Abstract
Hepatitis E Virus (HEV) is a zoonotic pathogen responsible for causing acute hepatitis in human, especially in developing countries. Diagnosis of HEV usually relies on the detection of antibodies mostly by enzyme-linked immunosorbent assay (ELISA). In the present study, we designed a new indirect ELISA (iELISA) based on a short recombinant peptide derived from the capsid protein (ORF2p) and demonstrated its potential for detecting human IgG against HEV genotype 3. The best polystyrene plate (Maxisorp®), optimal ORF2p coating antigen concentration (0,67μg/well) and primary antibody dilution (1:100) were determined. This iELISA showed a sensitivity of 91.4% and specificity of 95.9%. The comparison of our in house iELISA with a commercial assay (RecomWell, Mikrogen®) showed 94.25% of agreement and a kappa index of 0.88. The ORF2 recombinant ELISA was used to screen 780 blood donors for anti-HEV IgG and we found that 314 (40,25%) of these donors were IgG positive. This high prevalence of antibodies suggests, for the first time, that the Southern Brazil region might be endemic to Hepatitis E Virus genotype 3.
Introduction
Viral hepatitis stands up as a major public health issue worldwide and is caused by several different types of enterically and parenterally transmitted viruses. Hepatitis E (HE), for instance, caused by hepatitis E virus (HEV) is endemic in several developing African, Asian and South American countries [1] and autochthones cases are found at increasing and steadily frequency in developed countries [2]. The infection by HEV is usually unnoticed except in pregnant women and patients with liver-related problems [3]. However, a recent report indicated that the infection might become chronic mainly in immunocompromised individual such as kidney and liver-transplanted [4] and HIV-positive subjects [5]. In this scenario, one of the major question relates to the role HEV might take in causing chronic hepatitis in individual under immunosuppressive medication treatments [6] and recipients of blood derived product [7]. Thus, screening for anti-HEV antibodies or HEV RNA amongst blood donor should become mandatory.
HEV is a small icosahedral non-enveloped RNA virus with a single-stranded positive-sense genome with approximately 7.3 kilobases classified in the genus Hepevirus, Hepeviridae family [8]. The viral genome contains three open reading frames (ORFs) that encode the structural and nonstructural proteins. There are 4 well-known genotypes with distinct epidemiological distributions: genotypes 1 and 2 are epidemically found in Asia, Central and South America and in some African countries [9] and infect exclusively humans [10] whereas genotypes 3 and 4 are found mostly in Asia and developing countries and might cause infection in humans and animals, mainly domestic pigs [11] and wild boars [12]. Although there is a strong evidence of cross species transmission infection between human and pigs [13] the role of other animal species in the epidemiology of HEV infection remains to be evaluated.
In endemic countries the transmission of HEV occurs mostly by the oral-fecal route [14]; conversely, in developed countries, foodborne transmission [15], blood transfusion [7] and transplants of solid organs such as heart, lung, liver and kidney [16] are becoming major source of viral dissemination. The ingestion of undercooked contaminated pork meat and pork-related product might also constitute a potential source of infection [17]. However, the scarcity of data on these routes of infection hampers further evaluation on HEV epidemiology and the impact on public health. Nonetheless, the detection of anti-HEV antibodies or HEV nuclei acid amongst blood donors [18, 19] indicates that viral spread might be long occurring and the prevalence and incidence of HEV might be even higher than previously thought. In Brazil, HEV genotype 3 is commonly found in pig farms [20] and autochthonous human cases of HEV have already been described here [21], in Germany [22], France [23], Cambodia [24] and Israel [25] strengthening the zoonotic potential of HEV.
HE diagnosis is based mostly on the detection of anti-HEV antibodies (IgM, IgG and IgA) towards ORF-2 and ORF-3 encoded proteins [26]. Currently, there are commercial kits available to detect anti-HEV antibodies that differ in specificity and sensibility, mainly when used to HEV diagnosis in non-endemic countries [26, 27]. Recently, we expressed and characterized a recombinant protein based on the C-terminal of ORF-2 protein (ORF2p) from HEV genotype 3 and demonstrated its antigenic and diagnosis potential [28]. Here, we developed an indirect ELISA based on the recombinant ORF2p and screened blood samples from healthy blood donors to evaluate the prevalence of anti-HEV antibodies. We found a high prevalence of positive samples which indicates that the region might be potentially endemic to HEV.
Material and methods
Expression and purification of ORF2 recombinant protein
The recombinant HEV genotype 3 capsid protein was produced as previously described by our group [28]. Briefly, the plasmid pET20-His-Mbp-TEV-ORF2p was transformed into competent E. coli strain ER 2566 and induced with 100 mM isopropyl-β-D-thiogalactopyranoside (IPTG, Sigma). Culture media was centrifuged (8,000 × g, 1 h, 4 °C) and the bacteria pellet was suspended in NTA buffer (20mM NaH2PO4, 500 mM NaCl, 20 mM Imidazole, pH 8.0) and sonicated three times at 70 watts (Ultronique QR, Eco-sonics, Brazil). The bacteria lysate was then centrifuged (13.000 × g, 1 h, 4°C) and the supernatant was filtered (0.22 μM) to purify the protein by a two-step procedure, using the ÄKTA Pure® chromatography system (GE Healthcare, Germany) with HisTrap and Sepharose Q columns connected (both from GE Healthcare). The ORF2p was dialyzed in PBS and kept at -80°C until use.
Blood samples
The number of samples required for this study was calculated considering a prevalence of anti-HEV antibodies of 10%, a confidence interval of 95% and a precision of 2%. Then, seven hundred and eighty (780) samples were selected from the blood bank (Hemocentro de Passo Fundo—Hemopasso), in the state of Rio Grande do Sul, from March to October 2015. All samples were from healthy and suitable blood donors as required by the law number 1.075 from the Brazilian Ministry of Health. The study was approved by the Institutional Committee on Research Ethics (process number 849709).
ELISA assay design
Two commercially available polystyrene microplates (Maxisorp® and Polysorp®, Nunc, USA) were evaluated regarding the ability to adsorb the ORF2p. A total of 5 positive and 5 negative serum samples, previously evaluated by a commercial ELISA (RecomWell anti-HEV IgG, Mikrogen®, Germany) were tested in triplicates in three different plates. The plates were coated with ORF2p (5 μg/well) diluted in carbonate buffer (pH 9.6) at 4°C for 12 h and then washed three times with phosphate buffered saline 0.05% Tween (PBST, pH 7.2). Plate wells were blocked with PBST containing 3% bovine serum albumin (BSA) at 37°C for 2 h. Human serum diluted 1:100 in PBST 1% BSA was added and allowed to react for 1 h at 37°C. After washing, peroxidase conjugated goat anti-human IgG (Santa Cruz Biotecnology®, USA) diluted 1:5,000 in PBST 1% BSA was added and the plates were incubated as indicated above, followed by three washes and addition of substrate (3,3, 5,5’-tetrametilbenzidina + 0,06% H2O2). The plates were then incubated in the dark at 22 °C for 10 min and the reaction was stopped by adding 3 N HCl. Plates were read at 450 nm using a Synergy HI plate reader (BioTek®, USA).
The mean positive/negative (P/N) ratio and the mean within-plate percent coefficient of variation (CV%) were calculated for each type of plate as previously described [29]. The plates were then compared using an index obtained by dividing the P/N ratio by the CV%.
The optimal concentration of antigen was determined by titration in which twofold antigen dilution (final volume of 100 μl) was added to the plates and titrated against the positive and negative sera. The optimal antigen concentration was defined as the lowest antigen concentration that caused no significant changes in the OD obtained with the positive and negative sera.
Eight dilutions of the positive and negative serum samples were used to determine the ideal dilution to be used in the iELISA. Serum samples were diluted 1:100 to 1:1.000 in PBS in a 96-DeepWell™ plate (Nunc®, USA) and then 100μl of each dilution was transferred in duplicate to the sensitized plate wells (Maxisorp®, Nunc, USA). The optimal dilution was defined taken in consideration the mean absorption and the serum dilution as recommended previously [30]. Two diluents, PBST 1% BSA and PBST 1% skin milk were also analyzed.
Cut-off point determination
The iELISA cut-off point was set considering the relation between the optical density (OD) of the sample (ODS) and the OD of the negative control (ODNC). The ODNC was calculated using 50 serum samples that were free of anti-HEV IgG and IgM as determined by a commercial ELISA (RecomWell anti-HEV IgM and IgG (Mikrogen®, Germany) and free of HEV RNA [28]. Also, a total of 63 positive serum samples (anti-HEV IgG, determined by RecomWell kit) were included for the cut-off determination. The Receiver Operating Characteristic (ROC) curve analysis was performed to set the threshold taking into account the specificity and sensitivity of this new iELISA. The Area Under the Curve (AUC) analysis was used to evaluate the ability of this test to discriminate between those individuals that have IgG against HEV genotype 3 and those without any IgG antibody against the virus. The sample was considered positive when the relation ODS/ODNC was higher than 2.5.
Statistical analysis
The Kolmogorov-Smirnov test was used to determine the normal distribution of the data. The results were analyzed by Kruskal-Wallis or One Way Anova followed by Tukey post-test according to the data. Significant differences were considered when p < 0.05. All the statistics were performed using the GraphPad Prism software (GraphPad, USA).
Results
Microplates
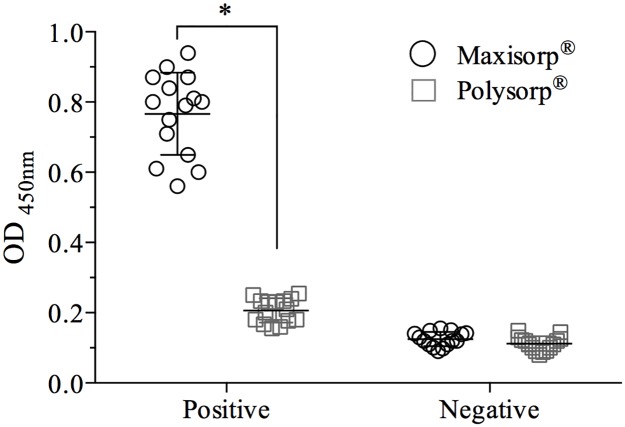
The Maxisorp® microplate adsorbed recombinant ORF2p more efficiently than the Polysorp® microplate. The mean OD of the positive control samples (0.76 ± 0.11) was 3.7 times higher (p = 0.0011) in the Maxisorp® plate (Fig 1). The mean of the ODNC were similar amongst plates (0.125 ± 0.021 in Maxisorp® and 0.121 ± 0.034 in Polysorp® microplate). The adsorption of ORF2p to the Maxisorp® was more constant (lower CV%) and allowed a better distinction between positive and negative samples (P/N ratio) compared to the Polysorp®. The P/N value normalized with its CV% obtained in the Maxisorp® microplate (1.24) was 4.27 times higher than that observed in the Polysorp® microplate (0.29) (Table 1).
Fig 1. Performance of ELISA Maxisorp® and Polysorp® microplates in the in house ELISA assay.
Plates were sensitized with recombinant capsid protein (5 μg/well) and reacted with 5 positive and 5 negative samples previously tested with the commercial kit. Significant differences (p<0.05) on the OD obtained with the same set of serum are indicated by asterisk (*).
Table 1. Performance of ELISA microplates.
Plates were sensitized with recombinant ORF-2 protein and tested with known positive (P) and negative (N) serum to determine the P/N relation and the percentile of the coefficient of variation (CV%). The index value was obtained by dividing the result of P/N by the CV%.
a Percentile of the coefficient of variation (CV) from positive samples tested in triplicates in each microplate.
b Index obtained by dividing the P/N ratio by the CV%. The microplates were classified by this index.
Optimal antigen concentration and primary serum dilution
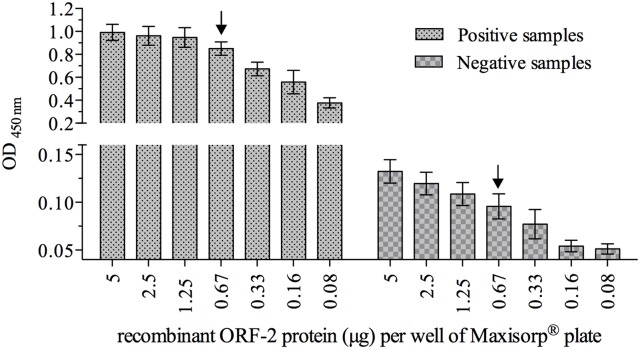
The ideal concentration of ORF2p was determined by twofold serial dilution using the Maxisorp® microplate and was adjusted at 0.67 μg/well (Fig 2). At this antigen concentration the mean OD readings obtained from the positive and negative control samples were similar to that obtained at the highest antigen concentration (5 μg/well) but significantly different (p = 0.0008) from the next concentration (0.33 μg/well). The remaining experiments were then performed using the ORF2p at 0.67 μg/well.
Fig 2. Optimal antigen concentration for the in house ELISA.
Recombinant capsid protein (5 μg) was twofold serially diluted on Maxisorp® microplate. Serum samples, previously determined to be positive (n = 5) or negative (n = 5) were evaluated. The data is represented as the mean ± SEM of the OD of each antigen dilution. The arrow indicates the optimal antigen concentration.
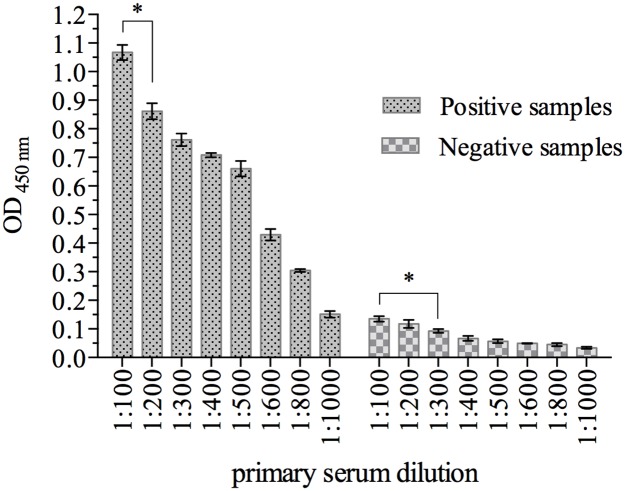
The optimal dilution of human sera to be used in the iELISA was determined taken into consideration the P/N OD relation. Positive control samples diluted 1:100 gave a significantly higher (p = 0.0001) OD reading mean than that obtained at the 1:200 dilution (Fig 3). At the same dilution (1:100), the mean OD of the negative control samples was below 0.15 and was similar at the 1:200 and 1:300 dilutions. A significantly (p = 0.0001) lower OD mean was obtained only when negative control samples were diluted more than 1:300. Thus, in the remaining experiments serum samples were diluted 1:100 to be evaluated.
Fig 3. Determination of the primary serum dilution.
Human serum samples known to be positive (n = 5) or negative (n = 5) were diluted as indicated in the figure and tested for their ability to bind the recombinant capsid protein (5 μg/well) adsorbed onto Maxisorp® ELISA microplates. The results are expressed as the mean ± SEM of the OD obtained at each serum dilution. The asterisk (*) indicates the lowest serum dilution with significant difference (p<0.05) to the immediately higher dilution within positive or negative samples.
Specificity and sensitivity of iELISA
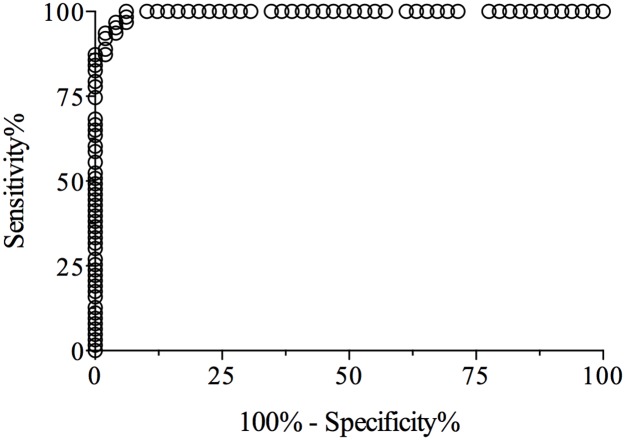
The cut-off value, specificity and sensitivity of the iELISA were determined by testing 50 negative and 63 positive human serum samples for antibodies against HEV. The ROC curve analysis of the iELISA data produced paired estimates of relative sensitivity and relative specificity at different cut-off values. A cut-off of OD ≥ 0.31 was recommended. At this cut-off value, the relative sensitivity and specificity estimates were 96.8% (95% confidence interval = 91.4% to 99.6%) and 95.9% (95% confidence interval = 88.7% to 99.5%), respectively. The Likelihood ratio at this cut-off was 42.78. The ROC curve (Fig 4) had an AUC value of 0.9955 (95% confidence interval = 0.988 to 1.002), which indicated a high level of accuracy for this iELISA. Based in this cut-off value we generated a relation between ODS (sample) and ODNC (negative control), and the sample was considered positive when the relation ODS/ODNC was higher than 2.5, as previously suggested [31].
Fig 4. Receiver Operating Characteristic (ROC) analysis.
The ROC curve was generated using the results obtained by analyzing 50 negative human serum samples and 63 positive human serum samples by the iELISA. The area under the ROC curve was of 0.9955.
Comparison between in house iELISA and commercial assay
The performance of the in house iELISA to detect anti-HEV IgG was compared with the commercial ELISA assay (RecomWell, Mikrogen®, Germany). We randomly selected 87 samples previously tested in the in house iELISA and evaluated them in duplicates using the commercial ELISA. The sample panel was composed by 49 positive and 38 negative sera. When analyzed in the commercial ELISA we found 50 positive and 37 negative samples (Table 2) resulting in 94,25% of agreement and a Kappa (K) index of 0.88 (strong/very good agreement).
Table 2. Agreement between the performance of the in house ELISA and the RecomWell (Mikrogen®) in detecting anti-HEV IgG.
Eight seven serum samples were randomly selected amongst the samples used in this study and tested in duplicates with the commercial ELISA kit to determine the presence of anti-HEV antibodies.
| Commercial ELISA kit | ||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| in house iELISA | Positive | 47 | 2 | 49 |
| Negative | 3 | 35 | 38 | |
| Total | 50 | 37 | 87 | |
Prevalence of anti-HEV IgG in blood donors
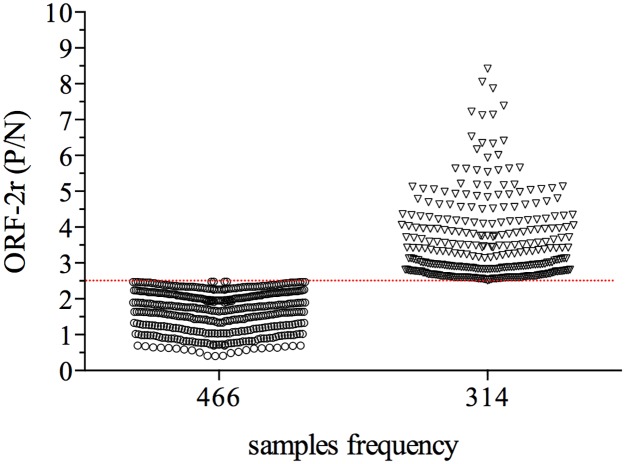
Aiming to evaluate the prevalence of anti-HEV IgG amongst blood donors we analyzed 780 samples collected from March to October (2015) at a major blood bank from north region of the Rio Grande do Sul State. We found that 314 (40.25%) samples had OD reading higher than the cut-off value that we set for the assay (Fig 5) and where thus considered positive to the presence of anti-HEV IgG antibodies. Amongst the positive samples, 90% had OD readings 2.5 to 5 times higher than the average obtained with the negative control samples and the remaining 10% of the positive samples had a P/N ratio higher than 5.
Fig 5. Prevalence of anti-HEV antibodies amongst blood donors.
The in house ELISA was performed using the ideal concentration of antigen and ideal serum dilution, with 780 blood samples. The results are expressed as the P/N ratio, as described in material and methods. Samples with a P/N ratio was ≥ 2.5. The cut-off value is represented by the dashed line.
Discussion
Hepatitis E is a neglected disease in Brazil and most developing countries and fatalities rates are of major concern to health authorities [31]. The course of HEV is usually asymptomatic and short lived [32] but in immunocompromised patients HEV might persist causing chronic infection and hepatic failure, neurological syndrome, renal injury and autoimmune hepatitis [32]. Furthermore, because HEV might be transmitted through blood transfusion [7], screening for anti-HEV antibodies in blood donors should help in preventing widespread dissemination of the virus, mainly in industrialized countries.
Recently, we described the production of a potential candidate antigen for the development of diagnosis to HEV [28]. The viral capsid protein, encoded by ORF 2 gene, is highly immunogenic and widely used in immunosorbent assays to detect anti-HEV antibodies in humans and animals [33–36]. Although different genotypes of HEV share common epitope within the ORF2 protein [37, 38], a better immunoreactivity in the ELISA platform is achieved when the antigens used to coat the plate belong to the same HEV genotype that is infecting the patients or animals tested [39]. Here, we describe the development of an indirect ELISA assay using a recombinant viral capsid protein suitable to detect anti-HEV antibodies in blood donors. We found that the characteristics of the microplate can affect the outcome of the assay. The MaxiSorp® microplate had a higher (p = 0.0011) antigen binding efficiency and a lower (p = 0.0021) CV compared to PolySorp® microplate. The MaxiSorp® microplate adsorbs molecules with hydrophilic and hydrophobic domains whereas PolySorp® microplates are more suitable to adsorb molecules with hydrophobic domains.
The optimal concentration of antigen was set at 0.67 μg/well; considering the maximal binding capacity/cm2 (0.5 μg of protein) and the final volume used in the assay (100 μl), the estimated saturation would occur with 0.42 μg of antigen. Indeed, in our study we found that with an antigen concentration of 0.33 μg/well the average OD value obtained with the positive control was significantly (p = 0.0008) lower compared to a higher antigen concentration (0.67 μg/well). The optimal antigen concentration in our study is higher than that reported previously [34] in which 0.25 μg/well of antigen was used. In similar studies, even lower antigen concentrations (15 ng and 30 ng/well) have been used in indirect ELISA to HEV [35, 38]. Considering that all studies evaluated the best microplate to be used in the assay, the differences in the amount of antigen used might be attributed to the system in which the recombinant protein was produced and with the size of the antigen used, that varied from approximately 30 KDa [28, 34] to 68 KDa [35, 38]. When antigens containing only the C-terminal domain of ORF2p are used, as reported here and by others [34], a higher antigen concentration is needed to obtain OD reading similar to that obtained when the entire ORF2p is used, indicating that the N-terminal domain of the protein contains important antigenic epitopes that should be useful for serological diagnosis. Furthermore, the ideal dilution of the primary antibody (1:100) in which the negative control samples had OD reading lower than 0.15, is similar to that reported with other iELISA assays for HEV [34, 35, 38].
The performance of our iELISA assay was compared to a commercial assay (RecomWell®) which is known for its higher sensitivity and widely use in epidemiological studies in areas endemic for HEV genotype 3 [40]. We found 94.25% of agreement between the results and a K value of 0.88, similar to the results reported previously by Jímenez de Oya (35) and slightly better when compared to the study published by Arankalle (36). It is worth noticing that the antigen we used was expressed in E. coli rather that in eukaryotic cells [35, 36] but still we obtained a high quality antigen suitable to be used in immunoassays.
In Brazil, antibodies to HEV genotype 3 were detected in pigs farms [20, 41, 42] and HEV was found in a case of human hepatitis [21] but blood related products were not evaluated yet for the presence of HEV RNA and limited is the information about the antibodies prevalence. In addition, because of the recent transfusion-transmitted HEV infection, blood products might pose a risk to immunocompromised or organ transplanted patient [40]. Thus, using our iELISA, we analyzed 780 blood samples from a major blood bank. Blood donors were from the North region of Rio Grande do Sul state, a region with a high density of pigs farms and high consumption of pig meat and related products. Amongst the samples analyzed, 314 (40.25%) had antibodies to HEV, a prevalence much higher than that reported recently in a neighboring state [43] or other countries. Even though differences in the ELISA settings and sensitivity of the assay might account for differences in the percentile of seropositive found amongst different studies [44] we credit the higher percentile of seropositive samples to characteristics inherent to the population studied and to the geographical localization. Endemic areas to HEV genotype 3 are found in France [19] and Cambodia [24] in which the prevalence of IgG antibodies to HEV was 52.5% and 28.5% amongst blood donors respectively. The population of our study resides within a region with high density of pig farms in which 15% of Brazilian pigs are produced [45]. Furthermore, more than 80% of the blood donors indicated that pig meat or related product are part of their everyday diet (data not shown) and this could account for the higher prevalence of anti-HEV antibodies, as already reported in other countries [19, 46].
In conclusion, we report the development of an iELISA based on the HEV genotype 3 recombinant capsid protein and found a high prevalence of antibodies to HEV amongst blood donors suggesting that the region might be endemic to HEV. Even though the prevalence of HEV RNA is low in blood samples from blood banks [18] our data points to the need to investigate the presence of viral RNA in samples with high titer aiming to minimize the possibility of horizontal transmission that could be harmful to immunocompromised patients.
Acknowledgments
We thank Jaqueline Mendes de Oliveira (Laboratório de Desenvolvimento Tecnológico em Virologia/IOC—Fundação Instituto Oswaldo Cruz, Rio de Janeiro, Brazil) for kindly reviewing this manuscript.
Data Availability
Data are available on the figshare repository (doi:10.6084/m9.figshare.4902104).
Funding Statement
The author RF received financial support from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, grant n° 485807/2013).
References
- 1.Aggarwal R. Hepatitis e: epidemiology and natural history. J Clin Exp Hepatol. 2013;3(2):125–33. 10.1016/j.jceh.2013.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pérez-Gracia MT, Suay-García B, García M, Mateos-Lindemann ML. Hepatitis E: latest developments in knowledge. Future Microbiology. 2016;11(6):789–808. [DOI] [PubMed] [Google Scholar]
- 3.Navaneethan U, Al Mohajer M, Shata MT. Hepatitis E and pregnancy: understanding the pathogenesis. Liver Int. 2008;28(9):1190–9. 10.1111/j.1478-3231.2008.01840.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamar N, Selves J, Mansuy JM, Ouezzani L, Peron JM, Guitard J, et al. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med. 2008;358(8):811–7. 10.1056/NEJMoa0706992 [DOI] [PubMed] [Google Scholar]
- 5.Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med. 2009;361(10):1025–7. 10.1056/NEJMc0903778 [DOI] [PubMed] [Google Scholar]
- 6.Pischke S, Gisa A, Suneetha PV, Wiegand SB, Taubert R, Schlue J, et al. Increased HEV seroprevalence in patients with autoimmune hepatitis. PLoS One. 2014;9(1):e85330 10.1371/journal.pone.0085330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khuroo MS, Kamili S, Yattoo GN. Hepatitis E virus infection may be transmitted through blood transfusions in an endemic area. J Gastroenterol Hepatol. 2004;19(7):778–84. 10.1111/j.1440-1746.2004.03437.x [DOI] [PubMed] [Google Scholar]
- 8.Emerson SU, Anderson D., Arankalle A., Meng X.-J., Purdy M., Schlauder G.G., Tsarev S.A. Hepevirus In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committe on the Taxonomy of Viruses. London: Elsevier Academic Press; 2005. p. 853–7. [Google Scholar]
- 9.Purcell RH, Emerson SU. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008;48(3):494–503. 10.1016/j.jhep.2007.12.008 [DOI] [PubMed] [Google Scholar]
- 10.Meng XJ. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet Microbiol. 2010;140(3–4):256–65. 10.1016/j.vetmic.2009.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seminati C, Mateu E, Peralta B, de Deus N, Martin M. Distribution of hepatitis E virus infection and its prevalence in pigs on commercial farms in Spain. Vet J. 2008;175(1):130–2. 10.1016/j.tvjl.2006.11.018 [DOI] [PubMed] [Google Scholar]
- 12.de Deus N, Peralta B, Pina S, Allepuz A, Mateu E, Vidal D, et al. Epidemiological study of hepatitis E virus infection in European wild boars (Sus scrofa) in Spain. Vet Microbiol. 2008;129(1–2):163–70. 10.1016/j.vetmic.2007.11.002 [DOI] [PubMed] [Google Scholar]
- 13.Meng XJ, Halbur PG, Shapiro MS, Govindarajan S, Bruna JD, Mushahwar IK, et al. Genetic and experimental evidence for cross-species infection by swine hepatitis E virus. J Virol. 1998;72(12):9714–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlauder GG, Mushahwar IK. Genetic heterogeneity of hepatitis E virus. J Med Virol. 2001;65(2):282–92. [DOI] [PubMed] [Google Scholar]
- 15.Yugo DM, Meng XJ. Hepatitis E virus: foodborne, waterborne and zoonotic transmission. Int J Environ Res Public Health. 2013;10(10):4507–33. 10.3390/ijerph10104507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pas SD, de Man RA, Mulders C, Balk AH, van Hal PT, Weimar W, et al. Hepatitis E virus infection among solid organ transplant recipients, the Netherlands. Emerg Infect Dis. 2012;18(5):869–72. 10.3201/eid1805.111712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng XJ. From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 2011;161(1):23–30. 10.1016/j.virusres.2011.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer C, Hofmann M, Danzer M, Hofer K, Kaar J, Gabriel C. Seroprevalence and Incidence of hepatitis E in blood donors in Upper Austria. PLoS One. 2015;10(3):e0119576 10.1371/journal.pone.0119576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansuy JM, Bendall R, Legrand-Abravanel F, Saune K, Miedouge M, Ellis V, et al. Hepatitis E virus antibodies in blood donors, France. Emerg Infect Dis. 2011;17(12):2309–12. 10.3201/eid1712.110371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.dos Santos DR, Vitral CL, de Paula VS, Marchevsky RS, Lopes JF, Gaspar AM, et al. Serological and molecular evidence of hepatitis E virus in swine in Brazil. Vet J. 2009;182(3):474–80. 10.1016/j.tvjl.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 21.Lopes Dos Santos DR, Lewis-Ximenez LL, da Silva MF, de Sousa PS, Gaspar AM, Pinto MA. First report of a human autochthonous hepatitis E virus infection in Brazil. J Clin Virol. 2010;47(3):276–9. 10.1016/j.jcv.2009.12.021 [DOI] [PubMed] [Google Scholar]
- 22.Pischke S, Behrendt P, Bock CT, Jilg W, Manns MP, Wedemeyer H. Hepatitis E in Germany—an under-reported infectious disease. Dtsch Arztebl Int. 2014;111(35–36):577–83. 10.3238/arztebl.2014.0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legrand-Abravanel F, Kamar N, Sandres-Saune K, Garrouste C, Dubois M, Mansuy JM, et al. Characteristics of autochthonous hepatitis E virus infection in solid-organ transplant recipients in France. J Infect Dis. 2010;202(6):835–44. 10.1086/655899 [DOI] [PubMed] [Google Scholar]
- 24.Nouhin J, Prak S, Madec Y, Barennes H, Weissel R, Hok K, et al. Hepatitis E virus antibody prevalence, RNA frequency, and genotype among blood donors in Cambodia (Southeast Asia). Transfusion. 2016. [DOI] [PubMed] [Google Scholar]
- 25.Erez-Granat O, Lachish T, Daudi N, Shouval D, Schwartz E. Hepatitis E in Israel: A nation-wide retrospective study. World J Gastroenterol. 2016;22(24):5568–77. 10.3748/wjg.v22.i24.5568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aggarwal R. Diagnosis of hepatitis E. Nat Rev Gastroenterol Hepatol. 2013;10(1):24–33. 10.1038/nrgastro.2012.187 [DOI] [PubMed] [Google Scholar]
- 27.Echevarria JM, Gonzalez JE, Lewis-Ximenez LL, Dos Santos DR, Munne MS, Pinto MA, et al. Hepatitis E virus infection in Latin America: a review. J Med Virol. 2013;85(6):1037–45. 10.1002/jmv.23526 [DOI] [PubMed] [Google Scholar]
- 28.de Almeida Ramos D, Miani M, Pandolfi R, Tondo L, Colli ML, Rosado Spilki F, et al. Production and characterization of a Brazilian candidate antigen for Hepatitis E Virus genotype 3 diagnosis. FEMS Microbiol Lett. 2016;363(5). [DOI] [PubMed] [Google Scholar]
- 29.Trottier YL, Wright PF, Lariviere S. Optimization and standardization of an enzyme-linked immunosorbent assay protocol for serodiagnosis of Actinobacillus pleuropneumoniae serotype 5. J Clin Microbiol. 1992;30(1):46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wright PF, Nilsson E, Van Rooij EM, Lelenta M, Jeggo MH. Standardisation and validation of enzyme-linked immunosorbent assay techniques for the detection of antibody in infectious disease diagnosis. Rev Sci Tech. 1993;12(2):435–50. [DOI] [PubMed] [Google Scholar]
- 31.WHO. Hepatitis E http://www.who.int/mediacentre/factsheets/fs280/en/ -: World Health Organization; 2015 [updated July 2015; cited 2015 August 2015].
- 32.Aggarwal R. Clinical presentation of hepatitis E. Virus Res. 2011;161(1):15–22. 10.1016/j.virusres.2011.03.017 [DOI] [PubMed] [Google Scholar]
- 33.Behrendt P, Bremer B, Todt D, Brown RJ, Heim A, Manns MP, et al. Hepatitis E Virus (HEV) ORF2 Antigen Levels Differentiate Between Acute and Chronic HEV Infection. J Infect Dis. 2016;214(3):361–8. 10.1093/infdis/jiw161 [DOI] [PubMed] [Google Scholar]
- 34.Pezzoni G, Caminiti A, Stercoli L, Grazioli S, Galletti G, Santi A, et al. Comparison of three in-house ELISAs for the detection of hepatitis E virus infection in pigs under field conditions. J Virol Methods. 2014;207:95–103. 10.1016/j.jviromet.2014.06.025 [DOI] [PubMed] [Google Scholar]
- 35.Jimenez de Oya N, Galindo I, Girones O, Duizer E, Escribano JM, Saiz JC. Serological immunoassay for detection of hepatitis E virus on the basis of genotype 3 open reading frame 2 recombinant proteins produced in Trichoplusia ni larvae. J Clin Microbiol. 2009;47(10):3276–82. 10.1128/JCM.00750-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arankalle VA, Lole KS, Deshmukh TM, Chobe LP, Gandhe SS. Evaluation of human (genotype 1) and swine (genotype 4)-ORF2-based ELISAs for anti-HEV IgM and IgG detection in an endemic country and search for type 4 human HEV infections. J Viral Hepat. 2007;14(6):435–45. 10.1111/j.1365-2893.2006.00801.x [DOI] [PubMed] [Google Scholar]
- 37.Ma H, Song X, Li Z, Harrison TJ, Zhang H, Huang W, et al. Varying abilities of recombinant polypeptides from different regions of hepatitis E virus ORF2 and ORF3 to detect anti-HEV immunoglobulin M. J Med Virol. 2009;81(6):1052–61. 10.1002/jmv.21484 [DOI] [PubMed] [Google Scholar]
- 38.Engle RE, Yu C, Emerson SU, Meng XJ, Purcell RH. Hepatitis E virus (HEV) capsid antigens derived from viruses of human and swine origin are equally efficient for detecting anti-HEV by enzyme immunoassay. J Clin Microbiol. 2002;40(12):4576–80. 10.1128/JCM.40.12.4576-4580.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma H, Song X, Harrison TJ, Zhang H, Huang W, Wang Y. Hepatitis E virus ORF3 antigens derived from genotype 1 and 4 viruses are detected with varying efficiencies by an anti-HEV enzyme immunoassay. J Med Virol. 2011;83(5):827–32. 10.1002/jmv.22032 [DOI] [PubMed] [Google Scholar]
- 40.Pas SD, Streefkerk RH, Pronk M, de Man RA, Beersma MF, Osterhaus AD, et al. Diagnostic performance of selected commercial HEV IgM and IgG ELISAs for immunocompromised and immunocompetent patients. J Clin Virol. 2013;58(4):629–34. 10.1016/j.jcv.2013.10.010 [DOI] [PubMed] [Google Scholar]
- 41.Vitral CL, Pinto MA, Lewis-Ximenez LL, Khudyakov YE, dos Santos DR, Gaspar AM. Serological evidence of hepatitis E virus infection in different animal species from the Southeast of Brazil. Mem Inst Oswaldo Cruz. 2005;100(2):117–22. [DOI] [PubMed] [Google Scholar]
- 42.da Costa Lana MV, Gardinali NR, da Cruz RA, Lopes LL, Silva GS, Caramori JG Junior, et al. Evaluation of hepatitis E virus infection between different production systems of pigs in Brazil. Trop Anim Health Prod. 2014;46(2):399–404. 10.1007/s11250-013-0503-3 [DOI] [PubMed] [Google Scholar]
- 43.Passos-Castilho AM, de Sena A, Geraldo A, Spada C, Granato CF. High prevalence of hepatitis E virus antibodies among blood donors in Southern Brazil. J Med Virol. 2016;88(2):361–4. 10.1002/jmv.24336 [DOI] [PubMed] [Google Scholar]
- 44.Wenzel JJ, Preiss J, Schemmerer M, Huber B, Jilg W. Test performance characteristics of Anti-HEV IgG assays strongly influence hepatitis E seroprevalence estimates. J Infect Dis. 2013;207(3):497–500. 10.1093/infdis/jis688 [DOI] [PubMed] [Google Scholar]
- 45.IBGE. Efetivo do rebanho suíno brasileiro no ano de 2015: IBGE; 2016 [updated 11/14/2016; cited 2016]. http://www.sidra.ibge.gov.br/bda/tabela/protabl.asp?c=3939&z=t&o=24&i=P.
- 46.Krumbholz A, Joel S, Dremsek P, Neubert A, Johne R, Durrwald R, et al. Seroprevalence of hepatitis E virus (HEV) in humans living in high pig density areas of Germany. Med Microbiol Immunol. 2014;203(4):273–82. 10.1007/s00430-014-0336-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available on the figshare repository (doi:10.6084/m9.figshare.4902104).