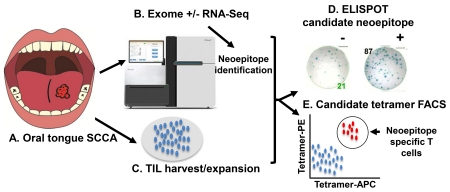
Abstract
The recent success of immunotherapies has demonstrated the potency of tumor-specific immune cells in mediating tumor rejection and generating durable tumor immunity. Our understanding of the scientific basis of these responses results from the confluence of a better comprehension of the cancer immunoediting process and the revolution in next generation sequencing of cancer genomes. Recent evidence suggests that T cell specificity for cancer cell expressed mutant proteins – termed neoantigens – is an important component of immune mediated tumor rejection. Improved neoantigen prediction algorithms have made it possible to predict and monitor immune responses to checkpoint inhibitors and adoptively transferred autologous lymphocytes and have enabled the development of tumor-specific therapeutic vaccines. Herein, we review the current research on cancer neoantigens in immunotherapies and its implications for the future of head and neck cancer management.
Keywords: neoepitope, next generation sequencing, immunogenomics
Graphical abstract
Introduction
“Further development in the area of specific immune responses may permit more meaningful measurements of tumor-specific reactions, thus yielding useful tools for immunodiagnosis as well as providing more effective and precise methods of immunotherapy.” From “Immunobiology of head and neck cancer: basic concepts”, HJ Wanebo (1979) [1]
Recent work on cancer exome based neoantigen identification promises to significantly contribute to the vision articulated in the above prescient quotation. There is now a body of literature showing that these “tumor-specific reactions” can be elicited, at least in part, by mutation-induced tumor neoantigens [2]. These tumor specific mutant antigens represent viable targets of the immune system underlying the recent successes in checkpoint based immunotherapies and have the potential for both greatly improved immune monitoring and the development of patient specific personalized vaccination [3]. The breakthrough of genomics-driven immunotherapies reflects the convergence of two major shifts in our thinking about neoplasia. First, basic studies in immunodeficient mouse cancer models, which led to the conceptualization of the “cancer immunoediting hypothesis” [4-6], have driven a renaissance in our understanding of the cancer-immune system interaction. Second, next generation sequencing technologies have revolutionized our understanding of the genomic complexities within cancer cells. In this review, we provide a historical context for antigenic specificity, review recent findings on cancer neoepitope identification, and discuss the implications for head and neck squamous cell carcinomas (HNSCC).
Historical Context - The Search for Specificity
“It would be as difficult to reject the right ear and leave the left ear intact as it is to immunize against cancer.” From “Immunity to transplantable tumors”, WH Woglom (1929)[7]
Our understanding of the host-tumor interaction has evolved significantly over the last several decades since the above quote on the prospects of cancer immunotherapy. Studies on specificity represent the critical milestones related to our progress in immunotherapy. Focusing on serologic responses, Lloyd Old and coworkers were among the first to concentrate on tumor specificity and the antitumoral immune response [8]. However, it was the seminal finding by Boon and colleagues of a melanoma protein, MZ2-E, recognized by tumor specific CD8+ T cells that finally confirmed the long held belief in specificity [9]. This finding that the immune system can detect discreet differences between cancer and normal cells dramatically changed the way we view the tumor-host interaction. This spurred an entire field of research devoted to the identification and characterization of these tumor signposts and also provided the fundamental context for understanding how the immune system shapes cancer growth and evolution.
Cancer Immunoediting
The cancer immunoediting hypothesis integrates the spectrum of tumor and immune interactions from initial elimination, to cancer equilibrium states and finally to the clinically evident escape phase of tumor growth (reviewed in [4-6]). We now recognize the critical role of the immune system in suppressing tumor growth based on seminal studies demonstrating that mice lacking IFNγ (IFNG) responsiveness, perforin (PRF1), or the capacity for adaptive immunity were significantly more susceptible to carcinogen-induced tumor development [10-14]. Developing tumors in immunocompetent mice face immune pressure that acts on the heterogeneity of the tumor cell population, eliminating immunogenic cells from the tumor mass and thereby selecting for a subset of cells that are resistant to immune attack. This immune pressure results in an “edited” tumor that is less immunogenic and therefore more resistant to immune-mediated rejection. Schreiber and colleagues found that tumors derived from Rag2−/− mice (recombination activating gene 2) deficient of antigen receptor rearranged B and T cells did not face this pressure as illustrated by the rejection of about half of these tumors upon re-transplantion into naïve immunocompetent mice. This is in contrast to “edited” tumors derived in wild type mice which all grow progressively upon re-transplantation into naïve mice. Thus, the immune system selects for tumor cells that are able to avoid immune recognition and elimination. Complementing these murine experiments, evidence that the immune system plays a crucial role in human cancer development arose from the finding that immunosuppressed patients were at significantly increased risk for developing malignancies, including HNSCCs (reviewed in [15]). Further, studies have found that immune infiltration and expression signatures of an adaptive immune response correlate with clinical outcomes in melanoma, colorectal cancer and other malignancies [16]. Thus, data from murine models and human cancer settings together support a role for ongoing immune responses to developing tumors.
Tumor Antigen Classification
Implicit in the immunoediting hypothesis is the idea that components of the immune system recognize the tumor as non-self. Four major categories of tumor antigens have been described: tumor-specific mutant antigens (TSMAs), tumor-associated antigens (TAAs), cancer-testis antigens (CTAs), and viral antigens. TSMAs are the byproduct of cancer-specific genomic alterations that result in protein changes and, independent of protein function, persist during tumor clonal expansion. As these neoantigens are not expressed in non-neoplastic tissue, they represent potent targets for immunotherapy because there is likely the least immunological tolerance to this class [17, 18]. An overview of TSMA processing and presentation is provided in Figure 1. TAAs are normal proteins, which have undergone posttranslational modification including phosphorylation and glycosylation, or are aberrantly expressed on cancer cells [19]. Because these are nonmutated proteins, T cells will frequently display partial tolerance to them, rendering these antigens less immunogenic than those that are absent entirely from the human genome [2]. CTAs are nonmutated proteins whose expression is typically restricted to germ cells but may become reactivated in cancer cells [20]. Finally, in virus-associated tumors such as HPV-associated oropharyngeal squamous cell carcinoma, circulating and tumor-infiltrating T cells may develop specificity for viral antigens presented on the tumor cell surface [21].
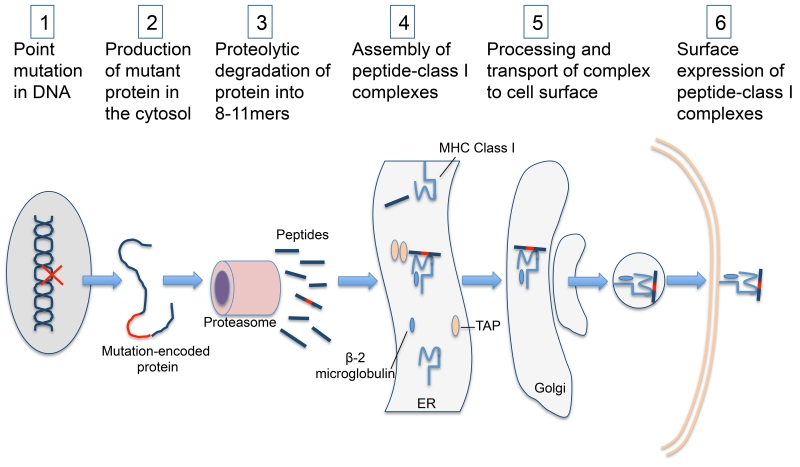
Figure 1.
Pathway of class I neoantigen presentation: Mutation-encoded proteins undergo proteolytic degradation in the cytosol and the mutation-derived peptide is loaded onto an MHC class I molecule in the ER with the assistance of TAP. This neoepitope-class I complex undergoes further processing and transport to the cell surface where it is accessible to neoantigen specific CD8+ T-cells (ER: endoplasmic reticulum. TAP: Transporter associated with antigen processing).
Classical Antigen Identification
Evidence identifying tumor specific antigens was for many years shown by indirect methods such as serologic responses and generation of tumor specific cytotoxic T cells (CTL). Early studies demonstrated that transplantation of a tumor cell line into immunocompetent mice could protect against re-challenge from the same cell line indicating a cell-line specific antigenicity [22]. This was further confirmed in the 1970s and 1980s with isolation of tumor-specific CTLs [23]. However, characterization of the actual tumor antigens remained largely elusive. The earliest techniques for antigen discovery involved the immunization of human cancer cells into rabbits and other animals and subsequent isolation of tumor-specific antibodies from the sera using complement fixation and agar gel immunoprecipitation [24]. This technique paved the way for human serological studies known as autologous typing but was limited both by the difficulty of routinely culturing tumor cells and by the lack of information it provided on the tumor antigens [24]. This deficiency was overcome by a technique known as serological analysis of recombinant cDNA expression libraries (SEREX) that used patient serum to screen cDNA expression libraries for immune targets [25]. The key steps entailed transfecting a library of tumor cDNA into a recipient cell line competent for antigen presentation, and evaluating patient sera for specificity to the transfected cells [26]. Positive “hits” were further delineated by transfecting fragments of the identified gene to further isolate the coding region responsible for the antigenic peptide. These approaches characterized a number of tumor antigens, but were mostly limited to epitopes resulting from activation of germline genes or from posttranscriptional or posttranslational alterations rather than mutation-derived antigens [27]
Numerous studies have looked in HNSCC to define the landscape of available immune targets. These studies have identified a number of CTAs and TAAs prevalent in HNSCCs by SEREX or using tumor infiltrating lymphocyte (TIL)-derived CTLs. These include CTAs from the MAGE family, SSX family, BAGE, SCP1 (SYCP1), PRAME and ENO1 [28-32]. Other HNSCC TAAs capable of inciting a lymphocytic response include art-4, and proteins from the Wnt, Frizzled and Kruppel families [33-36]. Detailed reviews of CTAs and TAAs in HNSCC have been published elsewhere [37, 38].
Although the above studies identified CTAs and TAAs, identification of TSMAs was more elusive. One exception to this was a study that used the CTL method and successfully identified a TSMA derived from a mutation in the CASP8 gene in a human oral cavity squamous cell carcinoma (OCSCC) [39]. As mutations in the CASP8 gene have been suggested to be driver mutations in HNSCCs [40, 41], this finding represents an example of an ideal immunotherapy target—a protein critical for tumorigenesis that can be targeted by directed immunotherapy.
The Next Generation - Enabling Technologies
The past decade has borne witness to a revolution in our understanding of the molecular basis of cancers due to the advent of next generation sequencing technologies. High throughput identification of tumor neoantigens has arisen as an extension of developments in cancer genome next-generation sequencing technologies. The seminal work of Vogelstein and Allison described a blueprint for defining TSMAs using next generation technologies and reordered the conventional passenger and driver mutation understanding of cancer genomes into a new hierarchy [18]. Whereas conventional thinking about cancer genomic changes focused on driver mutations that contribute to the hallmarks of cancer, Vogelstein and Allison instead proposed that all mutations are relevant for a given cancer depending on their respective affinity for HLA binding [18]. In this study, tumor neoantigens in breast and colorectal cancers were predicted from exome sequencing data [18]. Based on this in silico analysis, the authors proposed that neoantigens could be used to vaccinate patients, combined with immune stimulant therapy or represent targets of conventional targeted therapy, chemotherapy or radiation.
The actual steps involved in TSMA prediction start with next generation sequencing data to identify somatic mutations that generate a change in the amino acid sequence and verifying expression of the mutant gene at the RNA level. Following identification of mutation-encoded peptide regions of 8-11 amino acids in length, the next challenge is to filter these epitopes by affinity of binding to the major histocompatibility complex (MHC) molecule. Several tools are available to predict binding of an epitope to MHC including SYFPEITHI, the Immune Epitope Database and Analysis Resource (IEDB), and NetMHC [19]. Additionally, a recently described pipeline, Personalized Variant Antigens by Cancer Sequencing (pVac-Seq), is an automated workflow approach that integrates exome and RNA-sequence data to assemble and prioritize a list of candidate antigens [42]. Although most studies prioritize candidate epitopes based on the predicted binding affinity to the MHC molecule, others have proposed that addition of other features such as a change in the relative binding of mutant epitope to the wild-type epitope as well as the conformational stability of the peptide-MHC interaction can improve prediction [43-45]. Additionally, a variant allele frequency (VAF) cutoff is frequently applied since a recent study demonstrated that TIL reactivity to dominant (founding) clonal antigens are associated with a greater benefit from checkpoint blockade than does reactivity to sub-clonal neoantigens [46].
Determining MHC alleles is straightforward in inbred mouse strains but is more complex in humans due to the polymorphic nature of the human leukocyte antigen (HLA) resulting in over 6,500 unique HLA alleles [47]. Any given patient has at least 6 alleles that each may require different algorithms for predicting peptide binding. Although existing algorithms try to predict affinity for different alleles, they are still constrained by bias to certain alleles. An additional benefit of NGS approaches is that conventional HLA typing can be avoided and predicted from NGS data [47]. Rosenberg and colleagues described an in vitro approach to bypass the need to define HLA by generating tandem mini gene (TMG) constructs capable of screening candidate antigens [48]. DNA encoding each mutated amino acid and flanking amino acids are linked together and cloned into an expression vector [49]. Introduction of the TMG construct in vitro into a patient’s autologous antigen presenting cells (APCs) then results in expression on their native MHC molecule. Autologous TIL can then be incubated with these transfected APCs and screened for reactivity to candidate epitopes [49]. Although less commonly utilized due to poor variant calling results, another approach used to further focus and improve sequence data is to capture RNA from tumor specimens and use this to construct a cDNA library for sequencing [50].This technique focuses only on those mutations in coding exons and eliminates the need for further RNA-sequencing to eliminate non-expressed mutations [19].
Combining the sequencing data with prediction algorithms results in a list of candidate neoantigens that may be responsible for immune reactivity. Most studies that seek to validate these predictions further narrow the list of candidate neoantigens by in vitro autologous TIL reactivity assays, typically by enzyme-linked immunospot (ELISPOT), intracellular cytokine staining, or tetramer staining. Tetramers, MHC/peptide multimers bound to a fluorophore, have emerged as an extremely sensitive in vitro assay to identify antigen-specific T cell binding. Although tetramer production is labor intensive, Schumacher and colleagues have pioneered a high-throughput methodology involving UV-peptide exchange which enables a pre-made HLA molecule loaded with a photocleavable UV-sensitive peptide to exchange candidate epitopes in a matter of hours rather than weeks [51]. Large panels of patient specific neoepitope tetramers generated in this manner have been used to screen peripheral blood to identify reactive CD8+ T cells [52]. A pipeline for in vitro next-generation sequencing guided neoantigen discovery is provided in Figure 2. Ultimately, the only definitive evidence that a specific neoantigen is immunogenic and responsible for immune-mediated tumor rejection is via vaccination in either the prophylactic or therapeutic setting.
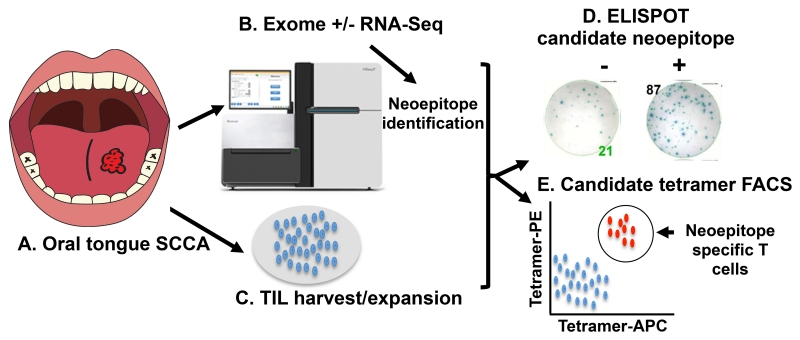
Figure 2.
Pipeline for clinical neoantigen identification. (A) A tumor biopsy is used for (B) sequencing via WES, WGS or cDNA cap-seq and predicted neoantigens are prioritized based on class I affinity. (C) PBMCs and/or tumor infiltrating lymphocytes are isolated from a tumor biopsy. (D) Peptides and/or tetramers are synthesized and TIL specificity is demonstrated by ELISPOT or (E) tetramer staining.
After Vogelstein and Allison’s initial theoretical description, it took another four years before the first genomics and bioinformatics approach would definitively identify a TSMA recognized by T cells. Two groups led by Ugur Sahin [53] and Robert Schreiber [54] independently used sequencing data and in silico neoepitope modeling to identify and validate TSMAs in murine B16-F10 melanoma and methylcholanthrene (MCA)-induced sarcoma cells, respectively. In the former study, predicted neoantigens were formulated into synthetic long peptide vaccines used to immunize mice in the protective and therapeutic setting [53]. In the latter study, it was found that a cell line of “unedited” MCA sarcomas developed in Rag2−/− mice would reject upon transplantation into immunocompetent mice. Using cDNA capture and deep sequencing, it was discovered that a CD8+ T cell response specific for an antigen encoded by the mutant spectrin β-2 gene was responsible for this immune-mediated tumor rejection [54]. In these studies, only a small subset of mutations encoded immunogenic epitopes and this immunogenicity was not correlated with oncologic relevance, gene structure, or subcellular location of the encoded protein [53, 54].
Genome-guided antigen discovery in human patients
With recent advances in immunotherapeutics, exome-guided studies to identify neoantigens have extended to human patients. Improvements in sequencing and bioinformatics have enabled the rapid identification of neoantigens and markedly reduced the time needed from tumor biopsy to screening of TIL and peripheral blood for neoantigen reactivity [44, 55]. Application of this neoantigen identification strategy to cancer patients has been performed either in the “standalone” setting or in the context of cell based or checkpoint inhibition therapies. Robbins, et al. were the first to use whole exome sequencing technology to identify neoantigen-specific TIL from patients with melanoma undergoing adoptive lymphocyte transfer [56]. Likewise, Schumacher and colleagues were the first to show that the high-throughput antigen discovery approach could be used to successfully detect T cell reactivity to a neoantigen in a patient following treatment with the anti-CTLA4 reagent ipilimumab [57]. Further, neoantigen specific CD8+ T cells were identified in patients with chronic lymphocytic leukemia who developed durable remission associated with antitumor immune responses following allogeneic hematopoietic stem cell transplantation [58]. Improvements in neoantigen identification techniques enabled Cohen et al. to show that CD8+ T cells specific for predicted neoantigens could be isolated from peripheral blood of melanoma patients who showed tumor responses to autologous TIL infusions [52].
Although substantial evidence supports the role of CD4+ T cells in cancer surveillance, prediction of MHC class II (MHC II) neoantigens has proved to be more challenging than class I due to the open peptide binding groove in MHC II which leads to significant variability in epitope prediction [19]. Nevertheless, several groups have successfully identified class II antigens in both mice and human tumors [59-61]. The most direct evidence of the potency of a class II neoantigen-specific T cell response is from a patient with metastatic cholangiocarcinoma who was enrolled in a TIL-based adoptive cellular transfer study [62]. The patient was found to have CD4+ TIL specific to an epitope from a mutation in the erbb2 interacting protein gene (ERBB2IP). After an initial transfer containing about 25% mutation-specific T cells, the patient developed some decrease in tumor size and disease stabilization but ultimately her cancer progressed. The patient underwent a subsequent adoptive transfer containing >95% pure mutation-specific CD4+ T cells and developed tumor regression which persisted at least 6 months. A complete list of next-generation sequencing guided neoantigen discovery studies is provided in Table 1.
Table 1.
Next-generation sequencing guided neoantigen discovery studies
| Author | Species/Tumor | CD4/CD8 | T cells | NGS | Epitope Filter | Validation Technique |
|---|---|---|---|---|---|---|
| Castle, CCR 2012 | Murine Melanoma | CD4/CD8 | spleen | WES | SIFT, POLYPHEN-2 |
SLP vaccine then ELISPOT of spleen for Ag reactivity |
| Matsushita, Nature 2012 |
Murine MCA sarcoma | CD8 | TIL, DLN, spleen |
cDNA CapSeq |
ANN | ELISA, ICS, tetramer staining, cDNA library screening |
| Robbins, Nat Med 2013 | Human melanoma | CD8 | TIL | WES | NetMHCPan2.4 | ELISPOT |
| Snyder, NEJM 2014 | Human Melanoma | CD8 | Blood | WES | NetMHC v3.4 | ICS, Tetramer |
| Gubin, Nature 2014 | Murine MCA sarcoma | CD8 | TIL, spleen |
cDNA CapSeq |
SMM, NetMHC NetMHCpan |
Tetramer staining, ELISPOT, ICS, Vaccination, |
| Tran, Science 2014 | Human metastatic cholangiocarcinoma |
CD4 | TIL | WES | TMG | ELISPOT, ELISA, ICS, TCR transduction of PBMCs |
| Wick, CCR 2014 | Human Ovarian Carcinoma |
CD8 | Ascites | WES | NetMHCpan 2.4 |
ELISPOT |
| Rajasagi, Blood 2014 | Human CLL | CD8 | Blood | WES | NetMHCpan 2.4 | ELISPOT, tetramer staining |
| Lu, CCR 2014 | Human Melanoma | CD8 | TIL | WES | TMG | ELISPOT, ELISA |
| Duan, J. Exp. Med 2014 | Murine MCA sarcomas | CD8 | DLN | cDNA-seq | NetMHC 3.0, DAI | Immunization then ICS, ELISPOT |
| Yadav, Nature 2014 | Murine colon adenocarcinoma |
CD8 | TIL, Blood, spleen |
WES | NetMHC-3.4, VAF>20%, M-S |
SLP vaccine then dextramer screen for Ag reactivity |
| Rizvi, Science 2015 | Human NSCLC | CD8 | Blood | WES | NetMHC | Multimer screening, ICS |
| Tran, Science 2015 | Human GI malignancies | CD4/CD8 | TIL | WES/WGS | TMG | ELISPOT, FACS |
| Carreno, Science 2015 | Human melanoma | CD8 | PBMC | WES | NetMHC 3.4, TMG, M-S |
Dendritic cell vaccine then ELISPOT, Dextramer staining |
| Cohen, JCI 2015 | Human melanoma | CD8 | TIL, PBMC |
WES | VAF>10%, IEDB | ELISA, tetramer staining |
| Robinson, Oncoimmunology 2015 |
Murine mesothelioma | CD8 | DLN, spleen |
RNA-seq | NetMHCpan 2.8 | ELISPOT |
| Linnemann, Nat. Med 2015 |
Human melanoma | CD4 | TIL | WES | RNA-seq only | Supernatant cytokine screening, ICS |
| McGranahan, Science 2016 |
Human NSCLC | CD8 | TIL | WES | NetMHCpan 2.8 | Multimer staining |
| Gros, Nat Med 2016 | Human Melanoma | CD8 | PBMCs | WES | TMG | ELISPOT, FACS analysis |
WES: Whole exome sequencing. SLP: Synthetic long peptide. Ag: Antigen. DLN: Draining lymph node. ANN: The Artificial Neural Network Algorithm. ICS: Intracellular cytokine staining. TMG: Tandem mini-gene. TCR: T cell receptor. PBMCs: peripheral blood mononuclear cells. MCA: Methylcholanthrene. SMM: The Stabilized Matrix Method Algorithm. VAF: Variant allele frequency. M-S: Mass spectroscopy. NSCLC: Non-small cell lung cancer. GI: gastrointestinal. WGS: Whole genome sequencing. FACS: Fluorescence-activated cell sorting. IEDB: Immune Epitope Database
Neoantigen identification has taken on a more prominent role as recent studies have suggested that neoantigen-specific T cells are key mediators of the antitumoral effect of checkpoint blocking monoclonal antibodies (mAbs) targeting CTLA-4 (CTLA4), PD-1 (PDCD1) or PDL-1 (CD274) [63, 64]. Additionally, numerous studies where genomics-based antigen discovery was applied to samples from checkpoint therapy clinical trials have found that the clinical benefit of checkpoint blockade is associated with the presence of neoantigen-specific T cells [65-67].
Tumor neoantigen content and patient outcomes
The above neoantigen studies have shed significant light on classification of patients as either responders or non-responders with respect to checkpoint inhibition and other immunotherapeutic efforts. In one study of patients with non-small cell lung cancer (NSCLC) treated with pembrolizumab, an anti-PD-1 mAb, a high neoantigen load conferred an improved progression-free survival of 14.5 months compared to 3.5 months in the low neoantigen load cohort [65]. A natural prediction from this finding is that the mutational burden of a given tumor will be directly correlated with immune responsiveness. In fact, studies have found that tumors with a greater number of nonsynonymous mutations are more likely to possess a higher load of mutation-derived neoepitopes and that immune responses to these epitopes correlate with clinical response [68]. Therefore, tumors with increased genomic instability, which are more likely to develop high numbers of mutations, should correspondingly express a greater number of neoantigens and be more likely to respond to checkpoint blockade. This was recently confirmed in a study of pembrolizumab in colorectal cancer patients with or without mismatch repair (MMR) deficiency, which results in a 10-100 fold increase in the number of somatic mutations. Patients with MMR deficiency had an objective response rate to checkpoint blockade of 40% (4/10) and disease control rate of 90% (9/10) whereas the MMR proficient group had an objective response rate of 0% (0/18) and a disease control rate of 11% (2/18) [69]. This finding was further corroborated by a study of patients with recurrent glioblastoma multiforme (GBM) with biallelic mismatch repair deficiency (bMMRD), which confers an average of 17,740 mutations per tumor [70]. Two siblings with GBM with bMMRD were treated with nivolumab, an anti PD-1 inhibitor, and demonstrated remarkable clinical and radiologic response [70]. However, studies to date suggest that the production of a neoantigen that elicits immune response is a stochastic event equivalent to a “neoantigen lottery” [2]. Therefore, high affinity neoepitopes can be identified in tumors with lower mutational loads. As HNSCCs frequently bear a high mutational burden, they should regularly contain neoantigens capable of immune recognition [2].
A role for personalized vaccine therapy
One of the ultimate goals of high throughput neoantigen identification is the development of patient-specific targeted vaccine therapies. Historically, outcomes with cancer vaccines have been disappointing due to poor vaccine design or an inability to overcome local immunosuppressive factors in the tumor microenvironment [17]. Several phase I and phase II clinical trials of cancer vaccines have demonstrated small improvements in disease-free survival, but overall they have been less effective in obtaining durable responses than other immunotherapies [71]. Studies of vaccines in HPV-negative HNSCC to date have focused on therapeutic vaccines using CTAs or TAAs linked to an adjuvant to facilitate APC uptake, antigen processing and presentation for antigen-specific responses [72]. Recently, a phase II clinical trial was conducted to evaluate the efficacy of a multipeptide vaccine containing the CTA and TAAs LY6K, CDCA1, and IMP3 in patients with locally advanced recurrent and/or metastatic HNSCC [73]. Relative to a control group receiving supportive care only, overall survival was improved (4.9 months to 3.5 months) and one patient demonstrated complete response. When patients had demonstrable CTL responses, the benefit was greater. However, as discussed above, partial tolerance to CTAs and TAAs may underly the relatively minor improvement in survival due to ineffective vaccination [74, 75].
Utilization of personalized cancer vaccines based on tumor-specific neoantigens has been proposed to surmount this tolerance barrier. In preclinical models, therapeutic vaccination with synthetic long peptides of predicted neoepitopes have induced tumor control and rejection in both the preventative and therapeutic settings [3, 53]. The decreased time required to complete exome sequencing and in silico neoantigen prediction promises to make therapeutic patient-specific vaccination a reality in a clinically relevant time frame. Therapeutic neoantigen vaccines have begun entering phase I clinical trials in a small number of malignancies including glioblastoma (NCT02510950, NCT02287428), melanoma (NCT01970358), colon cancer (NCT01885702) and breast cancer (NCT02348320, NCT02427581). In these trials, patient biopsies are sequenced and analyzed for neoantigen prediction. The predicted neoantigen candidates with the highest affinity are then formulated into either a peptide, or dendritic cell vaccine and delivered as a patient specific vaccine for promotion of anti-tumor responses. Response monitoring is performed by detection of neoantigen-specific T cells in the peripheral blood. A single phase I trial to treat advanced metastatic melanoma in three patients with a therapeutic, tumor-specific neoantigen vaccine was recently completed [76]. In this trial, the top seven neoantigen candidates per tumor with detectable expression by cDNA-capture sequencing were formulated into a dendritic cell vaccine. Pre- and post- vaccine TIL reactivity assays demonstrated successful expansion of the neoantigen-specific T cell population, from one neoantigen per patient in the pre-vaccinated TIL to three neoantigens per patient post-vaccination, thereby validating the feasibility of this approach [76]. This first attempt at patient specific vaccination shows that not all predicted neoantigens induce an immune response. Thus, one goal of effective prediction algorithms in the future is to allow for improved selection and prioritization of neoepitopes for inclusion in vaccines.
Vaccine Approaches in HPV-associated HNSCC
Although no neoantigen vaccine trials have begun in HNSCC, viral protein vaccines in HPV-associated HNSCC provide a framework for the efficacy of tumor-specific vaccines. In contrast to TSMAs, viral proteins represent shared tumors antigens and can therefore be targeted by vaccines without requiring tumor sequencing. Note that the TSMA approach focuses on therapeutic vaccination whereas the commonly used HPV vaccines represent a preventative methodology. The latter public health approach relies on two independent FDA-approved HPV vaccines, the bivalent HPV 16/18 (Cevarix®, Glaxo Smith Kline Biologicals) and the quadrivalent HPV 6/11/16/18 (Gardasil™, Merck Sharp and Dohme) that protect against the HPV strains responsible for the majority of HPV-related oropharyngeal squamous cell carcinoma. These vaccines target the L1 capsid protein using virus-like particles and prevent viral infection and subsequent cancer development. Given the dramatic efficacy of these vaccines in reducing cervical cancer (>95%) and other mucosal malignancies, there is an expectation that they will reduce the incidence of oropharyngeal squamous cell carcinoma although their efficacy is still unknown [77].
In addition to preventative vaccines, there are several ongoing therapeutic vaccine clinical trials for HPV-positive oropharyngeal squamous cell carcinoma. These trials build on earlier studies, which found that multiple HPV viral proteins including E2, E6, E7 and L1 are highly immunogenic antigens and that an HPV-specific immune response can be associated with disease clearance [78, 79]. One such approach involves using a natural adjuvant of live attenuated Listeria monocytogenes that secretes the listeriolysin protein fused to the HPV16 E7 protein [80] (ADXS11-001, NCT02002182). A second trial utilizes a vaccine targeting the p16/Ink4a protein (NCT01462838). Unfortunately, most therapeutic vaccine trials to date have been limited in efficacy due in part to an inability to overcome local immunosuppressive pathways [81]. Therefore, several current trials combine a therapeutic viral antigen vaccine with checkpoint inhibitors with the expectation that the two therapies will be synergistic [80].
These vaccine studies above provide a framework for optimal immunotherapy combinations for patients with advanced HNSCC who frequently face a poor prognosis and are in need of novel approaches. The average mutation rate in HNSCC was found to be 15.2 and 14.4 somatic exonic mutations among a panel of 617 cancer-associated genes in HPV negative and HPV positive tumors, respectively [82].This high mutational burden is likely to provide an array of potential antigenic targets for the immune system. Analysis of patient outcomes and correlative studies from preclinical and phase I trials with neoantigen and viral vaccines will hopefully provide insight into how to best utilize immunotherapeutics to approach these hard to treat patients.
Our institution is currently enrolling patients in a phase II window-of-opportunity plus post-operative treatment intensification clinical trial with the anti-PD1 antibody pembrolizumab, in conjunction with surgery and adjuvant chemoradiation for patients with locally advanced HPV-negative HNSCC (NCT02296684). Patients with surgically resectable HPV-negative stage III or IV primary HNSCC are treated with a single neoadjuvant dose of pembrolizumab 14-21 days prior to surgical resection. If the patient’s tumor displays pathologic high-risk features (such as extracapsular extension or positive margins) they will receive up to 6 more doses of pembrolizumab following standard adjuvant chemoradiation. The primary endpoints are a reduction in the local regional recurrence and distant metastasis rate by 15 percent from the historical 35% rate at the one-year time point. Additionally, we will evaluate how the mutational landscape, neoantigen load, and immune cell infiltrate predicts responsiveness to checkpoint therapy. One aim of this correlative work is to better characterize those neoantigens which induce a T cell response with the goal of providing a framework for development of neoantigen vaccines in HNSCC.
Conclusion
It is now well established that the immune system can identify mutation-derived antigens on cancer cells and that these neoantigens represent viable targets for immunotherapies. These findings help to fulfill the long search for specificity in tumor-host interactions. The mutational landscape of HNSCC supports the potential for a range of immunotherapeutic targets for integration in the clinical setting. Although early results from checkpoint-based HNSCC therapeutics are encouraging, outstanding questions remain regarding the broad efficacy of these approaches in HNSCC and how they should be incorporated into the clinical treatment paradigm. Emerging correlative and clinical results from Phase II/III trials in both the primary and recurrent/metastatic setting will provide needed scientific knowledge and clinical direction for future approaches.
Highlights.
The search for specificity of immune responses to cancer has a long history extending from serologic testing, to expression based screening to identify tumor antigens and finally to next generation sequencing technologies to identify mutation-derived neoantigens.
The success of checkpoint immunotherapies has highlighted the integral role of tumor neoantigens in mediating therapeutic benefit and these data may lead to better stratification in future trials.
Tumor neoantigen prediction and validation is continuing to evolve and may ultimately translate to better vaccination protocols for head and neck cancers.
Acknowledgements
Work in Dr. Uppaluri’s laboratory is supported by NIH/NIDCR (DE024403), the NCCN and the V Foundation. He is the PI of an investigator initiated clinical trial funded by Merck Incorporated.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest statement
None declared
References
- [1].Wanebo HJ. Immunobiology of head and neck cancer: basic concepts. Head & neck surgery. 1979;2:42–55. doi: 10.1002/hed.2890020107. [DOI] [PubMed] [Google Scholar]
- [2].Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348:69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- [3].Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–81. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nature immunology. 2002;3:991–8. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- [5].Dunn GP, Old LJ, Schreiber RD. The immunobiology of cancer immunosurveillance and immunoediting. Immunity. 2004;21:137–48. doi: 10.1016/j.immuni.2004.07.017. [DOI] [PubMed] [Google Scholar]
- [6].Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annual review of immunology. 2004;22:329–60. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- [7].Woglom WH. Immunity to Transplantable Tumours. Cancer review. 1929;4:129–214. [Google Scholar]
- [8].Old LJ. Cancer immunology: the search for specificity--G. H. A. Clowes Memorial lecture. Cancer research. 1981;41:361–75. [PubMed] [Google Scholar]
- [9].van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde BJ, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Journal of immunology (Baltimore, Md : 1950) 2007;178:2617–21. [PubMed] [Google Scholar]
- [10].Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–11. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- [11].Smyth MJ, Thia KY, Street SE, Cretney E, Trapani JA, Taniguchi M, et al. Differential tumor surveillance by natural killer (NK) and NKT cells. The Journal of experimental medicine. 2000;191:661–8. doi: 10.1084/jem.191.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Smyth MJ, Thia KY, Street SE, MacGregor D, Godfrey DI, Trapani JA. Perforin-mediated cytotoxicity is critical for surveillance of spontaneous lymphoma. The Journal of experimental medicine. 2000;192:755–60. doi: 10.1084/jem.192.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Street SE, Cretney E, Smyth MJ. Perforin and interferon-gamma activities independently control tumor initiation, growth, and metastasis. Blood. 2001;97:192–7. doi: 10.1182/blood.v97.1.192. [DOI] [PubMed] [Google Scholar]
- [14].Street SE, Trapani JA, MacGregor D, Smyth MJ. Suppression of lymphoma and epithelial malignancies effected by interferon gamma. The Journal of experimental medicine. 2002;196:129–34. doi: 10.1084/jem.20020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Uppaluri R, Dunn GP, Lewis JS., Jr. Focus on TILs: prognostic significance of tumor infiltrating lymphocytes in head and neck cancers. Cancer immunity. 2008;8:16. [PMC free article] [PubMed] [Google Scholar]
- [16].Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nature reviews Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- [17].Hacohen N, Fritsch EF, Carter TA, Lander ES, Wu CJ. Getting personal with neoantigen-based therapeutic cancer vaccines. Cancer immunology research. 2013;1:11–5. doi: 10.1158/2326-6066.CIR-13-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, et al. Epitope landscape in breast and colorectal cancer. Cancer research. 2008;68:889–92. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- [19].Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. The Journal of clinical investigation. 2015;125:3413–21. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Whitehurst AW. Cause and consequence of cancer/testis antigen activation in cancer. Annual review of pharmacology and toxicology. 2014;54:251–72. doi: 10.1146/annurev-pharmtox-011112-140326. [DOI] [PubMed] [Google Scholar]
- [21].Albers A, Abe K, Hunt J, Wang J, Lopez-Albaitero A, Schaefer C, et al. Antitumor Activity of Human Papillomavirus Type 16 E7-Specific T Cells against Virally Infected Squamous Cell Carcinoma of the Head and Neck. Cancer research. 2005;65:11146–55. doi: 10.1158/0008-5472.CAN-05-0772. [DOI] [PubMed] [Google Scholar]
- [22].Klein G, Sjogren HO, Klein E, Hellstrom KE. Demonstration of resistance against methylcholanthrene-induced sarcomas in the primary autochthonous host. Cancer research. 1960;20:1561–72. [PubMed] [Google Scholar]
- [23].Herin M, Lemoine C, Weynants P, Vessiere F, Van Pel A, Knuth A, et al. Production of stable cytolytic T-cell clones directed against autologous human melanoma. International journal of cancer Journal international du cancer. 1987;39:390–6. doi: 10.1002/ijc.2910390320. [DOI] [PubMed] [Google Scholar]
- [24].Old LJ, Chen YT. New paths in human cancer serology. The Journal of experimental medicine. 1998;187:1163–7. doi: 10.1084/jem.187.8.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sahin U, Tureci O, Schmitt H, Cochlovius B, Johannes T, Schmits R, et al. Human neoplasms elicit multiple specific immune responses in the autologous host. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:11810–3. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–7. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- [27].Vormehr M, Diken M, Boegel S, Kreiter S, Türeci Ö , Sahin U. Mutanome directed cancer immunotherapy. Current Opinion in Immunology. 2016;39:14–22. doi: 10.1016/j.coi.2015.12.001. [DOI] [PubMed] [Google Scholar]
- [28].Cuffel C, Rivals JP, Zaugg Y, Salvi S, Seelentag W, Speiser DE, et al. Pattern and clinical significance of cancer-testis gene expression in head and neck squamous cell carcinoma. International journal of cancer Journal international du cancer. 2011;128:2625–34. doi: 10.1002/ijc.25607. [DOI] [PubMed] [Google Scholar]
- [29].Atanackovic D, Blum I, Cao Y, Wenzel S, Bartels K, Faltz C, et al. Expression of cancer-testis antigens as possible targets for antigen-specific immunotherapy in head and neck squamous cell carcinoma. Cancer biology & therapy. 2006;5:1218–25. doi: 10.4161/cbt.5.9.3174. [DOI] [PubMed] [Google Scholar]
- [30].Tsai ST, Chien IH, Shen WH, Kuo YZ, Jin YT, Wong TY, et al. ENO1, a potential prognostic head and neck cancer marker, promotes transformation partly via chemokine CCL20 induction. European journal of cancer (Oxford, England : 1990) 2010;46:1712–23. doi: 10.1016/j.ejca.2010.03.018. [DOI] [PubMed] [Google Scholar]
- [31].Szczepanski MJ, DeLeo AB, Luczak M, Molinska-Glura M, Misiak J, Szarzynska B, et al. PRAME expression in head and neck cancer correlates with markers of poor prognosis and might help in selecting candidates for retinoid chemoprevention in pre-malignant lesions. Oral oncology. 2013;49:144–51. doi: 10.1016/j.oraloncology.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, de Smet C, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6:199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- [33].Kawano K, Gomi S, Tanaka K, Tsuda N, Kamura T, Itoh K, et al. Identification of a new endoplasmic reticulum-resident protein recognized by HLA-A24-restricted tumor-infiltrating lymphocytes of lung cancer. Cancer research. 2000;60:3550–8. [PubMed] [Google Scholar]
- [34].Kass ES, Greiner JW, Kantor JA, Tsang KY, Guadagni F, Chen Z, et al. Carcinoembryonic antigen as a target for specific antitumor immunotherapy of head and neck cancer. Cancer research. 2002;62:5049–57. [PubMed] [Google Scholar]
- [35].Rhee CS, Sen M, Lu D, Wu C, Leoni L, Rubin J, et al. Wnt and frizzled receptors as potential targets for immunotherapy in head and neck squamous cell carcinomas. Oncogene. 2002;21:6598–605. doi: 10.1038/sj.onc.1205920. [DOI] [PubMed] [Google Scholar]
- [36].Heubeck B, Wendler O, Bumm K, Schäfer R, Müller-Vogt U, Häusler M, et al. Tumor-associated antigenic pattern in squamous cell carcinomas of the head and neck-Analysed by SEREX. European Journal of Cancer. 2013;49:e1–e7. doi: 10.1016/j.ejca.2005.09.036. [DOI] [PubMed] [Google Scholar]
- [37].Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer immunology, immunotherapy : CII. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Allen CT, Judd NP, Bui JD, Uppaluri R. The clinical implications of antitumor immunity in head and neck cancer. The Laryngoscope. 2012;122:144–57. doi: 10.1002/lary.21913. [DOI] [PubMed] [Google Scholar]
- [39].Mandruzzato S, Brasseur F, Andry G, Boon T, van der Bruggen P. A CASP-8 mutation recognized by cytolytic T lymphocytes on a human head and neck carcinoma. The Journal of experimental medicine. 1997;186:785–93. doi: 10.1084/jem.186.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pickering CR, Zhang J, Yoo SY, Bengtsson L, Moorthy S, Neskey DM, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer discovery. 2013;3:770–81. doi: 10.1158/2159-8290.CD-12-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, et al. pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome medicine. 2016;8:11. doi: 10.1186/s13073-016-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Duan F, Duitama J, Al Seesi S, Ayres CM, Corcelli SA, Pawashe AP, et al. Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. The Journal of experimental medicine. 2014;211:2231–48. doi: 10.1084/jem.20141308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].van Buuren MM, Calis JJ, Schumacher TN. High sensitivity of cancer exome-based CD8 T cell neo-antigen identification. Oncoimmunology. 2014;3:e28836. doi: 10.4161/onci.28836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes in humans. Cancer immunology research. 2014;2:522–9. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016 doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Schellens IM, Hoof I, Meiring HD, Spijkers SN, Poelen MC, van Gaans-van den Brink JA, et al. Comprehensive Analysis of the Naturally Processed Peptide Repertoire: Differences between HLA-A and B in the Immunopeptidome. PloS one. 2015;10:e0136417. doi: 10.1371/journal.pone.0136417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–8. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lu Y-C, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, et al. Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clinical Cancer Research. 2014;20:3401–10. doi: 10.1158/1078-0432.CCR-14-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cabanski CR, Magrini V, Griffith M, Griffith OL, McGrath S, Zhang J, et al. cDNA hybrid capture improves transcriptome analysis on low-input and archived samples. The Journal of Molecular Diagnostics. 2014;16:440–51. doi: 10.1016/j.jmoldx.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Rodenko B, Toebes M, Hadrup SR, van Esch WJ, Molenaar AM, Schumacher TN, et al. Generation of peptide-MHC class I complexes through UV-mediated ligand exchange. Nature protocols. 2006;1:1120–32. doi: 10.1038/nprot.2006.121. [DOI] [PubMed] [Google Scholar]
- [52].Cohen CJ, Gartner JJ, Horovitz-Fried M, Shamalov K, Trebska-McGowan K, Bliskovsky VV, et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. The Journal of clinical investigation. 2015;125:3981–91. doi: 10.1172/JCI82416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Castle JC, Kreiter S, Diekmann J, Lower M, van de Roemer N, de Graaf J, et al. Exploiting the mutanome for tumor vaccination. Cancer research. 2012;72:1081–91. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- [54].Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 2012;482:400–4. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Fritsch EF, Rajasagi M, Ott PA, Brusic V, Hacohen N, Wu CJ. HLA-binding properties of tumor neoepitopes in humans. Cancer immunology research. 2014;2:522–9. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nature medicine. 2013;19:747–52. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2013;31:e439–42. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood. 2014;124:453–62. doi: 10.1182/blood-2014-04-567933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Linnemann C, van Buuren MM, Bies L, Verdegaal EM, Schotte R, Calis JJ, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nature medicine. 2015;21:81–5. doi: 10.1038/nm.3773. [DOI] [PubMed] [Google Scholar]
- [60].Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature. 2015;520:692–6. doi: 10.1038/nature14426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Schumacher T, Bunse L, Pusch S, Sahm F, Wiestler B, Quandt J, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512:324–7. doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- [62].Tran E, Turcotte S, Gros A, Robbins PF, Lu Y-C, Dudley ME, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–5. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nature reviews Cancer. 2012;12:252–64. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4275–80. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. The New England journal of medicine. 2014;371:2189–99. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–11. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome research. 2014;24:743–50. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine. 2015;372:2509–20. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bouffet E, Larouche V, Campbell BB, Merico D, de Borja R, Aronson M, et al. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016 doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- [71].Melero I, Gaudernack G, Gerritsen W, Huber C, Parmiani G, Scholl S, et al. Therapeutic vaccines for cancer: an overview of clinical trials. Nature reviews Clinical oncology. 2014;11:509–24. doi: 10.1038/nrclinonc.2014.111. [DOI] [PubMed] [Google Scholar]
- [72].DeLeo AB, Whiteside TL. Development of multi-epitope vaccines targeting wild-type sequence p53 peptides. Expert review of vaccines. 2008;7:1031–40. doi: 10.1586/14760584.7.7.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Yoshitake Y, Fukuma D, Yuno A, Hirayama M, Nakayama H, Tanaka T, et al. Phase II clinical trial of multiple peptide vaccination for advanced head and neck cancer patients revealed induction of immune responses and improved OS. Clinical Cancer Research. 2015;21:312–21. doi: 10.1158/1078-0432.CCR-14-0202. [DOI] [PubMed] [Google Scholar]
- [74].Sensi M, Anichini A. Unique tumor antigens: evidence for immune control of genome integrity and immunogenic targets for T cell-mediated patient-specific immunotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:5023–32. doi: 10.1158/1078-0432.CCR-05-2682. [DOI] [PubMed] [Google Scholar]
- [75].Melief CJ, van Hall T, Arens R, Ossendorp F, van der Burg SH. Therapeutic cancer vaccines. The Journal of clinical investigation. 2015;125:3401–12. doi: 10.1172/JCI80009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–8. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Kreimer AR. Prospects for prevention of HPV-driven oropharynx cancer. Oral oncology. 2014;50:555–9. doi: 10.1016/j.oraloncology.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Gildener-Leapman N, Lee J, Ferris RL. Tailored immunotherapy for HPV positive head and neck squamous cell cancer. Oral oncology. 2014;50:780–4. doi: 10.1016/j.oraloncology.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Bourgault Villada I, Moyal Barracco M, Ziol M, Chaboissier A, Barget N, Berville S, et al. Spontaneous regression of grade 3 vulvar intraepithelial neoplasia associated with human papillomavirus-16-specific CD4(+) and CD8(+) T-cell responses. Cancer research. 2004;64:8761–6. doi: 10.1158/0008-5472.CAN-04-2455. [DOI] [PubMed] [Google Scholar]
- [80].Massarelli E, Ferrarotto R, Glisson BS. New Strategies in Human Papillomavirus-Related Oropharynx Cancer: Effecting Advances in Treatment for a Growing Epidemic. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:3821–8. doi: 10.1158/1078-0432.CCR-14-1329. [DOI] [PubMed] [Google Scholar]
- [81].Badoual C, Sandoval F, Pere H, Hans S, Gey A, Merillon N, et al. Better understanding tumor-host interaction in head and neck cancer to improve the design and development of immunotherapeutic strategies. Head & neck. 2010;32:946–58. doi: 10.1002/hed.21346. [DOI] [PubMed] [Google Scholar]
- [82].Seiwert TY, Zuo Z, Keck MK, Khattri A, Pedamallu CS, Stricker T, et al. Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:632–41. doi: 10.1158/1078-0432.CCR-13-3310. [DOI] [PMC free article] [PubMed] [Google Scholar]