Abstract
The Fatty acids (FAs) metabolism is suggested to play a pivotal role in the development of lung cancer, we explored that by conducting pathway-based analysis. We performed a meta-analysis of published datasets of six genome wide association studies (GWASs) from the Transdisciplinary Research in Cancer of the Lung (TRICL) consortium, which included 12,160 cases with lung cancer and 16,838 cancer-free controls. A total of 30,722 single-nucleotide polymorphisms (SNPs) from 317 genes relevant to FA metabolic pathways were identified. An additional dataset from the Harvard Lung Cancer Study with 984 cases and 970 healthy controls was also added to the final meta-analysis. In the initial meta-analysis, 26 of 28 SNPs that passed false discovery rate multiple tests were mapped to the CYP4F3 gene. Among the 26 top ranked hits, was a proxy SNP, CYP4F3 rs4646904 (P = 8.65×10−6, FDR = 0.018), which is suggested to change splicing pattern/efficiency and to be associated with gene expression levels. However, after adding data of rs4646904 from the Harvard GWAS, the significance in combined analysis was reduced to P=3.52×10−3 [odds ratio (OR)=1.07, 95% confidence interval (95%CI)=1.03-1.12]. Interestingly, the small Harvard dataset also pointed to the same direction of the association in subgroups of smokers (OR = 1.07) and contributed to a combined OR of 1.13 (95%CI = 1.06-1.20, P=6.70×10−5). The results suggest that a potentially functional SNP in CYP4F3 (rs4646904) may contribute to the etiology of lung cancer, especially in smokers. Additional mechanistic studies are warranted to unravel the potential biological significance of the finding.
Keywords: lung cancer, fatty acid metabolism, tumor markers, genome wide association study
1. Introduction
Lung cancer is the leading cause of cancer deaths in the United States, with an estimated 224,210 new cancer cases and 159,260 deaths in 2014 [1]. Surgery is more effective for the early-stage lung cancer without metastasis, when the tumor can be completely removed. Unfortunately, the disease is typically asymptomatic, until it is in the late stages, and the best chance for surgical treatment is usually lost. Consequently, the mortality is high: the five-year survival rate for lung cancer is only 4.0% for patients with distant tumors, compared with 54.0% for patients with localized tumors [2]. Chest radiograph is a common approach for detecting lung cancer, but the annual screening with chest radiograph has not reduced lung cancer mortality, compared with usual care [3]. Although low-dose computed tomography (CT) screening is a promising method for detecting lung cancer in individuals at high risk [4, 5], the treatment is expensive, and it exposes patients to some extra doses of radiation. Besides, the low-dose CT screening has been associated with a high rate of false positive results, with ~95% of benign lung nodules that are challenging for radiologists to identify [4]. Therefore, there is still a demand for new less-invasive but more efficient biomarkers in the risk assessment and early detection of lung cancer.
Cancer cells often share the attributes of metabolic abnormalities, such as perturbation in the energy metabolism of glucose, glutamine and lipids [6, 7]. Fatty acids (FAs), an important subgroup of lipids, have emerged as a recent focus of cancer research. FAs are synthesized de novo to continually provide lipids for energy production, cell membrane regeneration and lipid modification of proteins to meet the excessive bioenergetics and structural demands of highly proliferating cancer cells. Additionally, FAs and their derivatives are also important signaling molecules that may affect many fundamental cell processes, including cellular survival, proliferation, migration, angiogenesis and therapy resistance [8]. Furthermore, the metabolites of FAs could augment survival [9–11] and enhance adhesion [12] in lung cancer cells, and thus promote progression and metastasis and induce angiogenesis in human lung cancer [13–16].
More recently, increasing evidence suggests that the FAs are potential biomarkers for monitoring development and progression of lung cancer [17–20]. For instance, in serum samples from 55 patients with lung cancer and 165 similar pulmonary patients without known cancer, Liu et al. found that free FAs and their metabolites demonstrated good sensitivity and specificity for the identification of adenocarcinoma of the lung [17]. Similarly, other studies demonstrated that FAs from erythrocyte total lipids might be used as diagnostic biomarkers of lung adenocarcinoma, squamous cell carcinoma, and small cell lung cancer [18, 19]. Most recently, Zhang et al. revealed that serum unsaturated free FAs could be used as potential biomarkers for early detection and disease progression of lung cancer [20]. Consequently, we hypothesized that genetic variants in genes involved in the FA synthesis, degradation, metabolism and transport were potential susceptibility factors for lung cancer.
The aim of the present study was to assess the associations between single-nucleotide polymorphisms (SNPs) in the FAs metabolic pathways and risk of lung cancer. We first conducted a meta-analysis of six lung cancer genome wide association studies (GWASs) within the Transdisciplinary Research in Cancer of the Lung (TRICL) consortium [21–23], and we added additional data from another GWAS from Harvard University [24] for the identified significant SNPs that were corrected by the false discovery rate (FDR) for multiple testing correction and with potential functions in bioinformatics analyses [25, 26].
2. Methods and Materials
2.1 Study populations and genotyping
The detailed information about the study participants is presented in Supplemental Table 1, and all the participants were of European descendent. A written informed consent was obtained from each participant in the GWASs, and the present study followed the study protocols approved by the institutional review board for each of the participating institutions.
2.2 Initial meta-analysis
The present study started from the combined genotyping and imputation dataset of six previously published GWASs of lung cancer from the TRICL consortium. Within the TRICL consortium [21], 12,160 lung cancer cases and 16,838 controls from Europe and North America participated in the GWASs[22]. All the cases and controls were at least frequency-matched on age and sex. The participants were described in previous publications: The University of Texas MD Anderson Cancer Center (MDACC) GWAS [27], the Institute of Cancer Research (ICR) GWAS [28], the National Cancer Institute (NCI) GWAS [21], the International Agency for Research on Cancer (IARC) GWAS [27], Samuel Lunenfeld Research Institute study (SLRI) GWAS [22], Toronto, and The Helmholtz-Gemeinschaft Deutscher Forschungszentren Lung Cancer GWAS, Germany (GLC) [22].
For the TRICL GWASs, the overwhelming majority of patients had non-small cell lung cancer, leaving only a few with small cell lung cancer. Genotyping data were from combined datasets of different platforms of Illumina HumanHap 317, 317+240S, 370Duo, 550, 610 or 1M arrays [21]. These datasets were imputed for all original scans for over 10 million SNPs using the 1000 Genomes Project (phase I integrated release 3, March 2012) as the reference by using IMPUTE2 v2.1.1, MaCH v1.0 or minimac (version 2012.10.3) software. The quality control process was detailed in previously published reports [21, 22].
2.3 Additional data for meta-analysis
An additional independent GWAS dataset of a Caucasian population was provided by the Harvard Lung Cancer Susceptibility Study [24]. For the Harvard GWAS, for which details of participant recruitment have been described previously [24], genotyping data was derived from 1000 cases and 1000 controls using Illumina Humanhap610-Quad arrays. Cases were patients aged >18 years, with newly diagnosed, histologically confirmed primary non-small cell lung cancer. Controls were healthy non-blood-related family members and friends of patients with cancer or with cardiothoracic conditions undergoing surgery.
Unqualified samples were excluded, if they had (i) overall genotype completion rates <95%; (ii) gender discrepancies; (iii) unexpected duplicates or probable relatives (based on pairwise identity by state value, PI_HAT in PLINK>0.185); (iv) heterozygosity rates >6 times the standard deviation from the mean; or (v) individuals evaluated to be of non-Caucasians [using the HapMap release 23 including Japanese in Tokyo, Japan (JPT), Han Chinese in Bejing, China (CHB), Utah Residents (CEPH) with Northern and Western Ancestry (CEU) and Yoruba in Ibadan, Nigeria (YRI) populations as a reference]. Unqualified SNPs were excluded, when they (i) were not mapped on autosomes; (ii) had a call rate <95% in all GWAS samples; (iii) had minor allele frequency (MAF) <0.01; or (iv) had a genotype distribution deviated from those expected by Hardy-Weinberg equilibrium (P < 1.0×10−6). After applying pre-specified quality control, the Harvard genotype data were available for 984 cases and 970 controls.
2.4 Gene and variant selection
To identify relevant genes of interest, we used two electronic databases, Gene Cards [29] and MSigDB [30] to search for FA metabolism-related genes. Specifically, keyword searches using “fatty acid” in pathway & interaction section and “cancer” in disorder section in the Gene Cards and one heading “fatty acid” in the MSigDB. As a result, 317 genes located on autosomal chromosomes were identified from the FA biosynthesis, metabolism and degration pathways (Supplementary Table 2 and Supplementary Figure 1). Genotyped, imputed common SNPs (minor allele frequency ≥ 0.05) within these genes or their ± 2kb flanking regions were selected for association analysis. Hence, 30,722 SNPs in the FA metabolic pathways had been extracted from the TRICL GWASs and used for further analysis.
2.5 Statistics
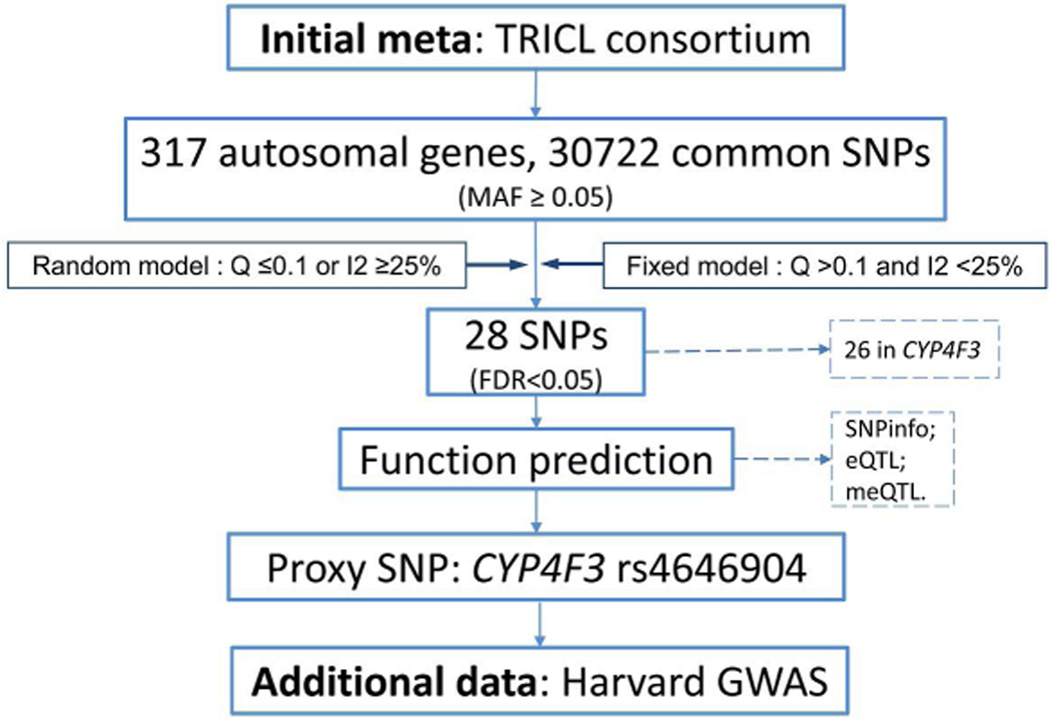
Meta-analysis was first performed among the six GWASs from the TRICL consortium. Briefly, the association between each SNP and lung cancer risk was assessed by unconditional logistic regression using an additive genetic model of the risk allele. The Cohran’s Q statistic to test for heterogeneity and the I2 statistic to quantify the proportion of the total variation due to heterogeneity were calculated [31]. Fixed-effects models were applied, when there was no heterogeneity among the datasets (based on the criteria P > 0.10 and I2 < 25%); otherwise, random-effects models were applied [32]. The Benjamini and Hochberg’s false discovery rate (FDR) method was used for correction of the multiple comparisons[33]. Associations were considered significant, if an FDR value was less than 0.05. For the top-hit variants of interest, we then focused on those SNPs with potential functional as predicted by the online prediction tools: SNPinfo [25] and pfSNP [34]. Linear regression analysis was used to test expression quantitative trait loci (eQTL) associations with data obtained from the RNA sequencing project of the 1000 Genomes Project samples [conducted by Genetic European Variation in Health and Disease Consortium (GEUVADIS) [35]. Gene expression values that were more than three standard deviations from the mean were considered as outliers[36]. Finally, we tested the association between predicted functional SNP and lung susceptibility in the Harvard GWAS [24], and subgroup analyses stratified by histology types (squamous and adenocarcinoma) and smoking status (smoker and non-smoker) were also performed. LocusZoom was used to produce regional association plots [37]. All statistical analyses were carried out by SAS software (version 9.1.3; SAS Institute, Cary, NC, USA) or R (2.6.0) unless specified otherwise. The analysis flow chart is present in Figure 1.
Figure 1.
Analysis flow chart of the present study.
3. Results
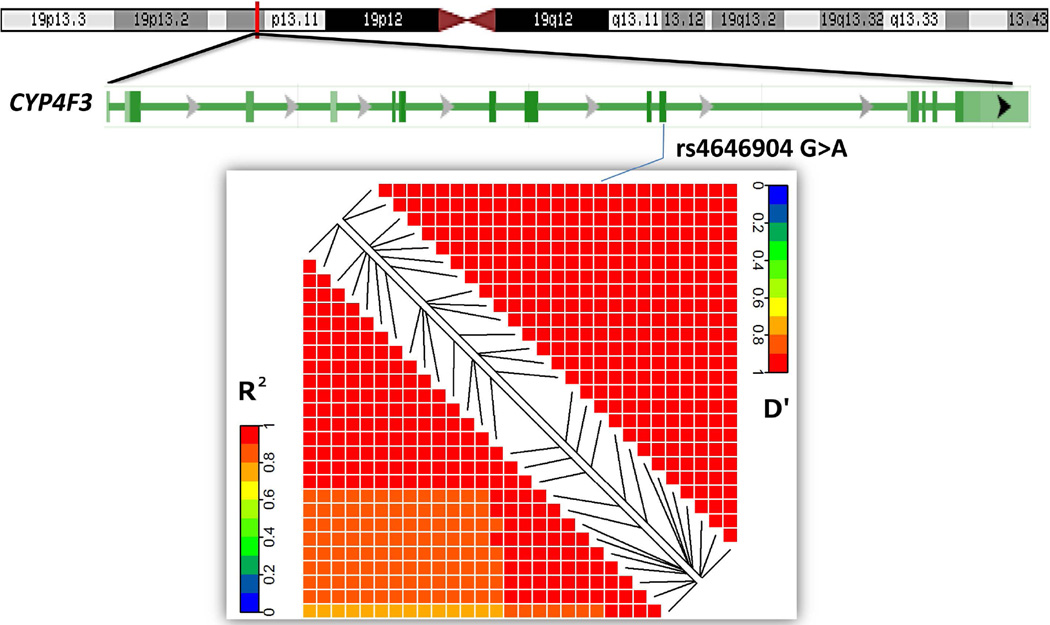
The six GWASs from the TRICL consortium consisted of 12,160 cases and 16,838 controls of European ancestry (Supplementary Table 1). The associations between SNPs of genes involved in the FA metabolic pathways and lung cancer risk in the TRICL consortium are shown in Supplementary Figure 2 (the Manhattan plot). Among 30,722 SNPs, 1,609 were nominally associated with lung cancer susceptibility at P < 0.05. Of these, as detailed in Table 1, 28 SNPs in three genes reached the preset statistical thresholds (FDR < 0.05). Among these 28 SNPs, two SNPs in TNF (rs1800628 G>A) and GPX5 (rs116260720 C>A) were located in the previously identified and reported lung cancer susceptibility major histocompatibility complex (MHC) region; therefore, they were not further pursued in further analyses. The other 26 SNPs were mapped within the CYP4F3 gene region on chromosome 19. Supplementary Figure 3 shows the regional association plots of CYP4F3. All the top-hits (FDR < 0.05 and P < 10−4) in the CYP4F3 region were in high linkage disequilibrium (LD, r2 > 0.8, Figure 2) based on the hg19/1000 Genome European populations.
Table 1.
Associations between SNPs in the fatty acid pathways and NSCLC risk with FDR-P<0.05
| SNP | Gene | Chr | Position | Allele* | TRICL |
Harvard |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EAF | Q† | I2 | Effects ‡ | OR (95%CI) | P | FDR | EAF | OR (95%CI) | P | |||||
| rs1800628 | TNF | 6 | 31546850 | G/A | 0.11 | 0.541 | 0 | ++++++ | 1.18 (1.11–1.24) | 5.46×10−9 | 0.0002 | 0.09 | 1.14 (0.99–1.32) | 0.071 |
| rs4646521 | CYP4F3 | 19 | 15769872 | G/T | 0.35 | 0.672 | 0 | ++++++ | 1.09 (1.05–1.13) | 3.83×10−6 | 0.018 | 0.37 | 0.99 (0.91–1.07) | 0.774 |
| rs10405638 | CYP4F3 | 19 | 15764870 | C/T | 0.36 | 0.478 | 0 | ++++-+ | 1.09 (1.05–1.12) | 7.59×10−6 | 0.018 | 0.38 | 0.99 (0.92–1.08) | 0.888 |
| rs4646904§ | CYP4F3 | 19 | 15763721 | G/A | 0.36 | 0.488 | 0 | ++++-+ | 1.09 (1.05–1.13) | 8.65×10−6 | 0.018 | 0.37 | 0.99 (0.92–1.08) | 0.884 |
| rs2102959 | CYP4F3 | 19 | 15767392 | A/G | 0.36 | 0.573 | 0 | ++++-+ | 1.08 (1.05–1.12) | 1.07×10−5 | 0.018 | 0.38 | 0.99 (0.91–1.07) | 0.779 |
| rs10401516 | CYP4F3 | 19 | 15760577 | C/T | 0.36 | 0.552 | 0 | ++++-+ | 1.08 (1.05–1.12) | 1.17×10−5 | 0.018 | 0.38 | 0.99 (0.92–1.08) | 0.895 |
| rs4646514 | CYP4F3 | 19 | 15769270 | A/C | 0.36 | 0.562 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.27×10−5 | 0.018 | 0.38 | 0.98 (0.90–1.06) | 0.633 |
| rs4646515 | CYP4F3 | 19 | 15769379 | G/C | 0.36 | 0.562 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.27×10−5 | 0.018 | 0.38 | 0.98 (0.90–1.06) | 0.615 |
| rs718258 | CYP4F3 | 19 | 15766419 | G/A | 0.36 | 0.564 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.28×10−5 | 0.018 | 0.38 | 0.99 (0.91–1.07) | 0.807 |
| rs4646520 | CYP4F3 | 19 | 15769775 | A/G | 0.36 | 0.558 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.33×10−5 | 0.018 | 0.38 | 0.98 (0.90–1.06) | 0.599 |
| rs10404163 | CYP4F3 | 19 | 15769805 | A/T | 0.36 | 0.558 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.33×10−5 | 0.018 | 0.38 | 0.98 (0.90–1.06) | 0.599 |
| rs4646513 | CYP4F3 | 19 | 15769253 | C/A | 0.36 | 0.561 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.34×10−5 | 0.018 | 0.38 | 0.98 (0.90–1.06) | 0.639 |
| rs2073600 | CYP4F3 | 19 | 15757643 | C/T | 0.36 | 0.475 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.51×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.983 |
| rs2072598 | CYP4F3 | 19 | 15757528 | A/G | 0.36 | 0.472 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.53×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.990 |
| rs2280750 | CYP4F3 | 19 | 15757428 | T/A | 0.36 | 0.473 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.54×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.992 |
| rs2283611 | CYP4F3 | 19 | 15759828 | A/G | 0.36 | 0.461 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.54×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.990 |
| rs2283609 | CYP4F3 | 19 | 15759764 | C/T | 0.36 | 0.462 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.54×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.990 |
| rs2077040 | CYP4F3 | 19 | 15755840 | T/C | 0.36 | 0.474 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.55×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.990 |
| rs2077080 | CYP4F3 | 19 | 15755392 | G/C | 0.36 | 0.474 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.55×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.990 |
| rs4808346 | CYP4F3 | 19 | 15758931 | T/C | 0.36 | 0.462 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.55×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.990 |
| rs8107976 | CYP4F3 | 19 | 15753683 | A/G | 0.36 | 0.481 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.55×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.988 |
| rs2203998 | CYP4F3 | 19 | 15754893 | A/G | 0.36 | 0.477 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.56×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.990 |
| rs8106799 | CYP4F3 | 19 | 15753759 | G/A | 0.36 | 0.48 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.56×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.990 |
| rs2204000 | CYP4F3 | 19 | 15755112 | G/A | 0.36 | 0.479 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.56×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.986 |
| rs2203999 | CYP4F3 | 19 | 15754975 | C/T | 0.36 | 0.479 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.56×10−5 | 0.018 | 0.37 | 1.00 (0.92–1.08) | 0.986 |
| rs4646512 | CYP4F3 | 19 | 15764220 | C/T | 0.36 | 0.454 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.58×10−5 | 0.018 | 0.38 | 1.00 (0.92–1.08) | 0.948 |
| rs116260720 | GPX5 | 6 | 28502794 | C/A | 0.22 | 0.335 | 12.5 | +++++- | 1.10 (1.05–1.14) | 1.65×10−5 | 0.018 | 0.21 | 1.11 (1.00–1.22) | 0.043 |
| rs4646516 | CYP4F3 | 19 | 15769449 | A/G | 0.36 | 0.568 | 0 | ++++-+ | 1.08 (1.04–1.12) | 1.67×10−5 | 0.018 | 0.38 | 0.98 (0.90–1.06) | 0.639 |
NSCLC: non-small cell lung cancer; EAF, effect allele frequency; FDR, false discovery rate; TRICL, Transdisciplinary Research in Cancer of the Lung;
reference allele / effect allele;
fixed effect models were used when no heterogeneity was found between studies (Q>0.10 and I2 <25.0%); otherwise, random effect models were used;
means positive association, and-means negative association;
online function prediction tool, SNPinfo [25], found that rs4646904 might influence splicing efficiency of CYP4F3;
SNPs in bold were putative functional and were not located in the major histocompatibility complex region.
Figure 2.
Gene structure of the CYP4F3 gene and linkage disequilibrium plot of 26 SNPs mapped to CYP4F3 and passed false discovery rate multiple tests.
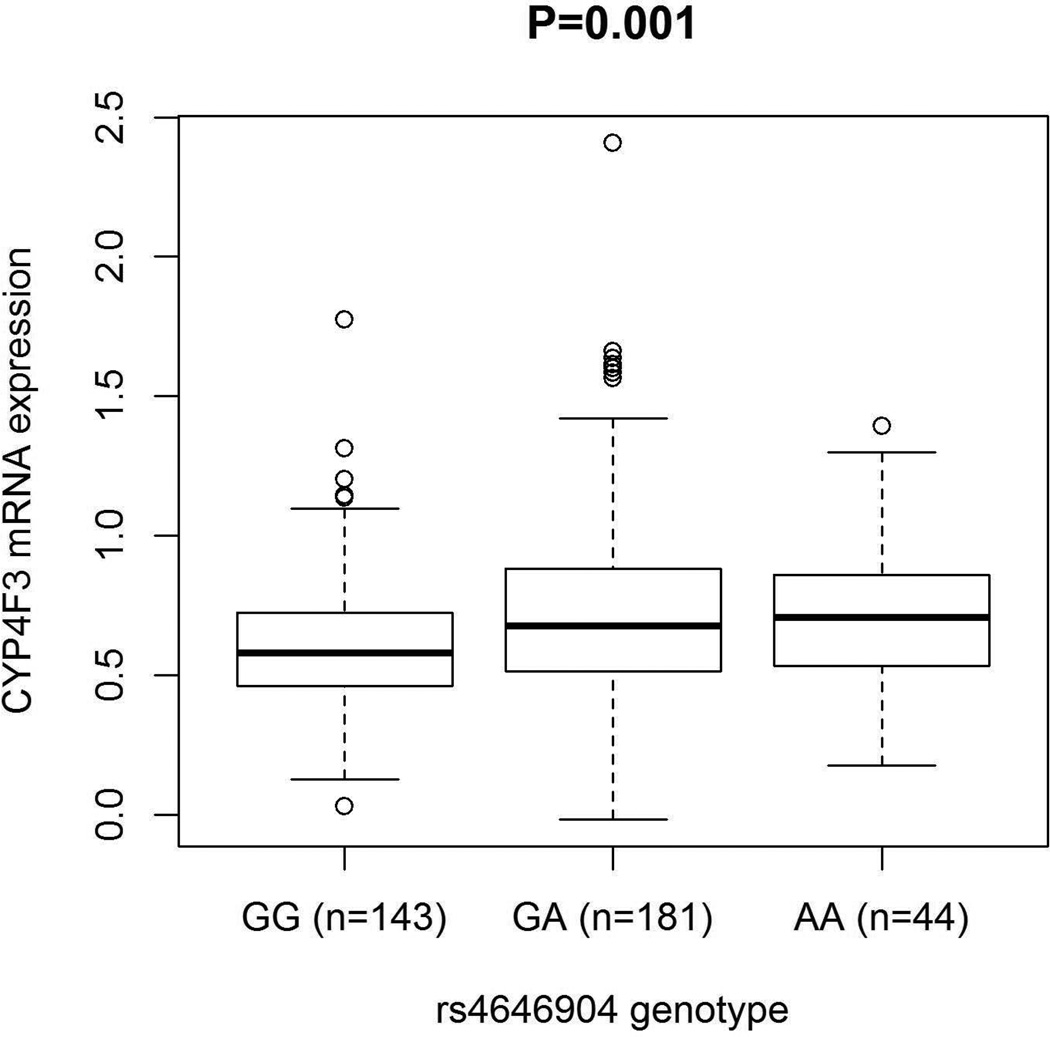
Using the online tool SNPinfo and pfSNP, the CYP4F3 rs4646904 G>A SNP was putatively functional, because it is predicted to influence exonic splicing efficiency of CYP4F3. We then performed an eQTL analysis to evaluate the mRNA expression levels of CYP4F3 by the genotypes of rs4646904. The GEUVADIS RNA sequencing of the 1000 Genomes Project has only a combined normalized transcriptome and genome sequencing data by performing mRNA and small RNA sequencing on 465 lymphoblastoid cell lines derived from five populations: CEU, Finns (FIN), British (GBR), Toscani (TSI) and Yoruba (YRI). To keep populations consistent and comparable with the present study, we only used data of 373 samples from European descendants (i.e., TSI, GBR, FIN and CEU). Besides, six values for CYP4F3 mRNA expression levels were considered an outlier and removed (Supplementary Figure 4). Gene expression differences among genotypes were examined using a regression model in which the association between gene expression and genotypes was considered additive, assuming that a trend by the number of variant alleles exists. As shown in Figure 3, the number of the variant A allele was shown to be associated with higher expression levels of CYP4F3 (Padditive = 0.001), compared with the common G allele.
Figure 3.
Expression quantitative trait loci (eQTL) association of CYP4F3 rs4646904. The eQTL analyses were preforemed in addtive model. We used RNA-seq data from 368 non-hispanic European individuals, which are part of the 1000 Genome Project.
Therefore, we chose rs4646904 as the tagSNP of the CYP4F3 region. In the meta-analysis of the TRICL consortium, there was no heterogeneity observed among the six GWASs, with I2 of 0 and the Q-test P value of 0.488. In combined analysis, we found that the per-unit increase of the variant A allele was associated with 1.09-fold increased risk of lung cancer [95% confidence interval (95% CI) = 1.05-1.13, P = 8.65×10−6, Table 1].
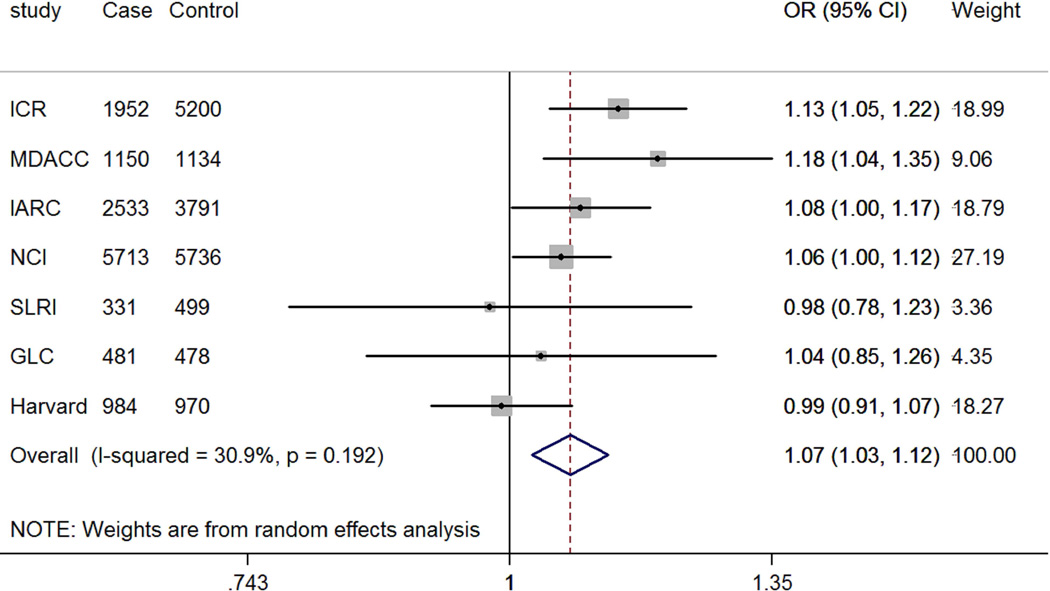
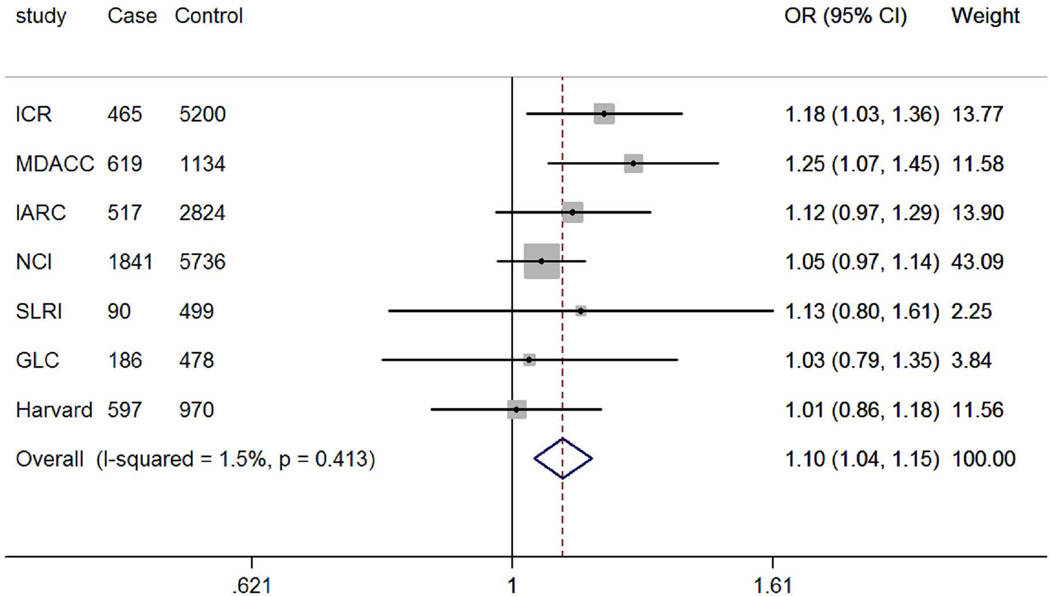
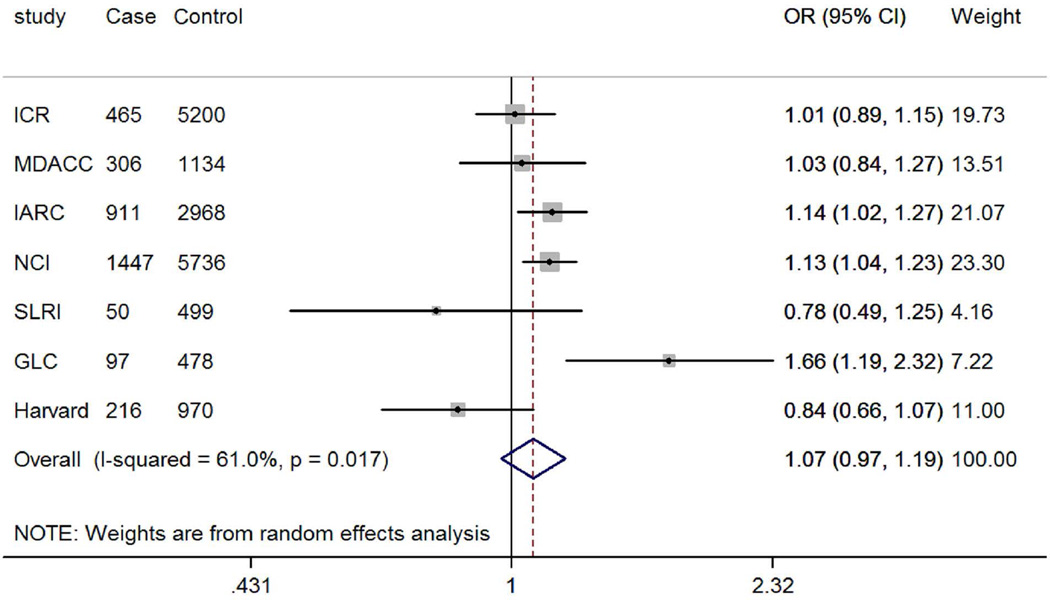
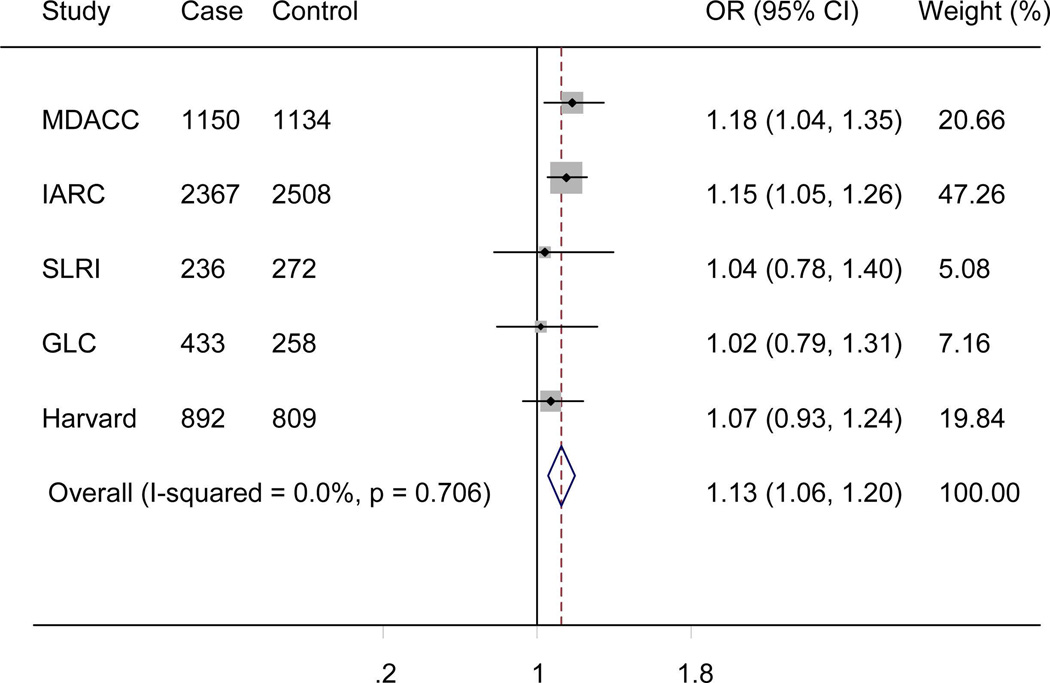
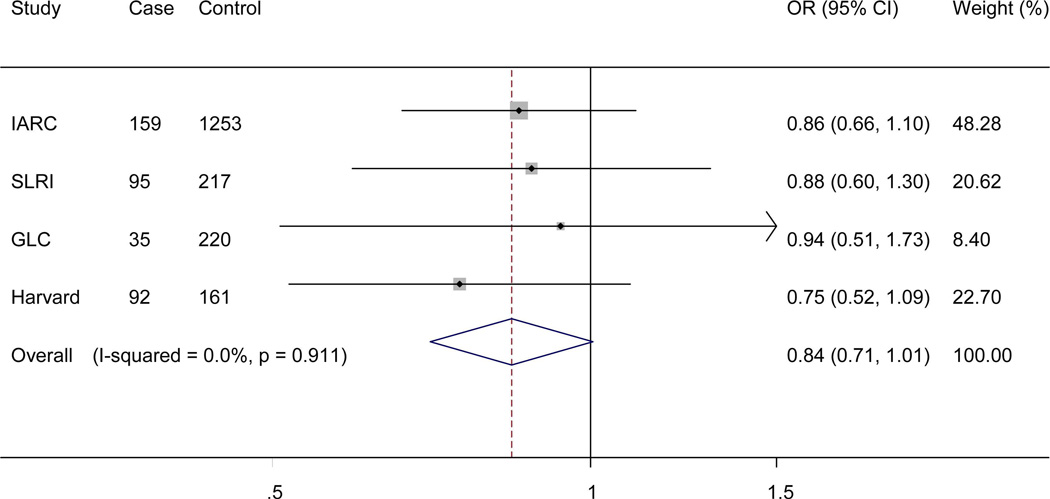
The genotyping data of the associated rs4646904 SNP was available in the Harvard GWAS (984 patients and 970 controls, minor allele frequency = 0.37, Table 1). When this dataset was added to the TRICL datasets, the summary effect estimate obtained from the expanded meta-analysis was 1.07 (95% CI =1.03-1.12, P = 3.52×10−3). For this analysis, we used in a random-effects model, because the effect estimate displayed a moderate degree of heterogeneity with I2 = 30.9% and the Q test P = 0.192 (Figure 4). When stratified by lung tumor types, the combined effect was 1.10 (95% CI = 1.04-1.15, P = 8.55×10−4I2 = 1.5%, Q-test P = 0.413) in a fixed-effects model (with a smaller I2 value) for adenocarcinomas and 1.07 (95% CI = 0.97-1.19, P = 0.212, I2 = 61.0%, Q-test P = 0.017) for squamous carcinomas in a random-effects model (with a larger I2 value). We also examined the expected association of rs4646904 with risk of lung cancer in the 10,059 smokers (5078 cases and 4981 controls) from MDACC, IARC, SLRI, GLC and Harvard GWASs. Consistently, a significant effect of the rs4646904 A allele on lung cancer risk was observed in the combined meta-analysis (OR = 1.13, 95%CI= 1.06-1.20, P = 6.7×10−5I2 = 0.0%, Q-test P = 0.706). However, among 2232 non-smokers (381 cases and 1851 controls), the result failed to reach significance (OR = 0.84, 95%CI= 0.71-1.01, P = 0.063, I2 = 0.0%, Q-test P = 0.911). Tests for heterogeneity indicated that the carcinogenesis effect of the rs4646904 A allele was predominantly limited to smokers (I2 = 89.7%, Q-test P = 0.002).
Figure 4.
Forest plot of A allele effect of CYP4F3 rs4646904 in all cases (Panel A), adenocarcinoma (Panel B), Squamous cell carcinoma (Panel C), smokers (Panel D), non-smokers (Panel E) from GWASs [the Institute of Cancer Research (ICR) GWAS, the MD Anderson Cancer Center (MDACC) GWAS, the International Agency for Research on Cancer (IARC) GWAS, the National Cancer Institute (NCI) GWAS, the Samuel Lunenfeld Research Institute study (SLRI) GWAS, German Lung Cancer Study (GLC) and Harvard lung cancer study (Harvard)].
3. Discussion
Deregulation of the FA metabolic pathways has been implicated in many cancers [38–40]. In the present study, we explored associations between genetic variants in the FA metabolic pathways and risk of lung cancer. We examined all typed and imputed SNPs of the genes involved in the FA metabolic pathways from six published GWASs of lung cancer within the TRICL consortium. We also tried to find additional support from the Harvard GWAS. In the meta-analyses of the TRICL GWASs, the predicted functional SNP, CYP4F3 rs4646904G > A, showed a significant association with lung cancer risk, but this association was not significant in the Harvard Lung Cancer Susceptibility Study; however, this SNP remained significant in the final combined analysis as well as in subgroups of squamous cancer and smokers.
The CYP4F3 gene contains 14 exons and 13 introns and encodes a member of the cytochrome P450 superfamily of enzymes. The CYP4F3 pre-messenger RNA is spliced into two mature transcripts (i.e., CYP4F3B and CYP4F3A). Human CYP4F3s are the main catalysts in the oxidation of FAs: they can ω-hydroxylate a variety of long chains and very long chains as well as saturated, unsaturated and branched chain FAs, vitamins with long alkyl side chains, and the physiologically important prostaglandins and hydroeicosatetraenoic acids[41]. Additionally, the ability of CYP4F3 to ω-hydroxylate both pro- and anti-inflammatory leukotrienes (i.e., LTB4) indicates that they may function in both the activation and resolution phases of the inflammatory response[42]. The CYP4F3 rs4646904G > A variant was a novel, potentially functional SNP that had the smallest P value in the present meta-analysis of the TRICL GWASs. Although CYP4F3 rs4646904G>A is a synonymous variant, it is located within an exon splicing enhancer (ESE)[25], which has discrete sequences within exons that promote both constitutive and regulated splicing[43]. Efficient splicing has limited tolerance of mutations in the ESEs, even if they have no effect on protein coding[44]. Additionally, ChIP-seq data indicate that rs4646904 is located in the enhancer region containing histone modification marks of H3k09me3, H3k27me3 and H3k09me3[45]. Cigarette smoking is the major cause of cancers of the respiratory tract, and CYP4F3 was found to be up-regulated in human airway epithelium in healthy current smokers, compared with those of never smokers [46–48], which suggests that CYP4F3 may be induced by smoking and thus contributes to carcinogenesis. The finding that the rs4646904 A variant allele was associated with significant higher gene expression levels than the G allele is consistent with the findings in other cancers, in which unregulated expression levels of CYP4F3s were found in pancreatic ductal adenocarcinoma, compared to benign lesions[49], as well as related with hepatocyte differentiation status [50] and progression of HCV-associated hepatocellular carcinoma [51].
Despite the underlying biological plausibility supporting this lung cancer-associated rs4646904 in the meta-analysis of the TRICL consortium with 12,160 cases and 16,838 controls, the relatively small Harvard GWAS dataset with 984 cases and 970 controls did not provide a further support for the association, but the combined meta-analysis revealed a slight effect with P < 0.05. Nevertheless, according to a PASS [52] power calculation, the Harvard GWAS had a mere power of 0.18 to detect an OR of 1.1. In contrast, the combined meta-analysis of the TRICL and Harvard GWASs with a total of 13,144 cases and 17,808 controls achieved a power of 0.98 for detecting an OR of 1.1, which suggest that rs4646904 may have a very small but genuine effect on lung cancer risk. Interestingly, the rs4646904-cancer risk association was significant in subgroups with squamous cancer and smokers but not adenocarcinoma and non-smokers, and there were high heterogeneity among the results of these subgroups, indicating that there might be an interaction between smoking and rs4646904 [53]. It may be because the exposure to cigarette smoke may negatively affect the synthesis of n−3 long-chain polyunsaturated fatty acid from the precursor in mammary gland cells [54]. Hence, rs4646904 might be biomarker for lung cancer risk in smokers.
It should be noted that there were limitations of the present study. First, none of the CYP4F3 variants was identified in previous TRICL GWASs as significant at a GWAS threshold (P < 10−7). Indeed, recent GWASs have identified at least 10 independent loci, and, as a result, only a small fraction of heritability could be explained by these SNPs. The challenge remains to identify the many additional common risk loci that are expected to have smaller genetic effects [55]. The pathway-based analysis, such as in the present study, reduces the significance level of P values based on the number SNPs examined in genes in the studied pathway. Therefore, such a pathway analysis with integration of association results with gene expression should be considered a complementary approach [56, 57] to the GWAS analyses. Second, we are aware that the observed effects may vary by body mass index or sex, but such data were not available from the TRICL consortium for us to further examine the risk modification. Thus, further analyses including body mass index and sex are warranted to improve our understanding of the apparent heterogeneity of effects. Third, because we only included non-Hispanic white populations, the generalizability to other ethnic populations needs further investigation.
In conclusion, this combined meta-analysis of six GWASs from the TRICL consortium and Harvard GWAS identified a potentially predictive functional marker (CYP4F3 rs4646904) for lung cancer risk in Caucasian populations, especially in smokers.
Supplementary Material
Acknowledgments
Jieyun Yin was sponsored by the China Scholarship Council for studying at Duke University. As Duke Cancer Institute members, QW and KO acknowledge support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: NIH CA014236). QW was also supported by a start-up fund from Duke Cancer Institute, Duke University Medical Center).
TRICL
This work was supported by the Transdisciplinary Research in Cancer of the Lung (TRICL) Study, U19-CA148127 on behalf of the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network. The Toronto study was supported by Canadian Cancer Society Research Institute(020214), Ontario Institute of Cancer and Cancer Care Ontario Chair Award to RH The ICR study was supported by Cancer Research UK (C1298/A8780 and C1298/A8362—Bobby Moore Fund for Cancer Research UK) and NCRN,HEAL and Sanofi-Aventis. Additional funding was obtained from NIH grants (5R01CA055769, 5R01CA127219, 5R01CA133996, and 5R01CA121197). The Liverpool Lung Project (LLP) was supported by The Roy Castle Lung Cancer Foundation, UK. The ICR and LLP studies made use of genotyping data from the Wellcome Trust Case Control Consortium 2 (WTCCC2); a full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Sample collection for the Heidelberg lung cancer study was in part supported by a grant (70–2919) from the Deutsche Krebshilfe. The work was additionally supported by a Helmholtz-DAAD fellowship (A/07/97379 to MNT) and by the NIH (U19CA148127). The KORA Surveys were financed by the GSF, which is funded by the German Federal Ministry of Education, Science, Research and Technology and the State of Bavaria. The Lung Cancer in the Young study (LUCY) was funded in part by the National Genome Research Network (NGFN), the DFG (BI576/2-1; BI 576/2-2), the Helmholtzgemeinschaft (HGF) and the Federal office for Radiation Protection (BfS: STSch4454). Genotyping was performed in the Genome Analysis Center (GAC) of the Helmholtz Zentrum Muenchen. Support for the Central Europe, HUNT2/Tromsø and CARET genome-wide studies was provided by Institut National du Cancer, France. Support for the HUNT2/Tromsø genome-wide study was also provided by the European Community (Integrated Project DNA repair, LSHG-CT- 2005–512113), the Norwegian Cancer Association and the Functional Genomics Programme of Research Council of Norway. Support for the Central Europe study, Czech Republic, was also provided by the European Regional Development Fund and the State Budget of the Czech Republic (RECAMO, CZ.1.05/2.1.00/03.0101). Support for the CARET genome-wide study was also provided by grants from the US National Cancer Institute, NIH (R01 CA111703 and UO1 CA63673), and by funds from the Fred Hutchinson Cancer Research Center. Additional funding for study coordination, genotyping of replication studies and statistical analysis was provided by the US National Cancer Institute (R01 CA092039). The lung cancer GWAS from Estonia was partly supported by a FP7 grant (REGPOT245536), by the Estonian Government (SF0180142s08), by EU RDF in the frame of Centre of Excellence in Genomics and Estoinian Research Infrastructure’s Roadmap and by University of Tartu (SP1GVARENG). The work reported in this paper was partly undertaken during the tenure of a Postdoctoral Fellowship from the IARC (for MNT). The Environment and Genetics in Lung Cancer Etiology (EAGLE), the Alpha-Tocopherol, Beta-Carotene Cancer Prevention Study (ATBC), and the Prostate, Lung, Colon, Ovary Screening Trial (PLCO) studies and the genotyping of ATBC, the Cancer Prevention Study II Nutrition Cohort (CPS-II) and part of PLCO were supported by the Intramural Research Program of NIH, NCI, Division of Cancer Epidemiology and Genetics. ATBC was also supported by US Public Health Service contracts (N01-CN-45165, N01-RC-45035 and N01-RC-37004) from the NCI. PLCO was also supported by individual contracts from the NCI to the University of Colorado Denver (NO1-CN-25514), Georgetown University (NO1-CN-25522), Pacific Health Research Institute (NO1-CN-25515),Henry Ford Health System (NO1-CN-25512), University of Minnesota(NO1-CN-25513), Washington University(NO1-CN-25516), University of Pittsburgh (NO1-CN-25511), University of Utah (NO1-CN-25524), Marshfield Clinic Research Foundation (NO1-CN-25518), University of Alabama at Birmingham (NO1-CN-75022, Westat, Inc. NO1-CN-25476), University of California, Los Angeles (NO1-CN-25404). The Cancer Prevention Study II Nutrition Cohort was supported by the American Cancer Society. The NIH Genes, Environment and Health Initiative (GEI) partly funded DNA extraction and statistical analyses (HG-06-033-NCI-01 andRO1HL091172-01), genotyping at the Johns Hopkins University Center for Inherited Disease Research (U01HG004438 and NIH HHSN268200782096C) and study coordination at the GENEVA Coordination Center (U01 HG004446) for EAGLE and part of PLCO studies. Funding for the MD Anderson Cancer Study was provided by NIH grants (P50 CA70907, R01CA121197, R01CA127219, U19 CA148127, R01 CA55769, K07CA160753) and CPRIT grant (RP100443). Genotyping services were provided by the Center for Inherited Disease Research (CIDR). CIDR is funded through a federal contract from the NIH to The Johns Hopkins University (HHSN268200782096C). The Harvard Lung Cancer Study was supported by the NIH (National Cancer Institute) grants CA092824, CA090578, and CA074386.
Abbreviations
- FA
Fatty acid
- GWAS
genome-wide association study
- TRICL
Transdisciplinary Research in Cancer of the Lung
- CYP4F3
cytochrome P450, family 4, subfamily F, polypeptide 3
- SNPs
single nucleotide polymorphisms
- FDR
false discovery rate
- OR
odds ratio
- CI
confident interval
- eQTL
expression quantitative trait loci
Footnotes
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA: a cancer journal for clinicians. 2014;64(1):9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Surveillance, Epidemiology, and End Results (SEER) Program ( www.seer.cancer.gov) SEER*Stat Database: Mortality - All COD, Aggregated With State, Total U.S. <Katrina/Rita Population Adjustment>, National Cancer Institute, DCCPS, Surveillance Research Program, Surveillance Systems Branch, released April 2013. 1969–2010 Underlying mortality data provided by NCHS ( www.cdc.gov/nchs)
- 3.Oken MM, Hocking WG, Kvale PA, et al. Screening by chest radiograph and lung cancer mortality: the Prostate, Lung, Colorectal, and Ovarian (PLCO) randomized trial. Jama. 2011;306(17):1865–1873. doi: 10.1001/jama.2011.1591. [DOI] [PubMed] [Google Scholar]
- 4.National Lung Screening Trial Research T. Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. The New England journal of medicine. 2011;365(5):395–409. doi: 10.1056/NEJMoa1102873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach PB, Mirkin JN, Oliver TK, et al. Benefits and harms of CT screening for lung cancer: a systematic review. Jama. 2012;307(22):2418–2429. doi: 10.1001/jama.2012.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 7.DeBerardinis RJ, Thompson CB. Cellular metabolism and disease: what do metabolic outliers teach us? Cell. 2012;148(6):1132–1144. doi: 10.1016/j.cell.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000;16(3):202–208. doi: 10.1016/s0899-9007(99)00266-x. [DOI] [PubMed] [Google Scholar]
- 9.Paige M, Saprito MS, Bunyan DA, Shim YM. HPLC quantification of 5-hydroxyeicosatetraenoic acid in human lung cancer tissues. Biomedical chromatography : BMC. 2009;23(8):817–821. doi: 10.1002/bmc.1191. [DOI] [PubMed] [Google Scholar]
- 10.Zajdel A, Wilczok A, Tarkowski M. Toxic effects of n-3 polyunsaturated fatty acids in human lung A549 cells. Toxicology in Vitro. 2015 doi: 10.1016/j.tiv.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Yao Q, Fu T, Wang LU, et al. Role of autophagy in the omega-3 long chain polyunsaturated fatty acid-induced death of lung cancer A549 cells. Oncology letters. 2015;9(6):2736–2742. doi: 10.3892/ol.2015.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulbricht B, Henny H, Horstmann H, Spring H, Faigle W, Spiess E. Influence of 12(S)-hydroxyeicosatetraenoic acid (12(S)-HETE) on the localization of cathepsin B and cathepsin L in human lung tumor cells. European journal of cell biology. 1997;74(3):294–301. [PubMed] [Google Scholar]
- 13.Panigrahy D, Greene ER, Pozzi A, Wang DW, Zeldin DC. EET signaling in cancer. Cancer metastasis reviews. 2011;30(3–4):525–540. doi: 10.1007/s10555-011-9315-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Nickkho-Amiry M, McVey R, Holland C. Peroxisome proliferator-activated receptors modulate proliferation and angiogenesis in human endometrial carcinoma. Molecular cancer research : MCR. 2012;10(3):441–453. doi: 10.1158/1541-7786.MCR-11-0233. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Sorenson AL, Poczobutt J, et al. Activation of PPARgamma in myeloid cells promotes lung cancer progression and metastasis. PloS one. 2011;6(12):e28133. doi: 10.1371/journal.pone.0028133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svensson RU, Parker SJ, Eichner LJ, et al. Inhibition of acetyl-CoA carboxylase suppresses fatty acid synthesis and tumor growth of non-small-cell lung cancer in preclinical models. Nature medicine. 2016;22(10):1108–1119. doi: 10.1038/nm.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J, Mazzone PJ, Cata JP, et al. Serum free Fatty Acid biomarkers of lung cancer. Chest. 2014;146(3):670–679. doi: 10.1378/chest.13-2568. [DOI] [PubMed] [Google Scholar]
- 18.de Castro J, Rodriguez MC, Martinez-Zorzano VS, Sanchez-Rodriguez P, Sanchez-Yague J. Erythrocyte fatty acids as potential biomarkers in the diagnosis of advanced lung adenocarcinoma, lung squamous cell carcinoma, and small cell lung cancer. American journal of clinical pathology. 2014;142(1):111–120. doi: 10.1309/AJCP1QUQQLLT8BLI. [DOI] [PubMed] [Google Scholar]
- 19.Sanchez-Rodriguez P, Rodriguez MC, Sanchez-Yague J. Identification of potential erythrocyte phospholipid fatty acid biomarkers of advanced lung adenocarcinoma, squamous cell lung carcinoma, and small cell lung cancer. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36(7):5687–5698. doi: 10.1007/s13277-015-3243-3. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y, He C, Qiu L, et al. Serum unsaturated free Fatty acids: potential biomarkers for early detection and disease progression monitoring of non-small cell lung cancer. Journal of Cancer. 2014;5(8):706–714. doi: 10.7150/jca.9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Timofeeva MN, Hung RJ, Rafnar T, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Human molecular genetics. 2012;21(22):4980–4995. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, McKay JD, Rafnar T, et al. Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nature genetics. 2014;46(7):736–741. doi: 10.1038/ng.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park SL, Fesinmeyer MD, Timofeeva M, et al. Pleiotropic associations of risk variants identified for other cancers with lung cancer risk: the PAGE and TRICL consortia. Journal of the National Cancer Institute. 2014;106(4):dju061. doi: 10.1093/jnci/dju061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhai R, Yu X, Wei Y, Su L, Christiani DC. Smoking and smoking cessation in relation to the development of co-existing non-small cell lung cancer with chronic obstructive pulmonary disease. International journal of cancer Journal international du cancer. 2014;134(4):961–970. doi: 10.1002/ijc.28414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic acids research. 2009;37(Web Server issue):W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grundberg E, Meduri E, Sandling JK, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. American journal of human genetics. 2013;93(5):876–890. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung RJ, McKay JD, Gaborieau V, et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452(7187):633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Broderick P, Webb E, et al. Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nature genetics. 2008;40(12):1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Safran M, Dalah I, Alexander J, et al. GeneCards Version 3: the human gene integrator. Database : the journal of biological databases and curation. 2010 doi: 10.1093/database/baq020. 2010:baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell systems. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penegar S, Wood W, Lubbe S, et al. National study of colorectal cancer genetics. British journal of cancer. 2007;97(9):1305–1309. doi: 10.1038/sj.bjc.6603997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hedges LV, Vevea JL. Fixed-and random-effects models in meta-analysis. Psychological methods. 1998;3(4):486. [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995:289–300. [Google Scholar]
- 34.Wang J, Ronaghi M, Chong SS, Lee CG. pfSNP: an integrated potentially functional SNP resource that facilitates hypotheses generation through knowledge syntheses. Human mutation. 2011;32(1):19–24. doi: 10.1002/humu.21331. [DOI] [PubMed] [Google Scholar]
- 35.Lappalainen T, Sammeth M, Friedlander MR, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proceedings of the National Academy of Sciences. 2001;98(1):31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fradet V, Cheng I, Casey G, Witte JS. Dietary omega-3 fatty acids, cyclooxygenase-2 genetic variation, and aggressive prostate cancer risk. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15(7):2559–2566. doi: 10.1158/1078-0432.CCR-08-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen PL, Ma J, Chavarro JE, et al. Fatty acid synthase polymorphisms, tumor expression, body mass index, prostate cancer risk, and survival. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28(25):3958–3964. doi: 10.1200/JCO.2009.27.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoeft B, Linseisen J, Beckmann L, et al. Polymorphisms in fatty-acid-metabolism-related genes are associated with colorectal cancer risk. Carcinogenesis. 2010;31(3):466–472. doi: 10.1093/carcin/bgp325. [DOI] [PubMed] [Google Scholar]
- 41.Hardwick JP. Cytochrome P450 omega hydroxylase (CYP4) function in fatty acid metabolism and metabolic diseases. Biochemical pharmacology. 2008;75(12):2263–2275. doi: 10.1016/j.bcp.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Corcos L, Lucas D, Le Jossic-Corcos C, et al. Human cytochrome P450 4F3: structure, functions, and prospects. Drug metabolism and drug interactions. 2012;27(2):63–71. doi: 10.1515/dmdi-2011-0037. [DOI] [PubMed] [Google Scholar]
- 43.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nature reviews Genetics. 2011;12(10):683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 44.Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(18):6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosenbloom KR, Sloan CA, Malladi VS, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic acids research. 2013;41(Database issue):D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carolan BJ, Harvey BG, Hackett NR, O'Connor TP, Cassano PA, Crystal RG. Disparate oxidant gene expression of airway epithelium compared to alveolar macrophages in smokers. Respiratory research. 2009;10:111. doi: 10.1186/1465-9921-10-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sridhar S, Schembri F, Zeskind J, et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC genomics. 2008;9:259. doi: 10.1186/1471-2164-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pickett G, Seagrave J, Boggs S, Polzin G, Richter P, Tesfaigzi Y. Effects of 10 cigarette smoke condensates on primary human airway epithelial cells by comparative gene and cytokine expression studies. Toxicological sciences : an official journal of the Society of Toxicology. 2010;114(1):79–89. doi: 10.1093/toxsci/kfp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gandhi AV, Saxena S, Relles D, et al. Differential expression of cytochrome P450 omega-hydroxylase isoforms and their association with clinicopathological features in pancreatic ductal adenocarcinoma. Annals of surgical oncology. 2013;20(Suppl 3):S636–S643. doi: 10.1245/s10434-013-3128-x. [DOI] [PubMed] [Google Scholar]
- 50.Madec S, Cerec V, Plee-Gautier E, et al. CYP4F3B expression is associated with differentiation of HepaRG human hepatocytes and unaffected by fatty acid overload. Drug metabolism and disposition: the biological fate of chemicals. 2011;39(10):1987–1996. doi: 10.1124/dmd.110.036848. [DOI] [PubMed] [Google Scholar]
- 51.Tsunedomi R, Iizuka N, Hamamoto Y, et al. Patterns of expression of cytochrome P450 genes in progression of hepatitis C virus-associated hepatocellular carcinoma. International journal of oncology. 2005;27(3):661–667. [PubMed] [Google Scholar]
- 52.Hintze J. PASS 11. NCSS, LLC Kaysville, Utah, USA. 2011 [Google Scholar]
- 53.Altman DG, Bland JM. Interaction revisited: the difference between two estimates. Bmj. 2003;326(7382):219. doi: 10.1136/bmj.326.7382.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marangoni F, Colombo C, De Angelis L, et al. Cigarette smoke negatively and dose-dependently affects the biosynthetic pathway of the n− 3 polyunsaturated fatty acid series in human mammary epithelial cells. Lipids. 2004;39(7):633–637. doi: 10.1007/s11745-004-1276-5. [DOI] [PubMed] [Google Scholar]
- 55.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature reviews Genetics. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 56.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nature reviews Genetics. 2010;11(12):843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 57.Wei S, Niu J, Zhao H, et al. Association of a novel functional promoter variant (rs2075533 C>T) in the apoptosis gene TNFSF8 with risk of lung cancer--a finding from Texas lung cancer genome-wide association study. Carcinogenesis. 2011;32(4):507–515. doi: 10.1093/carcin/bgr014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.