Abstract
Tcf1 is essential for T-cell development; however, it remains controversial if β-catenin, a known co-activator of Tcf1, has a role. Tcf1 is expressed in multiple isoforms in T-lineage cells, with the long isoforms interacting with β-catenin through an N-terminal domain. Here we specifically ablated Tcf1 long isoforms in mice (called p45−/−) to abrogate β-catenin interaction. Whereas thymic cellularity was diminished in p45−/− mice, transition of thymocytes through the maturation stages was unaffected, with no overt signs of developmental blocks. p45−/− thymocytes showed increase in apoptosis and alterations in transcriptome, but these changes were substantially more modest than thymocytes lacking all Tcf1 isoforms. These data indicate that Tcf1-β-catenin interaction is necessary for promoting thymocyte survival to maintain thymic output. Rather than being dominant negative regulators, Tcf1 short isoforms are adequate in supporting transition of maturation steps and proper expression of most Tcf1 target genes in developing thymocytes.
Introduction
T lymphocytes constitute the cellular branch of the adaptive immune responses and are essential for clearing infections by viruses, bacteria, and parasites. T cells are generated in the thymus, following step-wise maturation stages. The earliest step is the seeding of hematopoietic stem cell-derived progenitors in the thymus, including the early thymic progenitors (ETPs). ETPs undergo T cell lineage specification and commitment steps within the CD4−CD8− double negative (DN) stages. Based on CD44 and CD25 expression, DN thymocytes can be distinguished into four sequentially developing subsets, CD44+CD25− DN1, CD44+CD25+ DN2, CD44−CD25+ DN3, and CD44−CD25− DN4 cells. DN4 cells then pass through CD8+ immature single positive (ISP) stage and become CD4+CD8+ double positive (DP) thymocytes. After vigorous negative and positive selection, the DP thymocytes make lineage choice decisions to become either CD4+ or CD8+ single positive cells. Each of these maturation steps is orchestrated by multiple transcription factors in different combinations (1, 2).
T cell factor 1 (Tcf1) has been known as a transcription factor acting downstream of the Wnt pathway. It can interact with β-catenin coactivator, which is post-translationally regulated and stabilized by Wnt and prostaglandin-derived signals (3, 4). Tcf1 critically regulates several stages of thymic T cell development. At the very early step, Tcf1 is potently upregulated by Notch signaling and contributes to specification of early progenitors to the T cell lineage (5, 6). Whereas Tcf1 is not absolutely essential for TCRβ locus rearrangements at the DN3 stage, it facilitates maturation of DN to DP thymocytes (7). In recent studies, we have demonstrated that Tcf1 is important for directing DP thymocytes to the CD4+ T cell lineage by acting upstream of Thpok (8). Although not required for the CD8+ T cell lineage decision, Tcf1 utilizes its intrinsic histone deacetylase (HDAC) activity to suppress the expression of CD4+ lineage-associated genes and hence establish CD8+ T cell identity (9).
Due to differential promoter usage and alternative splicing, multiple Tcf1 isoforms can be detected in T cells (10). All isoforms contain a C-terminal high mobility group (HMG) DNA binding domain and a newly discovered HDAC domain (9). The Tcf1 long isoforms (p45 and p42) contain a unique N-terminal β-catenin-binding domain, while the Tcf1 short isoforms (p33 and p30) lack this domain and hence cannot interact with β-catenin. Most of the loss-of-function studies of Tcf1 ablated all Tcf1 isoforms. The Tcf1 short isoforms have been considered as dominant negative regulators of the full-length Tcf1 protein; yet their physiological roles in T cell development have not been elucidated. In this report, we performed structure-function analysis of Tcf1 protein in developing thymocytes, by ablating the β-catenin-binding domain or the HMG DNA-binding domain (Fig. 1A).
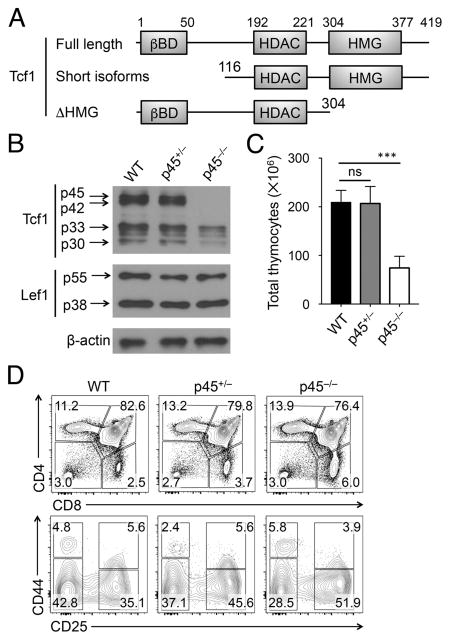
Figure 1. Impact of loss of Tcf1 long isoforms on T cell development.
(A) Diagram showing the functional domains of Tcf1 full-length protein, N-terminus truncated short isoforms, and C-terminus HMG domain truncation. Numbers denote boundaries of functional domains or truncation points.
(B) Detection of Tcf1 and Lef1 isoforms by immunoblotting. Cell lysates were extracted from total thymocytes of WT, p45+/− or p45−/− mice, and immunoblotted with anti-Tcf1, Lef1 or β-actin antibody. Representative data from ≥3 experiments are shown.
(C) Total thymic cellularity (n ≥ 4 from at least 4 experiments).
(D) Thymic maturation stages. Lin− thymocytes were analyzed for CD4 and CD8 expression, and the frequency of DN, DP, CD4+ and CD8+ thymic subsets is shown (top). Lin− DN thymocytes were further analyzed for CD44 and CD25 expression, and the frequency of DN1-DN4 subsets are shown (bottom). Data are representative from 4 experiments (n ≥ 4). ns, not statistically significant; ***, p<0.001 by Student’s t test.
Materials and Methods
Mice and CRISPR/Cas9-mediated genome editing
Tcf7fl/fl and p45−/− mice were generated previously (10, 11). To generate Tcf1ΔHMG mutant mice, we use the CRISPR/Cas9 approach (12) to insert a stop codon in Tcf7 exon 8 after the codon encoding Val304. Briefly, a single guide RNA (sgRNA: GAGGGGTTTCTTGATGAC) was cloned into an sgRNA vector using OriGene’s gRNA Cloning Services (Rockville, MD), and was then used as a template to synthesize sgRNAs using the MEGAshortscript T7 Kit (Life Technologies). Cas9 mRNA was in vitro transcribed using the mMESSAGE mMACHINE T7 Ultra Kit (Life Technologies) with plasmid MLM3613 (Addgene #42251) as a template. Next, Cas9 mRNA (100 ng/μl) was mixed with sgRNA (50 ng/μl) and 100 ng/μl oligos (TCT CAT CTC CTT CAT GTA AAG CAT GAA CGC ATT GAG GGG TTT CTT GAT CTA GAC TGG CTT CTT AGC CTC CTT CTC TGC CTT GGG TTC TGC CTG TGT TTT), and microinjected into the cytoplasm of fertilized eggs collected from C57BL/6N inbred mice. The injected zygotes were surgically implanted into the oviducts of pseudo-pregnant foster mothers, and offspring was genotyped using PCR and DNA sequencing. All mice analyzed were 5–9 weeks of age, and both genders are used without randomization or blinding. All mouse experiments were performed under protocols approved by the Institutional Animal Use and Care Committees of the University of Iowa and the NHLBI.
Flow cytometry and apoptosis assay
Single-cell suspensions were prepared from the thymus and surface stained as described (7). The fluorochrome-conjugated antibodies were as follows: anti-TCRβ (H57-597), anti-CD8 (53–6.7), anti-CD4 (RM4–5), anti-CD44 (IM7), anti-CD62L (MEL-14), anti-CD25 (PX61.5) (all from eBiosciences). Cell apoptosis was detected with Annexin V-PE Apoptosis Detection Kit (BD Biosciences). Hoechst 33258 was used to exclude dead cells so as to specifically measure Annexin V+ apoptotic cells. Data were collected on a FACSVerse (BD Biosciences) and were analyzed with FlowJo software (TreeStar).
Immunoblotting
Cell lysates were prepared from the total thymocytes and probed with anti-Tcf1 (C46C7), anti-Lef1 (C18A7) (both from Cell Signaling Technology) or anti-β-actin (I-19; Santa Cruz Biotechnology). The Tcf1 antibody is produced by immunizing animals with a synthetic peptide corresponding to a region surrounding Leu158 of human Tcf1 protein, recognizes all Tcf1 isoforms and would detect Tcf1ΔHMG truncated protein if it were translated and stably expressed.
RNA-Seq and data analysis
DN3 thymocytes were sorted from control, p45−/−, or Vav-Cre+Tcf7fl/fl (Tcf1−/−) mice, and total RNA was extracted as described (7). cDNA libraries were prepared and sequenced on Illumina HiSeq2000 in single-read mode with the read length of 50 nt. The single end fastq reads were assessed for quality using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), adaptor and low-quality regions are trimmed using Trimmomatic. Reads were then aligned to the mm9 mouse reference sequence using Tophat2. Read counts were then obtained using FeatureCounts (13). Differential expression was determined using DESeq2 using a custom R script (14). Heatmaps are created using standard R tools and plotting the log2 regularized log transformation. The RNA-seq data are deposited at the Gene Expression Omnibus under accession number GSE92323 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?&acc=gse92323).
Results and Discussion
Tcf1 short isoforms adequately support thymic maturation transitions
Tcf1 proteins are encoded by the Tcf7 gene, with long isoforms produced from transcripts initiated at exon 1 and short isoforms from transcripts initiated at exon 3. Previously, we have generated a Tcf1-EGFP reporter mouse strain, where we inserted IRES-EGFP cassette into the first intron of Tcf7 gene (11). In front of the cassette, we placed an En2 splice acceptor which forced the splicing of exon 1 to this cassette instead of exon 2. Mice homozygous for the Tcf1-EGFP reporter alleles thus failed to produce Tcf1 long isoforms, p45 and p42, but retained the short isoforms, p33 and p30, albeit at lower levels than WT or heterozygotes (Fig. 1B); for simplicity, this strain is called p45−/− herein. Importantly, Lef1 proteins (in two isoforms) were not detectably affected in p45−/− thymocytes (Fig. 1B).
Loss of Tcf1 long isoforms reduced the total thymic cellularity by approximately 60% (Fig. 1C). By phenotypic analysis, p45−/− mice showed similar profiles of thymic maturation stages as WT or p45+/− mice, with a modest decrease in the DP frequency and concurrent mild increase in the frequency of CD4+ and CD8+ single positive thymocytes (Fig. 1D, S1A). Due to the reduction in total thymic cellularity, all thymic subsets in p45−/− mice were reduced in numbers (Fig. S1A). As for early stages, p45−/− DN thymocytes showed a modest but consistent decrease in the DN2 subset, in both frequency and numbers, whereas other DN subsets were similar among WT, p45+/− and p45−/− mice (Fig. 1D, S1B). For non-T lineage thymocytes, thymic B cells, dendritic cells, natural killer cells and group 1 innate lymphoid cells were detected at increased frequencies in p45−/− mice, but their absolute counts were not significantly altered (Fig. S1C–E). It is known that complete deletion of all Tcf1 isoforms diminishes total thymic cellularity to < 5 million cells and results in several incomplete developmental blocks at the DN1 and/or DN3 stages (15) (also see below). Our analysis of p45−/− mice suggest that Tcf1 short isoforms, even at reduced levels, are adequate to support the normal transition among thymocyte maturation stages. Nevertheless, Tcf1 long isoforms remain important for optimal thymic cellularity.
Tcf1 long isoforms are necessary for thymocyte survival
Germline targeting of Tcf1 has detrimental effects on T cell development (15, 16). We previously established a Tcf7fl/fl strain (10) and crossed with Vav-Cre to specifically ablate all Tcf1 isoforms in hematopoietic cells. The Vav-Cre+Tcf7fl/fl mice (Tcf1−/−) showed a phenocopy of the germline-targeted strain, exhibiting greatly reduced thymic cellularity (Fig. 2A), strong block at the DN stage, corresponding reduction in DP thymocytes, and skewed CD4+ to CD8+ T cell lineage (Fig. 2B, S2A). Within the DN population, Tcf1−/− mice showed loss of DN2 and accumulation of DN1 and DN3 thymocytes; and the Tcf1−/− DN1 cells exhibited an aberrant upregulation of CD25 (Fig. 2B, S2A), consistent with previous reports (15).
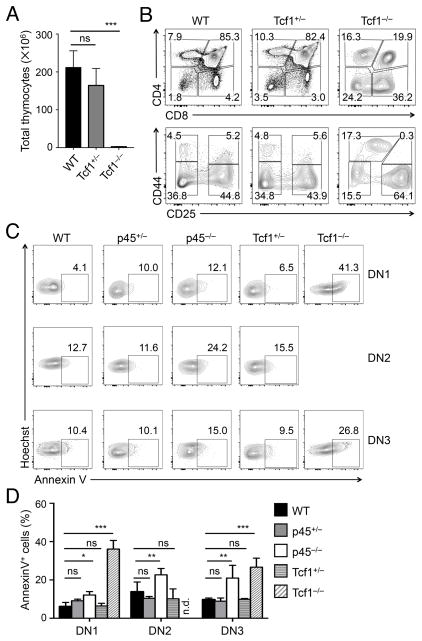
Figure 2. Loss of Tcf1 long isoforms modestly affects thymocyte survival.
(A)–(B) Impact of loss of Tcf1 all isoforms on T cell development. Thymocytes from WT, Tcf1+/− and Tcf1−/− mice were isolated and enumerated for total thymic cellularity (A, n ≥ 5 from at least 5 experiments). Lin− thymocytes were analyzed for DN, DP, CD4+ and CD8+ populations (top panels in B), and the Lin− DN cells were further analyzed for DN1-DN4 subsets (bottom in B). Contour plots are representative from 5 experiments (n ≥ 5).
(C)–(D) Early thymocyte survival in the absence of Tcf1 long isoforms or all isoforms. Lin− DN thymocytes from mice of indicated genotypes were surface-stained to identify DN1–DN3 subsets, followed by Hoechst 33258 and Annexin V staining. After excluding Hoechst+ dead cells, the viable cells were analyzed for Annexin V expression. The percentages of Annexin V+ cells are shown in representative contour plots (C), and cumulative data are in bar graphs (D) (n ≥ 3 from at least 3 experiments). n.d., not reliably detected. ns, not statistically significant; *, p<0.05; **, p<0.01; ***, p<0.001 by Student’s t test.
A prominent role of Tcf1 is to ensure survival of early thymocytes at the DN and DP stages (17, 18). In line with previous reports, ablation of all Tcf1 isoforms/proteins caused strong apoptosis of DP, CD4+ and mature CD8+ single positive thymocytes (Fig. S2B). In addition, all Tcf1−/− DN subsets were more apoptotic than controls (Fig. 2C–D). On the other hand, p45−/− thymocytes did show increase in apoptosis (except for DP thymocytes), but at much more modest levels compared with Tcf1−/− cells, whereas p45+/− and Tcf1+/− thymocyte survival was not detectably different from WT cells (Fig. 2C–D, S2B). Collectively, these data indicate that Tcf1 long isoforms remain important for supporting thymocyte survival, which at least partly account for the reduced numbers of thymocytes at each developmental stage.
Loss of Tcf1 long isoforms minimally affects the DN3 transcriptome
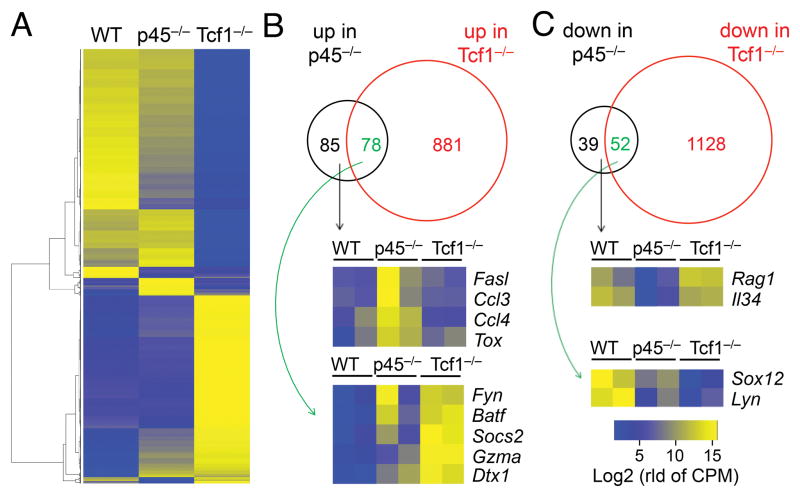
We next investigated how deficiency in Tcf1 long isoforms or all Tcf1 isoforms affected gene expression. Because Tcf1−/− mice showed a strong block at the DN3 stage, and both p45−/− and Tcf1−/− DN3 cells showed increased apoptosis, we focused on DN3 thymocytes for RNA-Seq analysis, which showed good reproducibility between two replicates (Fig. S2C). By a setting of ≥ 1.5 fold expression changes and adjusted p value < 0.05, we identified all differentially expressed genes (DEGs) in p45−/− versus control and Tcf1−/− versus control comparisons. Unsupervised clustering analysis revealed that Tcf1−/− cells caused strong transcriptomic changes, whereas only a small portion of such changes was observed in p45−/− DN3 thymocytes (Fig. 3A). Among the 1,044 upregulated genes, only 163 (15.6%) was upregulated in p45−/− DN3 cells (Fig. 3B). Functional annotation showed that Fasl (encoding Fas ligand) was among the 85 uniquely upregulated in p45−/− DN3 cells, and Gzma (encoding granzyme A) was induced in both p45−/− and Tcf1−/− DN3 cells (Fig. 3B). Another interesting gene is Dtx1 (encoding Deltex 1 downstream of the Notch signaling pathway), which was upregulated in both p45−/− and Tcf1−/− DN3 cells, consistent with a role of Tcf1 in terminating Notch signals in committed T lineage cells (19) (Fig. 3B). Among the 1,219 downregulated genes, only 91 (7.5%) were downregulated in p45−/− DN3 cells (Fig. 3C). Interestingly, Rag1 expression was among the 39 uniquely downregulated genes in p45−/− DN3 cells (Figure 3C), albeit the physiological impact remains to be determined. Overall, deficiency in Tcf1 long isoforms had limited impact on the DN3 transcriptome compared with loss of all Tcf1 isoforms/proteins. This observation held true when DEGs were identified using more stringent criteria (Fig. S2D). These data further suggest that the Tcf1 short isoforms are adequate to support most of Tcf1 target gene expression in developing thymocytes.
Figure 3. Loss of Tcf1 long isoforms minimally affects DN3 transcriptome.
(A) Heatmap showing differentially regulated genes (DEGs) in p45−/− and Tcf1−/− DN3 thymocytes. DN3 thymocytes were sort-purified from WT, p45−/−, or Tcf1−/− mice (each in two replicates) and analyzed by RNA-Seq. DEGs were identified using DESeq2 under permissive criteria (≥1.5 fold expression changes and p<0.05), and the average of two replicates in each genotype were used in cluster analysis.
(B)–(C) Venn diagrams showing upregulated (B) and downregulated (C) genes in p45−/− and Tcf1−/− DN3 thymocytes. Select genes of interest that are differentially expressed in p45−/− cells are shown in heatmaps. In color scale, “rld of CPM” stands for “regularized log transform of counts per million”.
Deletion of the HMG domain destabilizes Tcf1 protein
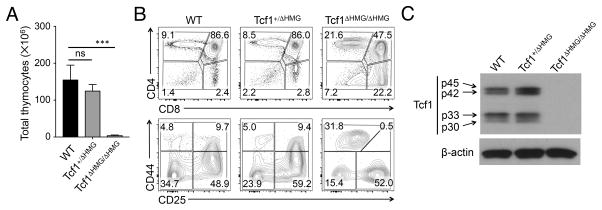
Recently we have demonstrated that Tcf1 has intrinsic HDAC activity, which is mapped between the β-catenin-binding domain and HMG binding domain (9). We aimed to determine if the Tcf1 HDAC activity can be dissociated from its DNA binding capacity. To this end, we used the CRISPR/Cas9 approach to insert a stop codon after Val304 in Tcf7 exon 8, thus leading to truncation of the HMG domain at the C-terminus (Fig. 1A, called Tcf1ΔHMG allele herein). Whereas the heterozygotes had similar thymic cellularity as littermate control, Tcf1ΔHMG/ΔHMG mice showed greatly reduced total thymocyte numbers (Fig. 4A). T cell development in Tcf1ΔHMG/ΔHMG mice showed a block at the DN stage, with concomitant reduction in DP cells and a skewed CD4+/CD8+ ratio (Fig. 4B, S2E). Furthermore, the DN cells in Tcf1ΔHMG/ΔHMG mice exhibited accumulation of DN1 and DN3 in addition to loss of DN2 cells (Fig. 4B, S2E). These profiles were remarkably similar to Tcf1−/− mice (compared with Fig. 2). By immunoblotting with a Tcf1 antibody that is generated with a peptide surrounding human Tcf1 Leu158 as an immunogen, we found that Tcf1ΔHMG/ΔHMG thymocytes were devoid of any Tcf1 protein (Fig. 4C). On the mRNA level, unexpectedly, Tcf1 transcripts were greatly reduced in Tcf1ΔHMG/ΔHMG thymocytes, and were even lower than those in Tcf1−/− cells (Fig. S2F). Therefore, the absence of all Tcf1 proteins in Tcf1ΔHMG/ΔHMG mice might be ascribed to at least two possibilities: 1) insertion of a stop codon in exon 8 destabilized the mRNA, and 2) any protein translated from the diminished amounts of mutant transcripts was not stable. This model does not achieve the intended dissociation of Tcf1 HDAC activity from DNA binding, but nonetheless suggests that unnatural mutation(s) in the HMG domain strongly impairs the stability of Tcf1 mRNA and/or protein.
Figure 4. Impact of truncating the HMG domain in Tcf1 on T cell development.
(A) Thymic cellularity in WT, Tcf1+/ΔHMG, and Tcf1ΔHMG/ΔHMG mice (n ≥ 4 from 4 experiments). ns, not statistically significant; ***, p<0.001 by Student’s t test.
(B) Thymic maturation stages. Lin− thymocytes were analyzed for DN, DP, CD4+ and CD8+ populations (top panels in B), and the Lin− DN cells were further analyzed for DN1-DN4 subsets (bottom in B). Contour plots are representative from 4 experiments (n ≥ 4).
(C) Detection of Tcf1 by immunoblotting. Cell lysates were extracted from total thymocytes of control, Tcf1+/ΔHMG, and Tcf1ΔHMG/ΔHMG mice, and immunoblotted with anti-Tcf1 or β-actin antibody. Representative data from ≥ 3 experiments are shown.
One unique feature of Tcf1 long isoforms is their ability to interact with the co-activator β-catenin. Forced expression of β-catenin extends thymocyte survival (20). However, conditional targeting of β-catenin neither enhanced thymocyte death (21) nor strongly diminished thymic cellularity (22). One possible reason is that the existing β-catenin-targeted mouse strains retained a truncated form of β-catenin protein in hematopoietic cells (23), and the truncated protein might be partially functional. γ-catenin is a homolog of β-catenin and interacts with Tcf4 at a region right next to the β-catenin binding domain (24). γ-catenin is expressed in T cells and its interaction with Tcf1 should be limited to the long isoforms (22). Therefore, our new targeting approach in p45−/− mice ensured complete abrogation of Tcf1 interaction with either β- or γ-catenin. Our data thus suggest that one major function of Tcf1-β-catenin interaction is to control thymocyte life span and sustain the output of mature T cells from the thymus. It should be noted that in addition to a complete loss of Tcf1 long isoforms, p45−/− thymocytes expressed Tcf1 short isoform proteins at a reduced level. The reduced amount of Tcf1 short isoforms could have contributed to the modest increase in apoptosis of p45−/− thymocytes, and further suggest that Tcf1-β-catenin interaction may also have a role in the proposed positive feed-forward regulation of Tcf1 gene transcription (6). Nonetheless, the latter possibility remains an integral part of Tcf1-β-catenin complex-mediated regulation of thymocyte survival.
Tcf1 short isoforms have been considered to be dominant negative regulators or non-functional (6, 25). It was previously showed that a Tcf1 p45 transgene can partly restore thymocyte numbers and rectify T cell developmental defects in Tcf1-deficient mice, but a Tcf1 p33 transgene failed to do so (17). In this study, instead of complementing Tcf1 deficiency using a transgene, we perturbed generation of Tcf1 long isoforms in the Tcf7 gene locus. Since Tcf1 can potentially regulate its own expression via a positive feed-forward loop (6), the expression of Tcf1 short isoforms appeared to be lower in p45−/− thymocytes (Fig. 1B). In spite of the diminished expression, the Tcf1 isoforms were adequate to support the developing thymocytes to traverse through each step without causing detectable blocks. In addition, Tcf1 short isoforms adequately support normal expression of majority (>85%) of Tcf1-dependent genes in DN3 thymocytes. Our findings thus suggest that the Tcf1 short isoforms are essential regulators of T cell maturation in the thymus rather than being dominant negative. Since Tcf1 has critical roles in regulating mature CD4+ and CD8+ T cell responses (4), it would be of interest to further investigate if Tcf1 short isoforms are adequate in directing generation of central memory CD8+ T cell and/or differentiation of follicular helper CD4+ T cells.
Supplementary Material
Acknowledgments
We thank the University of Iowa Flow Cytometry Core facility (J. Fishbaugh and H. Vignes) for cell sorting, Igor Antoshechkin (California Institute of Technology) for RNA-Seq.
This study is supported by grants from the NIH (AI112579, AI115149, AI119160, and AI121080 to H.-H. X.) and the US Department of Veteran Affairs (I01 BX002903 to H.-H. X.). C.L. is supported by the intramural research program of NHLBI, NIH.
Footnotes
The authors declare no conflict of interests.
References
- 1.Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nature reviews Immunology. 2014;14:529–545. doi: 10.1038/nri3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- 3.Staal FJ, Sen JM. The canonical Wnt signaling pathway plays an important role in lymphopoiesis and hematopoiesis. Eur J Immunol. 2008;38:1788–1794. doi: 10.1002/eji.200738118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue HH, Zhao DM. Regulation of mature T cell responses by the Wnt signaling pathway. Annals of the New York Academy of Sciences. 2012;1247:16–33. doi: 10.1111/j.1749-6632.2011.06302.x. [DOI] [PubMed] [Google Scholar]
- 5.Germar K, Dose M, Konstantinou T, Zhang J, Wang H, Lobry C, Arnett KL, Blacklow SC, Aifantis I, Aster JC, Gounari F. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A. 2011;108:20060–20065. doi: 10.1073/pnas.1110230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber BN, Chi AW, Chavez A, Yashiro-Ohtani Y, Yang Q, Shestova O, Bhandoola A. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu S, Zhou X, Steinke FC, Liu C, Chen SC, Zagorodna O, Jing X, Yokota Y, Meyerholz DK, Mullighan CG, Knudson CM, Zhao DM, Xue HH. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity. 2012;37:813–826. doi: 10.1016/j.immuni.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinke FC, Xue HH. From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunol Res. 2014;59:45–55. doi: 10.1007/s12026-014-8545-9. [DOI] [PubMed] [Google Scholar]
- 9.Xing S, Li F, Zeng Z, Zhao Y, Yu S, Shan Q, Li Y, Phillips FC, Maina PK, Qi HH, Liu C, Zhu J, Pope RM, Musselman CA, Zeng C, Peng W, Xue HH. Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nature immunology. 2016;17:695–703. doi: 10.1038/ni.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinke FC, Yu S, Zhou X, He B, Yang W, Zhou B, Kawamoto H, Zhu J, Tan K, Xue HH. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nature immunology. 2014;15:646–656. doi: 10.1038/ni.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Q, Li F, Harly C, Xing S, Ye L, Xia X, Wang H, Wang X, Yu S, Zhou X, Cam M, Xue HH, Bhandoola A. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nature immunology. 2015;16:1044–1050. doi: 10.1038/ni.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schilham MW, Wilson A, Moerer P, Benaissa-Trouw BJ, Cumano A, Clevers HC. Critical involvement of Tcf-1 in expansion of thymocytes. J Immunol. 1998;161:3984–3991. [PubMed] [Google Scholar]
- 16.Verbeek S, Izon D, Hofhuis F, Robanus-Maandag E, te Riele H, van de Wetering M, Oosterwegel M, Wilson A, MacDonald HR, Clevers H. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature. 1995;374:70–74. doi: 10.1038/374070a0. [DOI] [PubMed] [Google Scholar]
- 17.Ioannidis V, Beermann F, Clevers H, Held W. The beta-catenin--TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nature immunology. 2001;2:691–697. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 18.Wang R, Xie H, Huang Z, Ma J, Fang X, Ding Y, Sun Z. T cell factor 1 regulates thymocyte survival via a RORgammat-dependent pathway. J Immunol. 2011;187:5964–5973. doi: 10.4049/jimmunol.1101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu S, Xue HH. TCF-1 mediates repression of Notch pathway in T lineage-committed early thymocytes. Blood. 2013;121:4008–4009. doi: 10.1182/blood-2013-01-477349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Z, Xie H, Ioannidis V, Held W, Clevers H, Sadim MS, Sun Z. Transcriptional regulation of CD4 gene expression by T cell factor-1/beta-catenin pathway. J Immunol. 2006;176:4880–4887. doi: 10.4049/jimmunol.176.8.4880. [DOI] [PubMed] [Google Scholar]
- 21.Xu Y, Banerjee D, Huelsken J, Birchmeier W, Sen JM. Deletion of beta-catenin impairs T cell development. Nature immunology. 2003;4:1177–1182. doi: 10.1038/ni1008. [DOI] [PubMed] [Google Scholar]
- 22.Jeannet G, Boudousquie C, Gardiol N, Kang J, Huelsken J, Held W. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci U S A. 2010;107:9777–9782. doi: 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeannet G, Scheller M, Scarpellino L, Duboux S, Gardiol N, Back J, Kuttler F, Malanchi I, Birchmeier W, Leutz A, Huelsken J, Held W. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood. 2008;111:142–149. doi: 10.1182/blood-2007-07-102558. [DOI] [PubMed] [Google Scholar]
- 24.Miravet S, Piedra J, Miro F, Itarte E, Garcia de Herreros A, Dunach M. The transcriptional factor Tcf-4 contains different binding sites for beta-catenin and plakoglobin. J Biol Chem. 2002;277:1884–1891. doi: 10.1074/jbc.M110248200. [DOI] [PubMed] [Google Scholar]
- 25.Yu Q, Sharma A, Oh SY, Moon HG, Hossain MZ, Salay TM, Leeds KE, Du H, Wu B, Waterman ML, Zhu Z, Sen JM. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nature immunology. 2009;10:992–999. doi: 10.1038/ni.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.