Abstract
Amebiasis is one of the twenty major causes of disease in Mexico; however, the diagnosis is difficult due to limitations of conventional microscopy-based techniques. In this study, we analyzed stool samples using polymerase chain reaction-denaturing gradient gel electrophoresis (PCR-DGGE) to differentiate between Entamoeba histolytica (pathogenic) and E. dispar (non-pathogenic). The target for the PCR amplification was a small region (228 bp) of the adh112 gene selected to increase the sensitivity of the test. The study involved 62 stool samples that were collected from individuals with complaints of gastrointestinal discomfort. Of the 62 samples, 10 (16.1%) were positive for E. histolytica while 52 (83.9%) were negative. No sample was positive for E. dispar. These results were validated by nested PCR-RFLP (restriction fragment length polymorphism) and suggest that PCR-DGGE is a promising tool to differentiate among Entamoeba infections, contributing to determine the specific treatment for patients infected with E. histolytica, and therefore, avoiding unnecessary treatment of patients infected with the non-pathogenic E. dispar.
Keywords: Amebiasis, Neglected diseases, Diagnostic, PCR, DGGE, adh112 gene
Introduction
One of the major health problems in developing countries is amebiasis. In 1997, the World Health Organization declared this disease as the third leading cause of death due to parasitic infections (1,2). Currently, amebiasis is still a serious public health problem because parasitic infections are commonly neglected, particularly in populations that lack hygienic measures and clean drinking water (1,3). In Mexico, amebiasis is one of the twenty major causes of disease; its incidence rate in 2000 was 1,353.43 per 100,000 (1,4,5).
Amebiasis is caused by the parasite Entamoeba histolytica, including both intestinal and extra-intestinal infections. This parasite can be present in sewage and contaminated water. According to its cell cycle, it can exist in two forms: trophozoites and cysts (6,7). There is a second species with identical morphological characteristics to those described for E. histolytica called Entamoeba dispar; however, the biochemical, immunological and genetic data indicate that E. dispar is non-pathogenic (8). The life cycle in both species is the same. The infection begins with the ingestion of cysts from water or food contaminated with fecal matter. In the small intestine occurs the excystation and the trophozoites emerge. The trophozoites colonize the large intestine and adhere to the colonic mucosa (6,9). Only the not encysted trophozoites of E. histolytica acquire invasiveness. By the action of proteases, hyaluronidases and mucopolysaccharidases E. histolytica erodes the mucosa producing ulcers and may even reach the submucosa. The adhesive interaction of the trophozoites with the surface of host cells is determinant for the invasion of human tissues, cytotoxic activity, and severity of the disease (10). Primary molecules involved in the intestinal invasion process of E. histolytica are the Gal/Gal NAc lectin and EhCPADH112 (124 kDa) complex (11,12). This complex is formed by the genes: i) Ehcp112, encoding a cysteine protease (50 kDa), and ii) Ehadh112, encoding an adhesin (75 kDa). Some studies have analyzed the molecular role of EhCPADH112 in E. histolytica, but it has not been identified in E. dispar (13).
The laboratory diagnosis of amebiasis is usually based on microscopy, immunological methods and polymerase chain reaction (PCR). The occurrence of non-pathogenic species (particularly E. dispar) causes a confusing scenario for a correct diagnosis of intestinal amebiasis and E. histolytica is often inaccurately reported or diagnosed (8,14 –16). The denaturing gradient gel electrophoresis (DGGE) is a well-established tool for molecular microbiology; this method makes possible the electrophoretic separation of DNA fragments on the basis of differences in nucleotide composition rather than their size (17,18).
To address the need for a reliable diagnostic test of amebiasis caused by E. histolytica in human stools, we developed a polymerase chain reaction–denaturing gradient gel electrophoresis (PCR–DGGE), which differentiates E. histolytica from E. dispar, as it is highly specific and sensitive to these two species (more specific than other techniques such as ELISA). The PCR primers were based on a conserved portion of the adh112 gene spanning a region with substitutions that allowed the differentiation between pathogen and non-pathogen species. To our knowledge, there are no studies using DGGE as a molecular identification technique to determine the prevalence of E. histolytica in Mexico.
Material and Methods
Sample details
The study involved 62 stool samples collected from patients who presented gastrointestinal complaints and attended the "Maximiliano Dorantes" Health Center. They were all examined for intestinal parasites using coproparasitoscopic studies (for multiple ova and parasites); we used the method of Faust with subsequent staining with Lugol solution in order to find cysts (19,20). We used E. histolytica DNA as a positive control, which was obtained from a monogenic culture of strain HM-1 IMSS donated by the Molecular Diagnostic Laboratory, Department of Experimental Pathology CINVESTAV. Likewise, DNA of E. dispar was also used as a positive control; this was donated by the Department of Experimental Medicine, Faculty of Medicine UNAM. The coproparasitoscopic tests were performed in triplicate.
DNA extraction
The DNA of controls (E. histolytica and E. dispar) as well as the 62 studied samples were obtained directly from stools stored at –20°C, by means of Entamoeba cysts mechanical lysis, using zircon beads of 0.01 mm in diameter and then using the Wizard® Genomic DNA Purification Kit (Promega, USA) following the manufacturer's recommendations. As negative controls, we used human DNA extracted from whole blood of volunteer donors. We also utilized the Wizard® Genomic DNA Purification Kit to isolate DNA from white blood cells. The DNA samples were stored at –20°C until analyzed.
Bioinformatic analysis for primer design
In order to differentiate the orthologous adh112 gene of E. dispar from the adhesin of E. histolytica, a basic local alignment search tool (BLAST) was performed using the sequence reported by Garcia-Rivera et al. (21) obtained from GenBank database with accession number: AF127375 (E. histolytica). Taking this as a target sequence, it was aligned using the BLAST tool in the genome database of all organisms to find highly conserved regions. E. dispar showed 93% sequence identity with the adhesin gene from E. histolytica (accession No.: AANV02000421).
According to the high similarity between the two sequences, it was necessary to select a part of the sequence containing five differences for at least one nucleotide, because the DGGE is sensitive enough to detect differences of a single base between two sequences. The BioEdit (22) and GeneDoc (http://www.psc.edu/biomed/genedoc) softwares were used to interpret multiple alignments and manual adjustments. The alignment of E. histolytica and E. dispar sequences allowed the selection of a 228 bp-region containing five single base differences throughout the sequence (from base 916 to 1144) to be further amplified by PCR (Figure 1).
Figure 1. Alignment sequence between the adh112 gene fragments from E. histolytica and E. dispar. Five differences between sequences are highlighted.

An in silico analysis to examine the complementarity within the sequence of each primer was performed using the Oligonalyzer program (http://www.idtdna.com/Home/Home.aspx). The designed primers also amplified another species of amoeba: E. nuttalli; however, its selected 228 bp-region did not contain the five single base differences observed in E. histolytica and E. dispar, so it was not included in our analysis. Moreover, E. nuttalli is non-pathogenic for humans and therefore, not important for our study (23). In order to achieve greater sensitivity in the detection by DGGE between E. histolytica and E. dispar, a GC-clamp was added to the forward primer: 5′-CGCCCGCCGCGCGGCCGCGGCCGGCCGGGGGCACGCGGCG-3′ (24), changing the total size of the fragment to be amplified from 228 to 268 bp.
PCR amplification of adh111 gene
We tested various quantities of DNA (25, 50, and 100 ng) for PCR amplification and decided to use 100 ng of DNA. For the first amplification, a reaction volume of 25 µL comprised: 4 µL of 10xPCR buffer (Invitrogen, USA), 3.2 µL of MgCl2 (50 mM; Invitrogen, USA), 1 µL of dNTP's mix (10 mM each; Invitrogen), 0.3 µL of each primer (40 µM), 0.2 µL (5 U/µL) of Accu Prime™ TaqDNA Polymerase High Fidelity (Invitrogen, USA), 2 µL (100 ng) of DNA and 14 µL of sterile deionized water. Finally, 2 drops of mineral oil were added. The amplification program of DNA started with 2 min of denaturation at 94°C, followed by 40 cycles of 60 s at 92°C of denaturation, primer annealing for 60 s at 47°C and extension for 90 s at 72°C. The final extension was at 72°C for 7 min. We used the primers: Fw 5′-GCA GAA AAA AAT AAT AAT AAC-3′ and Rv 5′-TTC ATT TGT TTT ACT TTC A-3′. After the first PCR, 5 µL of the amplified product were used for a second amplification under the same conditions mentioned above, using the primers: Fw 5′-CGC CCG CCG CGC GGC CGC GGC CGG CCG GGG GCA CGC GGC GGC AGA AAA AAA TAA TAA TAA C-3′ and Rv 5′-TTC ATT TGT TTT ACT TTC A-3′. Both amplifications were done in triplicate. PCR amplification products were verified on 1.6% (w/v) agarose gel using electrophoresis containing ethidium bromide.
Denaturing gradient gel electrophoresis
The PCR products were subjected to DGGE with 10 and 30% linear denaturing gradients of urea and formamide in a 10% polyacrylamide gel (Promega, USA). DGGE was performed with a 10–30% denaturing gradient adding 210 µL of 10% ammonium persulfate and 10 µL of tetramethylethylenediamine (TEMED). The electrophoresis was pre-run in 1xTAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA, pH 8.0) at constant 200 V during 15 min and at 60°C using the Bio-Rad (USA) D-Code TM Universal Mutation Detection System. The final conditions for electrophoresis gels were 5 h and 30 min at 130 V; the presence of PCR products was visualized by gel staining with 1 µg/mL of ethidium bromide for 2 min at room temperature and photographed by UV transillumination.
Nested PCR-RFLP of SSU rRNA gene
For the first amplification, a reaction volume of 25 µL comprised: 2.5 µL of 10x PCR buffer (Invitrogen), 2 µL of MgCl2 (50 mM; Invitrogen), 0.64 µL of dNTP's mix (10 mM each; Invitrogen), 0.6 µL of each primer (40 µM), 0.2 µL (5 U/µL) of de Accu Prime™ TaqDNA Polymerase High Fidelity (Invitrogen), 2 µL (100 ng) of DNA, and 16.5 µL of sterile deionized water. Finally, 2 drops of mineral oil were added. The DNA amplification program started with 2 min of denaturation at 94°C, followed by 40 cycles of 60 s at 92°C of denaturation, primer annealing for 60 s at 47°C and extension for 90 s at 72°C; the final extension was at 72°C for 7 min. The primers used in this PCR were: Fw 5′-TTT GTA TTA GTA CAA A-3′; Rv 5′-GTA [A/G]TA TTG ATA TAC T-3′. Later, we used 4 µL of the first PCR product as a template for the nested PCR reaction with 4 µL of 10x PCR buffer (Invitrogen), 3.2 µL of MgCl2 (50 mM) (Invitrogen), 1 µL of dNTP's mix (10 mM each) (Invitrogen), 1 µL of each primer (40 µM), 0.2 µL (5 U/µL) of Accu Prime™ TaqDNA Polymerase High Fidelity (Invitrogen), 10.6 µL of sterile deionized water and 2 drops of mineral oil. The nested PCR was performed as described above using the first PCR conditions, except for the annealing temperature, which changed to 62°C and others primers were used: Selective for E. histolytica (Fw 5′-TTT AGA AAC AAT GCT TCT CT-3′ and Rv 5′- AAT GGC CAA TTC ATT CAA TG-3′) and selective for E. dispar (Fw 5′-AGT GGC CAA TTT ATG TAA GT-3′ and Rv 5′-TTT AGA AAC AAT GTT TCT TC-3′). Both amplifications were done in triplicate. The amplified products were stained with ethidium bromide after electrophoresis on a 1.6 % agarose gel. Positive and negative control reactions were included with each batch of samples analyzed by nested PCR. The nested PCR products of both E. histolytica and E. dispar showed approximately 874 bp fragments which correspond to small ribosomal RNA subunit (SSU rRNA) gene. These products were digested with the restriction endonuclease DraI or Sau96I (5 U/µL; BIOLabs, New England) during 16 h at 37°C according to the manufacturer's instructions. The RFLP-digested product was visualized by loading 5 µL of sample on a 1.6 % agarose gel containing ethidium bromide.
Results
Coproparasitoscopic exam of stool samples
All samples were tested for E. histolytica and E. dispar using the Faust coproparasitoscopic exam. Of the 62 stool samples screened, 18 were positive for E. histolytica, 22 were positive for either Escherichia coli, E. nana, Giardia lamblia, or Ascaris lumbricoides cysts and negative for Entamoeba. Finally, 22 samples were negative for any parasite. No sample was positive for E. dispar.
Specificity and sensitivity of PCR methods
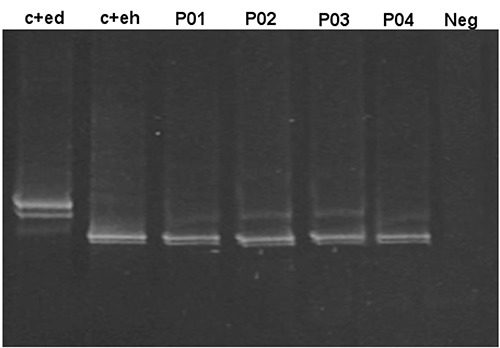
We found that the designed primers were specific for the expected fragment of 268 pb. This amplicon was observed in stool samples and positive controls (E. histolytica and E. dispar). Figure 2 shows a visible amplicon of 268pb in the analyzed samples. Although other amplicons were detected in the stool samples and negative controls, it should be noted that they were not found in the positive controls.
Figure 2. Stained agarose gel with products amplified by PCR with designed primers from samples containing E. histolytica or E. dispar. Lane 1, molecular weight marker; lanes 2–4, clinical samples; lane 5, DNA positive control for E. histolytica (c+eh); lane 6, DNA positive control for E. dispar (c+ed); and lane 7, negative control (human DNA from whole blood).
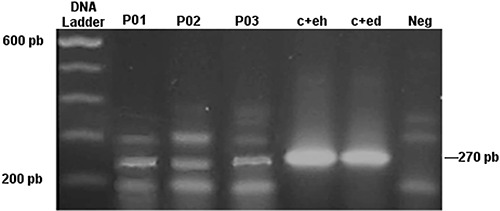
Clinical evaluation of PCR-DGGE-adh112 assay
Once the DNA was isolated from stool samples, the primers were selected and concentrations standardized, the denaturant conditions were fully optimized, then DGGE gels were run (Figure 3). Out of the 62 samples, 10 (16.1%) were positive for E. histolytica, while 52 (83.9%) were negative. No sample was found to be positive for E. dispar.
Figure 3. Denaturing gradient gel electrophoresis analysis of stool samples for the identification of E. histolytica and E. dispar. Lane 1, positive control of E. dispar; lane 2, positive control of E. histolytica; lanes 3–6, clinical samples; lane 7, negative control.
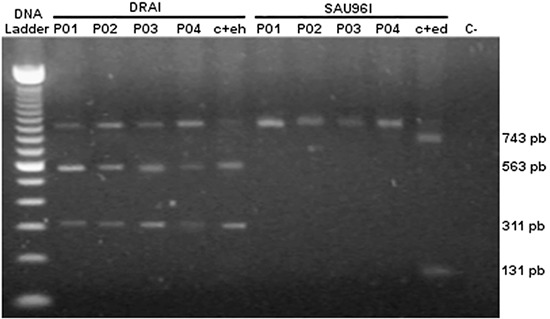
Nested PCR-RFLP of SSU rRNA gene
The RFLP pattern for E. histolytica showed 563 bp and 311 bp fragments and an undigested 874 bp fragment, whereas that for E. dispar showed 743 bp and 131 bp fragments (Figure 4). The nested PCR-RFLP was positive for E. histolytica in 10 (16.1%) stool samples and 52 (83.9%) samples were negative. These results are similar to what we observed with the PCR-DGGE technique, that is, the positive samples for E. histolytica by PCR-DGGE were also positive by PCR-RFLP. No sample was found to be positive for E. dispar. The sensitivity and specificity of both, PCR-DGGE and PCR-RFLP were 100%. The 95%CI values were also estimated and used to evaluate the sensitivity (95%CI=65.55–99.08) and specificity (95%CI=91.43–99.82) of PCR-DGGE technique.
Figure 4. Nested PCR-RFLP (restriction fragment length polymorphism) of SSU rRNA gene for the identification of E. histolytica and E. dispar. Lane 1, molecular weight marker; lanes 2–5, DraI digested PCR products; lane 6, DNA positive control for E. histolytica (c+eh); lane 7–10, Sau96I digested PCR products; lane 11, DNA positive control for E. dispar (c+ed); and lane 12, negative control.
Discussion
The identification of Entamoeba spp. has always been controversial and microscopy is usually used to diagnose protozoa in stool samples. However, this method is unable to differentiate E. histolytica from the morphologically identical non-pathogenic species such as E. dispar (8,14). Therefore, the WHO recommends the development and application of new methods for a specific diagnosis of E. histolytica infection (2). The present study describes a new PCR-DGGE strategy for species-specific detection and differentiation of E. histolytica and E. dispar DNA using stool samples. Several methods including isoenzyme analysis, antibody or antigen detection tests, immunochromatographic assays and real-time PCR have been used for an accurate detection of E. histolytica and E. dispar However, high cost limits their use in underdeveloped countries and, additionally, some yield false negative results (25 –28).
Another alternative for differentiating the species is using PCR. This technique demonstrates exceptional sensitivity and specificity compared with microscopy (8,14). This research is the first of its kind in Mexico, and was designed to detect and differentiate E. histolytica from E. dispar using a fragment of the adh112 gene, which presents five differences in single bases when comparing both species. The advantage of the PCR-DGGE-adh112 method over the PCR and restriction enzyme digestion method has to do with the reliability of the results. The electrophoretic pattern obtained in PCR and restriction enzyme digestion are based on the nucleotides size, and it can be difficult to see the differences and similarities that could exist among numerous samples (29). In contrast, DGGE is sensitive enough to detect differences of a single base. With the PCR-DGGE-adh112 the DNA fragments move through polyacrylamide gels containing a linear gradient of denaturing agents, resulting in a partially denatured molecule observed in the gel with distinctive electrophoretic pattern (the change in the sequence causes a change in the pattern of run), which gives greater effectiveness to the test and no likelihood of false positives or false negatives (17,24).
In Mexico, there is a lack of knowledge about the epidemiology of E. histolytica infection, although amebiasis has been considered for many years as a major health problem in the country (4,30). By using PCR-DGGE-adh112 we found the mono infection with E. histolytica to be 16.1%. No sample was found to be positive for E. dispar. These results were validated using another technique (nested PCR-RFLP of SSU rRNA gene) considered by several authors as the gold standard for the differential diagnosis between E. histolytica and E. dispar (31 –34). Our results were consistent with those observed by PCR-RFLP. Our PCR-DGGE-adh112 results showed sensitivity and specificity of 100%, indicating that it is a useful and reliable test to specifically detect E. histolytica in stool samples. PCR-DGGE has the advantage of identifying and differentiating E. histolytica and E. dispar, which is not possible using microscopy or ELISA (8,35). It should be noted that the differentiation of pathogenic E. histolytica from the morphologically identical E. dispar is important for the clinical management of patients.
In conclusion, the present study reports a new PCR-DGGE technique for species-specific detection and differentiation of E. histolytica and E. dispar DNA in stool samples. This technique could become an alternative or a complementary diagnostic tool.
References
- 1.Ximénez C, Morán P, Rojas L, Valadez A, Gómez A. Reassessment of the epidemiology of amebiasis: state of the art. Infect Genet Evol. 2009;9:1023–1032. doi: 10.1016/j.meegid.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous Amoebiasis. Wkly Epidemiol Rec. 1997;72:97–99. [PubMed] [Google Scholar]
- 3.Stauffer W, Abd-Alla M, Ravdin JI. Prevalence and incidence of Entamoeba histolytica infection in South Africa and Egypt. Arch Med Res. 2006;37:266–269. doi: 10.1016/j.arcmed.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Caballero-Salcedo A, Viveros-Rogel M, Salvatierra B, Tapia-Conyer R, Sepulveda-Amor J, Gutierrez G, et al. Seroepidemiology of amebiasis in Mexico. Am J Trop Med Hyg. 1994;50:412–419. doi: 10.4269/ajtmh.1994.50.412. [DOI] [PubMed] [Google Scholar]
- 5.Ramos F, Morán P, González E, García G, Ramiro M, Gómez A, et al. High prevalence rate of Entamoeba histolytica asymptomatic infection in a rural Mexican community. Am J Trop Med Hyg. 2005;73:87–91. [PubMed] [Google Scholar]
- 6.Begum S, Quach J, Chadee K. Immune evasion mechanisms of Entamoeba histolytica: progression to disease. Front Microbiol. 2015;6:1394. doi: 10.3389/fmicb.2015.01394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sepulveda B. Amebiasis: host-pathogen biology. Rev Infect Dis. 1982;4:1247–1253. doi: 10.1093/clinids/4.6.1247. [DOI] [PubMed] [Google Scholar]
- 8.Fotedar R, Stark D, Beebe N, Marriott D, Ellis J, Harkness J. Laboratory diagnostic techniques for Entamoeba species. Clin Microbiol Rev. 2007;20:511–532. doi: 10.1128/CMR.00004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luna-Nácar M, Navarrete-Perea J, Moguel B, Bobes RJ, Laclette JP, Carrero JC. Proteomic study of Entamoeba histolytica trophozoites, cysts, and cyst-like structures. PLoS One. 2016;11:e0156018. doi: 10.1371/journal.pone.0156018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aguirre Garcia M, Gutierrez-Kobeh L, Lopez Vancell R. Entamoeba histolytica: adhesins and lectins in the trophozoite surface. Molecules. 2015;20:2802–2815. doi: 10.3390/molecules20022802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ralston KS, Petri WA. The ways of a killer: how does Entamoeba histolytica elicit host cell death? Essays Biochem. 2011;51:193–210. doi: 10.1042/bse0510193. [DOI] [PubMed] [Google Scholar]
- 12.Ralston KS, Petri WA., Jr Tissue destruction and invasion by Entamoeba histolytica . Trends Parasitol. 2011;27:254–263. doi: 10.1016/j.pt.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Betanzos A, Javier-Reyna R, García-Rivera G, Baãuelos C, González-Mariscal L, Schnoor M, et al. The EhCPADH112 complex of Entamoeba histolytica interacts with tight junction proteins occludin and claudin-1 to produce epithelial damage. PLoS One. 2013;8:e65100. doi: 10.1371/journal.pone.0065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanyuksel M, Petri WA., Jr Laboratory diagnosis of amebiasis. Clin Microbiol Rev. 2003;16:713–729. doi: 10.1128/CMR.16.4.713-729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramos F, Morán P, González E, García G, Ramiro M, Gómez A, et al. Entamoeba histolytica and Entamoeba dispar prevalence infection in a rural Mexican community. Exp Parasitol. 2005;110:327–330. doi: 10.1016/j.exppara.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 16.Khairnar K, Parija SC, Palaniappan R. Diagnosis of intestinal amoebiasis by using nested polymerase chain reaction-restriction fragment length polymorphism assay. J Gastroenterol. 2007;42:631–640. doi: 10.1007/s00535-007-2080-6. [DOI] [PubMed] [Google Scholar]
- 17.Ercolini D. PCR-DGGE fingerprinting: novel strategies for detection of microbes in food. J Microbiol Methods. 2004;56:297–314. doi: 10.1016/j.mimet.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Righetti PG, Gelfi C. Analysis of clinically relevant, diagnostic DNA by capillary zone and double-gradient gel slab electrophoresis. J Chromatogr A. 1998;806:97–112. doi: 10.1016/S0021-9673(97)00304-X. [DOI] [PubMed] [Google Scholar]
- 19.Chaves A, de Alcantara OS, Carvalho OS, dos Santos JS. [Comparative study of Lutz, Kato-Katz and modified Faust coprologic methods] Rev Saúde Pública. 1979;13:348–352. doi: 10.1590/s0034-89101979000400010. [DOI] [PubMed] [Google Scholar]
- 20.Faust EC Parasitologic surveys in Cali, Departamento del Valle, Colombia. I. Incidence and morphologic characteristics of strains of Entamoeba histolytica . Am J Trop Med Hyg. 1958;7:4–15. doi: 10.4269/ajtmh.1958.7.4. [DOI] [PubMed] [Google Scholar]
- 21.García-Rivera G, Rodríguez MA, Ocádiz R, Martínez-López MC, Arroyo R, González-Robles A, et al. Entamoeba histolytica a novel cysteine protease and an adhesin form the 112 kDa surface protein. Mol Microbiol. 1999;33:556–568. doi: 10.1046/j.1365-2958.1999.01500.x. [DOI] [PubMed] [Google Scholar]
- 22.Hall TA. BioEdit: a user-friendly biological sequence alignment editor andanalysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 23.Tachibana H, Yanagi T, Feng M, Bandara KB, Kobayashi S, Cheng X, et al. Isolation and molecular characterization of Entamoeba nuttalli strains showing novel isoenzyme patterns from wild toque macaques in Sri Lanka. J Eukaryot Microbiol. 2016;63:171–180. doi: 10.1111/jeu.12265. [DOI] [PubMed] [Google Scholar]
- 24.Sheffield VC, Cox DR, Lerman LS, Myers RM. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci U S A. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calderaro A, Piergianni M, Buttrini M, Montecchini S, Piccolo G, Gorrini C, et al. MALDI-TOF mass spectrometry for the detection and differentiation of Entamoeba histolytica and Entamoeba dispar . PLoS One. 2015;10:e0122448. doi: 10.1371/journal.pone.0122448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavez-Munguia B, Martinez-Palomo A. High-resolution electron microscopical study of cyst walls of Entamoeba spp. J Eukaryot Microbiol. 2011;58:480–486. doi: 10.1111/j.1550-7408.2011.00576.x. [DOI] [PubMed] [Google Scholar]
- 27.Barnawi AB, Tonkal AM, Fouad MA, Al-Braiken FA. Detection of Entamoeba histolytica/dispar in stool specimens by using enzyme-linked immunosorbent assay in the population of Jeddah City, Saudi Arabia. J Egypt Soc Parasitol. 2007;37:143–150. [PubMed] [Google Scholar]
- 28.Spadafora LJ, Kearney MR, Siddique A, Ali IK, Gilchrist CA, Arju T, et al. Species-specific immunodetection of an Entamoeba histolytica cyst wall protein. PLoS Negl Trop Dis. 2016;10:e0004697. doi: 10.1371/journal.pntd.0004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noor Azian MY, Lokman Hakim S, Maslawaty MN. Use of molecular tools to distinguish Entamoeba histolytica and Entamoeba dispar infection among the aborigines in Cameron Highlands. Trop Biomed. 2006;23:31–36. [PubMed] [Google Scholar]
- 30.González CR, Isibasi A, Ortiz-Navarrete V, Paniagua J, García JA, Ramirez A, et al. Prevalence of antibodies against Entamoeba histolytica in Mexico measured by ELISA. Epidemiol Infect. 1995;115:535–543. doi: 10.1017/S0950268800058702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Troll H, Marti H, Weiss N. Simple differential detection of Entamoeba histolytica and Entamoeba dispar in fresh stool specimens by sodium acetate-acetic acid-formalin concentration and PCR. J Clin Microbiol. 1997;35:1701–1705. doi: 10.1128/jcm.35.7.1701-1705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haque R, Ali IK, Akther S, Petri WA., Jr Comparison of PCR, isoenzyme analysis, and antigen detection for diagnosis of Entamoeba histolytica infection. J Clin Microbiol. 1998;36:449–452. doi: 10.1128/jcm.36.2.449-452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katzwinkel-Wladarsch S, Loscher T, Rinder H. Direct amplification and differentiation of pathogenic and nonpathogenic Entamoeba histolytica DNA from stool specimens. Am J Trop Med Hyg. 1994;51:115–118. doi: 10.4269/ajtmh.1994.51.115. [DOI] [PubMed] [Google Scholar]
- 34.Clark CG, Diamond LS. Ribosomal RNA genes of 'pathogenic' and 'nonpathogenic' Entamoeba histolytica are distinct. Mol Biochem Parasitol. 1991;49:297–302. doi: 10.1016/0166-6851(91)90073-F. [DOI] [PubMed] [Google Scholar]
- 35.Zeyrek FY, Turgay N, Unver A, Ustün S, Akarca U, Töz S. Differentiation of Entamoeba histolytica/Entamoeba dispar by the polymerase chain reaction in stool samples of patients with gastrointestinal symptoms in the Sanliurfa Province. Turkiye Parazitol Derg. 2013;37:174–178. doi: 10.5152/tpd.2013.39. [DOI] [PubMed] [Google Scholar]


