Abstract
Background
Limited translational genomic research currently exists to guide the availability, comprehension, and appropriate use of personalized genomics in diverse general population subgroups. Melanoma skin cancers are preventable, curable, common in the general population, and disproportionately increasing in Hispanics.
Objective
Variants in the melanocortin-1 receptor (MC1R) gene are present in approximately 50% of the population, are major factors in determining sun sensitivity, and confer a 2-to-3-fold increase in melanoma risk in the general population, even in populations with darker skin. Therefore, feedback regarding MC1R risk status may raise risk awareness and protective behavior in the general population.
Methods
We are conducting a randomized controlled trial examining Internet presentation of the risks and benefits of personalized genomic testing for MC1R gene variants that are associated with increased melanoma risk. We will enroll a total of 885 participants (462 participants are currently enrolled), who will be randomized 6:1 to personalized genomic testing for melanoma risk versus waiting list control. Control participants will be offered testing after outcome assessments. Participants will be balanced across self-reported Hispanic versus non-Hispanic ethnicity (n=750 in personalized genomic testing for melanoma risk arm; n=135 in control arm), and will be recruited from a general population cohort in Albuquerque, New Mexico, which is subject to year-round sun exposure. Baseline surveys will be completed in-person with study staff and follow-up measures will be completed via telephone.
Results
Aim 1 of the trial will examine the personal utility of personalized genomic testing for melanoma risk in terms of short-term (3-month) sun protection and skin screening behaviors, family and physician communication, and melanoma threat and control beliefs (ie, putative mediators of behavior change). We will also examine potential unintended consequences of testing among those who receive average-risk personalized genomic testing for melanoma risk findings, and examine predictors of sun protection at 3 months as the outcome. These findings will be used to develop messages for groups that receive average-risk feedback. Aim 2 will compare rates of test consideration in Hispanics versus non-Hispanics, including consideration of testing pros and cons and registration of a decision to either accept or decline testing. Aim 3 will examine personalized genomic testing for melanoma risk feedback comprehension, recall, satisfaction, and cancer-related distress in those who undergo testing, and whether these outcomes differ by ethnicity (Hispanic vs non-Hispanic), or sociocultural or demographic factors. Final outcome data collection is anticipated to be complete by October 2017, at which point data analysis will commence.
Conclusions
This study has important implications for personalized genomics in the context of melanoma risk, and may be broadly applicable as a model for delivery of personalized genomic feedback for other health conditions.
Keywords: genetic testing, primary care, online health education, melanoma prevention, skin cancer risk, genetic risk communication
Introduction
Melanoma is a rapidly increasing and preventable cancer in the general population. Melanoma incidence rates have increased more rapidly than any other cancer in the past several decades [1,2]. Melanoma accounts for 70% of skin cancer deaths each year [3], and is currently the fourth most common cancer among men and sixth most common among women, both in the Unites States [3] and in New Mexico [4]. Among Hispanics, disproportionate increases in melanoma (particularly thicker tumors with poorer prognoses) have been documented in states with high levels of year-round sun exposure, such as California and Florida [5-9]. For example, in 2010 Rouhani and colleagues [5] compared data from the Florida Cancer Data System (FCDS) with national incidence rates from the Surveillance, Epidemiology, and End Results (SEER) Program. Male Hispanics from the FCDS had a 20% higher incidence rate of melanoma between 1992 and 2004, relative to SEER. Nationally, incidence rates continue to rise among people of lower socioeconomic status and among older men [9-11]. In nonwhites, melanoma results in greater morbidity and mortality due to the disease often being identified at later stages, and because of low physician and patient awareness that melanomas occur in these populations [5,8,12-15]. By 2060, Hispanics will comprise 29% of the US population, further increasing the public health significance of melanoma in Hispanics [16].
Ultraviolet radiation delivered via sunlight is the predominant modifiable cause of melanoma, with approximately 65-90% of melanomas caused by ultraviolet radiation [17-19]. As such, melanoma risk reduction recommendations include daily sun protection, such as sun exposure avoidance, use of hats and clothing, and use of sunscreen [20]. However, most individuals do not use sunscreen, wear protective clothing, or seek shade on a regular basis [21], and in the United States, large general population surveys show that approximately 35% of the population uses sunscreen consistently [20,22,23]. This behavior extends to Hispanics of varying skin types [24,25], and Hispanics in the United States have high sunburn rates [26].
Personalized genomic testing for melanoma risk may promote risk awareness and risk reduction in the general population. Variants of the melanocortin-1 receptor gene (MC1R) confer moderate melanoma and basal cell cancer risks in the general population [27]. This gene is located on the long arm of chromosome 16 and is related to cutaneous pigmentation (eg, fair skin, red hair) [28-37]. A great deal of accumulated evidence, including systematic analyses of candidate genes, genome wide-association studies, and a recent meta-analysis of 12 melanoma case-control studies involving 6000 individuals [38], has identified nine risk-increasing variants for melanoma with odds ratios ranging from 1.42 (95% CI 1.09-1.85) to 2.45 (95% CI 1.32-4.55) [39].
Importantly, variation in MC1R is associated with melanoma risk after adjustment for hair color and skin type [32-34,40-42]. As such, MC1R predicts melanoma risk in African-American [43], Spanish [44], and Mediterranean populations [34], with at least one study indicating that MC1R may confer greater risk on individuals with darker skin, compared to those with lighter skin [45]. Across Hispanic and non-Hispanic populations, approximately 50% of individuals have at least one risk variant [40,45]. This frequency is consistent across Europe [46]. Hispanics in Albuquerque, New Mexico have substantial Spanish ancestry [47,48], so we expect to find the frequency of at least one risk variant to be 50% across Hispanic and non-Hispanic study participants [44].
The translation of personalized genomics into real-world general population application is necessary [49] but understudied [50]. The sequencing of the human genome [51] and the isolation of high-risk mutations in tumor suppressor genes has led to the rapid development of clinically useful genetic testing strategies for various uncommon hereditary cancer syndromes. Psychosocial research has highlighted predictors and outcomes of genetic testing in high-risk families who present in specialized clinics and receive extensive genetic counseling [52,53], and is increasingly addressing the needs of diverse, high-risk individuals and families [54,55]. However, since most research has been conducted in the context of familial disease, it is not clear how the general population will respond to personalized genomics. A 2016 report from the National Academy of Sciences has highlighted the pressing need to address access issues in genomic medicine [56]. Despite this need, for-profit companies are already marketing and offering genetic testing directly to consumers [57-60]. This model has largely bypassed behavioral research that could ensure broad utility and reach of this technology through diverse populations, arguing for the time-sensitive need to develop an empirical basis to maximize the benefits and minimize the harms of genomic feedback, even as evidence for specific gene variants and panels inevitably shifts over time [59].
Communication and health behavior theories inform the anticipated impact of personalized genomic testing for melanoma risk. We propose that the personal utility of personalized genomic testing for melanoma risk can be best understood via enhanced communication regarding skin cancer risk with physicians and family, and individual belief processes (ie, arousal of health threat and threat control beliefs). Communication with family and physicians concerning skin cancer risk might be especially important among some individuals, if family opinions are prioritized within a collectivist culture and value system [61]. Protection Motivation Theory [62,63] proposes that individual beliefs, including heightened illness threat (beliefs about severity, susceptibility) and heightened risk information about skin cancer, lead to protective health behaviors when control beliefs are high. Control beliefs involve confidence to perform the behavior (self-efficacy) and confidence in the effectiveness of the behavior (response-efficacy). This study tests the role of personalized genomic testing for melanoma risk in influencing control and illness threat beliefs and communication, which are proposed mediators of behavioral outcomes.
We will examine the reach (defined as consideration of the pros and cons of testing and registration of test decision) of a feasible, generalizable channel with high dissemination potential: the Internet. We will also compare factors (Hispanic ethnicity, health literacy, health system distrust, sociocultural factors) that may differentially impact reach. Over the past decade, the rapid pace of discovery of risk-influencing genes and the use of the Internet as an important source of health information have evolved in parallel. However, the use of the Internet for health information drops sharply and directly with literacy levels, so we will assess health literacy as a moderator of reach in this study [64]. To date, uptake of Internet direct-to-consumer personalized genomic testing has generally been concentrated among white, highly-educated, and health-literate consumers [65]. This disproportionate access is a clear media justice issue, as the continuation of these trends [65-67] could widen health knowledge gaps [68] and the digital divide [69,70] in underserved populations, in the context of personalized genomics.
Based on these findings, prior work in primary care populations [71,72], as well as research examining barriers to participation among minority individuals in general cancer prevention trials [73,74], we anticipate that Hispanics may be less easily reached by personalized genomic testing for melanoma risk [75]. However, the Internet gives engaged individuals a direct method of accessing health information on a breadth of topics, and represents one of the most frequent reasons that individuals consult the Internet. A 2012 Pew Research Center Survey found that most general Internet users (66% of Hispanics; 73% of non-Hispanic whites) used the Internet to find health information [76]. A recent study indicated that Hispanics are highly receptive to online cancer information [77]. In the case of personalized genomics, the Internet could provide needed privacy for individuals to consider the benefits and drawbacks of testing. Indeed, for-profit companies have attempted to capitalize on this potential, and this may lead to disproportionate utilization among those who distrust (and may seek to bypass) the health system. Therefore, distrust of the health system, found to be highly relevant in minorities [78,79], may differentially impact reach. Finally, Hispanic sociocultural factors that are known to influence cancer prevention and screening activities [80], including cancer fatalism [81,82], an orientation to health that prioritizes the family over the individual [83], and specific misperceptions about skin cancer that are more common in Hispanics than non-Hispanics [15,84], may help us examine reasons for differential personalized genomic testing for melanoma risk reach. Examining these factors will help explain differences in reach by ethnic group, and thus provide critical direction in future personalized genomic testing for melanoma risk modifications for broad dissemination (see Figure 1).
Figure 1.
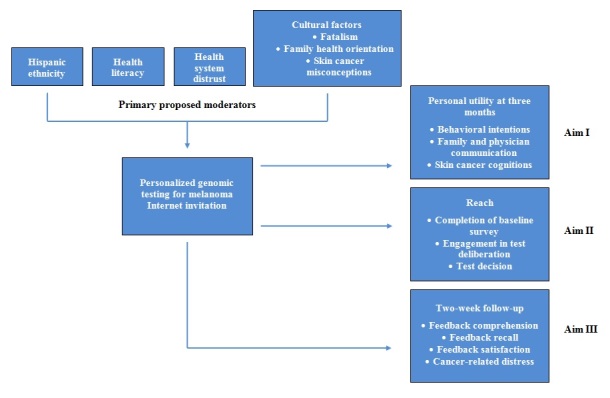
Conceptual model.
Specific Aims
This randomized controlled trial, “SOMBRA: Skin health Online for Melanoma: Better Risk Assessment” examines Internet presentation of the risks and benefits of personalized genomic testing for melanoma risk versus wait-list controls who are not offered testing. The study will compare personal utility and reach in a general population of English- or Spanish-speaking cohort in Albuquerque, New Mexico, which experiences year-round sun exposure.
In Aim 1, we will examine the personal utility of personalized genomic testing for melanoma risk in terms of short-term (3 months after testing) sun protection and skin screening (ie, behaviors), communication, and melanoma threat and control beliefs (ie, putative mediators of behavior change). Guided by Protection Motivation Theory [62], we hypothesize that behaviors and putative mediators will be higher in those who test, compared to those who decline testing or wait-list controls. Given that an important challenge of personal genomics involves the potential for those who receive negative genetic feedback to increase risky behaviors [85], we will also examine this potential unintended consequence of testing. To do so, we will conduct a subgroup analysis among those who receive average-risk personalized genomic testing for melanoma risk findings, and examine sun protection at 3 months as the outcome. Predictors will include baseline melanoma threat and control beliefs, melanoma risk factors, and demographics. These findings may be used in future studies to develop messages for groups that receive average-risk feedback, which accounts for large segments of those tested for moderate risk susceptibility factors across many diseases.
In Aim 2, we will examine differential reach of personalized genomic testing for melanoma risk between Hispanics and non-Hispanics, and potential explanations for any differential reach. Additional assessments of reach include baseline survey completion and the decision to pursue personalized genomic testing for melanoma risk testing. For the reasons listed above, we hypothesize that those who self-identify as Hispanic will show reduced reach, and that this reduction will in fact be the product of differences between Hispanics and non-Hispanics in relation to health literacy, health system distrust, and sociocultural factors [80], including cancer fatalism [81], family health orientation [83], and skin cancer misperceptions [15,84]. These results will inform future personalized genomic testing for melanoma risk modifications for Hispanics.
Finally, in Aim 3, among those who undergo testing we will examine test comprehension, recall and satisfaction, and cancer-related distress two weeks after test receipt. We will also examine whether these outcomes differ by ethnicity (Hispanic vs non-Hispanic) health literacy, health system distrust, sociocultural, or demographic factors.
Methods
Website Development and Usability Testing
Klein Buendel, Inc., a company specializing in health education programs and multimedia products in chronic disease prevention and control, provided the Web-based computer interface for the personalized genomic testing for melanoma risk education modules and testing invitation. Dr. David Buller (a study coinvestigator) is the Klein Buendel Research Director and an expert in skin cancer communications strategies. The Multiplex Study led by the National Human Genome Research Institute developed an Internet website where participants could opt to undergo genomic testing and risk feedback for common diseases, including the MC1R gene for melanoma risk, that was highly comprehensible, accurately interpreted, and did not increase distress in a primary care population [86,87]. We adapted these materials for our current study, and the modules are: (1) What genetic testing can and cannot tell you, (2) Skin cancer and genes, (3) Your rights if you take part in genetic research, and (4) Your decision to be tested or not. The website retains each of the four feasible Multiplex Study educational modules, with comprehension questions contained inside a website interface. The interface includes help files, navigation devices, and data collection code. The interface was tested and beta-tested by data professionals at Klein Buendel for stability and accuracy, and is hosted on secure data servers at Klein Buendel. Participants randomized to the personalized genomic testing for melanoma risk study arm view these materials via the Internet. To assess website usability, we conducted semistructured interviews (n=9) with English-speaking (n=8) and Spanish-speaking (n=1) primary care patients at 1209 Clinic, a University of New Mexico (UNM) General Internal Medicine Clinic that represents the primary recruitment site for the study. Any issue raised by at least one participant or study team member was evaluated for revision. Overall, participants found the website usable with 18 problems identified (eg, web pages with too much text, confusing wording, and unclear instructions). The research team developed solutions for these problems, which were confirmed with Klein Buendel before implementation.
Spanish Translation and Cognitive Interviews
We followed published guidance for translation and cognitive interviewing drawn from Translation, Review and Adjudication, Pretesting, and Documentation procedures [88,89]. First, the Memorial Sloan Kettering Cancer Center Linguistic and Cultural Competence Team (led by Mr. Javier Gonzalez and Dr. Francesca Gany, coinvestigators) within the Immigrant Health and Cancer Disparities Service provided Spanish translations and certificates of authenticity of all study materials, including: study invitation flyer, baseline survey, personalized genomic testing for melanoma risk Internet educational modules and corresponding knowledge surveys, buccal cell sample provision instructions, risk feedback comprehension assessment, 3-month telephone outcome survey, and consent forms. We internally reviewed the documents and provided fine-tuning.
Next, we conducted semistructured cognitive interviews (n=28) to assess the comprehension and acceptability of translated study materials with our target population (primary care Spanish-speaking Hispanic patients at 1209 Clinic at UNM) stratified across gender and education levels (≥high school, <high school). Bilingual interviewers administered only a portion of the materials to each participant to reduce patient burden and maximize completion with at least two patients viewing every item. If any issues were raised by at least one participant, a research assistant or the investigator panel (multidisciplinary team composed of experts in qualitative data analysis, linguistic translation, health and genetic literacy, and anthropology) evaluated the item for revision and labeled it as a problem [90]. Procedural details and results are reported elsewhere [91]. Most materials were comprehensible and acceptable, but 33 of 246 terms/concepts were not. These items were modified by the multidisciplinary team and retested. During this phase the team adopted the term skin cancer rather than melanoma in Internet and risk communication materials, due to the greater comprehensibility of this term in the translation and cognitive interviews.
Randomized Controlled Trial
In our ongoing randomized controlled trial, our bilingual Research Study Assistants approach primary care patients in UNM General Internal Medicine clinics with invitation flyers (English and Spanish) and National Cancer Institute skin cancer information for diverse skin types (available in English and Spanish versions; “ Anyone can get skin cancer ”). Patients are eligible for the study if they have been registered in any UNM clinic for at least six months, assigned a primary care physician in the UNM system, are aged ≥18 years, and are fluent in English or Spanish. We originally limited recruitment to the 1209 Clinic, but expanded to other UNM clinics to boost recruitment rates. If patients are eligible and interested in study participation, they complete the Baseline Survey (including an informed consent form) in-clinic via a semiprivate space with a Research Study Assistant who enters their responses on a dedicated study computer (tablet or laptop computer with wireless Internet access). In prior preliminary studies with UNM primary care patients, we found high levels of receptivity to skin cancer genomic information, yet higher skin cancer misconceptions than in the general population, making this an appropriate study context [84].
If patients are eligible but not interested in participating, we assess reasons for study refusal and ask them to complete a one-minute Refuser Survey (skin cancer risk perceptions, interest in genomic technologies, and demographics). Based on the Multiplex Study [86,87], we expect a 30% baseline survey response rate, for a total sample size of 885. After completion of the Baseline Survey, participants receive US $15 for their time and effort, and either a referral to consider personalized genomic testing for melanoma risk through a secure website or wait-list control (randomized 1:6; balanced across Hispanic vs non-Hispanic ethnicity; n=135 in control arm, n=750 in personalized genomic testing for melanoma risk arm). Hispanic ethnicity will be recorded by self-report. Trial design and reporting will adhere to The Consolidated Standards for Reporting Trials Statement [92,93]. Patients will choose Spanish or English study materials, and we will record their preference.
After completing the baseline survey in-clinic, all participants randomized to consider personalized genomic testing for melanoma risk are given an introductory letter inviting them to log onto the study website at their earliest convenience (preferably within the next month) to read the four educational modules regarding personalized genomic testing for melanoma risk, and to answer a series of questions regarding comprehension of, and satisfaction with, the content of each module. In section 4, participants register a test decision. Participants are only able to register a test decision if they have already read and completed the questions in the educational modules. Those who complete these steps receive a US $5 gift card for each educational module completed, for a total of US $15 in gift cards. Registration of a test decision (yes vs no) is our primary assessment of reach in this study. We expect a minimum of 30% of participants who complete the Baseline Survey to register a test decision, and that this will reach 50% in some subgroups, including those with higher literacy, and non-Hispanic subgroups [94]. Additional assessments of reach include completion of the baseline survey and decision to pursue personalized genomic testing for melanoma risk testing (yes vs no). Those who register a decision to proceed with testing will receive deoxyribonucleic acid (DNA) buccal cell test kits which will allow them to provide a saliva sample for genetic testing, postage prepaid envelopes, and instructions for buccal cell collection. Participants can return their kits at any point. Genetic counseling sessions are available at the participants’ request. In accordance with The Multiplex Study [72], we anticipate that 50% of those who consider personalized genomic testing for melanoma risk will return a saliva sample for genetic testing. Testing will be conducted on samples that are received by the lab and results will be mailed within one month.
Genomic DNA will be isolated from buccal cells using Oragene (Ottawa, ON, Canada). The Oragene kit generally provides at least 110 micrograms of high quality DNA. Standard polymerase chain reaction will be used to amplify the 951-nucleotide MC1R coding region. All amplified products will be sequenced on an ABI Prism 3100 (Applied Biosystems, Foster City, CA) using BigDye Terminators (Applied Biosystems) according to manufacturer’s specifications. Sequencing primers are:
5’-TCGTCTTCAGCACTCTCTTC-3’
5’-TTTAAGGCCAAAGCCCTGGT-3’
5’-AACCTGCACTCACCCATGTA-3’
5’-CTGCAGGTGATCACGTCAAT-3’
MC1R chromatograms will be read with the aid of Sequencher software version 4.05 (Gene Codes Corp., Ann Arbor, MI) and/or SeqScape software versions 1.0 to 2.1.1 (Applied Biosystems). These data will be read independently by two reviewers. This procedure is standard and has been used in most recent studies examining DNA isolation. We will sequence the coding region of MC1R in participants’ germline DNA and identify all variants. MC1R variants are typically classified as their risk for red hair (“R” variant) and low risk (“r” variant). The MC1R variants identified will be characterized as: (1) nonsynonymous or synonymous, (2) coding or noncoding, and (3) MC1R variants that are strongly associated with melanoma risk and represented as V60L, D84E, V92M, R142H, R151C, I155T, R160W, R163Q, and D294H; the nine variants that are associated with melanoma, regardless of skin type [41]. Genotype definitions used in these analyses are adapted from work exploring the association between MC1R variants and melanoma risk [39]. We estimate that 50% of participants will receive findings concerning at least one MC1R risk variant [40,45].
To provide risk feedback, molecular genotypes are combined into two categories: (1) average-risk feedback, the presence of no MC1R risk alleles associated with risk of melanoma; or (2) higher risk feedback, the presence of at least one MC1R risk allele, which is associated with risk of developing melanoma (range of 1.47 [95% CI 1.17-1.84] to 2.74 [95% CI 1.53-4.89]). We employ state-of-the-art methods of risk communication used with high comprehension in the Multiplex Study [86,87]. These materials include verbal and picture displays of risk information [95,96], given that people often neglect base rates for a disease and have difficulty understanding joint probabilities and shifting denominators [97]. We will also provide written information to clarify the bottom line information, given that individuals tend to rely on the gist of the information [97,98].
All participants who undergo personalized genomic testing for melanoma risk will receive a follow-up call two weeks after results are mailed to them, to assess result comprehension and potential distress (Risk Feedback Comprehension Assessment). All participants who complete this assessment will receive a US $5 gift card. Based on prior literature documenting low levels of distress in individuals undergoing genetic testing for high-risk mutations [99], and those found to carry Cyclin-Dependent Kinase Inhibitor 2A mutations indicating high melanoma risk [100], we expect low levels of distress regarding personalized genomic testing for melanoma risk feedback. Those who report high distress will be referred for follow-up by Clinic Director Dr. Jessica Bigney, who addresses distress issues in the UNM clinic.
All participants who complete Baseline Assessments (whether tested or not) are contacted by telephone after 3 months. Participants who complete the follow-up survey receive a US $15 gift card. See Figure 2 for study flow. Measures are outlined in Multimedia Appendix 1.
Figure 2.
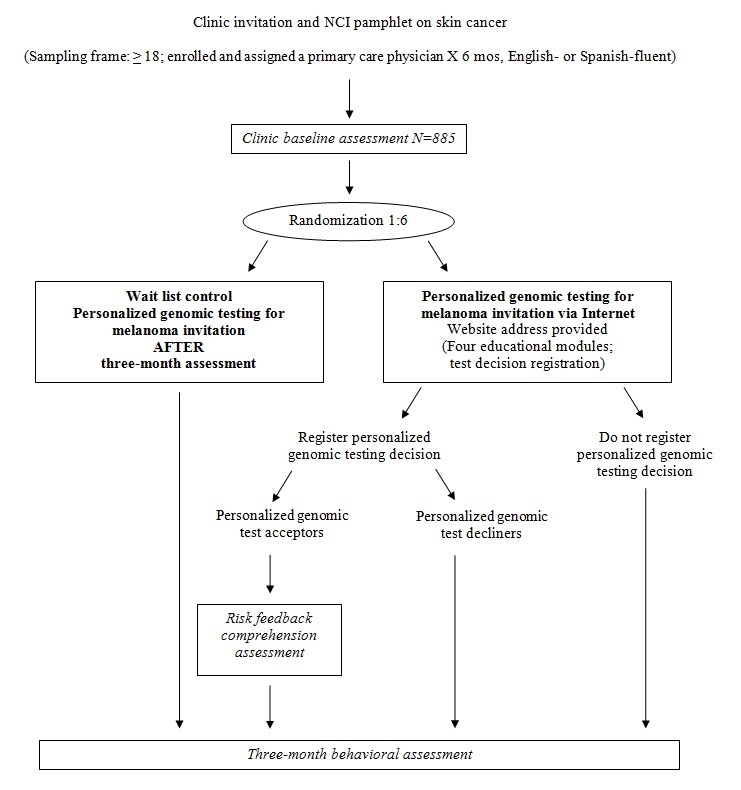
Study flow. PGT-M: personalized genomic testing for melanoma.
Data Analysis
We will examine four aspects of data quality and distributional assumptions: (1) data skewness, kurtosis, and parametric assumptions; (2) intention-to-treat principles; (3) missing data considerations; and (4) control of potentially inflated type-1 errors due to multiple statistical tests. We assume that up to 20% of the respondents will be unreachable at follow-up; missing assessments may be amenable to imputation by several techniques [101,102].
We will use a series of regression analyses to examine the personal utility of personalized genomic testing for melanoma risk. For these analyses, the dependent variables will be: short-term (3-month) sun protection and skin screening (ie, behaviors); communication; and skin cancer threat and control beliefs (ie, putative mediators of behavior change) [62,63,103]. Given guidelines regarding the importance of consistent use of sunscreen [3], patient-reported sunscreen use frequency will be dichotomized (frequent or more vs sometimes or less) to indicate consistent versus inconsistent use. Our expected approximate sample size for this analysis will be 708, assuming 20% attrition of our original 885 participants. Personalized genomic testing for melanoma risk uptake status will be categorized into three groups: (1) those who undergo personalized genomic testing for melanoma risk (acceptors), (2) those who decline personalized genomic testing for melanoma risk (decliners), and (3) those who are not offered personalized genomic testing for melanoma risk (controls). A logistic regression model will be used for sun protection outcomes that are dichotomous (eg, consistent use vs inconsistent use) as a function of personalized genomic testing for melanoma risk uptake. The baseline outcome assessments will be entered as covariates to provide control for ceiling and floor effects. Primary moderators will be considered, including ethnicity (Hispanic vs non-Hispanic), health literacy, health system distrust, sociocultural factors (cancer fatalism, family health orientation, skin cancer misconceptions), and high-risk versus average-risk personalized genomic testing for melanoma risk feedback, as well as demographics and skin cancer risk factors. Average ultraviolet index over the 3-month assessment time period will be considered as a covariate to provide control over seasonal variations in Albuquerque. The hypothesis is supported if there is a significant difference in sunscreen use between personalized genomic testing for melanoma risk acceptors and personalized genomic testing for melanoma risk decliners or controls, such that acceptors show greater sunscreen adherence.
Decreased sun protection may be an unintended consequence of testing among those who receive average-risk personalized genomic testing for melanoma risk feedback [85]. To examine this possibility, we will conduct a subgroup analysis among those who receive average-risk personalized genomic testing for melanoma risk feedback, and examine sun protection outcomes at 3 months as the outcome. Predictors will include baseline skin cancer threat and control beliefs, melanoma risk factors, demographics, and sociocultural factors (health literacy and health system distrust, cancer fatalism, family health orientation, and skin cancer misconceptions). Given the expected sample size for the analysis (n=60; participants who undergo testing and receive average-risk personalized genomic testing for melanoma risk feedback), we will use univariate analyses to guide predictor selection. Nonparametric statistics will be considered when appropriate to guard against violations of parametric assumptions in this restricted sample.
We will examine differential reach of personalized genomic testing for melanoma risk across Hispanics and non-Hispanics, and potential explanations for any differential reach. Reach is defined as registration of a personalized genomic testing for melanoma risk test decision, either accepting or declining testing (dichotomous outcome; test decision or no test decision). Additional assessments of reach include baseline survey completion and decision to pursue personalized genomic testing for melanoma risk testing. Only participants randomized to the personalized genomic testing for melanoma risk arm–those who are offered personalized genomic testing for melanoma risk–will be included in this analysis. Our sample size for this analysis will be 600, assuming 20% attrition of the original 750 who are offered personalized genomic testing for melanoma risk. We aim to offer putative explanations, such as differences in health literacy or skin cancer misconceptions concerning why Hispanics offered personalized genomic testing for melanoma risk may be less likely to register a personalized genomic testing for melanoma risk decision. This analysis will involve a moderation framework [104,105], such that reduced reach in Hispanics is moderated by one or more third variables (eg, skin cancer misconceptions). We will use a logistic regression modeling framework to address Aim 2. A standard requirement in moderation analysis [105] entails two sequential statistical findings: (1) there should first be a statistically significant Hispanic effect in Model 1; and (2) after adjusting for the moderator of interest in Model 2, the previously significant main Hispanic effect will no longer be significant. This approach may be applied to a variable coding Hispanic (yes vs no) and a moderator variable such as skin cancer misconceptions. This analysis will be applied to other putative explanations of why Hispanics might be less likely to register a personalized genomic testing for melanoma risk decision.
Among personalized genomic testing for melanoma risk test acceptors, we will examine (two weeks after test result receipt) test comprehension, recall, satisfaction, and distress. Our sample size for this analysis will be approximately 90, given that we expect to reach 80% (90/114) of those who undergo personalized genomic testing for melanoma risk testing, and thus receive personalized genomic testing for melanoma risk feedback. Based on the Multiplex Study [86,87], we anticipate that personalized genomic testing for melanoma risk feedback will be read by at least 80% of participants who undergo personalized genomic testing, and that at least 80% will correctly recall and accurately interpret their results. We anticipate that most participants (>95%) will report low levels of distress, including nervousness, testing regret, fear, and confusion. We will examine these outcomes using bivariate statistics, and examine differences across ethnicity, health literacy, health system distrust, and sociocultural factors.
Statistical Power
Regarding personal utility in Aim 1, we hypothesize that there will be higher rates of sunscreen use in personalized genomic testing for melanoma risk test acceptors, compared to personalized genomic testing for melanoma risk decliners or controls. We predict that those who accept personalized genomic testing for melanoma risk will have higher levels of sunscreen use (65% regular sunscreen use, consistent with rates of sunscreen use in those with melanoma risk factors [106]), compared to 35% sunscreen use in decliners or controls (consistent with rates of sunscreen use in the general population [20,22,23]). We estimated the statistical power in personal utility between personalized genomic testing for melanoma risk accepters compared to decliners or controls. The comparison between a 65% versus 35% difference in sunscreen use was carried out using Cohen’s method [107]. An estimated 65% versus 35% contrast translates to an arcsine-transformed effect size index of 0.61 [107], which yields a statistical power of 99.7% in a hypothesis test of these two proportions between personalized genomic testing for melanoma risk acceptors (an estimated n=90 after 20% attrition) and decliners (n=509 after 20% attrition), at a two-sided test with a tail probability of 0.01 (lower than the conventional 0.05 tail probability to reserve power for subset analyses).
Regarding the outcome of reach in Aim 2, we hypothesize that Hispanics will show reduced reach, but that differences in health literacy, health system distrust, and sociocultural factors (cancer fatalism, family health orientation, skin cancer misconceptions) will explain these findings. This method involves testing a moderation relationship in a logistic regression model. To estimate the statistical power, we ran 400 simulated logistic regression models, assuming Hispanics at 50% of the sample, and that within the Hispanic group high skin cancer misconceptions would be associated with a 0.38 odds ratio in personalized genomic testing for melanoma risk test registration; high skin cancer misconceptions would be associated with lower personalized genomic testing for melanoma risk registration. When converted to Cohen d, this 0.38 odds ratio translates to an effect size of -0.54, which Cohen considers a medium effect size. Based on the Multiplex Study [86], we estimate a minimum of 30% of participants will register a personalized genomic testing for melanoma risk decision among Hispanics, and a higher reach of 50% among non-Hispanics. This 30% versus 50% difference translates to a Cohen effect size of 0.50 [107]. We estimate 81% statistical power to detect a medium effect size at a conventional two-sided type-1 error rate of 5% for a moderator analysis.
In sum, we have adequate power for personal utility and reach to (1) ensure adequate representation of individuals with low health literacy, (2) ensure robust protection against missing data, and (3) ensure sufficient statistical power to detect moderation. If the effect size is larger than estimated, such as a 30% versus 80% difference and a Cohen effect size of 0.65, then we would have power to spare for additional comparisons.
Results
To date, 462 participants have been recruited to the study (203/462, 43.9% Hispanic; 222/462, 48.1% non-Hispanic white; 356/462, 77.1% female; mean age=54) and randomized 1:6 to usual care or the personalized genomic testing for melanoma risk offer. Final outcome data collection is anticipated to be complete by October 2017, at which point data analyses will commence.
Discussion
This study is one of the first population-based efforts to widen the reach of personal genomics in real-world settings. Along with other work examining ways to maximize the use of the Internet to bring emerging technologies [108] (including genomics [109]) to the general population, our research will use genomic information to raise melanoma risk awareness and prevention and control behaviors for this rapidly increasing cancer, which is extremely hard to treat when diagnosed beyond stage 1. We use a rigorous randomized controlled trial design, which increases the rigor of the proposal by comparing those who undergo personalized genomic testing for melanoma risk testing to: (1) those who have declined, and (2) those who have not been offered personalized genomic testing for melanoma risk testing. We use an established and feasible approach to Web-based communication regarding skin cancer genetic testing, and measure utility and reach using a real-world approach by which the general population may realistically access it (Internet personalized genomic testing for melanoma risk invitation). We have identified a highly diverse population for assessment in a geographical location that is exposed to year-round sun exposure.
The current proposal will examine potential unintended consequences of actual genetic testing, by directly examining those who receive personalized genomic testing for melanoma average-risk results to identify predictors of behavior change in this group. Examination of this question will have important implications for personalized genomics in the context of melanoma risk, and will be broadly applicable as a model for delivery of personalized genomic feedback for other health conditions.
Study limitations include the fact that we do not assess participants for Internet literacy, and do not include detailed assessments regarding context of sun exposure (occupational, recreational). Finally, we did not employ blinding of study condition among study staff.
Conclusions
Our findings will have important implications for personalized genomics in the context of melanoma risk, and will be broadly applicable as a model for delivery of personalized genomic feedback for other conditions in this population. We plan future work to expand personalized genomic testing for melanoma risk to include other melanoma risk and protective markers, and to expand risk stratification to multiple levels as the literature on genetic factors in skin cancer unfolds.
Acknowledgments
The SOMBRA study is supported by a research grant from the National Cancer Institute at the National Institutes of Health, which is a part of the United States Government (Grant # 1R01CA181241-01A1). Grant reviews are available in Multimedia Appendix 2.
Abbreviations
- DNA
deoxyribonucleic acid
- FCDS
Florida Cancer Data System
- MC1R
melanocortin-1 receptor
- SEER
Surveillance, Epidemiology, and End Results
- SOMBRA
Skin health Online for Melanoma: Better Risk Assessment
- UNM
University of New Mexico
Study measures.
Peer-review reports from National Cancer Institute grant.
CONSORT publication form.
Footnotes
Conflicts of Interest: Dr. David Buller is the Research Director of Klein Buendel.
References
- 1.Jemal A, Devesa SS, Hartge P, Tucker MA. Recent trends in cutaneous melanoma incidence among whites in the United States. J Natl Cancer Inst. 2001 May 02;93(9):678–83. doi: 10.1093/jnci/93.9.678. [DOI] [PubMed] [Google Scholar]
- 2.Ries LA, Melbert D, Krapcho M, Mariotto A, Miller BA, Feuer EJ, Clegg L, Horner MJ, Howlader N, Eisner MP, Reichman M, Edwards BK. SEER Cancer Statistics Review, 1975-2004. Bethesda, MD: National Cancer Institute; 2007. [2017-01-30]. http://seer.cancer.gov/csr/1975_2004/ [Google Scholar]
- 3.American Cancer Society . Cancer Facts & Figures. Atlanta, GA: American Cancer Society; 2017. [2017-02-07]. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2017/cancer-facts-and-figures-2017.pdf . [Google Scholar]
- 4.New Mexico Tumor Registry Top 10 Cancers in New Mexico, 2008-2012. 2017. [2017-02-07]. http://nmtrweb.unm.edu/statistics/top10_20jan2015.pdf .
- 5.Rouhani P, Pinheiro PS, Sherman R, Arheart K, Fleming LE, Mackinnon J, Kirsner RS. Increasing rates of melanoma among nonwhites in Florida compared with the United States. Arch Dermatol. 2010 Jul;146(7):741–6. doi: 10.1001/archdermatol.2010.133.146/7/741 [DOI] [PubMed] [Google Scholar]
- 6.Cockburn MG, Zadnick J, Deapen D. Developing epidemic of melanoma in the Hispanic population of California. Cancer. 2006 Mar 01;106(5):1162–8. doi: 10.1002/cncr.21654. doi: 10.1002/cncr.21654. [DOI] [PubMed] [Google Scholar]
- 7.Hu S, Parmet Y, Allen G, Parker DF, Ma F, Rouhani P, Kirsner RS. Disparity in melanoma: a trend analysis of melanoma incidence and stage at diagnosis among whites, Hispanics, and blacks in Florida. Arch Dermatol. 2009 Dec;145(12):1369–74. doi: 10.1001/archdermatol.2009.302.145/12/1369 [DOI] [PubMed] [Google Scholar]
- 8.Hu S, Soza-Vento RM, Parker DF, Kirsner RS. Comparison of stage at diagnosis of melanoma among Hispanic, black, and white patients in Miami-Dade County, Florida. Arch Dermatol. 2006 Jun;142(6):704–8. doi: 10.1001/archderm.142.6.704.142/6/704 [DOI] [PubMed] [Google Scholar]
- 9.Pollitt RA, Clarke CA, Swetter SM, Peng DH, Zadnick J, Cockburn M. The expanding melanoma burden in California hispanics: importance of socioeconomic distribution, histologic subtype, and anatomic location. Cancer. 2011 Jan 01;117(1):152–61. doi: 10.1002/cncr.25355. doi: 10.1002/cncr.25355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linos E, Swetter SM, Cockburn MG, Colditz GA, Clarke CA. Increasing burden of melanoma in the United States. J Invest Dermatol. 2009 Jul;129(7):1666–74. doi: 10.1038/jid.2008.423. http://linkinghub.elsevier.com/retrieve/pii/S0022-202X(15)34426-2 .S0022-202X(15)34426-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zell JA, Cinar P, Mobasher M, Ziogas A, Meyskens FL, Anton-Culver H. Survival for patients with invasive cutaneous melanoma among ethnic groups: the effects of socioeconomic status and treatment. J Clin Oncol. 2008 Jan 01;26(1):66–75. doi: 10.1200/JCO.2007.12.3604.26/1/66 [DOI] [PubMed] [Google Scholar]
- 12.Byrd-Miles K, Toombs EL, Peck GL. Skin cancer in individuals of African, Asian, Latin-American, and American-Indian descent: differences in incidence, clinical presentation, and survival compared to Caucasians. J Drugs Dermatol. 2007 Jan;6(1):10–6. [PubMed] [Google Scholar]
- 13.Pichon LC, Corral I, Landrine H, Mayer JA, Adams-Simms D. Perceived skin cancer risk and sunscreen use among African American adults. J Health Psychol. 2010 Nov;15(8):1181–9. doi: 10.1177/1359105310364177.1359105310364177 [DOI] [PubMed] [Google Scholar]
- 14.Robinson JK, Joshi KM, Ortiz S, Kundu RV. Melanoma knowledge, perception, and awareness in ethnic minorities in Chicago: recommendations regarding education. Psychooncology. 2011 Mar;20(3):313–20. doi: 10.1002/pon.1736. [DOI] [PubMed] [Google Scholar]
- 15.Roman C, Lugo-Somolinos A, Thomas N. Skin cancer knowledge and skin self-examinations in the Hispanic population of North Carolina: the patient's perspective. JAMA Dermatol. 2013 Jan;149(1):103–4. doi: 10.1001/2013.jamadermatol.263.1557757 [DOI] [PubMed] [Google Scholar]
- 16.Colby S, Ortman J. Population Estimates and Projections. United States Census Bureau; 2015. [2017-03-10]. Projection of the size and composition of the U.S. population: 2014 to 2060 https://www.census.gov/content/dam/Census/library/publications/2015/demo/p25-1143.pdf . [Google Scholar]
- 17.Armstrong BK, Kricker A. How much melanoma is caused by sun exposure? Melanoma Res. 1993 Dec;3(6):395–401. doi: 10.1097/00008390-199311000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Dal H, Boldemann C, Lindelöf B. Does relative melanoma distribution by body site 1960-2004 reflect changes in intermittent exposure and intentional tanning in the Swedish population? Eur J Dermatol. 2007;17(5):428–34. doi: 10.1684/ejd.2007.0242.ejd.2007.0242 [DOI] [PubMed] [Google Scholar]
- 19.Whiteman DC, Whiteman CA, Green AC. Childhood sun exposure as a risk factor for melanoma: a systematic review of epidemiologic studies. Cancer Causes Control. 2001 Jan;12(1):69–82. doi: 10.1023/a:1008980919928. [DOI] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention (CDC) Sunburn prevalence among adults--United States, 1999, 2003, and 2004. MMWR Morb Mortal Wkly Rep. 2007 Jun 01;56(21):524–8. https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5621a2.htm .mm5621a2 [PubMed] [Google Scholar]
- 21.Kasparian NA, McLoone JK, Meiser B. Skin cancer-related prevention and screening behaviors: a review of the literature. J Behav Med. 2009 Oct;32(5):406–28. doi: 10.1007/s10865-009-9219-2. [DOI] [PubMed] [Google Scholar]
- 22.Coups EJ, Manne SL, Heckman CJ. Multiple skin cancer risk behaviors in the U.S. population. Am J Prev Med. 2008 Feb;34(2):87–93. doi: 10.1016/j.amepre.2007.09.032.S0749-3797(07)00655-1 [DOI] [PubMed] [Google Scholar]
- 23.Hay J, Coups EJ, Ford J, DiBonaventura M. Exposure to mass media health information, skin cancer beliefs, and sun protection behaviors in a United States probability sample. J Am Acad Dermatol. 2009 Nov;61(5):783–92. doi: 10.1016/j.jaad.2009.04.023. http://europepmc.org/abstract/MED/19596487 .S0190-9622(09)00507-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coups EJ, Stapleton JL, Hudson SV, Medina-Forrester A, Goydos JS, Natale-Pereira A. Skin cancer screening among Hispanic adults in the United States: results from the 2010 National Health Interview Survey. Arch Dermatol. 2012 Jul;148(7):861–3. doi: 10.1001/archdermatol.2012.615. http://europepmc.org/abstract/MED/22801634 .1158554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coups EJ, Stapleton JL, Hudson SV, Medina-Forrester A, Natale-Pereira A, Goydos JS. Sun protection and exposure behaviors among Hispanic adults in the United States: differences according to acculturation and among Hispanic subgroups. BMC Public Health. 2012 Nov 15;12:985. doi: 10.1186/1471-2458-12-985. http://europepmc.org/abstract/MED/23153104 .1471-2458-12-985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saraiya M, Hall HI, Uhler RJ. Sunburn prevalence among adults in the United States, 1999. Am J Prev Med. 2002 Aug;23(2):91–7. doi: 10.1016/s0749-3797(02)00461-0.S0749379702004610 [DOI] [PubMed] [Google Scholar]
- 27.Udayakumar D, Tsao H. Moderate- to low-risk variant alleles of cutaneous malignancies and nevi: lessons from genome-wide association studies. Genome Med. 2009 Oct 27;1(10):95. doi: 10.1186/gm95. https://genomemedicine.biomedcentral.com/articles/10.1186/gm95 .gm95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudru V, Laud K, Avril M, Minière A, Chompret A, Bressac-de PB, Demenais F. Melanocortin-1 receptor (MC1R) gene variants and dysplastic nevi modify penetrance of CDKN2A mutations in French melanoma-prone pedigrees. Cancer Epidemiol Biomarkers Prev. 2005 Oct;14(10):2384–90. doi: 10.1158/1055-9965.EPI-04-0777. http://cebp.aacrjournals.org/cgi/pmidlookup?view=long&pmid=16214921 .14/10/2384 [DOI] [PubMed] [Google Scholar]
- 29.Duffy DL, Box NF, Chen W, Palmer JS, Montgomery GW, James MR, Hayward NK, Martin NG, Sturm RA. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum Mol Genet. 2004 Feb 15;13(4):447–61. doi: 10.1093/hmg/ddh043.ddh043 [DOI] [PubMed] [Google Scholar]
- 30.Flanagan N, Healy E, Ray A, Philips S, Todd C, Jackson IJ, Birch-Machin MA, Rees JL. Pleiotropic effects of the melanocortin 1 receptor (MC1R) gene on human pigmentation. Hum Mol Genet. 2000 Oct 12;9(17):2531–7. doi: 10.1093/hmg/9.17.2531. [DOI] [PubMed] [Google Scholar]
- 31.Goldstein AM, Landi MT, Tsang S, Fraser MC, Munroe DJ, Tucker MA. Association of MC1R variants and risk of melanoma in melanoma-prone families with CDKN2A mutations. Cancer Epidemiol Biomarkers Prev. 2005 Sep;14(9):2208–12. doi: 10.1158/1055-9965.EPI-05-0321. http://cebp.aacrjournals.org/cgi/pmidlookup?view=long&pmid=16172233 .14/9/2208 [DOI] [PubMed] [Google Scholar]
- 32.Kanetsky PA, Ge F, Najarian D, Swoyer J, Panossian S, Schuchter L, Holmes R, Guerry D, Rebbeck TR. Assessment of polymorphic variants in the melanocortin-1 receptor gene with cutaneous pigmentation using an evolutionary approach. Cancer Epidemiol Biomarkers Prev. 2004 May;13(5):808–19. http://cebp.aacrjournals.org/cgi/pmidlookup?view=long&pmid=15159314 .13/5/808 [PubMed] [Google Scholar]
- 33.Kennedy C, ter Huurne J, Berkhout M, Gruis N, Bastiaens M, Bergman W, Willemze R, Bavinck JN. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol. 2001 Aug;117(2):294–300. doi: 10.1046/j.0022-202x.2001.01421.x. http://linkinghub.elsevier.com/retrieve/pii/S0022-202X(15)41327-2 .S0022-202X(15)41327-2 [DOI] [PubMed] [Google Scholar]
- 34.Landi MT, Kanetsky PA, Tsang S, Gold B, Munroe D, Rebbeck T, Swoyer J, Ter-Minassian M, Hedayati M, Grossman L, Goldstein AM, Calista D, Pfeiffer RM. MC1R, ASIP, and DNA repair in sporadic and familial melanoma in a Mediterranean population. J Natl Cancer Inst. 2005 Jul 06;97(13):998–1007. doi: 10.1093/jnci/dji176.97/13/998 [DOI] [PubMed] [Google Scholar]
- 35.Matichard E, Verpillat P, Meziani R, Gérard B, Descamps V, Legroux E, Burnouf M, Bertrand G, Bouscarat F, Archimbaud A, Picard C, Ollivaud L, Basset-Seguin N, Kerob D, Lanternier G, Lebbe C, Crickx B, Grandchamp B, Soufir N. Melanocortin 1 receptor (MC1R) gene variants may increase the risk of melanoma in France independently of clinical risk factors and UV exposure. J Med Genet. 2004 Feb;41(2):e13. doi: 10.1136/jmg.2003.011536. http://jmg.bmj.com/cgi/pmidlookup?view=long&pmid=14757863 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sturm RA. Skin colour and skin cancer - MC1R, the genetic link. Melanoma Res. 2002 Oct;12(5):405–16. doi: 10.1097/00008390-200209000-00001. [DOI] [PubMed] [Google Scholar]
- 37.Valverde P, Healy E, Sikkink S, Haldane F, Thody AJ, Carothers A, Jackson IJ, Rees JL. The Asp84Glu variant of the melanocortin 1 receptor (MC1R) is associated with melanoma. Hum Mol Genet. 1996 Oct;5(10):1663–6. doi: 10.1093/hmg/5.10.1663. [DOI] [PubMed] [Google Scholar]
- 38.Raimondi S, Sera F, Gandini S, Iodice S, Caini S, Maisonneuve P, Fargnoli MC. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer. 2008 Jun 15;122(12):2753–60. doi: 10.1002/ijc.23396. doi: 10.1002/ijc.23396. [DOI] [PubMed] [Google Scholar]
- 39.Pasquali E, García-Borrón JC, Fargnoli MC, Gandini S, Maisonneuve P, Bagnardi V, Specchia C, Liu F, Kayser M, Nijsten T, Nagore E, Kumar R, Hansson J, Kanetsky PA, Ghiorzo P, Debniak T, Branicki W, Gruis NA, Han J, Dwyer T, Blizzard L, Landi MT, Palmieri G, Ribas G, Stratigos A, Council ML, Autier P, Little J, Newton-Bishop J, Sera F, Raimondi S, M-SKIP Study Group MC1R variants increased the risk of sporadic cutaneous melanoma in darker-pigmented Caucasians: a pooled-analysis from the M-SKIP project. Int J Cancer. 2015 Feb 01;136(3):618–31. doi: 10.1002/ijc.29018. doi: 10.1002/ijc.29018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanetsky PA, Panossian S, Elder DE, Guerry D, Ming ME, Schuchter L, Rebbeck TR. Does MC1R genotype convey information about melanoma risk beyond risk phenotypes? Cancer. 2010 May 15;116(10):2416–28. doi: 10.1002/cncr.24994. doi: 10.1002/cncr.24994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanetsky PA, Rebbeck TR, Hummer AJ, Panossian S, Armstrong BK, Kricker A, Marrett LD, Millikan RC, Gruber SB, Culver HA, Zanetti R, Gallagher RP, Dwyer T, Busam K, From L, Mujumdar U, Wilcox H, Begg CB, Berwick M. Population-based study of natural variation in the melanocortin-1 receptor gene and melanoma. Cancer Res. 2006 Sep 15;66(18):9330–7. doi: 10.1158/0008-5472.CAN-06-1634. http://cancerres.aacrjournals.org/cgi/pmidlookup?view=long&pmid=16982779 .66/18/9330 [DOI] [PubMed] [Google Scholar]
- 42.Palmer JS, Duffy DL, Box NF, Aitken JF, O'Gorman LE, Green AC, Hayward NK, Martin NG, Sturm RA. Melanocortin-1 receptor polymorphisms and risk of melanoma: is the association explained solely by pigmentation phenotype? Am J Hum Genet. 2000 Jan;66(1):176–86. doi: 10.1086/302711. https://linkinghub.elsevier.com/retrieve/pii/S0002-9297(07)62245-9 .S0002-9297(07)62245-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.John PR, Makova K, Li W, Jenkins T, Ramsay M. DNA polymorphism and selection at the melanocortin-1 receptor gene in normally pigmented southern African individuals. Ann N Y Acad Sci. 2003 Jun;994:299–306. doi: 10.1111/j.1749-6632.2003.tb03193.x. [DOI] [PubMed] [Google Scholar]
- 44.Scherer D, Nagore E, Bermejo JL, Figl A, Botella-Estrada R, Thirumaran RK, Angelini S, Hemminki K, Schadendorf D, Kumar R. Melanocortin receptor 1 variants and melanoma risk: a study of 2 European populations. Int J Cancer. 2009 Oct 15;125(8):1868–75. doi: 10.1002/ijc.24548. doi: 10.1002/ijc.24548. [DOI] [PubMed] [Google Scholar]
- 45.Demenais F, Mohamdi H, Chaudru V, Goldstein AM, Newton Bishop JA, Bishop DT, Kanetsky PA, Hayward NK, Gillanders E, Elder DE, Avril MF, Azizi E, van Belle P, Bergman W, Bianchi-Scarrà G, Bressac-de PB, Calista D, Carrera C, Hansson J, Harland M, Hogg D, Höiom V, Holland EA, Ingvar C, Landi MT, Lang JM, Mackie RM, Mann GJ, Ming ME, Njauw CJ, Olsson H, Palmer J, Pastorino L, Puig S, Randerson-Moor J, Stark M, Tsao H, Tucker MA, van der Velden P, Yang XR, Gruis N, Melanoma GC. Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: a GenoMEL study. J Natl Cancer Inst. 2010 Oct 20;102(20):1568–83. doi: 10.1093/jnci/djq363. http://europepmc.org/abstract/MED/20876876 .djq363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Torre C, Garcia-Casado Z, Martínez-Escribano JA, Botella-Estrada R, Bañuls J, Oliver V, Mercader P, Azaña JM, Frias J, Nagore E. Influence of loss of function MC1R variants in genetic susceptibility of familial melanoma in Spain. Melanoma Res. 2010 Aug;20(4):342–8. doi: 10.1097/CMR.0b013e32833b159d. [DOI] [PubMed] [Google Scholar]
- 47.Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Ann Hum Genet. 2004 Mar;68(Pt 2):139–53. doi: 10.1046/j.1529-8817.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- 48.Klimentidis YC, Miller GF, Shriver MD. Genetic admixture, self-reported ethnicity, self-estimated admixture, and skin pigmentation among Hispanics and Native Americans. Am J Phys Anthropol. 2009 Apr;138(4):375–83. doi: 10.1002/ajpa.20945. [DOI] [PubMed] [Google Scholar]
- 49.Green ED, Guyer MS, National Human Genome Research Institute Charting a course for genomic medicine from base pairs to bedside. Nature. 2011 Feb 10;470(7333):204–13. doi: 10.1038/nature09764.nature09764 [DOI] [PubMed] [Google Scholar]
- 50.Khoury MJ, Clauser SB, Freedman AN, Gillanders EM, Glasgow RE, Klein WM, Schully SD. Population sciences, translational research, and the opportunities and challenges for genomics to reduce the burden of cancer in the 21st century. Cancer Epidemiol Biomarkers Prev. 2011 Oct;20(10):2105–14. doi: 10.1158/1055-9965.EPI-11-0481. http://cebp.aacrjournals.org/cgi/pmidlookup?view=long&pmid=21795499 .1055-9965.EPI-11-0481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001 Feb 15;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 52.Lerman C, Trock B, Rimer BK, Jepson C, Brody D, Boyce A. Psychological side effects of breast cancer screening. Health Psychol. 1991;10(4):259–67. doi: 10.1037//0278-6133.10.4.259. [DOI] [PubMed] [Google Scholar]
- 53.Patenaude AF, Guttmacher AE, Collins FS. Genetic testing and psychology. New roles, new responsibilities. Am Psychol. 2002 Apr;57(4):271–82. [PubMed] [Google Scholar]
- 54.Kinney AY, Gammon A, Coxworth J, Simonsen SE, Arce-Laretta M. Exploring attitudes, beliefs, and communication preferences of Latino community members regarding BRCA1/2 mutation testing and preventive strategies. Genet Med. 2010 Feb;12(2):105–15. doi: 10.1097/GIM.0b013e3181c9af2d. http://europepmc.org/abstract/MED/20061960 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gammon AD, Rothwell E, Simmons R, Lowery JT, Ballinger L, Hill DA, Boucher KM, Kinney AY. Awareness and preferences regarding BRCA1/2 genetic counseling and testing among Latinas and non-Latina white women at increased risk for hereditary breast and ovarian cancer. J Genet Couns. 2011 Dec;20(6):625–38. doi: 10.1007/s10897-011-9376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Addie Siobhan, Olson Steve, Beachy Sarah H., Rapporteurs. Roundtable on Translating Genomic-Based Research for Health. Board on Health Sciences Policy. Health and Medicine Division. National Academics of Sciences, Engineering, and Medicine . Applying an Implementation Science Approach to Genomic Medicine: Workshop Summary. Washington DC: National Academies Press; 2016. p. 978. [PubMed] [Google Scholar]
- 57.Geransar R, Einsiedel E. Evaluating online direct-to-consumer marketing of genetic tests: informed choices or buyers beware? Genet Test. 2008 Mar;12(1):13–23. doi: 10.1089/gte.2007.0024. [DOI] [PubMed] [Google Scholar]
- 58.Gollust SE, Wilfond BS, Hull SC. Direct-to-consumer sales of genetic services on the Internet. Genet Med. 2003;5(4):332–7. doi: 10.1097/01.GIM.0000076972.83711.48. http://europepmc.org/abstract/MED/12865763 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khoury MJ, McBride CM, Schully SD, Ioannidis JP, Feero WG, Janssens AC, Gwinn M, Simons-Morton DG, Bernhardt JM, Cargill M, Chanock SJ, Church GM, Coates RJ, Collins FS, Croyle RT, Davis BR, Downing GJ, Duross A, Friedman S, Gail MH, Ginsburg GS, Green RC, Greene MH, Greenland P, Gulcher JR, Hsu A, Hudson KL, Kardia SL, Kimmel PL, Lauer MS, Miller AM, Offit K, Ransohoff DF, Roberts JS, Rasooly RS, Stefansson K, Terry SF, Teutsch SM, Trepanier A, Wanke KL, Witte JS, Xu J, Centers for Disease Control and Prevention The Scientific Foundation for personal genomics: recommendations from a National Institutes of Health-Centers for Disease Control and Prevention multidisciplinary workshop. Genet Med. 2009 Aug;11(8):559–67. doi: 10.1097/GIM.0b013e3181b13a6c. http://europepmc.org/abstract/MED/19617843 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams-Jones B. Where there's a web, there's a way: commercial genetic testing and the Internet. Community Genet. 2003;6(1):46–57. doi: 10.1159/000069538.CMG06046 [DOI] [PubMed] [Google Scholar]
- 61.Hallenbeck J. High context illness and dying in a low context medical world. Am J Hosp Palliat Care. 2006;23(2):113–8. doi: 10.1177/104990910602300208. [DOI] [PubMed] [Google Scholar]
- 62.Robberson M, Rogers R. Beyond fear appeals: negative and positive persuasive appeals to health and self-esteem. J Appl Social Pyschol. 1988 Mar;18(3):277–287. doi: 10.1111/j.1559-1816.1988.tb00017.x. [DOI] [Google Scholar]
- 63.Rogers RW. Cognitive and physiological processes in attitude change: a revised theory of protection motivation. In: Cacioppo JT, Petty RE, editors. Social Psychophysiology: A Sourcebook. New York: Guilford Press; 1983. pp. 153–176. [Google Scholar]
- 64.Kutner M, Greenberg E, Jin Y, Paulsen C. National Center of Education Statistics. Washington, DC: 2006. Oct, [2017-03-07]. The health literacy of America's adults: results from the 2003 National Assessment of Adult Literacy http://files.eric.ed.gov/fulltext/ED493284.pdf . [Google Scholar]
- 65.Bloss CS, Ornowski L, Silver E, Cargill M, Vanier V, Schork NJ, Topol EJ. Consumer perceptions of direct-to-consumer personalized genomic risk assessments. Genet Med. 2010 Sep;12(9):556–66. doi: 10.1097/GIM.0b013e3181eb51c6. [DOI] [PubMed] [Google Scholar]
- 66.Pagán JA, Su D, Li L, Armstrong K, Asch DA. Racial and ethnic disparities in awareness of genetic testing for cancer risk. Am J Prev Med. 2009 Dec;37(6):524–30. doi: 10.1016/j.amepre.2009.07.021.S0749-3797(09)00605-9 [DOI] [PubMed] [Google Scholar]
- 67.Suther S, Kiros G. Barriers to the use of genetic testing: a study of racial and ethnic disparities. Genet Med. 2009 Sep;11(9):655–62. doi: 10.1097/GIM.0b013e3181ab22aa.00125817-200909000-00010 [DOI] [PubMed] [Google Scholar]
- 68.Viswanath K, Nagler RH, Bigman-Galimore CA, McCauley MP, Jung M, Ramanadhan S. The communications revolution and health inequalities in the 21st century: implications for cancer control. Cancer Epidemiol Biomarkers Prev. 2012 Oct;21(10):1701–8. doi: 10.1158/1055-9965.EPI-12-0852. http://cebp.aacrjournals.org/cgi/pmidlookup?view=long&pmid=23045545 .21/10/1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lorence D, Park H. Group disparities and health information: a study of online access for the underserved. Health Informatics J. 2008 Mar;14(1):29–38. doi: 10.1177/1460458207086332.14/1/29 [DOI] [PubMed] [Google Scholar]
- 70.Lorence DP, Park H, Fox S. Racial disparities in health information access: resilience of the Digital Divide. J Med Syst. 2006 Aug;30(4):241–9. doi: 10.1007/s10916-005-9003-y. [DOI] [PubMed] [Google Scholar]
- 71.Hall MJ, Manne SL, Myers RE, Keenan EM, Balshem AM, Weinberg DS. Predictors of patient uptake of colorectal cancer gene environment risk assessment. Genome Med. 2012;4(11):92. doi: 10.1186/gm393. https://genomemedicine.biomedcentral.com/articles/10.1186/gm393 .gm393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McBride CM, Alford SH, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Characteristics of users of online personalized genomic risk assessments: implications for physician-patient interactions. Genet Med. 2009 Aug;11(8):582–7. doi: 10.1097/GIM.0b013e3181b22c3a. http://europepmc.org/abstract/MED/19606049 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grann VR, Jacobson JS, Troxel AB, Hershman D, Karp J, Myers C, Neugut AI. Barriers to minority participation in breast carcinoma prevention trials. Cancer. 2005 Jul 15;104(2):374–9. doi: 10.1002/cncr.21164. doi: 10.1002/cncr.21164. [DOI] [PubMed] [Google Scholar]
- 74.Maxwell AE, Bastani R, Vida P, Warda US. Strategies to recruit and retain older Filipino-American immigrants for a cancer screening study. J Community Health. 2005 Jun;30(3):167–79. doi: 10.1007/s10900-004-1956-0. http://europepmc.org/abstract/MED/15847243 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.James RD, Yu J, Henrikson NB, Bowen DJ, Fullerton SM, Health Disparities Working Group Strategies and stakeholders: minority recruitment in cancer genetics research. Community Genet. 2008;11(4):241–9. doi: 10.1159/000116878.000116878 [DOI] [PubMed] [Google Scholar]
- 76.Fox S, Duggan M. Pew Research Center Internet, Science & Tech. Washington, DC: Pew Research Center’s Internet & American Life Project; 2013. [2017-01-30]. Health Online 2013 http://pewinternet.org/Reports/2013/Health-online/Summary-of-Findings.aspx, [Google Scholar]
- 77.Selsky C, Luta G, Noone A, Huerta EE, Mandelblatt JS. Internet access and online cancer information seeking among Latino immigrants from safety net clinics. J Health Commun. 2013;18(1):58–70. doi: 10.1080/10810730.2012.688248. http://europepmc.org/abstract/MED/23066874 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Armstrong K, Putt M, Halbert CH, Grande D, Schwartz JS, Liao K, Marcus N, Demeter MB, Shea J. The influence of health care policies and health care system distrust on willingness to undergo genetic testing. Med Care. 2012 May;50(5):381–7. doi: 10.1097/MLR.0b013e31824d748b. http://europepmc.org/abstract/MED/22473221 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bernhardt JM, Lariscy RA, Parrott RL, Silk KJ, Felter EM. Perceived barriers to Internet-based health communication on human genetics. J Health Commun. 2002;7(4):325–40. doi: 10.1080/10810730290088166. [DOI] [PubMed] [Google Scholar]
- 80.Marin G, Sabogal F, Marin B, Otero-Sabogal R, Perez-Stable E. Development of a short acculturation scale for Hispanics. Hisp J Behav Sci. 1987 Jun;9(2):183–205. doi: 10.1177/07399863870092005. [DOI] [Google Scholar]
- 81.Espinosa de Los Monteros K, Gallo LC. The relevance of fatalism in the study of Latinas' cancer screening behavior: a systematic review of the literature. Int J Behav Med. 2011 Dec;18(4):310–8. doi: 10.1007/s12529-010-9119-4. http://europepmc.org/abstract/MED/20953916 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Magai C, Consedine N, Conway F, Neugut A, Culver C. Diversity matters: unique populations of women and breast cancer screening. Cancer. 2004 Jun 01;100(11):2300–7. doi: 10.1002/cncr.20278. doi: 10.1002/cncr.20278. [DOI] [PubMed] [Google Scholar]
- 83.Pasick RJ, Barker JC, Otero-Sabogal R, Burke NJ, Joseph G, Guerra C. Intention, subjective norms, and cancer screening in the context of relational culture. Health Educ Behav. 2009 Oct;36(5 Suppl):91S–110S. doi: 10.1177/1090198109338919. http://europepmc.org/abstract/MED/19805793 .36/5_suppl/91S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Torres S, Ramos M, Leverence R, Bowen D, Berwick M, Hay J. A pilot study of skin cancer risk reduction behaviors, cancer communication, and skin cancer beliefs in hispanics. Calif J Health Promot. 2014;12(1):95–100. ISSN: 1545-8717 http://www.cjhp.org/Volume12Issue1_2014/documents/Formatted_CJHP07092013_Torres.pdf. [Google Scholar]
- 85.McBride CM, Koehly LM, Sanderson SC, Kaphingst KA. The behavioral response to personalized genetic information: will genetic risk profiles motivate individuals and families to choose more healthful behaviors? Annu Rev Public Health. 2010;31:89–103. doi: 10.1146/annurev.publhealth.012809.103532. [DOI] [PubMed] [Google Scholar]
- 86.Kaphingst KA, McBride CM, Wade C, Alford SH, Reid R, Larson E, Baxevanis AD, Brody LC. Patients' understanding of and responses to multiplex genetic susceptibility test results. Genet Med. 2012 Jul;14(7):681–7. doi: 10.1038/gim.2012.22. http://europepmc.org/abstract/MED/22481132 .gim201222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaphingst KA, McBride CM, Wade C, Alford SH, Brody LC, Baxevanis AD. Consumers' use of web-based information and their decisions about multiplex genetic susceptibility testing. J Med Internet Res. 2010 Sep 29;12(3):e41. doi: 10.2196/jmir.1587. http://www.jmir.org/2010/3/e41/ v12i3e41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Berrigan D, Forsyth BH, Helba C, Levin K, Norberg A, Willis GB. Cognitive testing of physical activity and acculturation questions in recent and long-term Latino immigrants. BMC Public Health. 2010 Aug 13;10:481. doi: 10.1186/1471-2458-10-481. http://europepmc.org/abstract/MED/20707902 .1471-2458-10-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harkness J, Edwards B, Braun M, Johnson TD, Lyberg L, Mohler PP, Pennell B, Smith T. Survey Methods in Multicultural, Multinational, and Multiregional Contexts. Hoboken, NJ: Wiley; 2010. [Google Scholar]
- 90.Willis G. Cognitive interviewing: a tool for improving questionnaire design. Thousand Oaks, CA: Sage Publications; 2005. [Google Scholar]
- 91.Rodríguez VM, Robers E, Zielaskowski K, Javier GC, Hunley K, Kaphingst KA, Guest DD, Sussman A, Meyer White KA, Schwartz MR, Greb J, Talamantes Y, Bigney J, Berwick M, Hay JL. Translation and adaptation of skin cancer genomic risk education materials for implementation in primary care. J Community Genet. 2017 Jan;8(1):53–63. doi: 10.1007/s12687-016-0287-z. http://europepmc.org/abstract/MED/27924449 .10.1007/s12687-016-0287-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davidson KW, Goldstein M, Kaplan RM, Kaufmann PG, Knatterud GL, Orleans CT, Spring B, Trudeau KJ, Whitlock EP. Evidence-based behavioral medicine: what is it and how do we achieve it? Ann Behav Med. 2003 Dec;26(3):161–71. doi: 10.1207/S15324796ABM2603_01. [DOI] [PubMed] [Google Scholar]
- 93.Schulz KF, Altman DG, Moher D. CONSORT 2010 Statement: Updated guidelines for reporting parallel group randomised trials. J Clin Epidemiol. 2010 Aug;63(8):834–40. doi: 10.1016/j.jclinepi.2010.02.005. https://linkinghub.elsevier.com/retrieve/pii/S0895-4356(10)00079-X .S0895-4356(10)00079-X [DOI] [PubMed] [Google Scholar]
- 94.Hensley-Alford S, McBride CM, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Participation in genetic testing research varies by social group. Public Health Genomics. 2011;14(2):85–93. doi: 10.1159/000294277. https://www.karger.com?DOI=10.1159/000294277 .000294277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brewer NT, Tzeng JP, Lillie SE, Edwards AS, Peppercorn JM, Rimer BK. Health literacy and cancer risk perception: implications for genomic risk communication. Med Decis Making. 2009;29(2):157–66. doi: 10.1177/0272989X08327111.0272989X08327111 [DOI] [PubMed] [Google Scholar]
- 96.Lipkus IM. Numeric, verbal, and visual formats of conveying health risks: suggested best practices and future recommendations. Med Decis Making. 2007;27(5):696–713. doi: 10.1177/0272989X07307271.0272989X07307271 [DOI] [PubMed] [Google Scholar]
- 97.Reyna VF, Lloyd FJ, Whalen P. Genetic testing and medical decision making. Arch Intern Med. 2001 Nov 12;161(20):2406–8. doi: 10.1001/archinte.161.20.2406.ied10002 [DOI] [PubMed] [Google Scholar]
- 98.Reyna VF, Lloyd FJ, Brainerd C. Memory, development, and rationality: An integrative theory of judgment and decision making. In: Schneider S, Shanteau J, editors. Emerging perspectives on judgment and decision research. Cambridge: Cambridge University Press; 2003. pp. 201–245. [Google Scholar]
- 99.Coyne JC, Benazon NR, Gaba CG, Calzone K, Weber BL. Distress and psychiatric morbidity among women from high-risk breast and ovarian cancer families. J Consult Clin Psychol. 2000 Oct;68(5):864–74. [PubMed] [Google Scholar]
- 100.Bergenmar M, Hansson J, Brandberg Y. Family members' perceptions of genetic testing for malignant melanoma--a prospective interview study. Eur J Oncol Nurs. 2009 Apr;13(2):74–80. doi: 10.1016/j.ejon.2008.12.003.S1462-3889(08)00155-5 [DOI] [PubMed] [Google Scholar]
- 101.Graham JW, Hofer SM, Donaldson SI, MacKinnon DP, Schafer JL. Analysis with missing data in prevention research. In: Bryant KJ, Windle MT, West SH, editors. The science of prevention: methodological advances from alcohol and substance abuse research. Washington, D.C: American Psychological Association; 1997. [Google Scholar]
- 102.Schafer J, Olsen M. Multiple imputation for multivariate missing data problems: a data analyst's perspective. Multivariate Behav Res. 1998 Oct 01;33(4):545–571. doi: 10.1207/s15327906mbr3304_5. [DOI] [PubMed] [Google Scholar]
- 103.Rogers E. Diffusion of Innovations, 5th Edition. New York: Free Press; 2003. [Google Scholar]
- 104.Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol. 1986 Dec;51(6):1173–82. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 105.MacKinnon D. Introduction to statistical mediation analysis. New York: Lawrence Erlbaum Associates/Taylor & Francis Group; 2008. [Google Scholar]
- 106.Geller AC, Emmons KM, Brooks DR, Powers C, Zhang Z, Koh HK, Heeren T, Sober AJ, Li F, Gilchrest BA. A randomized trial to improve early detection and prevention practices among siblings of melanoma patients. Cancer. 2006 Aug 15;107(4):806–14. doi: 10.1002/cncr.22050. doi: 10.1002/cncr.22050. [DOI] [PubMed] [Google Scholar]
- 107.Cohen J. Statistical power analysis for the behavioral sciences. Hillsdale, NJ: L. Erlbaum Associates; 1988. [Google Scholar]
- 108.Eng TR. Emerging technologies for cancer prevention and other population health challenges. J Med Internet Res. 2005 Jul 01;7(3):e30. doi: 10.2196/jmir.7.3.e30. http://www.jmir.org/2005/3/e30/ v7i3e30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shaer O, Nov O, Okerlund J, Balestra M, Stowell E, Ascher L, Bi J, Schlenker C, Ball M. Informing the design of direct-to-consumer interactive personal genomics reports. J Med Internet Res. 2015 Jun;17(6):e146. doi: 10.2196/jmir.4415. http://www.jmir.org/2015/6/e146/ v17i6e146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Glanz K, Yaroch AL, Dancel M, Saraiya M, Crane LA, Buller DB, Manne S, O'Riordan DL, Heckman CJ, Hay J, Robinson JK. Measures of sun exposure and sun protection practices for behavioral and epidemiologic research. Arch Dermatol. 2008 Feb;144(2):217–22. doi: 10.1001/archdermatol.2007.46.144/2/217 [DOI] [PubMed] [Google Scholar]
- 111.Ersig AL, Williams JK, Hadley DW, Koehly LM. Communication, encouragement, and cancer screening in families with and without mutations for hereditary nonpolyposis colorectal cancer: a pilot study. Genet Med. 2009 Oct;11(10):728–34. doi: 10.1097/GIM.0b013e3181b3f42d. http://europepmc.org/abstract/MED/19707152 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hay J, Shuk E, Zapolska J, Ostroff J, Lischewski J, Brady M, Berwick M. Family communication patterns after melanoma diagnosis. J Fam Commun. 2009 Oct 09;9(4):209–232. doi: 10.1080/15267430903182678. [DOI] [Google Scholar]
- 113.Hay J, Ostroff J, Martin A, Serle N, Soma S, Mujumdar U, Berwick M. Skin cancer risk discussions in melanoma-affected families. J Cancer Educ. 2005;20(4):240–6. doi: 10.1207/s15430154jce2004_13. [DOI] [PubMed] [Google Scholar]
- 114.Hay J, Kaphingst KA, Baser R, Li Y, Hensley-Alford S, McBride CM. Skin cancer concerns and genetic risk information-seeking in primary care. Public Health Genomics. 2012;15(2):57–72. doi: 10.1159/000330403. https://www.karger.com?DOI=10.1159/000330403 .000330403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yennurajalingam S, Parsons HA, Duarte ER, Palma A, Bunge S, Palmer JL, Delgado-Guay MO, Allo J, Bruera E. Decisional control preferences of Hispanic patients with advanced cancer from the United States and Latin America. J Pain Symptom Manage. 2013 Sep;46(3):376–85. doi: 10.1016/j.jpainsymman.2012.08.015.S0885-3924(12)00495-2 [DOI] [PubMed] [Google Scholar]
- 116.Witte K, Cameron KA, McKeon JK, Berkowitz JM. Predicting risk behaviors: development and validation of a diagnostic scale. J Health Commun. 1996;1(4):317–41. doi: 10.1080/108107396127988. [DOI] [PubMed] [Google Scholar]
- 117.Waters EA, Hay JL, Orom H, Kiviniemi MT, Drake BF. “Don't know” responses to risk perception measures: implications for underserved populations. Med Decis Making. 2013 Feb;33(2):271–81. doi: 10.1177/0272989X12464435. http://europepmc.org/abstract/MED/23468476 .33/2/271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Weinstein ND. Unrealistic optimism about susceptibility to health problems. J Behav Med. 1982 Dec;5(4):441–60. doi: 10.1007/BF00845372. [DOI] [PubMed] [Google Scholar]
- 119.Aiken LS, West SG, Woodward CK, Reno RR, Reynolds KD. Increasing screening mammography in asymptomatic women: evaluation of a second-generation, theory-based program. Health Psychol. 1994 Nov;13(6):526–38. doi: 10.1037//0278-6133.13.6.526. [DOI] [PubMed] [Google Scholar]
- 120.Bandura A. Social foundations of thought and action: a social cognitive theory. Englewood Cliffs, NJ: Prentice-Hall; 1986. [Google Scholar]
- 121.Hay JL, Baser R, Weinstein ND, Li Y, Primavera L, Kemeny MM. Examining intuitive risk perceptions for cancer in diverse populations. Health Risk Soc. 2014 Jan 01;16(3):227–242. doi: 10.1080/13698575.2014.911822. http://europepmc.org/abstract/MED/24999304 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hay JL, Ramos M, Li Y, Holland S, Brennessel D, Kemeny MM. Deliberative and intuitive risk perceptions as predictors of colorectal cancer screening over time. J Behav Med. 2016 Feb;39(1):65–74. doi: 10.1007/s10865-015-9667-9. http://europepmc.org/abstract/MED/26280754 .10.1007/s10865-015-9667-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hay JL, Oliveria SA, Dusza SW, Phelan DL, Ostroff JS, Halpern AC. Psychosocial mediators of a nurse intervention to increase skin self-examination in patients at high risk for melanoma. Cancer Epidemiol Biomarkers Prev. 2006 Jun;15(6):1212–6. doi: 10.1158/1055-9965.EPI-04-0822. http://cebp.aacrjournals.org/cgi/pmidlookup?view=long&pmid=16775183 .15/6/1212 [DOI] [PubMed] [Google Scholar]
- 124.Chew LD, Griffin JM, Partin MR, Noorbaloochi S, Grill JP, Snyder A, Bradley KA, Nugent SM, Baines AD, Vanryn M. Validation of screening questions for limited health literacy in a large VA outpatient population. J Gen Intern Med. 2008 May;23(5):561–6. doi: 10.1007/s11606-008-0520-5. http://europepmc.org/abstract/MED/18335281 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chew LD, Bradley KA, Boyko EJ. Brief questions to identify patients with inadequate health literacy. Fam Med. 2004 Sep;36(8):588–94. http://www.stfm.org/fmhub/fm2004/September/Lisa588.pdf . [PubMed] [Google Scholar]
- 126.Shea JA, Micco E, Dean LT, McMurphy S, Schwartz JS, Armstrong K. Development of a revised Health Care System Distrust scale. J Gen Intern Med. 2008 Jun;23(6):727–32. doi: 10.1007/s11606-008-0575-3. http://europepmc.org/abstract/MED/18369678 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Powe BD. Cancer fatalism among elderly Caucasians and African Americans. Oncol Nurs Forum. 1995 Oct;22(9):1355–9. [PubMed] [Google Scholar]
- 128.National Cancer Institute. 2007. [2017-01-30]. Health Information National Trends Survey https://hints.cancer.gov/
- 129.Horowitz M, Wilner N, Alvarez W. Impact of Event Scale: a measure of subjective stress. Psychosom Med. 1979 May;41(3):209–18. doi: 10.1097/00006842-197905000-00004. [DOI] [PubMed] [Google Scholar]
- 130.Joseph S. Psychometric evaluation of Horowitz's Impact of Event Scale: a review. J Trauma Stress. 2000 Jan;13(1):101–13. doi: 10.1023/A:1007777032063. [DOI] [PubMed] [Google Scholar]
- 131.Thewes B, Meiser B, Hickie IB. Psychometric properties of the Impact of Event Scale amongst women at increased risk for hereditary breast cancer. Psychooncology. 2001;10(6):459–68. doi: 10.1002/pon.533.10.1002/pon.533 [DOI] [PubMed] [Google Scholar]
- 132.Nelson DE, Kreps GL, Hesse BW, Croyle RT, Willis G, Arora NK, Rimer BK, Viswanath KV, Weinstein N, Alden S. The Health Information National Trends Survey (HINTS): development, design, and dissemination. J Health Commun. 2004;9(5):443–60; discussion 81. doi: 10.1080/10810730490504233.3F44L3KHM6U0HRED [DOI] [PubMed] [Google Scholar]
- 133.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Abeni D, Boyle P, Melchi CF. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Eur J Cancer. 2005 Jan;41(1):28–44. doi: 10.1016/j.ejca.2004.10.015. https://linkinghub.elsevier.com/retrieve/pii/S0959-8049(04)00832-9 .S0959-8049(04)00832-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Study measures.
Peer-review reports from National Cancer Institute grant.
CONSORT publication form.


