Abstract
Focal adhesions (FAs) are important mediators of cell-substrate interactions. One of their key functions is the transmission of forces between the intracellular acto-myosin network and the substrate. However, the relationships between cell traction forces, FA architecture, and molecular forces within FAs are poorly understood. Here, by combining Förster resonance energy transfer (FRET)-based molecular force biosensors with micropillar-based traction force sensors and high-resolution fluorescence microscopy, we simultaneously map molecular tension across vinculin, a key protein in FAs, and traction forces at FAs. Our results reveal strong spatiotemporal correlations between vinculin tension and cell traction forces at FAs throughout a wide range of substrate stiffnesses. Furthermore, we find that molecular tension within individual FAs follows a biphasic distribution from the proximal (towards the cell nucleus) to distal end (towards the cell edge). Using super-resolution imaging, we show that such a distribution relates to that of FA proteins. On the basis of our experimental data, we propose a model in which FA dynamics results from tension changes along the FAs.
Keywords: Mechanobiology, molecular force sensor, focal adhesion, micropillar substrate, rigidity sensing, cell traction force, vinculin
Introduction
Cells’ ability to sense and respond to the mechanical cues from their microenvironment in the form of geometry, topography, and stiffness,1,2 is crucial for understanding cell function3 and major processes in cancer4 and development.5 Focal adhesions (FAs) are important mediators of force transmission from the external environment to the cell’s interior and vice versa.6 They are macromolecular assemblies comprising trans-membrane receptors and cytoplasmic adapters that serve as physical links between the intracellular contractile acto-myosin network and the extracellular matrix (ECM).1,7 However, due to their complex architecture,8,9 it remains unclear how actomyosin forces are transmitted to the ECM through and distributed within these protein assemblies.10 Several studies have revealed key roles of FA proteins such as talin,11 vinculin,12–14 and paxillin15 in ECM stiffness sensing, or mechanosensing. In addition, single molecule studies have revealed that some of these proteins respond to forces through conformational changes16,17 that may facilitate the recruitment of other proteins and potentially the clustering of FA constituents. Vinculin is a key protein which binds to both talin and actin8,18 in the FAs and also regulates the recruitment of several adhesion factors.18–20 In addition, vinculin functions like a molecular clutch that takes part in ECM traction generation, FA maturation, and turnover.21 Along this line, several studies have revealed a complex relationship between cell traction forces and FA dynamics. For example, Balaban et al. showed that the size of FAs scales with the traction force.22 A later study revealed that such correlation applies only to growing FAs.23 Cell migration also adapts to substrate stiffness.24,25 A Förster resonance energy transfer (FRET) based tension sensor inserted between the head and tail domains of vinculin molecules revealed that FA-localized vinculin molecules bore tension.26 This tension appears to share a complex relationship with the spatial distribution of FAs and cell traction forces,27 which may arise from the multiple possible arrangements of proteins within large clusters at FAs and affect FA dynamics.28 Hence, a better understanding of FA regulation by traction forces requires combined knowledge of FA internal forces at molecular scales with external traction forces at FAs in a wide range of conditions.
In the present study, we addressed this challenge by simultaneously using FRET-based biosensors of vinculin tension and micro-force sensor arrays (µFSA)29,30 to determine the relationship between molecular forces within FAs, FA architecture, and traction forces exerted at the cell-substrate interface (Figure 1). The use of µFSA of allowed us to vary the substrate stiffness by changing the length the micropillars and thus to determine a correlation between cell traction forces and molecular tensions over a broad range of stiffnesses. Interestingly, examination of protein recruitment to micropillars allowed us to observe distinct regions within a FA with respect to the pillar surface as confirmed by structured illumination microscopy (SIM). We further showed that FA anchoring correlated with a non-monotonic tension profile along the FA length. Altogether, these data reveal that molecular tensions within individual FAs not only correlate with traction forces and substrate stiffness but also exhibit sub-FA variations that follow FA architecture and anchoring to the underlying substrate. This behavior supports the idea that molecular tension variations may be harnessed by different cellular functions through various mechanosensing pathways at distinct length scales.
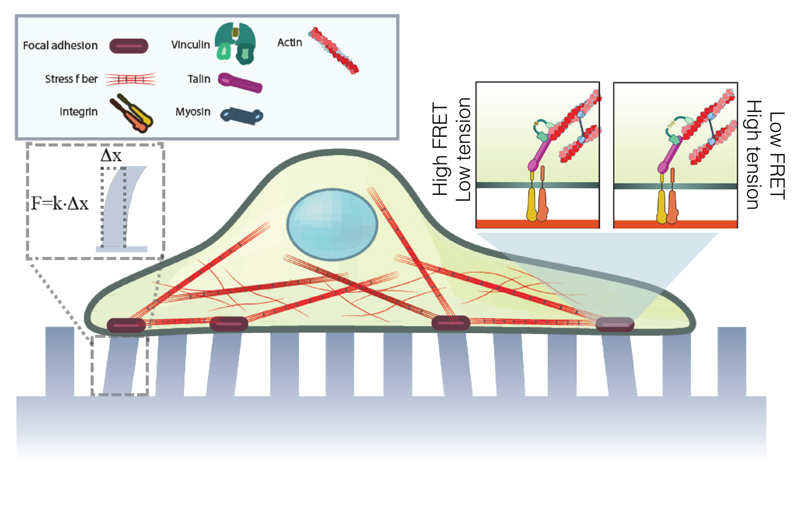
Figure 1.
Schematics showing the experimental approach. Cells expressing FRET-based vinculin tension sensor, VinTS, were plated on micropillar substrates. Deflection of micropillar tops and the FRET index of VinTS reveal the cell traction forces and vinculin molecular tensions, respectively, as indicated in the inset.
Results
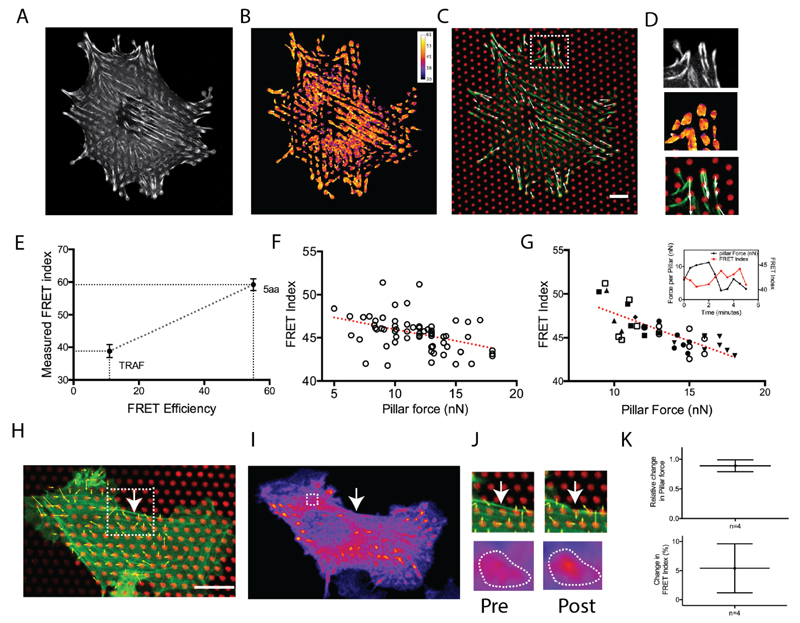
To simultaneously measure intra-FA molecular forces and traction forces at FAs, we expressed a FRET biosensor of vinculin tension26 in vinculin-deficient mouse embryonic fibroblast cells (MEF), and plated them on µFSA whose surface was coated with dye-conjugated fibronectin (Figure 1). Using image segmentation based on fluorescence intensity, we isolated FAs to measure an average FRET index for individual FAs. In order to convert FRET index to FRET efficiency and further infer molecular tensions (see discussion), we measured the FRET index of two FRET standards,31 mTFP-TRAF-Venus and mTFP-5aa-Venus, expressed in MEF cells (Figure 2E). FRET index of the biosensor is inversely related to the tension across it with lower FRET index indicating higher tension across the vinculin molecule. We also measured traction forces of individual FAs from the pillar deflections32 (Figure 2A-D). From a plot of FRET index as a function of force on pillar per FA, we observed a decrease in FRET index as the force on pillar increased (Figure 2F). As a control, we showed that a vinculin TS construct lacking the actin-binding tail did not, however, exhibit a FRET index dependent on traction force (Supporting Information Figure S1). This result demonstrated that the magnitude of traction forces correlates with tension across vinculin among a population of FAs at a given time. We then analyzed the temporal variations of molecular tensions and traction forces. Again, we observed a similar anti correlation between temporal fluctuations of FRET index and pillar force for individual FAs (Figure 2G). In principle, traction forces may vary as a function of individual protein tensions but may also depend on the amount of protein within FAs. To assess this, we measured the integrated intensity of segmented FAs, as a proxy for the amount of vinculin (Supporting Information Figure S2). No correlation was found between vinculin amount and traction forces for FAs of similar size (length = 2.5 ± 0.4 µm). This result supports the hypothesis that the change in cell traction forces relies more on tension changes across vinculin than in protein density.
Figure 2.
(A) MEF cells transfected with VinTS. (B) Corresponding FRET index image. (C) Pillars are coated with fluorescent fibronectin (red). VinTS expression is shown is green. Calculated force vectors are represented by arrows (white). Scale bar = 10 µm. (D) Closer view of FAs in the inset in C. (E) Calibration of VinTS FRET sensor using mTFP-5aa-venus (5aa; five amino acid linker) and mTFP-TRAF-venus (TRAF; 229-amino acid tumor necrosis factor receptor-associated factor) as standards. (n ≥ 12 cells). Error bars represent s.e. of mean. (F) Forces at FAs as calculated from pillar deflection and corresponding average FRET index. Each circle corresponds to a FA. The red line represents a linear fit. (Correlation Coefficient R= -0.442, n ≥ 8 cells in two independent experiements). (G) Temporal correlation of force and FRET index at FAs. Different symbols correspond to the measurement at one FA and associated pillar at different time points (time interval = 1 minute). Inset: Typical example of temporal variation of pillar force (black line) and corresponding FRET index(red line). (H) REF52 cell on micropillars showing actin (green), fibronectin (red), and calculated force vectors (white). Scale bar = 10 µm. (I) FRET channel image of FAs for the corresponing cell. Terminal FA for the stress fiber is shown inside dashed white box. The white arrow indicates the ablation point. (J) Top panel: pre and post-ablation of stress fiber in the inset in (H). Bottom panel: FRET channel intensity of corresponding FA showing increased FRET post-ablation. (K) Top panel: Relative change in pillar force (|Fpost-Fpre|/Fpre). Bottom panel: Relative change in FRET index (|FRETpost-FRETpre|/FRETpre). Error bars represent s.e. of mean. The relaxation of force at the FA is associated with the increase in the FRET index indicates a relaxation of molecular tension within the FAs.
Cell traction forces are generated by acto-myosin contractility and transmitted to the substrate through stress fibers and FAs.1 As cells apply forces, both the actin cytoskeleton24 and FAs25 are remodeled. We thus investigated vinculin tension and cell traction forces in response to laser ablation of acto-myosin contractile stress fibers.33,34 To do so, we selectively ablated individual stress fibers and measured changes in pillar deflection. Simultaneously, we measured the change in FRET index of the FAs associated with the corresponding fiber before and after ablation (Figure 2H-J). Our measurements on those FAs revealed that forces on the pillar instantaneously decreased and the average FRET index of the associated FA increased after stress fiber ablation (Figure 2K). These data confirm the close relationship between molecular tensions and traction forces but also show that vinculin tension responds to stress fiber perturbations.
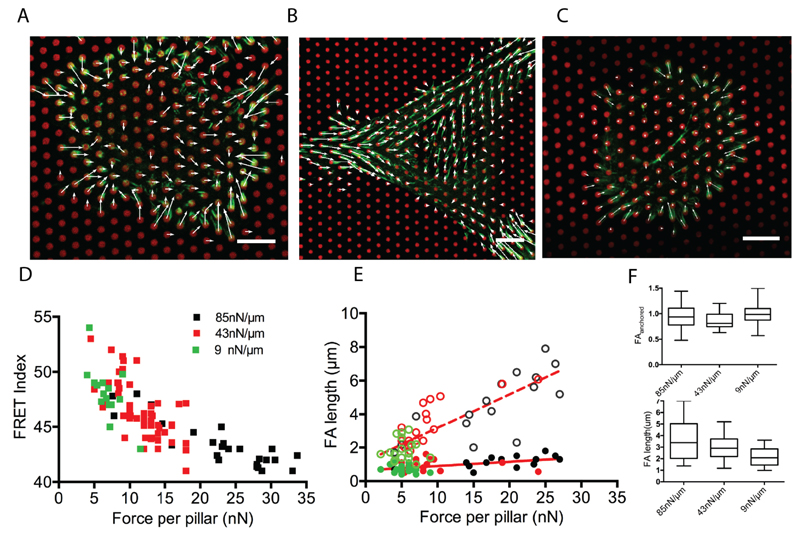
To further investigate the relationship between cell-traction forces and molecular tension, we varied the stiffness of µFSA by changing the height of the pillars.35 Numerous studies have shown that cell contractility3,31,36 as well as FA size and dynamics are affected by substrate stiffness.25,37 Moreover, talin tension increases with substrate stiffness.38 In order to probe the effect of substrate stiffness on the relationship between vinculin molecular tension and traction forces, we used micropillars of three different stiffnesses: 85, 43, and 9 nN/µm (Figure 3A-C). Consistent with previous results,32 we observed a decrease in average force per pillar with decreasing substrate stiffness (Figure 3D). More importantly, the correlation between vinculin tension and traction forces remains over the whole range of tested stiffnesses (Figure 3D). Even though lower forces were measured on average on softer substrates, vinculin tension within FA remained the same for pillar experiencing similar forces regardless of substrate stiffness. Altogether, these results confirm the correlation between molecular tensions and cell traction forces and show that this correlation is stiffness-independent.
Figure 3.
MEF cells transfected with vinTS (green) plated on fibronectin coated µFSAs (red) of different stiffnesses (A) 85, (B) 43, and (C) 9nN/µm. Scale bars = 10 µm. White arrows indicate traction forces. (D) Correlation of force per pillar with average FRET index of corresponding FAs at different substrate stiffnesses. (n≥ 6 cells for each substrate stiffness). (E) Length of FAs as a function of force on pillars. The total and the anchored lengths of the FAs are represented by the open and closed circle, respectively. Dashed and solid red lines are linear fits for the total length and the anchored part, respectively. Green, red and black symbols correspond to 9, 43 and 85 nN/µm substrate stiffness respectively. n≥ 12 FAs in minimum of three cells for each stifnesess. (F) Length of FAs for substrates of different stiffnesses. Total length and anchored part of FAs are shown in bottom and top panels, respectively (The boxes represent the data from the 25th and 75th percentile. n≥ 20 FAs for each substrate stiffness in three independent experiments).
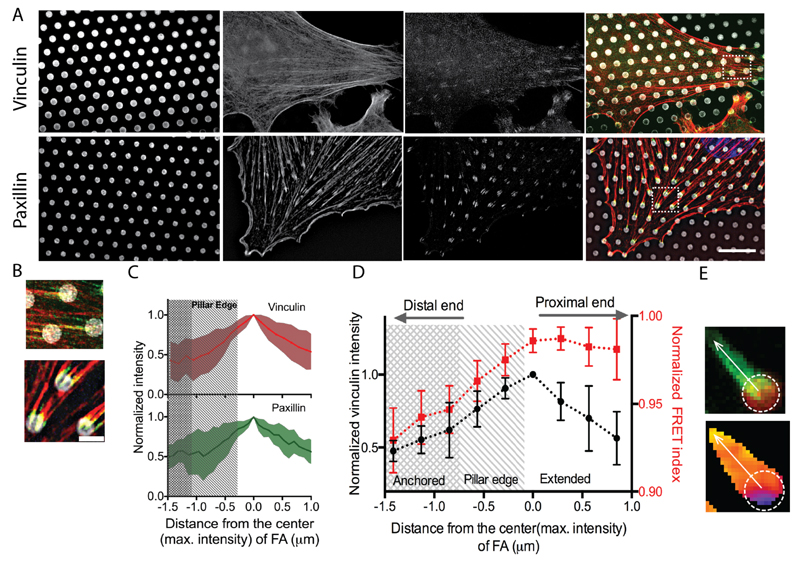
Previous experiments have shown that the distribution of traction forces varied along the FA, exhibiting a maximum in the region between the center and the distal tip (toward the cell periphery).25 Furthermore the distribution of proteins within individual FAs was reported to be inhomogeneous, exhibiting maximum recruitment at the center of the FAs. In addition, recent studies using super-resolution microscopy showed that paxillin recruitment extended beyond the region of contact between FAs and the substrate.39 In view of this, we next sought to determine protein recruitment and tension at the sub-FA scale. To do so, we first analyzed the fluorescence intensity of vinculin along the adhesion. We observed that only a part of the vinculin staining overlapped the micropillars while a substantial part overhung from the pillars. We then investigated the distribution of vinculin as a function of pillar stiffness. Surprisingly, the length of the anchored part on the pillar tops remained unchanged over the range of stiffnesses we tested even though traction forces increased with stiffness. Conversely, the overhanging length increased with substrate stiffness. (Figure 3F). In order to obtain higher resolution details, we performed structured illumination microscopy (SIM) on REF52 cells immunostained for vinculin and paxillin (see Methods). SIM images for both vinculin and paxillin confirmed that FAs extend beyond the pillars (Figure 4A-C). The intensity distribution of both vinculin and paxillin followed a non-monotonic profile including first an increase from the distal end and then a slight decrease from roughly the center of the FA (Figure 4C and Figure S3).
Figure 4.
(A) 3D-SIM micrographs of REF52 cell on micropillars showing, from left to right, the micropillar tops, actin filaments, vinculin and the merged image respectively (top panel). Bottom panel shows the micropillar tops, actin filaments, paxillin, and the merged image. Scale bar = 10 μm. (B) Enlarged views of FAs in the insets in (A), showing, vinculin(top) and paxillin(bottom). Scale bar = 2μm. (C) Intensity profile (mean±1SD) of vinculin (red) and paxillin (green) along the length of a FA (n≥ 12). (D) Normalized intensity and FRET index profile along the length of a FA showing the inhomogeneous distribution of vinculin intensity and the molecular tension. Both intensity and FRET index are normalized to the maximum values observed in individual FAs. Error bars represent the s.e of mean. n=9. (E) (top) A zoomed-in view of FA (green) on micropillar (red) and (bottom) corresponding FRET index image where the position of the pillar and the line scan path have been indicated.
Consequently, we measured the distribution of FRET index within individual FAs. We observed an increase of FRET index along the FA from the distal tip to the proximal tip following vinculin and paxillin recruitment profiles up to the pillar edge from where the index remained rather constant inwards, which is in contrast with the decrease of vinculin and paxillin recruitment (Figure 4D,E). This shows that the forces experienced by vinculin molecules are the highest where their relative amount is the lowest within the region in contact with the substrate. This is consistent with a model whereby vinculin molecules share a load per unit area as long as they are in a substrate-contacting region. In contrast, there is no correlation in the overhanging, low-force region although the length of this suspended part scales with the traction force (Figure 3E).
Discussion
While earlier studies have quantified the relationships between cell traction forces, FA areas,39 substrate stiffness, and the distribution of cell traction forces at the sub-FA scale, it is not clear how molecular tensions within FAs relate to cell traction forces and distribute within FAs. FAs are protein clusters that can vary in composition, molecular and supra-molecular conformations, and size.37 Thus, how traction forces distribute across a population of FA proteins is not straightforward.
Here, by combining the use of micropillar substrates with a FRET-based molecular tension sensor, we could simultaneously map the traction forces associated with individual FAs with molecular tensions within FA-localized vinculin molecules. This allowed us to assess molecular tension within FAs and cell traction forces from the sub-FA scale to the cell scale, and for a wide range of substrate stiffnesses. We found a spatiotemporal correlation between cell traction forces and molecular tensions across FAs of whole cells, both dependent on stress fiber integrity. Altogether these data show that the tension across vinculin maps synchronously to the changes in the traction force at the FAs.
Using previously published FRET efficiency to molecular tension calibration26 (about 0.16 pN per unit FRET Efficiency in the linear regime) and FRET index to FRET Efficiency calibration (Figure 2, about 0.5 index unit per Efficiency unit), we estimate that the largest molecular tension differential observed for cells on substrate stiffness spanning about an order of magnitude reaches about 4 pN, which is within the detectable range of tensions of this sensor (Figure 3E). Assuming actual molecular tensions do not significantly exceed the detectable range, traction forces can thus be used as a quantitative proxy for evaluating these tensions, and reciprocally. Moreover, within this same stiffness range the traction force differential reached about 30nN. Assuming anchored FA surface area lying around 1µm2 (Figure 3F), this is equivalent to a force surface density differential of about 3pN/(10nm)2, a change that proteins densely packed in a parallel fashion can easily accommodate, given the molecular tension differential of vinculin we measured (see above) and that other Fa proteins such as talin also bear tension in parallel. Interestingly, the FRET index of VinTS appeared somewhat higher than its tailless counterpart on the softest substrate (Figure S3). As the sensor has previously been seen to be sensitive to compression,40 we suspect that at very low to no cytoskeletal tension on soft substrates, the tail of VinTS is sufficient to hinder the sensor and retrieve slightly higher FRET index than the tailless control. The phenomenon was previously exploited as such to measure compression of glycocalyx proteins.41 While this probably leads to an underestimation of the actual cytoskeletal force exerted on the protein in our context, this does not affect the overall trend. Future experiments may benefit from a control construct impaired for actin-binding but retaining most of its tail.
At the scale of individual FAs, we consistently found that the intensity of both paxillin and vinculin are maximum at the central region of the FAs. In addition, we show that tension across vinculin is maximal at FA distal tip and decreases away from it within the substrate contacting region. A recent study using FRET-based tension sensor linked to integrin-binding RGD peptides showed that the integrins with highest tension within FAs colocalize more with paxillin recruitment center of mass than with that of other FA proteins.42 Another study, using traction force microscopy (TFM), had shown that the traction force peak localized distally within a few µm away from the paxillin recruitment maximum.25 The apparent mismatch between sub-FA traction force and molecular tension profiles suggests that the local molecular density, molecular orientations, and interactions may have significant impact on the traction force distribution over FA proteins. Nevertheless, the regularization step inherent to TFM method may limit its ability to retrieve traction force profiles sharp enough to match that of molecular tensions so that interpreting their comparison requires caution.
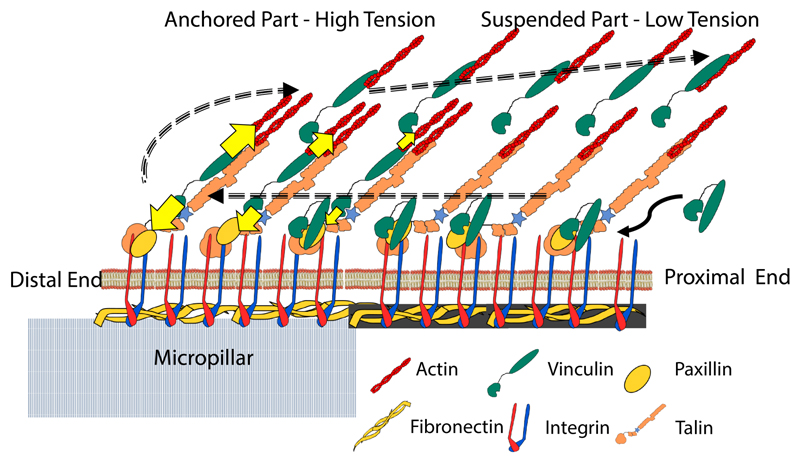
Previous studies had shown the treadmilling of FAs43,44 whereby FA proteins incorporate into FAs from the proximal edge and detach from the substrate at the distal edge. More recently, vinculin was shown to exhibit a more closed conformation and closer to the substrate localization associated with paxillin when at the distal end, and a more open conformation at the proximal end when associated with talin and actin.45 Here, we show a biphasically increasing tension from the proximal to distal edge. In this framework, we propose that vinculin incorporates with minimal tension at the FA proximal end in the lower paxillinrich layer. As it treadmills toward the FA distal edge, vinculin binds to talin and actin, its conformation opens, and its tension increases, consistently with a molecular clutch model featuring a gradient of clutch engagement along the FA length. As tension increases, vinculin increasingly detaches from the lower layer and is carried inward and upward by the actin retrograde flow in a tension-less open conformation in a motion reminiscent of sticky tape peeling (Figure 5). Of note, a theoretical model of flexible molecular linker peeling between a cell and its substrate had predicted a width of a few 100nm for the adhesion belt, the region over which linkers exhibit a stretch gradient at the cell periphery,46 a length scale similar to that of the vinculin tension gradient in the present study. Finally, a lag between tension release and conformation closing could explain the orthogonal gradients of tension and conformation. In this context, the overhanging region of FAs may contain newly incorporated proteins not yet involved in force transmission in a lower layer as well as older proteins detached from the lower layer and carried by the actin retrograde flow. Importantly, overhanging FAs are not an exclusive feature of micropillar substrates and have been also observed over non-adhesive regions of flat, micropatterned substrates.47 It is likely that such interfaces between adhesive and non-adhesive regions may better reflect the heterogeneous nature of native cellular environments than continuously flat and adhesive substrates. Therefore, we expect this model can generally be applied to understand the cellular adhesion structures in vivo and shed more light towards the understanding of the cell migration in complex three-dimensional porous environments.
Figure 5.
A model describing the structure of FA on the micropillar top wherein one part of the FA is anchored and a significant portion of the FA is suspended from the pillar. The dashed arrows describe the hypothesized travel of vinculin during FA treadmilling. Intra-molecular tension is higher in the anchored part whereas the protruding part experiences relatively lower tension correlating with the lack of substrate attachment. The shaded fibronectin in the protruding part represents the fibrils formed by the cell (see Figure S4).
Methods
Preparation and Calibration of the micropillar substrates
Preparation
The preparation of micropillar substrate has been described in detail elsewhere.29 In brief, the micropillar substrates were prepared from Polydimethylsiloxane (PDMS, Sylgard 184, Dow-Corning) elastomer by replica molding from silicon wafers. The substrate was characterized by using Scanning Electron Microscopy (SEM). The pillars are arranged in hexagonal arrays with an inter-pillar distance of 2 µm. The pillar diameter was 2 µm and height was 5 µm. With such a geometry we obtained substrate stiffness of 43 nN/µm. For facilitating cell adhesion, the pillar tops were coated with fibronectin using micro contact printing. For that, flat PDMS stamps were incubated with fibronectin (50 µg/ml unconjugated fibronectin and 5µg/ml dye-conjugated fibronectin, ATTO647N, Sigma-Aldrich) for one hour in room temperature. The stamps were then dried and placed on UV/Ozone-treated micropillar substrates for 5 minutes. To restrict the cell adhesion only to the pillar tops, we added 0.2% Pluronics-F127 solution to the substrate for 1 hour, which passivated the sides of the pillar. The substrate was then rinsed gently with PBS and subsequently immersed in cell culture medium.
Calibration
To obtain reproducible Young’s Modulus E= 2MPa, we adopted a consistent procedure for PDMS preparation where in a fixed ratio of cross-linker to base polymer (1:10) and the curing time (80°C for 2 hours) were maintained for all our experiments. After obtaining the dimensions of the pillars from SEM images, we used Finite Element Method (FEM) to calculate the stiffness of the pillars.29
Cell Biology
Cell Culture and Transfection
Vinculin deficient mouse embryonic fibroblasts were a gift from Christian Grashoff. Both MEF and REF52 cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin, and 100 µg/ml L-glutamine. To label the actin filaments, cells were transfected with RFP-Ftractin. VinculinTS was a gift from Martin Schwartz (Adgene plasmid # 26019). We used electroporation (Nucleofactor, Lonza) for all our transfections.
Immunostaining
For immunostaing, cells were first allowed to spread on micropillars for sufficient time (~3 hours) and then subsequently fixed (4% formaldehyde in PBS, 10 min) at room temperature, permeabilized (0.1% Triton-X , 5 min), blocked ( 1% BSA in PBS, 1 h), and rinsed with PBS. Phalloidin-TRITC was used for actin staining, and for cellular fibronectin we used anti-fibronectin antibody (Abcamab18265).
Confocal Microscopy and FRET imaging
Live cell imaging was performed after 2 hours on a confocal microscope (Zeiss LSM-710, Carl Zeiss, Germany). For FRET measurement, we used 458nm laser to excite the donor. The fluorescent signal from the sensor was collected with a spectral detector in 13 continuous bands from 465nm to 582nm. This contained both the emission from donor and the acceptor. Subsequently using PixFret plugin48 with ImageJ we deduced the FRET index at desired ROI. After background correction, the plugin calculates the FRET index by measuring the intensity ratio between the donor channel and the FRET channel, for example, FRET Index = IFRET / (Idonor + IFRET). The index (F) to Efficiency (E) relationship is extracted from the measured F of TRAF and 5aa and their known E using linear interpolation: E=(F-F5aa)*(E5aa-ETRAF)/(F5aa-FTRAF)+E5aa.
To measure the pillar deflection the fibronectin-dye on the pillar top was imaged concurrently on a different track using a separate detector. From the pillar image, individual pillar deflection was calculated using a home-built plugin in imageJ. For a particular FA, the average FRET index was measured. Laser ablation experiments where performed by using a Zeiss LSM 780 NLO microscope (Carl Zeiss, Germany) coupled to a pulsed NIR laser Ti:Sapphire crystal laser (Coherent Chameleon Ultra, Santa Clara, CA, USA). We used several excitation wavelength 458nm (For VinTS sensor donor excitation), 561nm (PSS-laser) for Actin visualization, and 633nm (HeNe-laser) for micropillar top visulization. For ablation experiments NIR laser was tuned at 800 nm (power: 65%-100%). The system included a 34-Channel GaAsP detector (GaAsp-array with 32 PMTs and two flanking single PMTs). A plan c-apochromat ×63 (NA = 1.2) water immersion objective was used for all experiments. Local temperature was monitored and maintained at 37°C (PECON, Germany).
Structured Illumination Microscopy (SIM)
For 3D-SIM, we used fixed samples immunostained for actin or cellular fibronectin. FAs were visualized with YFP-paxillin, which was stably expressed in REF52 cells. The nucleus was stained with Hoechst. The equipment consisted of ELYRA PS.1 Zeiss microscope coupled with an EMCCD camera (Andor iXon 885, 1004×1002, pixel size 8µm, QE=65%). We used excitation lasers of 405nm (for nucleus), 488nm(for FA), 561 nm (for actin or cellular fibronectin), and 641nm (for micropillar tops). Structured illumination grids at three different angles and five different phases were used for image acquisition. A Zeiss PLAN-APO, 63x, NA 1.40 (Zeiss) objective was used for all our experiments. 3-D reconstruction was done using IMARIS. (Bitplane AG, Switzerland).
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website.
Acknowledgments
The authors thank group members from IJM and MBI for helpful discussions. We acknowledge C. Grashoff for his generous gift of MEF cells and fruitful discussions. The authors would also like to thank MBI Microfabrication (Gianluca Grenci and Mohammed Ashraf), Fabrice Licata from the Institut Universitaire d’Hematologie (Saint-Louis Hospital, France) for his help in ablation experiments and acknowledge the IJM Imagoseine Imaging Facility, member of the France BioImaging infrastructure supported by the French National Research Agency (ANR-10-INBS-04, « Investments of the future »). Financial supports from the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement n° 617233, the Agence Nationale de la Recherche “PillarCell” (ANR 13-NANO-0011) and the USPC-NUS collaborative program are gratefully acknowledged. NT and NB are supported by the Agence Nationale de la Recherche (ANR-10-BLAN-1515) and the France-BioImaging infrastructure (ANR-10-INBS-04).
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
References
- (1).Geiger B, Spatz JP, Bershadsky AD. Nat Rev Mol Cell Biol. 2009;10(1):21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- (2).Vogel V, Sheetz M. Nat Rev Mol Cell Biol. 2006;7(4):265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- (3).Engler AJ, Sen S, Sweeney HL, Discher DE. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- (4).Butcher DT, Alliston T, Weaver VM. Nat Rev Cancer. 2009;9(2):108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Heisenberg C-P, Bellaïche Y. Cell. 2013;153(5):948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- (6).Gardel ML, Schneider IC, Aratyn-Schaus Y, Waterman CM. Annu Rev Cell Dev Biol. 2010;26(1):315–333. doi: 10.1146/annurev.cellbio.011209.122036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Hynes RO. Cell. 2002:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- (8).Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, Hess HF, Waterman CM. Nature. 2010;468(7323):580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Zaidel-Bar R, Itzkovitz S, Ma’ayan A, Iyengar R, Geiger B. Nat Cell Biol. 2007;9(8):858–867. doi: 10.1038/ncb0807-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ladoux B, Nicolas A. Rep Prog Phys. 2012;75(11):116601. doi: 10.1088/0034-4885/75/11/116601. [DOI] [PubMed] [Google Scholar]
- (11).Margadant F, Chew LL, Hu X, Yu H, Bate N, Zhang X, Sheetz M. PLoS Biol. 2011;9(12) doi: 10.1371/journal.pbio.1001223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Dumbauld DW, Lee TT, Singh A, Scrimgeour J, Gersbach CA, Zamir EA, Fu J, Chen CS, Curtis JE, Craig SW, García AJ. Proc Natl Acad Sci U S A. 2013;110(24):9788–9793. doi: 10.1073/pnas.1216209110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Goldmann WH, Auernheimer V, Thievessen I, Fabry B. Cell Biology International. 2013:397–405. doi: 10.1002/cbin.10064. [DOI] [PubMed] [Google Scholar]
- (14).Liu Z, Bun P, Audugé N, Coppey-Moisan M, Borghi N. Integr Biol. 2016;8(6):693–703. doi: 10.1039/c5ib00307e. [DOI] [PubMed] [Google Scholar]
- (15).Zaidel-Bar R, Milo R, Kam Z, Geiger B. J Cell Sci. 2007;120(Pt 1):137–148. doi: 10.1242/jcs.03314. [DOI] [PubMed] [Google Scholar]
- (16).Moore SW, Roca-Cusachs P, Sheetz MP. Developmental Cell. Elsevier Inc; 2010. pp. 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Science. 2009;323(5914):638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Carisey A, Tsang R, Greiner AM, Nijenhuis N, Heath N, Nazgiewicz A, Kemkemer R, Derby B, Spatz J, Ballestrem C. Curr Biol. 2013;23(4):271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Carisey A, Ballestrem C. Eur J Cell Biol. 2011;90(2-3):157–163. doi: 10.1016/j.ejcb.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hernández-Varas P, Berge U, Lock JG, Strömblad S. Nat Commun. 2015 May;6:7524. doi: 10.1038/ncomms8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Thievessen I, Thompson PM, Berlemont S, Plevock KM, Plotnikov SV, Zemljic-Harpf A, Ross RS, Davidson MW, Danuser G, Campbell SL, Waterman CM. J Cell Biol. 2013;202(1):163–177. doi: 10.1083/jcb.201303129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Balaban N, Schwarz U, Riveline D. Nat cell. 2001 May;3 doi: 10.1038/35074532. [DOI] [PubMed] [Google Scholar]
- (23).Stricker J, Aratyn-Schaus Y, Oakes PW, Gardel ML. Biophys J. 2011;100(12):2883–2893. doi: 10.1016/j.bpj.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Trichet L, Le Digabel J, Hawkins RJ, Vedula SRK, Gupta M, Ribrault C, Hersen P, Voituriez R, Ladoux B. Proc Natl Acad Sci U S A. 2012;109(18):6933–6938. doi: 10.1073/pnas.1117810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Cell. 2012;151(7):1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, Schwartz MA. Nature. 2010;466(7303):263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Chang C, Kumar S. J Cell Sci. 2013;126(Pt 14):3021–3030. doi: 10.1242/jcs.119032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ladoux B, Mege RM, Trepat X. Trends Cell Biol. 2016;26(6):420–433. doi: 10.1016/j.tcb.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Gupta M, Kocgozlu L, Sarangi BR, Margadant F, Ashraf M, Ladoux B. Biophysical Methods in Cell Biology. Vol. 125. Elsevier Ltd; 2015. pp. 289–308. [DOI] [PubMed] [Google Scholar]
- (30).du Roure O, Saez A, Buguin A, Austin RH, Chavrier P, Silberzan P, Ladoux B. Proc Natl Acad Sci U S A. 2005;102(7):2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Day RN, Booker CF, Periasamy A. J Biomed Opt. 13(3):031203. doi: 10.1117/1.2939094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Gupta M, Sarangi BR, Deschamps J, Ne-matbakhsh Y, Callan-Jones A, Margadant F, Mège R-M, Lim CT, Voituriez R, La-doux B. Nat Commun. 2015;6:7525. doi: 10.1038/ncomms8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hotulainen P, Lappalainen P. J Cell Biol. 2006;173(3):383–394. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Kumar S, Maxwell IZ, Heisterkamp A, Polte TR, Lele TP, Salanga M, Mazur E, Ingber DE. Biophys J. 2006;90(10):3762–3773. doi: 10.1529/biophysj.105.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Saez A, Buguin A, Silberzan P, Ladoux B. Biophys J. 2005;89(6):L52–L54. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Mitrossilis D, Fouchard J, Pereira D, Postic F, Richert A, Saint-Jean M, Asnacios A. Proc Natl Acad Sci U S A. 2010;107(38):16518–16523. doi: 10.1073/pnas.1007940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Geiger T, Zaidel-Bar R. Curr Opin Cell Biol. 2012;24(5):562–568. doi: 10.1016/j.ceb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- (38).Austen K, Ringer P, Mehlich A, Chrostek-Grashoff A, Kluger C, Klingner C, Sabass B, Zent R, Rief M, Grashoff C. Nat Cell Biol. 2015;17(12):1597–1606. doi: 10.1038/ncb3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Van Hoorn H, Harkes R, Spiesz EM, Storm C, Van Noort D, Ladoux B, Schmidt T. Nano Lett. 2014;14(8):4257–4262. doi: 10.1021/nl5008773. [DOI] [PubMed] [Google Scholar]
- (40).Rothenberg KE, Neibart SS, LaCroix AS, Hoffman BD. Cell Mol Bioeng. 2015;8(3):364–382. [Google Scholar]
- (41).Paszek MJ, DuFort C, Rossier O, Bainer R, Mouw JK, Godula K, Hudak JE, Lakins JN, Wijekoon AC, Cassereau L, Rubashkin MG. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Morimatsu M, Mekhdjian AH, Chang AC, Tan SJ, Dunn AR. Nano Lett. 2015;15(4):2220–2228. doi: 10.1021/nl5047335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ballestrem C, Hinz B, Imhof BA, Wehrle-Haller B. J Cell Biol. 2001;155(7):1319–1332. doi: 10.1083/jcb.200107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Digman MA, Brown CM, Horwitz AR, Mantulin WW, Gratton E. Biophys J. 2008;94(7):2819–2831. doi: 10.1529/biophysj.107.104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Case LB, Baird MA, Shtengel G, Campbell SL, Hess HF, Davidson MW, Waterman CM. Nat Cell Biol. 2015;17(7):880–892. doi: 10.1038/ncb3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Garrivier D, Décavé E, Bréchet Y, Bruckert F, Fourcade B. Eur Phys J E. 2002;8(1):79–97. doi: 10.1140/epje/i2002-10010-8. [DOI] [PubMed] [Google Scholar]
- (47).Théry M, Pépin A, Dressaire E, Chen Y, Bornens M. Cell Motil Cytoskeleton. 2006;63(6):341–355. doi: 10.1002/cm.20126. [DOI] [PubMed] [Google Scholar]
- (48).Feige JN, Sage D, Wahli W, Desvergne B, Gelman L. Microsc Res Tech. 2005;68(1):51–58. doi: 10.1002/jemt.20215. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.