Abstract
In the kidney, the epithelial sodium channel (ENaC) regulates blood pressure through control of sodium and volume homeostasis, and in the lung, ENaC regulates the volume of airway and alveolar fluids. ENaC is a heterotrimer of homologous α-, β- and γ-subunits, and assembles in the endoplasmic reticulum (ER) before it traffics to and functions at the plasma membrane. Improperly folded or orphaned ENaC subunits are subject to ER quality control and targeted for ER-associated degradation (ERAD). We previously established that a conserved, ER lumenal, molecular chaperone, Lhs1/GRP170, selects αENaC, but not β- or γ-ENaC, for degradation when the ENaC subunits were individually expressed. We now find that when all three subunits are co-expressed, Lhs1-facilitated ERAD was blocked. To determine which domain–domain interactions between the ENaC subunits are critical for chaperone-dependent quality control, we employed a yeast model and expressed chimeric α/βENaC constructs in the context of the ENaC heterotrimer. We discovered that the βENaC transmembrane domain was sufficient to prevent the Lhs1-dependent degradation of the α-subunit in the context of the ENaC heterotrimer. Our work also found that Lhs1 delivers αENaC for proteasome-mediated degradation after the protein has become polyubiquitinated. These data indicate that the Lhs1 chaperone selectively recognizes an immature form of αENaC, one which has failed to correctly assemble with the other channel subunits via its transmembrane domain.
Introduction
Endoplasmic reticulum-associated degradation (ERAD) targets polypeptides that are misfolded or mutated, proteins that are subject to regulated degradation, and subunits in multimeric complexes that are orphaned for degradation by the cytosolic 26S proteasome [1–4]. ERAD can be divided into four steps [5–7]: recognition, polyubiquitination, retrotranslocation and degradation. ERAD substrates are initially recognized and targeted for degradation by molecular chaperones and chaperone-like lectins that reside either in the cytosol or in the endoplasmic reticulum (ER) lumen [8,9]. These chaperones and lectins recruit the machinery — most notable the E3 ubiquitin ligases — that polyubiquitinate the substrate, marking it for degradation [10]. Next, the ubiquitinated protein is retrotranslocated from the ER to the cytosol with energy provided by the conserved Cdc48/p97 complex [11–14], and degraded by the cytosolic 26S proteasome [15].
Epithelial sodium channel (ENaC) is a heterotrimeric integral membrane protein found primarily in the aldosterone-sensitive distal nephron and in epithelial cells lining the airway and in the lung alveoli. In the kidney, ENaC maintains extracellular fluid volume balance, controls blood pressure and facilitates potassium excretion [16–19]. ENaC is transcriptionally regulated by hormones, such as aldosterone, which bind mineralocorticoid receptors in the nephron [20]. ENaC residence at the cell surface is also controlled by endocytosis, which is triggered by physiological signals [21]. Each ENaC subunit, α, β and γ, has a large extracellular loop, two transmembrane segments and short cytosolic N- and C-termini [22]. Even though the ENaC subunits share ~40% identity, they are subject to differential modification and regulation [16,23]. For example, in some cell types, the β- and γ-ENaC subunits are expressed constitutively and degraded by ERAD. Only after aldosterone-induced αENaC expression can the channel assemble and traffic to the plasma membrane of epithelial cells where it reabsorbs sodium [18,24,25].
Mutations that affect ENaC expression at the apical membrane are linked to abnormal blood pressure. For example, ENaC gain-of-function mutations in Liddle’s syndrome lead to early-onset high blood pressure with hypokalemia. This phenomenon arises due to defects in ubiquitin-mediated endocytosis, resulting in a high number of functional channels at the plasma membrane, an increase in channel open probability and excess sodium reabsorption [26]. In contrast, loss-of-function mutations lead to the premature degradation of ENaC and result in pseudohypoaldosteronism type I, which presents with low blood pressure, hyperkalemia and renal salt wasting [27]. Altered ENaC function has also been linked to the pathogenesis of cystic fibrosis lung disease. Specifically, there is evidence that the cystic fibrosis transmembrane conductance regulator (CFTR) negatively regulates ENaC, and that hyperabsorption of sodium in the absence of functional CFTR contributes to airway surface liquid volume depletion, which exacerbates the cystic fibrosis disease phenotype [28–32].
Even when all three ENaC subunits are present, a significant fraction of the channel is still degraded by the ERAD pathway [33–35]. These data are in line with numerous reports that a significant fraction of wild-type (WT), polytopic membrane proteins fold inefficiently and are subject to ERAD [36]. Alternatively, ER-associated quality control may provide a means (along with endocytosis) to regulate ENaC levels [37,38]. Indeed, a growing number of substrates in both yeast and mammals are proteins whose steady-state levels are regulated by ERAD [39–43].
To begin to define how ENaC is subject to ER quality control, and more specifically how it is targeted for ERAD, we established a yeast expression system in which each of the three subunits could be individually expressed in various mutant backgrounds. We demonstrated that the ER lumenal Hsp40 chaperones, Scj1 and Jem1, as well as Cdc48, the small heat shock proteins and the E3 ubiquitin ligases, Hrd1 and Doa10, target αENaC for degradation at the ER [44,45]. More recently, we reported that the ER lumenal chaperone, Lhs1, also helps target αENaC, but not the β- or γ-subunits, for ERAD. Consistent with these data, overexpression of GRP170, which is the mammalian Lhs1 homolog, promoted αENaC turnover in human cells [46]. Lhs1/GRP170 is an Hsp70-like chaperone with both nucleotide- and substrate-binding domains and functions as a nucleotide exchange factor (NEF) for the lumenal Hsp70, Kar2/BiP [47–50]. However, Lhs1 also posseses protein ‘holdase’ activity and prevents misfolded protein aggregation in vitro; Lhs1 holdase activity requires neither ATP binding nor BiP association [51]. Accordingly, we found that αENaC degradation is BiP-independent and that Lhs1-dependent degradation was ATP-independent. These data strongly suggest that αENaC degradation employs Lhs1’s holdase activity [46]. In addition, Lhs1 and GRP170 preferentially target unglycosylated and thus immature forms of αENaC, consistent with the chaperone acting as a mediator of ER quality control.
In the present study, we have mapped how Lhs1 selects αENaC for Lhs1-dependent ERAD and defined where in the ERAD pathway the chaperone acts. First, we developed a yeast expression system so that the α-, β- and γ-ENaC subunits can be co-expressed. We then demonstrated that heterotrimer assembly blocks Lhs1-dependent degradation of αENaC. These data provide the first evidence that a molecular chaperone differentially recognizes monomeric versus heterotrimic ENaC subunits. Second, we constructed chimeric α/βENaC species to determine that intersubunit interactions via the αENaC transmembrane domains (TMDs) prevent Lhs1-dependent degradation. These data are consistent with the predicted, extensive interactions among the TMDs of the three ENaC subunits [52,53,54]. Therefore, Lhs1 either directly recognizes the unassembled αENaC TMDs, or interactions between the TMDs trigger a structural change in the extracellular (ECL) loop that is recognized by Lhs1. Third, we demonstrate that Lhs1 acts after the subunit has been polyubiquitinated, which is consistent with Lhs1 helping target ER-resident, ubiquitinated αENaC to the proteasome.
Experimental procedures
Yeast strains and growth conditions
Yeast strains were propagated at 26°C using established methods, and media preparation and transformations were performed as previously published unless otherwise noted [55]. The WT yeast strain was BY4742. The BY4742 and Δlhs1 strains were obtained from Open Biosystems (Thermo Scientific). The absence of Lhs1 in the Δlhs1 strain was confirmed by western blot analysis and by phenotypes associated with the loss of this protein [46].
Plasmid construction and molecular techniques
The constitutive expression of a C-terminally HA epitope-tagged form of the ENaC subunits was previously described [45]. The HA-tagged αENaC was removed from the pRS426GPD vector and ligated into pRS425GPD using EcoRI and ClaI. To introduce alternate epitope tags in the β- and γ-subunits, pRS426GPD-βENaC-13myc was created by PCR overlap extension, as previously described [56], using pRSGPD426-βENaC-HA [45] and pFA6-13myc-kanMX [57] as templates. The products were then digested and ligated into the BamHI and EcoRI sites of pRS426GPD. An NheI site was inserted between the βENaC fragment and the 13myc fragment. pRS425GPD-γENaC-V5 was made by PCR amplifying pRS426GPD-γENaC-HA [45] with a C-terminal primer containing the V5 sequence. The DNA fragment was next ligated into pRS425GPD using the HindIII and SpeI sites. Previous work indicated that epitope tags at these positions had no effect on channel function [35].
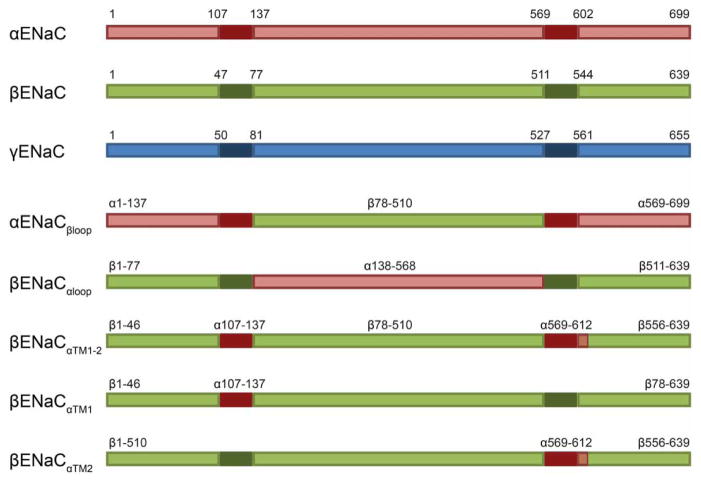
Chimeric ENaC constructs were also constructed using PCR overlap extension [56] and are depicted in Figure 1. Because a crystallographic structure for ENaC has not been achieved, the structural borders for these constructs are based on published predictive analyses of the degenerin family member, ASIC [52–54]. All constructs were subjected to DNA sequence analysis to confirm their identities.
Figure 1. Chimeric ENaC constructs used in the present study.
Diagrams of αENaC (red), βENaC (green) and γENaC (blue) segments are shown. The solid darker regions correspond to the TMDs. Relative amino acid positions for the TMDs and domain boundaries used for chimeric constructs are indicated. The marked residues correspond to published predictive analyses [54]. The chimeric constructs βENaCαTM1–2 and βENaCαTM2 contain an additional ~10 amino acids C-terminal to the TM2 border (as illustrated).
Protein degradation assays
Cycloheximide chase analyses to measure the stabilities of the ENaC subunits were performed as published previously [45]. Cell lysates from chase samples were generated using alkaline lysis followed by trichloroacetic acid (TCA) precipitation [58], and protein fractions in sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) sample buffer [0.325 M Tris (pH 6.8), 10% SDS, 5% β-mercaptoethanol and 0.25 mg/ml bromophenol blue] were immediately resolved by SDS–PAGE before western blot analysis. The αENaC subunit was detected using anti-HA-horseradish peroxidase (HRP) (clone 3F10; Roche Applied Science) at a dilution of 1:5000. The βENaC, βENaCαloop, αENaCβloop, βENaCαTM1–2, αENaCβTM1–2, βENaCαTM1 and βENaCαTM2 constructs were probed using anti-myc antiserum (Clontech). The anti-myc primary antibody was then detected with anti-mouse monoclonal IgG HRP-conjugated secondary antibody (Cell Signaling Technology). γENaC was detected with anti-V5 antibody (Novex), and the V5 primary antibody was detected with anti-mouse monocolonal IgG-conjugated secondary antibody (Cell signaling Technology). Western blots were also probed with anti-glucose-6-phosphate dehydrogenase antiserum (G6P; Sigma), which served as a loading control. The anti-G6P primary antibody was detected with a donkey HRP-conjugated anti-rabbit IgG secondary antibody (GE Healthcare). All images of western blots for the present study were obtained on a BioRAD Universal Hood II Imager, and data were analyzed using ImageJ software [Version 1.49U (100)]. P-values for all experiments, which were conducted with a minimum of six different yeast transformants, were calculated using a Student’s t-test and are indicated in the figure legends.
Other biochemical analyses
Endoglycosidase H assays were completed by harvesting 1 ml of cells at an attenuance (D)600 = 1. Cell lysates were obtained as described above by alkaline lysis and TCA precipitation, and resuspended in SDS sample buffer. Samples were treated in the presence or absence of Endoglycosidase H (Roche) for 2 h at 37°C according to the manufacturer’s instructions and subsequently analyzed by SDS–PAGE and western blotting as described above.
For co-immunoprecipitations, WT yeast (BY4742) transformed with the indicated plasmids were grown overnight in selective medium, and 20 ml of cells at a D600 = ~0.5 cells were harvested. The pellet was resuspended in 500 μl of lysis buffer [1% n-dodecyl-β-D-maltoside (DDM), 150 mM NaCl, 50 mM Tris–HCl (pH = 7.4), 1 mM PMSF (phenylmethylsulfonyl fluoride), 2 μg/ml leupeptin, 0.1 μg/ml pepstatin A and one Roche mini-EDTA-free complete protease inhibitor tablet per 7 ml buffer] plus glass beads and was agitated four times on a Vortex mixer for 1 min. The lysate was cleared by centrifugation at 5000 × g for 5 min in a microfuge, and the supernatant was placed in a new tube with 300 μl of lysis buffer and either 25 μl of HA-conjugated agarose beads (Roche) or unconjugated sepharose beads as a control. Reactions were incubated at 4°C for 3 h and then washed twice with lysis buffer and twice with a buffer equivalent to lysis buffer but lacking detergent. The samples were resuspended in 25 μl of SDS sample buffer, divided in half and run in duplicate SDS–polyacryl-amide gels followed by western blotting. Immunoblots were probed with anti-HA HRP or anti-V5 as described above. Nitrocellulose membranes were stripped with 0.1 M glycine (pH 2.0) for 1 h, reblocked [50 mM Tris (pH = 7.4), 150 mM NaCl, 2% nonfat dry milk, 1% Tween-20 and 5 mM NaN3] and probed with anti-Sec61 (raised against peptide LVPGFSDLM and purified by Cocalico Biologicals, Stevens, PA) or anti-myc antibodies. Anti-Sec61 primary antibody was detected with a donkey HRP-conjugated anti-rabbit IgG secondary antibody (GE Healthcare). Images were obtained as described above. The antibodies used for these immunoprecipitations have been extensively used and are well characterized, and no cross-reactivity has been observed.
Sucrose gradients were performed essentially as previously described [59]. Briefly, yeast strains were grown overnight in selective media, and 35 ml of D600 = 0.8 of cells were collected, washed and resuspended in 10 mM Tris–HCl (pH 7.5), 1 mM EDTA (ethylenediaminetetraacetic acid) and 10% sucrose, and then disrupted by agitation with glass beads. Lysates were cleared by centrifugation at 5000 × g for 5 min in a microfuge and were layered on top of 11 ml of 30–70% sucrose gradient. The gradients were centrifuged at 100 000 × g in a Beckman SW41 rotor for 14 h at 4°C, and fractions were collected from the top of the tube. Next, the fractions were analyzed by SDS–PAGE and western blotting. Immunoblots were probed with anti-HA HRP, anti-V5, anti-Myc, anti-Sec61 and anti-Pma1 (Abcam) antibodies. Each antibody was detected as described above.
For carbonate extraction, yeast strains were grown overnight in selective media, and 75 ml of D600 = 0.5 cells were collected. The cells were resuspended in 600 μl of IP buffer [20 mM HEPES (pH = 7.4), 50 mM KOAc, 2 mM EDTA, 0.1 M sorbitol, 1 mM DTT (dithiothreitol), 1 mM PMSF, 2 μg/ml leupeptin, 0.1 μg/ml pepstatin A and protease inhibitor tablet (Complete Mini, EDTA-free, Roche)] plus glass beads and were subjected to agitation on a Vortex mixer four times for 1 min with 1 min incubations on ice. The lysate was removed from the glass beads and placed in a clean tube, and cleared by centrifugation at 2500 × g for 3 min. The supernantant fraction was then removed and membranes were pelleted by centrifugation at 14 000 × g for 20 min at 4°C. The membrane pellet was resuspended in 500 μl of IP buffer to wash and again subjected to centrifugation at 14 0000 × g for 10 min at 4°C. Next, the supernatant was removed and the membrane pellet was washed with 100 μl of Buffer 88 [20 mM HEPES (pH 6.8), 150 mM potassium acetate, 5 mM magnesium acetate, 250 mM sorbitol and the protease inhibitors described above] and then resuspended in 100 μl of Buffer 88 and split into 40 μl samples, which were treated with either 1 ml of 0.1 M Na2CO3 or 1 ml of Buffer 88 as a control. The samples were incubated on ice for 30 min and subjected to centrifugation at 50 000 × g for 1 h at 4°C. The supernatant was removed and set aside for TCA precipitation, and the pellet was resuspended in 500 μl of the appropriate buffer (either containing or lacking Na2CO3) and subjected to centrifugation at 600 000 × g for 10 min at 4°C. This supernatant was discarded and the combined pellets were resuspendend in 35 μl of SDS sample buffer using a mechanical pestle. A total of 100 μl of 50% TCA was then added to each supernatant fraction, and the samples were incubated on ice for 15 min followed by centrifugation at 140 000 × g for 10 min at 4°C. Next, the supernatant was aspirated and the pellet was resuspended in 35 μl of SDS sample buffer using a mechanical pestle. All samples were incubated at 37°C for 10 min before analysis by SDS–PAGE and western blotting. Immunoblots were probed with anti-HA HRP, anti-Sec61 and anti-Pdi1 antibodies. The anti-Pdi1 primary antibody (a kind gift from Dr Vlad Denic, Harvard University) was detected using donkey HRP-conjugated anti-rabbit IgG secondary antibody (GE Healthcare).
For detergent solubility assays, 4 l cultures of BY4742 and Δlhs1 yeast strains expressing pRS426GPD-αENaC-HA or pRS426GPD-ΔGαENaC-HA [46] were grown to a D600 of 1.5 in selective medium. The cultures were then shifted to 37°C for 1 h in a shaking water bath, and the cells were harvested at 4°C by centrifugation for 5 min at 3000 × g. Once harvested, yeast ER-enriched microsomes were purified using a previously described large-scale technique [60]. Solubilization assays were performed by adding 30 μl of Buffer 88, the indicated concentration of DDM and ER-enriched microsomes. Each sample contained a final concentration of 0.5 mg/ml of protein, as determined spectrophotometrically (A280). Following the addition of the ER-enriched microsomes, the samples were incubated at room temperature (~21°C) for 30 min and then centrifuged for 10 min at 18 000 × g at 4°C. The supernatant was removed and dispensed into an Eppendorf tube, which contained 5× SDS–PAGE sample buffer. The pellet was resuspended in an equal final volume of 1× sample buffer by pipetting. The samples were then incubated at 37°C for 30 min followed by centrifugation for 1 min at 13 000 × g, and subjected to SDS–PAGE and western blotting with anti-HA or anti-Sec61 antisera as described above. The data were analyzed as described above.
In vitro ubiquitination assay
Yeast cytosol was purified from WT (BY4742) yeast cells that were incubated at 39°C for 2 h according to a previously published technique [60], except that yeast cells were lysed six times for 1 min using a cold mortar and pestle with the continuous addition of liquid nitrogen. ER-enriched microsomes were purified from either WT or Δlhs1 strains expressing pRS426GPD-ΔGαENAC-HA, as described above. In vitro reactions contained 1 mg/ml yeast cytosol, 1 mg/ml ER-derived microsomes and an ATP-regenerating system (1 mM ATP, 40 M creatine phosphate and 0.2 mg/ml creatine phosphokinase in Buffer 88). A negative control contained apyrase (0.02 units/reaction) in place of the ATP-regenerating system. In vitro reactions were incubated at room temperature for 10 min prior to the addition of 125I-ubiquitin to a final concentration of 1.5 mg/ml. The samples were then incubated at 37°C for 45 min. A total of 125 μl of a 1.25% SDS Stop solution [50 mM Tris–Cl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1.25% SDS, 1 mM PMSF, 1 μg/ml leupeptin, 0.5 μg/ml pepstatin A and 10 mM N-ethylmaleimide (NEM)] was added to the reactions, which were then incubated at 37°C for 30 min. Next, 400 μl of a Triton solution [50 mM Tris–Cl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 2% Triton X-100, 1 mM PMSF, 1 μg/ml leupeptin, 0.5 μg/ml pepstatin A and 10 mM NEM], 30 μl of a 50/50 protein A Sepharose slurry in the manufacturer’s suggested buffer and 2.5 μl of anti-HA (Roche, mouse) antibody were added to each reaction, and samples were immunoprecipitated overnight at 4°C on a rotator. The samples were then washed three times in an IP wash buffer [50 mM Tris–Cl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.2% SDS and 10 mM NEM], and 40 μl of sample buffer was added. Finally, the precipitated proteins were incubated at 37°C for 30 min and were resolved in duplicate by SDS–PAGE. One gel was subjected to western blotting and probed with anti-HA HRP antibody, as described above. A second gel was dried on filter paper and subjected to phosphoimager analysis. Data were analyzed and quantified using a Typhoon FLA 7000 and Image J Software.
Results
The expression of associated partners blocks the Lhs1-dependent degradation of αENaC
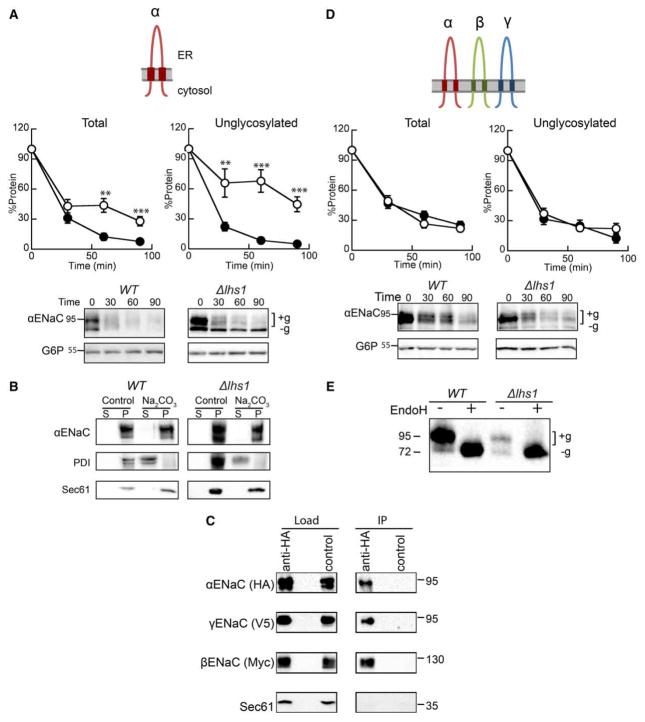
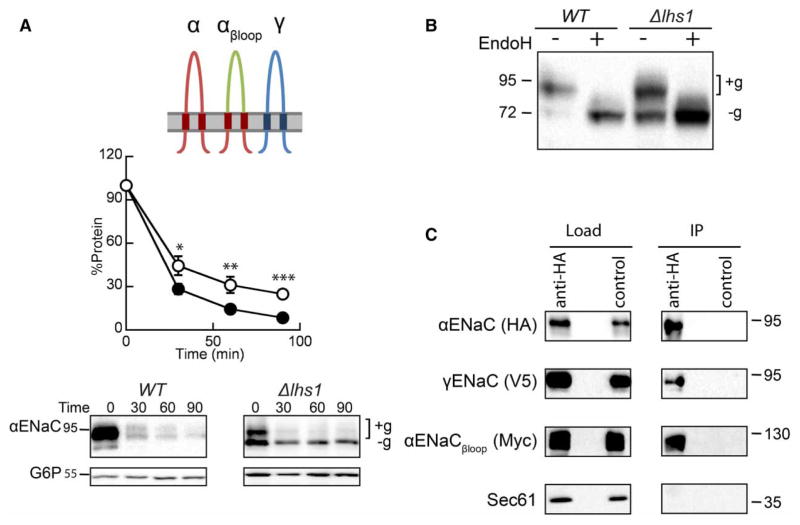
The ER lumenal molecular chaperone, Lhs1, targets the orphaned αENaC subunit for ERAD. As shown in Figure 2A and as previously demonstrated [46], αENaC is stabilized in a yeast strain lacking Lhs1 (Δlhs1) in comparison with WT (BY4742) yeast when protein levels are monitored following the addition of cycloheximide. Moreover, stabilization of the unglycosylated form of αENaC (‘−g’, Δlhs1 in Figure 2A) is magnified (compare ‘Total’ with ‘Unglycoslated’), whereas the upper, glycosylated band (+g) is unaffected. This dominant, lower molecular mass species serves as a convenient readout for Lhs1-dependent degradation of αENaC since it is absent in WT yeast. A significant portion of the overall ENaC pool is also unglycoyslated. For example, ~20% of steady-state αENaC is unglycosylated in WT yeast [46], and greater levels of unglycosylated ENaC are observed in some higher cell types [61–63]; therefore, the mechanism underlying the biosynthesis and quality control of this fraction of ENaC protein is biologically relevant. Because Lhs1 also plays a role in the translocation of some proteins [47–50], we confirmed that αENaC is membrane-integrated in both WT and Δlhs1 yeast after treatment with sodium carbonate (Figure 2B) as was the ER membrane protein, Sec61. In contrast, the soluble ER lumenal protein, protein disulfide isomerase (PDI), was released into the supernatant. Therefore, the differences we observe between WT and Δlhs1 are unlikely to be due to a translocation defect.
Figure 2. Expression of the three ENaC subunits eliminates Lhs1-targeted ERAD.
Cycloheximide chase reactions were performed as described in Experimental Procedures using WT (filled circles) and Δlhs1 (open circles) yeast strains transformed with plasmids engineered for the expression of either (A) αENaC-HA or (D) αENaC-HA, βENaC-13myc and γENaC-V5 simultaneously. Chase reactions were performed with cells shifted to 37°C, and lysates were resolved by SDS–PAGE and proteins were immunoblotted with anti-HA antisera (αENaC) and with anti-G6P as a loading control. βENaC and γENaC expression and degradation were confirmed, and the levels of degradation mirrored the levels when expressed alone and showed no Lhs1 dependence (data not shown). Glycosylated (+g) and unglycosylated (−g) species are indicated. Data represent the mean of 6–9 experiments, ±SEM. **P < 0.01, ***P ≤ 0.0001. (B) Cellular membranes from WT or Δlhs1 yeast expressing αENaC-HA were treated with Na2CO3 or buffer (control) and subjected to centrifugation analysis. Pellet and supernatant fractions were obtained and immunoblotted for ENaC or the control proteins, PDI (soluble, ER lumenal) and Sec61 (ER membrane), as described in Experimental Procedures. (C) Cell lysates from WT yeast expressing αENaC-HA, βENaC-13myc and γENaC-V5 were subjected to immunoprecipitation with anti-HA agarose resin or sepharose (control), and proteins were immunoblotted with anti-HA (αENaC), anti-myc (βENaC), anti-V5 (γENaC) or anti-Sec61 as a control. Samples equal to 1% of immunoprecipitated material were also immunoblotted (load). (E) Cell lysates from WT or Δlhs1 yeast expressing α ENaC-HA, βENaC-13myc and γENaC-V5 were treated with Endoglycosidase H (Endo H). Anti-HA immunoblots are shown.
As observed with numerous multimeric proteins, ENaC function and cellular trafficking are presumed to require assembly of the heterotrimeric channel in the ER. For example, in the kidney, the β- and γ-ENaC subunits are constitutively expressed and targeted for ERAD [18,34,35]. Only when the expression of the α-subunit is induced in response to aldosterone, can the channel assemble in the ER and then traffic and function at the cell surface [24,25]. Therefore, we asked how the presence of the other subunits affects the role of Lhs1 in αENaC degradation. To this end, a new yeast expression system for the heterotrimeric channel was developed. Differentially epitope-tagged α-, β- and γ-ENaC subunits (αENaC-HA, βENaC-13myc and γENaC-V5) were co-expressed in WT (BY4742) yeast. To confirm that the ENaC subunits assemble, we performed co-immunoprecipitations under nondenaturing conditions. As described in the Experimental Procedures section, αENaC-HA was immunoprecipitated with anti-HA agarose beads, and immunoblots were performed to detect each of the subunits. In contrast to an abundant ER membrane protein, Sec61, both the β- and γ-subunits co-precipitated with αENaC (Figure 2C). These data strongly suggest that the trimeric ENaC channel assembles in yeast. This result is also consistent with an earlier report using an alternate yeast ENaC expression system [64].
Next, the stability of αENaC was measured in WT and Δlhs1 yeast co-expressing βENaC-13myc and γENaC-V5 by cycloheximide chase analysis (Figure 2D). We observed that αENaC is somewhat more stable when the β and γENaC subunits are co-expressed than when expressed alone (~20% of protein remained after 90 min compared with <10% in Figure 2A). These results are in accordance with those seen in higher cell systems. For example, the half-lives of the ENaC subunits approximately double in oocytes expressing all three subunits [35]. However, the overall degradation rate remains quite efficient. We also determined by sucrose density centrifugation that the majority of ENaC remains ER-retained (see Supplementary Figure S1). These data are also in line with observations in higher cells [33–35] and are consistent with the poor assembly efficiency observed for numerous polytopic membrane proteins (see Introduction). In addition, the presence of all three subunits eliminated the Lhs1-dependent degradation of αENaC (Figure 2D). Importantly, the prominent unglycosylated species we observed when αENaC was expressed alone is no longer present (compare Δlhs1 in Figure 2A versus D). Consistent with these data, EndoH digestion of protein lysates obtained from the experiment in Figure 2D confirmed that the lower molecular mass band is the unglycosylated protein (Figure 2E). For each experiment, we also confirmed β- and γ-ENaC expression by western blot, but no effect was observed on the stabilities of these subunits (data not shown; also see below). Taken together, these results suggest that channel oligomerization occludes a recognition motif in αENaC or that channel oligomerization alters αENaC structural elements that are necessary for Lhs1-targeted ERAD.
Expression of both the β- and γ-ENaC subunits is required to eliminate the Lhs1-dependent degradation of αENaC
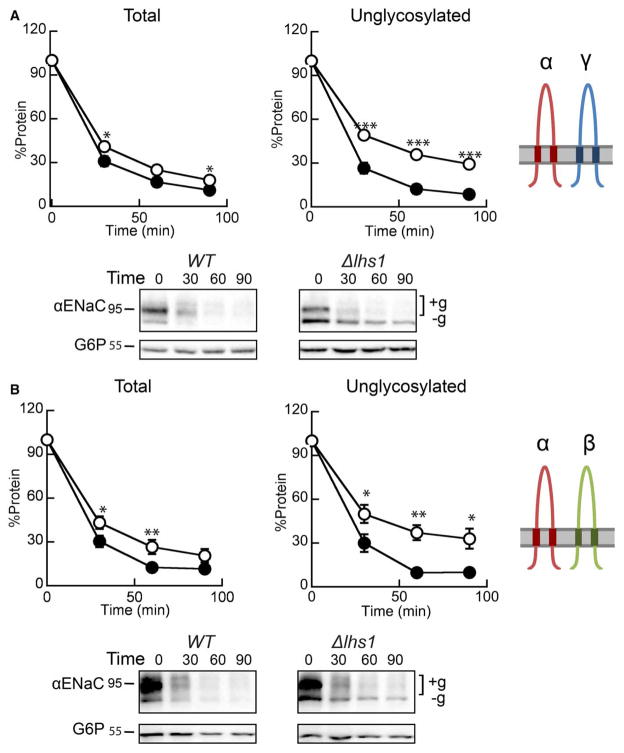
We next asked whether expression of both the β- and γ-ENaC subunits was necessary to eliminate the Lhs1-dependent ERAD of αENaC. To answer this question, either the α- and γ-ENaC subunits (Figure 3A) or the α- and β-ENaC subunits (Figure 3B) were co-expressed in the WT and Δlhs1 yeast strains. As illustrated in Figure 3A,B, αENaC was nearly absent after 90 min in the WT strain, but was partially stabilized in the Δlhs1 strain co-expressing either βENaC or γENaC. The presence of a lower molecular mass species at 72 kDa (−g) that resists degradation in Δlhs1 yeast co-expressing αENaC and γENaC (Figure 3A) or αENaC and βENaC (Figure 3B) is consistent with the heightened selection of unglycosylated αENaC by Lhs1 [46]. This species is also observed when αENaC is expressed alone (see Figure 2A). When the unglycosylated protein (−g) is quantified separately from the total protein, a stronger Lhs1 dependence is observed, whereas the degradation of the glycosylated αENaC protein (+g) is unaffected (not shown). Therefore, co-expression of either the β- or γ-ENaC subunit reduces Lhs1-dependent degradation of the ENaC α-subunit (compare ‘Total’ protein in Figure 2A with ‘Total’ protein in Figure 3A,B), but only the presence of both of the other subunits completely blocks the Lhs1-supported degradation of αENaC.
Figure 3. The expression of both βENaC and γENaC blocks Lhs1-dependent degradation of αENaC.
Cycloheximide chase reactions were performed as described in the Experimental Procedures section in WT (filled circles) and Δlhs1 (open circles) yeast strains transformed with plasmids engineered to express either (A) αENaC-HA and γENaC-V5 or (B) αENaC-HA and βENaC-13myc. Lysates were prepared and resolved, and proteins were immunoblotted with anti-HA antisera (αENaC) and with anti-G6P as a loading control. Glycosylated (+g) and unglycosylated (−g) species are indicated. The expression of βENaC and γENaC was confirmed by western blot (data not shown). Data represent the means of 7–12 experiments, ±SEM. *P < 0.05, **P < 0.01, ***P < 0.0001.
Lhs1-dependent degradation of αENaC is unaffected by interactions with extracellular domains of the α- and β-ENaC subunits
Based on the data presented above, we hypothesized that disrupting crucial subunit–subunit interactions would restore Lhs1-dependent ERAD of the α-subunit and result in a reappearance of the stable unglycosylated species. Although experiments presented in Figure 3 suggest that interactions between α- and γ-ENaC are also critical for evading Lhs1-targeted ERAD, based on preliminary data, we chose to focus our investigation on the interactions between the α- and β-ENaC subunits. Therefore, to systematically test which domains of βENaC subvert Lhs1-dependent ERAD, a series of chimeric α/βENaC constructs was synthesized (see Figure 1). Our strategy was to insert αENaC domains into the analogous domains in βENaC and then co-express the chimeric proteins with αENaC and γENaC. We surmised that this strategy would allow us to identify which intersubunit domain interactions block αENaC degradation. It is important to note that αENaC on its own can trimerize, traffic to the plasma membrane and act as a sodium-conducting channel at ~10% efficiency of the heterotrimeric channel in higher cells [65]. Therefore, we anticipated that the chimeric proteins would assemble with αENaC.
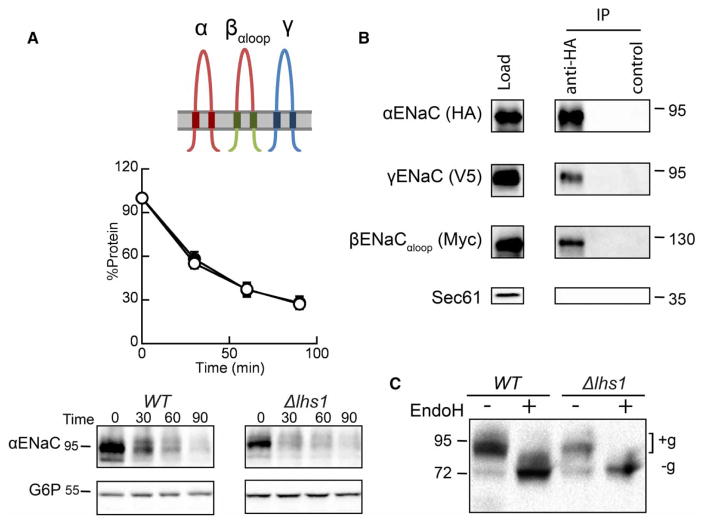
Because the ECL accounts for ~75% of the mass of each subunit and provides extensive interaction interfaces with the other subunits [22,52–54,66–68], we first tested whether ECL interactions stabilize the α-subunit. Moreover, the ECL resides within the ER lumen and might interact with ER lumenal chaperones, such as Lhs1. Thus, we predicted that αENaC degradation would again be Lhs1-dependent when co-expressed with γENaC and βENaCαloop, a construct in which the ECL loop of βENaC is replaced with that from αENaC (Figures 1 and 4A). Surprisingly, there was no significant difference between the degradation of αENaC when co-expressed with βENaCαloop and γENaC in a WT and an Δlhs1 strain (Figure 4A). In fact, the degradation profile was identical with that observed when αENaC was co-expressed with the WT β- and γ-ENaC subunits (Figure 2D). Subunit–subunit associations were retained as both βENaCαloop and γENaC were precipitated with αENaC (Figure 4B), suggesting that assembly of the α- and β-ENaC ECLs is not sufficient to confer stabilization (see below). Moreover, the prominent lower molecular mass, unglycosylated protein species that we observed when αENaC was expressed alone (Figure 2A) was largely absent (Figure 4A,C).
Figure 4. The β-subunit ECL has no effect on the Lhs1-dependent degradation of αENaC.
(A) Cycloheximide chase reactions were performed as described in the Experimental Procedures section in WT (filled circles) and Δlhs1 (open circles) yeast strains transformed with plasmids engineered for the expression of αENaC-HA, βENaCαloop-13myc and γENaC-V5. Lysates were prepared, and resolved proteins were immunoblotted with anti-HA antisera (αENaC) and with anti-G6P as a loading control. The expression of βENaCαloop and γENaC were confirmed by western blot (data not shown). Data represent the means of 12–14 experiments, ±SEM. (B) Cell lysates from WT yeast expressing αENaC-HA, βENaCαloop-13myc and γENaC-V5 were prepared, and resolved proteins were subjected to immunoprecipitation with either anti-HA agarose resin or sepharose (control) and immunoblotted with anti-HA (αENaC), anti-myc (βENaCαloop), anti-V5 (γENaC) or anti-Sec61 as a control. Samples equal to 1% of immunoprecipitated material were also immunoblotted (load). (C) Cell lysates from WT or Δlhs1 yeast expressing αENaC-HA, βENaCαloop-13myc and γENaC-V5 were treated with Endo H. Anti-HA immunoblots are shown. Glycosylated (+g) and unglycosylated (−g) species are indicated.
The βENaC transmembrane segments are sufficient to prevent Lhs1 selection and degradation of αENaC within the ENaC heterotrimer
We next investigated whether replacing the TMD and cytosolic domains of βENaC with those from αENaC would restore α-subunit degradation. As shown in Figure 5A, when the αENaCβloop chimera was expressed with the WT α- and γENaC subunits, Lhs1-dependent degradation of αENaC was again evident. Notably, the degradation profile of αENaC when co-expressed with αENaCβloop and γENaC in the Δlhs1 strain resembled that observed when αENaC was expressed alone (Figure 2A). Furthermore, a lower molecular mass, unglycosylated (−g) (Figure 5A) species was again preferentially stabilized, whereas the higher molecular mass, glycosylated (+g) species was not. As in Figure 4, αENaC was core glycosylated (Figure 5B) and the αENaCβloop protein was immunoprecipitated with αENaC, as was γENaC (Figure 5C). Therefore, although αENaCβloop still associates with αENaC, either the recognition motif for Lhs1-targeted ERAD remains exposed, or the structural changes in αENaC required to evade Lhs1-targeted degradation are absent when the βENaC transmembrane and cytosolic domains are replaced.
Figure 5. Inserting the α-subunit TMDs and N- and C-termini into βENaC restores Lhs1-dependent degradation of αENaC.
(A) Cycloheximide chase reactions were performed as described in the Experimental Procedures section in WT (filled circles) and Δlhs1 (open circles) yeast strains transformed with plasmids engineered for the expression of αENaC-HA, αENaCβloop-13myc and γENaC-V5. Lysates were prepared, and resolved proteins were immunoblotted with anti-HA antisera (αENaC) and with anti-G6P as a loading control. The expression of αENaCβloop and γENaC were confirmed by western blot (data not shown). Glycosylated (+g) and unglycosylated (−g) species are indicated. Data represent the means of 11–12 experiments, ±SEM. *P < 0.05, **P ≤ 0.01, ***P < 0.001. (B) Cell lysates from WT or Δlhs1 yeast expressing αENaC-HA, αENaCβloop-13myc and γENaC-V5 were treated with Endo H. Anti-HA immunoblots are shown. (C) Cell lysates from WT yeast expressing αENaC-HA, αENaCβloop-13myc and γENaC-V5 were subjected to immunoprecipitation with either anti-HA agarose resin or sepharose (as a control), and resolved proteins were immunoblotted with anti-HA (αENaC), anti-myc (αENaCβloop), anti-V5 (γENaC) or anti-Sec61 as a control. Samples equal to 1% of immunoprecipitated material were also immunoblotted (load).
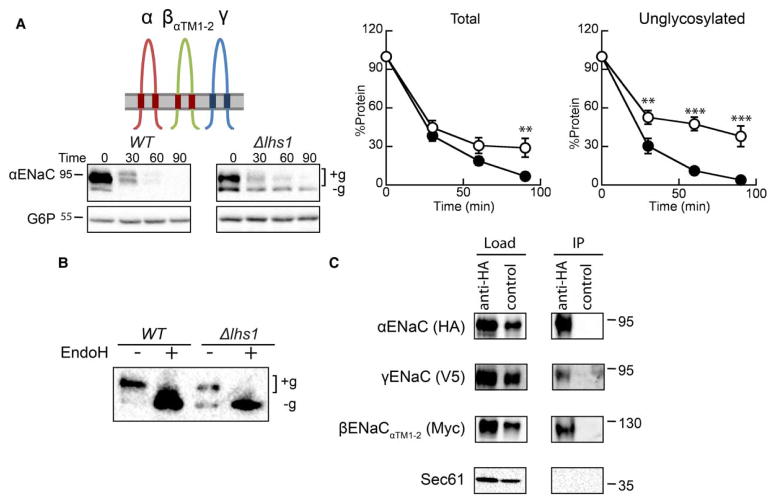
To further refine which domain(s) in βENaC prevent αENaC turnover, we next co-expressed a chimeric ENaC construct composed of βENaC with the two transmembrane segments from αENaC (βENaCαTM1–2; Figures 1 and 6A). In other words, the presence of the α- and β-ENaC TMDs may be required to assemble the ENaC heterotrimer in a native conformation and block Lhs1-targeted degradation. Therefore, we predicted that co-expressing βENaCαTM1–2 with the α- and γ-ENaC subunits would restore ERAD, and that degradation would be compromised in the Δlhs1 mutant strain. As hypothesized, αENaC was more stable in the Δlhs1 strain than in a WT yeast strain, and the stable, unglycosylated αENaC band (−g) was again prominent (Figure 6A,B). Moreover, as previously observed ([46] and see above), the unglycosylated species was significantly more stable, whereas the degradation rate of the glycosylated (+g) species was unchanged. Importantly, the presence of the αENaC TMDs in the context of βENaC did not inhibit interactions among the channel subunits (Figure 6C). These data suggest that only productive interactions between the α- and β-ENaC TMDs subvert Lhs1-dependent degradation.
Figure 6. Inserting the α-subunit TMDs into βENaC is sufficient to restore Lhs1-dependent degradation of αENaC.
(A) Cycloheximide chase reactions were performed as described in the Experimental Procedures section in WT (filled circles) and Δlhs1 (open circles) yeast strains transformed with plasmids designed to express αENaC-HA, γENaC-V5 and βENaCαTM1–2-13myc. Lysates were prepared, and resolved proteins were immunoblotted with anti-HA antisera (αENaC) and with anti-G6P as a loading control. The expression of the myc-tagged and γENaC proteins were confirmed by western blot (data not shown). Glycosylated (+g) and unglycosylated (−g) species are indicated. Data represent the means of 8–11 experiments, ±SEM. **P < 0.01, ***P < 0.001. (B) Cell lysates from WT or Δlhs1 yeast expressing αENaC-HA, βENaCαTM1–2-13myc and γENaC-V5 were treated with Endo H. Anti-HA immunoblots are shown. (C) Cell lysates from WT yeast expressing αENaC-HA, αENaCβTM1–2-13myc and γENaC-V5 were subjected to immunoprecipitation with either anti-HA agarose resin or sepharose (control) and immunoblotted with anti-HA (αENaC), anti-myc (βENaCαTM1-2), anti-V5 (γENaC) or anti-Sec61 as a control. Samples equal to 1% of immunoprecipitated material were also immunoblotted (load).
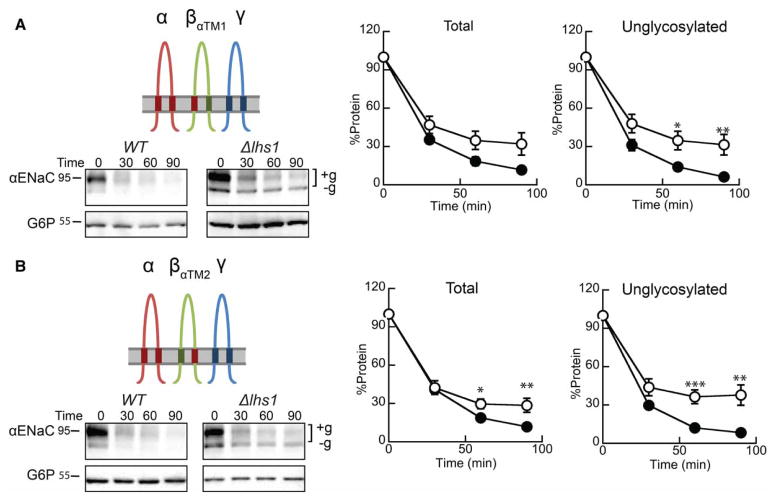
We then asked whether the βENaC TMD1 or TMD2 is sufficient to slow αENaC degradation. βENaC constructs with either TMD1 or TMD2 were replaced with the corresponding αENaC TMD (βENaCαTM1 or βENaCαTM2; Figures 1 and 7A,B), and the chimeric subunits were co-expressed with the α- and γ-ENaC subunits in WT or Δlhs1 yeast. The results of the subsequent cycloheximide chase analysis show that replacing either βENaC TMD restored Lhs1-dependent degradation to the co-expressed αENaC subunit (Figure 7A,B). Again, the unglycosylated protein was preferentially stabilized, and a prominent lower molecular mass species was observed. Therefore, evasion of Lhs1-targeted ERAD requires the interaction of both βENaC TMDs with αENaC.
Figure 7. Inserting the α-subunit TMD1 or TMD2 into βENaC restores Lhs1-dependent degradation of αENaC.
Cycloheximide chase reactions were performed as described in the Experimental Procedures section in WT (filled circles) and Δlhs1 (open circles) yeast strains transformed with plasmids designed to express αENaC-HA, γENaC-V5 and (A) βENaCαTM1-13myc or (B) βENaCαTM2-13myc. Lysates were prepared and resolved proteins were immunoblotted with anti-HA antisera (αENaC) and with anti-G6P as a loading control. The expression of myc-tagged and γENaC proteins were confirmed by western blot (data not shown). Glycosylated (+g) and unglycosylated (−g) species are indicated. Data represent the means of 8–11 experiments, ±SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
As noted above, Lhs1 also plays a role in the translocation of select proteins, so we confirmed that swapping the TMDs between αENaC and βENaC did not affect membrane integration, as assessed by carbonate extraction. Similar to our observation for WT αENaC (Figure 2B), each of the ENaC chimeras remained in the pellet fraction after treatment with carbonate (data not shown), implying that at least one of the TMDs is membrane-integrated (see Models in Discussion).
Ubiquitinated αENaC accumulates in yeast ER membranes lacking Lhs1
The experiments described in the preceding sections define structural elements within αENaC, namely the TMDs, which allow the protein to escape ER quality control in the absence of Lhs1. The results also confirm our previous observation that Lhs1 preferentially targets unglycosylated αENaC for degradation [46]. It is important to reiterate that variable glycosylation of the ENaC subunits occurs in higher cell systems [62,63], and we previously showed that the mammalian homolog of Lhs1, GRP170, preferentially binds unglycosylated αENaC in HEK293 cells [46].
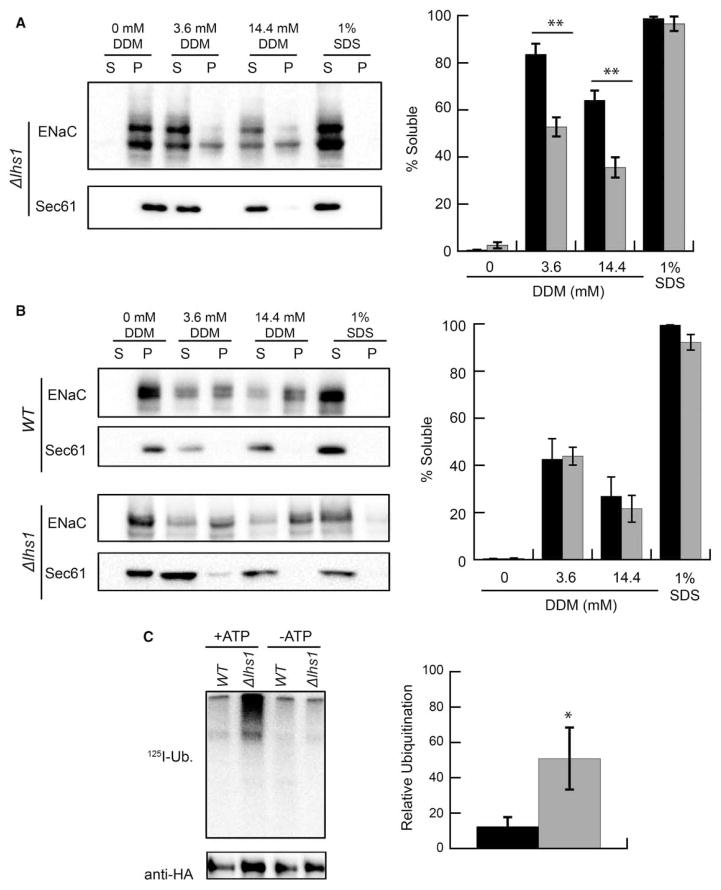
To begin to understand how Lhs1 targets unglycosylated αENaC for degradation, we tested the hypothesis that unglycosylated αENaC is less soluble than the glycosylated species. Therefore, the Lhs1 and GRP170 holdase activities might facilitate ERAD since N-linked glycans can increase the solubility of misfolded proteins [69–71]. αENaC was expressed in Δlhs1 yeast, which allows for the formation of both the glycosylated and unglycosylated αENaC species. ER-enriched microsomes were then prepared and subjected to treatment with either the nonionoic detergent, DDM, or the ionic detergent, SDS, and also subjected to centrifugation to obtain a pellet (insoluble) and supernatant (soluble) fraction (see Experimental Procedures). Consistent with our hypothesis, more glycosylated (higher molecular mass) than unglycosylated (lower molecular mass) protein was found in the soluble fraction, and a greater percentage of the unglycosylated protein remained in the insoluble pellet fraction when treated with DDM (Figure 8A). In the absence of detergent, αENaC resided only in the pellet fraction, but when treated with SDS, it was exclusively in the supernatant fraction. Based on these data, we hypothesized that Lhs1 increases the solubility of the unglycosylated αENaC species, and expressed an αENaC construct lacking the six N-linked glycosylation sites, ΔGαENaC, in either WT or Δlhs1 yeast. We previously showed that ΔGαENaC degradation was significantly more dependent on Lhs1 than WT αENaC [46]. Contrary to our prediction, however, the presence or absence of Lhs1 had no effect on ΔGαENaC solubility (Figure 8B).
Figure 8. Lhs1 acts after αENaC ubiquitination.
(A) ER-enriched microsomes were prepared as described in Experimental Procedures from Δlhs1 yeast transformed with the αENaC-HA expression vector. The microsomes were treated with DDM or SDS, and the solution was centrifuged at 18 000 × g. The isolated proteins were immunoblotted with anti-HA antisera (αENaC) and with anti-Sec61, which served as an ER-resident membrane protein control. The relative amounts of the glycosylated (black bars) and unglycosylated (gray bars) proteins were quantified. Data represent the means of four experiments ±SEM. **P < 0.01. (B) ER-enriched microsomes were prepared from WT and Δlhs1 yeast transformed with an expression vector for αENaC that lacks the N-linked glycosylation sites, ΔGαENaC-HA. A solubility assay was then performed as described in A. The solubility of ΔGαENaC-HA in microsomes from WT (black bars) and Δlhs1 (gray bars) yeast is indicated. Data represent the means of four experiments ±SEM. (C) ER-enriched microsomes from WT (black bar) or Δlhs1 (gray bar) yeast expressing ΔGαENaC-HA were subjected to an in vitro ubiquitination assay as described in Experimental Procedures. Levels of ubiquitination were corrected to the relative amount of ENaC (anti-HA signal) present in each lane. Data represent the means of 6–9 experiments relative to WT ubiquitination ratios, ±SEM, *P < 0.05.
Ubiquitination targets proteins for degradation by the cytosolic 26S proteasome and is required for the hydrolysis of nearly every ERAD substrate [3,15]. Hrd1 and Doa10 are the E3 ligases most commonly associated with ERAD in yeast [72], and consistent with this fact, we showed that αENaC degradation and ubiquitination are facilitated by these ligases [45]. Based on its presence in the ER lumen and noted effects on ENaC degradation, we reasoned that Lhs1 acts upstream of substrate ubiquitination and anticipated that αENaC ubiquitination would decrease in Δlhs1 versus WT yeast. To this end, we quantified the amount of ubiquitinated αENaC in ER-derived microsomes using an established in vitro ubiquitination assay; the assay has been used to define the requirements for the ubiquitination of several ERAD substrates, including αENaC [45,60,73]. In brief, purified ER microsomes from WT or Δlhs1 yeast expressing ΔGαENaC were prepared and incubated with yeast cytosol, 125I-ubiquitin, and either an ATP-regenerating system or apyrase (-ATP). We used ΔGαENaC instead of WT αENaC because Lhs1 preferentially targets unglycosylated αENaC for degradation ([46] and see above), so this substrate provides a more robust signal. After ΔGαENaC immunoprecipitation and autoradiography, we found that deletion of Lhs1 resulted in the accumulation of approximately four times more ubiquitinated ΔGαENaC than in WT yeast (Figure 8C). Contrary to our expectations, these results are in accordance with Lhs1 acting downstream from αENaC ubiquitination. Therefore, in contrast with lumenal Hsp40 molecular chaperones that facilitate αENaC ubiquitination [45], Lhs1 appears to identify the ubiquitinated substrate and/or helps target ubiquitinated αENaC to the proteasome. Consistent with Lhs1 acting downstream of channel ubiquitination, ~50% less αENaC was retrotranslocated from isolated ER membranes prepared from Δlhs1 versus WT cells in an established in vitro assay ([60,73]; data not shown). In fact, a recent paper from the Tsai Laboratory suggested that GRP170 promotes the retrotranslocation of the Null Hong Kong variant of α-1 antitrypsin (NHK) [74], which would occur downstream of substrate ubiquitination. A model that takes into account the potential role of Lhs1 in the pathway for the ERAD of αENaC is presented below.
Discussion
Our work begins to unravel how ENaC channel assembly affects ER quality control decisions made by a conserved, ER lumenal molecular chaperone, Lhs1. First, we demonstrate that the presence of the other ENaC subunits blocks Lhs1-dependent αENaC degradation. Second, we found that βENaC TMDs hinder the Lhs1-targeted degradation of αENaC. And third, we observed that Lhs1 acts downstream of αENaC ubiquitination in the ERAD pathway.
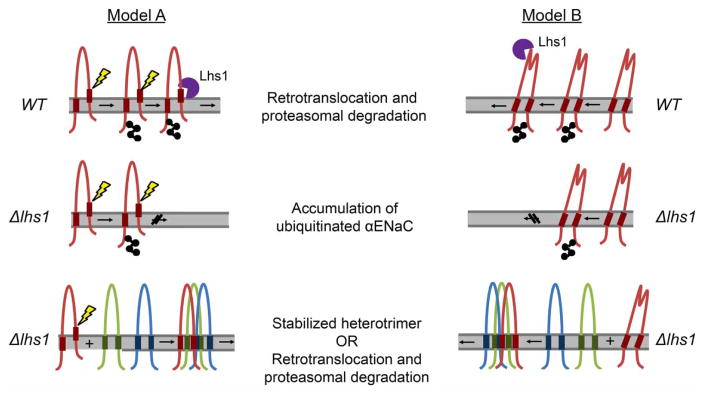
How does Lhs1 differentially distinguish monomeric αENaC versus αENaC in the context of the trimeric channel? One explanation is that oligomerization simply occludes an Lhs1-binding site on the αENaC monomer, which eliminates the ability of the chaperone to identify the subunit. We disfavor this explanation because βENaCαTM1–2 and other chimeric constructs continue to assemble with αENaC (see, for example, Figures 5–7), but do not eliminate Lhs1-targeted ERAD. We propose that this association is mediated either through the ECLs or through the WT γENaC subunit that is also co-expressed. Therefore, the Lhs1-binding site(s) must still be available for chaperone recognition. Instead, we propose two alternate models (Figure 9) for recognition of the ubiquitinated αENaC protein by Lhs1. First, in the absence of the β- and γ-subunits, the transmembrane domains of αENaC may poorly integrate into the membrane and expose a ‘degron’ [75,76] that is recognized directly or indirectly by Lhs1 (Model A). Second, associations between the αENaC TMDs and the β- and γ-subunit TMDs may facilitate a conformational change in the ECL that allows evasion of Lhs1-targeted ERAD (Model B).
Figure 9. Selection of αENaC for Lhs1-facilitated ER-associated degradation.
Models A and B describe the selection of monomeric αENaC (red) for Lhs1-targeted degradation and the evasion of ERAD by the ENaC heterotrimer. In Model A, one αENaC TMD fails to stably integrate into the membrane, which exposes TMD residues to the ER lumen (depicted with a yellow lightning blot) and the ER quality control machinery. Lhs1 (purple) either directly or indirectly recognizes the exposed TMD as a degron and targets αENaC for ERAD. When all three subunits are present, interactions between the αENaC TMDs with the βENaC (green) and γENaC (blue) TMDs stabilize the αENaC TMD within the membrane and thwart Lhs1-dependent ERAD. Model B predicts a conformational change associated with ENaC heterotrimerization. Here, Lhs1 binds to the ECL of monomeric αENaC, targeting it for ERAD. However, oligomerization and association between the three ENaC subunit TMDs result in a conformational change that is transmitted through the wrist domain to the αENaC ECL. This relay hinders Lhs1-dependent degradation.
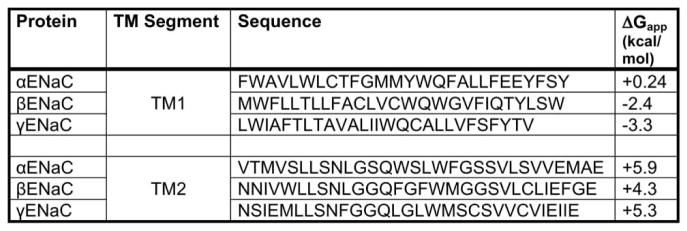
Our first model predicts that one or both of the αENaC TMDs fail to efficiently integrate into the lipid bilayer in the absence of the βENaC TMD. A study by von Heijne and colleagues reported that >30% of TMDs inefficiently integrate into the membrane because they contain marginally hydrophobic TMDs [77]. The integration of these TMDs requires the presence of other stabilizing TMDs [78–81]. To determine whether the αENaC TMDs poorly integrate, we performed free energy calculations for membrane insertion (http://dgpred.cbr.su.se/index.php?p=home). The results shown in Figure 10 predict that the integration of the αENaC TMDs is less favorable than that of the β- and γ-ENaC subunits. In fact, the ΔGapp for helix insertion is positive for both αENaC TMD1 and TMD2, whereas the ΔGapp for helix insertion is positive for only one TMD in the β- and γENaC subunits (Figure 10). Therefore, one of the αENaC TMDs may slip into the ER lumen, allowing for direct recognition by Lhs1. However, when βENaC is co-expressed, the βENaC TMDs may stabilize the αENaC TMDs in the lipid bilayer (Model A, Figure 9). Similar phenomena have been described for other multimeric proteins. For example, the Na+/K+ ATPase is composed of an α-subunit that contains 10 TMDs and a β-subunit that contains just 1 TMD. In the absence of the β-subunit, the α-subunit is targeted for ERAD [82–85]. Specifically, TMD7 of the α-subunit inefficiently integrates into the membrane and instead slips into the cytoplasm, exposing a degradation signal that resides in the loop between TM7 and TM8 [83,85]. The exposed degradation signal is recognized by the quality control machinery and the protein is targeted for ERAD. In addition, unpaired charges within the TMD in another well-characterized ERAD substrate, TCRα, act as a degron unless the TCRα-binding partner, CD3δ, is present. Charge pairing between the TMDs of TCRα and CD3δ allows for stable expression of both proteins [86–88]. In the absence of its binding partners, TCRα slips entirely into the ER lumen where the ER lumenal chaperones, BiP and calnexin/calreticulin, facilitate retrotranslocation and degradation [89,90].
Figure 10. ENaC transmembrane sequences and predicted membrane insertion free energies.
The amino acid sequence for the predicted first and second αENaC, βENaC and γENaC TMDs is indicated. The apparent free energy of insertion was calculated for each TMD using http://dgpred.cbr.su.se/index.php?p=home.
Our second model predicts that association between the ENaC TMDs induces a conformational change in the extracellular/ER lumenal loop that precludes recognition by the ER quality control machinery, e.g. Lhs1 (Model B, Figure 10). Structural models of ENaC based on the crystal structure of the ENaC family member, ASIC [52–54], predict that the TMDs of αENaC are connected to the ECL by a short linker region known as the ‘wrist’ domain [91]. Structure–function and modeling studies of ENaC and other degenerin family members determined that binding of extracellular factors to the ECL induces movements that are transferred through the wrist domain to the channel pore, and — therefore — the TMDs [52,54,92–94]. For example, extracellular sodium and laminar sheer stress act on the ECL to alter the sodium-conducting properties of the ENaC pore, and mutations in the wrist domain alter channel regulation by sodium and sheer stress [91]. Thus, it is reasonable to posit that changes in the TMDs relay conformational changes to the ECL. In turn, conformational changes induced by TMD assembly may prevent Lhs1 binding and, formally, promote recognition by factors that stabilize ENaC. A thorough test of these models will systematically be investigated in future studies.
Another question is why unglycosylated αENaC is stabilized in the absence of Lhs1, and why the human homolog, GRP170, preferentially associates with the unglycosylated αENaC species in HEK293 cells [46]. The Perlmutter laboratory reported that tunicamycin treatment, which blocks N-linked glycosylation, increased GRP170 binding to another ERAD substrate, α1-antitrypsin Z [95]. In pulse-chase experiments, we found that the ratio of unglycosylated protein to glycosylated protein immediately after labeling was similar in the WT and Δlhs1 strains and that the glycosylated and unglycosylated proteins possess unique turnover rates (data not shown). We hypothesize that during ER quality control, Lhs1/GRP170 acts downstream of the ER localized lectins, which recognize and target glycosylated proteins for degradation [9]. Therefore, unglycosylated αENaC that evades detection by the lectins is targeted for ERAD by Lhs1/GRP170. In the future, it will also be important to assess whether the Lhs1/GRP170 chaperones affect the stability of other soluble and integral membrane ERAD substrates, whether the holdase or NEF [74] activity favors different unassembled monomers in protein complexes, and whether Lhs1/GRP170 recognizes distinct motifs in αENaC (Figure 9).
Based on the growing interest in identifying chemical modulators of molecular chaperones [96], a clarification of the mechanism of Lhs1/GRP170-dependent degradation may lead to therapeutic targets for patients with mutations that affect ENaC folding and/or degradation. In addition, our data predict that polymorphisms in the gene encoding GRP170 will compromise salt/water balance and blood pressure regulation. The expansion of genomic databases corresponding to patients with hypertension (for example, http://www.bioguo.org/CADgene/index.php, http://bws.iis.sinica.edu.tw/THOD and VAHC (Veterans Administration Healthcare System) [97]) will allow us to test this hypothesis.
Supplementary Material
Acknowledgments
Funding
This work was funded by the National Institute of Health [grants K01DK090195 to T.M.B., DK051391 to T.R.K., GM75061 to J.L.B., DK061296 to G.M.P. and DK79307 (University of Pittsburgh George O’Brien Kidney Research Center) to T.R.K. and J.L.B.] and CF Foundation Student Traineeship, PRESTO15H0, to G.M.P.
We thank Drs Patrick Needham, Rebecca Hughey, Arohan Subramanya, Ossama Kashlan, Allyson O’Donnell, Chris Guerriero, Vlad Denic and Linda Hendershot for helpful discussions, reagents and/or technical assistance.
Abbreviations
- CFTR
cystic fibrosis conductance regulator
- DDM
n-dodecyl-β-D-maltoside
- ECL
extracellular loop
- ENaC
epithelial sodium channel
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- HRP
horseradish peroxidase
- IP
immunoprecipitation
- NEF
nucleotide exchange factor
- NEM
N-ethylmaleimide
- PDI
protein disulfide isomerase
- SDS–PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- TCA
trichloroacetic acid
- TCR
T cell receptor
- TM
transmembrane segment
- TMD
transmembrane domain
- WT
wild type
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Author Contribution
T.M.B. conceived the idea, conducted experiments and wrote the paper. A.S.J. and M.E.Y. conducted experiments, and A.S.J. also contributed to writing the manuscript. G.M.P. performed the solubility assays and in vitro ubiquitination experiments. E.C. conducted the pilot studies that form the basis for this project. T.R.K. provided intellectual input and edited the manuscript. J.L.B. provided intellectual input and assisted in writing and editing the manuscript.
References
- 1.Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amm I, Sommer T, Wolf DH. Protein quality control and elimination of protein waste: the role of the ubiquitin-proteasome system. Biochim Biophys Acta. 2014;1843:182–196. doi: 10.1016/j.bbamcr.2013.06.031. [DOI] [PubMed] [Google Scholar]
- 3.Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hampton RY, Garza RM. Protein quality control as a strategy for cellular regulation: lessons from ubiquitin-mediated regulation of the sterol pathway. Chem Rev. 2009;109:1561–1574. doi: 10.1021/cr800544v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ismail N, Ng DTW. Have you HRD? Understanding ERAD is DOAble! Cell. 2006;126:237–239. doi: 10.1016/j.cell.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 7.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hebert DN, Molinari M. Flagging and docking: dual roles for N-glycans in protein quality control and cellular proteostasis. Trends Biochem Sci. 2012;37:404–410. doi: 10.1016/j.tibs.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rutkevich LA, Williams DB. Participation of lectin chaperones and thiol oxidoreductases in protein folding within the endoplasmic reticulum. Curr Opin Cell Biol. 2011;23:157–166. doi: 10.1016/j.ceb.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 10.Claessen JHL, Kundrat L, Ploegh HL. Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 2012;22:22–32. doi: 10.1016/j.tcb.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hitchcock AL, Krebber H, Frietze S, Lin A, Latterich M, Silver PA. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol Biol Cell. 2001;12:3226–3241. doi: 10.1091/mbc.12.10.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 15.Raasi S, Wolf DH. Ubiquitin receptors and ERAD: a network of pathways to the proteasome. Semin Cell Dev Biol. 2007;18:780–791. doi: 10.1016/j.semcdb.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Bhalla V, Hallows KR. Mechanisms of ENaC regulation and clinical implications. J Am Soc Nephrol. 2008;19:1845–1854. doi: 10.1681/ASN.2008020225. [DOI] [PubMed] [Google Scholar]
- 17.Kashlan OB, Kleyman TR. Epithelial Na(+) channel regulation by cytoplasmic and extracellular factors. Exp Cell Res. 2012;318:1011–1019. doi: 10.1016/j.yexcr.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder PM. Minireview: regulation of epithelial Na+ channel trafficking. Endocrinology. 2005;146:5079–5085. doi: 10.1210/en.2005-0894. [DOI] [PubMed] [Google Scholar]
- 19.Soundararajan R, Lu M, Pearce D. Organization of the ENaC-regulatory machinery. Crit Rev Biochem Mol Biol. 2012;47:349–359. doi: 10.3109/10409238.2012.678285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blazer-Yost BL, Liu X, Helman SI. Hormonal regulation of ENaCs: insulin and aldosterone. Am J Physiol. 1998;274(5 Pt 1):C1373–C13739. doi: 10.1152/ajpcell.1998.274.5.C1373. [DOI] [PubMed] [Google Scholar]
- 21.Eaton DC, Malik B, Bao HF, Yu L, Jain L. Regulation of epithelial sodium channel trafficking by ubiquitination. Proc Am Thorac Soc. 2010;7:54–64. doi: 10.1513/pats.200909-096JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canessa CM, Merillat AM, Rossier BC. Membrane topology of the epithelial sodium channel in intact cells. Am J Physiol. 1994;267(6 Pt 1):C1682–C1690. doi: 10.1152/ajpcell.1994.267.6.C1682. [DOI] [PubMed] [Google Scholar]
- 23.Weisz OA, Wang JM, Edinger RS, Johnson JP. Non-coordinate regulation of endogenous epithelial sodium channel (ENaC) subunit expression at the apical membrane of A6 cells in response to various transporting conditions. J Biol Chem. 2000;275:39886–39893. doi: 10.1074/jbc.M003822200. [DOI] [PubMed] [Google Scholar]
- 24.Asher C, Wald H, Rossier BC, Garty H. Aldosterone-induced increase in the abundance of Na+ channel subunits. Am J Physiol. 1996;271(2 Pt 1):C605–C611. doi: 10.1152/ajpcell.1996.271.2.C605. [DOI] [PubMed] [Google Scholar]
- 25.Masilamani S, Kim GH, Mitchell C, Wade JB, Knepper MA. Aldosterone-mediated regulation of ENaC α, β, and γ subunit proteins in rat kidney. J Clin Invest. 1999;104:R19–R23. doi: 10.1172/JCI7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, et al. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 27.Grunder S, Firsov D, Chang SS, Jaeger NF, Gautschi I, Schild L, et al. A mutation causing pseudohypoaldosteronism type 1 identifies a conserved glycine that is involved in the gating of the epithelial sodium channel. EMBO J. 1997;16:899–907. doi: 10.1093/emboj/16.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobbs CA, Da Tan C, Tarran R. Does epithelial sodium channel hyperactivity contribute to cystic fibrosis lung disease? J Physiol. 2013;591:4377–4387. doi: 10.1113/jphysiol.2012.240861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mall M, Grubb BR, Harkema JR, O’Neal WK, Boucher RC. Increased airway epithelial Na+ absorption produces cystic fibrosis-like lung disease in mice. Nat Med. 2004;10:487–493. doi: 10.1038/nm1028. [DOI] [PubMed] [Google Scholar]
- 30.Gentzsch M, Dang H, Dang Y, Garcia-Caballero A, Suchindran H, Boucher RC, et al. The cystic fibrosis transmembrane conductance regulator impedes proteolytic stimulation of the epithelial Na+ channel. J Biol Chem. 2010;285:32227–32232. doi: 10.1074/jbc.M110.155259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ismailov II, Awayda MS, Jovov B, Berdiev BK, Fuller CM, Dedman JR, et al. Regulation of epithelial sodium channels by the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 1996;271:4725–4732. doi: 10.1074/jbc.271.9.4725. [DOI] [PubMed] [Google Scholar]
- 32.Yan W, Samaha FF, Ramkumar M, Kleyman TR, Rubenstein RC. Cystic fibrosis transmembrane conductance regulator differentially regulates human and mouse epithelial sodium channels in Xenopus oocytes. J Biol Chem. 2004;279:23183–23192. doi: 10.1074/jbc.M402373200. [DOI] [PubMed] [Google Scholar]
- 33.Malik B, Schlanger L, Al-Khalili O, Bao HF, Yue G, Price SR, et al. ENaC degradation in A6 cells by the ubiquitin-proteosome proteolytic pathway. J Biol Chem. 2001;276:12903–12910. doi: 10.1074/jbc.M010626200. [DOI] [PubMed] [Google Scholar]
- 34.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, et al. Regulation of stability and function of the epithelial Na+ channel (ENaC) by ubiquitination. EMBO J. 1997;16:6325–6336. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valentijn JA, Fyfe GK, Canessa CM. Biosynthesis and processing of epithelial sodium channels in Xenopus oocytes. J Biol Chem. 1998;273:30344–30351. doi: 10.1074/jbc.273.46.30344. [DOI] [PubMed] [Google Scholar]
- 36.Needham PG, Brodsky JL. How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: the early history of ERAD. Biochim Biophys Acta. 2013;1833:2447–2457. doi: 10.1016/j.bbamcr.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kolb AR, Buck TM, Brodsky JL. Saccharomyces cerivisiae as a model system for kidney disease: what can yeast tell us about renal function? Am J Physiol Renal Physiol. 2011;301:F1–11. doi: 10.1152/ajprenal.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotin D, Staub O. Role of the ubiquitin system in regulating ion transport. Pflugers Arch. 2011;461:1–21. doi: 10.1007/s00424-010-0893-2. [DOI] [PubMed] [Google Scholar]
- 39.Foresti O, Ruggiano A, Hannibal-Bach HK, Ejsing CS, Carvalho P. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. eLife. 2013;2:e00953. doi: 10.7554/eLife.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olzmann JA, Richter CM, Kopito RR. Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc Natl Acad Sci USA. 2013;110:1345–1350. doi: 10.1073/pnas.1213738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X, Tukachinsky H, Huang CH, Jao C, Chu YR, Tang HY, et al. Processing and turnover of the Hedgehog protein in the endoplasmic reticulum. J Cell Biol. 2011;192:825–838. doi: 10.1083/jcb.201008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zettl M, Adrain C, Strisovsky K, Lastun V, Freeman M. Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell. 2011;145:79–91. doi: 10.1016/j.cell.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hampton RY. Proteolysis and sterol regulation. Annu Rev Cell Dev Biol. 2002;18:345–378. doi: 10.1146/annurev.cellbio.18.032002.131219. [DOI] [PubMed] [Google Scholar]
- 44.Kashlan OB, Mueller GM, Qamar MZ, Poland PA, Ahner A, Rubenstein RC, et al. Small heat shock protein αA-crystallin regulates epithelial sodium channel expression. J Biol Chem. 2007;282:28149–28156. doi: 10.1074/jbc.M703409200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buck TM, Kolb AR, Boyd C, Kleyman TR, Brodsky JL. The ER associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol Biol Cell. 2010;21:1047–1058. doi: 10.1091/mbc.E09-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buck TM, Plavchak L, Roy A, Donnelly BF, Kashlan OB, Kleyman TR, et al. The Lhs1/GRP170 chaperones facilitate the endoplasmic reticulum-associated degradation of the epithelial sodium channel. J Biol Chem. 2013;288:18366–18380. doi: 10.1074/jbc.M113.469882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baxter BK, James P, Evans T, Craig EA. SSI1 encodes a novel Hsp70 of the Saccharomyces cerevisiae endoplasmic reticulum. Mol Cell Biol. 1996;16:6444–6456. doi: 10.1128/MCB.16.11.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Craven RA, Egerton M, Stirling CJ. A novel Hsp70 of the yeast ER lumen is required for the efficient translocation of a number of protein precursors. EMBO J. 1996;15:2640–2650. [PMC free article] [PubMed] [Google Scholar]
- 49.Steel GJ, Fullerton DM, Tyson JR, Stirling CJ. Coordinated activation of Hsp70 chaperones. Science. 2004;303:98–101. doi: 10.1126/science.1092287. [DOI] [PubMed] [Google Scholar]
- 50.Tyson JR, Stirling CJ. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 2000;19:6440–6452. doi: 10.1093/emboj/19.23.6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Keyzer J, Steel GJ, Hale SJ, Humphries D, Stirling CJ. Nucleotide binding by Lhs1p is essential for its nucleotide exchange activity and for function in vivo. J Biol Chem. 2009;284:31564–31571. doi: 10.1074/jbc.M109.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 53.Gonzales EB, Kawate T, Gouaux E. Pore architecture and ion sites in acid-sensing ion channels and P2X receptors. Nature. 2009;460:599–604. doi: 10.1038/nature08218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kashlan OB, Kleyman TR. ENaC structure and function in the wake of a resolved structure of a family member. Am J Physiol Renal Physiol. 2011;301:F684–F696. doi: 10.1152/ajprenal.00259.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics. A Cold Spring Harbor Laboratory Course Manual. 1997. Cold Spring Harbor Laboratory Press; Plainview, New York: 1997. [Google Scholar]
- 56.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 57.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. <953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, et al. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sullivan ML, Youker RT, Watkins SC, Brodsky JL. Localization of the BiP molecular chaperone with respect to endoplasmic reticulum foci containing the cystic fibrosis transmembrane conductance regulator in yeast. J Histochem Cytochem. 2003;51:545–548. doi: 10.1177/002215540305100417. [DOI] [PubMed] [Google Scholar]
- 60.Nakatsukasa K, Brodsky JL. In vitro reconstitution of the selection, ubiquitination, and membrane extraction of a polytopic ERAD substrate. Methods Mol Biol. 2010;619:365–376. doi: 10.1007/978-1-60327-412-8_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prince LS, Welsh MJ. Cell surface expression and biosynthesis of epithelial Na+ channels. Biochem J. 1998;336(Pt 3):705–710. doi: 10.1042/bj3360705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prince LS, Welsh MJ. Effect of subunit composition and Liddle’s syndrome mutations on biosynthesis of ENaC. Am J Physiol. 1999;276(6 Pt 1):C1346–C1351. doi: 10.1152/ajpcell.1999.276.6.C1346. [DOI] [PubMed] [Google Scholar]
- 63.Snyder PM, Cheng C, Prince LS, Rogers JC, Welsh MJ. Electrophysiological and biochemical evidence that DEG/ENaC cation channels are composed of nine subunits. J Biol Chem. 1998;273:681–684. doi: 10.1074/jbc.273.2.681. [DOI] [PubMed] [Google Scholar]
- 64.Gupta SS, Canessa CM. Heterologous expression of a mammalian epithelial sodium channel in yeast. FEBS Lett. 2000;481:77–80. doi: 10.1016/S0014-5793(00)01977-3. [DOI] [PubMed] [Google Scholar]
- 65.Canessa CM, Schild L, Buell G, Thorens B, Gautschi I, Horisberger JD, et al. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 66.Renard S, Lingueglia E, Voilley N, Lazdunski M, Barbry P. Biochemical analysis of the membrane topology of the amiloride-sensitive Na+ channel. J Biol Chem. 1994;269:12981–12986. [PubMed] [Google Scholar]
- 67.Snyder PM, McDonald FJ, Stokes JB, Welsh MJ. Membrane topology of the amiloride-sensitive epithelial sodium channel. J Biol Chem. 1994;269:24379–24383. [PubMed] [Google Scholar]
- 68.Kashlan OB, Adelman JL, Okumura S, Blobner BM, Zuzek Z, Hughey RP, et al. Constraint-based, homology model of the extracellular domain of the epithelial Na+ channel α subunit reveals a mechanism of channel activation by proteases. J Biol Chem. 2011;286:649–660. doi: 10.1074/jbc.M110.167098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferris SP, Jaber NS, Molinari M, Arvan P, Kaufman RJ. UDP-glucose:glycoprotein glucosyltransferase (UGGT1) promotes substrate solubility in the endoplasmic reticulum. Mol Biol Cell. 2013;24:2597–2608. doi: 10.1091/mbc.E13-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu C, Ng DT. Glycosylation-directed quality control of protein folding. Nat Rev Mol Cell Biol. 2015;16:742–752. doi: 10.1038/nrm4073. [DOI] [PubMed] [Google Scholar]
- 71.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, et al. Essentials of Glycobiology. 2. Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]
- 72.Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. J Cell Biol. 2014;204:869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inoue T, Tsai B. The Grp170 nucleotide exchange factor executes a key role during ERAD of cellular misfolded clients. Mol Biol Cell. 2016;27:1650–1662. doi: 10.1091/mbc.E16-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 76.Varshavsky A. Naming a targeting signal. Cell. 1991;64:13–15. doi: 10.1016/0092-8674(91)90202-A. [DOI] [PubMed] [Google Scholar]
- 77.Hessa T, Meindl-Beinker NM, Bernsel A, Kim H, Sato Y, Lerch-Bader M, et al. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- 78.Lu Y, Xiong X, Helm A, Kimani K, Bragin A, Skach WR. Co- and posttranslational translocation mechanisms direct cystic fibrosis transmembrane conductance regulator N terminus transmembrane assembly. J Biol Chem. 1998;273:568–576. doi: 10.1074/jbc.273.1.568. [DOI] [PubMed] [Google Scholar]
- 79.Ota K, Sakaguchi M, von Heijne G, Hamasaki N, Mihara K. Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol Cell. 1998;2:495–503. doi: 10.1016/S1097-2765(00)80149-5. [DOI] [PubMed] [Google Scholar]
- 80.Öjemalm K, Halling KK, Nilsson I, von Heijne G. Orientational preferences of neighboring helices can drive ER insertion of a marginally hydrophobic transmembrane helix. Mol Cell. 2012;45:529–540. doi: 10.1016/j.molcel.2011.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kida Y, Ishihara Y, Fujita H, Onishi Y, Sakaguchi M. Stability and flexibility of marginally hydrophobic-segment stalling at the endoplasmic reticulum translocon. Mol Biol Cell. 2016;27:930–940. doi: 10.1091/mbc.E15-09-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beguin P, Hasler U, Beggah A, Horisberger JD, Geering K. Membrane integration of Na,K-ATPase α-subunits and β-subunit assembly. J Biol Chem. 1998;273:24921–24931. doi: 10.1074/jbc.273.38.24921. [DOI] [PubMed] [Google Scholar]
- 83.Beguin P, Hasler U, Staub O, Geering K. Endoplasmic reticulum quality control of oligomeric membrane proteins: topogenic determinants involved in the degradation of the unassembled Na,K-ATPase α subunit and in its stabilization by β subunit assembly. Mol Biol Cell. 2000;11:1657–1672. doi: 10.1091/mbc.11.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Beggah A, Mathews P, Beguin P, Geering K. Degradation and endoplasmic reticulum retention of unassembled α- and β-subunits of Na, K-ATPase correlate with interaction of BiP. J Biol Chem. 1996;271:20895–20902. doi: 10.1074/jbc.271.34.20895. [DOI] [PubMed] [Google Scholar]
- 85.Beggah AT, Beguin P, Bamberg K, Sachs G, Geering K. β-subunit assembly is essential for the correct packing and the stable membrane insertion of the, H,K-ATPase α-subunit. J Biol Chem. 1999;274:8217–8223. doi: 10.1074/jbc.274.12.8217. [DOI] [PubMed] [Google Scholar]
- 86.Bonifacino JS, Cosson P, Shah N, Klausner RD. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bonifacino JS, Suzuki CK, Klausner RD. A peptide sequence confers retention and rapid degradation in the endoplasmic reticulum. Science. 1990;247:79–82. doi: 10.1126/science.2294595. [DOI] [PubMed] [Google Scholar]
- 88.Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-M. [DOI] [PubMed] [Google Scholar]
- 89.Feige MJ, Behnke J, Mittag T, Hendershot LM. Dimerization-dependent folding underlies assembly control of the clonotypic αβT cell receptor chains. J Biol Chem. 2015;290:26821–26831. doi: 10.1074/jbc.M115.689471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feige MJ, Hendershot LM. Quality control of integral membrane proteins by assembly-dependent membrane integration. Mol Cell. 2013;51:297–309. doi: 10.1016/j.molcel.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shi S, Carattino MD, Kleyman TR. Role of the wrist domain in the response of the epithelial sodium channel to external stimuli. J Biol Chem. 2012;287:44027–44035. doi: 10.1074/jbc.M112.421743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baconguis I, Gouaux E. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature. 2012;489:400–405. doi: 10.1038/nature11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li T, Yang Y, Canessa CM. Interaction of the aromatics Tyr-72/Trp-288 in the interface of the extracellular and transmembrane domains is essential for proton gating of acid-sensing ion channels. J Biol Chem. 2009;284:4689–4694. doi: 10.1074/jbc.M805302200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi S, Ghosh DD, Okumura S, Carattino MD, Kashlan OB, Sheng S, et al. Base of the thumb domain modulates epithelial sodium channel gating. J Biol Chem. 2011;286:14753–14761. doi: 10.1074/jbc.M110.191734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schmidt BZ, Perlmutter DH. Grp78, Grp94, and Grp170 interact with α1-antitrypsin mutants that are retained in the endoplasmic reticulum. Am J Physiol Gastrointest Liver Physiol. 2005;289:G444–G455. doi: 10.1152/ajpgi.00237.2004. [DOI] [PubMed] [Google Scholar]
- 96.Kondoh Y, Osada H. High-throughput screening identifies small molecule inhibitors of molecular chaperones. Curr Pharm Des. 2013;19:473–492. doi: 10.2174/138161213804143743. [DOI] [PubMed] [Google Scholar]
- 97.Salem RM, Pandey B, Richard E, Fung MM, Garcia EP, Brophy VH, et al. The VA hypertension primary care longitudinal cohort: electronic medical records in the post-genomic era. Health Informatics J. 2010;16:274–286. doi: 10.1177/1460458210380527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.