphocytes (TILs) was noted, and immunohistochemical staining for programmed death‐ligand 1 (PD‐L1) was negative (DAKO Inc., Carpenteria CA, Clone 22C3). (C): Appearance of tumor from patient 2 reveals a more conventional colonic adenocarcinoma, which was moderately differentiated with cribriform growth and irregular glands. (D): While histologically dissimilar from patient 1, here too, the presence of TILs was insignificant and PD‐L1 staining was negative. Immunohistochemical staining for the mismatch repair proteins was retained in both specimens (not shown). All images taken at ×200 total magnification.
Patient #2
The second case is a 56‐year‐old man with untreated hepatitis C, prior intravenous heroin use, and metastatic rectal cancer. He initially presented at age 55 with weight loss, rectal pain, and bleeding. CT scan demonstrated a large centrally necrotic rectal mass, tumor thrombus occluding the left portal vein, as well as multiple perirectal abscesses. Biopsy of the rectal mass done by flexible sigmoidoscopy revealed invasive adenocarcinoma with extensive necrosis (Fig. 1). The patient subsequently underwent diagnostic laparoscopy with diverting colostomy and incision and drainage of abscesses. He was treated with concurrent capecitabine and radiation. Follow‐up imaging showed decrease in the size of the rectal mass as well as progression of portal vein thrombosis and suspicion of new carcinomatosis. He was then started on capecitabine and oxaliplatin (XELOX) and panitumumab. Follow‐up imaging demonstrated partial response, and he remains on this regimen at the time of writing. Genomic profiling of the primary rectal tumor revealed mutations in BRCA2, ATM, PIK3CA, APC, TP53, POLE, RAF1, ABL2, NF1, CTNNB1, FBXW7, NTRK2, PTEN, ACVR1B, ARID1A, ATRX, CHD2, CUL3, IRF2, KDM5A, KEL, LRP1B, MAP2K4, PBRM1, PIK3R1, PRDM1, PRKDC, SETD2, TET2, XPO1, and an additional 118 variants of uncertain significance. His tumor was RAS wild‐type, MSS, and had an extremely high TMB, with 208.43 mutations per mega base.
Molecular Tumor Board
Genotyping Results and Interpretation of the Molecular Results
These two cases were discussed at a Montefiore Medical Center Molecular Tumor Board in September 2016. This novel entity characterized by a POLE mutation and an ultramutated and MSS phenotype was identified, and consideration of immunotherapy or experimental options was recommended based on the findings that are reviewed below.
In these two cases, tumor specimens from formalin fixed paraffin embedded tissue were sent to a Clinical Laboratory Improvement Amendments certified laboratory for genomic profiling using a next‐generation sequencing (NGS) panel, which covers the coding region of 315 “cancer‐related” genes plus introns from 28 genes known to be altered in cancer (FoundOne). In addition to accurate determination of base pair substitutions, in/dels, rearrangements, and copy number changes, microsatellite status and TMB are also defined. Measurement of microsatellite instability (MSI) is done by assessing indel characteristics at 114 homopolymer repeat loci in or near the targeted gene regions. TMB is extrapolated to the whole tumor genome after measuring somatic mutations in the sequenced genes. Mutation rates ≥20/Mb, 6–19/Mb, and ≤5/Mb are reported as high, intermediate, and low, respectively.
The genomic testing for both index patients revealed the samples were MSS but categorized as high TMB. In fact, the mutation rate for both patients was remarkably increased (116/Mb in patient 1 and 208.43/Mb in patient 2), consistent with an ultramutated phenotype. The Cancer Genome Atlas Network (TCGA) 2012 examined 224 colorectal cancer (CRC) tumors with exome capture DNA sequencing and identified two subsets of tumors based on mutation rate [1]. Eighty‐four percent of cases had a mutation rate <8.24/Mb and the remaining 16% had mutation rates >12/Mb, classified as nonhypermutated and hypermutated, respectively. Thirty hypermutated tumors had complete data sets, and, of these, 23 were found to be MSI‐high. The seven hypermutated cases that were not MSI‐high, which accounted for the six cases with the highest mutation rates, all contained missense mutations in the proofreading exonuclease domain of DNA polymerase ɛ (POLE). Among other mutations identified in our two index patients, both contained a POLE mutation. The POLE mutation reported, p.Val411Leu, is the same for both of our patients, indicating the encoded amino acid at position 411 has changed from a valine to a leucine. Mutation allele frequencies of the POLE mutation were 10% in patient 1 and 24% in patient 2. In both cases, allele frequencies were consistent with a clonal mutation.
Patterns of genes that were recurrently mutated in the TCGA data were also characterized. Fifteen expressed genes were found to be recurrently mutated in the hypermutated CRC and 17 in the nonhypermutated cancers. The patterns and frequency of mutations were significantly different between the two groups. For example, TP53 and APC were mutated significantly more frequently in the nonhypermutated tumors, whereas TGFBR2 was more frequently mutated in the hypermutated cases. The mutational profile of hypermutated tumors with MLH1 silencing and MSI‐high demonstrated significantly higher rates of frameshift mutations in 28 assessed genes with long mononucleotide repeats in their coding sequences [1]. Further analysis of the CRCs with POLE exonuclease domain mutations (EDM) in the TCGA data demonstrated an increase in all types of base substitutions. C:G>T:A was the most common mutation type, but compared to other cancers the relative increase in G:C>T:A was most dramatic [2].
Functional and Clinical Significance of a POLE Mutation in CRC
High‐fidelity DNA replication in human cells is safeguarded by two pivotal mechanisms: proofreading by the DNA polymerases themselves and DNA mismatch repair (MMR). The chances of new mutations arising during genome replication are thereby drastically limited by the accuracy of DNA replicases in base selection, aided by nearly instant proofreading via built‐in exonuclease function and subsequent MMR for errors escaping proofreading. Germline effects in these genes affecting DNA repair can lead to unique phenotypes characterized by cancer susceptibility syndromes. On the other hand, somatic alterations can provide a recurrent mechanism of carcinogenesis via the accumulation of large number of mutations [3].
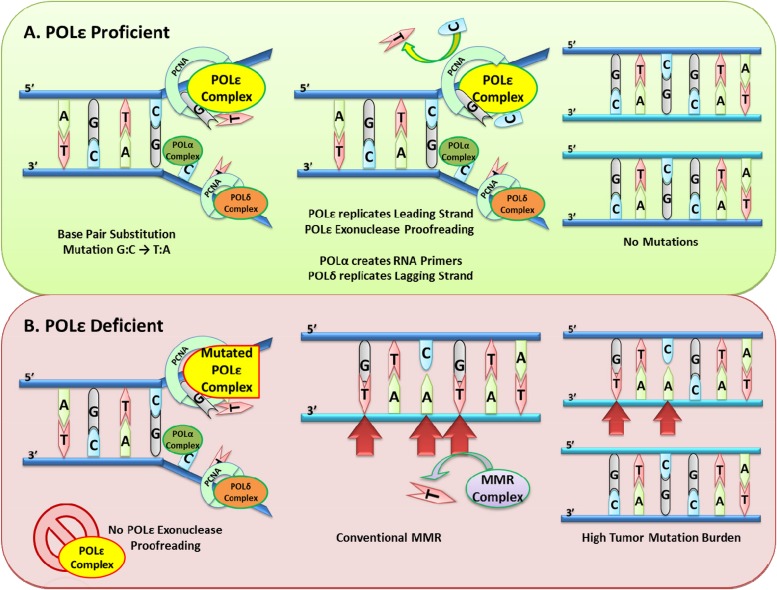
MutS and MutL are proteins which are critical to initiation of mismatch repair and are often mutated in hereditary nonpolyposis colorectal cancer. MMR begins when the MutS heterodimer complex (α or β) recognizes and binds mismatched DNA or insertion/deletion loops (IDL) on the daughter strand of replicating DNA (Fig. 2). The MutS DNA complex then recruits the MutL heterodimer (consisting of MLH1 and PMS2). A third complex, proliferating cell nuclear antigen and replication factor C, binds MutL and serves as a DNA clamp, activating the endonuclease function of PMS2. Simultaneously, exonuclease 1 function is also activated for excision of the mismatched DNA. DNA ligase then rejoins the replacement nucleotide(s) of the excised mismatch [4].
Figure 2.
Biochemical basis of POLE deficiency.
Abbreviations: MMR, mismatch repair; PCNA, proliferating cell nuclear antigen; POL α, DNA polymerase α; POL δ, DNA polymerase δ; POL ɛ, DNA polymerase ɛ.
Heterozygous mutations in any of the four MMR genes, MLH1, MSH2, MSH6, and PMS2, contribute to a state termed MSI. MSI is characterized by deficiency in repair of base‐base mismatches and/or IDL, leading to missense or frameshift mutations, as well as by frequent insertion and deletions especially at homopolymer and dinucleotide repeats. MSI is commonly associated with hereditary nonpolyposis CRC and a high DNA mutational burden in associated cancers ranging from 10 to 100 mutations per Mb [4], [5].
Distinct from MMR, proofreading during DNA replication is performed by the exonuclease domains of DNA polymerase δ (Pol δ) and Pol ɛ, which are critical components of the normal DNA replication process that is carried out by several interacting polymerases. Polymerase α initiates DNA synthesis, then Pol ɛ and Pol δ take over replication of the leading and lagging strands of eukaryotic DNA, respectively. During replication, DNA polymerase α (Pol α) and Pol ɛ, via their exonuclease function, detect and replace mismatched bases in the leading strand by checking against the methylated parent strand [4], [6], [7]. This proofreading function improves replication fidelity by 100‐fold or so.
Despite high levels of evolutionary conservation in the exonuclease domain of both genes, the polymerases are susceptible to germline and somatic EDM. POLE‐mutated, MSS patients present with high tumor burden due to deficiency in the 3′ to 5′ proofreading functions in the exonuclease domain of the polymerase. Recurrent “hotspot” mutations have been identified in the exonuclease domain of Pol ɛ, and the majority are located along the DNA‐binding pocket—most commonly affecting the P286 amino acid residue. Others, including residue 411, are not predicted to directly interact with DNA. However, in biochemical studies, this clearly leads to deficient proofreading function. Mutations such as V411L are seen repeatedly in these cancers, including our two cases, suggesting an indirect effect on the proofreading function of the exonuclease domain.
Although mutations in POLE can be corrected by MMR, the substantial mutation burden simply overwhelms the MMR pathway and demonstrates the characteristic multifaceted oncogenic progression [4], [7], [8]. In addition, many POLE‐deficient tumors harbor additional mutations in MMR genes, possibly further contributing to the excessive mutation rate. Overall, the very high mutation rate leads to an average of 5,000 coding sequence mutations in POLE cases just in the coding sequence. Many of the recurrent mutations include nonsense changes in APC, MSH6, p53, KRAS, and PIK3CA that are often of types and positions other than common hotspots and might be contributing to overall carcinogenesis as so‐called “mini‐drivers.”
In 2010, Yoshida et al. reported the first POLE mutation in a human colorectal adenocarcinoma [9]. Since then, multiple germline and somatic mutations in POLD1 and POLE have been reported, predominantly in endometrial cancer and CRC. Data from The TCGA reported POLE mutations in approximately 3% of colorectal and 7% of endometrial cancers [1]. Genomic profiling found that the majority of tumors with these POLE mutations demonstrate very high TMB, often exceeding 100 mutations per Mb. Ultramutated tumors make up 6.4% of low‐grade and 17.4% of high‐grade endometrioid endometrial cancers (ECs) [10]. As discussed, the specific alterations are distinct from those typically seen in cancers with MSI [1]. Rare germline mutations in POLD1 and POLE have been described in patients with polymerase proofreading‐associated polyposis, a clinical phenotype quite similar to that of MMR deficiency [11], [12]. While germline POLD1 mutations might be involved in familial cases, there is little evidence to suggest that somatic POLD1 mutations act as a driver of spontaneous CRC, as seen in POLE mutations.
Enhanced immunogenicity and improved clinical outcomes have been reported in endometrial cancer patients with POLE mutations [13]. Recent data suggest similar findings in CRC with POLE mutations. Cox regression analysis on pooled data from three clinical trials and multiple patient cohorts detected 66 POLE mutations in 6,517 (1.0%) CRC samples [14]. POLE mutations were associated with young age, male sex, right‐sided tumor location, early disease stage, and absence of MMR deficiency. Furthermore, POLE mutations were associated with a reduced risk of disease recurrence (hazard ratio [HR] 0.34, p = .006), and this association was stronger than that seen with MMR deficiency, which is known to carry a more favorable prognosis, particularly in early‐stage CRC. Immunohistochemical studies also demonstrated increased tumor‐infiltrating lymphocytes in tumors with POLE mutations compared with MMR‐proficient tumors, but this finding was not statistically different when compared with MMR‐deficient tumors. Interestingly, a smaller study by Stenzinger et al. did not identify a clear association between POLE mutations and clinical outcomes in 431 CRC patients with MSS disease [15].
Potential Strategies to Target the Pathway
A phase II study of pembrolizumab in 41 patients with advanced cancers concluded that MMR deficiency predicts higher response rates [16]. Eleven patients had MMR‐deficient CRC, 21 patients had MMR‐proficient CRC, and 9 patients had MMR‐deficient non‐CRC. Both primary endpoints were significant: the immune‐related objective response rate and 20‐week immune‐related progression‐free survival were 40% and 78%, respectively for MMR‐deficient CRC and 0% and 11% for MMR‐proficient CRC. Non‐CRC with MMR‐deficient tumors had similar results to the cohort of MMR‐deficient CRC: 71% immune‐related objective response rate and 67% immune‐related progression free survival rate. Of note, a high mutation burden was associated with prolonged progression‐free survival [16].
Indeed, several tumor types with high TMB have recently been shown to have better response rates with immune checkpoint inhibitors. The increased tumor‐infiltrating lymphocytes observed in MSI‐high CRC are themselves a sign of immune activation, and it has been hypothesized that the increased mutational burden seen in these tumors could predict immunogenicity and response to immune checkpoint blockade. It is estimated that POLE mutated tumors on average have 15‐fold more neoantigens than MSI tumors and 100‐fold more than typical MSS tumors [17], [18].
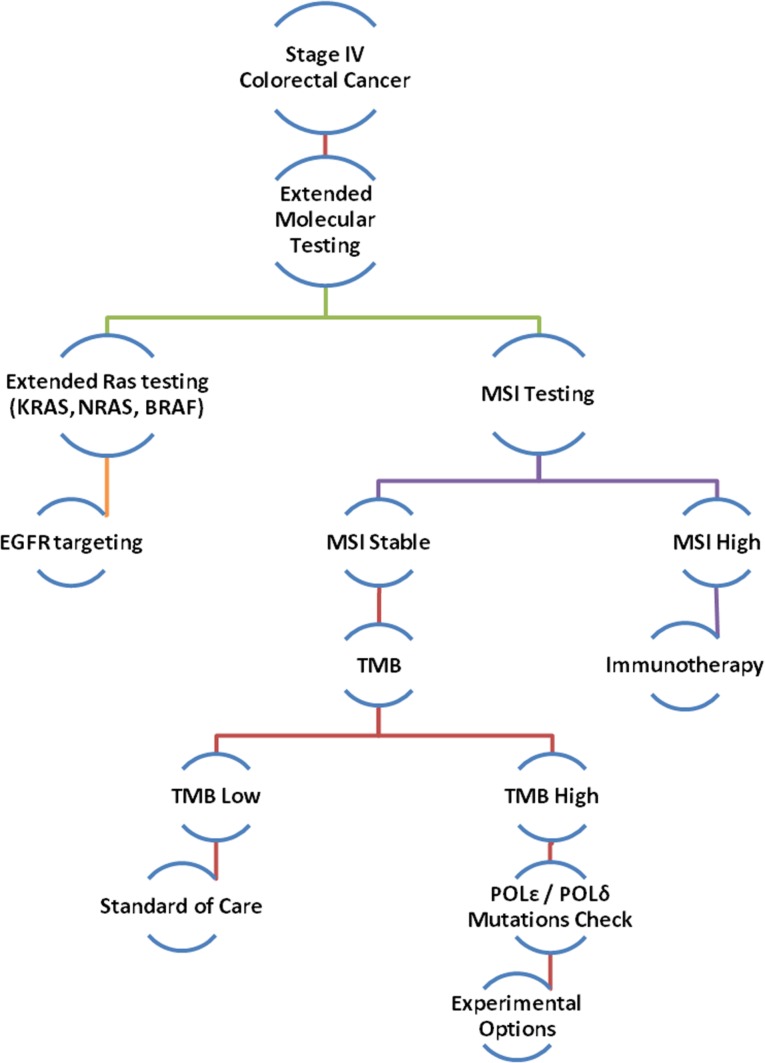
With evidence to support improved response to immune checkpoint blockade in tumors with high mutation rates, it is logical to hypothesize similar responses in the subset of cancers with POLE mutations leading to an ultramutated MSS phenotype. There are ongoing clinical trials addressing this question; however, identifying such cases with this potentially meaningful genomic abnormality poses a challenge. Interestingly, a recent study demonstrated that screening for MMR status could be done very reliably using NGS profiling and cutoffs for mutational load [19]. Out of 224 CRC tumors that underwent NGS profiling (341 gene panel), 100% of the 193 tumors with <20 mutations were MMR proficient. Twenty‐eight of the 31 tumors with >20 mutations were MMR deficient, and the remaining three had distinctly higher mutation rates and all harbored the POLE P286R mutation. Given that POLE mutations are found in a small but relevant portion of CRC, we propose an algorithm that includes screening for such mutations when performing genomic profiling on these tumors (Fig. 3). Clinical trials targeting programmed cell death 1/programmed death‐ligand 1 (PD‐L1) in a variety of malignancies with POLE mutations are ongoing (NCT02912572, NCT02899793, and NCT02658279).
Figure 3.
Proposed algorithm for expanded molecular testing for stage IV colorectal cancer.
Abbreviations: EGFR, epidermal growth factor receptor; MSI, microsatellite instability; POL δ, DNA polymerase δ; POL ɛ, DNA polymerase ɛ; TMB, tumor mutation burden.
As discussed, tumors with POLE EDM display characteristic increases in base substitutions. However, there is evidence that these mutations do not all lead to one unique clinical, pathologic, or molecular phenotype. Ahn et al. examined 28 MSS early‐onset CRC with whole exome sequencing and identified 6 hypermutated cases, 4 of which had the same POLE P286R mutation and 2 of which had POLE mutations outside the exonuclease domain [6]. Findings were validated with an expanded cohort of 83 MSS early‐onset CRC, which identified 6 tumors with the same POLE P286 mutation. No significant difference was seen in the immune profiles of tumor cells and tumor‐infiltrating lymphocytes with respect to POLE mutation status. It would not be surprising that such high rates of mutation might lead to a variety of alterations and drivers of tumorigenesis. This heterogeneity may also prove to be a challenge when applying treatment strategies. Despite the promise of immunotherapy in highly mutated tumors, not all such cancers will respond to checkpoint inhibitor therapy. In our first case, in which no response to immunotherapy was observed, the lack of PD‐L1 staining might suggest primary poor immune recognition, possibly calling for combination immunotherapy for such cases. Alternatively, the exceedingly high mutation rate could lead to subclonal alterations, allowing for rapid adaptation, clonal selection, and subsequent immune evasion.
A potential targeted treatment strategy to consider involves the concept of “synthetic lethality.” In a tumor with an underlying defect in DNA repair such as POLE mutation, targeting a separate DNA repair mechanism may lead to preferential death of tumor cells. This approach has been investigated using poly(ADP)ribose polymerase (PARP) inhibitors in BRCA gene‐mutated ovarian and breast cancers [20], [21]. A phase II study with the PARP inhibitor olaparib did not demonstrate activity in advanced CRC patients with MSS or MSI‐high tumors [22]. Nonetheless, POLE‐deficient cancers encompass a different mechanism of failed DNA repair, and targeted therapies such as these may prove successful in the future.
Author Contributions
Conception/Design: Balazs Halmos, Jennifer Chuy
Provision of study material or patients: Enrico Castellucci, Jennifer Chuy, Yitzhak Goldstein
Collection and/or assembly of data: Tianfang He, Enrico Castellucci, Yitzhak Goldstein
Data analysis and interpretation: Enrico Castellucci, Tianfang He, D. Yitzchak Goldstein, Balazs Halmos, Jennifer Chuy
Manuscript writing: Enrico Castellucci, Tianfang He, D. Yitzchak Goldstein, Balazs Halmos, Jennifer Chuy
Final approval of manuscript: Enrico Castellucci, Tianfang He, D. Yitzchak Goldstein, Balazs Halmos, Jennifer Chuy
Disclosures
The authors indicated no financial relationships.
References
- 1.Cancer Genome Atlas Network , Muzny DM, Bainbridge MN et al. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Palles C, Cazier JB, Howarth KM. Germline mutations in the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet 2013;45: 136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ghosal G, Chen J. DNA damage tolerance: A double‐edged sword guarding the genome. Transl Cancer Res 2013;2:107–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li SK, Martin A. Mismatch repair and colon cancer: Mechanisms and therapies explored. Trends Mol Med 2016;22:274–289. [DOI] [PubMed] [Google Scholar]
- 5. Martin SA, Lord CJ, Ashworth A. Therapeutic targeting of the DNA mismatch repair pathway. Clin Cancer Res 2010;16:5107–5113. [DOI] [PubMed] [Google Scholar]
- 6. Ahn SM, Ansari AA, Kim J et al. The somatic POLE P286R mutation defines a unique subclass of colorectal cancer featuring hypermutation, representing a potential genomic biomarker for immunotherapy. Oncotarget 2016;7:68638–68649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edwards S, Li CM, Levy DL et al. Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion. Mol Cell Biol 2003;23:2733–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Briggs S, Tomlinson I. Germline and somatic polymerase ɛ and δ mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol 2013;230:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yoshida R, Miyashita K, Inoue M et al. Concurrent genetic alterations in DNA polymerase proofreading and mismatch repair in human colorectal cancer. Eur J Hum Genet 2011;19:320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Network , Kandoth C, Schultz N et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wimmer K, Beilken A, Nustede R et al. A novel germline POLE mutation causes an early onset cancer prone syndrome mimicking constitutional mismatch repair deficiency. Fam Cancer 2017;16:67–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bellido F, Pineda M, Aiza G et al. POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: Review of reported cases and recommendations for genetic testing and surveillance. Genet Med 2016;18:325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meng B, Hoang LH, McIntyre JB et al. POLE exonuclease domain mutation predicts long progression‐free survival in grade 3 endometrioid carcinoma of the endometrium. Gynecol Oncol 2014;134:15–19. [DOI] [PubMed] [Google Scholar]
- 14. Domingo E, Freeman‐Mills L, Rayner E et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: A retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol 2016;1:207–216. [DOI] [PubMed] [Google Scholar]
- 15. Stenzinger A, Pfarr N, Endris V et al. Mutations in POLE and survival of colorectal cancer patients–Link to disease stage and treatment. Cancer Med 2014;3:1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Le DT, Uram JN, Wang H et al. PD‐1 blockade in tumors with mismatch‐repair deficiency. N Engl J Med 2015;372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Howitt BE, Shukla SA, Sholl LM et al. Association of polymerase e‐mutated and microsatellite‐instable endometrial cancers with neoantigen load, number of tumor‐infiltrating lymphocytes, and expression of PD‐1 and PD‐L1. JAMA Oncol 2015;1:1319–1323. [DOI] [PubMed] [Google Scholar]
- 18. Shinbrot E, Henninger EE, Weinhold N et al. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res 2014;24:1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stadler ZK, Battaglin F, Middha S et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in next‐generation sequencing panels. J Clin Oncol 2016;34:2141–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Banerjee S, Kaye S. PARP inhibitors in BRCA gene‐mutated ovarian cancer and beyond. Curr Oncol Rep 2011;13:442–449. [DOI] [PubMed] [Google Scholar]
- 21. Rios J, Puhalla S. PARP inhibitors in breast cancer: BRCA and beyond. Oncology (Williston Park) 2011;25:1014–1025 [PubMed] [Google Scholar]
- 22. Leichman L, Groshen S, O'Neil BH et al. Phase II study of Olaparib (AZD‐2281) after standard systemic therapies for disseminated colorectal cancer. The Oncologist 2016;21:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]