Abstract
Circadian coordination of metabolism, physiology, and behaviour is found in all living kingdoms. Clock genes are transcriptional regulators, and their rhythmic activities generate daily rhythms in clock-controlled genes which result in cellular and organismal rhythms. Insects provide numerous examples of rhythms in behaviour and reproduction, but less is known about control of metabolic processes by circadian clocks in insects. Recent data suggest that several pathways involved in protecting cells from oxidative stress may be modulated by the circadian system, including genes involved in glutathione (GSH) biosynthesis. Specifically, rhythmic expression of the gene encoding the catalytic subunit (Gclc) of the rate-limiting GSH biosynthetic enzyme was detected in Drosophila melanogaster heads. The aim of this study was to determine which clocks in the fly multi-oscillatory circadian system are responsible for Gclc rhythms. Genetic disruption of tissue-specific clocks in D. melanogaster revealed that transcriptional rhythms in Gclc mRNA levels occur independently of the central pacemaker neurons, because these rhythms persisted in heads of behaviourally arrhythmic flies with a disabled central clock but intact peripheral clocks. Disrupting the clock specifically in glial cells abolished rhythmic expression of Gclc, suggesting that glia play an important role in Gclc transcriptional regulation, which may contribute to maintaining homeostasis in the fly nervous system.
Keywords: Central pacemaker, circadian clock, circadian rhythms, glutathione biosynthesis, peripheral clocks
Introduction
Circadian rhythms regulate many physiological, metabolic, and behavioural functions with an approximately 24 h periodicity. This way of molecular timekeeping has likely evolved in organisms to provide for optimal survival in a diurnally changing environment. In both insects and mammals, circadian regulation is achieved by a negative feedback loop consisting of transcriptional activators and repressors, which among insects, are best understood in Drosophila melanogaster (Hardin & Panda, 2013). The core clock genes are period (per), timeless (tim), Clock (Clk), and cycle (cyc). CLK and CYC induce transcription of per and tim mRNA. As PER and TIM proteins accumulate in the early night, they form heterodimers and repress CLK-CYC activity, thus leading to the suppression of their own transcription. This suppression eventually subsides when PER and TIM are degraded, starting another clock cycle over again. The clock feedback loops are cell autonomous and are known to operate in many different cell types.
The multi-oscillatory circadian system consists of a “central clock” and cell autonomous clocks in peripheral tissues (Tomioka et al., 2012). In flies, the central pacemaker neurons comprise several dozen of lateral and dorsal neurons, which control different aspects of rest/activity rhythms (Rieger et al., 2006). In addition, circadian oscillators are present in retinal photoreceptors, olfactory and gustatory sensory neurons, glia, and most other tissues. These clocks are called peripheral, as their function is not necessary for behavioural rest/activity rhythms (Glossop & Hardin, 2002), but rather for the control of tissue-specific rhythmic processes (Tomioka et al., 2012). These rhythmic processes are initiated by transcriptional regulation of clock-controlled genes (CCGs). Several genome-wide studies have shown that a substantial number of genes exhibit circadian expression in heads of Drosophila melanogaster (Keegan et al., 2007; Rodriguez et al., 2013; Hughes et al., 2012) and other insects (Leming et al., 2014) but functional significance of oscillatory CCG expression is poorly understood.
Recent studies have suggested that circadian clocks have a role in regulating oxidative stress responses. Flies with a null mutation in the core clock gene per show increased susceptibility to hydrogen peroxide (Krishnan et al., 2008), and their aging is associated with accelerated neurodegeneration in the brain and reduced lifespan following hyperoxia exposure (Krishnan et al., 2009, 2012). This role of the circadian system appears to be conserved, as clock-deficient mice have elevated oxidative damage and accelerated aging symptoms (Kondratova & Kondratov, 2012).
Glutathione (GSH) is an essential molecule for defence against toxins and oxidative insult. GSH levels have previously been demonstrated to oscillate in heads of wild type D. melanogaster but not in per01 or cyc01 clock mutants (Beaver et al., 2012). The first, rate-limiting reaction in GSH biosynthesis is catalyzed by the holoenzyme glutathione cysteine ligase. Glutathione cysteine ligase is composed of two subunits: the catalytic subunit encoded by Gclc, and the modulatory subunit encoded by Gclm. Both Gclc and Gclm are rhythmically expressed with peak expression at night (Beaver et al., 2012). The peak in Gclc is lost in cyc01 mutants, while expression is constitutively high in per01; this is typical of clock-controlled genes, due to loss of CLK-CYC activation and loss of PER-TIM repression, respectively. A previous genome-wide search for clock-controlled genes also showed cycling Gclc RNA, and revealed that CLK is bound periodically to the promoter region of the Gclc gene via E-boxes, which are the binding sites for CLK/CYC (Abruzzi et al., 2011; Rodriguez et al., 2013). In addition, Gene Ontology analysis found glutathione metabolism to be a category enriched in the dataset of cycling transcripts (Rodriguez et al., 2013). Together, these results provide strong evidence that GSH production is clock-regulated.
The present study investigated whether GSH-related rhythms in fly heads are generated by the central pacemaker, which controls locomotor activity rhythms, or by other peripheral clocks in the fly head. The focus of the present study was on Gclc, because changes in Gclc mRNA levels alone can affect overall glutathione cysteine ligase activity (Lu, 2009), and manipulations in Gclc levels have been shown to have a greater effect on overall GSH levels than Gclm manipulations (Luchak et al., 2007). Results of the current study show that in fly heads, Gclc rhythms do not depend on the central pacemaker, but persist cell-autonomously in peripheral clocks of the nervous system, specifically in glial cells.
Materials and methods
Fly rearing and strains
Drosophila melanogaster were raised on a standard yeast (35g L−1), cornmeal (50g L−1), and molasses (5%) diet at 25 ± 1°C, under a 12 h light/12 h dark (LD 12:12 h) regimen. Flies were exposed to fluorescent light of luminous energy 8 ± 2 μmol m−2 s−1.
To abolish clock function selectively in central clock cells, the Drosophila binary UAS/GAL4 system was used (Brand & Perrimon, 1993). The UAS-cycΔ construct encodes a dominant negative version of the CYC protein, which disrupts the clock mechanism when expressed in target cells (Tanoue et al., 2004). Flies carrying UAS-cycΔ were crossed with Pdf-Gal4, driving expression in both small ventral lateral neurons (s-LNvs) and large ventral lateral neurons (l-LNvs) (Kaneko, 1998), or with cry-Gal4-39, driving expression in the majority of central pacemaker neurons (Grima et al., 2004). These three fly lines were backcrossed for 8 generations to w1118.
To maintain clock function specifically in central clock cells, per01 7.2.2d;;ry506 transgenic flies containing a 7.2kb section of DNA from the per genomic region were used. This fragment excludes most of the promotor region, the 5′ UTR, and part of the first intron of the per gene, yet it is sufficient for rescuing behavioural rhythmicity and clock function in lateral pacemaker neurons (Frisch et al., 1994). The control line was per01;13.2.2e;ry506 flies carrying a 13.2kb genomic DNA fragment that includes the 7.2kb section mentioned above, as well as an additional 4.2kb of regulatory sequences upstream of the per gene. These per01 13.2.2e flies have clock function rescued in all clock cells (Zerr et al., 1990).
eya2 flies without eyes were obtained from the Bloomington Stock Center (stock #2285). Two different cyc-RNAi lines were used in this study to reduce expression of the core clock gene cyc. Bloomington Stock Center stock #42563 is referred to here as cyc-RNAi-sh because it encodes a short hairpin of the cyc sequence. The other cyc-RNAi line from the National Institute of Genetics (stock #8727R-1) is referred to as cyc-RNAi-lh, as it carries a longer cyc-matching hairpin. To abolish clock function in glia, neurons, or both, the cyc-RNAi lines were crossed to the glial driver loco-Gal4 (Bloomington stock #26883), neuronal driver elav-Gal4 (Robinow & White, 1991), or all clock cell driver tim-Gal4 (Kaneko & Hall, 2000), respectively. These three driver lines were backcrossed for 8 generations to w1118.
Locomotor activity analysis
Locomotor activity was measured using Trikinetics Drosophila Activity Monitors DAM2 or DAM5, (Waltham, Massachusetts). Activity counts were taken in 15 min bins for 3 days in LD followed by 7 days in constant darkness (DD). A quantitative measure of rhythmicity in DD was obtained using the fast Fourier Transform (FFT) along with chi-squared periodogram analysis (ClockLab version 2.72, Actimetrics, Wilmette, Illinois). Individuals with a FFT ≥ 0.04 at a period near 24 h or 12 h and a periodogram amplitude peak breaking the 99% confidence line were deemed rhythmic.
Quantitative real-time PCR
Mated flies were separated 1–2 days after emergence, and 5 day old males were used for all experiments. Flies were collected every 4 h over 24 h in LD 12:12 h. Each sample of 50 heads was separated using 710 μm and 425 μm diameter stainless steel sieves frozen with liquid nitrogen, and homogenized in TRI Reagent (Sigma-Aldrich, St. Louis, Missouri) using a Kontes handheld motor and pestle. The RNA was treated with rDNase I (Takara, Otsu, Shiga, Japan), which was removed with a phenol/chloroform extraction, followed by ethanol/sodium acetate precipitation. cDNA was synthesized using the Bio-Rad iScript cDNA synthesis kit (Hercules, California). Real-time PCR was performed with Bio-Rad iTaq SYBR Green Supermix with Rox (Hercules, California) on an Applied Biosystems Step-One Plus real-time machine. Primers were obtained from Integrated DNA Technology (Coralville, Iowa). All primers used in this study had efficiency > 96%, and their sequences are as follows: rp49 forward 5′ GCCCAGCATACAGGCCCAAG 3′, rp49 reverse 5′ AAGCGGCGACGCACTCTGTT 3′; robl forward 5′ AATCCAGAGCCACAAAGGTG 3′, robl reverse 5′ AGTGTTGTCCAGCGTGGATT 3′; tim forward 5′ GTGCTTCTGCTGAGGCGTTTCAAT 3′, tim reverse 5′ GGCGAATGGTTTGACATCCACCAA 3′; Gclc forward 5′ ATGACGAGGAGAATGAGCTG 3′, Gclc reverse 5′ CCATGGACTGCAAATAGCTG 3′. RNA levels were normalized to rp49 or robl (Ling & Salvaterra, 2011) and analyzed using the 2−∆∆CT method. Statistics were calculated using GraphPad Prism 6 (San Diego, California).
Results
Transcriptional rhythms of Gclc persist in fly heads when the central clock is disrupted
Gclc, the gene encoding the catalytic subunit comprising the glutathione cysteine ligase holoenzyme, has been previously shown to display significant transcriptional rhythms in heads of young CantonS (Beaver et al., 2012) and w1118 D. melanogaster (Klichko et al., 2015). To probe the mechanism generating these rhythms, this study investigated whether they are controlled systemically by central pacemaker neurons, or in cell autonomously in peripheral oscillators.
In the first experiment, tim and Gclc mRNA was measured around the clock in heads of flies with disrupted central clock function, but intact peripheral clocks. The most important central clock neurons are small ventral lateral neurons (LNvs) expressing Pdf. Therefore, the Pdf-Gal4 driver combined with a dominant-negative version of cyc (UAS-cycΔ) was used to disable the central clock; these flies are hereon referred to as Pdf>cycΔ. Locomotor activity monitoring showed that 87% of these flies became arrhythmic, whereas 94% of cycΔ>+ control flies remained rhythmic (Table 1). Two-way ANOVA with factors being genotype and time of day show significant differences between peak and trough expression in tim (P < 0.0001, Fig. 1A) and Gclc mRNA levels (P < 0.0001, Fig. 1B) in these behaviourally arrhythmic flies, with similar phase and amplitude as in control flies. Even with a disabled central clock, high-amplitude cycling of the core clock gene tim is expected, because the disrupted Pdf clock cells consist of only 16 LNvs in the brain (Helfrich-Forster, 1998). The bulk of tim gene expression in fly heads comes from circadian oscillators in retinal photoreceptors and glial cells (Cheng & Hardin, 1998; Ng et al., 2011); therefore the clock disruption in the central clock would not be detectable by qRT-PCR.
Table 1.
Locomotor activity statistics in adult Drosophila melanogaster with genetically disrupted circadian clocks.
| Genotype | n | % Rhythmic | Avg Period (h) ± SEM | Avg FFT± SEM |
|---|---|---|---|---|
| Pdf-Gal4/UAS-cycΔ | 16 | 12.50 | 23.67± 0.00 | 0.025± 0.006 |
| UAS-cycΔ/+ | 18 | 94.44 | 23.73± 0.03 | 0.108± 0.008 |
| cry-Gal4-39/UAS-cycΔ | 17 | 0 | N/A | 0.007± 0.001 |
| cry-Gal4-39/+ | 14 | 100 | 24.21± 0.09 | 0.099± 0.010 |
| per01 7.2.2d;;ry506 | 11 | 63.64 | 24.50± 0.14 | 0.063± 0.022 |
| per01;13.2.2e;ry506 | 11 | 90.91 | 23.43± 0.10 | 0.084± 0.014 |
Fig. 1.
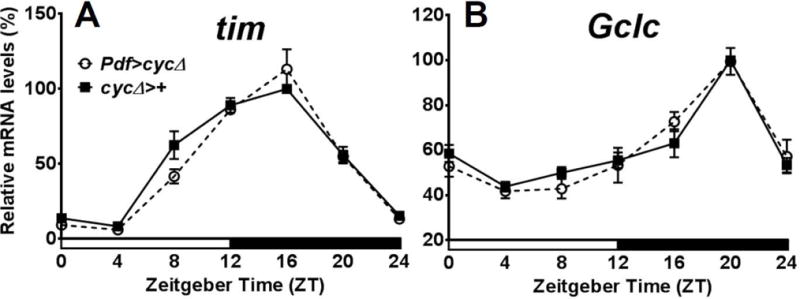
Expression of the genes tim and Gclc in Drosophila melanogaster with clock function disrupted in PDF-positive pacemaker neurons. tim (A) and Gclc (B) mRNA show similar rhythms in heads of flies with clock disrupted in PDF neurons (Pdf>cycΔ) as in the control (cycΔ>+). Levels are normalized to the reference gene rp49. Values are reported as percent of peak expression in the control and represent mean of 3 independent biological replicates ± SEM. Two-way ANOVA with Bonferonni post-test showed no significance difference in tim or Gclc expression between the two genotypes at each time point, but within each genotype, a significant difference between peak and trough expression (P < 0.0001 for both genotypes in tim; P < 0.0001 for both genotypes in Gclc).
A second driver, cry-Gal4-39, was combined with UAS-cycΔ (cry-39>cycΔ) to disable the clock in a larger number of central clock cells. cry-Gal4-39 is active in additional groups of dorsal central pacemaker neurons (Klarsfeld et al., 2004). Again, in both central clock disabled flies and controls, Gclc mRNA levels remained rhythmic in fly heads (P < 0.0001 between peak and trough), as well as tim levels (P < 0.0001, Fig. 2A). Locomotor activity rhythms were abolished in cry-39>cycΔ flies (Fig. 2B, Table 1), confirming that central clock neurons were not functioning. These experiments demonstrate that peripheral clocks in the head can maintain rhythm of Gclc mRNA in the absence of a functioning central circadian clock.
Fig. 2.
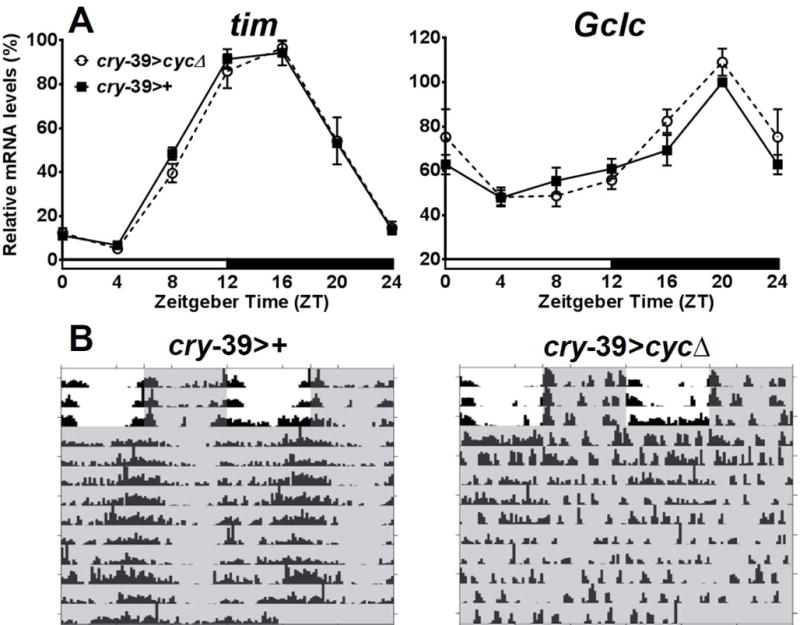
Expression of tim and Gclc in Drosophila melanogaster with a disrupted central clock. (A) Both tim and Gclc mRNA expression profiles are similar in cry-39>cycΔ and cry-39>+ control. Levels are normalized to the reference gene robl. Values are reported as percent of peak expression in the control and represent mean of 3 biological replicates ± SEM. Two-way ANOVA with Bonferonni post-test showed no significant difference in tim or Gclc expression between the two genotypes at each time point, but within each genotype, a significant difference between peak and trough expression (P<0.0001 for both genotypes in tim; P < 0.0001 for both genotypes in Gclc). (B) Representative examples of locomotor activity in cry-39>+ and cry-39>cycΔ. Shaded areas represent periods of darkness.
Gclc rhythms are absent in flies with disrupted peripheral clocks but functioning central clocks
If the observed Gclc rhythms originate from peripheral clock cells, then abolishing clock function in these cells should cause these rhythms to disappear, even if the central clock is functional. To test this prediction, Gclc profiles were measured in per01 7.2.2d flies, which have rescued clock function only in central pacemaker neurons and restored locomotor activity rhythms (Frisch et al., 1994). For the control, per01 13.2.2e flies with rescued clock function in all central and peripheral clocks were used (Zerr et al., 1990). In this experiment, per01 13.2.2e displayed rhythmic tim and Gclc expression as expected, while in per017.2.2d flies, expression of both genes was constitutively high and arrhythmic (Fig. 3A), similar as in non-rescued per01 mutants (Beaver et al., 2012). Locomotor activity monitoring confirmed that 91% of the per0113.2.2e were behaviourally rhythmic, as well as a majority (64%) of the per01 7.2.2d flies (Fig. 3B, Table 1).
Fig. 3.
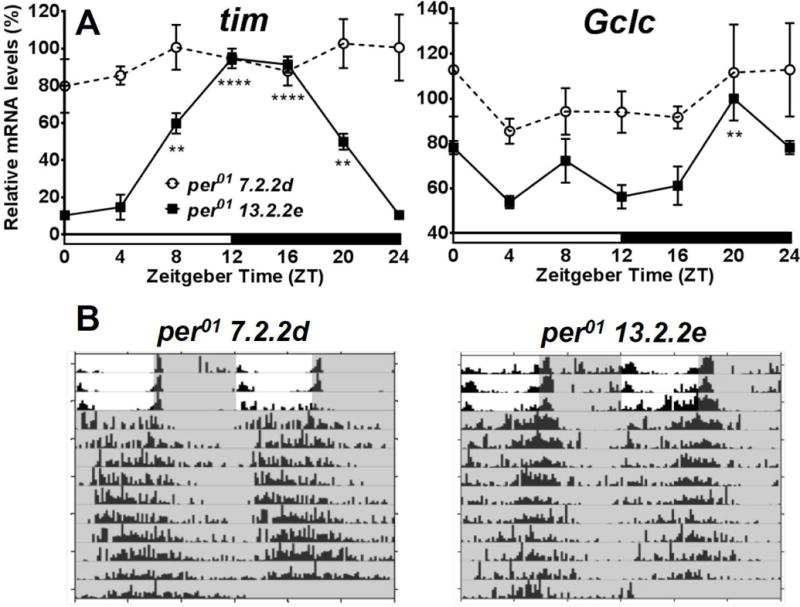
Expression of tim and Gclc expression in heads of per01 background Drosophila melanogaster with rescued clock function in central pacemaker or all head clocks. (A) mRNA expression of tim and Gclc in per01 7.2.2d flies with a rescued central clock, and per01 13.2.2e flies with per rescued in all clock cells. Levels are normalized to the reference gene rp49. Values are reported as percent of peak expression in the control and represent mean of 3 independent biological replicates ± SEM. Stars indicate a significant difference from the trough expression of each genotype, analyzed by two-way ANOVA with Bonferonni post-test (*P < 0.05, **P < 0.01, ****P < 0.0001). (B) Representative actograms showing rhythmic locomotor activity in both genotypes. Shaded areas represent periods of darkness.
Removing photoreceptors does not disrupt Gclc rhythms in the head
Having established that peripheral clocks are responsible for Gclc mRNA rhythms, the next step was to determine whether specific cell types are regulating rhythmic Gclc expression. Peripheral clocks function in retinal photoreceptor cells, glia, and some sensory neurons. When sampling mRNA levels from the whole head, a large part of clock gene expression comes from photoreceptor cells of the compound eyes (Cheng & Hardin, 1998). To determine if Gclc rhythms observed in whole heads are generated by retinal photoreceptor oscillators, eyes absent (eya2) mutants, which are missing the compound eyes, were tested around the clock. Even without the peripheral oscillators in the eyes, heads of eya2 flies show rhythmic profiles of tim, and importantly, they also retained rhythmic expression of Gclc mRNA (P<0.01), as shown in Fig. 4.
Fig. 4.
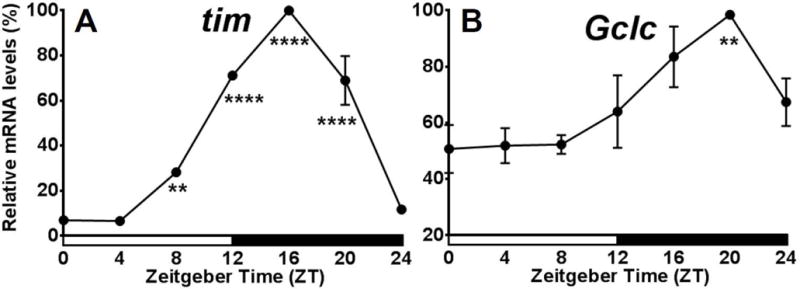
Expression profiles of tim and Gclc in eya2 Drosophila melanogaster. mRNA levels of tim (A) and Gclc (B) in heads of eya2 flies. Levels are normalized to the reference gene rp49. Values are reported as percent of peak expression and represent mean of 3 independent biological replicates ± SEM. Stars indicate a significant difference from the trough using one-way ANOVA with Bonferonni post-test (**P < 0.01, ****P < 0.0001).
Disrupting clock in glial cells abolishes Gclc rhythms in the head
Since genetic removal of photoreceptors did not abolish Gclc rhythms in the whole head, the question of which other peripheral clocks could be responsible for this rhythm remained. It is known that not only neurons but also glial cells possess the clock mechanism (Ng et al., 2011) and express several genes rhythmically (Jackson et al., 2015). In addition, Gclc is listed as a transcript that is enriched in glia (Huang et al., 2015). Therefore, it was investigated how disruption of only neuronal or only glial clocks would affect the Gclc expression pattern in whole heads. Since expression of cycΔ via tim-Gal4 is lethal, two different cyc-RNAi lines, which have been shown to be effective in cell type specific knockdown of cyc (Karpowicz et al., 2013) were used in the following experiments. First, it was verified that the expression of the target gene cyc was significantly reduced when either of the cyc-RNAi lines were driven with tim-Gal4. Indeed, average cyc mRNA was significantly reduced by more than 50% compared to controls (Fig. 5A). This decrease in cyc expression also significantly reduced tim mRNA levels at the peak expression time point. In tim>cyc-RNAi-sh, peak tim expression at ZT16 was significantly lower (P<0.05) compared to both control genotypes (Fig. 5B). Similarly, peak tim expression was also significantly lower (P<0.01) in tim>cyc-RNAi-lh flies compared to both control groups (Fig. 5B). These data, together with significantly reduced locomotor rhythmicity (not shown) suggest that both cyc-RNAi lines are effective in disrupting the clock mechanism.
Fig. 5.
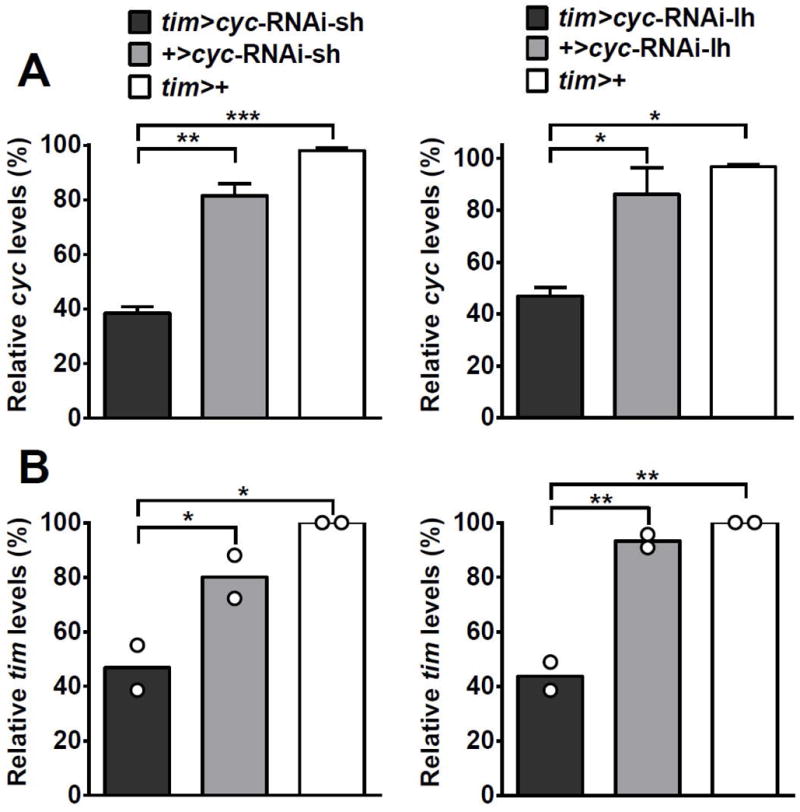
Verification of cyc mRNA knockdown in heads of Drosophila melanogaster expressing either cyc-RNAi-sh or cyc-RNAi-lh via the tim-Gal4 driver. (A) Relative expression of overall cyc mRNA is significantly reduced in flies expressing cyc-RNAi-sh or cyc-RNAi-lh driven by tim-Gal4 compared to controls. Values are reported as percent expression compared to the tim>+ control and represent mean of 4 biological replicates ± SEM. (B) Relative expression of tim mRNA is significantly reduced at the peak time point in flies expressing cyc-RNAi-sh or cyc-RNAi-lh driven by tim-Gal4 compared to respective controls. Values are reported as percent of peak expression in the tim>+ control. Bars represent mean of 2 biological replicates, with dots showing individual replicate values. Levels are normalized to the reference gene rp49. Mean expression levels were compared using one-way ANOVA. Stars indicate significant differences (*P < 0.05, **P < 0.01, ***P < 0.001).
To investigate whether reduced levels of cyc in neuronal or glial clocks would affect the Gclc expression pattern in whole heads, cyc expression was reduced first in all clock cells via tim-Gal4. Gclc expression in tim>cyc-RNAi-sh and tim>cyc-RNAi-lh flies showed significantly reduced peak levels compared to respective controls (Fig. 6A), whereas trough levels were not significantly different, except between tim>cyc-RNAi-sh and +>cyc-RNAi-sh (P < 0.05, Fig 6A). On the other hand, reducing levels of cyc in neurons only via elav-Gal4 did not have such an effect. Both elav>cyc-RNAi-sh and elav>cyc-RNAi-lh flies did not show significantly different expression patterns in Gclc mRNA compared to respective controls (Fig. 6B). In contrast to what was seen with neuronal clock disruption, reducing cyc in all glial cells via loco-Gal4 resulted in a significant (P < 0.01) decrease in the Gclc expression at the peak time point of ZT20. This decrease was observed in both loco>cyc-RNAi-sh and loco>cyc-RNAi-lh flies compared to their respective controls (Fig. 6C). Both cyc-RNAi lines when driven by loco-Gal4 showed no statistical difference between the peak and trough time points providing evidence that the loss of clock function in glia abolish rhythm in Gclc mRNA expression to a similar degree as disruption of all clocks in the fly head.
Fig. 6.
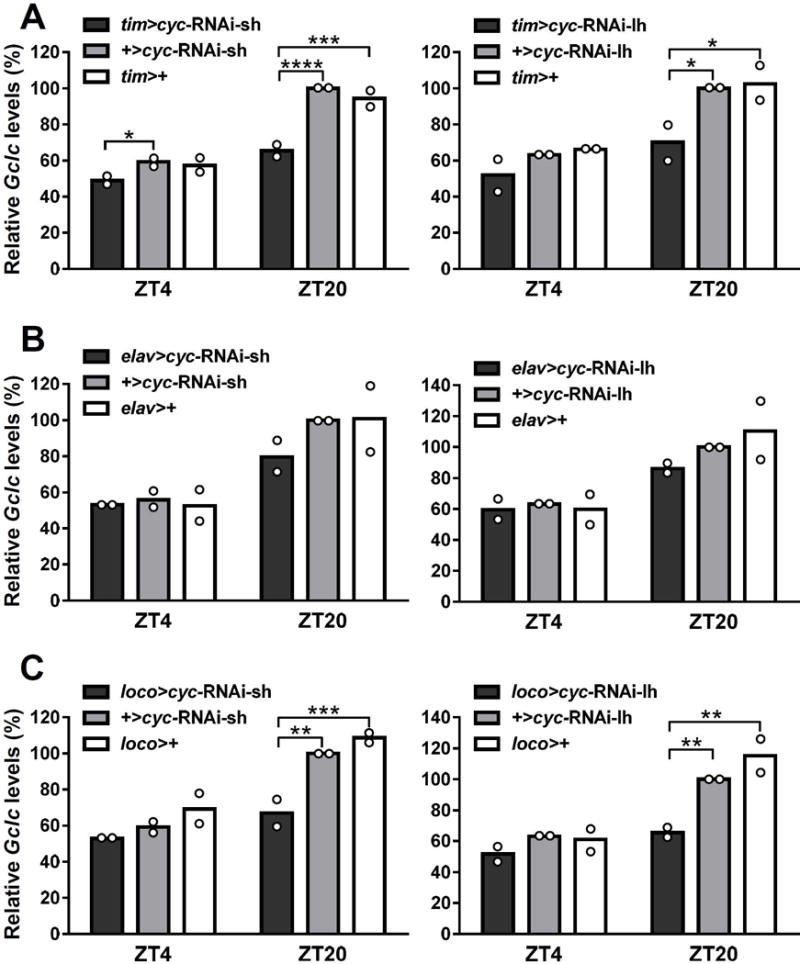
Gclc mRNA profiles in heads of Drosophila melanogaster flies with clock disrupted in all clock cells, neurons only, or glia only. Expression of Gclc in flies expressing cyc-RNAi-sh or cyc-RNAi-lh driven by (A) tim-Gal4, (B) elav-Gal4, or (C) loco-Gal4. Levels of Gclc mRNA are normalized to the reference gene rp49. Each bar represents mean of 2 biological replicates, with dots showing individual replicate values. Mean expression levels at ZT4 (trough) and ZT20 (peak) were compared between genotypes using two-way ANOVA. Values are reported as percent of peak expression in the responder-only control. Stars indicate a significant difference (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Discussion
Due to the power of D. melanogaster genetic tools (Duffy, 2002), it is possible to disable and rescue clocks in specific cells of the brain. This allows for the investigation of which clock-harbouring cells in the multi-oscillatory circadian system are responsible for the rhythm in Gclc mRNA expression. The central pacemaker of the Drosophila circadian clock is made up of three groups of dorsal neurons, and two groups of lateral neurons (LNs) (Lin et al., 2004). Ventral LNs are important for the generation of rest/activity rhythms, and also release the neuropeptide pigment dispersing factor (PDF). In D. melanogaster and other insects, PDF is required to maintain locomotor rhythms in constant darkness, as well as for synchronization of autonomous rhythms in many neurons expressing the PDF receptor (Renn et al., 1999; Lin et al., 2004; Shafer et al., 2008; Im & Taghert, 2010). However, disabling the clock in Pdf-positive cells via expression of cycΔ did not impede the rhythmic expression of Gclc. The cry-Gal4-39 driver is expressed in both dorsal neurons and LNvs (Yoshii et al., 2010). Expression of cycΔ in these cells via cry-Gal4-39 abolished locomotor activity rhythms; however, the rhythm in Gclc expression with a peak at ZT20 still occurred with no difference from the control flies. Together, these data suggest that the central clock, which controls behavioural rhythms, is not responsible for the rhythmic control of Gclc expression. Additional evidence supporting this idea came from investigating flies in a per-null background carrying constructs that rescue per expression in the central pacemaker (Frisch et al., 1994) or all per-expressing cells (Zerr et al., 1990). Flies with rescue of per in all clock cells were confirmed to have rhythmic expression of Gclc mRNA. However, flies with per rescued only in central pacemaker neurons lacked rhythmic expression of Gclc, despite most exhibiting rest/activity rhythms.
Peripheral clocks function in many cell types in the brain including photoreceptor cells of the compound eyes, other sensory neurons, and glia. The profile of Gclc was measured in eya2 flies, which are missing the compound eyes. These flies displayed rhythmic expression of Gclc, suggesting that a different clock-containing cell type in central brain may be responsible for Gclc expression.
Glial cells (or glia) consist of multifunctional cell types that play major roles in nervous system development, defence, and functioning. In the adult brain, glia cells provide nutrients, remove waste products, and provide neurotransmitter precursors (Jackson et al., 2015). It has been shown that Gclc mRNA is enriched in astrocyte-like glia in adult Drosophila (Huang et al., 2015). The present study demonstrates that disruption of the circadian clock machinery in glial cells via two different cyc-RNAi constructs abolished the rhythm in Gclc mRNA expression in whole heads, while disruption of neuronal clocks did not have this effect. It has long been known that glial cells express PER (Ewer et al., 1992). A more recent study reported that glial cells exclusively express ebony under circadian control, which is essential for maintaining locomotor activity rhythms (Suh & Jackson, 2007). Glial cells have also been shown to play a role in daily cell morphology changes that occur in the Drosophila visual system (Gorska-Andrzejak, 2013). In studies of mammalian cell cultures, it was demonstrated that astrocyte glia not only generate GSH, but also release it into the extracellular space (Dringen & Hirrlinger, 2003). Neurons cultured with glial cells contain higher levels of GSH than those cultured alone, possibly through glial contribution of GSH precursors (Dringen & Hirrlinger, 2003).
Glutathione biosynthesis is the rate-limiting factor in other aspects of glutathione metabolism pathways such as glutathione-S-transferase (GST) activity, which is also rhythmic in flies (Hooven et al., 2009) and mosquitos (Balmert et al., 2014). The circadian regulation of genes involved in GSH production and utilization may also explain time-of-day differences in pesticide resistance found in flies (Hooven et al., 2009), mosquitos (Balmert et al., 2014), cockroaches (Lin et al., 2014), and other insects. Whilst many insect studies report rhythms in GST expression or activity in response to pesticide challenge, rhythmic Gclc expression has been reported so far in Drosophila melanogaster (Beaver et al., 2012) and the mosquito Aedes aegypti (Leming et al., 2014).
Glutathione biosynthesis is important for cellular defence against reactive oxygen species (ROS) inter- and extracellularly. The daily rhythms in Gclc may help coordinate ROS defence with increases in GSH production (Patel et al., 2014). The present study demonstrates that the central clock and other neurons are dispensable for the Gclc expression rhythms observed in whole heads. These results suggest that the circadian clock in glia generate the daily rhythm of Gclc expression in the adult Drosophila brain.
Acknowledgments
We thank Drs. P. Hardin, P. Karpowicz, and R. Jackson, and the Bloomington Drosophila Stock Center for fly stocks. Research reported in this publication was supported by the National Institute of Aging of the National Institutes of Health under award number R01 AG045830 to JMG. DML was supported by the NSF IGERT in Aging Sciences Program at OSU.
References
- Abruzzi KC, Rodriguez J, Menet JS, et al. Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes & Development. 2011;25:2374–2386. doi: 10.1101/gad.178079.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmert NJ, Rund SS, Ghazi JP, et al. Time-of-day specific changes in metabolic detoxification and insecticide resistance in the malaria mosquito Anopheles gambiae. Journal of Insect Physiology. 2014;64:30–39. doi: 10.1016/j.jinsphys.2014.02.013. [DOI] [PubMed] [Google Scholar]
- Beaver LM, Klichko VI, Chow ES, et al. Circadian regulation of glutathione levels and biosynthesis in Drosophila melanogaster. PLoS One. 2012;7:e50454. doi: 10.1371/journal.pone.0050454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Hardin PE. Drosophila photoreceptors contain an autonomous circadian oscillator that can function without period mRNA cycling. Journal of Neuroscience. 1998;18:741–750. doi: 10.1523/JNEUROSCI.18-02-00741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dringen R, Hirrlinger J. Glutathione pathways in the brain. Journal of Biological Chemistry. 2003;384:505–516. doi: 10.1515/BC.2003.059. [DOI] [PubMed] [Google Scholar]
- Duffy JB. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- Ewer J, Frish B, Hamblen-Coyle MJ, et al. Expression of the period clock gene within different cell types in the brain of Drosophila adults and mosaic analysis of these cells’ influence on circadian behavioral rhythms. Journal of Neuroscience. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch B, Hardin PE, Hamblen-Coyle MJ, et al. A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Hardin PE. Central and peripheral circadian oscillator mechanisms in flies and mammals. Journal of Cell Science. 2002;115:3369–3377. doi: 10.1242/jcs.115.17.3369. [DOI] [PubMed] [Google Scholar]
- Gorska-Andrzejak J. Glia-related circadian plasticity in the visual system of Diptera. Frontiers in Physiology. 2013;4:36. doi: 10.3389/fphys.2013.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, Rouyer F. Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature. 2004;431:869–873. doi: 10.1038/nature02935. [DOI] [PubMed] [Google Scholar]
- Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Current Opinion in Neurobiology. 2013;23:724–731. doi: 10.1016/j.conb.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C. Robust circadian rhythmicity of Drosophila melanogaster requires the presence of lateral neurons: a brain-behavioral study of disconnected mutants. Journal of Comparative Physiology A. 1998;182:435–453. doi: 10.1007/s003590050192. [DOI] [PubMed] [Google Scholar]
- Hooven LA, Sherman KA, Butcher S, Giebultowicz JM. Does the clock make the poison? Circadian variation in response to pesticides. PLoS One. 2009;4:e6469. doi: 10.1371/journal.pone.0006469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Ng FS, Jackson FR. Comparison of larval and adult Drosophila astrocytes reveals stage-specific gene expression profiles. G3 (Bethesda) 2015;5:551–558. doi: 10.1534/g3.114.016162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Grant GR, Paquin C, et al. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Research. 2012;22:1266–1281. doi: 10.1101/gr.128876.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SH, Taghert PH. PDF receptor expression reveals direct interactions between circadian oscillators in Drosophila. Journal of Comparative Neurology. 2010;518:1925–1945. doi: 10.1002/cne.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson FR, Ng FS, Sengupta S, et al. Glial cell regulation of rhythmic behavior. Methods Enzymology. 2015;552:45–73. doi: 10.1016/bs.mie.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M. Neural substrates of Drosophila rhythms revealed by mutants and molecular manipulations. Current Opinion in Neurobiology. 1998;8:652–658. doi: 10.1016/s0959-4388(98)80095-0. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. Journal of Comparative Neurology. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Karpowicz P, Zhang Y, Hogenesch JB, et al. The circadian clock gates the intestinal stem cell regenerative state. Cell Reports. 2013;3:996–1004. doi: 10.1016/j.celrep.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan KP, Pradhan S, Wang JP, Allada R. Meta-analysis of Drosophila circadian microarray studies identifies a novel set of rhythmically expressed genes. PLoS Computational Biology. 2007;3:e208. doi: 10.1371/journal.pcbi.0030208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarsfeld A, Malpel S, Michard-Vanhee C, et al. Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila. Journal of Neuroscience. 2004;24:1468–1477. doi: 10.1523/JNEUROSCI.3661-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klichko VI, Chow ES, Kotwica-Rolinska J, et al. Aging alters circadian regulation of redox in Drosophila. Frontiers in Genetics. 2015;6:83. doi: 10.3389/fgene.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratova AA, Kondratov RV. The circadian clock and pathology of the ageing brain. Nature Reviews Neuroscience. 2012;13:325–335. doi: 10.1038/nrn3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Davis AJ, Giebultowicz JM. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochemical and Biophysical Research Communications. 2008;374:299–303. doi: 10.1016/j.bbrc.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Kretzschmar D, Rakshit K, et al. The circadian clock gene period extends healthspan in aging Drosophila melanogaster. Aging. 2009;1:937–948. doi: 10.18632/aging.100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Rakshit K, Chow ES, et al. Loss of circadian clock accelerates aging in neurodegeneration-prone mutants. Neurobiology of Disease. 2012;45:1129–1135. doi: 10.1016/j.nbd.2011.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leming MT, Rund SS, Behura SK, et al. A database of circadian and diel rhythmic gene expression in the yellow fever mosquito Aedes aegypti. BMC Genomics. 2014;15:1128. doi: 10.1186/1471-2164-15-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Stormo GD, Taghert PH. The neuropeptide pigment-dispersing factor coordinates pacemaker interactions in the Drosophila circadian system. Journal of Neuroscience. 2004;24:7951–7957. doi: 10.1523/JNEUROSCI.2370-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Lee CM, Huang JH, Lee HJ. Circadian regulation of permethrin susceptibility by glutathione S-transferase (BgGSTD1) in the German cockroach (Blattella germanica) Journal of Insect Physiology. 2014;65:45–50. doi: 10.1016/j.jinsphys.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Ling D, Salvaterra PM. Robust RT-qPCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS One. 2011;6:e17762. doi: 10.1371/journal.pone.0017762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SC. Regulation of glutathione synthesis. Molecular Aspects of Medicine. 2009;30:42–59. doi: 10.1016/j.mam.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchak JM, Prabhudesai L, Sohal RS, et al. Modulating longevity in Drosophila by over- and underexpression of glutamate-cysteine ligase. Annals of the New York Academy of Sciences. 2007;1119:260–273. doi: 10.1196/annals.1404.000. [DOI] [PubMed] [Google Scholar]
- Ng FS, Tangredi MM, Jackson FR. Glial cells physiologically modulate clock neurons and circadian behavior in a calcium-dependent manner. Current Biology. 2011;21:625–634. doi: 10.1016/j.cub.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SA, Velingkaar NS, Kondratov RV. Transcriptional control of antioxidant defense by the circadian clock. Antioxidants & Redox Signaling. 2014;18:2997–3006. doi: 10.1089/ars.2013.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, et al. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/s0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Rieger D, Shafer OT, Tomioka K, Helfrich-Forster C. Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. Journal of Neuroscience. 2006;26:2531–2543. doi: 10.1523/JNEUROSCI.1234-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow S, White K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. Journal of Neurobiology. 1991;22:443–461. doi: 10.1002/neu.480220503. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Tang CH, Khodor YL, et al. Nascent-Seq analysis of Drosophila cycling gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E275–284. doi: 10.1073/pnas.1219969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, et al. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh J, Jackson FR. Drosophila ebony activity is required in glia for circadian regulation of locomotor activity. Neuron. 2007;55:435–447. doi: 10.1016/j.neuron.2007.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanoue S, Krishnan P, Krishnan B, et al. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Current Biology. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Tomioka K, Uryu O, Kamae Y, et al. Peripheral circadian rhythms and their regulatory mechanism in insects and some other arthropods: a review. Journal of Comparative Physiology B. 2012;182:729–740. doi: 10.1007/s00360-012-0651-1. [DOI] [PubMed] [Google Scholar]
- Yoshii T, Hermann C, Helfrich-Forster C. Cryptochrome-positive and -negative clock neurons in Drosophila entrain differentially to light and temperature. Journal of Biological Rhythms. 2010;25:387–398. doi: 10.1177/0748730410381962. [DOI] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, Siwicki KK. Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. Journal of Neuroscience. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]


