Abstract
The totality of microbial genomes in the gut exceeds the size of the human genome, having around 500-fold more genes that importantly complement our coding potential. Microbial genes are essential for key metabolic processes, such as the breakdown of indigestible dietary fibres to short-chain fatty acids, biosynthesis of amino acids and vitamins, and production of neurotransmitters and hormones. During the last decade, evidence has accumulated to support a role for gut microbiota (analysed from faecal samples) in glycaemic control and type 2 diabetes. Mechanistic studies in mice support a causal role for gut microbiota in metabolic diseases, although human data favouring causality is insufficient. As it may be challenging to sort the human evidence from the large number of animal studies in the field, there is a need to provide a review of human studies. Thus, the aim of this review is to cover the current and future possibilities and challenges of using the gut microbiota, with its capacity to be modified, in the development of preventive and treatment strategies for hyperglycaemia and type 2 diabetes in humans. We discuss what is known about the composition and functionality of human gut microbiota in type 2 diabetes and summarise recent evidence of current treatment strategies that involve, or are based on, modification of gut microbiota (diet, probiotics, metformin and bariatric surgery). We go on to review some potential future gut-based glucose-lowering approaches involving microbiota, including the development of personalised nutrition and probiotic approaches, identification of therapeutic components of probiotics, targeted delivery of propionate in the proximal colon, targeted delivery of metformin in the lower gut, faecal microbiota transplantation, and the incorporation of genetically modified bacteria that express therapeutic factors into microbiota. Finally, future avenues and challenges for understanding the interplay between human nutrition, genetics and microbial genetics, and the need for integration of human multi-omic data (such as genetics, transcriptomics, epigenetics, proteomics and metabolomics) with microbiome data (such as strain-level variation, transcriptomics, proteomics and metabolomics) to make personalised treatments a successful future reality are discussed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00125-017-4278-3) contains peer-reviewed but unedited supplementary material including a slideset of the figures for download, which is available to authorised users.
Keywords: 16S sequencing, Faecal microbiome, Genetics, Glycaemic control, Gut microbiome, Metagenomics, Metformin, Personalised nutrition, Probiotics, Review, Type 2 diabetes
Introduction
Accumulating evidence indicates the importance of the gut microbiota in the development of a multitude of complex human diseases, including type 2 diabetes. The intimate coevolution of the microbiota with mammalian hosts has created a coherent symbiotic relationship that directs and contributes to a plethora of essential physiological functions, such as energy metabolism, metabolic signalling, formation of the immune system, and regulation of integrity and mobility of the gut barrier. A paradigm shift in the research of gut microbiota in health and disease came after the development of PCR-based techniques for the characterisation and quantification of bacterial composition via sequencing of bacterial genes. In contrast to cloning-, blotting- or cultivation-based analyses, sequencing techniques enable a much more comprehensive mapping of bacteria, alongside investigation of their abundance in samples of interest, which in human gut microbiota studies have almost exclusively been faeces specimens. Thus, throughout this review, the term ‘gut microbiota’ refers to faecal microbiota. The two main approaches for next-generation sequencing of the microbiome are 16S ribosomal RNA gene amplicon sequencing and shotgun metagenomics sequencing, both of which have their strengths and weaknesses (see summary text box: gut microbiome analysis) [1]. The sequencing methods used in the studies discussed in this review are listed in ESM Table 1. Whilst considering the findings discussed in this review, we would like the readers to note that, in addition to the type of sequencing methods used, the results of gut microbiome analyses are affected by the methods used for sampling and storage and DNA extraction, the approaches and pipelines used for taxonomical classification of bacterial sequences, and the bioinformatics and statistical approach applied for data integration and analysis [1, 2].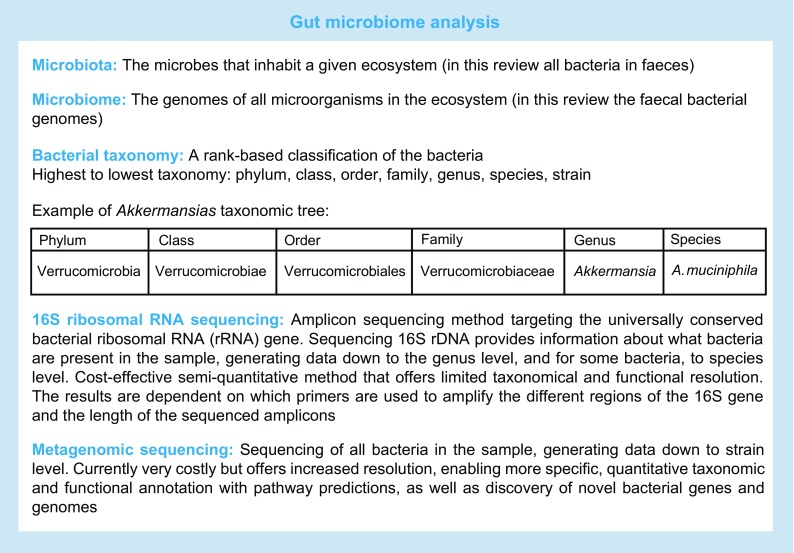
What factors impact on the gut microbiota?
During 2016 the first two population-based gut microbiota studies were published. In an attempt to obtain a comprehensive picture of the factors that correlate with the human gut microbiota, Falony et al investigated the association between 503 clinical-, health- and lifestyle-related variables and the diversity and composition of gut microbiota among 1106 Belgian individuals and also in a replication cohort (Dutch LifeLines DEEP; n = 1135) [3]. They observed that stool consistency had the largest effect size on gut microbiota, while medication-, diet- and health-related variables majorly contributed to the variance in gut microbiota. Zhernakova et al performed a more detailed analysis of the Dutch LifeLines DEEP cohort using metagenomics and reported that, of the 207 exogenous and intrinsic host factors investigated, 60% correlated significantly with gut microbiota (including 31 intrinsic factors, 12 diseases, 19 drug groups, four smoking categories and 60 dietary factors). Collectively, these factors explained 18.7% of the inter-individual variation in microbial composition [4]. These studies demonstrate that a wide range of health and environment factors associate with changes in the composition and functionality of the gut microbiota.
A role for the gut microbiota in diabetes prevention and treatment
From an evolutionary perspective, the rapid increase in the prevalence of type 2 diabetes worldwide is related to rapid environmental changes that have had a negative impact on risk factors of diabetes, such as dietary habits and sedentary time. Although the composition of gut microbiota is affected by genetic and environmental factors, unless marked environmental changes occur, it is rather stable in adult (‘middle-aged’) individuals. Nonetheless, inter-individual variation in gut microbial composition is remarkably high. Given the plethora of biological functions that are regulated by the gut microbiota, and that its composition is dependent on adjustable factors (e.g. diet or drugs), new evidence for the potential use of the modifiable capacity of the gut microbiota in the development of better prevention and treatment options for type 2 diabetes are emerging.
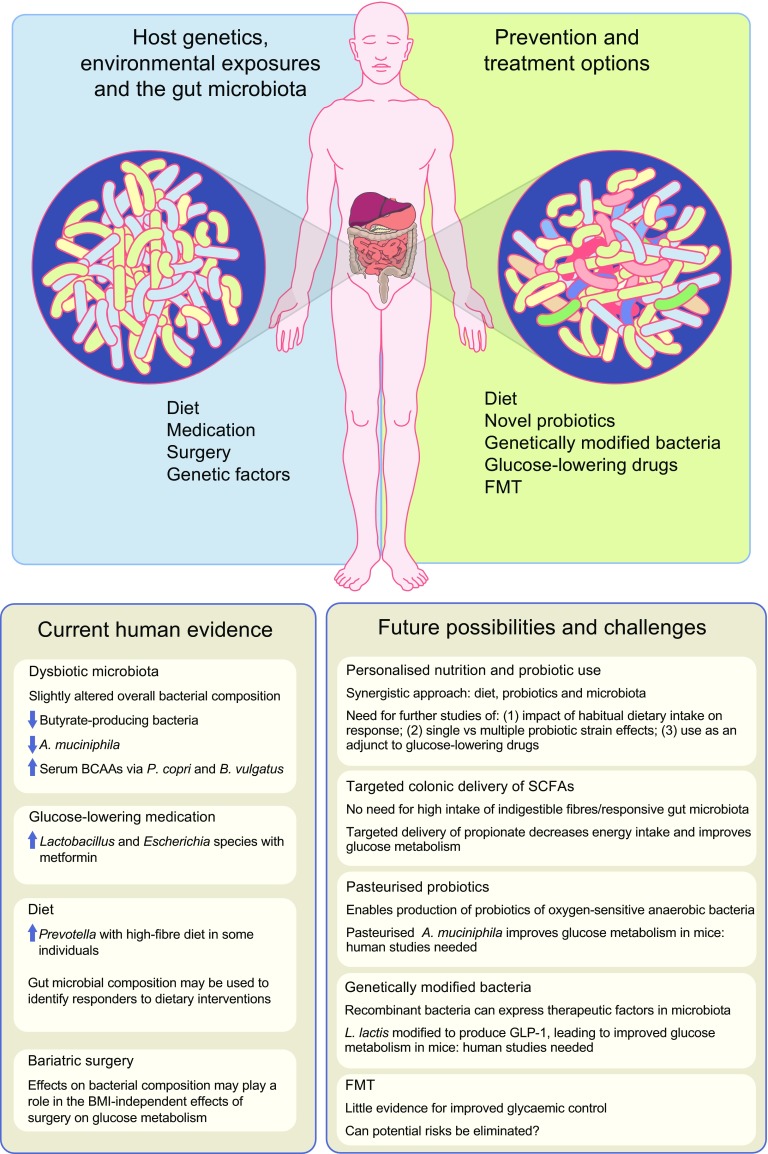
In a review in 2007, Cani et al were the first to mention gut microbiota in the context of type 2 diabetes [5]. To date, a search of PubMed (www.ncbi.nlm.nih.gov/pubmed/, accessed 16 March 2017) using ‘type 2 diabetes (or hyperglycaemia) and gut microbiota (or gut microbiome)’ produces a list of over 600 papers that have been published in this area, of which almost 50% are reviews. Yet, despite there being some excellent (recent) reviews on this topic [6], it is challenging to identify the human evidence from the much larger number of animal studies in the field. Similarly, separating the findings related to obesity or inflammation from those related to glycaemic traits is not straightforward. Therefore, this review focuses on human studies and, when possible, on BMI-independent findings, aiming to converge the current and future possibilities and challenges of using the gut microbiota, and its plastic nature, in preventive and treatment strategies for type 2 diabetes (Fig. 1).
Fig. 1.
The gut microbiome as a prevention and treatment target for hyperglycaemia in type 2 diabetes: current evidence from human studies and future possibilities. Research has aimed to investigate how host genetics and internal and external environmental exposures (e.g. diet, medication, surgery) affect the gut microbiota, and how this information, along with information on gut bacterial composition, may be used to develop prevention and treatment strategies for personalised medicine in type 2 diabetes. Current human evidence has revealed that individuals with type 2 diabetes have a slightly altered overall bacterial composition, including decreased abundance of butyrate-producing bacteria and those with prediabetes/type 2 diabetes have a decreased abundance of A. muciniphila. Furthermore, in individuals with insulin resistance, serum BCAAs are shown to be elevated in insulin resistance and gut microbiota are thought to be a significant contributor, with P. copri and B. vulgatus being the main producers of BCAAs. Interestingly, glucose-lowering treatments, such as metformin, can alter overall bacterial composition e.g. by increasing abundance of Lactobacillus and Escherichia species. Diet has also been shown to cause short- and long-term alteration in gut microbial richness and composition. For example, high fibre intake increases Prevotella abundance. However, response to diet is varied, with some individuals being non-responders to dietary interventions. In line with this, gut microbial composition may provide a tool for the identification of individuals who are expected to benefit from dietary interventions. Further, evidence from bariatric surgery suggests that its effects on bacterial composition in the gut may contribute to the BMI-independent effects on glucose metabolism after surgery. Together, these findings open doors for novel future possibilities for the prevention and treatment of type 2 diabetes; however, the implementation of these is associated with many challenges. For example, a personalised nutrition approach (a synergistic approach involving diet, probiotics and microbiota) may be developed to improve glycaemic control. However, prior to implementing this strategy, there is a need for prospective follow-up studies, studies investigating the influence of habitual dietary intake on responsiveness to personalised nutrition, and large long-term randomised controlled trials (RCTs) to investigate single vs multiple probiotic strain effects and the use of probiotics as adjuncts to glucose-lowering drugs. Targeted colonic delivery of SCFAs may also be used to beneficially alter glycaemic control, without the need for high consumption of indigestible fibres (and for responsive gut microbiota, which may not be available in diabetes). In line with this, targeted delivery of propionate has been shown to decrease energy intake and improve glucose metabolism, making this a promising approach. Pasteurised probiotics provide another potential therapy in diabetes; these enable use of oxygen-sensitive anaerobic bacteria in diabetes treatment. For example, pasteurised A. muciniphila has been shown to improve glucose metabolism in mice. However, human studies are needed. Likewise, genetically modified bacteria can be designed to express therapeutic factors and incorporated into the microbiota. For example, L. Lactis (a ‘safe’ bacteria) has been genetically modified to produce GLP-1 and, hence, improve glucose metabolism in mice; however, human studies are lacking. Finally, FMT has been shown to be effective in the treatment of severe forms of C. difficile infection (for which there are high mortality rates if not treated). However, evidence is required to ensure that it can be used to improve glycaemic control in diabetes and that the potential risks of this therapeutic approach can be eliminated
Gut microbiota composition in type 2 diabetes and insulin resistance
The first study investigating the differences between human gut microbiota in individuals with type 2 diabetes and control participants was conducted by Larsen et al in 2010. Significant differences in the composition of microbiota at the phylum level were observed between the groups using a principal component analysis [7]. Thereafter, other studies have reported BMI-independent alterations in the gut microbiota of participants with type 2 diabetes, and although there has been a lack of consensus concerning which bacteria are significantly altered in type 2 diabetes, a common observation has been a decreased abundance of butyrate-producing bacteria with this condition [7–10]. More recently, Forslund et al challenged the question of whether type 2 diabetes-associated changes in the gut microbiota can be separated from those induced by commonly used blood glucose-lowering drugs [11]. They found that a large part of the previously observed differences in the gut microbiota in diabetes could indeed be explained by metformin treatment. In line with previous studies [10, 12], Forslund and colleagues found that individuals with type 2 diabetes who were not treated with metformin had less bacteria from genera known to produce butyrate as compared with control participants without diabetes. Meanwhile, they also elucidated that the previously reported increase in Lactobacillus species among type 2 diabetes individuals results from metformin treatment [11]. Metformin-treated individuals were also observed to have increased abundance of Escherichia species in the gut, suggested to potentially contribute to the adverse gastrointestinal effects of metformin.
Another recent study integrated data on insulin resistance, the gut microbiome and the fasting serum metabolome of 277 Danish individuals without diabetes to investigate whether the gut microbiome impacts upon insulin resistance-associated metabolic signatures. The serum metabolome of individuals with insulin-resistance was characterised by an increased potential for branched-chain amino acid (BCAA) biosynthesis and reduced potential for BCAA transport into bacterial cells [13]. The finding that BCAAs tightly co-varied with fasting serum metabolites of microbial origin, and that BCAA synthesis was driven by a few specific bacterial species (Prevotella copri and Bacteroides vulgatus; ‘top producers’ of BCAA), suggested that the gut microbiota could be an important source of elevated BCAAs in insulin resistance [13, 14].
One specific bacteria that has been the focus of several obesity and type 2 diabetes studies is the mucus-colonising Akkermansia muciniphila. Zhang et al found A. muciniphila to be decreased in individuals with prediabetes (impaired glucose tolerance and/or impaired fasting glucose) and newly diagnosed type 2 diabetes, and suggested that low abundance of this bacteria could be a biomarker for glucose intolerance [8]. More recently, A. muciniphila was observed to be decreased prior to the onset of type 2 diabetes in twins [9], while high abundance was associated with a healthier metabolic status in overweight/obese humans, as well as with greater improvements in glucose homoeostasis and body composition after energy restriction [15].
Current prevention and treatment strategies for hyperglycaemia in type 2 diabetes by gut microbiota modification
Modification of gut microbiota by diet and prebiotics
A number of studies have demonstrated that the gut microbial richness and composition are, to a large extent, modulated by diet [16, 17]; there is increasing evidence that insufficient consumption of indigestible carbohydrates may have led to a loss of bacterial species in the human gut that are reliant on these substrates and, as a consequence, there is decreased production of their fermentation end-products, short-chain fatty acids (SCFA) [18]. Epidemiological studies have been consistent in showing inverse association between dietary fibre and the incidence of type 2 diabetes, and dietary fibre and whole grains have been shown to increase the diversity of the human gut microbiota [19, 20]. In several studies, high fibre intake has been shown to associate with increased levels of the bacterial genus Prevotella [16, 17, 21]. Further, Kovatcheva-Datchary et al observed that improvements in postprandial glucose and insulin response after a 3-day intervention with barley kernel bread were dependent on the enrichment of P. copri in the microbiota of participants, and this enrichment was functionally connected to increased efficiency to digest complex polysaccharides in the barley kernel bread. Interestingly, for those whose glucose metabolism was not improved following the intervention, their gut microbiota was found to be neither enriched with P. copri nor was its potential to ferment complex polysaccharides increased following the trial. These findings suggest that analysis of the gut microbiota could be used to gain an understanding of individual responses to dietary interventions [22].
The therapeutic effects of metformin are mediated by gut microbiota
The primary site of action and the main glucose-lowering effects of metformin have been debated. However, important novel information has now emerged demonstrating that the primary effects of the drug may reside in the gut; the pharmacological action of metformin was observed to include alterations in bile acid recirculation and gut microbiota, resulting in enhanced glucagon-like polypeptide-1 (GLP-1) secretion [23]. Further, a meta-analysis of 199 individuals with type 2 diabetes and 544 control participants without diabetes, stratified by treatment regiments, found that metformin had a significant effect on gut microbiota composition, such that individuals with type 2 diabetes who were on metformin could be identified based on compositional changes in their gut microbiota [11]. Functionally, the gut microbiota of metformin-treated individuals had increased potential to produce butyrate and propionate, while the untreated participants had enrichment of microbial genes involved in the degradation of glycine and tryptophan, and depletion of genes involved in threonine and arginine degradation. This is particularly interesting since glycine has previously been associated with insulin sensitivity [24], and glycine supplementation has been suggested as a treatment for glutathione deficiency and has been shown to improve insulin sensitivity in type 2 diabetes [25]. Further, increased abundance of Escherichia in metformin-treated individuals has been functionally attributed to the enrichment of virulence factors and genes associated with gas metabolism, indicating involvement of this genus in the intestinal adverse effects of metformin. The work by Forslund et al thus suggests potential microbiota-mediated mechanisms behind both the therapeutic and adverse effects of metformin [11].
Additional evidence and mechanistic insights supporting a predominant action of metformin in the lower gut have recently been provided by clinical studies testing the response to delayed-release metformin (MetDR) in healthy volunteers and participants with type 2 diabetes [26, 27]. The delayed release of this drug relies on a pH-dependent dissolution of the enteric coat of the tablet; this first occurs at pH 6.5 in the distal intestine, where the density of L cells is high, to maximise the gut-based mechanisms of the drug. Pharmacokinetic and dose-ranging studies have demonstrated that MetDR has a glucose-lowering efficacy comparable with that of extended- release metformin but with a 40% increase in potency, thus allowing for lower doses of MetDR to be used [27]. In line with this, two randomised trials demonstrated that MetDR resulted in glucose-lowering effects comparable with immediate-release metformin, despite significantly lower systemic exposure [26]. Large-scale Phase III clinical trials are now needed, as well as studies in individuals with renal impairment, to fully elucidate the efficacy and safety of MetDR. In addition, whether MetDR affects the gut microbiota in a similar manner to immediate-release metformin is currently unknown and must be investigated.
Bariatric surgery remodels the gut microbiota composition
Bariatric surgery leads to fast and major weight loss, and many individuals with type 2 diabetes achieve normal glucose and insulin regulation within days after surgery, suggesting a BMI-independent effect [28]. Bariatric surgery leads to many important physiological alterations (for a recent review see [29]) including both short- and long-term changes in gut microbiota composition [29, 30]. Proteobacteria have been found to be increased in the microbiota of operated individuals and elevations in plasma bile acids and alterations in bile acid composition have been linked to the compositional changes of microbiota [30]. Both the changes in Proteobacteria and bile acids following bariatric surgery have been suggested to contribute to the BMI-independent surgical effects on glucose metabolism [28]. However, the precise molecular mechanisms underlying the many physiological effects of bariatric surgery still need to be determined and whether such information can lead to development of novel non-surgical interventions for the treatment of type 2 diabetes remains to be seen.
Towards novel prevention and treatment strategies for type 2 diabetes: possibilities and challenges offered or inspired by gut microbiota studies
Personalised nutrition, probiotics and targeted delivery of propionate
Inter-individual differences in the composition of gut microbiota have been shown to result in individualised responses to dietary fibre interventions [22, 31] and analysis of gut microbial composition has been proposed as a tool for the identification of individuals who are expected to benefit from dietary interventions [32–34]. A promising future approach could use a synergistic approach involving both diet and microbiota in the prevention and treatment of type 2 diabetes. However, prospective follow-up studies that use gut microbiota composition as a strategy to identify individuals who would benefit from a certain dietary intervention are obligatory to provide evidence for the effectiveness of this approach. In addition, studies investigating the influence of habitual dietary fibre intake on an individual’s responsiveness (and the responsiveness of their gut microbiota) to probiotic intervention are needed [35]. A further challenge will be to perform large, long-term randomised controlled trials to investigate the effect of single vs multiple probiotic strains on glycaemic control and to determine their usefulness as an adjunct to glucose-lowering drugs, whilst elucidating how such effects may be mediated by changes in gut microbiota ecology.
The increasing evidence for a beneficial role of SCFAs in appetite control and energy homeostasis has inspired the development of methods to increase colonic SCFA levels without the need for high consumption of indigestible fibres and, hence, a responsive gut microbiota for their effective fermentation. A novel targeted delivery system was recently developed based on an inulin-propionate ester capable of releasing gram quantities of propionate in the proximal colon [36]. Such targeted delivery of propionate was demonstrated to acutely reduce energy intake and ameliorate long-term weight gain in overweight adults [37]. In addition, 24 weeks of supplementation with the inulin-propionate ester significantly improved acute insulin secretion and beta cell function, suggesting that increasing colonic propionate improves glucose metabolism, both via acute elevations in GLP-1 and peptide YY (PYY) [36] and by long-term stimulatory actions of propionate on beta cells [37].
Pasteurisation and identification of the therapeutic components of probiotics
The use of A. muciniphila in humans is hampered by its high sensitivity to oxygen and its requirement for mucus-based medium that is incompatible with human administration. To circumvent these hurdles, in a recent study, Plovier et al grew this bacterium on a synthetic medium where mucus was replaced with compounds appropriate for human administration. Further, Plovier and colleagues were able to produce a non-replicative, pasteurised preparation of A. muciniphila [38], which was surprisingly found to have an enhanced capacity to improve glucose metabolism in mouse models of obesity and diabetes. Further, a specific outer membrane protein of A. muciniphila, Amuc_1100, was found to partly recapitulate the bacteria’s beneficial effects. Preliminary human data indicated that, following 2 weeks of treatment, when grown on the synthetic medium both the pasteurised and the live bacterium were safe for human administration [38]. However, the efficacy of A. muciniphila preparations as a potential therapeutic tool for diabetes remains to be shown.
Faecal microbiota transplantation
Faecal microbiota transplantation (FMT) is a technique where case or control microbiota is transplanted into germ-free mice to study the effect of faecal microbiota independent of other environmental and microbial interactions. FMT is commonly used to prove causality, however, whether causality can be effectively determined via this method is debated [39]. For example, only a subset of human gut microorganisms is capable of colonising the gut of germ-free mice [39, 40] and, in particular, many potential butyrate-producing genera may be poorly transferred [39]. Nonetheless, the fact that individuals with severe Clostridium difficile infection are successfully treated with FMT has led researchers to hypothesise that transplantation of healthy faecal microbiota could cure metabolic disease. Testing this hypothesis in mice has provided promising results [41]. In the first human study of 18 individuals, FMT from lean individuals to men with the metabolic syndrome resulted in significant improvements in peripheral insulin sensitivity along with an increase in butyrate-producing bacteria in the recipient’s faecal microbiota [42], which is in line with observational studies where butyrate-producing bacteria have been observed to be enriched in the faeces of healthy individuals [12]. However, we must emphasise that this study was small, it did not report data on glucose levels during the intervention and it has not yet been replicated. Moreover, not all participants responded to FMT, again raising the question as to why some individuals are responsive while others are not [22]. Thus, the current evidence for FMT as a therapeutic tool to improve glycaemic control and/or insulin sensitivity is very limited. In addition, this method is still very new and, hence, much more research must be devoted to FMT to explore the potential risks related to its impact on the vast physiological functions regulated by gut microbiota and to eliminate the threat of transplantation of pathogenic bacteria.
Genetically modified microbiota
One novel strategy to alter the microbiome is to incorporate genetically modified bacteria that express therapeutic factors into microbiota. Recently, a recombinant Lactococcus lactis strain that was genetically modified to produce GLP-1 was shown to stimulate insulin secretion and improve glucose tolerance in mice [6]. L. lactis is used in many fermented food products and is generally regarded as safe and, despite the lack of human evidence for its ability to treat diabetes by acting as a host for the production, secretion and delivery of incretins (or other compounds) directly to the intestine, recombinant bacteria that produce IL-10 have been shown to be safe in a Phase I clinical trial in participants with Crohn’s disease [43]. Other studies in mice have demonstrated that therapeutic compounds produced by genetically modified colonic bacteria can have additional therapeutic effects in tissues beyond the intestinal tract, such as the liver [44]. Compared with wild-type probiotics, the use of genetically modified gut microbiota has clear advantages as it enables selection of an effective ‘coloniser’ as the carrier bacteria, and production of a desired therapeutic compound. Further, the recombinant bacteria could be designed to respond to specific temporal signals, like dietary factors [44].
Multi-omics in gut microbiota research
During the last decade genome-wide association studies (GWAS) have revolutionised our understanding of genetic factors affecting multifactorial diseases in humans, including identification of over 120 loci that associate with type 2 diabetes or glycaemic traits. The gut microbiome essentially complement our coding potential and have also been named our ‘second genome’ [45]. Indeed, based on metagenomic sequencing data of faecal samples from 1267 individuals, a catalogue of human faecal microbiome was created including 9,879,896 genes, which is almost 500 times the number of human genes (~20,000). However, each sample contained an average of 762,665 bacterial genes, thus representing around 40 times more genes as compared with the human genome [45]. In 2014, Goodrich et al assessed the heritability of the human gut microbiota in a twin study and demonstrated that human genetic variation influences the microbial phenotype [46]. More recently, the same research group tripled the sample size to 1126 twin pairs and compared the differences in heritability of 945 widely shared bacterial taxa in monozygotic vs dizygotic twin pairs. Their findings indicated that 8.8% of all taxa have heritability estimates of greater than 0.2, meaning that at least 20% of the variance in the abundance of these 8.8% of taxa could be attributed to genetic differences. Christensenellaceae was identified as the most heritable taxa [47]. The first human GWAS of gut microbiota composition observed a significant association between variation in the gene PLD1, which has been associated with obesity in African-American individuals, and the abundancy of Akkermansia [48]. Another GWAS found nine SNPs to be associated with specific taxa, and replicated an earlier finding that an interaction between the LCT gene and Bifidobacteria was dependent on dairy intake [49]. This study provided the first example of how dietary intake can modify the association between a genetic variant related to metabolic disease and the abundance of specific gut bacteria. For GWAS, the size of these studies is very small and, thus, much better powered studies are warranted. This is especially true for GWAS in gut microbiota research since, compared with GWAS in other areas, there are a vast number of variables (i.e. a huge array of possible microbial phenotypes), increasing the multitude of tests that are required [50].
Another future opportunity in gut microbiota research is microbial metagenomic GWAS, which, so far, has only been used in the study of pathogenic microorganisms [51]. Although a number of analytical advances are required to handle the unique features of microbial genomics, such an approach could provide exciting new insights into the effects of microbial variants and, if integrated with human genomic data, how they may be dependent on the human genetic background. Arguably, for microbiota-based personalised treatments to become a successful future reality, integration of human multi-omic data (such as genetics, transcriptomics, epigenetics, proteomics and metabolomics) with microbiome data (such as strain-level variation, transcriptomics, proteomics and metabolomics) in large, longitudinal cohorts will most likely be required [52].
Conclusion
The WHO has predicted type 2 diabetes to be the seventh leading cause of death by 2030 and there is an urgent need for the development of more efficient prevention and treatment strategies. The metabolic potential of the gastrointestinal tract and its microbiota are increasingly recognised as promising targets to improve glycaemic control and treat type 2 diabetes. Importantly, recent data has demonstrated that the glucose-lowering effects of metformin are mediated by changes in the composition and function of gut microbiota. Several potential gut-targeting glucose-lowering treatment strategies are now emerging and show initial promise, yet a better understanding of the mechanisms underlying these effects in humans is required. So far, human studies in this field have been limited in size and the results have often been inconclusive and difficult to interpret. Therefore, to fully elucidate the modifiable capacity of the gut microbiota and its potential for use in the prevention and treatment of type 2 diabetes, large longitudinal and interventional studies are required. Another great challenge for the future is the personalisation of nutrition to allow for individual dietary advice in diabetes, taking into account gut microbiota, drug use, genetics and relevant environmental factors (here probiotics might provide an opportunity to enhance the beneficial health effects of diet or glucose-lowering drugs). However, methodologies must be developed to enable integration of multi-omics data of both human and microbial origin for this to happen. In addition, many other methodological challenges remain in this area of research, from appropriate standardisation of the DNA-extraction methods used, to functional characterisation of bacterial genes with unknown functions and improvement of the statistical and bioinformatic processing of this complicated data. Hence, advancing our knowledge on the role of gut microbiota in type 2 diabetes and using this knowledge to enable a personalised medicine approach for the treatment and management of type 2 diabetes will presumably require long, painstaking work with no scope for short cuts.
Electronic supplementary material
(PPTX 225kb)
(PDF 106 kb)
Abbreviations
- BCAA
Branched-chain amino acid
- FMT
Faecal microbiota transplantation
- GLP-1
Glucagon-like polypeptide-1
- GWAS
Genome-wide association study
- MetDR
Delayed-release metformin
- SCFA
Short-chain fatty acid
Funding
MO-M is a recipient of grants from the Swedish Research Council, the Swedish Heart and Lung Foundation, the Novo Nordic Foundation, the Swedish Diabetes Foundation, the Region Skåne (Skåne University Hospital) and the European Research Council (Consolidator grant no. 649021, Orho-Melander).
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript.
Contribution statement
Both authors were responsible for drafting the article and revising it critically for important intellectual content. Both authors approved the version to be published.
References
- 1.Jovel J, Patterson J, Wang W, et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol. 2016;7:459. doi: 10.3389/fmicb.2016.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodrich JK, Di Rienzi SC, Poole AC, et al. Conducting a microbiome study. Cell. 2014;158:250–262. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 4.Zhernakova A, Kurilshikov A, Bonder MJ, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cani PD, Neyrinck AM, Fava F, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50:2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 6.Arora T, Backhed F. The gut microbiota and metabolic disease: current understanding and future perspectives. J Intern Med. 2016;280:339–349. doi: 10.1111/joim.12508. [DOI] [PubMed] [Google Scholar]
- 7.Larsen N, Vogensen FK, van den Berg FW, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS One. 2010;5:e9085. doi: 10.1371/journal.pone.0009085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang X, Shen D, Fang Z, et al. Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One. 2013;8:e71108. doi: 10.1371/journal.pone.0071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yassour M, Lim MY, Yun HS, et al. Sub-clinical detection of gut microbial biomarkers of obesity and type 2 diabetes. Genome Med. 2016;8:17. doi: 10.1186/s13073-016-0271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 11.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin J, Li Y, Cai Z, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490:55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen HK, Gudmundsdottir V, Nielsen HB, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 14.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dao MC, Everard A, Aron-Wisnewsky J, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 16.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20:779–786. doi: 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tap J, Furet JP, Bensaada M, et al. Gut microbiota richness promotes its stability upon increased dietary fibre intake in healthy adults. Environ Microbiol. 2015;17:4954–4964. doi: 10.1111/1462-2920.13006. [DOI] [PubMed] [Google Scholar]
- 20.Martinez I, Lattimer JM, Hubach KL, et al. Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J. 2013;7:269–280. doi: 10.1038/ismej.2012.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovatcheva-Datchary P, Nilsson A, Akrami R, et al. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab. 2015;22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Napolitano A, Miller S, Nicholls AW, et al. Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One. 2014;9:e100778. doi: 10.1371/journal.pone.0100778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gall WE, Beebe K, Lawton KA, et al. α-Hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;5:e10883. doi: 10.1371/journal.pone.0010883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekar S, Chandrasekaran A, Rao U, Sastry TP. Comparison of some of the physicochemical characteristics of type 2 diabetic and normal human bones: a sample study. J Diabetes Complicat. 2011;25:187–192. doi: 10.1016/j.jdiacomp.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 26.DeFronzo RA, Buse JB, Kim T, et al. Once-daily delayed-release metformin lowers plasma glucose and enhances fasting and postprandial GLP-1 and PYY: results from two randomised trials. Diabetologia. 2016;59:1645–1654. doi: 10.1007/s00125-016-3992-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buse JB, DeFronzo RA, Rosenstock J, et al. The primary glucose-lowering effect of metformin resides in the gut, not the circulation: results from short-term pharmacokinetic and 12-week dose-ranging studies. Diabetes Care. 2016;39:198–205. doi: 10.2337/dc15-1531. [DOI] [PubMed] [Google Scholar]
- 28.Kaska L, Sledzinski T, Chomiczewska A, Dettlaff-Pokora A, Swierczynski J. Improved glucose metabolism following bariatric surgery is associated with increased circulating bile acid concentrations and remodeling of the gut microbiome. World J Gastroenterol. 2016;22:8698–8719. doi: 10.3748/wjg.v22.i39.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evers SS, Sandoval DA, Seeley RJ. The physiology and molecular underpinnings of the effects of bariatric surgery on obesity and diabetes. Annu Rev Physiol. 2016;79:313–334. doi: 10.1146/annurev-physiol-022516-034423. [DOI] [PubMed] [Google Scholar]
- 30.Tremaroli V, Karlsson F, Werling M, et al. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22:228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lampe JW, Navarro SL, Hullar MA, Shojaie A. Inter-individual differences in response to dietary intervention: integrating omics platforms towards personalised dietary recommendations. Proc Nutr Soc. 2013;72:207–218. doi: 10.1017/S0029665113000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korpela K, Flint HJ, Johnstone AM, et al. Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PLoS One. 2014;9:e90702. doi: 10.1371/journal.pone.0090702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Chatelier E, Nielsen T, Qin J, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 34.Salonen A, Lahti L, Salojarvi J, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014;8:2218–2230. doi: 10.1038/ismej.2014.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Healey G, Brough L, Butts C, Murphy R, Whelan K, Coad J. Influence of habitual dietary fibre intake on the responsiveness of the gut microbiota to a prebiotic: protocol for a randomised, double-blind, placebo-controlled, cross-over, single-centre study. BMJ Open. 2016;6:e012504. doi: 10.1136/bmjopen-2016-012504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chambers ES, Viardot A, Psichas A, et al. Effects of targeted delivery of propionate to the human colon on appetite regulation, body weight maintenance and adiposity in overweight adults. Gut. 2015;64:1744–1754. doi: 10.1136/gutjnl-2014-307913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pingitore A, Chambers ES, Hill T, et al. The diet-derived short chain fatty acid propionate improves beta-cell function in humans and stimulates insulin secretion from human islets in vitro. Diabetes Obes Metab. 2016;19:257–265. doi: 10.1111/dom.12811. [DOI] [PubMed] [Google Scholar]
- 38.Plovier H, Everard A, Druart C, et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2016;23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 39.Zhang L, Bahl MI, Roager HM, et al. Environmental spread of microbes impacts the development of metabolic phenotypes in mice transplanted with microbial communities from humans. ISME J. 2016;11:676–690. doi: 10.1038/ismej.2016.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodman AL, Kallstrom G, Faith JJ, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011;108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khoruts A, Sadowsky MJ. Understanding the mechanisms of faecal microbiota transplantation. Nat Rev Gastroenterol Hepatol. 2016;13:508–516. doi: 10.1038/nrgastro.2016.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vrieze A, Van Nood E, Holleman F, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913-916 e917. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 43.Braat H, Rottiers P, Hommes DW, et al. A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin Gastroenterol Hepatol. 2006;4:754–759. doi: 10.1016/j.cgh.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, Guo L, Zhang Y, et al. Incorporation of therapeutically modified bacteria into gut microbiota inhibits obesity. J Clin Invest. 2014;124:3391–3406. doi: 10.1172/JCI72517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Jia H, Cai X, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 46.Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodrich JK, Davenport ER, Beaumont M, et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davenport ER, Cusanovich DA, Michelini K, Barreiro LB, Ober C, Gilad Y. Genome-wide association studies of the human gut microbiota. PLoS One. 2015;10:e0140301. doi: 10.1371/journal.pone.0140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonder MJ, Kurilshikov A, Tigchelaar EF, et al. The effect of host genetics on the gut microbiome. Nat Genet. 2016;48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 50.Turpin W, Espin-Garcia O, Xu W, et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet. 2016;48:1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- 51.Power RA, Parkhill J, de Oliveira T. Microbial genome-wide association studies: lessons from human GWAS. Nat Rev Genet. 2017;18:41–50. doi: 10.1038/nrg.2016.132. [DOI] [PubMed] [Google Scholar]
- 52.Franzosa EA, Hsu T, Sirota-Madi A, et al. Sequencing and beyond: integrating molecular ‘omics’ for microbial community profiling. Nat Rev Microbiol. 2015;13:360–372. doi: 10.1038/nrmicro3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPTX 225kb)
(PDF 106 kb)