Abstract
The Pacific oyster, Crassostrea gigas, was introduced to Europe for aquaculture purposes, and has had a rapid and unforeseen northward expansion in northern Europe. The recent dramatic increase in number of C. gigas populations along the species’ northern distribution limit has questioned the efficiency of Skagerrak as a dispersal barrier for transport and survival of larvae. We investigated the genetic connectivity and possible spreading patterns between Pacific oyster populations on the southern Norwegian coast (4 localities) and Swedish and Danish populations by means of DNA microsatellite analysis of adult oysters, and by simulating larvae drift. In the simulations we used a 3D oceanographic model to explore the influence of recent climate change (1990–2010) on development, survival, and successful spreading of Danish and Swedish Pacific oyster larvae to Norwegian coastal waters. The simulations indicated adequate temperature conditions for development, survival, and settlement of larvae across the Skagerrak in warm years since 2000. However, microsatellite genotyping revealed genetic differences between the Norwegian populations, and between the Norwegian populations and the Swedish and Danish populations, the latter two populations being more similar. This patchwork pattern of genetic dissimilarity among the Norwegian populations points towards multiple local introduction routes rather than the commonly assumed unidirectional entry of larvae drifted from Denmark and Sweden. Alternative origins of introduction and implications for management, such as forecasting and possible mitigation actions, are discussed.
Introduction
The Pacific oyster, Crassostrea gigas, was repeatedly introduced to Europe for aquaculture purposes in the second half of the 20th Century (see [1] for a review), and has established wild populations in the Black Sea, the Mediterranean Sea and along the Atlantic European coasts, to Scandinavia [2]. Temperature conditions north of France were erroneously thought inappropriate for natural reproduction, and the species was actively introduced for aquaculture purposes in the Netherlands [3], Germany [4], Denmark [5], Sweden [6], and Norway [7]. Climate changes and broader eco-physiological tolerances of the species than first supposed [8] are proposed to be the cause of the recently rapid northward expansion of the species from the Wadden Sea to Sweden [3]. From 2007 the species was found in high densities along the Swedish coast [9] and it has been hypothesized that the species has been introduced to Sweden from Denmark through spreading of larvae with coastal currents [10, 11]. In 2005 the species was observed for the first time in the wild on the northern side of the Skagerrak coast, in Norwegian coastal areas [9, 12], presumably due to larva drifted from “parent populations on the Continent” [11]. The repeated introduction of the species to several countries in northern Europe, and multiple documented accounts of its spreading from locations where it has been introduced [13], makes it important to elucidate the processes that cause the observed rapid expansion of the species distribution along its northern distribution limit in Europe. Two mechanisms of introduction have been hypothesized for this region: 1) natural dispersal of larvae across country borders and 2) post-introduction dispersal from local populations founded through other introduction pathways such as i.e. aquaculture, shipping (ballast water, hull fouling), and live trade (live seafood, bait). Moreover, the changing sea temperature conditions within this region [14] may have caused temporal differences in the dispersal abilities of C. gigas along its northern distribution limit.
Crassostrea gigas was introduced to Europe from source populations in either Japan or Canada, which are shown to be genetically similar [1]. However, recent DNA studies of C. gigas in Europe identify two genetically distinct groups, a northern and a southern. Genetic studies of samples from the south of France to Sweden [15], the south of France to the Wadden Sea [16], samples within the Wadden Sea [17] and samples within the British Isles [1], all indicate two main genetic groups. The two groups seem to be separated by one border in the Wadden Sea and another border within southern UK (Fig 1). The southern group (France, southwestern England, The Netherlands, southern Wadden Sea) with high genetic diversity, was genetically similar to populations from Canada and Japan, whereas the northern group (northern Wadden Sea, Germany, Denmark, Sweden, Ireland and eastern England), with low genetic diversity [1, 15], has, to our knowledge, no genetically matching populations elsewhere in the world. This is consistent with the history of multiple introductions of the species from Canada and Japan to southern Europe, forming a genetically diverse southern group, whereas most of the introductions we are aware of, to the countries belonging to the northern group, come from the UK (see Fig 1 and references). Based on this, the UK appears to be the key source for the Pacific oyster populations within the northern group.
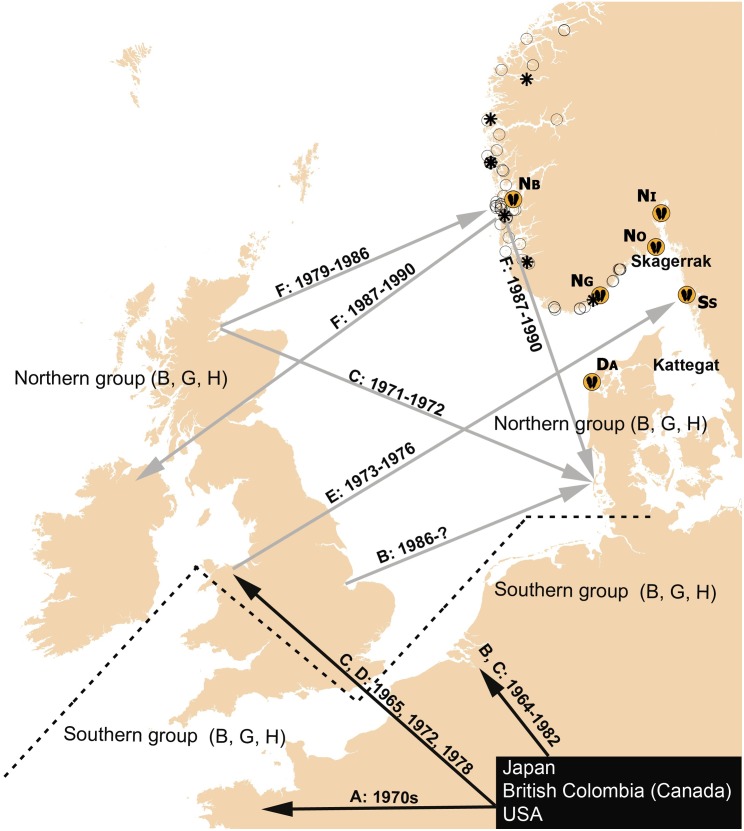
Fig 1. Sampling overview and simplified introduction history.
Crassostrea gigas aquaculture introduction pathways in Europe (A[23], B[17], C[3], D[24], E[9] & F[7]) and the genetic differenciation boundary between a documented southern and northern genetic group delineated by a dotted line (B[17], G[15] & H[1]). The six C. gigas collection sites used in this study are indicated by the oyster symbole (See Table 1 for details). For Norway, valid and withdrawn aquaculture licenses for Ostrea edulis (http://www.fiskeridir.no/register/akvareg/?m=utl_lok&s=1; 20. May 2014) and C. gigas (Directorate of Fisheries) are indicated by open circles and stars, respectively. The map is produced using ESRIs GIS software ArcMap v 10.4.1 (www.esri.com), and the country dataset GISCO NUTS 2010.
Temperature is a critical factor for C. gigas larvae development and survival [18]. Maturity and spawning in summer demand temperature above 16–20°C for several days [19, 20]. In warmer water the larvae grow faster [21], the planktonic phase is shorter and a higher proportion of the larvae are successfully metamorphosed [22]. Recent global warming has likely increased the chance of spawning, recruitment, and survival in established populations at the outer edge of its present distribution, accelerating the species proliferation rate and spread to new areas.
Since feral populations of C. gigas were first observed in Norwegian waters in 2005 [9, 12], the number of known Pacific oyster localities has increased dramatically and the species is at present observed at 435 sites along the Norwegian coastline in Skagerrak and the North Sea (http://artskart.artsdatabanken.no/default.aspx, downloaded 26. February 2017. Some of the 516 observations (81) were duplicates, reported at the same site). This rapid expansion of the species in northern Europe has raised a concern for further uncontrolled northwards expansion through massive larvae supply across Skagerrak from southern countries. This would cause severe problems for any mitigation actions against further northward spread of the species. In this study we used genetic analysis to investigate the origin of 4 established C. gigas populations along the Norwegian coast. We expect that if the main origin of the Norwegian populations is larvae dispersal from Swedish and Danish populations, then these populations would be genetically similar. Alternatively, if the origin is from post-introduction dispersal from local populations founded through other origins (e.g. aquaculture, shipping, or live trade), we expect these populations to be genetically different. We also examined what influence recent climate change and temperature conditions might have on dispersal of oyster larvae from Swedish and Danish populations, using a 3D oceanographic model, modelled sea water temperature for the region for selected years, and known temperature thresholds for larval development, spawning, and survival.
Materials and methods
Sampling and DNA preparation
A total of 262 individuals of Crassostrea gigas oysters sampled from six Scandinavian populations in 2010 (Table 1 and Fig 1), were analyzed. As the Pacific oyster is an invasive species considered to be a threat to marine ecosystems in all three countries, permission was not required before sampling. The field studies did not involve endangered or protected species. Shell length varied between 6.5 and 18 cm with an average length of 10.9 cm among sampling locations, implying that all individuals were adults and probably from multiple generations. Oyster mantle samples were collected in 15 ml cap tubes and preserved in ethanol 96% (the ethanol was changed once). We used a new simple DNA preparation protocol without any purification step [25]. Briefly, individual tissue samples were washed in deionized water and about 5 mg mantle tissue was transferred to 100 μl 0.3% SDS with 2 μl proteinase K, incubated at 65°C for 10 min followed by 98°C inactivation for 2 min. The lysates were further diluted 10−2 in Tris EDTA buffer (Fluka, Chemie GmbH, Switzerland) prior to performing PCR.
Table 1. Sample information.
| Country | Region | Code | Date sampled | Sample size | WGS84 DD Lat. / Long. |
|---|---|---|---|---|---|
| Norway | Bergen Espevik | NB | June 2012 | 12 | 59.9159 / 5.64759 |
| Norway | South Norway Grimstad | NG | December 2011 | 50 | 58.292 / 8.517 |
| Norway | Inner Oslo fjord Sætrepollen | NI | May 2012 | 50 | 59.68449 / 10.53466 |
| Norway | Outer Oslo fjord Hui | NO | May 2012 | 50 | 59.11554 / 10.35547 |
| Sweden | Smalsund | SS | August 2011 | 50 | 58.30262 / 11.36911 |
| Denmark | Agger Tange | DA | August 2011 | 50 | 56.75923 / 8.24432 |
Country, region, sample code and size, and geographic position (decimal degrees) of the sampling sites.
Microsatellite genotyping
PCR amplifications were performed using a CFX96 thermocycler (BioRad, Hercules, CA, USA) in 10 μl reaction volume containing 5 μl iProof mastermix (Bio-Rad), primers (Eurofins MWG, Ebersberg, Germany) were used in two optimized (see Results) multiplex reactions with primer concentrations as indicated in Table 2; Bovine Serum Albumin (BSA) 0.1 μg/μl (VWR, 2 μg/μl) and 2.5 μl sample. Reaction volume was completed with sterile deionised water. Multiplex PCR amplifications were optimized and carried out under the following conditions: a denaturing step for 1 min at 98°C, followed by 35 cycles of 98°C for 15 s, 55°C for 30 s and 72°C for 30 s. Multiplex PCR plates, each with either 4 or 2 different dyes (Table 2), were mixed and diluted by transferring 5 μl from each well to a plate prefilled with 100 μl deionized water per well. From this dilution plate 1.2 μl per sample was transferred to the run plate prefilled with 10 μl HiDi Formamide (Applied Biosystems, Foster City, CA, USA) and 40% strength orange standard (MCLAB, San Francisco, CA, USA). PCR product sizes were determined using a 3730XL DNA analyzer (Applied Biosystems) and scored using GeneMapper software version 4.0 (Applied Biosystems).
Table 2. Genotyping of 262 Crassostrea gigas individuals using six microsatellite loci in two multiplex PCR.
| Locus | Repeat motif | F & R primer sequences 5’-3’ | Dye | Conc. (μM) | Size range (bp) | NA | N | HO | HE | Primer Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| L10* | AG |
GGTCAATTCAAAGTCAATTTCCC CATGTTTTCCCTTGACTGATCC |
FAM | 0.15 | 109–173 | 29 | 262 | 0.86 | 0.90 | [51] |
| Cgsili44* | (AG)7AAA(GA)4 / 25 |
TGGCATTTCATGGTTAATTT TGTTGTATGAAATGTCGGAA |
ATTO 565 | 0.075 | 337–363 | 12 | 259 | 0.60 | 0.83 | [52] |
| HSat1 & HSat2R (AMY)** | TC |
ACCGGTATTGCCCGAGTTACAA AGTTAGGCATCCCCCATTGTTC |
FAM | 0.1 | 196–238 | 27 | 262 | 0.83 | 0.89 | [53, 54] |
| L48** | GA |
TCAAACCATCTGCTCGTCTACG TCCGAAAATCCAGGAATACCGG |
Yakima Yellow | 0.2 | 96–158 | 26 | 262 | 0.86 | 0.90 | [51] |
| CGE009** | AG |
TTCGTTGAAGGTGACAAGTG GCATTTTGGGATGAACAGA |
ATTO 565 | 0.05 | 102–126 | 8 | 262 | 0.66 | 0.71 | [55] |
| CG49** | GT |
CATCAGGGGTAAATTAAAGTAAGC CCACAGACGATTTCATATATCCTG |
ATTO 550 | 0.05 | 128–184 | 26 | 259 | 0.60 | 0.86 | [56] |
* Duplex PCR reaction ** Fourplex PCR reaction, number of alleles (NA), number of individuals that amplified (N), observed heterozygosity (HO), expected heterozygosity (HE).
Genetic diversity and structure
Genotyping results were analyzed with Micro-Checker v2.3.3 [26] to identify potential inconsistencies and errors (e.g., null alleles and large allele drop-out). All incidences identified by Micro-Checker were chromatographically inspected before proceeding with further analyses. GenAlEx software v6.5 [27] was used to report overall observed (HO) and expected (HE) heterozygosity. Genepop v4.2 [28, 29] was used to report observed (HO) and expected (HE) heterozygosity for each locus within each sampling location. The number of observed alleles (NA) and calculated allelic richness (AR), compensating for less individuals in NB (Rarefaction option), at each locus within each location was assessed using HP-RARE [30]. Independence among loci was tested by linkage disequilibrium (LD) and Hardy-Weinberg equilibrium (HWE) was calculated to identify loci and populations departing from theoretical equilibrium of allele frequencies, using the Arlequin software v3.5.1.3 [31]. Calculations of statistical significance were corrected for multiple tests according to the B-Y FDR method [32]. A multivariate Discriminant Analysis of Principal Components (DAPC) was used to resolve genetic connectivity between populations through sequential clustering and model selection. The DAPC was performed in R v3.3.2 [33] using the adegenet package [34] and the sampling locations as prior groups. The genetic relationship between samples, at the population level, was evaluated according to Chords distances (DCE) [35], calculated and bootstrapped 2000 times with MSA v4.05 [36], and presented in a neighbour joining (NJ) tree [37] using the PHYLIP v3.68 software package [38] and SPLITSTREE v4.0 [39]. The significance of splits in the NJ tree were evaluated according to Hillis & Bull [40], i.e., all splits > 70% are considered statistically significant. Calculated pairwise DCE’s were also used to generate a Principal Coordinates Analysis (PCoA) in GenAlEx v6.5 [27]. Pairwise genetic differentiation was estimated by calculating the fixation index, FST [41], and the statistical significance of the differences between populations was tested by 10,000 permutations of individuals between samples using MSA v4.05.
Larvae dispersal and survival simulations
Simulation of larvae dispersal and survival to settlement was performed with an open source, numerical 3D oceanographic model (ROMS) [42] with a spatial resolution of 800 m [43]. ROMS has shown accurate results when compared with field observations [44–46] and is a widely used model at both local and global scale (myroms.org). We focused on the influence of recent climate change by performing the simulations for 6 years (1990, 1998, 2002, 2006, 2007 and 2010) representing the climate since the 90s, one cold (2007), two warm (2002 and 2006) and one moderately warm year (2010). For survival, the simulated larvae have to had experienced 225 recruitment degree days [47], and the temperature at the landing site had to be ≥ 18°C (according to Mann, Burreson [48]). Due to the high reproduction capacity (several million larvae per spawning individual), true individual-based modelling was impossible. Hence we used the super-individual approach suggested by Scheffer, Baveco [49], where each modelled individual represents a large number of actual individuals. From each of 44 locations equally distributed along the Danish and Swedish coastline, 7 simulated larvae were released between 1 and 14 August (1 larvae every second day, i.e. 7*44 = 308 larvae per year [50]), and their floating path, experienced degree days and temperature at the landing sites were recorded. This could represent one viable super-individual from each of seven individuals in a small colony on each location. We did not include any behavior or random walk approach, but chose to distribute the time of release within the two first weeks of August, known to be the most relevant period for oyster spawning [12]. The number of landed larvae in Swedish and Norwegian coastal areas was counted within coastal grid cells of 50x50 km resolution for each simulated year, by summing the landed larvae within all the 800 m cells that fall within each of the coarser grid cells.
Results
Genetic diversity
A total of eight microsatellite markers [15] were initially tested using a Norwegian oyster sample (NO). Two of these microsatellites, CG108 and Cgsili29, failed to amplify and were not further used. The remaining six markers were first tested in simplex PCR to determine optimal annealing temperatures, and thereafter tested in combinations using various primer concentrations and cycling conditions for multiplex PCR testing. Both MgCl2 and BSA were also tested as PCR helpers for multiplex optimization. Successful conditions were found for a fourplex and duplex PCR used in this study as described in the methods and Table 2.
Among the six microsatellite loci analysed, four amplified for all samples while CG49 and Cgsili44 each amplified in 259 out of 262 samples (Table 2 and S1 Appendix). All loci were polymorphic and the total number of alleles detected per locus varied from eight to 33 (Table 2 and S1 Appendix). Populations did not differ markedly in allelic richness, with average number of alleles ranging between 7.19, for NB which also had the lowest analysed sample size (n = 12), to10.35 for NG (Tables 1 and 3). The number of private alleles ranged from none at sampling location NB to eight at SS (Table 3). When the four Norwegian sampling locations are compared to a compiled Swedish and Danish sample, private alleles are 25 and 26 respectively. Genetic diversity was homogenous among the populations with the highest expected heterozygosity found in population NG (0.89), the lowest in NB (0.83), and highest and lowest observed heterozygosity in NB (0.76) and NI (0.71) respectively (Table 3), showing that NB conforms closest to HWE.
Table 3. Summary statistics of the six genotyped microsatellites loci in the six analysed sampling locations.
| Location | Locus | Mean | |||||
|---|---|---|---|---|---|---|---|
| L10 | Cgsili44 | AMY | L48 | CGE009 | CG49 | ||
| NB | |||||||
| NA | 7 | 5 | 12 | 7 | 5 | 8 | 7.3 |
| AR | 6.83 | 5.00 | 11.49 | 6.83 | 5.00 | 8.00 | 7.19 |
| PA | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HO | 0.92 | 0.50 | 0.92 | 0.92 | 0.83 | 0.45 | 0.76 |
| HE | 0.81 | 0.80 | 0.91 | 0.83 | 0.81 | 0.80 | 0.83 |
| p | NS | NS | NS | NS | NS | ** | |
| NG | |||||||
| NA | 23 | 8 | 20 | 23 | 7 | 16 | 16.2 |
| AR | 13.36 | 7.30 | 11.65 | 13.70 | 5.54 | 10.57 | 10.35 |
| PA | 0 | 0 | 0 | 2 | 0 | 2 | 0.67 |
| HO | 0.80 | 0.60 | 0.84 | 0.90 | 0.58 | 0.64 | 0.73 |
| HE | 0.95 | 0.85 | 0.91 | 0.95 | 0.76 | 0.91 | 0.89 |
| p | NS | *** | NS | NS | ** | ** | |
| NO | |||||||
| NA | 21 | 9 | 16 | 18 | 7 | 15 | 14.3 |
| AR | 11.64 | 7.36 | 10.67 | 10.31 | 5.21 | 8.79 | 9.00 |
| PA | 0 | 0 | 0 | 0 | 1 | 0 | 0.17 |
| HO | 0.72 | 0.76 | 0.78 | 0.96 | 0.68 | 0.58 | 0.75 |
| HE | 0.90 | 0.86 | 0.88 | 0.89 | 0.72 | 0.86 | 0.85 |
| p | *** | NS | NS | NS | NS | *** | |
| NI | |||||||
| NA | 25 | 9 | 19 | 21 | 6 | 13 | 15.5 |
| AR | 13.52 | 7.39 | 10.68 | 12.51 | 5.43 | 9.00 | 9.76 |
| PA | 1 | 0 | 0 | 0 | 0 | 1 | 0.33 |
| HO | 0.98 | 0.44 | 0.74 | 0.82 | 0.64 | 0.62 | 0.71 |
| HE | 0.95 | 0.85 | 0.88 | 0.93 | 0.72 | 0.87 | 0.87 |
| p | NS | *** | * | NS | NS | *** | |
| SS | |||||||
| NA | 22 | 12 | 24 | 20 | 7 | 18 | 17.2 |
| AR | 13.43 | 7.24 | 12.68 | 12.00 | 5.13 | 10.16 | 10.11 |
| PA | 1 | 1 | 3 | 1 | 0 | 2 | 1.33 |
| HO | 0.84 | 0.66 | 0.90 | 0.86 | 0.60 | 0.56 | 0.74 |
| HE | 0.94 | 0.81 | 0.93 | 0.93 | 0.69 | 0.89 | 0.87 |
| p | NS | NS | NS | NS | *** | *** | |
| DA | |||||||
| NA | 24 | 11 | 18 | 21 | 7 | 17 | 16.3 |
| AR | 12.89 | 7.73 | 10.68 | 11.65 | 5.01 | 9.68 | 9.61 |
| PA | 0 | 0 | 0 | 0 | 0 | 3 | 0.50 |
| HO | 0.92 | 0.60 | 0.80 | 0.72 | 0.64 | 0.74 | 0.74 |
| HE | 0.94 | 0.85 | 0.90 | 0.92 | 0.62 | 0.89 | 0.86 |
| p | NS | ** | NS | * | NS | NS | |
Sampling locations (in bold); NA—number of alleles, AR—allelic richness according to the rarefaction method, PA—private alleles, HO—observed heterozygosity, HE—expected heterozygosity, p—the p-value from the Hardy-Weinberg equilibrium (HWE) test. Hardy-Weinberg after B-Y FDR adjustment (K = 15) significance levels:
* - 5%, ** - 1%, *** - 0.1% and NS -not significant.
Only one pair of loci (CGE009 & CG49) was identified as significantly linked by the LD analysis (B-Y FDR adjusted α = 0.00984). However, since this non-random association among alleles was not consistent among sampling locations and only identified in NO, loci CGE009 and CG49 were retained for further analysis. Significant departure from HWE was identified in 13 of the 36 tests (B-Y FDR adjusted α = 0.01198, Table 3). However, no locus showed significant departure from HWE in all sampled locations.
Population genetic structure
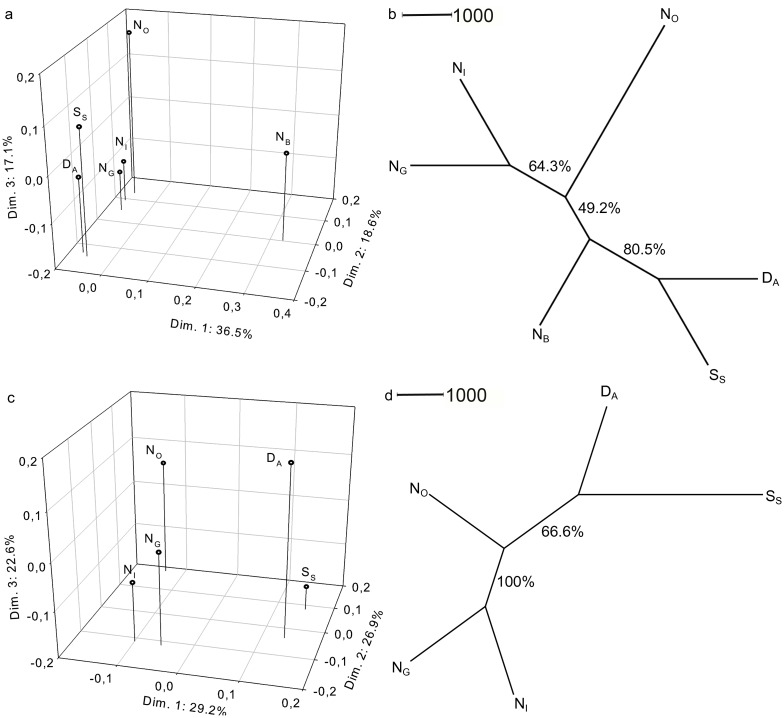
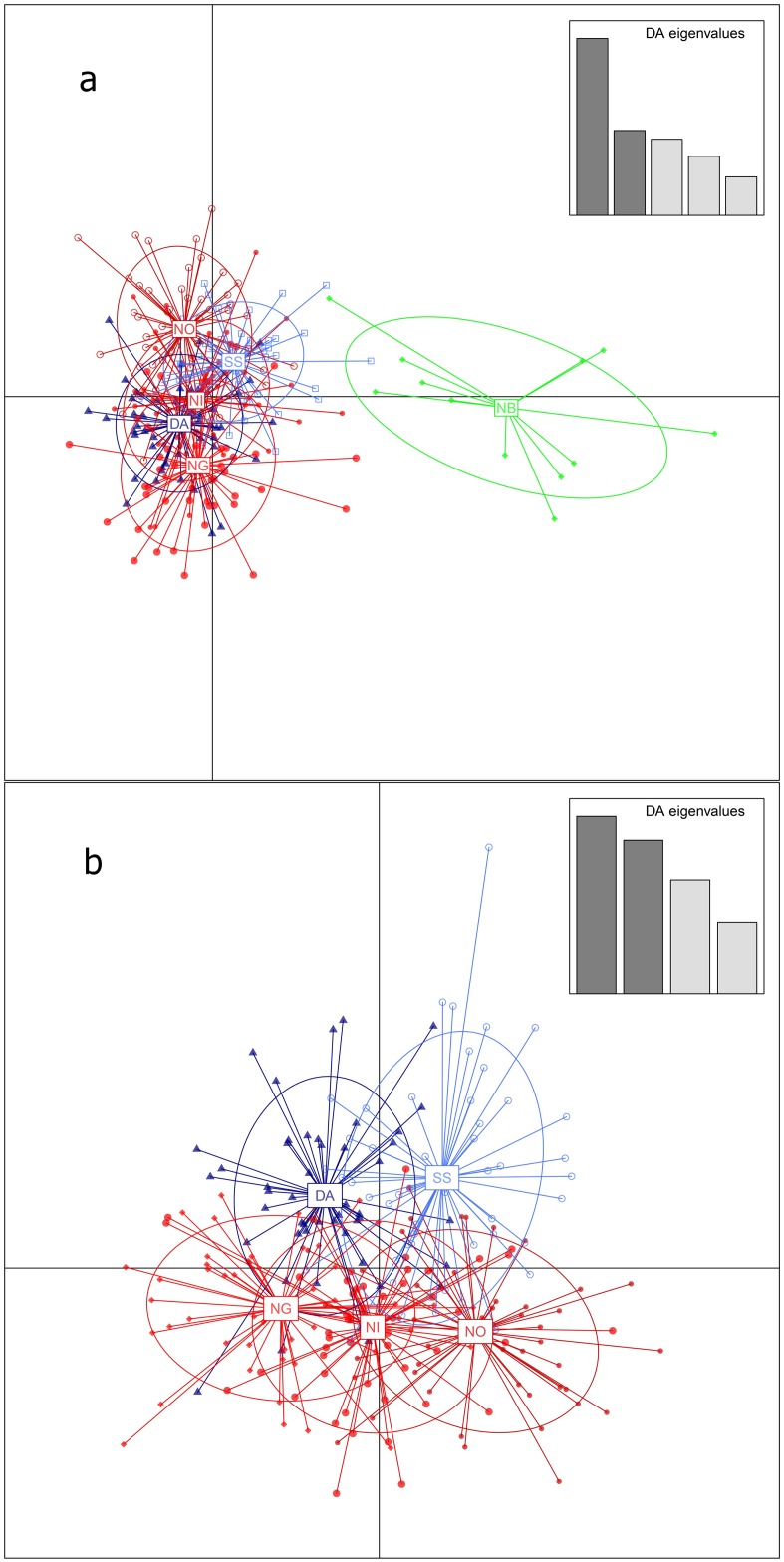
The PCoA analysis of all samples (Fig 2A) separated location NB (at the Norwegian west coast) from the remaining samples along the first dimension representing 36.5% of the total variance (72.2%). The remaining samples clustered into two groups along the second dimension, representing 18.6% of the total variance in the data set. The two clusters were made up of the Swedish (SS) and the Danish (DA) population in one group and the remaining Norwegian populations (NI, NO and NG) in the other (Fig 2A). The third dimension separated outer Oslo fjord (NO) from the NI and NG samples, representing 17.1% of the variance. The same clustering pattern was also shown by the NJ tree (Fig 2B). In the NJ tree, sampling locations DA and SS were significantly split from the remaining sample locations. The NB sample occurred at an intermediate position within the NJ tree, between the other Norwegian localities (i.e. NO, NI and NG), and the foreign countries localities (i.e. DA and SS, Fig 2B). For both the PCoA and the NJ tree, removal of location NB caused clustering of the remaining samples into three groups: 1) NO, 2) NI and NG, and 3) SS and DA (Fig 2c and 2d). The DAPC analyses, with all sampling locations included, showed a clear separation of NB from the remaining samples, primarily separated by the first principal component (Fig 3A). The second and third principal components did not vary in information value based on DA eigenvalues. With the removal of NB a stronger tendency for structuring between DA and SS versus the remaining Norwegian samples occurred (NI, NO, and NG, Fig 3B). Despite overlapping of individuals, the population ellipses for the Norwegian samples did not cross the centers of the DA and SS samples. The separation of sampling location NB was supported by high and significant FST values when compared with the remaining sampling locations (Table 4). Among the remaining sampling locations, the pairwise FST values showed non-significant genetic differences, except for the outer Oslo fjord location (NO) versus the Danish and Swedish populations (DA and SS, respectively, Table 4). Hence all the statistical analyses indicate genetic differentiation.
Fig 2. Genetic distance.
Similarities and differences among Norwegian (NB, NO, NI, NG), Swedish (SS) and Danish (DA) Pacific oyster (Crassostrea gigas) populations visualized by Chords distance [57] in a Principal Coordinate analysis (a and c) and Neighbour Joining tree plot (b and d). Based on all sampled locations (a and b), and for all locations except location NB (c and d), to explore and visualize the genetic distances without location NB that act as an outlier in the data set. Overview of the sampled oyster locations and abbreviations are given in Table 1.
Fig 3. Discriminant Analysis of Principal Components (DAPC).
Scatter plot with (a) and without (b) location NB in the analysis. Sampling locations are internally connected with lines to the center of each ellipses. The Danish and Swedish samples are indicated by blue colors (DA, dark blue and SS, light blue), the Norwegian outlier location (NB, green) is differentiated from the remaining Norwegian samples (NI, NO, and NG) represented by red color.
Table 4. Pairwise comparisons of FST (below diagonal) among the six sampled Pacific oyster locations with tested statistical significance between pairs (above diagonal).
| NB | DA | NG | NI | SS | NO | |
| NB | *** | *** | *** | *** | *** | |
| DA | 0.049 | NS | NS | NS | *** | |
| NG | 0.031 | 0.002 | NS | NS | NS | |
| NI | 0.042 | 0.006 | 0.002 | NS | NS | |
| SS | 0.043 | 0.004 | 0.004 | 0.006 | *** | |
| NO | 0.046 | 0.009 | 0.006 | 0.004 | 0.010 |
Sample abbreviations are explained in Table 1.
*** - 0.1% and NS -not significant.
Changes in dispersal and survival of Danish and Swedish oyster larvae
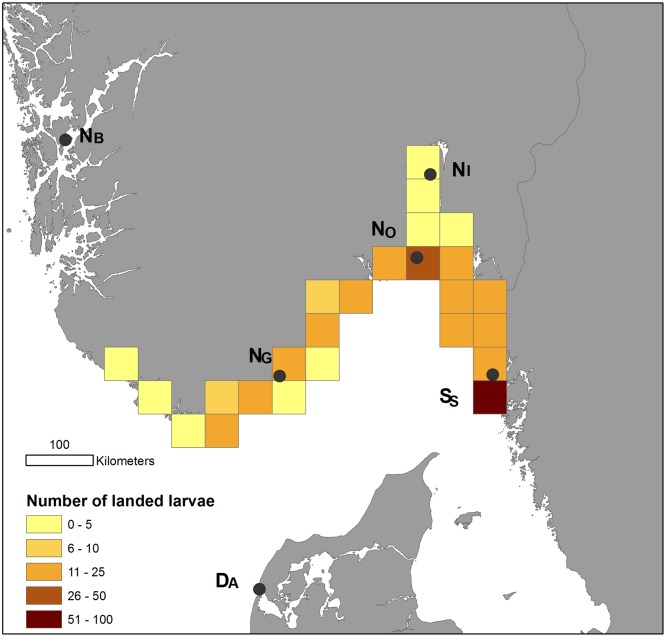
The simulation results showed that the water temperatures were too cold for the oyster larvae to develop and settle in Norwegian coastal waters in the 1990s (1990 and 1998), but warm summers since 2000 had adequate temperatures for development and survival of transported larvae across the Skagerrak (Table 5 and Fig 4). In the warmest year, 2002, a high fraction (36%) of the released larvae landed in Norwegian coastal waters, whereas in the following cold and moderately warm years (2007 and 2010) only 2–6% of the released larvae experienced sufficient water temperatures to successfully develop and settle on the Norwegian coast. In the two warm years (2002 and 2006), the simulated larvae could reach beyond the limit of the Skagerrak region and into the North Sea region. The hot spot for receiving the highest supply of oyster larvae along the Norwegian coast (i.e. the 50x50 km grid cell with the largest number of landed oyster larvae, Fig 4) included the sampling site at Hui in the Outer Oslo fjord (NO).
Table 5. Overview of simulated number and fraction of the Pacific oyster (Crassostrea gigas) larvae landed in total and within Norwegian coastal waters provided ocean climate in six different years.
| Year | 1990 | 1998 | 2002 | 2006 | 2007 | 2010 | In total |
|---|---|---|---|---|---|---|---|
| n released | 308 | 308 | 308 | 308 | 308 | 308 | 1848 |
| n landed | 0 | 0 | 192 | 95 | 38 | 44 | 369 |
| Fraction landed | 0 | 0 | 0.62 | 0.31 | 0.12 | 0.14 | 0.20 |
| n in Norway | 0 | 0 | 111 | 43 | 7 | 20 | 181 |
| Fraction in Norway | 0 | 0 | 0.36 | 0.14 | 0.02 | 0.06 | 0.10 |
Fig 4. Simulation of larval dispersal.
The spatial distribution of the 369 landed Pacific oyster (Crassostrea gigas) larvae in Swedish and Norwegian coastal waters in total for the simulated years (1990, 1998, 2002, 2006, 2007, 2010), summed per coastal grid cell (50x50 km). Number of landed larvae (super-individuals) per grid cell is shown (see legend). The location and names of the sampled DNA stations in this study are indicated (black circles, cf. Table 1). For simulation details see [50]. Reprinted from Rinde et al. 2016 under a CC BY license, with permission from NIVA, original copyright 2016. The map is produced using ESRIs GIS software ArcMap v 10.4.1 (www.esri.com), and the country dataset GISCO NUTS 2010.
Discussion
The analyses of the Pacific oyster (Crassostrea gigas) samples showed that the studied Norwegian populations were genetically different from the Danish and the Swedish populations. This contradicts the hypothesis that the Norwegian populations mainly origin from natural dispersal of larvae drifting from established populations in Denmark and Sweden [11].
Among the studied Norwegian Skagerrak C. gigas populations, sampling location NO differed genetically from the studied Swedish and Danish populations although it is located within the hot spot area for larvae supply as indicated by the larvae drift simulations. However, the identification of this area as a hotspot area for supply of larvae is otherwise supported by being the only area in Norway with oyster reef formation in 2015 [58]. The genetic difference identified between NO and the Swedish (SS) and Danish populations (DA) concurs with unpublished genetic studies [10] showing high similarity between Swedish and Danish populations, whereas the population at Hui (NO) differed from the studied Swedish and Danish populations. The remaining two Norwegian Skagerrak populations (NI and NG), located on each side of NO, form a cluster differentiated from both SS/DA and NO. This patchwork of dissimilar populations across Skagerrak separating the five studied populations into three groups, NI/NG and NO on the Norwegian side and SS/DA on the opposite side of Skagerrak, is unlikely to be caused by natural dispersal from the SS/DA-group to Norway. Indeed, genetic discontinuity is observed among three populations along a contiguous stretch of the Norwegian coast (400 km), and only 126 km separates the genetically different populations NI and NO. In contrast, the Kattegat Sea separates the Swedish and Danish populations, they are 500 km apart, and yet they form a homogeneous cluster, also different from the three Norwegian Skagerrak populations. Moreover, the larvae drift simulations indicated only two warm years (2002 and 2006) with significant gene flow events across the Skagerrak area since 2000. These two years have the highest summer sea temperatures since measurements on the south coast of Norway (Flødevigen Research station, see www.imr.no) started in 1924. Considering the aquaculture history of C. gigas in Norway in the 70 – 80s until 2010 [7, 9], when all licenses for Pacific oyster aquaculture in Norway were revoked (Pers. comm. Directorate of Fisheries) it seems more likely that the identified genetic differences separating NI/NG from NO and from SS/DA are a consequence of multiple introduction events such as from aquaculture or shipping activities. On the other hand, this does not preclude the possible existence of other populations originating from larval drift across the Skagerrak.
The differentiation and independency of the studied Norwegian samples towards the Danish and Swedish samples is furthermore supported by the presence of private alleles in both groups in high and almost equal numbers (25 and 26, respectively). In order for the Norwegian populations to be a result of a frontier/range expansion (drift) scenario, as known from terrestrial [59] and marine organisms [60, 61], the Norwegian Skagerrak group should have had lower genetic diversity, i.e. less private alleles, than the Swedish and Danish populations. Even among the Norwegian populations private alleles occur, providing further evidence of a lack of a uniform population structure, and pointing towards multiple introductions from separate sources of origin.
The larvae drift simulations indicated that the water temperatures in the Skagerrak were too cold for larvae development, survival, and settlement to support successful natural dispersal of Danish and Swedish larva until 2000. Since 2000, summer temperatures have several years been sufficiently high for such natural dispersal and successful crossing of the Skagerrak barrier into Norwegian coastal areas. However, the lack of genetic similarity between the population at Hui (NO), which is situated within the hot spot area for landing of foreign C. gigas larvae in the simulation study, and the studied Danish and Swedish oyster populations, indicates that so far there has been low success rate of this pathway. However, other genetic studies [10] have revealed some similarities between one Norwegian population (approximately 20 km north of NG) that was not included in this study, and another Swedish population, which indicates the possible occurrence of successful recruitment of Swedish oyster larvae in Norwegian waters. Future climate change with rising summer temperatures is likely to increase the risk of C. gigas larvae dispersal [62]. Analysis of sea surface temperature data along the Swedish Skagerrak coast [50] suggests a 125 km northwards displacement of the 19°C temperature isocline in August. This implies a northward shift of the summer temperature needed to enable Pacific oyster spawning in wild populations. This will further push the distribution range of the Pacific oyster northward into previously unfavourable areas/ecosystems, as previously documented for other species [63]. Our findings of a theoretical possible increased supply of foreign C. gigas larvae in recent and future years clearly indicate a need to monitor and investigate the newly established populations of C. gigas along the Norwegian coast to assess the connectivity link across the Skagerrak area. There are many factors that may cause high pre- or post-settlement mortality of drifting C. gigas larva (e.g. predation, starvation, etc.) and that could counteract successful dispersal and colonization across the Skagerrak. In addition, selection imposed by strong environmental gradients, such as the temperature gradient in the studied region, promotes adaptive differentiation [64]. Local adaptation of earlier introduced C. gigas in Norwegian waters would imply that the local genotypes would have higher fitness than genotypes from foreign habitats [65]. Accordingly, recently landed foreign C. gigas larvae, would have lower chances of survival than locally adapted oysters.
The simulation model does not include any other mortality factors than the influence of temperature on the larvae’s possibility to develop successfully during the planktonic phase, and sufficient temperature for the larvae to survive at the landing site. This implies that the predicted rate of success of transported Pacific oyster larvae, is likely to be higher than the real success rate. Other mortality factors in the planktonic phase (e.g. starving and predation), when settling (finding suitable substrate), and post settlement (including spatial competition with other species), will all reduce the larvae’s chances of successful spreading. Hence the predicted rate of successful spreading in the two warm years, are likely to be higher than the actual rate because of these limitations, further reducing the possibility of connectivity between the populations.
The genetic differences found among the Norwegian Pacific oyster populations suggests that multiple introductions may have occurred along the coast. This could involve previous aquaculture activities or other introduction pathways such as shipping activities and live trade. Unfortunately, no aquaculture sources were included in the present analysis, so its role as a potential source of introduction cannot be established. Introduction of oysters to ports by shipping is possible since C. gigas larvae and adults have been found in ballast water and on ship hulls, respectively [66]. The Norwegian Skagerrak coast houses some large ports with high shipping activities and hence the potential for this introduction pathway exists [67]. Spreading of C. gigas from aquaculture sites has occurred in several countries [13, 68]. In Norway aquaculture licenses for both native (Ostrea edulis) and the invasive oyster species (C. gigas) (Directorate of Fisheries, http://www.fiskeridir.no/register/akvareg/?m=utl_lok&s=1) have been given, indicating that introduction for aquaculture purposes is another plausible introduction pathway. Indeed, the Norwegian population from Bergen (NB, western Norway) was collected in the vicinity of a former aquaculture site (Espevik, Tysnes) for C. gigas, for which the origin of the imported larvae was reported to be Scotland [7]. This population, NB, showed strong genetic differentiation from the other studied populations. The possible aquaculture origins for the remaining sampled populations in Norway are difficult to establish from literature. Despite strong restrictions on the import of molluscs for cultivation purposes in Norway in 1986 [9], these aquaculture licenses were still assigned until 2001. The restrictions may have reduced the likelihood of recent repetitive aquaculture introductions.
Although being genetically different, the studied Norwegian oyster populations had low genetic diversity. This agrees with other studies [1, 15, 17] indicating a general pattern of low genetic diversity in the north. Among these studies [15] used the same six microsatellites as this study. Few differences in mean allelic richness were shown for all the analyzed sampling locations (Table 3), except for the westernmost sampling location, Bergen (NB). Despite the relatively low number of analyzed individuals, the low allelic richness of the Bergen population could be due to a founder effect or subsequent bottleneck effects [69–71]. However, a bottleneck analysis [72, 73] using the two-phase mutation model with default settings did not identify any bottleneck events in the analyzed samples (data not shown). Moreover, the simulation study indicates low chances for natural dispersal of Danish and Swedish larvae so far along the Norwegian coastline as to Bergen given recent year’s climate. It therefore seems plausible that the Bergen population has adapted to local environmental conditions within the area since the 1970s when the species was introduced for aquaculture purposes. The clustering software STRUCTURE v2.3.4 [74] was run with and without NB (data not shown), to detect any population structure among the sampled populations. The program failed to show any structuring pattern. This concurs with previous C. gigas studies showing no population structuring except between the northern and the southern European populations [1] or between aquaculture and feral populations [75].
Initial introduction of C. gigas to Europe entirely originates from Japan, USA, and BC in Canada. Several studies [1, 15, 17] have demonstrated that the southern European populations genetically cluster with the introduction source populations. This indicates that the northern group has developed locally in Europe. As the history of introduction to the north (i.e. to Denmark, Norway and Sweden) mainly originate from the UK following the 1970s, it seems that genetic differentiation between the northern and southern group may have begun in the UK. Hybridization is known to be an evolution mechanism by which genetic variations may be swiftly introduced and fixed in populations, and in particular when species colonize new environments [76]. Considering that Crassostrea angulata has been introduced both prior to C. gigas, and in parallel into the UK [24], hybridization between the two species within the UK may have been possible during this spatial and temporal overlap. Hybridization is documented for C. gigas with C. angulata [77, 78], supporting that such an event may have occurred. Although considered as conspecific to C. gigas by some authors [79], C. angulata has been shown to be sufficiently genetically distinct to be a separate species [78, 80] and listed under the World Register of Marine Species (http://www.marinespecies.org/aphia.php?p=taxdetails&id=146900, July 2016).
This study contributes to the understanding of the genetic pattern of C. gigas in northern Europe and shows so far, low connectivity across the Skagerrak. Furthermore, it also demonstrates a likely future increase in successful dispersal of C. gigas larvae across the Skagerrak, as sea surface temperatures keep rising [14]. The current expansion might therefore temporarily be mitigated by reducing the density of the species in locations with suitable conditions for oyster growth and spawning, e.g. semi-enclosed bays, traditionally used for oyster aquaculture in Norway [7]. The suggested importance of aquaculture, shipping and import for live food as likely introduction pathways is highly relevant for nature management and implies the need to inform aquaculture industries and the public about the risk of introducing invasive species. The future risk of successful dispersal of Pacific oyster larvae from Danish and Swedish populations, due to climate change, emphasize the need for monitoring to detect any massive expansion as basis for targeted management of affected ecosystems. These conclusions may be extended to other invasive species with pelagic larvae stages also affected by climatic change.
Supporting information
(XLSX)
Acknowledgments
The genetic analysis was done as a part of the project “ALIEN OYSTER—distribution, population development and effects of the invasive Pacific oyster (Crassostrea gigas) in the Skagerrak”, funded by the Norwegian Research Council (Project no. 203792). The simulation study was funded by NIVA’s Strategic Institute Initiative ‘Climate effects from Mountains to Fjords’ (The Research Council of Norway, contract number 208279). We thank Hans Erik Karlsen (University of Oslo), Stein Mortensen (IMR), Lise Tveiten and Trine Dale (NIVA) for help with planning and conducting field work, and Karen Filbee-Dexter (NIVA) for useful comments and major improvements to the manuscript.
Data Availability
All relevant data are within the paper.
Funding Statement
The genetic analysis was done as a part of the project “ALIEN OYSTER – distribution, population development and effects of the invasive Pacific oyster (Crassostrea gigas) in the Skagerrak”, funded by the Norwegian Research Council (Project no. 203792). The simulation study was funded by NIVA’s Strategic Institute Initiative ‘Climate effects from Mountains to Fjords’ (The Research Council of Norway, contract number 208279). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lallias D, Boudry P, Batista FM, Beaumont A, King JW, Turner JR, et al. Invasion genetics of the Pacific oyster Crassostrea gigas in the British Isles inferred from microsatellite and mitochondrial markers. Biol Invasions. 2015; 17(9):2581–95. [Google Scholar]
- 2.Nehring S. NOBANIS—Invasive Alien Species Fact Sheet Crassostrea gigas Online Database of the European Network on Invasive Alien Species—NOBANIS wwwnobanisorg. 2011.
- 3.Drinkwaard AC. Introductions and developments of oysters in the North Sea area: a review. Helgolander Meeresun. 1999; 52(3–4):301–8. [Google Scholar]
- 4.Diederich S, Nehls G, Beusekom JEEv, Reise K. Introduced Pacific oysters (Crassostrea gigas) in the northern Wadden Sea: invasion accelerated by warm summers? Helgoland Marine Research. 2005; 59(2):97–106. [Google Scholar]
- 5.Jensen KR, Knudsen J. A summary of alien marine benthic invertebrates in Danish waters. Oceanol Hydrobiol Stud. 2005; 24(1):137–62. [Google Scholar]
- 6.Eklund U, Håkansson M, Haamer J. En undersökningom förutsättningarna för ostronodling vid svenska västkusten. Chalmers Tekniska Högskola och Göteborgs Universitet; 1977; No. B83:35. [Google Scholar]
- 7.Strand Ø, Vølstad JH. The molluscan fisheries and culture of Norway In: MacKenzie JL, Burrel V, Hobart WL, editors. The History, Present Condition, and Future of the Molluscan fisheries of North America and Europe: Dep Commer, NOAA Tech Rep NMFS; 1997. p. 7–24. [Google Scholar]
- 8.Strand A, Waenerlund A, Lindegarth S. High Tolerance of the Pacific Oyster (Crassostrea Gigas, Thunberg) to Low Temperatures. Journal of Shellfish Research. 2011; 30(3):733–5. [Google Scholar]
- 9.Wrange AL, Valero J, Harkestad LS, Strand O, Lindegarth S, Christensen HT, et al. Massive settlements of the Pacific oyster, Crassostrea gigas, in Scandinavia. Biol Invasions. 2010; 12(6):1453–8. [Google Scholar]
- 10.Bodvin T, Rinde E, Mortensen A. Faggrunnlag stillehavsøsters (Crassostrea gigas). Norway: Institute of Marine Research and Norwegian Institute for Water Research, 2014 Nr. 32–2014.
- 11.Gederaas L, Loennechen Moen T, Skjelseth S, Larsen L-K. Alien species in Norway—with the Norwegian Black List 2012. Trondheim, Norway: Te Norwegian Biodiversity Information Centre (NBIC); 2012. [Google Scholar]
- 12.Dolmer P, Holm MW, Strand A, Lindergarth S, Bodvin T, Norling P, et al. The invasive Pacific oyster, Crassostrea gigas, in Scandinavian coastal waters: A risk assessment on the impact in different habitats and climate conditions. Norway, Bergen: Institute of Marine Research, 2014 March. Report No.: 2/2014 Contract No.: 80190.
- 13.Andrews JD. A Review of Introductions of Exotic Oysters and Biological Planning for New Importations. Mar Fish Rev. 1980; 42(12):1–11. [Google Scholar]
- 14.IPCC. Climate Change 2013 The Physical Science Basis. Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Stocker TF, Qin D, Plattner G-K, Tignor M, Allen SK, Boschung J, et al., editors. Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press; 2013. 1535 p.
- 15.Rohfritsch A, Bierne N, Boudry P, Heurtebise S, Cornette F, Lapègue S. Population genomics shed light on the demographic and adaptive histories of European invasion in the Pacific oyster, Crassostrea gigas. Evol Appl. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meistertzheim AL, Arnaud-Haond S, Boudry P, Thebault MT. Genetic structure of wild European populations of the invasive Pacific oyster Crassostrea gigas due to aquaculture practices. Mar Biol. 2013; 160(2):453–63. [Google Scholar]
- 17.Moehler J, Wegner KM, Reise K, Jacobsen S. Invasion genetics of Pacific oyster Crassostrea gigas shaped by aquaculture stocking practices. J Sea Res. 2011; 66(3):256–62. [Google Scholar]
- 18.Fabioux C, Huvet A, Le Souchu P, Le Pennec M, Pouvreau S. Temperature and photoperiod drive Crassostrea gigas reproductive internal clock. Aquaculture. 2005; 250(1–2):458–70. [Google Scholar]
- 19.Dutertre M, Beninger PG, Barille L, Papin M, Haure J. Rising water temperatures, reproduction and recruitment of an invasive oyster, Crassostrea gigas, on the French Atlantic coast. Mar Environ Res. 2010; 69(1):1–9. 10.1016/j.marenvres.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 20.Ruiz C, Abad M, Sedano F, Garcia-Martin LO, López JLS. Influence of seasonal environmental changes on the gamete production and biochemical composition of Crassostrea gigas (Thunberg) in suspended culture in El Grove, Galicia, Spain. Journal of Experimental Marine Biology and Ecology. 1992; 155(2):249–62. [Google Scholar]
- 21.Ben Kheder R, Quéré C, Moal J, Robert R. Effect of nutrition on Crassostrea gigas larval development and the evolution of physiological indices. Part A: Quantitative and qualitative diet effects. Aquaculture. 2010; 305(1–4):165–73. [Google Scholar]
- 22.Rico-Villa B, Woerther P, Mingant C, Lepiver D, Pouvreau S, Hamon M, et al. A flow-through rearing system for ecophysiological studies of Pacific oyster Crassostrea gigas larvae. Aquaculture. 2008; 282(1–4):54–60. [Google Scholar]
- 23.Grizel H, Héral M. Introduction into France of the Japanese oyster (Crassostrea gigas). Journal du Conseil: ICES Journal of Marine Science. 1991; 47(3):399–403. [Google Scholar]
- 24.Utting SD, Spencer BE. Introductions of marine bivalve molluscs into the United Kingdom for commercial culture—case histories. ICES mar Sci Symp. 1992; 194:8. [Google Scholar]
- 25.Anglès d'Auriac MB, Norling P, Rinde E. A rapid and inexpensive DNA extraction protocol for oysters. Animal Genetics. 2016; 47(3):389–90. 10.1111/age.12417 [DOI] [PubMed] [Google Scholar]
- 26.Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004; 4(3):535–8. [Google Scholar]
- 27.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012; 28(19):2537–9. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raymond M, Rousset F. Genepop (Version-1.2)—Population-Genetics Software for Exact Tests and Ecumenicism. J Hered. 1995; 86(3):248–9. [Google Scholar]
- 29.Rousset F. genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour. 2008; 8(1):103–6. 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- 30.Kalinowski ST. HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 2005; 5(1):187–9. [Google Scholar]
- 31.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010; 10(3):564–7. 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 32.Narum SR. Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv Genet. 2006; 7(5):783–7. [Google Scholar]
- 33.R Core Team. R: A language and environment for statistical computing: R Foundation for Statistical Computing, Vienna, Austria; 2016. [cited 2017 March]. https://www.R-project.org/. [Google Scholar]
- 34.Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008; 24(11):1403–5. 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- 35.Ll Cavalli-Sforza, Edwards AWF. Phylogenetic analysis: models and estimation procedures. Evolution. 1967; 21(3):550–70. [DOI] [PubMed] [Google Scholar]
- 36.Dieringer D, Schlötterer C. MICROSATELLITE ANALYSER (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes. 2003; 3(1):167–9. [Google Scholar]
- 37.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987; 4(4):406–25. [DOI] [PubMed] [Google Scholar]
- 38.Felsenstein J. PHYLIP (Phylogeny Inference Package). Distributed by the author. Version 3.6 ed. USA, Seattle: Department of Genome Sciences, University of Washington; 2005.
- 39.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006; 23(2):254–67. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 40.Hillis DM, Bull JJ. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Systematic Biology. 1993; 42(2):182–92. [Google Scholar]
- 41.Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984; 38(6):1358–70. [DOI] [PubMed] [Google Scholar]
- 42.Haidvogel DB, Arango H, Budgell WP, Cornuelle BD, Curchitser E, Di Lorenzo E, et al. Ocean forecasting in terrain-following coordinates: Formulation and skill assessment of the Regional Ocean Modeling System. Journal of Computational Physics. 2008; 227(7):3595–624. [Google Scholar]
- 43.Albretsen J, Sperrevik AK, Staalstrøm A, Sandvik AD, Vikebø F, Asplin L. NorKyst800 Report No.1 User Manual and technical descriptions. Institute of Marine Research, 2011 43. Report No.: nr.2/2011.
- 44.LaCasce JH, Røed LP, Bertino L, Ådlandsvik B. CONMAN Technical Report No. 2: Analysis of model results. Oslo: met.no, 2007 May 18, 2007. Report No.: Report 5/2007.
- 45.Capet X, Mcwilliams JC, Mokemaker MJ, Shchepetkin AF. Mesoscale to submesoscale transition in the California current system. Part I: Flow structure, eddy flux, and observational tests. J Phys Oceanogr. 2008; 38(1):29–43. [Google Scholar]
- 46.Moore AM, Arango HG, Broquet G, Edwards C, Veneziani M, Powell B, et al. The Regional Ocean Modeling System (ROMS) 4-dimensional variational data assimilation systems Part II—Performance and application to the California Current System. Prog Oceanogr. 2011; 91(1):50–73. [Google Scholar]
- 47.Syvret M, FitzGerald A, Hoare P. Development of a Pacific Oyster Aquaculture Protocol for the UK. UK: Sea Fish Industry Authority, 2008.
- 48.Mann R, Burreson E, Baker P. The decline of the Virginia oyster fishery in Chesapeake Bay: considerations for the introduction of a non-endemic species, Crassostrea gigas (Thunberg, 1793). Journal of Shellfish Research. 1991; 10(2):379–88. [Google Scholar]
- 49.Scheffer M, Baveco JM, DeAngelis DL, Rose KA, van Nes EH. Super-individuals a simple solution for modelling large populations on an individual basis. Ecological Modelling. 1995; 80(2–3):161–70. [Google Scholar]
- 50.Rinde E, Hjermann DØ, Staalstom A. Larvae drift simulations of the Pacific oyster in Skagerrak—influence of climate change on larvae development, survival and dispersal. Oslo, Norway: NIVA, 2016 7016–2016 Contract No.: NIVA report 7016–2016.
- 51.Huvet A, Boudry P, Ohresser M, Delsert C, Bonhomme F. Variable microsatellites in the Pacific Oyster Crassostrea gigas and other cupped oyster species. Anim Genet. 2000; 31(1):71–2. [DOI] [PubMed] [Google Scholar]
- 52.Sauvage C, Boudry P, Lapegue S. Identification and characterization of 18 novel polymorphic microsatellite makers derived from expressed sequence tags in the Pacific oyster Crassostrea gigas. Mol Ecol Resour. 2009; 9(3):853–5. 10.1111/j.1755-0998.2009.02525.x [DOI] [PubMed] [Google Scholar]
- 53.Li RH, Li Q, Cornette F, Degremont L, Lapegue S. Development of four EST-SSR multiplex PCRs in the Pacific oyster (Crassostrea gigas) and their validation in parentage assignment. Aquaculture. 2010; 310(1–2):234–9. [Google Scholar]
- 54.Sellos D, Moal J, Degremont L, Huvet A, Daniel JY, Nicoulaud S, et al. Structure of amylase genes in populations of pacific cupped oyster (Crassostrea gigas): Tissue expression and allelic polymorphism. Mar Biotechnol. 2003; 5(4):360–72. 10.1007/s10126-002-0089-7 [DOI] [PubMed] [Google Scholar]
- 55.Yu H, Li Q. EST-SSR markers from the Pacific oyster, Crassostrea gigas. Mol Ecol Notes. 2007; 7(5):860–2. [Google Scholar]
- 56.Magoulas A, Gjetvaj B, Terzoglou V, Zouros E. Three polymorphic microsatellites in the Japanese oyster, Crassostrea gigas (Thunberg). Anim Genet. 1998; 29(1):69–70.9682462 [Google Scholar]
- 57.Cavalli-Sforza LL, Edwards AWF. Phylogenetic analysis. Models and estimation procedures. Am J Hum Genet. 1967; 19(3 Pt 1):233–57. [PMC free article] [PubMed] [Google Scholar]
- 58.Postmyr E. National action plan against pacific oyster—Crassostrea gigas. Miljødirektoratet, 2016 Contract No.: M-588.
- 59.Manier MK, Arnold SJ. Population genetic analysis identifies source-sink dynamics for two sympatric garter snake species (Thamnophis elegans and Thamnophis sirtalis). Mol Ecol. 2005; 14(13):3965–76. 10.1111/j.1365-294X.2005.02734.x [DOI] [PubMed] [Google Scholar]
- 60.Chen K, Ciannelli L, Decker MB, Ladd C, Cheng W, Zhou Z, et al. Reconstructing source-sink dynamics in a population with a pelagic dispersal phase. PLoS One. 2014; 9(5):e95316 10.1371/journal.pone.0095316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Munguia P. Role of sources and temporal sinks in a marine amphipod. Biol Lett. 2015; 11(2):20140864 10.1098/rsbl.2014.0864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rinde E, Tjomsland T, Hjermann DØ, Kempa M, Norling P, Kolluru VS. Increased spreading potential of the invasive Pacific oyster (Crassostrea gigas) at its northern distribution limit in Europe due to warmer climate. Mar Freshwater Res. 2016: [Google Scholar]
- 63.Swaegers J, Mergeay J, Therry L, Larmuseau MHD, Bonte D, Stoks R. Rapid range expansion increases genetic differentiation while causing limited reduction in genetic diversity in a damselfly. Heredity. 2013; 111(5):422–9. 10.1038/hdy.2013.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bible JM, Sanford E. Local adaptation in an estuarine foundation species: Implications for restoration. Biological Conservation. 2016; 193:95–102. [Google Scholar]
- 65.Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecology Letters. 2004; 7(12):1225–41. [Google Scholar]
- 66.Gollasch S. The Importance of Ship Hull Fouling as a Vector of Species Introductions into the North Sea. Biofouling. 2002; 18(2):105–21. [Google Scholar]
- 67.Bax N, Williamson A, Aguero M, Gonzalez E, Geeves W. Marine invasive alien species: a threat to global biodiversity. Marine Policy. 2003; 27(4):313–23. [Google Scholar]
- 68.Troost K. Causes and effects of a highly successful marine invasion: Case-study of the introduced Pacific oyster Crassostrea gigas in continental NW European estuaries. J Sea Res. 2010; 64(3):145–65. [Google Scholar]
- 69.Kinziger AP, Nakamoto RJ, Anderson EC, Harvey BC. Small founding number and low genetic diversity in an introduced species exhibiting limited invasion success (speckled dace, Rhinichthys osculus). Ecol Evol. 2011; 1(1):73–84. 10.1002/ece3.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Frankham R, Ballou JD, Briscoe DA. Introduction to conservation genetics. New York, USA: Cambridge University Press; 2002. [Google Scholar]
- 71.Hedrick PW. Genetics of populations. Sudbury MA, USA: Jones and Barnett; 2005. [Google Scholar]
- 72.Piry S, Luikart G, Cornuet J-M. Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J Hered. 1999; 90(4):502–3. [Google Scholar]
- 73.Luikart G, Cornuet J-M. Empirical Evaluation of a Test for Identifying Recently Bottlenecked Populations from Allele Frequency Data. Conserv Biol. 1998; 12(1):228–37. [Google Scholar]
- 74.Pritchard JK, Stephens M, Donnelly P. Inference of Population Structure Using Multilocus Genotype Data. Genetics. 2000; 155(2):945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kochmann J, Carlsson J, Crowe TP, Mariani S. Genetic Evidence for the Uncoupling of Local Aquaculture Activities and a Population of an Invasive Species—A Case Study of Pacific Oysters (Crassostrea gigas). J Hered. 2012; 103(5):661–71. 10.1093/jhered/ess042 [DOI] [PubMed] [Google Scholar]
- 76.Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004; 19(4):198–207. 10.1016/j.tree.2004.01.003 [DOI] [PubMed] [Google Scholar]
- 77.Leitão A, Chaves R, Santos S, Guedes-Pinto H, Boudry P. Interspecific hybridization in oysters: Restriction Enzyme Digestion Chromosome Banding confirms Crassostrea angulata × Crassostrea gigas F1 hybrids. Journal of Experimental Marine Biology and Ecology. 2007; 343(2):253–60. [Google Scholar]
- 78.Huvet A, Fabioux C, McCombie H, Lapegue S, Boudry P. Natural hybridization between genetically differentiated populations of Crassostrea gigas and C-angulata highlighted by sequence variation in flanking regions of a microsatellite locus. Mar Ecol-Prog Ser. 2004; 272:141–52. [Google Scholar]
- 79.Humphreys J, Herbert RJH, Roberts C, Fletcher S. A reappraisal of the history and economics of the Pacific oyster in Britain. Aquaculture. 2014; 428–429:117–24. [Google Scholar]
- 80.Batista F, Leitao A, Huvet A, Lapegue S, Heurtebide S, Boudry P. The taxonomic status and origin of the Portuguese oyster Crassostea angulata (Lamark, 1819). International Oyster Symposium; July 13–14; Tokyo, Japan2005.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
Data Availability Statement
All relevant data are within the paper.