Abstract
Collective migration of cells is of fundamental importance for a number of biological functions such as tissue development and regeneration, wound healing and cancer metastasis. The movement of cell groups consisting of multiple cells connected by cell-cell junctions depends on both extracellular and intercellular contacts. Epithelial cell assemblies are thus regulated by a cross-talk between cell-substrate and cell-cell interactions. Here, we investigated the onset of collective migration in groups of cells as they expand from few cells into large colonies as a function of extra-cellular matrix (ECM) protein coating. By varying the amount of extracellular matrix proteins (ECM) presented to the cells, we observe that the mode of colony expansion as well as their overall geometry is strongly dependent on substrate adhesiveness. On high ECM protein coated surfaces, cells at the edges of the colonies are well spread exhibiting large outward-pointing protrusive activity whereas cellular colonies display more circular and convex shapes on less adhesive surfaces. Actin structures at the edge of the colonies also show different organizations with the formation of lamellipodial structures on highly adhesive surfaces and a pluricellular actin cable on less adhesive ones. The analysis of traction forces and cell velocities within the cellular assemblies confirm these results. By increasing ECM protein density, cells exert higher traction forces together with a higher outward motility at the edges. Furthermore, tuning cell-cell adhesion of epithelial cell lines modified the mode of expansion of the colonies. Finally, we used a recently developed computational model to recapitulate the emergent experimental behaviors of expanding cell colonies and extract that the main observed differences are dependent on the different cell-substrate interactions. Overall, our data suggest that switching behaviors of epithelial cell assemblies results of a tug-of-war between friction forces at cell-substrate interface and cell-cell interactions.
Introduction
Important biological and pathophysiological phenomena, such as formation of tissues and organs during development, wound healing, and cancer metastasis, are multicellular processes requiring coordinated migration of cells as a group1–3. To achieve such coordination for both in vivo and in vitro conditions, cells require to interact with each other and cooperate over length scales involving multiple cells4–6. The adhesion and migration of single cells on 2D substrates rich in ECM requires to establish heterophilic adhesion to the substrate at the site of specialized protein complexes7. This allows cells to stabilize their protrusions such as lamellipodia and exert forces onto the substrate to propel forward in a mechanism termed cell crawling. This motility mode involves protrusion of lamellipodia at the leading edge and adhesion to ECM proteins such as fibronectin (FN) by transmembrane proteins (e.g. integrins). Based on this model, single cell adhesion and migration have been shown to depend on cell-matrix interactions including ECM adhesiveness and ligand density8, 9. Besides this well-established mode of migration, one additional major mechanism is relevant to cell translocation within tissues: the movement of cell groups, sheets, or strands consisting of multiple cells connected by cell-cell junctions10. The growth and migration of cell clusters over 2D surfaces also display similar types of protrusions at the edges, their cohesiveness being maintained by cell-cell junctions (CCJs)11. This cooperation thus relies on different types of interactions at cell-matrix and cell-cell interfaces. Indeed, the organization of multicellular assemblies in 2D and 3D environments has been shown to depend on the relative strength of these interactions which may be explained by a cross-talk between cell-matrix and cell-cell adhesions12–15. In epithelial cells, adherens junctions through homophilic interactions between E-cadherin proteins have been shown to be crucial to understand forces transmitted from one cell to its neighbors and as such, tissue cohesion16–19. During tissue migration and growth, CCJs experiment various intercellular stresses that include compressive, tensile and shear stresses20–23. Various experimental and theoretical models have been described to explain multicellular movements including contact inhibition of locomotion24, agent-based models25–27 and continuum multicellular approaches28–30. These models include the description of bulk cellular motions through cell-substrate interactions and intercellular tension for a continuous monolayer29, 31, 32. However, in many cases including wound healing, gap closure and also morphogenetic movements, epithelial tissues present discontinuities. In such situations, cellular movements and tension at the free edge together with bulk cellular rearrangements largely contribute to tissue dynamics4, 27, 29, 33–38. In this scenario that includes either epithelial expansion or gap closure into cell-free areas, collective migration is aided by the protrusive activity of leading cells 21, 39. Such cells are able to pull adjacent cells along and form finger-like structures. As such, the colonies’ edge presents fingering patterns40 with protrusive activity and actomyosin cables at the rear. Interestingly such contractile actomyosin cables play a major role during epithelial gap closure34–36, 41, 42 and are prominent in concave regions during tissue migration or wound closure. The assembly of actomyosin contractile cables plays an opposite role at convex regions where it limits the expansion of the tissue34, 36, 43. Taken together, the above considerations draw a complex picture of collective cell dynamics during epithelial gap closure, wound healing or tissue expansion driven by cell proliferation and migration that strongly depend on both biochemical and physical environmental factors5, 34, 44–47. Cohesive forces acting at the CCJ19, 48 as well as the tension exerted by supracellular actomyosin cables at the cell edge36 play a role in the maintenance of colony cohesion. Meanwhile, adhesiveness to the substrate and traction forces acting onto it through FAs seem to work in the opposite way to modify cell polarity and expand the tissue39, 49. This process is somewhat reminiscent of substrate wetting phenomena by liquid droplets that can be described by the relative contributions of adhesive properties of the liquids versus cohesive properties50. Such analogy has been indeed used to describe the spreading of cell aggregates51.
Even though single cell responses including their signaling and mechanical properties have been shown to be tightly coupled to the geometrical, chemical and physical interactions with the underlying substrate52–55, little is known about the coupling between substrate properties and regulation of colony shape, tissue expansion and migration for multicellular assemblies. Here, we investigate the emerging behaviors of expanding epithelial Madin-Darby canine kidney (MDCK) colonies in response to various substrate ECM coatings to vary cell-substrate interactions and thus determine their impact on tissue organization.
Results and discussion
Varying amounts of FN substrate coating leads to different degrees of spreading of epithelial cells colonies, in analogy to partial wetting of water droplets
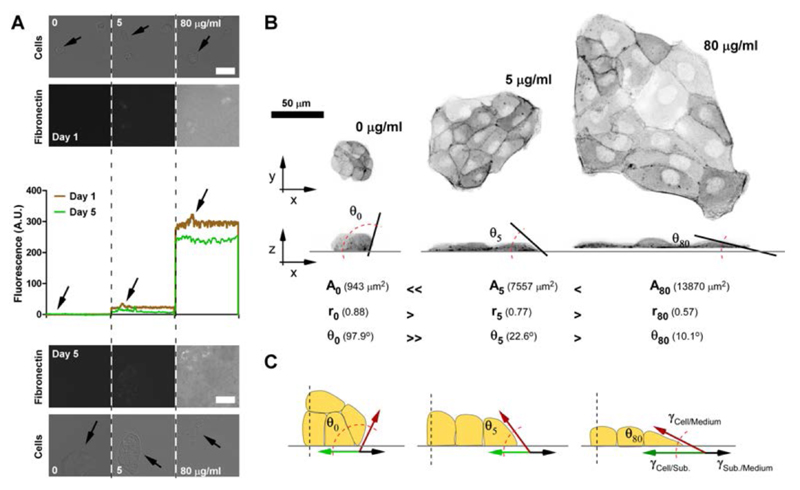
Previous studies reported that spreading of cell aggregates56 as well as coalescence of cells57 on varying ECM at short time scales (< 10 hrs) can be described by two parameters, i.e. the cohesive (Wcohesion ; Wc) and adhesive (Wadhesion ; Wa) energy of the cells. In order to alter adhesive properties of the cells on the substrate, thus changing Wa, we coated glass-bottom petri dishes (borosilicate) with varying amounts of FN. Fluorescence images revealed that 80 μg/ml provided for ample surface coating, whereas 5 μg/ml showed only limited covering and 0 μg/ml no coating (Fig. 1A). In all cases, we observed an overall homogenous protein coating of the substrates with inhomogenies being much smaller than the sell size (Suppl. Fig. 1). Importantly, surface coating did not markedly change after 5 days submerged in culture medium and in the presence of cells. Furthermore, cells did not significantly change the amount and the homogeneity of the protein coating. Under these conditions, we observed that small colonies of epithelial cells originating from few cells (< 2) and grown for three days adopted diverse shapes in response to different amounts of FN coating (Fig 1B). In particular, the overall aspect ratio of the cell colonies varied in all dimensions. Colonies on high FN (80 μg/ml) grew into large, flat and anisotropic patches, those at low FN (5 μg/ml) were rather round, small and tall. Even more dramatically, colonies tended to form three-dimensional spheroids in absence of FN (0 μg/ml). Interestingly, also the contact angle between the cell patches and the substrate changes as a function of the FN coating (Fig. 1B, x/z images), in a manner highly reminiscent to partial wetting of droplets on surfaces with varying affinity for the fluid. In the droplet case, the contact angle (θ) is a direct measure for the wettability parameter, S = Wa − Wc ; with Wa < Wc and S < 0 for partial wetting. The more negative S is, the less the droplet spreads, thus giving a larger θ. In our experiments, small θ were measured for colonies provided with high adhesion (80 μg/ml) and large θ (> 90°) for those deprived of it (0 μg/ml) (Fig. 1C and D). This indicates that increased FN concentration increases Wa and a less negative S (smaller θ). Under this condition, colonies spread into patches of larger area. Furthermore, colonies with higher Wa show less isotropic shapes (low roundness) indicating for indicating for a higher crawling activity.
Fig. 1.
Surface FN coating changes the aspect ratio of epithelial cell colonies. A. Top, physisorption of FN on glass substrate was measured by detection of fluorescently labeled FN (Cy5-FN). Middle, line profiles across the images show the fluorescence intensity values of the Cy5-FN. Bottom, FN coating after 5 days in culture with cells. FN coating was only marginally decreased after 5 days in culture. Presence of cells (arrows) caused only minor variations in the fluorescence intensity. (Scale bar = 50 μm). For direct visual comparison, acquisition and display setting are equal for all fluorescence images in A. B. Exemplary fluorescence images of colonies grown on different FN coating for 3 days. Colonies originated from few ( < 2) cells stably transfected with GFP-actin are shown. x/y (top) and x/z (side) views of cell colonies are shown. Top (x/y) views were used to measure area spread (A) and roundness (r - see text) of the colony. Side (x/z) views were used to measure contact angle (θ). Actual values for the colonies shown are reported in brackets. C. Schematic of contact angle as a function of FN concentration.
Dynamics of expansion of cell colonies depends on FN surface coating
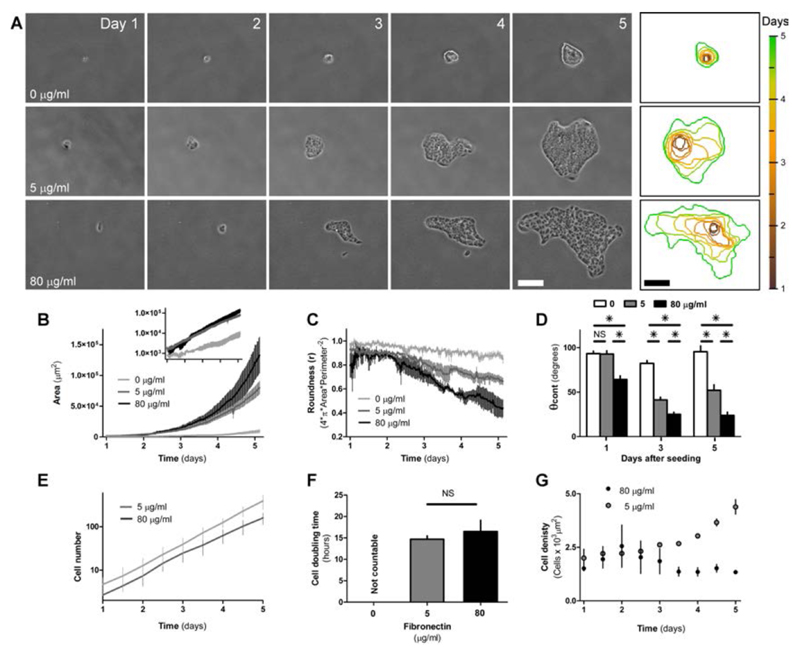
Despite previously mentioned analogies, spreading tissue colony is much more complex than a simple spreading of water droplet as it is driven far from equilibrium due to cellular activities. Further, the localization of activity (e.g. lamellipodia, actin cable) is variable depending on FN concentration and probably also time. To investigate these complexities, we visualized the expansion of colonies for 5 days over different ECM coating (Fig 2 A and Suppl. Movies 1 - 3). Interestingly, colonies on different FN amounts looked alike for the first two days when the number of cells was limited and clear differences appeared only after the second day. Potentially this was due to very high number of cells per units of area, which may favors high Wc. After the second day, the appearance of the colonies started to differ between the different FN coatings. This was reflected by an exponential increase in colonies’ area (Fig. 2 B). Rise in area correlated with the amount of FN absorbed onto the surfaces, with cells on high FN being able to spread into large patches, whereas colonies on 5 μg/ml had less pronounced enlargement. Conversely, cells with low adhesion (0 μg/ml) never really expanded onto the substrate, but rather aggregated into small three dimensional spheroids and remained round till the end of the process. In parallel to area changes, roundness of colonies provided with ECM declined after the third day (Fig. 2 C). This decline in roundness, which reflected more anisotropic shapes, was also correlated to FN concentrations. In particular, colonies on higher FN coating showed more anisotropic shapes (lower roundness) then on lower FN amounts. Differences in colonies’ area and shape could potentially result from differences in proliferation in the two different conditions58, 59. Contact angle of the cell colonies with the substrate, was also subject to changes over time (Fig. 2 D). One day after seeding, the colonies had the highest contact angle during the first day and only colonies at 80 μg/ml FN showed to be able to effectively spread onto the substrate. In the following days θ sensibly declined for colonies at 5 and 80 μg/ml FN but not for 0, which remainded rather high throughout the whole time of sampling. Contact angle of the cell colonies with the substrate, was also subject to changes over time (Fig. 2 D). One day after seeding, the colonies had the highest contact angle during the first day and only colonies at 80 μg/ml FN showed to be able to effectively spread onto the substrate. In the following days θ sensibly declined for colonies at 5 and 80 μg/ml FN but not for 0, which remainded rather high throughout the whole time of sampling. Despite these time-dependent changes, cell number grew exponentially by the same rate at both, 5 and 80 μg/ml FN (Fig. 2 F). This was reflected by a nearly identical doubling time (time take by the population to duplicate in number) of approximately 15 h in both cases (Fig. 2 E). This could be due to the fact that cell proliferation was at its maximum speed at this low confluence. Despite having the same proliferation rates, colonies on different FN had marked differences in the cell density (Fig. 2 F), during the process of expansion. In both cases, low and high FN, cell density increased between day 1 and 2. Thereafter, density of cells continued climbing at a lower rate for colonies cultured at 5 μg/ml FN, but it declined for those at 80 μg/ml FN resulting in colonies with large, flat cells. Taken together, these results suggest that active expansion dominates the development of colonies at high Wa leading to higher colony spreading (S).
Fig. 2.
Growth of MDCK colonies depends on FN concentrations. A. Time lapse shows the evolution of cell colonies over 5 days as a function of FN coating concentration. Colonies derived from 1 or 2 cells seeded at time 0. Perimeters of colonies at different time points are shown in the last column. For all images, horizontal scale bars = 100 μm; vertical scale bar = color-code of time points. Variation of colony area (B) and roundness (C) as a function of FN coating are plotted over time. Inset in B, plot of area over time in semi-logarithmic scale. Geometrical descriptor termed roundness is independent of the size of the patch and it would be 1 for a perfect circle and would tend to 0 for a highly scattered, anisotropic shape. D. Contact angle of colonies (θ) as a function of FN coating is plotted over time. 4 to 10 colonies per conditions have been analyzed. 8 different positions per each colony have been used to measure θ. E. Plot of cell number in semilog scale shows exponential increase of cell population over time. At 0 µg/ml FN, number of cells cannot be resolved due to very high density within the spheroid. F. Doubling time of cell population as a function of FN coating is calculated from E. G. Cell density as a function of time. For B, C, E, F and G, n = 3 - 7. For all images, error bars = standard error of the mean. Data are considered significantly different for p < 0.05 (unpaired Student’s t-test).
Cell motility and mode of collective migration within cell colonies depends on FN surface concentration
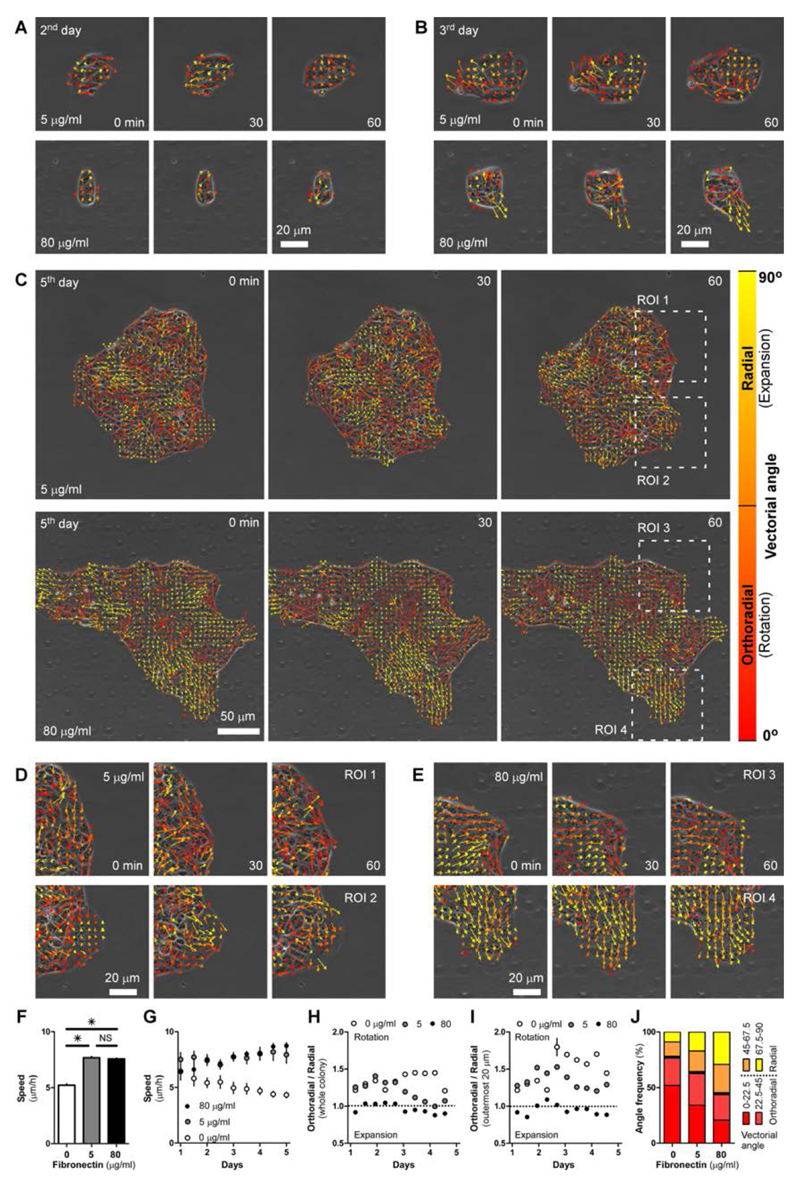
Cell proliferation fails to explain the different spreading behaviors at different FN coatings. Thus, we further investigated the effect of cell motility and collective behavior on the colony expansion process. Increasing amounts of ECM coating and adhesion of cells to the substrate proved to cause increased cell motility of single cells60 as well as in tissues38, 61. Also in our system, MDCK cells overexpressing Snail transcription factor, which down-regulates E-cadherin62 and induce single cell migration, showed increased motility at higher FN coating (Suppl. Fig. 2). To explore this aspect in our system, we analyzed the motility within the colonies by particle image velocimetry (PIV; Fig. 3 and Suppl. Movies 4 and 5). At early times, when the number of cells and the patch size was still small (second day), cells had a clear tendency to rotate for both FN conditions (Fig. 3 A) in a similar fashion as shown on confined micropatterned disks63–65. After the second day, this rotation continued in colonies on low FN coatings, but it was replaced by a predominantly outward movement for the higher FN concentration (Fig. 3 B - E). Besides the case of no FN coating where movement remains slow at all times (average speed = 5.21 ± 1.72 μm/h), average speed was low for the duration of this whole initial period for both FN coatings (day 1 to 3, average speed = 7.41 ± 2.17 μm/h for low FN and 6.98 ± 2.02 μm/h for high FN) and it increased only thereafter (day 4 and 5, average speed = 6.98 ± 2.50 μm/h for 5 μg/ml and 8.21 ± 1.23 μm/h for 80 μg/ml) (Fig. 3 F and G). During the fifth day, colonies on low FN continued to show a rotational movement (Fig. 3 C and D). Meanwhile, a radial outward motion became more visible for colonies on high FN (Fig. 3 C and E). This picture was confirmed by quantitative analysis of movement directionality (Fig. 3 H, I and J). Rotational movement during the second and third day was a common characteristic of all conditions (orthoradial/radial ratio > 1). After the second day, this indicator for rotational movement declined in both cases but the decline was more pronounced for high FN coating. Despite this decline, rotation remained prominent for colonies on low FN for the entire duration of the experiment (orthoradial component higher than radial one). Conversely, movement became more radially oriented (orthooradial/radial ratio < 1) at high FN, indicating that outward collective movement was driving the overall expansion of the tissue through cell crawling mechanism. Similarly, analysis of the orientation of the vectors shows that majority of vectors are orthoradial for 0 and 5 μg/ml FN and radial for 80 μg/ml. In our interpretation, rotational movement would be induced by high cohesion (high Wc) between the cells and/or lack of sufficient adhesion to the substrate (low Wa), and this was preventing outward cell migration and onset a rotation of the colony. In contrast, on higher FN, higher cell-surface friction would counterbalance the strong cell-cell cohesion. Therefore, cells were able to move and broaden the relative distance separating them from each other and allow spreading of the colony over the substrate. This expansion was further enhanced by the appearance of leader cells pulling neighbors into finger-like structures (Fig. 3 E, bottom). Interestingly, in rare cases leader cells moved too fast for their followers and detached from the colony (Suppl. Fig. 3 A and Movie 6) in a process that reflects the consequences of an extreme imbalance between cohesion and adhesion (with Wa > Wc). These results suggest that at low FN cells have limited capability of expansion due to high cohesion between cells (Wa preponderant over Wc). Thus, their motion is frustrated into a rotation63, 65, which further suggests a self-confining mechanism leading to high cell density and roundness. Thus, in absence of outward migration, expansion of colonies on low FN can mostly be attributed to cell proliferation rather than cell motility. On the other hand, cells within colonies on high FN showed high motility and freedom to collectively move outward (Wc preponderant over Wa). In the process, cells become flat and well spread out. Furthermore, appearance of leader cells seems to be the reason for highly anisotropic shapes of colonies on high FN concentration.
Fig. 3.
Particle Image Velocimetry (PIV) analysis of collective motion of cells. A to E. Merge of PIV quiver plot and phase contrast images show patterns of cell movement at three consecutive time points (Δt = 30 min) during the 2nd (A), 3rd (B), and 5th (C) day after seeding. For each image sequence, top row shows migration patterns of a colony growing over substrate coated with 5 μg/ml FN and bottom row demonstrate those with 80 μg/ml. Direction and magnitude of movement are represented by orientation and length of vectors. D and E. enlarged views from ROI in C. For all images from A to E, vectors are color-coded according to the angle respective to the center of mass of the colony. 90° angle are vectors radially oriented to the center of mass (expanding tissue), whereas 0° angle are vectors perpendicular to the radial direction (orthoradial – rotational movement of the tissue). F. Bar plot of speed magnitude distribution of colonies grown at different FN coating. Average speed of 3 independent colonies over the 4 days of observation is shown. G. Evolution of the speed magnitude over time. Average of 3 independent colonies is shown. H. Ratio between orthoradial and radial components of velocities within the whole colony plotted over time. For ratio above 1, tissue is mostly rotating; below 1 is mostly expanding. I. Plot of orthoradial/radial components within 20 µm from the edge of the colonies. J. Frequency (%) of angles of velocity vectors as compared to tissue center of mass. For H-J, average of 2 independent colonies is shown. For all panels, 90° is radial, 0° is orthoradial. Error bars = standard error of the mean. When not visible, SEM bar is smaller than symbol. Data are considered significantly different for p < 0.05 (unpaired Student’s t-test).
Actomyosin cable provides cohesiveness to the cell colony, whereas lamellipodia are responsible for adhesiveness and substrate wetting
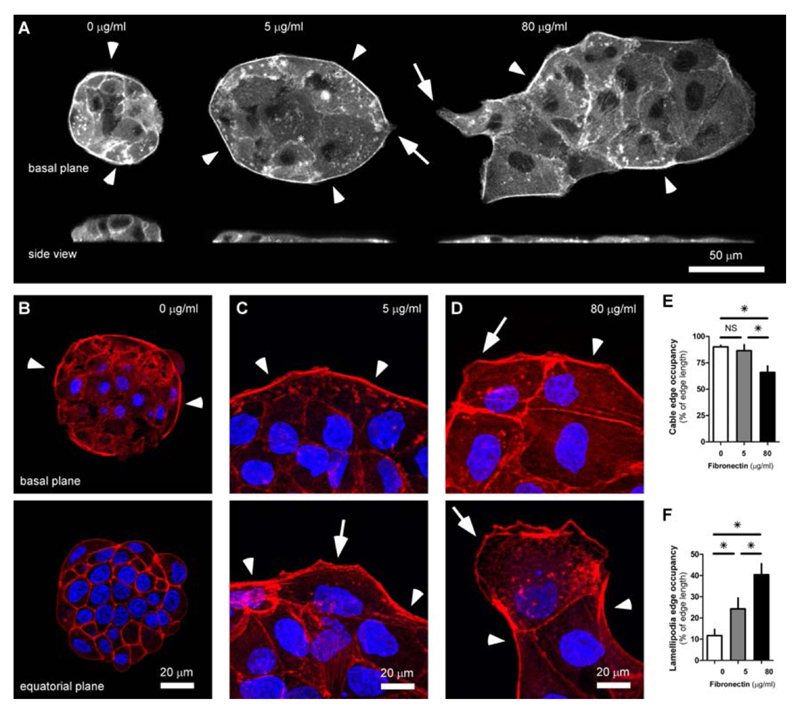
To investigate the nature of cohesive and adhesive mechanisms, we visualized actin structures at different FN coating. Previous studies have shown that epithelial cell migration in various situations can be regulated by acto-myosin contractility through purse-string mechanism and/or cell crawling activity35, 38, 66. We looked for actomyosin cable and lamellipodia distribution at the three different FN concentrations (Fig. 4). As previously mentioned, at 0 μg/ml FN cells did not spread, but formed 3D spheroids. At the base, these aggregates where confined by a thick actomyosin cable (Fig. 4 A and B bottom). However, contact with the substrate is very limited in this case and images take at the equatorial plane show that actin is mostly engaged at the cell-cell junction (Fig. 4 B bottom). For the case of 5 and 80 μg/ml, the most obvious difference was the presence of large stress fibers at the basal plane of colonies on high FN, that were virtually absent at lower FN coatings (Fig. 4 A - D). The actomyosin cable, which provides a strong barrier at the outer perimeter of the colony and inward forces at concave edges, thus improving the cohesiveness of rounder colonies38, appeared to be continuous over multiple cells at 5 μg/ml FN (Fig 4 A and C top). In contrast, the cable was frequently interrupted by the protrusive activity of small and large lamellipodia in colonies on high FN (Fig 4 A and D top). Lamellipodia and leader cells were clearly favored at high FN coating, whereas they appeared to be small at low FN (Fig. 4 A, C and D bottom). This scenario was confirmed by quantitative analysis of edge occupancy by the actomyosin cable (Fig. 4 E) and lamellipodia (Fig 4 F). The edge of colonies at 0 and 5 μg/ml was mostly limited by the actomyosin cable whereas those at 80 μg/ml had higher lamellipodia occupancy. Altogether, these results confirmed that the relative contributions of the pluricellular actin cable and the formation of lamellipodia-based protrusions could be tuned by cell-substrate adhesion and were important to control epithelial tissue organization.
Fig. 4.
Lamellipodia and actomyosin cable organization. A. GFP-actin staining at the basal plane (top) and from the side (bottom) show actin structure organization in colonies grown on different FN coating. Cells were grown on substrates coated with 0, 5 or 80 µg/ml FN for 3 days. B - D. High resolution exemplary images of F-actin structures (Phalloidin-TRITC, red) and nuclei (DAPI, blue) different FN coating. B. Basal plane (top) and equatorial plane (bottom) of a cell spheroid grown at 0 µg/ml FN. C and D. Basal plane of cell colonies grown at 5 and 80 µg/ml FN for 3 days. In C and D, top shows a region with prominent actomyosin cable. Bottom show a cell with lamellipodia with characteristics of a leader cell. In all panels, arrowheads indicate actomyosin cable, arrows lamellipodia. E and F. Plots show the percentage of colonies’ edge occupied by actomyosin cable (E) and lamellipodia (F). n= 2 for 0 μg/ml and = 3 for 5 and 80 μg/ml. bars = standard error of the mean. When not visible, SEM bar is smaller than symbol. Data are considered significantly different for p < 0.05 (unpaired Student’s t-test).
Tissue traction forces on different FN coatings
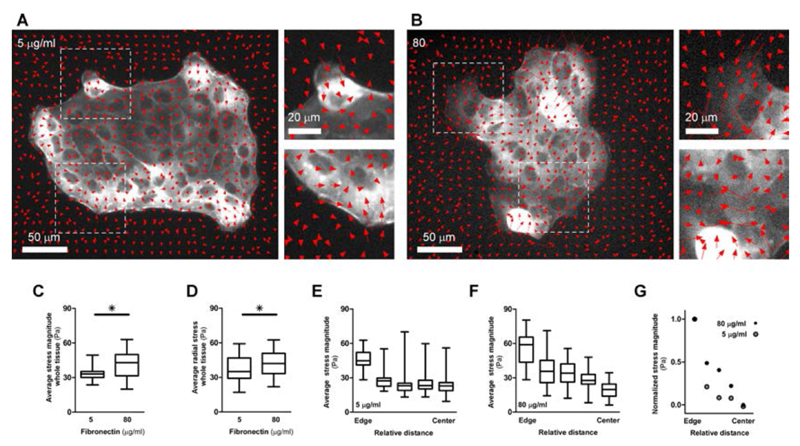
Next, we measured traction forces by traction force microscopy (TFM)67 on soft silicon gels coated with 5 and 80 μg/ml FN while monitoring actin structures to verify their contribution to adhesive and cohesive mechanisms. Despite differences in substrate rigidity and potentially in FN physisorption, colonies of cells grown on the soft gels demonstrated similar phenotypes as those grown on glass coverslips. Indeed, colonies of cells at low FN were more circular, with more actomyosin cable and less lamellipodia as compared to those grown on high FN (Fig. 5A and B, left and Suppl. Movies 7 and 8). Compatible with crawling mechanism, large inward pulling forces were observed at the site of lamellipodia (Fig. 5A and 5B, top right). Importantly, larger forces and larger lamellipodia were found at the edge of colonies grown at 80 μg/ml FN. On the other hand, converging force dipoles (Fig. 5A, bottom right) were found at the site of contractile actin cables acting tangentially to the tissue edges. This was particularly obvious at low FN coating. In contrast, at high FN, large converging force dipoles appeared within the tissue (Fig. 5B, bottom right). This was presumably due to prominent basal stress fibers within cells at high FN only (Fig. 4A). Overall, colonies on high FN were able to exert higher forces on the substrate as compared to low FN, thus unmasking a higher adhesion (higher Wa) with the surface (Fig. 5 C). In particular, inward radial force component (with respect to colony’s center of mass, inwards pointing is defined as positive) was higher for 80 μg/ml (Fig. 5 D). Finally, we measured the average force magnitudes as a function of the relative distance from the edge to the center (Fig. 5 E and F). In both cases, forces were more localized at the edge of the colonies. Interestingly, forces dropped more gradually from tissue edge to the innermost region for high FN than for low FN (Fig. 5G). This is in line with the fact that basal stress fibers were more pervasive in high FN, and these structures penetrated deep into the tissue colony, thus leading to high traction forces. Interestingly, less average traction force magnitudes in the tissue indicated a higher degree of cohesiveness between cells in the tissue for low FN. Conversely, high average force magnitudes and force dipoles within the tissue indicate for higher adhesiveness and more autonomous behavior for cells at high FN.
Fig. 5.
A. and B. Quiver plot of traction force vectors overlapped on GFP-Actin fluorescence images. Top right, enlarged views from left images show inward forces at lamellipodia regions. Bottom right and force dipoles caused by actomyosin cable (A) and stress fibers (B). C. Force magnitude averaged under whole tissue. D. Average of radial component of force with respect to center of mass (COM) – defined as positive for outwards radial component. E. and F. Average force magnitudes as function of relative distance from tissue edge. Distance is normalized by the size of the patch. G. Normalized traction forces are plotted versus the relative distance from the edge of the colony to mark the sharper drop in forces at low FN as compared to the more gradual drop at higher FN amount. For all experiments, n = 5 - 6. Error bars = standard error of the mean. Data are considered significantly different for p < 0.05 (unpaired in Student’s t-test).
Alteration of the adhesive and cohesive machineries changes the colonies’ wetting properties
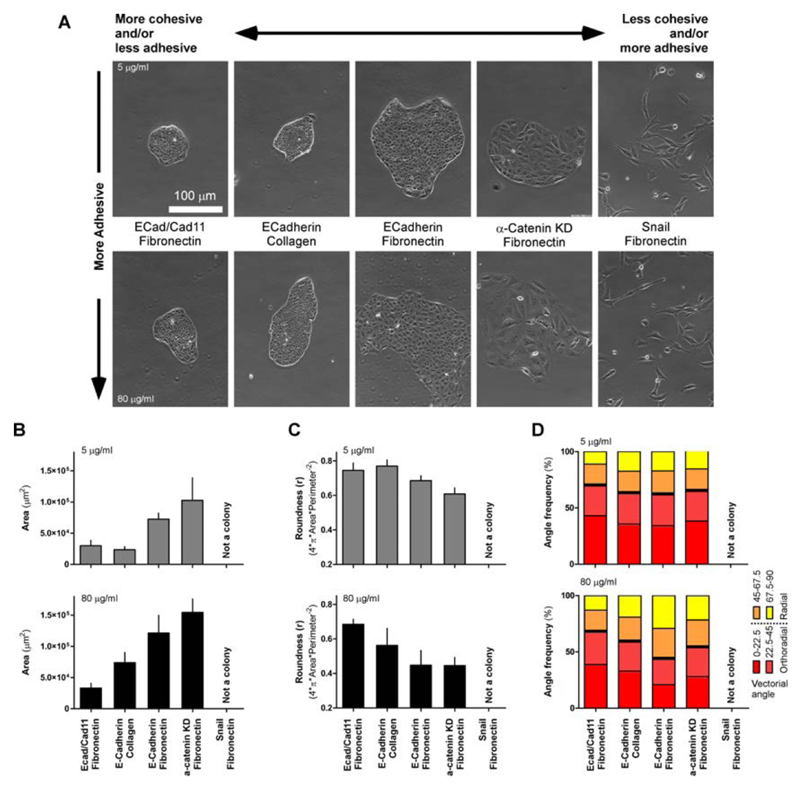
Our experiments showed two distinct phenotypes in response to changes in adhesion properties (Fig. 2 and 3). At low FN (low Wa), colonies were small, round and with a tendency to rotate. Conversely, at high FN (high Wa) they were large, anisotropic and with an outward cell motility driving cell expansion. The general aspect of actin structure and characteristics of traction forces on the substrate showed that the FN concentration modified the overall adhesive properties of the colonies and this correlated with cell motility. To further elucidate the role of ECM and CCJ in regulating adhesive and cohesive properties of the expanding tissue, we introduced collagen coating as the main support for cell adhesion and we used genetically modified MDCK cell lines to specifically alter Wa and Wc, respectively (Fig. 6). In particular, we used MDCK cells overexpressing an additional cadherin, cadherin 11 (osteoblastic cadherin), which together with the other constitutively-expressed cadherins could promote strong interaction between cells, and MDCK α-catenin knock down (α-catenin KD) cell line. α-catenin is involved in the mechanical regulation of CCJ68, 69 and reduces the cohesiveness of colonies5. Furthermore, we used MDCK cells overexpressing the transcription factor Snail that induce epithelial-to-mesenchymal transition (EMT) by down-regulating E-cadherin 62. Our experiments demonstrated that by introducing collagen colonies grew into smaller and rounder patches as compared to their respective FN (Fig. 6 A, B and C) indicating for a less adhesive phenotype. In our interpretation, this was mostly due to the lower ability of cell to perform 2D crawling on this type of substrate70. Indeed, on collagen fewer leader cells and lamellipodia appeared even at high protein concentrations. Similarly, cell expressing cadherin 11 also showed limited ability to expand. This is consistent with higher cohesiveness of the tissue. In both cases, analysis of the direction of motion by PIV showed that colonies with cadherin 11 and those grown on collagen were characterized by rotational movement (Fig. 6 D). On the other hand, α-catenin KD cells had very poor cohesiveness and grew into larger patches as compared to wild-type cells. These cells started to behave in a less collective manner (Suppl. Movies 9 and 10). This was particularly evident at high FN coating and analysis of the motion showed that cells were moving with no preferential direction. Finally, Snail cells did not form a colony, but crawled as single isolated cells. Overall these data suggest that a similar collective behavior can be obtained by reducing cells-substrate adhesion (e.g. collagen instead of fibronectin) as well as by increasing their cohesiveness (e.g. overexpression of cadherin 11).
Fig. 6.
Variations of ECM or cell-cell junction proteins changes the phenotypes of the colony. A. Representative images of cell colonies grown for 5 days. Colonies with different modifications of the CCJ apparatus were grown on 5 (top row) or 80 μg/ml (bottom) of either fibronectin or collagen coated substrates. Specific conditions for each column are as indicated. Standard conditions (wild type E-Cadherin based junction on FN substrate) are taken from Fig. 2. Day 5 was chosen as end point condition. B and C. Plots of colonies area and roundness after 5 days of expansion. Data in B and C are significantly different in a two-ways ANOVA analysis (p < 0.05). (Ecad/Cad11-Fibronectin = 6 and 6 independent colonies for both protein concentration; E-Cadherin-Collagen = 8 and 6 ind. col. for 5 and 80 μg/ml, respectively; E-Cadherin-Fibronectin = 7 and 3 ind. col. for 5 and 80 μg/ml, respectively; a-catenin KD-Fibronectin = 8 and 11 ind. col. for 5 and 80 μg/ml, respectively Error bars = standard error of the mean. D. Frequency (%) of angles of velocity vectors as compared to tissue center of mass. 90° is radial, 0° is orthoradial. Average of 2 independent colonies for each condition is shown.
Mathematical simulation of colony expansion
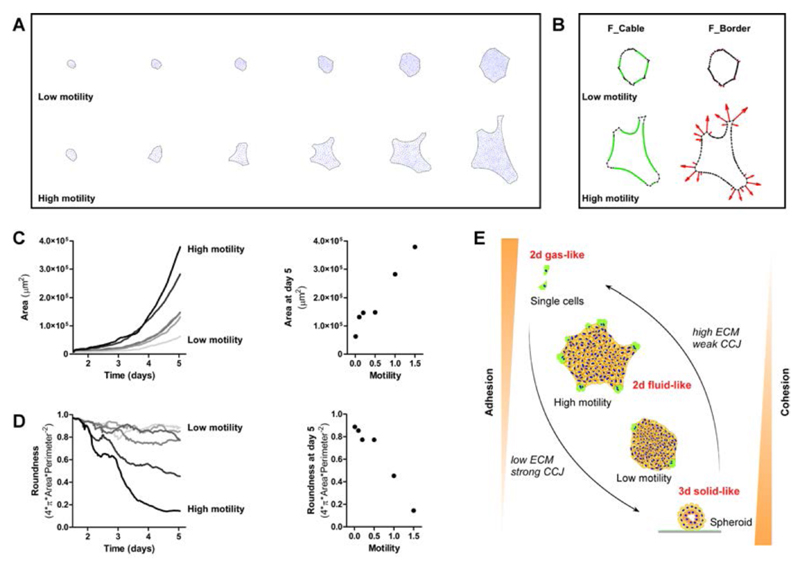
We next applied a recently developed theoretical model for the dynamics of confluent cellular layers, to describe the growth and shapes of the colonies. In this model (for details see61 and table S1) the dynamics of the cells within the bulk of the layer are determined by the following ingredients27: cells undergo a random motion (treated as a randomly oriented traction force, F_noise table S1), but interact with each other through effective binary potentials (F_interaction, Table S1). In addition, cells have a tendency to move in the same direction as their neighbors, which is the property that gives rise to collective migration effects. In the model this is described through a Vicsek-like orientational interaction that acts to create long-range collective motions (F_vicsek, Table S1). The interaction potential between the cells is such that they have a repulsive central part (soft-core), and attract when further apart (representing cell-cell adhesion) until some maximal interaction length |(maximal cell extension). The Vicsek-like orientational interaction is a standard technique to endow the cells with the property of preferring to move in the same direction as their neighbors, and is used to describe a variety of collective motion phenomena. In addition, we have that cells at the colony’s edge have the following additional forces: they form an outwards crawling force (due to lamellipodia), which grows linearly with the local convex curvature of the edge40 (F_border, Table S1). This positive feedback between the traction force and the edge cell shape drives the instability that spontaneously forms “leader-cells” at the tips of cellular fingers61. There is also the force of line tension that acts due to the acto-myosin cables (and membrane bending), mostly for cells along the colony’s edges that have concave (or very small convex) curvature. This line tension and acto-myosin cables act to contract the colony’s edge and straighten it(F_bending, F_cable, Table S1). For the detailed mathematical formalism and the numerical techniques used to calculate the evolution of the colony using this model (Fig.7) we refer the reader to the companion paper61.
Fig. 7.
Particle-based simulation and model. A. Time laps of in-silico evolution of expanding cell colonies as a function of varying the outwards (and curvature-dependent) motility of the edge cells( F_border, Table S1). This curvature-dependent force is representing the traction produced by the edge cells through the extension of lamellipodia beyond the monolayer edge .Dark outer-line indicates for colony boundary, blue dots represent the cells. B. Decomposition of the forces shows the relative contribution of each force component acting on the boundary: Green edges of the colony represent edge cells that feel the contractile force of the acto-myosin supracellular cable F_cable, while the red arrows indicate the outwards pointing lamellipodia-driven traction forces of the convex edge cells F_border (see Table S1 for explanation of forces terminology). C and D. Calculated dependence of the colonies area and shape on the outwards, curvature-dependent motility of the edge cells, respectively. E. Illustration shows the relation between cohesion/adhesion and the dynamics of expansion of a colony of cells.
In Fig. 7, we demonstrate that this model is able to explain qualitatively the observed changes due to different cell-substrate adhesion (Fig.2). We find that only increasing the motile force due to the outwards-directed lamellipodia of the edge cells (F_border), was sufficient to recreate the trend observed in the experiments, suggesting that the dominant effect of the fibronectin coating is on the crawling lamellipodia of edge cells. The decrease in roundness is explained in this model as being driven by the destabilization of the colony’s edge to the formation of fingering40, 61, while the actomyosin cables act to straighten the edges in-between the sharp corners that initiate leader cells and fingers (as observed in Fig. 4 A). The instability arises in our model due to the positive feedback between the convex shape of the edge cell and the amplitude of its outwards directed lamellipodia. This positive feedback can lead to the growth of small undulations at the colony’s edge, whereby edge cells move outwards and become leader cells at the highly convex tips. When the cell-cell cohesion is decreased, the model predicts that leader cells may detach at the convex corners of the colony (Suppl. Fig. 3D), as observed in the experiments (Suppl. Fig. 3A - C and Movie 6).
Conclusions
During development, tumor formation and metastasis and many other biological processes, the mode of expansion of a small group of cells into a large colony fundamentally influences the outcome of the process. In a biomechanical prospective, this can be analyzed as the result of the interplay between cell-cell cohesive (Wc) and cell-substrate adhesive (Wa) energies as shown for tumor progression in 3D environments71. Collective motion deployed during development to form tissue relies on a condition of partial spreading over the substrate (for S = Wa − Wc; with Wa < Wc and S < 0). The degree, speed and overall dynamic of this spreading depends on the relative importance of Wa and Wc. Similarly, metastatic processes typically take place when tumor cells lose cohesiveness and, due to high substrate adhesiveness or low cohesiveness, manage to separate from the tumor mass (for S = Wa − Wc; with Wa > Wc and S > 0). To investigate this process, we have manipulated adhesive properties of a small group of epithelial cells expanding over several days. Furthermore, we have interfered with tissue cohesiveness by genetically changing the strength of the CCJ machinery. Importantly, both manipulations showed similar phenotypes proving the duality of adhesion and cohesion (Fig. 7 E). In particular, colonies were large and well spread if adhesion was high (80 μg/ml FN) and/or cohesion low (α-catenin KD). Conversely, colonies were likely to be small and compact at high cohesion (cadherin 11) and/or low adhesion (5 or 0 μg/ml FN). This process seems to be mostly driven by changes in the type of cell motility and its relative directionality. Indeed, our analysis of actin organization reveled at higher adhesiveness promoted structures that enhanced migration and colony’s expansion – i.e. lamellipodia and leader cells. On the other hand, down regulation of adhesion promoted cohesive structures such as an actomyosin cable surrounding the colony. In line with previous reports56, 58, 72, our in vitro investigation demonstrates the importance of tissue cohesion and substrate adhesion to the biomechanics of multicellular groups.
Materials and Methods
Cell culture and reagents
MDCK (madin-darby canine kidney) cells were cultured in DMEM (Life Technologies) supplemented with 10% Fetal Bovine Serum (FBS; Life Technologies). Stably transfected GFP-Actin, α-Catenin KD, Cad11 MDCK cell lines have been kindly provided by Prof. J.W. Nelson (Stanford University, USA); Snail 1 MDCK cell line by Dr. Amparo Cano (IIB, CSIC-UAM, Spain)73. Cells were cultured at 37° C in a humidified incubator with 5% CO2 and subcultured every 2/3 days by Trypsin/EDTA (Life Technologies) method. Fibronectin (Sigma) was reconstituted in deionized water at a concentration of 1mg/ml. Dilution to working solution was also done in deionized water.
Preparation of cell seeding onto varying fibronectin concentration
600µl of varying concentration (5, 80µg/ml) of fibronectin solution was incubated for 1h at 37° C onto glass-bottom petri dish (IBIDI, Sciencewerke Pte Ltd.). Thereafter, excess of protein still in solution or weakly bound was washed away three times by deionized water. After washing and replacing pure water with culturing media, a total of 15-20 cells suspended in 300 μl of calcium free media were seeded. To prevent washout, cells were allowed to adhere to the substrate for 30-45 minutes in a cell culture incubator. Thereafter, 2ml of DMEM media supplemented with 10% FBS were added to provide cell with sufficient nutrients.
Live-cell imaging
Samples prepared as described in previous section were placed in BioStation CT imaging system (Nikon) 24 h after seeding. Dynamics of tissue expansion were visualized for four days by phase contrast microscopy using BioStation CT equipped with 4x, 10x and 20x phase objective. All live-cell experiments have been conducted at 37° C and 5% CO2. Three dimensional images of cell colonies were acquired using a laser scanning confocal microscope (Zeiss – LSM 710) equipped with a 40X water immersion objective.
Image segmentation and analysis
Image segmentation of phase contrast data was performed using in-house MATLAB software. The algorithm applies a range filter on the raw image. The range filter gives high response at region with large intensity variations and low response at region with homogeneous intensity. In our image, the large variation region corresponds to the cell colonies. The size of the range filter used varied from 3x3 to 5x5 pixels for images acquired with 4X objective and 7x7 to 13x13 for 10X. The range-filtered image was then thresholded and refined by morphological operations to generate a binary mask corresponding to the colonies. After segmentation, the contour of the cell colonies was extracted from the binary mask. Binary image was used to compute area and roundness. Cell number was accessed by manual count of cells nuclei. To measure velocity, Particle Image Velocimetry (PIV) analysis was performed using MatPIV 1.6.1 (http://www.mn.uio.no/math/english/people/aca/jks/matpiv/). Single-pass PIV with window size 64x64 pixels was used. The velocity vectors were further decomposed into radial and orthoradial components with respect to the colonies’ center of mass (COM). For lamellipodia and actomyosin cable occupancy, stably transfected cells have been imaged for 5 days. Sampling have been acquired every 24 h starting at day 1 (24 h). Cumulative length of lamellipodia was divided by the total perimeter of the colony and multiplied by 100 to obtain the % of the edge occupied by lamellipodia structures. Equivalents analysis has been performed for the actomyosin cable to obtain the % of edge occupied by the cable. For actin staining shown in Fig. B-D, PFA-fixed cells were incubated with Phalloidin-TRITC (1:100) for 15 min at room temperature.
Force measurements
Traction forces exerted on the substrate was assessed by deformation of a soft elastic substrate (Traction Force Microscopy, TFM)13, 41, 45. TFM was performed by detecting displacement of beads embedded onto the surface of a soft silicon elastomer. Briefly, CyA and CyB (Dow Corning) components were mixed in a 1:1 ratio, spin-coated on a IWAKI glass-bottom dish at 500 rpm for 1 min and cured at 80 °C for 2h (elastic modulus ~8 kPa). The substrate was silanized using a 5% solution of (3-Aminopropyl) trimethoxysilane (Sigma) in 100% ethanol for 15 min. Subsequently, red fluorescent beads (100 nm, Invitrogen) were diluted in deionized water (1:500) and added to the substrate. After incubation for 5 min, the substrates were washed with deionized water to remove loosely bound beads and dried for 15 mins at 80 °C. The beads were then passivated by incubating in TRISS 100mM for 30 mins. After removing TRISS, the substrates could be kept in incubator overnight. Substrates were coated with fibronectin at the concentration of 5 and 80µg/ml for 30 min at 37° C. Afterwards, substrates were incubated with 0.2% pluronics for 30 mins and washed with PBS. Thereafter, cells can be seeded as described in previous part. Live-cell imaging of tissue expansion and displacement of beads (traction force microscopy) was used by a fully automated Olympus IX81 inverted microscope equipped with air objective (20X). Analysis of forces by beads displacement was performed as previously described41, 74.
Mathematical modelling
We used here the theoretical model that we have recently developed to describe the collective dynamics of cellular monolayers61. The parameters used in the simulations shown here (Fig. 7) are defined in Table S1 [SI]. We simulated the effect of the varying ECM concentration as changing the maximal lamellipodia-driven, curvature-dependent crawling force, directed outwards, that can be produced by edge cells, which is the parameter Fmax in our model(which is the maximal value of F_border, Table S1). Larger values of this parameter are associated in the simulations to higher ECM concentration. In Fig. 7 we scaled the values of this parameter from the values fitted to previous experiments61, in the range of 0.001-2.5. This effect of the ECM concentration is also observed on the formation of fingers during wound-healing geometries, both in experiments and in our simulations (by changing the value of Fmax61). Due to the positive feedback between the shape (curvature) of the edge cell and its ability to produce large extending lamellipodia, our model describes naturally how edge cells can destabilize the colony’s edge and become leader cells at the highly convex tips.
Supplementary Material
Acknowledgments
The authors thank group members from MBI, William J. Nelson, Jacques Prost, Virgile Viasnoff and Alpha Yap for helpful discussions. The authors would also like to thank MBI Microfabrication (Gianluca Grenci and Mohammed Ashraf), MBI Science Communication Core (Andrew Wong and Steven Wolf) and MBI Microscopy Core (Felix Margadant, Wai Han Lau) for continuous support. The authors are grateful to A Cano and W. J. Nelson for their generous gift of MDCK cell lines. Financial supports from the Human Frontier Science Program (grant RGP0040/2012), the European Research Council under the European Union's Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement n° 617233, and the Mechanobiology Institute are gratefully acknowledged. B.L. acknowledges the Institut Universitaire de France. N.S.G is the incumbent of the Lee and William Abramowitz Professorial Chair of Biophysics, and would like to thank also the ISF grant 580/12 for support.
Footnotes
Author Contributions
A.R., B.L. designed research; A.R., A.P.L., T.B.S. and C.B. performed experiments; V.T. and N.G. conceived and implemented in silico simulations; A.R., V.T., T.B.S., A.P.L., H.T.O., C.B., R.M.M., C.T.L., N.G., B.L. contributed new reagents, modelling and computational tools; A.R., T.B.S., N.G. and B.L. wrote the paper; N.G. and B.L. supervised the project. All authors read the manuscript and commented on it.
References
- 1.Bertet C, Sulak L, Lecuit T. Nature. 2004;429:667–671. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Fernandez B, Campos I, Geiger J, Santos AC, Jacinto A. International Journal of Developmental Biology. 2009;53:1549–1556. doi: 10.1387/ijdb.072308bg. [DOI] [PubMed] [Google Scholar]
- 3.Friedl P, Locker J, Sahai E, Segall JE. Nature cell biology. 2012;14:777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- 4.Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vedula SRK, Leong MC, Lai TL, Hersen P, Kabla AJ, Lim CT, Ladoux B. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:12974–12979. doi: 10.1073/pnas.1119313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cetera M, Juan G, Oakes PW, Lewellyn L, Fairchild MJ, Tanentzapf G, Gardel ML, Horne-Badovinac S. Nat Commun. 2014;5 doi: 10.1038/ncomms6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ladoux B, Nicolas A. Reports on progress in physics Physical Society. 2012;75:116601. doi: 10.1088/0034-4885/75/11/116601. [DOI] [PubMed] [Google Scholar]
- 8.Huttenlocher A, Horwitz AR. Cold Spring Harbor perspectives in biology. 2011;3:a005074. doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palecek SP, Huttenlocher A, Horwitz AF, Lauffenburger DA. Journal of cell science. 1998;111(Pt 7):929–940. doi: 10.1242/jcs.111.7.929. [DOI] [PubMed] [Google Scholar]
- 10.Friedl P, Hegerfeldt Y, Tusch M. The International journal of developmental biology. 2004;48:441–449. doi: 10.1387/ijdb.041821pf. [DOI] [PubMed] [Google Scholar]
- 11.Vedula SRK, Ravasio A, Lim CT, Ladoux B. Physiology. 2013;28:370–379. doi: 10.1152/physiol.00033.2013. [DOI] [PubMed] [Google Scholar]
- 12.Guo WH, Frey MT, Burnham NA, Wang YL. Biophys J. 2006;90:2213–2220. doi: 10.1529/biophysj.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mertz AF, Che Y, Banerjee S, Goldstein JM, Rosowski KA, Revilla SF, Niessen CM, Marchetti MC, Dufresne ER, Horsley V. Proc Natl Acad Sci U S A. 2013;110:842–847. doi: 10.1073/pnas.1217279110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butcher DT, Alliston T, Weaver VM. Nature reviews Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Al-Kilani A, de Freitas O, Dufour S, Gallet F. Biophys J. 2011;101:336–344. doi: 10.1016/j.bpj.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ladoux B, Anon E, Lambert M, Rabodzey A, Hersen P, Buguin A, Silberzan P, Mege RM. Biophysical Journal. 2010;98:534–542. doi: 10.1016/j.bpj.2009.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chu YS, Thomas WA, Eder O, Pincet F, Perez E, Thiery JP, Dufour S. Journal of Cell Biology. 2004;167:1183–1194. doi: 10.1083/jcb.200403043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leckband DE, le Duc Q, Wang N, de Rooij J. Current opinion in cell biology. 2011;23:523–530. doi: 10.1016/j.ceb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Maitre JL, Berthoumieux H, Krens SF, Salbreux G, Julicher F, Paluch E, Heisenberg CP. Science. 2012;338:253–256. doi: 10.1126/science.1225399. [DOI] [PubMed] [Google Scholar]
- 20.Gov NS. In: Cell and Matrix Mechanics. Kaunas R, Zemel A, editors. ch.9. CRC Press; 2014. pp. 219–238. [Google Scholar]
- 21.Das T, Safferling K, Rausch S, Grabe N, Boehm H, Spatz JP. Nature cell biology. 2015;17:276–287. doi: 10.1038/ncb3115. [DOI] [PubMed] [Google Scholar]
- 22.Trepat X, Fredberg JJ. Trends in cell biology. 2011;21:638–646. doi: 10.1016/j.tcb.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaritsky A, Kaplan D, Hecht I, Natan S, Wolf L, Gov NS, Ben-Jacob E, Tsarfaty I. PLoS computational biology. 2014;10:e1003747. doi: 10.1371/journal.pcbi.1003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayor R, Carmona-Fontaine C. Trends in cell biology. 2010;20:319–328. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szabo B, Szollosi GJ, Gonci B, Juranyi Z, Selmeczi D, Vicsek T. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;74:061908. doi: 10.1103/PhysRevE.74.061908. [DOI] [PubMed] [Google Scholar]
- 26.Basan M, Elgeti J, Hannezo E, Rappel WJ, Levine H. Proc Natl Acad Sci U S A. 2013;110:2452–2459. doi: 10.1073/pnas.1219937110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sepulveda N, Petitjean L, Cochet O, Grasland-Mongrain E, Silberzan P, Hakim V. PLoS computational biology. 2013;9 doi: 10.1371/journal.pcbi.1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li B, Sun SX. Biophysical Journal. 2014;107:1532–1541. doi: 10.1016/j.bpj.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee P, Wolgemuth CW. PLoS computational biology. 2011;7 doi: 10.1371/journal.pcbi.1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchetti MC, Joanny JF, Ramaswamy S, Liverpool TB, Prost J, Rao M, Simha RA. Reviews of Modern Physics. 2013;85 [Google Scholar]
- 31.Gov NS. HFSP journal. 2009;3:223–227. doi: 10.2976/1.3185785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabla AJ. J R Soc Interface. 2012;9:3268–3278. doi: 10.1098/rsif.2012.0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson CM, Jean RP, Tan JL, Liu WF, Sniadecki NJ, Spector AA, Chen CS. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11594–11599. doi: 10.1073/pnas.0502575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klarlund JK. Proc Natl Acad Sci U S A. 2012;109:15799–15804. doi: 10.1073/pnas.1210992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brugues A, Anon E, Conte V, Veldhuis JH, Gupta M, Colombelli J, Munoz JJ, Brodland GW, Ladoux B, Trepat X. Nature Physics. 2014;10:684–691. doi: 10.1038/nphys3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vedula SRK, Peyret G, Cheddadi I, Chen T, Brugues A, Hirata H, Lopez-Menendez H, Toyama Y, de Almeida LN, Trepat X, Lim CT, Ladoux B. Nat Commun. 2015;6:6111. doi: 10.1038/ncomms7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rausch S, Das T, Soine JR, Hofmann TW, Boehm CH, Schwarz US, Boehm H, Spatz JP. Biointerphases. 2013;8:32. doi: 10.1186/1559-4106-8-32. [DOI] [PubMed] [Google Scholar]
- 38.Ravasio A, Cheddadi I, Chen T, Pereira T, Ong HT, Bertocchi C, Brugues A, Jacinto A, Kabla AJ, Toyama Y, Trepat X, et al. Nat Commun. 2015;6:7683. doi: 10.1038/ncomms8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reffay M, Parrini MC, Cochet-Escartin O, Ladoux B, Buguin A, Coscoy S, Amblard F, Camonis J, Silberzan P. Nature cell biology. 2014;16:217–223. doi: 10.1038/ncb2917. [DOI] [PubMed] [Google Scholar]
- 40.Mark S, Shlomovitz R, Gov NS, Poujade M, Grasland-Mongrain E, Silberzan P. Biophysical Journal. 2010;98:361–370. doi: 10.1016/j.bpj.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vedula SRK, Hirata H, Nai MH, Brugues A, Toyama Y, Trepat X, Lim CT, Ladoux B. Nature Materials. 2014;13:87–96. doi: 10.1038/nmat3814. [DOI] [PubMed] [Google Scholar]
- 42.Cochet-Escartin O, Ranft J, Silberzan P, Marcq P. Biophys J. 2014;106:65–73. doi: 10.1016/j.bpj.2013.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacinto A, Martin P. Curr Biol. 2001;11:R705–707. doi: 10.1016/s0960-9822(01)00414-6. [DOI] [PubMed] [Google Scholar]
- 44.Angelini TE, Hannezo E, Trepat X, Fredberg JJ, Weitz DA. Physical Review Letters. 2010;104 doi: 10.1103/PhysRevLett.104.168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trepat X, Wasserman MR, Angelini TE, Millet E, Weitz DA, Butler JP, Fredberg JJ. Nature Physics. 2009;5:426–430. [Google Scholar]
- 46.Tamada M, Perez TD, Nelson WJ, Sheetz MP. The Journal of cell biology. 2007;176:27–33. doi: 10.1083/jcb.200609116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ng MR, Besser A, Danuser G, Brugge JS. The Journal of cell biology. 2012;199:545–563. doi: 10.1083/jcb.201207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. Proc Natl Acad Sci U S A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paszek MJ, Nastaran Z, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettinger D, Hammer DA, et al. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 50.de Gennes P-G, Brochard-Wyart F, Quere D. Capillarity and Wetting Phenomena: Drops, Bubbles, Pearls, Waves (Book) Springer-Verlag; New York: 2004. [Google Scholar]
- 51.Gonzalez-Rodriguez D, Guevorkian K, Douezan S, Brochard-Wyart F. Science. 2012;338:910–917. doi: 10.1126/science.1226418. [DOI] [PubMed] [Google Scholar]
- 52.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425–1428. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 53.Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA, Horwitz AF. Nature. 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 54.Gupta M, Sarangi BR, Deschamps J, Nematbakhsh Y, Callan-Jones A, Margadant F, Mege RM, Lim CT, Voituriez R, Ladoux B. Nature Communications. 2015;6 doi: 10.1038/ncomms8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu YJ, Le Berre M, Lautenschlaeger F, Maiuri P, Callan-Jones A, Heuze M, Takaki T, Voituriez R, Piel M. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 56.Douezan S, Guevorkian K, Naouar R, Dufour S, Cuvelier D, Brochard-Wyart F. Proc Natl Acad Sci U S A. 2011;108:7315–7320. doi: 10.1073/pnas.1018057108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan PL, Foty RA, Kohn J, Steinberg MS. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:4323–4327. doi: 10.1073/pnas.071615398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang S, Ingber DE. Nature cell biology. 1999;1:E131–138. doi: 10.1038/13043. [DOI] [PubMed] [Google Scholar]
- 59.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 60.McCarthy JB, Palm SL, Furcht LT. The Journal of cell biology. 1983;97:772–777. doi: 10.1083/jcb.97.3.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tarle V, Ravasio A, Hakim V, Gov NS. Integrative biology : quantitative biosciences from nano to macro. 2015 doi: 10.1039/c5ib00092k. [DOI] [PubMed] [Google Scholar]
- 62.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, de Herreros AG. Nature cell biology. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 63.Doxzen K, Vedula SRK, Leong MC, Hirata H, Gov N, Kabla AJ, Ladoux B, Lim CT. Integrative Biology. 2013 doi: 10.1039/c3ib40054a. [DOI] [PubMed] [Google Scholar]
- 64.Wan LQ, Ronaldson K, Park M, Taylor G, Zhang Y, Gimble JM, Vunjak-Novakovic G. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:12295–12300. doi: 10.1073/pnas.1103834108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Deforet M, Hakim V, Yevick HG, Duclos G, Silberzan P. Nat Commun. 2014;5:3747. doi: 10.1038/ncomms4747. [DOI] [PubMed] [Google Scholar]
- 66.Grasso S, Hernandez JA, Chifflet S. American journal of physiology Cell physiology. 2007;293:C1327–1337. doi: 10.1152/ajpcell.00001.2007. [DOI] [PubMed] [Google Scholar]
- 67.Munevar S, Wang YL, Dembo M. Biophysical Journal. 2001;80:1744–1757. doi: 10.1016/s0006-3495(01)76145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. Nature cell biology. 2010;12:533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 69.Thomas WA, Boscher C, Chu YS, Cuvelier D, Martinez-Rico C, Seddiki R, Heysch J, Ladoux B, Thiery JP, Mege RM, Dufour S. The Journal of biological chemistry. 2013;288:4957–4969. doi: 10.1074/jbc.M112.403774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Olivero DK, Furcht LT. Invest Ophth Vis Sci. 1993;34:2825–2834. [PubMed] [Google Scholar]
- 71.Butcher DT, Alliston T, Weaver VM. Nature Reviews Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Foty RA, Steinberg MS. Developmental Biology. 2005;278:255–263. doi: 10.1016/j.ydbio.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 73.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. Nature cell biology. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 74.Martiel JL, Leal A, Kurzawa L, Balland M, Wang I, Vignaud T, Tseng Q, Thery M. Methods in cell biology. 2015;125:269–287. doi: 10.1016/bs.mcb.2014.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.