Abstract
Organ-on-a-chip has emerged as a powerful platform with widespread applications in biomedical engineering, such as pathology studies and drug screening. However, the fabrication of organ-on-a-chip is still a challenging task due to its complexity. For an integrated organ-on-a-chip, it may contain four key elements, i.e., a microfluidic chip, live cells/microtissues that are cultured in this chip, components for stimulus loading to mature the microtissues, and sensors for results readout. Recently, bioprinting has been used for fabricating organ-on-a-chip as it enables the printing of multiple materials, including biocompatible materials and even live cells in a programmable manner with a high spatial resolution. Besides, all four elements for organ-on-a-chip could be printed in a single continuous procedure on one printer; in other words, the fabrication process is assembly free. In this paper, we discuss the recent advances of organ-on-a-chip fabrication by bioprinting. Light is shed on the printing strategies, materials, and biocompatibility. In addition, some specific bioprinted organs-on-chips are analyzed in detail. Because the bioprinted organ-on-a-chip is still in its early stage, significant efforts are still needed. Thus, the challenges presented together with possible solutions and future trends are also discussed.
Organ-on-a-chip emerges as a powerful tool in biomedical applications, where live microtissues are cultured in a microfluidic chip in vitro to mimic specific functions of an organ or even multiorgans. Usually, a fully integrated organ-on-a-chip involves four key elements, i.e., a microfluidic chip, live microtissues that are cultured in this chip, components for stimulus loading to mature the microtissues, and also sensors for results readout. Due to its remarkable advantages, such as high throughput, high efficiency, and excellent capability of mimicking in vivo microenvironment, organ-on-a-chip has shown great potential in pathology studies and screening applications.1 Although applications are appealing, the corresponding fabrication techniques are left behind, especially methods that enable integrated organs-on-chips are lacking.
Thus far, various methods, e.g., photolithography and soft lithography, have been adopted to fabricate different organs-on-chips, such as breathing lung-on-a-chip, liver-on-a-chip, and tumor-on-a-chip.2–6 Howbeit, these methods are not satisfying as they require multi-step lithographic processes, masks, and dedicated equipment, which make the fabrication expensive and time consuming. Moreover, these methods are only capable of fabricating microfluidic chip itself, while the other elements (i.e., microtissues, stimulus loading components, and results-readout sensors) require further processes. Recently, bioprinting has been used for organ-on-a-chip fabrication. As a straightforward method, it is based on layer-by-layer printing and thus capable of printing various materials to build 3D complex constructs. Besides, it enables the rapid prototyping and customized design, and is thus considered as one of the most promising candidate for organ-on-a-chip fabrication.
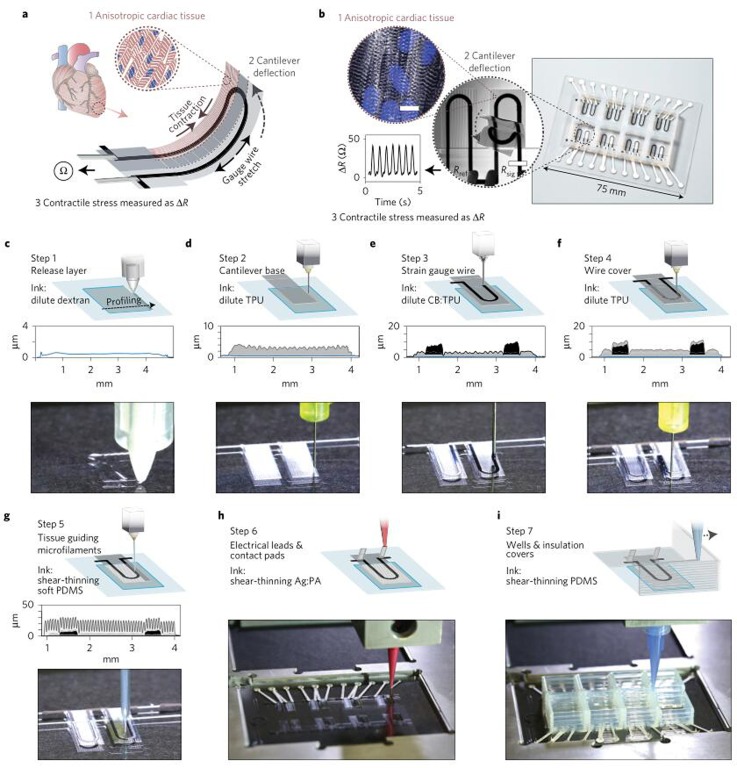
As a good example, Lewis and Parker's research group from Harvard University recently reported in Nature Materials the design and fabrication of a cardiac organ-on-a-chip by bioprinting.7 Using a customized bioprinter, the authors sequentially printed multiple functional materials in a programmable manner. The printed parts were integrated into a comprehensive chip for the purpose of testing contractile force and studying drug response of cardiac tissues (Fig. 1(a)). The main part of this chip was an array of wells performing as cell incubators that were fenced with polydimethylsiloxane (PDMS) walls. Inside each well, a layer of flexible filament with TPU-CB-TPU (TPU-thermoplastic polyurethane, CB-carbon black nanoparticles mixed with TPU) sandwich structure was printed (Fig. 1(b)). The embedded CB line whose electrical conductivity is affected by the strain can be used to measure the contractile forces of cells that were cultured on the cantilever. To ease the readout of results, the CB wire was linked outside by a printed conductive Ag line. The entire device was printed by multiple nozzles in a single continuous procedure (Figs. 1(c)–1(i)). Such a cardiac organ-on-a-chip enabled the real time monitoring of cellular behaviors in vitro over a long time. This chip was then employed to study the effect of two model drugs (verapamil and isoproterenol) on the beating of cardiac microtissues, where the beating frequency and strength were observed and recorded directly with this chip.
FIG. 1.
The fabrication process of cardiac organ-on-a-chip by bioprinting. (a) Working mechanism of the chip. The contraction of cardiac tissues would cause the cantilever deformation and thus the gauge wire stretch. Eventually, the contractile stress can be detected by measuring the electrical resistant. (b) The printed chip together with the obtained results. (c)–(i) Detailed printing procedures for chip fabrication. Reprinted with permission from Lind et al., Nat. Mater. 16, 303–308 (2017). Copyright 2016 Nature Publishing Group.
The success of using bioprinting as a tool to fabricate the integrated cardiac organ-on-a-chip is inspiring for biomedical engineers and other researchers. The main obstacle for organ-on-a-chip fabrication is that it is more than constructing a chip but also creating a proper microenvironment for cells. With the improvement in printing technique and also the advances of biocompatible materials, bioprinting makes it a reality. For instance, Lewis and her colleagues printed grooved microstructures as small as tens of microns, with the purpose of engineering and functionalizing the cardiac tissues. Moreover, the sensors that characterize the microtissues' behavior were integrated on this chip that eases the results readout. The success of this chip may inspire more researchers to devote to the field of organ-on-a-chip which could mimic the complexity of human organs and interactions between different organs. These chips hold great potential in high-throughput drug screening and thus have drawn attention from pharmacy companies. Lewis et al. integrated eight-wells in this chip, yet more wells could be realized with ease in the future to improve the efficiency of drug screening.7
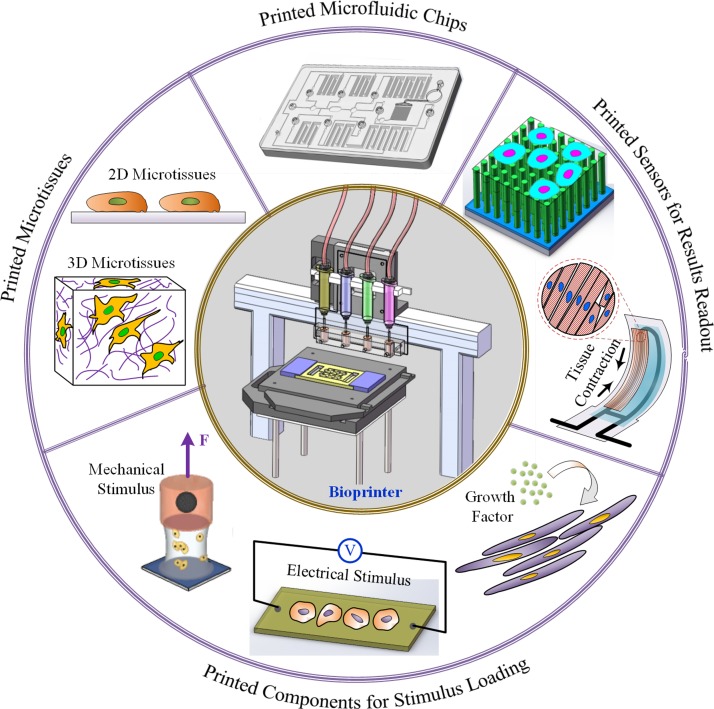
For an integrated organ-on-a-chip, four key elements are involved during the fabrication process, i.e., a microfluidic chip; 2D/3D microtissues that are cultured in the chip; components for stimulus loading; and sensors for monitoring the physiological behavior of microtissues and for results readout (see Fig. 2). In Lewis and Parker's method, a microfluidic chip, microscopic and macroscopic structures for maturing tissues, and result readout sensors are integrated onto the chip by bioprinting. As to the microtissues, they were formed on the chip after it was removed from the printer, through self-assembly guided by printed microstructures. By using the approach, Lewis et al. were able to overcome limitations relating to transport and off-the-shelf use of the chips. However, for thicker and more complex tissue models that require positioning of multiple cell types, it would be convenient to perform chip sterilization and cell seeding on the same printer.
FIG. 2.
Bioprinting the key elements of integrated organ-on-a-chip. Besides a fine bioprinter, four key elements are involved during the fabrication process, i.e., microfluidic chips; 2D/3D microtissues that are cultured in the chip; components for stimulus loading, the stimulus could be biophysical (e.g., electrical, mechanical, and thermal) or biochemical (e.g., growth factor) ones with the purpose to mature and functionalize the microtissues; and sensors for monitoring the physiological behavior of microtissues and for results readout.
The bioprinted organ-on-a-chip makes a significant step towards the fabrication of practical organ-on-a-chip; nonetheless, lots of work are still needed to advance the chip up to the commercial level. We would consequently discuss the state of art and future trends of 3D bioprinted organs-on-chips. For the first element of organs-on-chips (i.e., microfluidic chips), 3D bioprinting has been widely adopted for the fabrication due to its merits such as versatility and assembly-free.8 Thus far, numerous 3D printing strategies have been developed, such as micro-extrusion, inkjet, and laser-assistant printing.9 Among them, micro-extrusion printing is commonly used for microfluidic chips fabrication.6,10 Besides, stereolithography (SLA) and fused deposition modeling (FDM) also have great potential for fabricating microfluidic chips. Currently, some printers that enable microfluidic chips are even commercially available, such as Fluidic Factory 3D Printer from Dolomite. For the 3D printed microfluidic chips, it would be favorable if the materials are transparent, which could ease the observation and optical measurement.
The second element of organ-on-a-chip is the microtissues that are cultured in the microfluidic chips, which could be 2D or 3D. For microtissues bioprinting, usually, cells are encapsulated with some biocompatible materials (e.g., hydrogels) in order to prevent the mechanical damage. As to the printing strategy, micro-extrusion and inkjet bioprinting are capable of printing microtissues and they have already been widely used for various tissues fabrication. As to SLA and FDM, they are powerful in printing microfluidic chips, which has been mentioned above; however, they may encounter some challenges in cell printing. Perhaps, one solution is combining them together with other printing strategies. For instance, one could use FDM for the microfluidic chips and micro-extrusion (or inkjet) bioprinting for the microtissues. To the best of our knowledge, howbeit, such a hybrid technology has not been reported yet. So far, 2D microtissues are commonly used in organ-on-a-chip. Compared with the 2D case, the 3D microtissues can better mimic the in vitro situation, and of course is more complicated. For 3D microtissues, how to print the extracellular matrix (ECM) and microfluidic channels mimicking perfused vasculature is still a challenge. Recently, fabrication of microfluidic chips and microtissues by bioprinting has received much attention. For more information, one can refer to the review paper by Cho et al.9
Another key element of organ-on-a-chip is stimulus loading component to make the microtissues functional mimic specific functions of live organs. Usually, the development of live microtissues necessitates proper microenvironment involving certain stimulus. The stimulus could be biochemical (e.g., growth factor) or biophysical (e.g., electrical, mechanical, and thermal) ones with the purpose to mature and functionalize the microtissues.11 For the biochemical stimulus, growth factors are commonly printed together with cell laden hydrogels. One advantage of 3D bioprinting is that it enables the precision positioning of multiple materials (including cells, hydrogels, and growth factors) in a sequential order with a high spatial resolution. As to the mechanical stimulus, one trend is printing actuators on chips and applying stimulus (e.g., electrical, mechanical, and thermal) on cells after printing to make them mature and functional. It has been known that mechanical stimulus can significantly affect the cells' (e.g., cardiomyocyte) alignment and thus the cells proliferation, spreading. Li et al. have mixed magnetic particles with cell laden hydrogels, and thus a noncontact force can be imposed upon the cells by applying a magnetic field. An alternative is to print actuators with smart materials that are capable to change their shapes by tuning the environment conditions (termed as 4D bioprinting).12 Such actuators could be used to impose extern stimulus (e.g., mechanical forces) to the microtissues on organ-on-a-chip.
Last but not least, an integrated organ-on-a-chip shall comprise of sensors to monitor the physiological behavior of microtissues and to read out results. For instance, Lewis et al. embedded an electronic sensor on the chip to detect the contractile forces of cells that were cultured in the chip. An alternative is to fabricate the chips with transparent materials and then observe the cells' behavior by optical microscopy. For instance, Chen's group has used micro-post assays to measure the contraction force of microtissues via observing the deflection of pillars.13 A more common method is to stain the microtissues and then observe the physiological behavior with/without disassembling the chips. Compared with these methods, the merits of an embedded sensor are that the results are facility free and immediately available. Despite the significant advantages, embedded sensors on organ-on-a-chip are still rare as the fabrication is challenging for bioprinting. Perhaps the techniques for printing Micro-Electro-Mechanical System (MEMS) could be helpful in solving the embedded sensors fabrication. One issue that should be paid attention is the biocompatibility as some materials for electronic sensors are toxic to living cells.
In conclusion, bioprinted organ-on-a-chip is the trend of practical organ-on-a-chip and has opened a new era for biomedical engineering. For organ-on-a-chip, the first two elements (i.e., microfluidic chips and microtissues) have received broad attention and much work has been done. However, for the other two elements (i.e., components for stimulus loading and sensors for results read out), they are still in the early stage and more input is needed. Besides, the storage and transportation of organ-on-a-chip are also challenging, which however are necessary for their off-the-shelf use. One possible solution is vitrifying the chips rapidly for storage and then defrosting them before using, as inspired by the cryopreservation of oocytes in clinics.14
Acknowledgments
We acknowledge the financial support from the National Natural Science Foundation of China (Grant Nos. 51605377, 11522219, 11372243, and 51375371). Q.Y. also acknowledges the partial support from National Engineering Laboratory for Highway Maintenance Equipment (Chang'an University) (Grant No. 310825161103).
References
- 1. Reardon S., Nature 523, 266 (2015). 10.1038/523266a [DOI] [PubMed] [Google Scholar]
- 2. Huh D., Matthews B. D., Mammoto A., Montoya-Zavala M., Hsin H. Y., and Ingber D. E., Science 328, 1662 (2010). 10.1126/science.1188302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Torisawa Y.-S., Spina C. S., Mammoto T., Mammoto A., Weaver J. C., Tat T., Collins J. J., and Ingber D. E., Nat. Methods 11, 663 (2014). 10.1038/nmeth.2938 [DOI] [PubMed] [Google Scholar]
- 4. Zhang B., Montgomery M., Chamberlain M. D., Ogawa S., Korolj A., Pahnke A., Wells L. A., Massé S., Kim J., and Reis L., Nat. Mater. 15, 669 (2016). 10.1038/nmat4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fan Y., Nguyen D. T., Akay Y., Xu F., and Akay M., Sci. Rep. 6, 25062 (2016). 10.1038/srep25062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee H. and Cho D.-W., Lab Chip 16, 2618 (2016). 10.1039/C6LC00450D [DOI] [PubMed] [Google Scholar]
- 7. Lind J. U., Busbee T. A., Valentine A. D., Pasqualini F. S., Yuan H., Yadid M., Park S.-J., Kotikian A., Nesmith A. P., Campbell P. H., Vlassak J. J., Lewis J. A., and Parker K. K., Nat. Mater. 16, 303 (2017). 10.1038/nmat4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dixon C., Lamanna J., and Wheeler A. R., Adv. Funct. Mater. 27, 1604824 (2017). 10.1002/adfm.201604824 [DOI] [Google Scholar]
- 9. Yi H.-G., Lee H., and Cho D.-W., Bioengineering 4, 10 (2017). 10.3390/bioengineering4010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Snyder J., Hamid Q., Wang C., Chang R., Emami K., Wu H., and Sun W., Biofabrication 3, 034112 (2011). 10.1088/1758-5082/3/3/034112 [DOI] [PubMed] [Google Scholar]
- 11. Li Y., Poon C. T., Li M., Lu T. J., Pingguan-Murphy B., and Xu F., Adv. Funct. Mater. 25, 5999 (2015). 10.1002/adfm.201502018 [DOI] [Google Scholar]
- 12. Gao B., Yang Q., Zhao X., Jin G., Ma Y., and Xu F., Trends Biotechnol. 34, 746 (2016). 10.1016/j.tibtech.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 13. Legant W. R., Pathak A., Yang M. T., Deshpande V. S., McMeeking R. M., and Chen C. S., Proc. Natl. Acad. Sci. 106, 10097 (2009). 10.1073/pnas.0900174106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi M., Ling K., Yong K. W., Li Y., Feng S., Zhang X., Pingguan-Murphy B., Lu T. J., and Xu F., Sci. Rep. 5, 17928 (2015). 10.1038/srep17928 [DOI] [PMC free article] [PubMed] [Google Scholar]