Abstract
Betulinic acid (BetA) is a plant-derived pentacyclic triterpenoid that exerts potent anti-cancer effects in vitro and in vivo. It was shown to induce apoptosis via a direct effect on mitochondria. This is largely independent of proapoptotic BAK and BAX, but can be inhibited by cyclosporin A (CsA), an inhibitor of the permeability transition (PT) pore. Here we show that blocking apoptosis with general caspase inhibitors did not prevent cell death, indicating that alternative, caspase-independent cell death pathways were activated. BetA did not induce necroptosis, but we observed a strong induction of autophagy in several cancer cell lines. Autophagy was functional as shown by enhanced flux and degradation of long-lived proteins. BetA-induced autophagy could be blocked, just like apoptosis, with CsA, suggesting that autophagy is activated as a response to the mitochondrial damage inflicted by BetA. As both a survival and cell death role have been attributed to autophagy, autophagy-deficient tumor cells and mouse embryo fibroblasts were analyzed to determine the role of autophagy in BetA-induced cell death. This clearly established BetA-induced autophagy as a survival mechanism and indicates that BetA utilizes an as yet-undefined mechanism to kill cancer cells.
Keywords: cancer, betulinic acid, autophagy, LC3, apoptosis, necroptosis
Betulinic acid (BetA) is a naturally occurring triterpenoid with potent cytotoxic effects on cancer cells.1, 2, 3, 4 It was initially proposed to have a direct effect on the mitochondria and to induce apoptosis in a BCL-2-dependent fashion.5, 6, 7 However, BCL-2 overexpression can only provide short-term protection and eventually these cells do succumb to apoptosis. In addition, BAK/BAX double deficiency does not protect against BetA-induced cell death,8 indicating that cell death ensues independently of the BCL-2 family. Mitochondrial outer membrane permeabilization (MOMP), however, is crucial as cyclosporin A (CsA) and bongkrekic acid, both inhibitors of the permeability transition pore (PT-pore), and as a consequence MOMP were able to protect BetA-treated cells from releasing cytochrome c and subsequent apoptosis.7, 8 Downstream of the mitochondria caspases are activated, but direct inhibition of caspase activity with the pan-caspase inhibitor zVAD.fmk (N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone) did not offer protection to BetA-induced cell death.4 Combined this indicates that BetA does induce apoptosis, but that additional mechanisms must exist by which cell death is induced by the compound.
Alternative cell death pathways include necrosis, necroptosis, lysosomal membrane permeabilization (LMP) and autophagy. Necrosis is a form of passive cell death that is induced upon strong insults such as mechanical injury of cells. Typical features include swelling, rupture of organelle membranes as well as the outer cell membrane and as a result the cell contents are released, often causing inflammation in vivo.9 Necroptosis, a highly regulated form of necrosis, can be induced by ligation of death receptors in the presence of caspase inhibitors.10 Necroptosis is regulated by distinct complexes called the necrosome or ripoptosome that include receptor interacting protein (RIP) kinases, FAS-associated death domain and caspase-8.10, 11 Necroptosis can be blocked by small molecules, such as necrostatin-1, that allosterically block the kinase activity of RIP1.12, 13
Lysosomal cell death is induced by a destabilization of the lysosomal membrane.14 The cell death depends on the leakage of lysosomal components into the cytoplasm, and the activity of cathepsins that can induce a necrotic-like or apoptotic cell death depending on the extent of the leakage. As such the induction of cell death is clearly distinct from necrosis and apoptosis; nevertheless, the execution phase is similar.
Autophagy is a highly conserved cellular proteolytic degradation process in which cytosolic components are first encapsulated by a membrane and subsequently degraded in the lysosome, thereby providing new building blocks for the cell. Autophagy is typically induced upon nutrient starvation. However, autophagy also involves sequestration of damaged cytoplasmic components and organelles, via induction of de novo double-membrane vesicles (autophagosomes) that surround the cargo. The autophagosome transports the cargo to the lysosome where fusion of the autophagosome and lysosome leads to degradation of the cargo (autophagic flux).15 Autophagy, even though initially regarded as a cell survival mechanism, has also been suggested to serve a role in inducing cell death. It may therefore represent a balancing mechanism between cell survival and cell death.16, 17, 18 Autophagy can target cytosolic components specific for degradation, known as specific autophagy. Specific autophagy can include ubiquitinated proteins, peroxisomes and mitochondria.19, 20, 21 Mitophagy involves the selective degradation of mitochondria and is among others used for clearance of damaged mitochondria.17, 22 Previously, BetA and a derivative of BetA, B10, have been shown to induce autophagosome formation in multiple myeloma cells and glioblastoma cells.23, 24 In these studies it was suggested that the autophagic flux was prevented, leading to an accumulation of undigested autophagosomes. Although both studies observed autophagy, the role of autophagy as a cell death mechanism was not addressed for BetA.23, 24
We hypothesized that autophagy/mitophagy is induced upon BetA treatment to clear the damaged mitochondria. We show that autophagy is massively induced in various BetA-treated tumor cells, but is prevented by CsA, suggesting that autophagy occurs downstream of the BetA-induced mitochondrial damage. With the use of knockout and knockdown studies of key regulators of the autophagy pathway, we demonstrate that autophagy serves as a rescue pathway and is not responsible for the cell death induced by BetA.
Results
BetA-induced cell death is independent of apoptosis
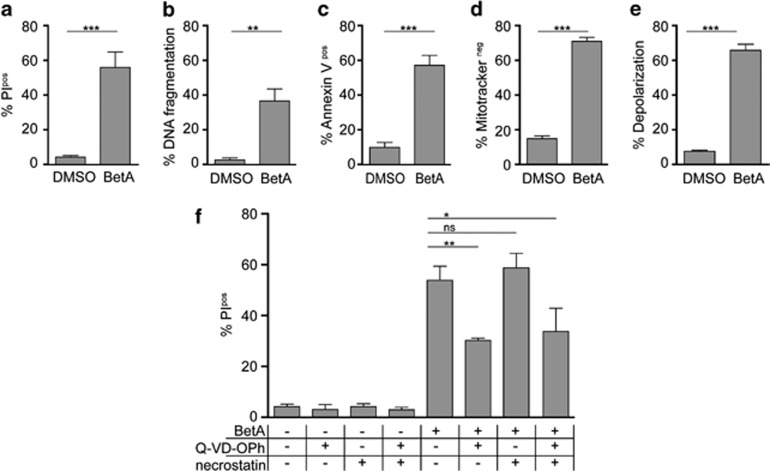
BetA induced a very potent form of cell death in HeLa cells that displayed features like cell membrane rupture (Figure 1a), apoptosis (Figures 1b and c) and mitochondrial depolarization (Figures 1d and e). The effect of BetA was, as shown before, concentration dependent, starting at 7.5 μg/ml and reaching a plateau at 10 μg/ml (Supplementary Figure 1). Previously we have shown, in Jurkat cells, that BetA-induced apoptosis is blocked by co-treatment with zVAD.fmk, but that this pan-caspase inhibitor does not prevent cell death as measured by propidium iodide (PI) exclusion.4 Similarly, cell death induced by BetA in HeLa cells was only partly blocked with a more potent caspase inhibitor Q-VD-OPh (Figure 1f), which did block apoptotic features like DNA fragmentation (results not shown). This indicates that caspase inhibition was effective, but not sufficient to block the induction of cell death. Thus, while apoptosis is induced by BetA, these findings indicate that alternative cell death pathways are activated as well. To study whether necroptosis serves as an alternative cell death mechanism induced by BetA, we made use of necrostatin, a specific inhibitor of this cell death mechanism. Either necrostatin alone or in combination with caspase inhibition failed to prevent cell death (Figure 1f). As a control for the action of necrostatin, U937 cells were used, which have been reported to undergo necroptosis upon tumor necrosis factor-α (TNF-α) treatment12 in the presence caspase inhibitors. In accordance with literature12 this indeed led to a significant induction necroptosis, which was prevented by necrostatin (Supplementary Figure 2). As this combination did not inhibit BetA-induced cell death, our data point to a cell death mechanism that is independent of caspases and necroptosis.
Figure 1.
BetA induces cell death independent of apoptosis and necroptosis. HeLa cells were subjected to 10 μg/ml BetA for 48 h after which cell death by PI exclusion (a), DNA fragmentation (b), phosphatidylserine (PS) exposure (c), and loss of mitochondrial activity by mitotracker orange (d) or JC-1 (e) were assessed (f) HeLa cells were pretreated for 1 h with or without 20 μM Q-VD-Oph and with or without 25 μM necrostatin-1 and subjected to 10 μg/ml BetA for 48 h after which cell death was assed via PI exclusion. Mean±S.D. of three independent experiments are shown, *P<0.05; **P< 0.01; ***P<0.001; NS, not significant
BetA induces autophagy
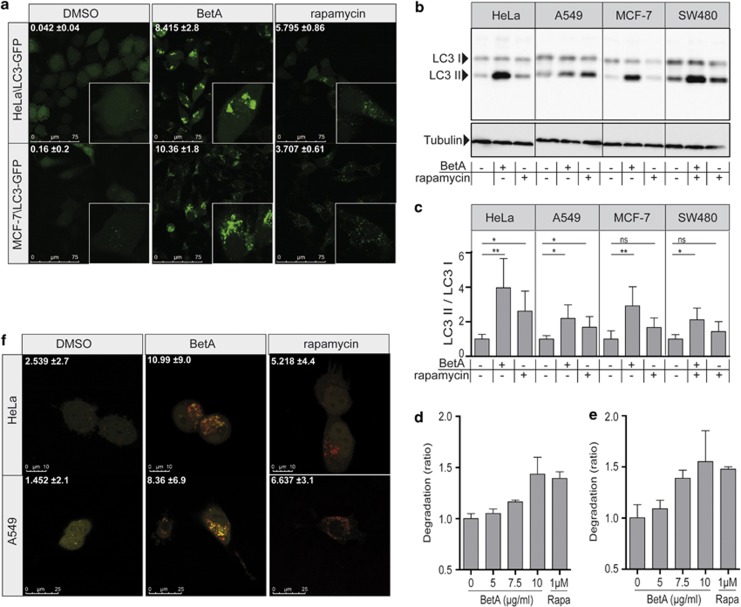
To determine whether autophagy is responsible for the cell death observed, LC3 conjugation was studied. Conjugated LC3 (LC3-II) is associated with autophagic membranes and therefore fusion of LC3 to green fluorescent protein (GFP) can be used to detect autophagosomes.25 In untreated cells, LC3–GFP is evenly dispersed in the cells, with a slight preference for the nucleus, whereas in cells where autophagy is induced, LC3 translocation is evident (Figure 2a).25 Both HeLa\LC3–GFP and MCF-7\LC3–GFP cells treated with the classical autophagy inducer rapamycin displayed a clear induction of punctuated LC3–GFP structures. Interestingly, BetA was even more potent in inducing autophagy, showing enlarged punctae (Figure 2a). To validate that BetA indeed induced autophagy, a separate measure to monitor the induction of autophagy was used, namely the processing of LC3-I to its PE-conjugated LC3-II using immunoblotting.26 Rapamycin treatment resulted in enhanced LC3-II levels in HeLa\LC3–GFP, A549, MCF-7\LC3–GFP and SW480\LC3-GFP cells as compared with their DMSO-treated counterparts (Figure 2b). Analyzing multiple independent assays consistently revealed this induction, but due to the variation in background levels of LC3-II it only reached significance in HeLa and A549 cells (Figure 2c). In contrast, BetA-induced LC3-II levels are significantly elevated in all cell lines and were clearly more pronounced as compared with rapamycin (Figures 2b and c), similar to the observations with LC3–GFP staining. The effect of BetA on autophagy occurs relatively rapid upon BetA treatment and required concentrations that are also exerting a toxic effect (7.5–10 μg/ml) (Supplementary Figures 3a and b).
Figure 2.
BetA induces autophagy. (a) HeLa\LC3–GFP cells and MCF-7\LC3–GFP cells were treated with 10 μg/ml BetA or 10 μM rapamycin and after 18 h cells were analyzed by confocal microscopy. Quantification of LC3 puncta was performed using Image J software. Mean of positive pixels/total pixel cell (%)±S.D. of three fields of view per sample are shown. (b) HeLa\LC3–GFP, A549, MCF-7\LC3–GFP and SW480\LC3–GFP were treated with 10 μg/ml BetA or 10 μM rapamycin for 18 h. LC3 processing was assessed via immunoblotting. Results for endogenous LC3 are shown. LC3-I (18 kDa) and LC3-II (16 kDa). Tubulin is used as a control. (c) Band intensity of western blot was quantified using Image J software. DMSO control is set to 1. Mean±S.D. of three independent experiments are shown, *P<0.05; **P< 0.01; NS, not significant. (d) MCF-7 cells were treated with the indicated concentrations of BetA or 1 μM rapamycin and after 14 h degradation of long-lived proteins was measured or (e) after 14 h medium was refreshed and degradation in the following 6 h was measured. Mean±S.D. of triplicates are shown. (f) HeLa and A549 cells with tandem fluorescent LC3 (mRFP–eGFP–LC3) were treated with 10 μg/ml BetA or 10 μM rapamycin and after 18 h cells were analyzed by confocal microscopy. Quantification of red LC3 puncta was performed using Image J software. Mean of positive pixels/total pixel cell (%)±S.D. of three fields of view per sample are shown
Both detection of LC3-II via immunoblotting and LC3–GFP translocation are measures for the amount of autophagosomes present at a certain time point, but do not provide information about the cause of this phenotype. It is possible that an increased number of autophagosomes is caused by an inhibition of basal autophagic flux rather than by induction of autophagy.27, 28 Previously, it was reported that BetA in KM3 cells dose-dependently induces the expression of LC3-II, but also of p62, a protein that is normally degraded during autophagy. This study suggested that BetA rather inhibited autophagic flux instead of inducing autophagy.24 To analyze the effect of BetA on p62 we studied its expression in HeLa cells. This confirmed a strong induction of p62 upon BetA treatment (Supplementary Figure 3c). However, this is due to induction of de novo synthesis and could be blocked with the addition of cycloheximide. The combination of cycloheximide and BetA did reveal degradation of p62, pointing to a functional flux (Supplementary Figure 3c). To better evaluate this autophagic flux, degradation of long-lived proteins was measured, which reportedly are at least in part degraded by autophagy.27, 28, 29 In agreement, rapamycin clearly induced degradation of these long-lived proteins after 14 h (Figure 2d). Moreover, degradation continued after this time point as was evident from analyzing the release of labeled amino acids in the time period between 14 and 20 h (Figure 2e). In contrast to the finding of Yang et al.24 that BetA prevents autophagic flux, BetA clearly increased the degradation of long-lived proteins after 14 h and the following 6 h after medium change. This was also more pronounced as compared to rapamycin-induced degradation (Figures 2d and e). Because degradation of long-lived proteins is not solely specific for autophagy and cannot discriminate between proteasomal- and autophagosomal degradation more specific assays using tandem RFP/eGFP-tagged LC3 as well as a tandem mCherry/GFP-tagged p62 were used.30 As RFP and mCherry are pH stable, they remain fluorescent after fusion of the autophagosomes with the lysosomal compartment, while eGFP fluorescence is lost. This allows detection of autophagic flux by simply observing the formation of red fluorescent lysosomes from green/red fluorescent autophagosomes. Using either LC3 (Figure 2f) or p62 (Supplementary Figure 3d) we observed that rapamycin as well as BetA induced a functional autophagic flux using. This confirms that BetA is a potent inducer of autophagy resulting in an enhanced autophagic flux.
Cyclosporin A blocks BetA-induced autophagy
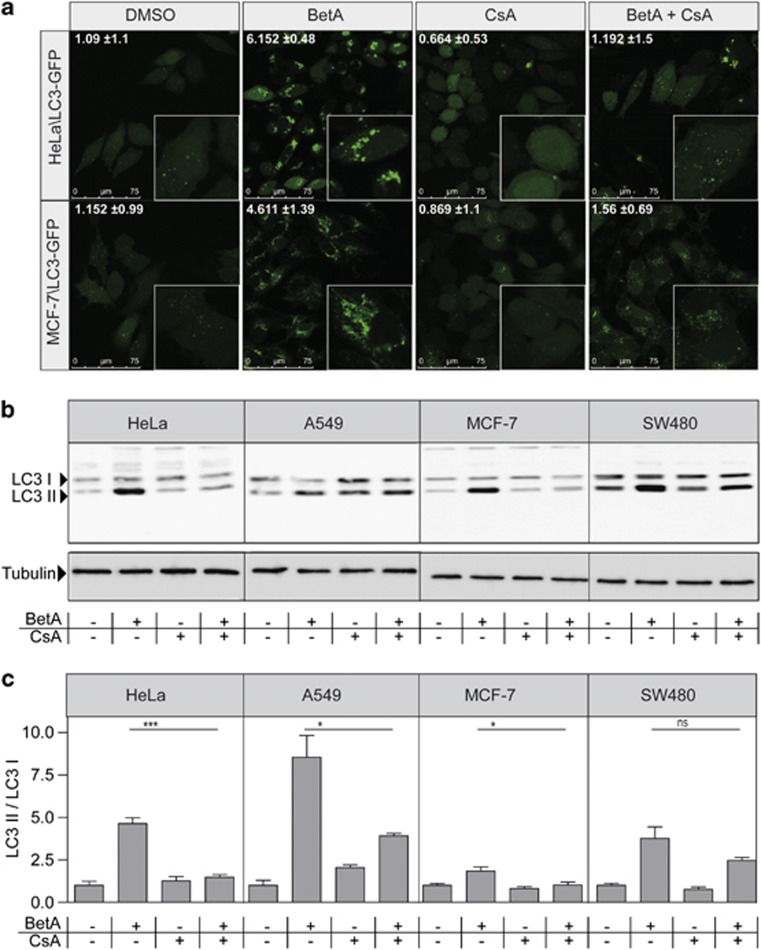
Permeability transition pore (PT-pore) opening results in membrane depolarization and leads to cytochrome c release and subsequent caspase activation.31 The proposed structure of the pore is formed by a voltage-dependent anion channel, adenine nucleotide translocator and cyclophilin D complex,32 and opening of the PT-pore can be blocked by inhibition of cyclophilin D using CsA.33 Previously we have shown that BetA-induced apoptosis and cytochrome c release proceeded in a PT-pore-dependent fashion.8 As CsA inhibits BetA-induced apoptotic features,8 the effect of CsA on BetA-induced autophagy was investigated. HeLa\LC3–GFP cells were treated with BetA alone or in combination with CsA and analyzed via confocal microscopy. Autophagosome formation was clearly inhibited in the presence of CsA (Figure 3a). A similar CsA dependency was observed in MCF-7\LC3 cells (Figure 3a). The effect of CsA was confirmed by immunoblotting for LC3 in BetA-treated HeLa, but also in other cancer lines (A549, MCF-7 and SW480). In all tested cell lines, BetA-induced formation of lipidated LC3-II was reduced when cells were pretreated with CsA (Figures 3b and c). These data suggest that BetA-induced autophagy is a consequence of mitochondrial damage triggered by BetA and can be prevented by inhibition of PT-pore opening.
Figure 3.
CsA inhibits BetA-induced autophagy. (a) HeLa\LC3–GFP cells and MCF-7\LC3–GFP cells were pretreated with 10 μg/ml CsA for 1 h and subjected to 10 μg/ml BetA for 18 h and analyzed by confocal microscopy. Quantification of LC3 puncta was perfomed using Image J software. Mean of positive pixels/total pixel cell (%)±S.D. of three fields of view per sample are shown. (b) HeLa\LC3-GFP, A549, MCF-7\LC3–GFP and SW480\LC3–GFP were pre–treated with 10 μg/ml CsA for one hour and subjected to 10 μg/ml BetA for 18 h. LC3 processing was assessed via immunoblotting. Results for endogenous LC3 are shown; LC3-I (18 kDa) and LC3-II (16 kDa). Tubulin is used as a control. (c) Band intensity of LC3 western blot was quantified using Image J software. DMSO control is set to 1. Mean±S.D. of three independent experiments are shown, *P<0.05; ***P<0.001; NS, not significant
Autophagy serves as a rescue pathway, not as an alternative cell death pathway in BetA-treated cells
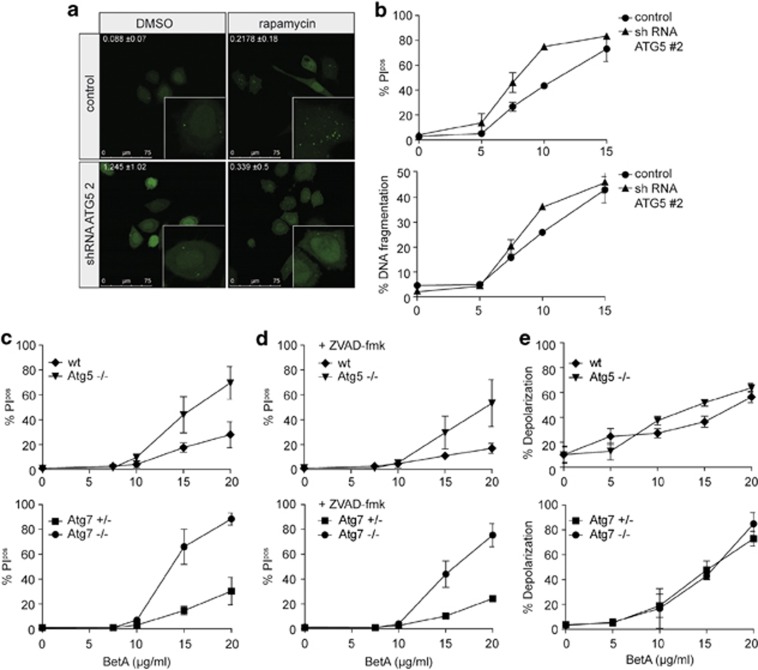
Autophagy, even though initially regarded as a cell survival pathway, can play a role in dying cells as well and has been suggested to serve as a balancing mechanism between cell survival and cell death.16, 17 Autophagy is detectable already at low BetA concentrations (7.5 μg/ml) and we reasoned that it is primarily induced as a rescue mechanism. However, it is feasible that at higher BetA concentrations autophagy is induced beyond a certain threshold, which shifts the balance from cell survival to cell death.17, 34 To test this hypothesis, we used cell lines with a deficiency in ATG5 or ATG7, which are crucial regulators in the induction of autophagy.35, 36 First we used retroviral shRNA against ATG5 in HeLa and HeLa\LC3–GFP cells. ATG5 knockdown levels of 77 and 86% were obtained with shRNA against ATG5 in HeLa and HeLa\LC3–GFP cells, respectively (Supplementary Figure 4a). The abrogation of autophagy in these cells was confirmed by confocal microscopy (Figure 4a). Importantly, when analyzing BetA-induced cell death in HeLa ATG5 knockdown cells, this was found, if anything, to be enhanced as compared to the cell death induced in control knockdown cells (Figure 4b). The increase in BetA-induced cell death in autophagy-impaired cells was also observed in MCF-7\LC3–GFP cells (Supplementary Figures 4a and b). To formally confirm that impaired autophagy enhances BetA-induced cell death, MEFs derived from Atg5 or Atg7 knockout mice were used. While these cells are completely autophagy deficient, they showed increased levels of BetA-induced cell death (Figure 4c) and Annexin V exposure (Supplementary Figure 4c). Interestingly, caspase inhibition in these autophagy-deficient cells did not prevent cell death either (Figure 4d). However, both autophagy-proficient and -deficient cells displayed a similar increase in mitochondrial depolarization (Figure 4e), suggesting that the initial mitochondrial insults are independent of the induction of autophagy. Combined these data indicate that autophagy serves primarily as a survival mechanism in BetA-treated cells and that BetA-induced cell death is independent of autophagy, necroptosis and caspases and follows an as yet-undefined pathway.
Figure 4.
Autophagy serves as a rescue pathway. (a) HeLa\LC3–GFP cells with pSuper empty (control) or ATG5 #2 knockdown were treated with 10 μM rapamycin for 6 h and analyzed with confocal microscopy. Representative pictures of two experiments are shown. Quantification of LC3 puncta was performed using Image J software. Mean of positive pixels/total pixel cell (%) ±S.D. of three fields of view per sample are shown. (b) HeLa cells with ATG5 #2 knockdown and the control were subjected to different concentrations of BetA for 48 h after which cell death was measured via PI exclusion and DNA fragmentation was assessed. Mean±S.D. of triplicates are shown. (c) Mouse embryonic fibroblasts lacking either ATG5 or ATG7 and their respective control cells were subjected to different concentrations of BetA, and cell death (24 h) using (c) PI exclusion was measured. (d) Mouse embryonic fibroblasts lacking either ATG5 or ATG7 and their respective control cells were subjected to different concentrations of BetA for 24 h in the presence or absence of 20 μM zVAD.fmk, and cell death using PI exclusion was measured. Mean±S.D. of three independent experiments are shown. (e) Loss of mitochondrial activity measured by JC-1 after 18 h was assessed. Mean±S.D. of three independent experiments are shown
Discussion
BetA is a promising anti-cancer agent with apoptosis-inducing effects that acts on the PT-pore in a BAX/BAK-independent fashion.8 Here we show that BetA highly efficiently induced autophagy in several cancer cells. Autophagic flux was found to be functional and led to enhanced degradation of long-lived proteins and degradation of GFP in tandem fluorescent LC3 and p62 proteins (Figure 2 and Supplementary Figure 3). Our observations are in contrast to previous findings by Yang et al.24 reporting that BetA inhibits autophagic flux in human multiple myeloma cells as measured by accumulation of p62 protein. Similar to BetA, B10, a semi-synthetic glycosylated derivative of BetA, induced cell death in both an apoptosis-dependent and apoptosis-independent fashion. In response to B10 treatment, LMP and subsequent release of lysosomal enzymes was observed in a human glioblastoma cell line, impairing autophagy in later stages.23 LMP results in the release of lysosomal hydrolases and cathepsins into the cytosol.37 Massive lysosomal leakage leads to uncontrolled necrosis, whereas minor LMP can activate either the intrinsic caspase-dependent apoptosis pathway or caspase-independent alternative cell death programs.38, 39, 40, 41, 42, 43 LMP could therefore also be a possible mechanism by which BetA induces cell death. However, in contrast to B10, which disrupts the autophagic flux, our data show that autophagy is enhanced after BetA treatment. It could well be that at later time points the flux is halted, potentially due to an overload of cargo. This could explain the relatively large LC3–GFP structures that are observed at later time points. Nevertheless, we show that the initial enhanced autophagic flux also serves a protective function as it delays the execution of cell death, while it does not appear to limit the mitochondrial depolarization. Blocking autophagy completely is detrimental to the cells confirming that the observed increase in autophagosomes and LC3-II is not simply due to a block in the flux.
An interesting connection between BetA-induced apoptosis and autophagy is the fact that both are efficiently inhibited by CsA, an inhibitor of the PT-pore8 (Figure 3) CsA has been reported to block mitophagy induced by loss of MOMP in rat hepatocytes.44, 45 Blocking of apoptosis and autophagy by CsA hints to a scenario in which both pathways are triggered by a common upstream event that is related to PT-pore opening. How this is achieved by BetA remains yet to be established. It is consistent though with the observations in autophagy-deficient cells, which do show a decrease in cell death, but not in mitochondrial defects. This suggests that the primary insult is independent of autophagy, but that the execution of cell death is dampened by an initial salvage response through the induction of autophagy. The fact that autophagy occurs relatively rapid upon BetA treatment is also in line with this notion.
Even though our results suggest that autophagy does not primarily serve as a cell death mechanism, it is conceivable that it plays an indirect role in the toxicity exerted by BetA. At higher concentration of BetA, autophagy may keep the balance up for a period of time, after which either the mitochondrial damage is too massive to be counteracted by autophagy or that lysosomal exhaustion occurs, which as a consequence will prevent further autophagic flux. This may then result in release of cytochrome c, activation of caspases and subsequent apoptosis. This is supported by the observations that BetA-induced cell death is higher in autophagy-deficient cells compared to control cells (Figure 4 and Supplementary Figure 4). Nevertheless, even under these conditions caspase inactivation is not sufficient to rescue the cells either, suggesting that the mitochondrial damage inflicted is not compatible with cellular survival.
Taken together our findings show that BetA-induced cell death is independent of caspases, necroptosis and autophagy and, thus, alternative cell death mechanisms must be involved.
This potentially explains the high tumoricidal efficacy of BetA, as it induces cell death that is not prevented by classical antiapoptotic mechanisms.
Materials and Methods
Chemicals/antibodies
Betulinic acid (BioSolution Halle, Halle, Germany, >99% purity) was dissolved in DMSO at 4 mg/ml and stored at −80 °C. PI and CsA were purchased from Sigma-Aldrich (St. Louis, MO, USA). Rapamycin was purchased from VWR (Amsterdam, The Netherlands) and C-14 valine (CFB75-50 μCi) from Amersham Biosciences (Amersham, UK). Q-VD-OPh was purchased from R&D Systems (Minneapolis, MN, USA). Necrostatin-1 and TNF-α were obtained from Enzo Life Sciences (Farmingdale, NY, USA). Anti-LC3 antibody (51520) was obtained from Abcam (Cambridge, UK). Anti-ATG5 antibody and anti-SQSTM1/p62 were from Cell Signaling Technology (Danvers, MA, USA). Anti-αTubulin antibody was from Santa Cruz Biotechnology (Dallas, TX, USA). Annexin V-APC and 7-AAD were from BD Biosciences (Franklin Lakes, NJ, USA). Mitotracker Orange CMTMRos and JC-1 were obtained from Life Technologies (Carlsbad, CA, USA).
Cell culture
HeLa cells, A549 cells, MCF-7 cells, SW480 and U937 cells were obtained from the ATCC (Manassas, VA, USA). MCF-7 cells overexpressing LC3–GFP were generated as described.46 HeLa cells and SW480 cells overexpressing LC3-GFP were made by stable transfection of plasmid pEGFP-C1-LC3, which was a gift from G. Kroemer (Institut Gustave Roussy, Villejuif, France). All LC3–GFP-overexpressing cell lines were cultured with 500 μg/ml G418 in order to select for LC3–GFP. ATG5 knockout and the corresponding control MEFs were kindly provided by N Mizushima (Department of Cell Biology, Okazaki, Japan)25 and the ATG7 knockout and corresponding control MEFs were obtained from M Komatsu (Department of Molecular Oncology, Tokyo, Japan).35
All cells were cultured in IMDM supplemented with 8% FCS, 2 mM L-glutamine, 40 U/ml penicillin and 40 μg/ml streptomycin.
RNA interference
Retroviral vectors pSuper ATG5 #2, pSuper ATG5 #3 and pSuper empty, a gift from S.W. Tait (Beatson Institute, Institute of Cancer Sciences, Glasgow, UK),47 were transfected into Phoenix Amphothrophic cells using the standard Ca2+-phosphate procedure. After 48 h, the virus containing medium was centrifuged at 1200 r.p.m. to purify virus from cell debris and supernatant was supplemented with 10 μg/ml polybrene. Target cells were plated at 70% confluence and allowed to attach overnight in standard medium. For infections, the culture medium was replaced with the virus containing medium and incubated for 8 h at 37 °C. Transduced cell populations were subsequently selected with 1–2 μg/ml puromycin, depending on cell type (2 μg/ml for HeLa cells and 1 μg/ml for MCF-7 cells). After 1 week of selection, expression of the targeted proteins was determined by western blot.
Cell death analysis
Cell death was determined by PI exclusion. In short, 50 000 cells were plated in a 24-well plate and allowed to attach overnight. Cells were treated for 48 h and harvested. The cell pellet was resuspended in 200 μl medium and stained with PI at 1 μg/ml just before measuring by flow cytometry. Samples were analyzed using FlowJo software (Tree Star Inc., Ashland, OR, USA). For quantification of apoptotic DNA fragmentation (Nicoletti assay) cells were resuspended in Nicoletti buffer48 supplemented with 50 μg/ml PI for 12 h, subsequent flow cytometric measurement of PI-stained nuclei was performed. Phosphatidylserine exposure during apoptosis was detected by annexin V assay. In short, 50 000 cells were plated in a 24-well plate and allowed to attach overnight. Cells were treated for 48 h and harvested. The cell pellet was stained with annexin V-APC and 7-AAD for 15 min at RT, subsequently flow cytometric measurement of annexin V and 7-AAD was performed.
Mitochondrial membrane potential analysis
Mitochondrial activity was measured with Mitotracker Orange CMTMRos. Treated cells were incubated for 30 min with 25 nM Mitotracker Orange at 37 °C. After staining, cells were washed and 7-AAD was added followed by cytometric analysis.
As a second mitochondrial membrane potential assay, JC-1 staining was used. Cells were stained for 30 min at RT with 4 μM JC-1. Depolarization was measured in FITC and PE channel by flow cytometry.
LC3 fluorescence microscopy
HeLa and MCF-7 cells overexpressing LC3–GFP were cultured on Poly-D-lysine-coated 24 mm diameter glass coverslips in six-well plates and treated with BetA, rapamycin and CsA for indicated time points and concentrations. The glass coverslips were mounted into a chamber of the microscope for live-cell imaging and Z-stack measurements and placed in a Leica DMI 6000 (TCS SP8) microscope (with adaptive focus, Motorized XY-Stage and SuperZ Galvo) and a case incubator at 37 °C. Samples were analyzed using Leica Las AF software. Quantification was performed with Image J49 generating a cut-off for the basal dispersed LC3 fluorescence, allowing for the quantification of fluorescence in the concentrated areas (LC3–GFP-associated autophagosomes).
Tandem fluorescence microscopy
HeLa and A549 cells were transiently transfected with tandem fluorescent LC3 (mRFP–eGFP–LC3, kindly provided by T Yoshimori30 using polyethylenimine (PEI) transfection (Brunschwig, The Netherlands). 24 h after transfection cells were treated with 10 μg/ml BetA or 10 μM rapamycin for 18 h. HeLa cells were transiently transfected with tandem fluoresecent p62 (pDest-mCherry-EGFP-p62, kindly provided by E Reits, AMC, The Netherlands), using PEI transfection. Twenty-four hours after transfection, cells were treated with 10 μg/ml BetA or 10 μM rapamycin for 12 h.
Degradation of long-lived proteins
MCF-7 cells were seeded in 12-well plates. The next day cells were labeled with C-14 valine (0.2 μCi/ml). After 24 h, a cold chase using cell culture medium only was performed before adding BetA or rapamycin for various time points. Supernatants and cells were collected separately and both were precipitated for 30 min on ice in 10% TCA (trichloroacetic acid). Precipitates were collected via centrifugation for 10 min at 10 000 r.p.m. and subsequently dissolved in 0.5 M NaOH. Radioactivity in supernatant and cell samples was measured and the ratio determined. The relative degradation ratio in the control cells was set to ‘one' and compared with the degradation ratios in BetA and rapamycin-treated samples.
Western blot analysis
Cells were lysed in 1 × RIPA lysis buffer (Thermo Fisher Scientific, Waltham, MA, USA) containing complete proteinase inhibitor (Roche, Penzberg, Germany) and subjected to protein quantification using a BCA kit (Pierce, Thermo Fisher Scientific, Waltham, MA, USA). A quantity of 25 μg protein per lane was applied for SDS-PAGE. Subsequent blotting was performed using a PVDF membrane (Amersham Biosciences, Amersham, UK). Membranes were blocked in 5% milk (blocking buffer) in TRIS phosphate-buffered saline solution containing TWEEN 20 (0.2%) (TBST-T) for 1 h at room temperature. Primary antibody incubations were performed in 1% milk/TBS-T (LC3 antibody, tubulin antibody) or 5% BSA/TBS-T (ATG5 antibody, p62 antibody and ERK1/2 antibody) overnight at 4 °C. Membranes were washed three times and incubated in blocking buffer for 1 h with a secondary HRP (horseradish peroxidase)-labeled antibody (anti-rabbit IgG (H+L) or anti-mouse IgG (H+L); Southern Biotech, Birmingham, AL, USA). For chemiluminescent visualization, Lumi-Light plus substrate (Roche, Penzberg, Germany) was used. Blots were analyzed by ImageQuant LAS4000. Band intensities were quantified using Image J software (NIH, Bethesda, MD, USA).
Statistical analysis
Statistical analyses were performed with Prism 5 (GraphPad Software, La Jolla, CA, USA) applying Student's t-test. Differences were considered significant with P-values <0.05 (*), <0.01 (**) and <0.001 (***).
Acknowledgments
We would like to thank G Kroemer (Institut Gustave Roussy, Villejuif, France) for the LC3–GFP construct, N Mizushima (Department of Cell Biology, Okazaki, Japan) and M Komatsu (Department of Molecular Oncology, Tokyo, Japan) for providing MEFs, SW Tait (Beatson Institute, Institute of Cancer Sciences Glasgow, UK) for the plasmids. The microscopy images were taken at the core facility Cellular Imaging/LCAM-AMC. We would like to thank DI Picavet for technical assistance, furthermore MF Bijlsma for his valuable scientific input and helpful discussions. This work was supported by a grant of the Stichting Nationaal Fonds tegen Kanker (SNFK), Amsterdam, The Netherlands.
Glossary
- ATG5
autophagy-related gene 5
- BetA
betulinic acid
- CsA
cyclosporin A
- GFP
green fluorescent protein
- LC3
light chain 3
- LMP
lysosomal membrane permeabilization
- MEFs
mouse embryonic fibroblasts
- MOMP
mitochondrial outer membrane permeabilization
- PI
propidium iodide
- PT-pore
permeability transition pore
- Q-VD-OPh
quinolyl-valyl-O-methylaspartyl-[2,6-difluorophenoxy]-methyl ketone
- RIP
receptor interacting protein
- TNF
tumor necrosis factor
- zVAD.fmk
N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by E Baehrecke
The authors declare no conflict of interest.
Supplementary Material
References
- Ehrhardt H, Fulda S, Fuhrer M, Debatin KM, Jeremias I. Betulinic acid-induced apoptosis in leukemia cells. Leukemia 2004; 18: 1406–1412. [DOI] [PubMed] [Google Scholar]
- Rzeski W, Stepulak A, Szymanski M, Sifringer M, Kaczor J, Wejksza K et al. Betulinic acid decreases expression of bcl-2 and cyclin D1, inhibits proliferation, migration and induces apoptosis in cancer cells. Naunyn Schmiedebergs Arch Pharmacol 2006; 374: 11–20. [DOI] [PubMed] [Google Scholar]
- Jung GR, Kim KJ, Choi CH, Lee TB, Han SI, Han HK et al. Effect of betulinic acid on anticancer drug-resistant colon cancer cells. Basic Clin Pharmacol Toxicol 2007; 101: 277–285. [DOI] [PubMed] [Google Scholar]
- Kessler JH, Mullauer FB, de Roo GM, Medema JP. Broad in vitro efficacy of plant-derived betulinic acid against cell lines derived from the most prevalent human cancer types. Cancer Lett 2007; 251: 132–145. [DOI] [PubMed] [Google Scholar]
- Fulda S, Friesen C, Los M, Scaffidi C, Mier W, Benedict M et al. Betulinic acid triggers CD95 (APO-1/Fas)- and p53-independent apoptosis via activation of caspases in neuroectodermal tumors. Cancer Res 1997; 57: 4956–4964. [PubMed] [Google Scholar]
- Fulda S, Susin SA, Kroemer G, Debatin KM. Molecular ordering of apoptosis induced by anticancer drugs in neuroblastoma cells. Cancer Res 1998; 58: 4453–4460. [PubMed] [Google Scholar]
- Fulda S, Scaffidi C, Susin SA, Krammer PH, Kroemer G, Peter ME et al. Activation of mitochondria and release of mitochondrial apoptogenic factors by betulinic acid. J Biol Chem 1998; 273: 33942–33948. [DOI] [PubMed] [Google Scholar]
- Mullauer FB, Kessler JH, Medema JP. Betulinic acid induces cytochrome c release and apoptosis in a Bax/Bak-independent, permeability transition pore dependent fashion. Apoptosis 2009; 14: 191–202. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P, Vanden Berghe T, Festjens N. Caspase inhibitors promote alternative cell death pathways. Sci STKE 2006; 2006: e44. [DOI] [PubMed] [Google Scholar]
- Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010; 11: 700–714. [DOI] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 2011; 43: 432–448. [DOI] [PubMed] [Google Scholar]
- Degterev A, Huang Z, Boyce M, Li Y, Jagtap P, Mizushima N et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat Chem Biol 2005; 1: 112–119. [DOI] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 2008; 4: 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aits S, Jaattela M. Lysosomal cell death at a glance. J Cell Sci 2013; 126(Pt 9): 1905–1912. [DOI] [PubMed] [Google Scholar]
- Parzych KR, Klionsky DJ An Overview of Autophagy: Morphology, Mechanism, and Regulation. Antioxid Redox Signal 2014; 20: 460–473. [DOI] [PMC free article] [PubMed]
- Saeki K, Yuo A, Okuma E, Yazaki Y, Susin SA, Kroemer G et al. Bcl-2 down-regulation causes autophagy in a caspase-independent manner in human leukemic HL60 cells. Cell Death Differ 2000; 7: 1263–1269. [DOI] [PubMed] [Google Scholar]
- Baehrecke EH. Autophagy: dual roles in life and death? Nat Rev Mol Cell Biol 2005; 6: 505–510. [DOI] [PubMed] [Google Scholar]
- Chen N, Karantza V. Autophagy as a therapeutic target in cancer. Cancer Biol Ther 2011; 11: 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H, Shvets E, Elazar Z. Biogenesis and cargo selectivity of autophagosomes. Annu Rev Biochem 2011; 80: 125–156. [DOI] [PubMed] [Google Scholar]
- Lee J, Giordano S, Zhang J. Autophagy, mitochondria, and oxidative stress: cross-talk and redox signalling. Biochem J 2012; 441: 523–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till A, Lakhani R, Burnett SF, Subramani S. Pexophagy: the selective degradation of peroxisomes. Int J Cell Biol 2012; 2012: 512721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 2007; 462: 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez P, Mader I, Tchoghandjian A, Enzenmuller S, Cristofanon S, Basit F et al. Impairment of lysosomal integrity by B10, a glycosylated derivative of betulinic acid, leads to lysosomal cell death and converts autophagy into a detrimental process. Cell Death Differ 2012; 19: 1337–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LJ, Chen Y, He J, Yi S, Wen L, Zhao J et al. Betulinic acid inhibits autophagic flux and induces apoptosis in human multiple myeloma cells in vitro. Acta Pharmacol Sin 2012; 33: 1542–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol 2004; 36: 2491–2502. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy 2007; 3: 542–545. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4: 151–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8: 445–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimore GE, Poso AR. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu Rev Nutr 1987; 7: 539–564. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3: 452–460. [DOI] [PubMed] [Google Scholar]
- Scorrano L, Ashiya M, Buttle K, Weiler S, Oakes SA, Mannella CA et al. A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell 2002; 2: 55–67. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell 2003; 112: 481–490. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Shimizu S. Role of the mitochondrial membrane permeability transition in cell death. Apoptosis 2007; 12: 835–840. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vicencio JM, Kepp O, Tasdemir E, Maiuri MC, Kroemer G. To die or not to die: that is the autophagic question. Curr Mol Med 2008; 8: 78–91. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005; 169: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T et al. The role of autophagy during the early neonatal starvation period. Nature 2004; 432: 1032–1036. [DOI] [PubMed] [Google Scholar]
- Petersen NH, Olsen OD, Groth-Pedersen L, Ellegaard AM, Bilgin M, Redmer S et al. Transformation-associated changes in sphingolipid metabolism sensitize cells to lysosomal cell death induced by inhibitors of Acid sphingomyelinase. Cancer Cell 2013; 24: 379–393. [DOI] [PubMed] [Google Scholar]
- Brunk UT, Neuzil J, Eaton JW. Lysosomal involvement in apoptosis. Redox Rep 2001; 6: 91–97. [DOI] [PubMed] [Google Scholar]
- Leist M, Jaattela M. Four deaths and a funeral: from caspases to alternative mechanisms. Nat Rev Mol Cell Biol 2001; 2: 589–598. [DOI] [PubMed] [Google Scholar]
- Guicciardi ME, Leist M, Gores GJ. Lysosomes in cell death. Oncogene 2004; 23: 2881–2890. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Jaattela M. Lysosomes and autophagy in cell death control. Nat Rev Cancer 2005; 5: 886–897. [DOI] [PubMed] [Google Scholar]
- Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene 2008; 27: 6434–6451. [DOI] [PubMed] [Google Scholar]
- Groth-Pedersen L, Jaattela M. Combating apoptosis and multidrug resistant cancers by targeting lysosomes. Cancer Lett 2013; 332: 265–274. [DOI] [PubMed] [Google Scholar]
- Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J 2001; 15: 2286–2287. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, He L, Lemasters JJ. Role of mitochondrial permeability transition pores in mitochondrial autophagy. Int J Biochem Cell Biol 2004; 36: 2463–2472. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T et al. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell 2007; 25: 193–205. [DOI] [PubMed] [Google Scholar]
- Colell A, Ricci JE, Tait S, Milasta S, Maurer U, Bouchier-Hayes L et al. GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 2007; 129: 983–997. [DOI] [PubMed] [Google Scholar]
- Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods 1991; 139: 271–279. [DOI] [PubMed] [Google Scholar]
- Hoogendijk AJ, Roelofs JJ, Duitman J, van Lieshout MH, Blok DC, van der Poll T et al. R-roscovitine reduces lung inflammation induced by lipoteichoic acid and Streptococcus pneumoniae. Mol Med 2012; 18: 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.