Abstract
The transfer of an acetyl group from acetyl-CoA to an acceptor amine is a ubiquitous biochemical transformation catalyzed by Gcn5-related N-acetyltransferases (GNATs). Although it is established that the reaction proceeds through a sequential ordered mechanism, the role of the acetyl group in driving the ordered formation of binary and ternary complexes remains elusive. Herein, we show that CoA and acetyl-CoA alter the conformation of the substrate binding site of an arylalkylamine N-acetyltransferase (AANAT) to facilitate interaction with acceptor substrates. However, it is the presence of the acetyl group within the catalytic funnel that triggers high affinity binding. Acetyl group occupancy is relayed through a conserved salt bridge between the P-loop and the acceptor binding site, and is manifested as differential dynamics in the CoA and acetyl-CoA-bound states. The capacity of the acetyl group carried by an acceptor to promote its tight binding even in the absence of CoA, but also its mutually exclusive position to the acetyl group of acetyl-CoA underscore its importance in coordinating the progression of the catalytic cycle.
Introduction
GNAT enzymes catalyze the acetylation of a wide range of primary amine acceptors from histone and ribosomal proteins, to small molecules like serotonin, dopamine and aminoglycoside antibiotics [1–3]. Thus, they are central regulators of diverse cellular processes such as gene transcription [4], time and seasonal adaptation [5–7] and metabolism [8, 9]. Structural analysis of GNAT family members guided primarily by crystallography has revealed a conserved core domain, made up of a mixed β sheet and flanking α helices, which contains four signature motifs (A-D) [1–3]. Strands β4 and β5, from motifs A and B, respectively, form the only parallel stretch of the mixed β sheet. A conserved β bulge in strand β4 results in partial separation of the β4- β5 stretch, creating a characteristic V-shaped catalytic cavity that interacts with the pantetheine arm and the acetyl group of acetyl-CoA. The amide nitrogen of the residue downstream of the β bulge forms a conserved hydrogen bond with the carbonyl of the acetyl group. In contrast to the pantetheine arm and the acetyl group which reach deep in the aforementioned cavity, the 3’-phosphate ADP is exposed at the surface of the protein, resulting in an overall bent conformation for acetyl-CoA. The highly conserved Q/R-X-X-G-X-G/A consensus sequence found at the loop downstream of β4 (P-loop) coordinates the pyrophosphate moiety of the 3’-phosphate ADP via hydrogen bonding of main-chain atoms. Although the mode of acetyl-CoA binding is highly conserved, the mode of interaction of GNATs with acceptor substrates is highly divergent due to their substantially different chemical nature [1–3].
Arylalkylamine N-acetyltransferases (AANATs) catalyze the transfer of an acetyl group to a diverse gamut of arylalkylamines, such as indolethylamines and phenylethylamines [10, 11]. In mammals, a single AANAT gene is expressed in the pineal gland and in the retina, where it is involved in the melatonin pathway [12], and retinal neurotransmission and detoxification [13, 14], respectively. AANAT, also known as serotonin N-acetyltransferase (SNAT), is the penultimate enzyme in the melatonin biosynthesis pathway and it catalyzes the N-acetylation of serotonin using acetyl-CoA as acetyl donor to produce N-acetylserotonin [15]. On the other hand, insects have multiple copies of AANATs that function beyond the regulation of the circadian system, particularly in neurotransmitter metabolism [16, 17] and sclerotization and pigmentation [18–21]. Structural studies on three AANATs identified in Aedes aegypti, revealed that the three enzymes share high structural homology and contain the signature motifs of the GNAT superfamily, but display distinct binding specificities with respect to the acceptor substrate [22]. Accumulating evidence from kinetic analyses of AANATs, has shown that they follow an ordered, ternary complex mechanism, where acetyl-CoA binding facilitates the interaction with the acceptors [15, 23–25]. Indeed, the structure of the prototypical AANAT, serotonin N-acetyltransferase, from Ovies aries (oaAANAT) in the free state and when in complex with a bisubstrate analog supports this mechanism, as the acceptor binding site is fully formed only after acetyl-CoA-induced conformational changes [26, 27]. These changes involve rearrangement of loops over the acetyl-CoA binding cavity as well as a notable loop-to-helix transition at the C-terminal end of helix α1. However, significant deviations from this model are observed. Comparison of the crystal structures of the highly homologous dopamine N-acetyltransferase from Drosophila melanogaster (DAT) solved in the free and acetyl-CoA-bound forms, revealed that the acetyl-CoA induced loop-to-helix transition at the C-terminal end of helix α1 observed for oaAANAT preexists in the free form of DAT [28]. Similarly, comparison of the crystal structure of free Aedes aegypti AANAT2 (aaAANAT2) to that of free or acetyl-CoA bound DAT shows the same distribution and three dimensional arrangement of secondary structure elements, suggesting that, again, the conformational changes observed for oaAANAT preexist in the free form of aaAANAT2 [22, 28]. Thus, even for closely related GNAT homologues as the AANAT subfamily members, a common, ternary complex kinetic mechanism can be driven by distinct underlying molecular events. In addition, more divergent models, such as global unfolded-to-folded transitions, are encountered when GNATs other than AANATs are considered [29, 30].
In the current study we used a combination of nuclear magnetic resonance spectroscopy (NMR), isothermal titration calorimetry (ITC) and homology modelling to follow the formation of binary and ternary complexes during the catalytic cycle of bmAANAT3, a small GNAT from Bombyx mori involved in the acetylation of a broad range of amines. Our data reveal that the acetyl group plays a central role in coordinating the ordered progression of AANATs through the binding and release events encountered during the ternary complex mechanism, by regulating the conformational properties of both, the donor and acceptor binding sites.
Materials and methods
Expression and purification of bmAANAT3
Full-length bmAANAT3 cloned into a pET28a vector (thrombin cleavage site and kanamycin resistance) was expressed in BL21(DE3) competent E. Coli cells (New England Biolabs) as a fusion protein with an N-terminal His-tag. Cell growth was performed at 37°C and protein expression was induced at an OD600 = 0.5 by the addition of 1 mM IPTG, for 5 hours. Cells were harvested by centrifugation at 4°C, resuspended in 50 mM Tris-HCl, pH = 8.0, 20 mM imidazole, 500 mM NaCl, 1 mM Phenylmethylsulfonyl fluoride and 5 mM β-mercaptoethanol and lysed by sonication. Cell debris were removed by centrifugation at 35,000 x g (4°C). The clarified lysate was loaded on a Ni2+ affinity column, washed with 10 column volumes of cell lysis buffer and eluted with 400 mM imidazole in the same buffer. Fusion bmAANAT3 was further purified through a Superdex-75 gel-filtration column using a 50 mM Tris-HCl pH = 8.0, 150 mM NaCl, 0.5 mM EDTA and 5 mM β-mercaptoethanol buffer. The purification polyhistidine tag was removed by incubating bmAANAT3 with biotinylated thrombin (2U/mg). 15N- or 13C/15N-labeled bmAANAT3 was prepared using the same expression and purification protocol, except that cells were grown in M9 media containing 15N-NH4Cl and 13C-glucose as the sole source of nitrogen and carbon, respectively.
NMR spectroscopy
All NMR spectra were acquired in 20 mM Tris, 100 mM KCl, 5 mM DTT, 0.5 mM EDTA and 7% D2O, at 30°C, using Varian direct drive 600 and 800 MHz spectrometers equipped with a cryogenic probe. {1H}-15N NOE experiments were acquired in an interleaved manner and with a recycling delay of 5 s. Spectra were processed using NMRPipe [31] and analyzed using Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). bmAANAT3 titrations with acetyl-CoA and CoA were performed at 30°C and at a protein concentration of 250 μM. The titration with acetyl-CoA progressed to 750 μM. Two titrations were performed with CoA, one in small increaments that progressed up to 1.75 mM for extracting the Kd of the interaction, and one in large increments that progressed to 8 M and was used to achieve > 99.6% saturation. The assignment of free bmAANAT3 has been reported previously and can be found at the BMRB under the accession number 26962 [32]. The assignment of the 15N-HSQC of bmAANAT3 in the CoA-bound state was performed by following the signal trajectories during the course of the titration. For the assignment of the 15N-HSQC of bmAANAT3 in the acetyl-CoA-bound state, the spectrum of the CoA-bound state was used. Titrations with substrate acceptors in the presence or absence of acetyl-CoA or CoA were performed at 22°C at a concentration of 120–150 μM, and progressed close to saturation.
Combined chemical shift changes were calculated using the following expression:
Dissociation constants were determined by global fitting of several residues to [33]:
where Δδobs is the incremental chemical shift change at each point of the titration, Δδmax is the maximum chemical shift change (at saturation), [P]t and [L]t are the total protein and ligand concentrations at each point of the titration and Kd is dissociation constant.
Isothermal titration calorimetry
ITC experiments were performed on a VP-ITC calorimeter (GE Healthcare) at 30 and 22°C for cofactors and acceptor substrates, respectively. bmAANAT3 was dialyzed against 20 mM Tris, 100 mM NaCl, 0.5 mM EDTA and 1 mM tris(2-carboxyethyl)phosphine and degassed. bmAANAT3 concentration was ~40 μM and the ligands at a 10- to 100-fold excess. Titrations included an initial 0.2-μl injection and were completed by 10–12 injections, with a 4 min time interval between each injection. The data were processed using Origin 7.0 (OriginLab Corporation) with the point of the initial injection excluded and using an one-site binding model.
Homology model
The structure of bmAANAT3 was generated with SWISS-MODEL [34] using the crystal structure of the arylalkylamine N-Acetyltransferase 2 from the yellow fever mosquito, Aedes aegypti (aaAANAT2, PDB: 4FD6) [22] as a template. aaAANAT2 is found in the same cluster of a phylogenetic tree of insect aaNATs as bmAANAT3 and exhibits 47% sequence identity and 62% sequence similarity to bmAANAT3.
Results and discussion
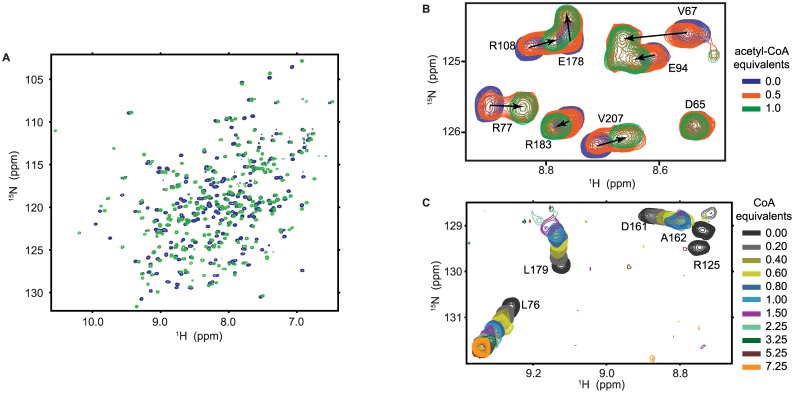
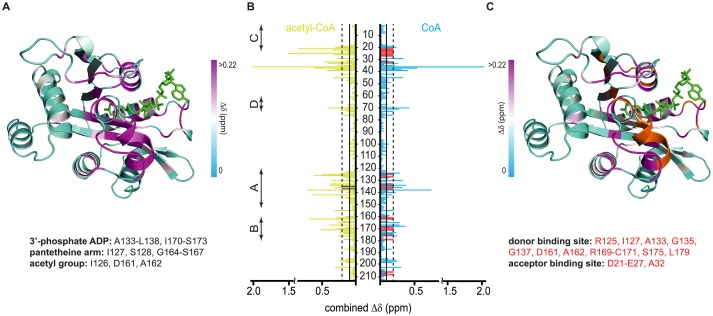
To characterize the mode of donor binding, we followed the titration of bmAANAT3 with acetyl-CoA and CoA by NMR. Addition of acetyl-CoA to 15N-labeled bmAANAT3 results in significant chemical shift perturbation (CSP) of a large number of signals (Figs 1A, 2A and 2B), suggesting that acetyl-CoA binding does not affect only residues in the vicinity of the binding site but induces conformational changes to distal regions. Changes in the 15N-HSQC spectrum of bmAANAT3 as a function of added acetyl-CoA occur on the slow-exchange regime of the NMR timescale, causing progressive disappearance of the signals of the free-state and the concomitant appearance of a new set of signals corresponding to the bound-state (Fig 1B). To discriminate between CSPs reporting direct binding from those reporting induced conformational changes, we took advantage of the highly conserved mode of acetyl-CoA binding to GNAT family members. Thus, we used the structure of DAT (20% identity and 50% similarity) to identify CSPs caused by direct acetyl-CoA binding. As expected, significant CSPs are observed for residues within the conserved motifs A and B (Fig 2A and 2B). Therefore, prominent CSPs that report induced conformational changes outside the acetyl-CoA binding site, span the end of motif C and the downstream sequence prior to motif D, and include amino acids L19-H42 (Fig 2A and 2B). Notably, this region corresponds to a segment of oaAANAT’s acceptor binding site which undergoes a loop-to-helix transition upon bisubstrate binding to allow for a high affinity interaction with the acceptor [26, 27].
Fig 1. Association of bmAANAT3 with acetyl-CoA and CoA followed by NMR.
(A) 15N-HSQC of bmAANAT3 in the free (blue) and acetyl-CoA-bound state (green). The very large number of signals affected by acetyl-CoA indicates that chemical shift perturbations do not solely report cofactor binding at the catalytic funnel, but also induced conformational changes to distal regions of the enzyme. (B) Expanded region of the 15N-HSQC of bmAANAT3 in the free-state (blue), after the addition of 0.5 (orange) or 1.0 (green) equivalents of acetyl-CoA, showing the concomitant disappearance and appearance of the signals for the free and bound states. (C) Expanded region from the NMR titration course of bmAANAT3 with CoA highlighting signals from residues that either do not exhibit any line broadening (L76) or broaden beyond detection at different points of the titration (R125, D161, A162 and L179).
Fig 2. Cofactor-mediated conformational remodeling of bmAANAT3 monitored by NMR.
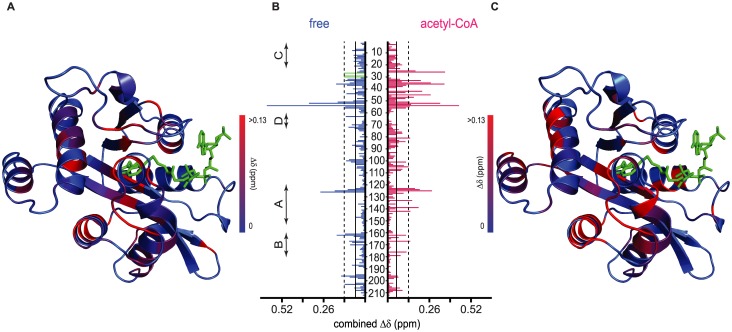
(A) acetyl-CoA induced CSPs mapped on the model of bmAANAT3. (B) CSPs as a function of primary sequence for the interaction of bmAANAT3 with acetyl-CoA (green) and CoA (cyan). CSPs greater than the mean or one SD above the mean are marked by continuous and broken lines, respectively. The conserved motifs of the GNAT superfamily are indicated on the left. Black bars in the acetyl-CoA portion of the plot mark G135 and G137 which are absent in the free state of the enzyme, but tentatively assigned in the bound state. Red bars in the CoA portion of the plot mark residues that become broadened beyond detection in the CoA-bound state. (C) CoA induced CSPs mapped on the model of bmAANAT3. In (A) and (C) the white region in the three-color gradient corresponds to the mean CSP, while the magenta region to greater than mean + σ. Residues highlighted in orange in (C) highlight residues that broaden beyond detection in the presence of CoA. The position of acetyl-CoA is highlighted in green, after aligning the bmAANAT3 model with the structure of acetyl-CoA-bound DAT. Residues that experience significant CSPs for acetyl-CoA and CoA are listed in (A), while, signals that broaden (beyond detection) upon CoA binding are listed in (C).
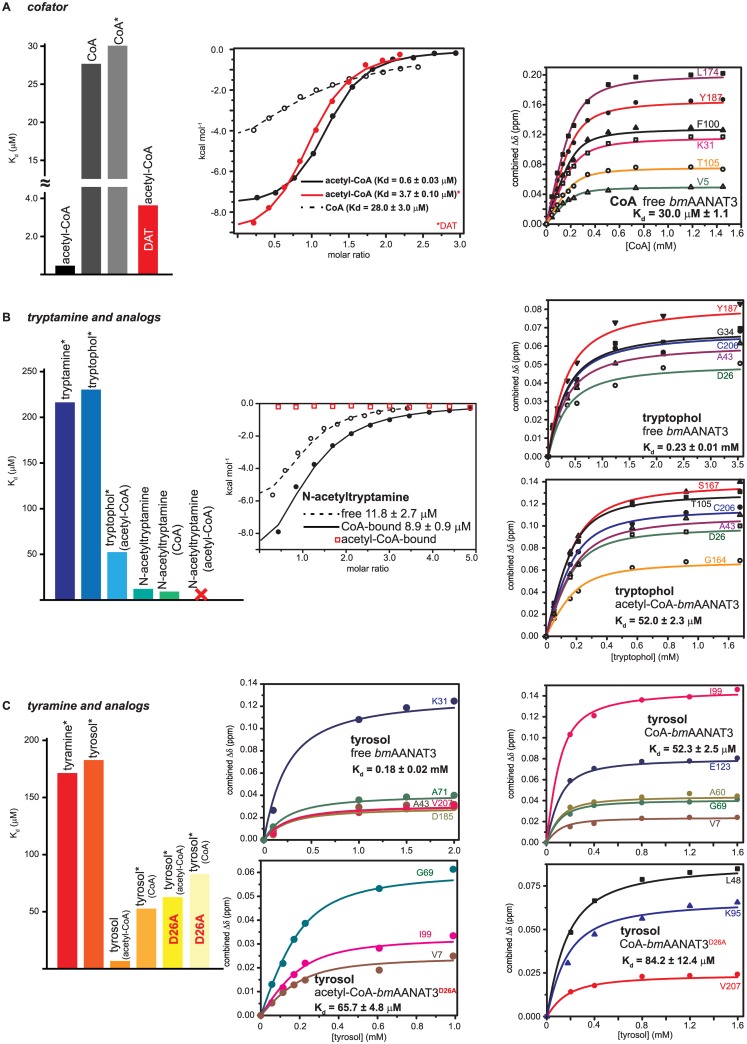
To elucidate how conformational changes at this region are linked to the formation of ternary complexes with acceptors, we generated a homology model of bmAANAT3 (S1A and S1B Fig). Overall, two major conformational variations are observed between the predicted bmAANAT3 structure and the prototypical structure of free oaAANAT, both of which occur at the acceptor binding site. First, helix α1 preexists as a long helical element, whereas in oaAANAT, helix α1 extends towards its C-terminal end only upon bisubstrate analog binding (S1C Fig). Second, residues 28–33 of bmAANAT3, that correspond to the C-terminus of oaAANAT’s loop1, form a short helical “plug” on top of the catalytic funnel (S1B Fig). In agreement with the NMR derived secondary structure (S1A Fig), heteronuclear {1H}-15N NOEs recorded for this region (S1D Fig) indicate a rigid backbone and further support the presence of a preformed long helix α1 and a helical plug in the unliganded state of bmAANAT3. These observations explain the significantly higher Kd of oaAANAT for acetyl-CoA (~242 μM) [35] as compared to that of bmAANAT3 (0.66 ± 0.03 μM, Fig 3) and account for the three-fold difference in KM [15, 36]. Hence, the observed CSPs for the L19-H42 segment do not report a loop-to-helix transition, but rather a subtle structural reorganization of helix α1 and the plug over the catalytic funnel. This observation is in line with the proposed ordered mechanism for bmAANAT3 and other GNATs, but highlights significant variations within the AANAT class. In this respect, bmAANAT3 resembles DAT both in terms of the thermodynamics of acetyl-CoA binding (Kd = 3.7 μM, Table 1 and Fig 3) and the steady-state kinetic constants [24, 28, 36].
Fig 3. An overview of dissociation constants measured in this study.
For those complexes that gave poor c-values in the ITC, Kd values were determined by NMR (marked by *). A summary of Kd values is presented in Table 1. (A) Determination of Kd values for the interaction of free bmAANAT3 with acetyl-CoA (black) and CoA (gray), and DAT with acetyl-CoA (red). The isotherms presented in the middle panel correspond to interaction of bmAANAT3 with acetyl-CoA and CoA (black) and to the interaction of DAT with acetyl-CoA (red). NMR was used to confirm the Kd of bmAANAT3 for CoA (right panel). (B) Determination of Kd values for the interaction of different cofactor-liganded states of bmAANAT3 with tryptamine and tryptamine analogs. The presence of acetyl-CoA in the catalytic funnel leads to an increase in the affinity for tryptophol, while an even more substantial increase is observed when an acetylated derivative of tryptamine (N-acetyltryptamine) is utilized. Binding of N-acetyltryptamine to bmAANAT3 is not sensitive to the presence of CoA, but it is abolished in the presence of acetyl-CoA. The isotherms presented in the middle panel correspond to the interaction of bmAANAT3 with N-acetyltryptamine in the free- or CoA-bound states (black) and in the acetyl-CoA-bound state (red). NMR was used to measure the Kd for the interaction of free- and acetyl-CoA-bound bmAANAT3 with tryptophol (right panel). (C) Determination of Kd values for the interaction of different cofactor-liganded states of bmAANAT3 and variants (D26A) with tyramine and tyramine analogs. The presence of CoA in the catalytic funnel leads to an increase in the affinity for tyrosol, while an even more substantial increase is observed in the presence of acetyl-CoA. The D26A mutation renders tyrosol binding insensitive to acetyl-group occupancy of the catalytic center, in a manner that tyrosol affinity of acetyl-CoA-bound bmAANAT3D26A is similar to that of CoA-bound bmAANAT3D26A or CoA-bound bmAANAT3. NMR was used to measure the Kd for the interaction of tyrosol with free- or CoA-bound bmAANAT3, and for acetyl-CoA- or CoA-bound bmAANAT3D26A.
Table 1. Dissociation constants measured for bmAANAT3, in μM.
| cofactor | acetyl-CoA | CoA | ||||
| 0.66 ± 0.03 3.70 ± 0.10a |
28 ± 3.0 | |||||
| tryptamine analogs | tryptamine | tryptophol | Tryptophol (acetyl-CoA) | N-acetyltryptamine | N-acetyltryptamine (CoA) | N-acetyltryptamine (acetyl-CoA) |
| 219 ± 8 | 230 ± 12 | 52 ± 2.40 | 11.8 ± 2.7 | 8.9 ± 0.9 | no binding | |
| tyramine analogs | tyramine | tyrosol | tyrosol (acetyl-CoA) | tyrosol (CoA) | tyrosolb(acetyl-CoA) | tyrosolb(CoA) |
| 169 ± 17 | 180 ± 21 | 6.45 ± 0.52 | 52.3 ± 2.5 | 65.7 ± 4.8 | 84.2 ± 12.4 | |
When bmAANAT3 was titrated with an acceptor in the presence of a cofactor (acetyl-Coa or CoA), the cofactor is shown in parenthesis.
a refers to the Kd measured for DAT under identical conditions to bmAANAT3.
b refers to the Kd measured for the bmAANAT3 variant D26A.
To further characterize the role of the acetyl group in the progression of the catalytic cycle from an activated acetyl-donor to the CoA product, we investigated the association of bmAANAT3 with CoA. Consistent with its higher Kd measured by ITC (28.00 ± 3.00 μM, Fig 3A), incremental additions of CoA cause a gradual shift of the signals from the free to the fully saturated state (Fig 1C). Evidently, signals that experience significant CSPs cluster to the same regions as for the titration with acetyl-CoA, and include residues from motif A and motif B (Fig 2B and 2C). This is in agreement with the available structural information for GNATs, where a conserved mode of binding is observed for the two cofactors. Nevertheless, CoA produces distinct conformational changes to bmAANAT3 as, for two sets of signals, a change in chemical shift is also associated with significant and incremental signal broadening, leading to complete signal loss (Figs 1C and 2B). Surprisingly, one set maps to the acetyl-CoA binding site and the second to the segment spanning the end of helix α1 and the plug, which partly comprises the acceptor binding site (Fig 2C), indicating that in the presence of CoA the cofactor and acceptor binding sites undergo exchange at a μs-ms timescale. The origin of exchange broadening is not chemical exchange due to CoA binding, since bmAANAT3 is fully saturated with the cofactor under these conditions (>99.6%). When 15N-HSQC signals from residues that do not experience line broadening but lie in the vicinity of the acetyl-CoA binding site were used to determine the Kd, there was a very good agreement between NMR and ITC (Fig 3A), confirming saturation with CoA. Therefore broadening is caused by conformational heterogeneity of bmAANAT3 in the CoA-bound state, suggesting that the acetyl group of acetyl-CoA (and its absence thereof) modulates the dynamic properties of the enzyme, both locally, throughout the donor binding site, and remotely at the acceptor binding site. Analysis of available crystal structures of GNATs in the presence of either of the two cofactors reveals that, overall, CoA exhibits higher conformational variability as compared to acetyl-CoA. In this respect, the B-factors of CoA atoms are consistently higher than the B-factors of the corresponding atoms of acetyl-CoA (S2 Fig). In addition, there are several structures of CoA-bound GNATs for which poor or no electron density is observed for the adenosine [37–39], pantothenic [40] and β-mercaptoethylamine [37, 40, 41] moieties, or for which multiple conformations are observed, particularly for the pantetheine [42] and β-mercaptoethylamine [43–45] moieties. The inherent mobility observed for CoA segments that are in contact with the enzymes suggests that the CoA-induced conformational heterogeneity observed for bmAANAT3 is a common characteristic among GNATs. Thus, although the ΔΔG for CoA and acetyl-CoA binding to bmAANAT3 (~-10.0 kJ/mol) indicates a simple additive contribution of the acetyl group to the overall binding affinity of acetyl-CoA as compared to CoA, in the absence of the acetyl group, motions are propagated throughout the cofactor binding site and ultimately to the substrate binding site.
The conformational heterogeneity of the acceptor binding site in the CoA-bound form suggests that acetyl-group occupancy in the catalytic funnel may control the sequential mechanism. To test this hypothesis, we characterized binary and ternary complexes of bmAANAT3 with acceptor substrates. To prevent acetyl group transfer in ternary complexes, we used tyrosol and tryptophol as model non-acceptor analogs of tyramine and tryptamine, respectively [15]. Both ligands give poor ITC isotherms with low c-values and thus NMR titrations were used to determine Kd values for binary complexes (Fig 3B and 3C). In agreement with an ordered kinetic mechanism, saturation occurs only in the presence of high acceptor excess (Kd values of 180 ± 21 and 230 ± 12 μM for tyrosol and tryptophol, respectively), indicating a weak acceptor-bmAANAT3 interaction. Mapping the observed CSPs on the model of bmAANAT3 (Fig 4) reveals that the most prominent changes occur at the following structural elements (i) the β-sheet, at a position of the catalytic funnel that coincides with the tryptamine moiety of the bisubstrate in the crystal structure of oaAANAT, (ii) the plug and the downstream loop, (iii) helix α2, (iv) the insect-specific helical insert and (v) the loop connecting the last two β-strands on top of the catalytic funnel. In the homology model of bmAANAT3 generated using the crystal structure of aaAANAT2, a large number of hydrophobic sidechains from these regions protrude towards the catalytic funnel forming a hydrophobic cage, which presumably encapsulates the arylalkyl acceptor (S3 Fig). A similar cluster of hydrophobic residues is also observed in the crystal structures of DAT in both the free- and acetyl-CoA-bound states [28]. In addition, a set of residues within the plug (E27, L29 and N30) exhibit significant line broadening or disappear at near saturating substrate concentrations (Fig 4). Thus, although the acceptor interacts weakly with the hydrophobic part of the catalytic funnel even in the context of binary acceptor complexes, the dynamic nature of the plug does not allow for a high affinity interaction.
Fig 4. Assembly of bmAANAT3 complexes with acceptor substrates monitored by NMR.
(A) Mapping of tryptophol induced CSPs to free bmAANAT3. The conserved motifs of the GNAT superfamily are indicated on the left. (B) CSPs as a function of primary sequence for the interaction of tryptophol with free- (blue) and acetyl-CoA-bound bmAANAT3 (red). CSPs greater than the mean or one SD above the mean are marked by continuous and broken lines, respectively. Green, open bars in the free-state mark residues that disappear in the substrate-bound state. (C) Mapping of tryptophol induced CSPs to acetyl-CoA bound bmAANAT3. The bisubstrate, CoA-S-acetyltryptamine (green), was modelled by aligning bmAANAT3 model to the structure of oaAANAT.
On the other hand, in the presence of acetyl-CoA the interaction with acceptors is significantly enhanced (Kd values of 6.45 ± 0.52 and 52 ± 2.40 μM, for tyrosol and tryptophol, respectively, Fig 3B and 3C). To identify the molecular origin of this strong, positive binding cooperativity (ΔΔG ~ -8 kJ/mol), we looked for differences in the NMR titrations of substrates in free and acetyl-CoA-bound bmAANAT3. The CSP patterns obtained from the two titrations are very similar (Fig 4), suggesting that the mode of substrate binding at the two states is similar. However, in the presence of acetyl-CoA, residues at the C-terminal end of helix α1 that either contribute directly to the substrate binding site (F22) or lie in the vicinity of the 3’-phosphate ADP moiety binding site (A25, D26), experience significantly higher chemical shift changes. Furthermore, the residues from the plug for which we have available assignment in the acetyl-CoA-bound state, do not show any line broadening. These results suggest that acetyl-CoA binding selectively stabilizes a conformation of the plug which is competent for a strong interaction with the substrate.
The differential effect of CoA and acetyl-CoA binding on the conformational properties of the acceptor binding site, implies that acetyl group occupancy of the catalytic cavity may act as an effector that modulates the affinity of the enzyme for acceptors during the ordered catalytic cycle. To test this hypothesis we studied the interaction of acceptors with CoA-bound bmAANAT3. Indeed, the affinity of tyrosol is significantly reduced as compared to the acetyl-CoA-bound state of bmAANAT3 (Kd of 52.3 ± 2.5 μM), although it is still higher than for the free enzyme. Hence, the extent of the binding cooperativity is highly dependent on the presence of the acetyl group on the cofactor and binding of CoA does not shift bmAANAT3 to an acceptor binding competent state to the same extent as acetyl-CoA does. Next, we utilized N-acetyltryptamine as an acetylated acceptor to test binding to free bmAANAT3 and to a binary complex with CoA. N-acetyltryptamine shows a 20-fold increase in affinity for the free protein as compared to the non-acetylated substrate (Kd = 11.8 ± 2.7 μM), which is only marginally improved in the presence of CoA (Kd = 8.9 ± 0.9 μM) (Fig 3). It must be noted that this large difference in Kd is not due to an erroneous comparison between the alcohol mimic and the natural amine, as the affinity of tyramine and tryptamine is comparable to that tyrosol and tryptophol for free bmAANAT3 (Fig 3). Thus, acetyl group occupancy of the catalytic site has a profound effect on the binding properties of the acceptor binding site even when the acetyl group is carried by the acceptor. Importantly, the position of the acetyl group of the acetylated acceptor is mutually exclusive to the position of the acetyl group of acetyl-CoA, as N-acetyltryptamine does not bind to bmAANAT3-acetyl-CoA (Fig 3B). This is expected to provide a mechanism for productive dissociation of the acetylated acceptor upon acetyl-CoA binding during the progression to a new catalytic cycle. In support of this hypothesis, comparison of available GNAT structures in the presence of acetyl-CoA and bisubstrates or acetylated acceptors, shows that the acetyl group occupies the same space and in some cases with a conserved hydrogen bond network [46–50].
As the most significant difference between the NMR titrations of bmAANAT3 with acetyl-CoA and CoA was the conformational heterogeneity of the C-terminus of helix α1 and the plug, we hypothesized that residues at the interface between this region and the P-loop encompass the molecular switch that couples acetyl group occupancy to the acceptor binding site. Comparison of the structure of DAT in the free and acetyl-CoA-bound state reveals that a salt-bridge between D46 and R153 is only formed in the presence of acetyl-CoA, while a salt bridge from residues of the same structural elements of oaAANAT (E54 and R131) is observed in the presence of a bisubstrate, but not in the free-state. To test our hypothesis, we mutated the corresponding residue, D26, of bmAANAT3 to alanine (S4 Fig). Our rationale was that disruption of a potential interaction with R134 from the P-loop in the acetyl-CoA-bound state would affect the substrate binding properties. Indeed, tyrosol binding to acetyl-CoA-bound bmAANAT3D26A, shows a 10-fold drop in affinity (Kd = 65.7 ± 4.8 μM), to the level of CoA-bound bmAANAT3 or CoA-bound bmAANAT3D26A (Kd = 84.2 ± 12.4 μM) (Fig 3C). Hence, this salt bridge comprises the structural switch that mediates the observed positive cooperativity by sensing acetyl group occupancy at the catalytic site, since when broken, acetyl group occupancy of the catalytic funnel is not relayed to the substrate binding site. This observation is also in agreement with the 20-fold higher Km,app observed for DATR153A for tyramine as compared to wild type DAT [24].
Conclusions
In summary, we find that the sequential progression between binary and ternary complexes formed during the catalytic cycle of an N-acetyltransferase is coordinated by the acetyl group occupancy in the catalytic funnel. When carried by acetyl-CoA, the acetyl group shifts the conformational ensemble of the acceptor binding site to a high affinity competent state, while when carried by the acceptor it allows for displacement of the acetylated acceptor upon acetyl-CoA binding. Our NMR data also show that the C-terminus of helix α1 of bmAANAT3 preexists in a helical conformation in the free-state of the enzyme and thus allows for a high affinity interaction with the cofactor, acetyl-CoA. Similar observations were made for the free-states of DAT and aaAANAT2, suggesting that this is a general mechanism for insect acetyltransferases [22, 28]. However, the highly conserved sequential mechanism observed for other GNATs suggests that this model may apply to other members of the superfamily.
Supporting information
(A) Primary sequence alignment of bmAANAT3 and of the template, aaAANAT2, generated using Jalview and color-coded according to amino acid properties. Invariant residues are marked with an asterisk. A comparison between the secondary structure elements predicted by homology modeling (noted as predicted SS) to those determined by analysis of HN, N, Cα, Cβ and C’ backbone chemical shifts of bmAANAT3 (noted as NMR SS) is shown under the alignment. Strands and helices are represented by arrows and cylinders, respectively, and named according to the GNAT nomenclature. Conserved motifs of the GNAT superfamily and amino acid numbering is shown above the alignment. (B) Orthogonal views of the homology model of bmAANAT3, where the conserved GNAT family fold is colored in red. The insect specific helical insert and the helical plug, are shown in cyan. The bisubstrate analog CoA-S-Acetyltryptamine (shown in green) was used to illustrate the position of acetyl-donor and -acceptor ligands in the catalytic funnel, after structural alignment of the model with the crystal structure of the binary complex of oaAANAT with a bisubstrate analog (PDBID: 1CJW). (C) Overlay of the bmAANAT3’s homology model (colored as in panel B) to the structures of oaAANAT in the free (yellow) and bisubstrate-bound state (blue). The bisubstrate analog is shown in green and loop1, which undergoes a disorder-to-helix transition upon bisubstrate binding is shown in gray. In the extended conformation, loop1 partially occupies binding of acetyl-CoA, while in the helical conformation it completes the substrate binding site. These differences may account for the higher Kd of oaAANAT for acetyl-CoA as well as for the higher KM observed, compared to bmAANAT3. (D) {1H}-15N heteronuclear NOEs, which are sensitive to ps-ns timescale dynamics of backbone atoms, reveal that the plug region (colored in magenta) acquires a helical conformation in solution.
(EPS)
A comparison between the B-Factors of acetyl-CoA and CoA atoms obtained from those GNAT crystal structures that have been determined with both acetyl-CoA and CoA. The PDB IDs for each set of structures is provided.
(EPS)
Two orthogonal views of the acceptor substrate binding cavity of the modelled bmAANAT3 structure. The tryptamine and CoA moieties of the bisubstrate analog CoA-S-acetyltryptamine are colored pink and yellow respectively, while hydrophobic residues that exhibit significant chemical shift perturbation are shown as green spheres.
(EPS)
Cartoon diagram of bmAANAT3 model highlighting the salt bridge between D26 located at the end of helix α1 and R134 located at the P-loop.
(EPS)
Acknowledgments
We thank Dr. Gary Daughdrill for access to the ITC instrumentation and Morgan Parries for assistance with sample preparations.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
I.G. would like to thank the University of South Florida for internal funds. D.J.M. acknowledges the National Institute of Health for support (R15-GM107864). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Salah Ud-Din AI, Tikhomirova A, Roujeinikova A. Structure and Functional Diversity of GCN5-Related N-Acetyltransferases (GNAT). Int J Mol Sci. 2016;17(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. 10.1146/annurev.biophys.29.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vetting MW, SdC LP, Yu M, Hegde SS, Magnet S, Roderick SL, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433(1):212–26. 10.1016/j.abb.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 4.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389(6649):349–52. 10.1038/38664 [DOI] [PubMed] [Google Scholar]
- 5.Arendt J. Significance of melatonin in humans. Nato Adv Sci Inst Se. 1995;277:165–71. [Google Scholar]
- 6.Klein DC, Roseboom PH, Coon SL. New light is shining on the melatonin rhythm enzyme: the first postcloning view. Trends Endocrinol Metab. 1996;7(3):106–12. [DOI] [PubMed] [Google Scholar]
- 7.Klein DC, Weller JL. Indole metabolism in the pineal gland: a circadian rhythm in N-acetyltransferase. Science. 1970;169(3950):1093–5. [DOI] [PubMed] [Google Scholar]
- 8.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–4. 10.1126/science.1179689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327(5968):1004–7. 10.1126/science.1179687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coon SL, Roseboom PH, Baler R, Weller JL, Namboodiri MA, Koonin EV, et al. Pineal serotonin N-acetyltransferase: expression cloning and molecular analysis. Science. 1995;270(5242):1681–3. [DOI] [PubMed] [Google Scholar]
- 11.Khalil EM, De Angelis J, Cole PA. Indoleamine analogs as probes of the substrate selectivity and catalytic mechanism of serotonin N-acetyltransferase. J Biol Chem. 1998;273(46):30321–7. [DOI] [PubMed] [Google Scholar]
- 12.Klein DC, Coon SL, Roseboom PH, Weller JL, Bernard M, Gastel JA, et al. The melatonin rhythm-generating enzyme: molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog Horm Res. 1997;52:307–57; discussion 57–8. [PubMed] [Google Scholar]
- 13.Zilberman-Peled B, Ron B, Gross A, Finberg JP, Gothilf Y. A possible new role for fish retinal serotonin-N-acetyltransferase-1 (AANAT1): Dopamine metabolism. Brain Res. 2006;1073–1074:220–8. [DOI] [PubMed] [Google Scholar]
- 14.Klein DC. Evolution of the vertebrate pineal gland: the AANAT hypothesis. Chronobiol Int. 2006;23(1–2):5–20. 10.1080/07420520500545839 [DOI] [PubMed] [Google Scholar]
- 15.De Angelis J, Gastel J, Klein DC, Cole PA. Kinetic analysis of the catalytic mechanism of serotonin N-acetyltransferase (EC 2.3.1.87). J Biol Chem. 1998;273(5):3045–50. [DOI] [PubMed] [Google Scholar]
- 16.Brodbeck D, Amherd R, Callaerts P, Hintermann E, Meyer UA, Affolter M. Molecular and biochemical characterization of the aaNAT1 (Dat) locus in Drosophila melanogaster: differential expression of two gene products. DNA Cell Biol. 1998;17(7):621–33. 10.1089/dna.1998.17.621 [DOI] [PubMed] [Google Scholar]
- 17.Sloley BD. Metabolism of monoamines in invertebrates: the relative importance of monoamine oxidase in different phyla. Neurotoxicology. 2004;25(1–2):175–83. 10.1016/S0161-813X(03)00096-2 [DOI] [PubMed] [Google Scholar]
- 18.Dai FY, Qiao L, Tong XL, Cao C, Chen P, Chen J, et al. Mutations of an arylalkylamine-N-acetyltransferase, Bm-iAANAT, are responsible for silkworm melanism mutant. J Biol Chem. 2010;285(25):19553–60. 10.1074/jbc.M109.096743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer KJ, Kanost MR, Hopkins TL, Jiang HB, Zhu YC, Xu RD, et al. Oxidative conjugation of catechols with proteins in insect skeletal systems. Tetrahedron. 2001;57(2):385–92. [Google Scholar]
- 20.Mun S, Noh MY, Dittmer NT, Muthukrishnan S, Kramer KJ, Kanost MR, et al. Cuticular protein with a low complexity sequence becomes cross-linked during insect cuticle sclerotization and is required for the adult molt. Sci Rep. 2015;5:10484 10.1038/srep10484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osanai-Futahashi M, Ohde T, Hirata J, Uchino K, Futahashi R, Tamura T, et al. A visible dominant marker for insect transgenesis. Nat Commun. 2012;3:1295 10.1038/ncomms2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han Q, Robinson H, Ding H, Christensen BM, Li J. Evolution of insect arylalkylamine N-acetyltransferases: structural evidence from the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A. 2012;109(29):11669–74. 10.1073/pnas.1206828109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dempsey DR, Carpenter AM, Ospina SR, Merkler DJ. Probing the chemical mechanism and critical regulatory amino acid residues of Drosophila melanogaster arylalkylamine N-acyltransferase like 2. Insect Biochem Mol Biol. 2015;66:1–12. 10.1016/j.ibmb.2015.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dempsey DR, Jeffries KA, Bond JD, Carpenter AM, Rodriguez-Ospina S, Breydo L, et al. Mechanistic and structural analysis of Drosophila melanogaster arylalkylamine N-acetyltransferases. Biochemistry. 2014;53(49):7777–93. 10.1021/bi5006078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dempsey DR, Jeffries KA, Handa S, Carpenter AM, Rodriguez-Ospina S, Breydo L, et al. Mechanistic and Structural Analysis of a Drosophila melanogaster Enzyme, Arylalkylamine N-Acetyltransferase Like 7, an Enzyme That Catalyzes the Formation of N-Acetylarylalkylamides and N-Acetylhistamine. Biochemistry. 2015;54(16):2644–58. 10.1021/acs.biochem.5b00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickman AB, Klein DC, Dyda F. Melatonin biosynthesis: the structure of serotonin N-acetyltransferase at 2.5 A resolution suggests a catalytic mechanism. Mol Cell. 1999;3(1):23–32. [DOI] [PubMed] [Google Scholar]
- 27.Hickman AB, Namboodiri MA, Klein DC, Dyda F. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 A resolution with a bisubstrate analog. Cell. 1999;97(3):361–9. [DOI] [PubMed] [Google Scholar]
- 28.Cheng KC, Liao JN, Lyu PC. Crystal structure of the dopamine N-acetyltransferase-acetyl-CoA complex provides insights into the catalytic mechanism. Biochem J. 2012;446(3):395–404. 10.1042/BJ20120520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freiburger LA, Baettig OM, Sprules T, Berghuis AM, Auclair K, Mittermaier AK. Competing allosteric mechanisms modulate substrate binding in a dimeric enzyme. Nat Struct Mol Biol. 2011;18(3):288–94. 10.1038/nsmb.1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norris AL, Serpersu EH. Ligand promiscuity through the eyes of the aminoglycoside N3 acetyltransferase IIa. Protein Sci. 2013;22(7):916–28. 10.1002/pro.2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6(3):277–93. [DOI] [PubMed] [Google Scholar]
- 32.Aboalroub AA, Zhang Z, Keramisanou D, Gelis I. Backbone resonance assignment of an insect arylalkylamine N-acetyltransferase from Bombyx mori reveals conformational heterogeneity. Biomol NMR Assign. 2017;11(1):105–9. 10.1007/s12104-017-9729-8 [DOI] [PubMed] [Google Scholar]
- 33.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog Nucl Magn Reson Spectrosc. 2013;73:1–16. 10.1016/j.pnmrs.2013.02.001 [DOI] [PubMed] [Google Scholar]
- 34.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014;42(Web Server issue):W252–8. 10.1093/nar/gku340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation. Cell. 2001;105(2):257–67. [DOI] [PubMed] [Google Scholar]
- 36.Battistini MR, O’Flynn, B. G., Shoji, C., Dillashaw, J.E., and Merkler, D.J. Bm-iAANAT3: Expression and Characterization of a Novel Arylalkylamine N-acyltransferases from Bombyx mori submitted. [DOI] [PMC free article] [PubMed]
- 37.Dorfmueller HC, Fang W, Rao FV, Blair DE, Attrill H, van Aalten DM. Structural and biochemical characterization of a trapped coenzyme A adduct of Caenorhabditis elegans glucosamine-6-phosphate N-acetyltransferase 1. Acta Crystallogr D Biol Crystallogr. 2012;68(Pt 8):1019–29. 10.1107/S0907444912019592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oda K, Matoba Y, Noda M, Kumagai T, Sugiyama M. Catalytic mechanism of bleomycin N-acetyltransferase proposed on the basis of its crystal structure. J Biol Chem. 2010;285(2):1446–56. 10.1074/jbc.M109.022277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung MN, Rangarajan E, Munger C, Nadeau G, Sulea T, Matte A. Crystal structure of TDP-fucosamine acetyltransferase (WecD) from Escherichia coli, an enzyme required for enterobacterial common antigen synthesis. J Bacteriol. 2006;188(15):5606–17. 10.1128/JB.00306-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burk DL, Ghuman N, Wybenga-Groot LE, Berghuis AM. X-ray structure of the AAC(6')-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members. Protein Sci. 2003;12(3):426–37. 10.1110/ps.0233503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurtado-Guerrero R, Raimi OG, Min J, Zeng H, Vallius L, Shepherd S, et al. Structural and kinetic differences between human and Aspergillus fumigatus D-glucosamine-6-phosphate N-acetyltransferase. Biochem J. 2008;415(2):217–23. 10.1042/BJ20081000 [DOI] [PubMed] [Google Scholar]
- 42.Bewley MC, Graziano V, Jiang J, Matz E, Studier FW, Pegg AE, et al. Structures of wild-type and mutant human spermidine/spermine N1-acetyltransferase, a potential therapeutic drug target. Proc Natl Acad Sci U S A. 2006;103(7):2063–8. 10.1073/pnas.0511008103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen JY, Liu L, Cao CL, Li MJ, Tan K, Yang X, et al. Structure and function of human Naa60 (NatF), a Golgi-localized bi-functional acetyltransferase. Sci Rep. 2016;6:31425 10.1038/srep31425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filippova EV, Shuvalova L, Minasov G, Kiryukhina O, Zhang Y, Clancy S, et al. Crystal structure of the novel PaiA N-acetyltransferase from Thermoplasma acidophilum involved in the negative control of sporulation and degradative enzyme production. Proteins. 2011;79(8):2566–77. 10.1002/prot.23062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maksimoska J, Segura-Pena D, Cole PA, Marmorstein R. Structure of the p300 histone acetyltransferase bound to acetyl-coenzyme A and its analogues. Biochemistry. 2014;53(21):3415–22. 10.1021/bi500380f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liszczak G, Goldberg JM, Foyn H, Petersson EJ, Arnesen T, Marmorstein R. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat Struct Mol Biol. 2013;20(9):1098–105. 10.1038/nsmb.2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hegde SS, Chandler J, Vetting MW, Yu M, Blanchard JS. Mechanistic and structural analysis of human spermidine/spermine N1-acetyltransferase. Biochemistry. 2007;46(24):7187–95. 10.1021/bi700256z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peneff C, Mengin-Lecreulx D, Bourne Y. The crystal structures of Apo and complexed Saccharomyces cerevisiae GNA1 shed light on the catalytic mechanism of an amino-sugar N-acetyltransferase. J Biol Chem. 2001;276(19):16328–34. 10.1074/jbc.M009988200 [DOI] [PubMed] [Google Scholar]
- 49.Majorek KA, Kuhn ML, Chruszcz M, Anderson WF, Minor W. Structural, functional, and inhibition studies of a Gcn5-related N-acetyltransferase (GNAT) superfamily protein PA4794: a new C-terminal lysine protein acetyltransferase from pseudomonas aeruginosa. J Biol Chem. 2013;288(42):30223–35. 10.1074/jbc.M113.501353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vetting MW, Bareich DC, Yu M, Blanchard JS. Crystal structure of RimI from Salmonella typhimurium LT2, the GNAT responsible for N(alpha)-acetylation of ribosomal protein S18. Protein Sci. 2008;17(10):1781–90. 10.1110/ps.035899.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Primary sequence alignment of bmAANAT3 and of the template, aaAANAT2, generated using Jalview and color-coded according to amino acid properties. Invariant residues are marked with an asterisk. A comparison between the secondary structure elements predicted by homology modeling (noted as predicted SS) to those determined by analysis of HN, N, Cα, Cβ and C’ backbone chemical shifts of bmAANAT3 (noted as NMR SS) is shown under the alignment. Strands and helices are represented by arrows and cylinders, respectively, and named according to the GNAT nomenclature. Conserved motifs of the GNAT superfamily and amino acid numbering is shown above the alignment. (B) Orthogonal views of the homology model of bmAANAT3, where the conserved GNAT family fold is colored in red. The insect specific helical insert and the helical plug, are shown in cyan. The bisubstrate analog CoA-S-Acetyltryptamine (shown in green) was used to illustrate the position of acetyl-donor and -acceptor ligands in the catalytic funnel, after structural alignment of the model with the crystal structure of the binary complex of oaAANAT with a bisubstrate analog (PDBID: 1CJW). (C) Overlay of the bmAANAT3’s homology model (colored as in panel B) to the structures of oaAANAT in the free (yellow) and bisubstrate-bound state (blue). The bisubstrate analog is shown in green and loop1, which undergoes a disorder-to-helix transition upon bisubstrate binding is shown in gray. In the extended conformation, loop1 partially occupies binding of acetyl-CoA, while in the helical conformation it completes the substrate binding site. These differences may account for the higher Kd of oaAANAT for acetyl-CoA as well as for the higher KM observed, compared to bmAANAT3. (D) {1H}-15N heteronuclear NOEs, which are sensitive to ps-ns timescale dynamics of backbone atoms, reveal that the plug region (colored in magenta) acquires a helical conformation in solution.
(EPS)
A comparison between the B-Factors of acetyl-CoA and CoA atoms obtained from those GNAT crystal structures that have been determined with both acetyl-CoA and CoA. The PDB IDs for each set of structures is provided.
(EPS)
Two orthogonal views of the acceptor substrate binding cavity of the modelled bmAANAT3 structure. The tryptamine and CoA moieties of the bisubstrate analog CoA-S-acetyltryptamine are colored pink and yellow respectively, while hydrophobic residues that exhibit significant chemical shift perturbation are shown as green spheres.
(EPS)
Cartoon diagram of bmAANAT3 model highlighting the salt bridge between D26 located at the end of helix α1 and R134 located at the P-loop.
(EPS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.