ABSTRACT
The diversity of the genetic code systems used by microbes on earth is yet to be elucidated. It is known that certain methanogenic archaea employ an alternative system for cysteine (Cys) biosynthesis and encoding; tRNACys is first acylated with phosphoserine (Sep) by O-phosphoseryl-tRNA synthetase (SepRS) and then converted to Cys-tRNACys by Sep-tRNA:Cys-tRNA synthase (SepCysS). In this study, we searched all genomic and metagenomic protein sequence data in the Integrated Microbial Genomes (IMG) system and at the NCBI to reveal new clades of SepRS and SepCysS proteins belonging to diverse archaea in the four major groups (DPANN, Euryarchaeota, TACK, and Asgard) and two groups of bacteria (“Candidatus Parcubacteria” and Chloroflexi). Bacterial SepRS and SepCysS charged bacterial tRNACys species with cysteine in vitro. Homologs of SepCysE, a scaffold protein facilitating SepRS⋅SepCysS complex assembly in Euryarchaeota class I methanogens, are found in a few groups of TACK and Asgard archaea, whereas the C-terminally truncated homologs exist fused or genetically coupled with diverse SepCysS species. Investigation of the selenocysteine (Sec)- and pyrrolysine (Pyl)-utilizing traits in SepRS-utilizing archaea and bacteria revealed that the archaea carrying full-length SepCysE employ Sec and that SepRS is often found in Pyl-utilizing archaea and Chloroflexi bacteria. We discuss possible contributions of the SepRS-SepCysS system for sulfur assimilation, methanogenesis, and other metabolic processes requiring large amounts of iron-sulfur enzymes or Pyl-containing enzymes.
KEYWORDS: biochemistry, bioinformatics, cysteine biosynthesis, genetic code, translation
IMPORTANCE
Comprehensive analyses of all genomic and metagenomic protein sequence data in public databases revealed the distribution and evolution of an alternative cysteine-encoding system in diverse archaea and bacteria. The finding that the SepRS-SepCysS-SepCysE- and the selenocysteine-encoding systems are shared by the Euryarchaeota class I methanogens, the Crenarchaeota AK8/W8A-19 group, and an Asgard archaeon suggests that ancient archaea may have used both systems. In contrast, bacteria may have obtained the SepRS-SepCysS system from archaea. The SepRS-SepCysS system sometimes coexists with a pyrrolysine-encoding system in both archaea and bacteria. Our results provide additional bioinformatic evidence for the contribution of the SepRS-SepCysS system for sulfur assimilation and diverse metabolisms which require vast amounts of iron-sulfur enzymes and proteins. Among these biological activities, methanogenesis, methylamine metabolism, and organohalide respiration may have local and global effects on earth. Taken together, uncultured bacteria and archaea provide an expanded record of the evolution of the genetic code.
INTRODUCTION
Two minor genetic code systems were discovered in methanogenic archaea a decade ago (1–3). In most organisms, Cys biosynthesis and Cys-tRNACys formation are carried out separately by a cysteine synthase and cysteinyl-tRNA synthetase (CysRS), respectively. However, methanogens employ a tRNACys-dependent Cys biosynthesis pathway (3). In these archaea, Cys-tRNACys is formed in a two-step process; first, O-phosphoserine (Sep) is acylated to tRNACys by SepRS and then Sep-tRNACys is converted by Sep-tRNA:Cys-tRNA synthase (SepCysS) to Cys-tRNACys (3–6). An additional component, SepCysE, stabilizes the SepRS⋅SepCysS⋅tRNACys ternary complex, but it is known to be present in class I methanogens only (4, 7). The class I methanogens are also exceptional among methanogens in that they encode selenocysteine (Sec), the 21st genetically encoded amino acid used in some archaea and many bacteria and eukaryotes (8, 9). The coupled biosynthesis and coding of Cys are considered as the original mechanism of Cys-tRNACys formation in the last common ancestor of archaea (3, 4) because archaeal CysRS genes appear to have multiple bacterial origins (10, 11) and bacterial CysRS is a highly evolved Cys-specific enzyme using a zinc atom to ensure specificity (12, 13). However, our knowledge is confined to well-studied lineages of cultured archaea, and it remains unclear whether the SepRS-SepCysS pathway is present outside the major Euryarchaeota clade, which includes class I, II, and III methanogens, methanotrophic archaea 1 (ANME-1), and Archaeoglobi (4, 14–16).
Pyrrolysine (Pyl), the 22nd genetically encoded amino acid, is charged to tRNAPyl by pyrrolysyl-tRNA synthetase (PylRS) (17, 18), which is specific for this unusual amino acid. PylRS is present in diverse bacteria and a few archaeal groups (19). PylRS is encoded by a single pylS gene in the Methanosarcinaceae, by the pylSn and pylSc gene, encoding the N- or C-terminal part, respectively, of PylRS in some anaerobic bacteria and “Candidatus Methanomethylicus sp. V1,” or by pylSc only in Methanomassiliicoccales (1, 19–21). The evolutionary pathways of the three types of PylRS remain unclear (19). Pyrrolysine biosynthesis genes (pylBCD), a tRNAPyl gene (pylT), and Pyl-utilizing methylamine methyltransferase genes (mtxBC) usually form a single gene cluster with the PylRS gene, which may have facilitated the horizontal gene transfer (HGT) of a Pyl-encoding system (22). The Pyl-utilizing methylamine:corrinoid methyltransferases (MtxB) transfer a methyl group from methylamines to their corrinoid protein partners (MtxC). The methyl group is then transferred to coenzyme M (CoM) in methanogens and possibly to CoM or tetrahydrofolate (THF) in bacteria (8, 23). Finally, the methyl group is released as methane by methyl-CoM reductase in methanogens and probably fuels anaerobic respiration in bacteria (8, 23).
In the last few years, analyses of genomic and metagenomic sequences have identified large numbers of novel bacterial and archaeal lineages. Some of these archaea are methanogens (21, 24–27). Most importantly, single-cell genomics and the composite-genome approach have dissected microbial dark matter (MDM) (28), the candidate phylum radiation (CPR) (29, 30), and the Asgard archaeal superphylum (31) by detecting and classifying uncultivated microbes (28, 29, 31–34). Progress in DNA sequence and de novo assembly technologies have led to the generation of larger genomic and metagenomic contigs encoding proteins. Phylogenetic studies of organisms based on protein sequences challenge traditional phylogenies based solely on rRNA sequences (29).
In this study, we assumed that the SepRS-SepCysS-SepCysE system might exist in diverse organisms whose genomic sequences were not available several years ago. We addressed (i) the distribution of the genes for SepRS, SepCysS, and SepCysE homologs outside the major Euryarchaeota groups and (ii) any relationships between RNA-dependent Cys biosynthesis and Sec- or Pyl-utilizing traits. In addition to investigating the genomic data, we investigated metagenomic protein sequence data, whose usage has been limited due to the low reliability of the data and the difficulty of inferring firm phylogenetic results. To overcome these problems, we performed a comprehensive survey of all metagenomic data sets in the IMG system (35) and at the NCBI rather than using an individual data set.
RESULTS
Identification of homologs of SepRS, SepCysS, and SepCysE.
In a preliminary search, we found SepRS genes in Hadesarchaea and MSBL1 (36, 37), “Candidatus Bathyarchaeota” (24, 38), and a few more groups of Euryarchaeota (26, 27) in the NCBI database. Furthermore, a SepRS-SepCysS operon was found in a metagenomic bin of a CPR bacterium, “Candidatus Parcubacteria” bacterium DG_74_2 (39). Although this “Ca. Parcubacteria” DG_74_2 bin is apparently composed of a few different genomes, including those of two “Ca. Parcubacteria” species, the SepRS-SepCysS operon is flanked by a typical “Ca. Parcubacteria” gene encoding a signal transduction histidine kinase. Thus, it is suggested that SepRS is not limited to the archaeal domain of life. Because the “Ca. Parcubacteria” DG_74_2 SepRS sequence (GenBank accession no. KPJ56532) differs from the methanogen SepRS sequences (about 40% similarity), we used it as query for the first round of genomic and metagenomic BLASTp searches. SepRS sequences that showed more than 40% similarity with the query were collected and grouped by similarity using Clustal X (40). Representative sequences of each group were subsequently used as queries for another run of metagenomic BLASTp to identify close relatives.
Corresponding/paired SepCysS genes are readily available in genomic sequences and metagenomic contig sequences, in which they exist in the vicinity of the SepRS gene. However, to identify the SepCysS genes paired with SepRSs obtained from different metagenomic contigs, the metagenomic contigs were binned based on GC contents and read depths. For precise phylogenetic inference, (i) some raw sequence data were used to connect neighboring contigs (41), (ii) binning of a single-cell genome and metagenomic contigs was performed in cases where both the cell and the DNA samples derived from the same sampling point, and (iii) rRNA and protein sequences were identified whenever possible. Binning was facilitated by an observation that similar organisms have similar SepRS and SepCysS genes and thrive in similar environments. In our analysis, we were able to pair most of the representative SepRS genes with one or two SepCysS genes.
(i) Occurrence of SepRS.
Our analysis shows that SepRS is widespread among uncultured archaea and bacteria (Fig. 1A and see Fig. S1 in the supplemental material). SepRS is present in four clades of archaea (Euryarchaeota, DPANN, TACK, and Asgard) and in Chloroflexi and a few other bacterial species (Fig. 1A; Fig. S1). As an exception, a truncated SepRS gene that lacks the C-terminal anticodon binding domain (SepRS-ΔC) exists in an uncultured Crystal Geyser groundwater (“Ca. Parcubacteria”) bacterium (Fig. 1A). SepRS is common in some lineages of archaea (Euryarchaeota methanogens and Archaeoglobi, Hadesarchaea/MSBL1, “Candidatus Altiarchaeales,” Crenarchaeota pJP 33/pSL50/pJP 41, and “Ca. Bathyarchaeota”), while it is sparsely distributed or appears to be absent in others. Within the same SepRS subgroup, SepRS phylogeny tends to show lineage specificity (Fig. 1A) (see reference 4 for the case of methanogens), indicating coevolution with the host organism for various time periods.
FIG 1 .
Distribution of SepRS, SepCysS, and SepCysE homologs in the prokaryotic domains of life. The bootstrap values (percentages) are shown for the unrooted maximum likelihood trees made with 100 replicates using MEGA 7. (A) Distribution of SepRS in archaea and bacteria. The archaeal species are (i) Rice cluster II or “Ca. Methanoflorentaceae” archaea (26); (ii) Thaumarchaeota AK59 archaea similar to clone AK59 locus tag AY555832 (all accession numbers are from GenBank unless otherwise specified) and clone 24Earc79 locus tag JN605031 (48, 82); (iii) “Ca. Bathyarchaeota” archaea (24, 38); (iv) three subgroups of uncultured Crenarchaeota (83) each similar to pSL50 gene U63342, pJP 33 gene L25300, and pJP 41 gene L25301, probably including “Ca. Verstraetearchaeota” archaea (21); (v) pMC2A209 archaea (49) similar to clone IAN1-71 locus tag AB175574 and clone ARC_OTU_72 locus tag KP091046 (47, 84); (vi) Hadesarchaea and MSBL1 archaea (36, 37); (vii) clone AK8/W8A-19 archaea similar to clone W8A-19 locus tag KM221272 (46) and clone AK8 locus tag AY555814 (48) or clone ARC_OTU_92 locus tag KP091068 (47); (viii) archaeon Odin LCB_4, “Ca. Lokiarchaeota” archaea, and a few unknown species in the Asgard superphylum (9, 31, 33); (ix) a Crystal Geyser groundwater (“Ca. Woesearchaeota”) archaeon most similar to archaeon GW2011_AR9 (85); (x) “Ca. Micrarchaeota” (Eury4AB group) archaea most similar to clone C1AA1CA10 locus tag GU127467 (86, 87); (xi) “Ca. Altiarchaeales” archaea (88–91); (xii) Crystal Geyser groundwater archaea (91), one of which is similar to clone SCA130 locus tag EU735580 (92); (xiii) locus tag Z7ME43 or Theionarchaea archaea (93); (xiv) Arc 1 group or “Ca. Methanofastidiosa” archaea (27); and (xv) unknown groups of archaea. The SepRS-harboring bacterial species are the “Ca. Parcubacteria” DG_74_2 bin bacterium (39), a putative deltaproteobacterium (CG bacterium no. 3), and Chloroflexi (probably Dehalococcoides) bacteria (34). SepRS sequences were classified into three clades and a few orphans. A few representative genetic loci of clade I SepRS genes are shown below the tree. As indicated, modern Crenarchaeota, including Thermofilum pendens, lack SepRS. TRAX belongs to translin superfamily proteins. FADS and RFK denote FAD synthase and riboflavin kinase, respectively. (B) Distribution of SepCysS in the prokaryotic domains of life. SepCysS sequences were classified into seven clades. (C) Distribution of SepCysE in three groups of selenocysteine-encoding archaea. (D) Multiple-alignment analysis of SepCysE homologs based on the crystal structure of M. jannaschii SepCysE (PDB accession no. 3wkr). The SepCysSn peptide is either encoded as a split gene preceding the SepCysS gene or N-terminally fused with SepCysS (SepCysSN).
Full trees for SepRS and SepCysS. Download FIG S1, PDF file, 0.1 MB (149KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
SepRS genes form three major clades (Fig. 1A). SepRS clade I is the largest and probably arose from a common ancestral operon containing the gene encoding an archaeal translin-associated protein X (TRAX) homolog (42) (Fig. 1A; Fig. S2). Assuming that the recently published (29) phylogenetic tree of life is mostly true, SepRS clade I represents relatively modern lineages of Euryarchaeota and the TACK and Asgard superphyla, whereas SepRS clades II and III represent more-ancient lineages of archaea (DPANN, “Ca. Altiarchaeales,” and Z7ME43/Theionarchaea) as well as bacteria. However, the evolutionary relationship of the three SepRS clades and the archaeal lineages remains unclear, notably because of an HGT event of SepRS in “Ca. Bathyarchaeota” and rapid evolution of SepRS in putative Euryarchaeota archaea (Fig. 1A). A late-branching “Ca. Bathyarchaeota” archaeon, BA2 (38), has a Hadesarchaea-type SepRS, which is different from other “Ca. Bathyarchaeota” SepRSs (Fig. 1A). The latter represent the most-diverged SepRS (or SepRS-like) genes, which were not paired with SepCysS genes by our contig binning. They may belong to the rapidly evolving groups of Euryarchaeota (Fig. 1A) (27, 43), which explains the extent of divergence.
Genetic loci of SepRS and SepCysS genes and bacterial tRNACys genes. Bacterial tRNACys species are shown with residue 37 marked with a red circle. Download FIG S2, PDF file, 0.5 MB (606.5KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(ii) Occurrence of SepCysS.
Classification of the collected SepCysS sequences revealed that, in addition to the three known SepCysS clades (here, clades I, III, and VII) (10), four other clades exist (here, clades II, IV, V, and VI) (Fig. 1B; Fig. S1). Residues critical for the Sep-to-Cys conversion are conserved in all SepCysS species, with only two exceptions. While residues involved in pyridoxal-phosphate (PLP) binding are conserved in all collected SepCysSs, the Cys residues involved in persulfide formation (6) are missing in these two cases, thereby suggesting that these SepCysS proteins might employ a different mechanism for sulfur transfer (see below).
SepCysS phylogeny shows a very low correlation with the SepRS phylogeny. There are two plausible explanations for it. (i) Some archaea have two copies of SepCysS genes (of the same clade or different clades) that are shared within the same subgroup of archaea (Fig. 1B; Fig. S1 and S2) (10, 44). Likewise, it is possible that a second SepCysS gene copy was excluded from our analysis due to incomplete genome sequencing and contig binning. Importantly, in our genome and metagenome analyses, no additional copies of SepRS genes were identified, nor were any SepCysS genes found in the complete genomes lacking SepRS. (ii) Because SepCysS shows less tRNA specificity than SepRS (45), SepCysS genes may be more prone to HGT than SepRS genes. The occurrence of a SepCysS gene duplication in some Euryarchaeota methanogens (Fig. S1) implies that an additional gene copy may enhance RNA-dependent cysteine biosynthesis under certain conditions.
(iii) Occurrence of SepCysE homologs.
SepCysE genes are present in a few selenocysteine-encoding archaea other than class I methanogens (Fig. 1C; Fig. S3). SepCysE is present either in an operon with SepCysS in AK8/W8A-19 group archaea (46–49) or separately in a BOG (Asgard) archaeon (Fig. 1B and C; Fig. S2 and S3). In the archaeal domain, the Sec utilization trait was found within the Euryarchaeota class I methanogens (8) and two Asgard superphylum members (31) (“Candidatus Lokiarchaeota” [9] and Thorarchaeota [50]). The AK8/W8A-19 group archaea and the BOG (Asgard) archaeon share four selenoproteins (SPS, HdrA, VhuD, and VhuU) with Methanopyrus, Methanococcus, and “Ca. Lokiarchaeota” (9), whereas the AK8/W8A-19 SPS proteins are split in two fragments (Fig. S3). Our findings lend support to the hypotheses that the archaeal Sec-encoding system and the SepRS-SepCysS-SepCysE system emerged prior to the divergence of class I methanogens and “Ca. Lokiarchaeota” (9) and prior to the divergence of class I, II, and III methanogens (4), respectively. However, the possibility of HGT events after the division of these archaeal groups cannot be excluded.
Cooccurrence of SepRS-SepCysS-SepCysE and Sec-encoding systems in the clone AK8 W8A-19 group archaea and the BOG (Asgard) archaeon. See Table S1 for other members. Download FIG S3, PDF file, 0.4 MB (507KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Binning and naming of genomic and metagenomic contigs. Download TABLE S1, DOCX file, 0.03 MB (39.2KB, docx) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Some archaeal genomes contain homologs of the N-terminal helix-turn-helix domain of SepCysE (Fig. 1B and D). This homolog is present as an additional domain fused to SepCysS (some of the clade VII SepCysSs) or encoded as a split gene in front of clade SepCysS genes (clade VII SepCysSs and a few clade I and VI SepCysS genes) (Fig. 1B and D; Fig. S1). This SepCysE homolog was named “SepCysSn” when encoded by a separate gene or “SepCysSN” when fused to SepCysS (Fig. 1D).
The genetic loci of SepRS and SepCysS.
The genetic loci and genes accompanying SepRS and SepCysS genes support the protein sequence-based phylogenies (Fig. S2). (i) Bacteria (and an archaeon) share the SepRS-SepCysS operon. In two cases, bacterial tRNACys is found in an operon with either SepRS-ΔC or the second copy of SepCysS (Fig. S2). (ii) “Ca. Bathyarchaeota” archaeon BA2 has a compact operon encoding tRNACys, SepCysSn, SepCysS, and SepRS. This operon is widespread among marine sediment archaea, possibly because it is so amenable to HGT. (iii) Clade VII SepCysS genes are often associated with a tRNACys gene, a few sulfur metabolism genes, and a small gene which was annotated to encode tRNA-Thr-editing domain (ED). As shown in Fig. S4, tRNA-Thr-ED is a homolog of the editing domain of archaeal threonyl-tRNA synthetase (ThrRS-R) (51) and the editing domain of archaeal transediting ThrRS-ED protein (52) (see Text S1 in the supplemental material). The tRNA-Thr-ED proteins of Euryarchaeota methanogens form a clade distinct from those of MSBL1/Hadesarchaea (Fig. S4B and C). The SepCysS proteins associated with these tRNA-Thr-ED proteins are distributed in the same manner (methanogens’ clade and MSBL1/Hadesarchaea clade) in our clade VII SepCysS phylogeny (Fig. 1B).
Supplemental text about tRNA-Thr-ED proteins. Download TEXT S1, DOCX file, 0.01 MB (18.2KB, docx) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification and analysis of homologs of the editing domain of archaeal ThrRS. (A) Crystal structure of the editing domain of ThrRS from M. jannaschii with l-Ser3AA (a substrate analog) (PDB accession no. 4RRF). (B) Phylogeny of tRNA-Thr-ED homologs and the editing domains of archaeal ThrRS (ThrRS-R) and ThrRS-ED. The bootstrap values (percentages) are shown for the unrooted maximum likelihood tree made with 100 replicates using MEGA 7. The tRNA-Thr-ED proteins associating with clade VII SepCysS (shown in the red box) are very similar to the archaeal ThrRS (ThrRS-R) and ThrRS-ED proteins. d-Tyrosyl-tRNATyr deacylase (DTD), used as an outgroup, was previously known as the closest homolog of the editing domain of ThrRS-R/ThrRS-ED. The full tree is shown below the compressed tree. (C) The SepCysS-associating tRNA-Thr-ED proteins from class II/III methanogens and rice cluster II methanogens have an additional C-terminal cysteine-rich motif, while Hadesarchaea MSBL1 proteins have no extension. Download FIG S4, PDF file, 0.3 MB (398.7KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Idiosyncrasies of bacterial SepRS and SepCysS.
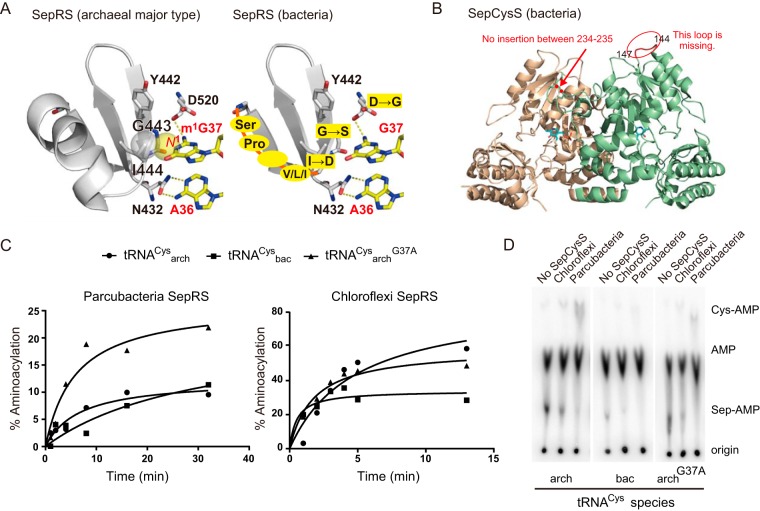
Bacterial SepRSs are highly distant from the well-studied methanogen SepRS gene (Fig. 1A). Bacterial SepCysSs, on the other hand, have a close evolutionary relationship with methanogens’ SepCysSs (Fig. 1B). Therefore, bacterial SepRS and SepCysS genes may have different archaeal origins and formed an operon after the branching of class I and class II and III methanogens. It is apparent from the multiple alignments of SepRS sequences that bacterial SepRS lacks a small motif involved in archaeal tRNACys recognition (53, 54) (Fig. 2A). As this motif binds methylated guanosine 37, an identity determinant in methanogen SepRS systems (53–55), it appears that the N1-methyl modification of G37 does not contribute to bacterial SepRS⋅tRNACys recognition. This is consistent with the fact that bacteria lack methyltransferase Trm5, which catalyzes m1G37 formation in archaeal tRNACys species (54, 56–58). Our structural models of bacterial SepRSs based on the Archaeoglobus fulgidus SepRS⋅tRNACys (PDB accession no. 2du3) crystal structure (59) show that a hydrophilic residue (mostly Asp) replaces the hydrophobic Ile444 within the enzyme’s anticodon binding domain. In A. fulgidus and methanogen SepRSs, Ile444 might be involved in m1G37 recognition (Fig. 2A). In addition, the vicinal helix of Ile444 is replaced with a short loop in bacterial SepRSs (Fig. 2A).
FIG 2 .
Bacterial SepRS and SepCysS and “Ca. Parcubacteria” tRNACys species. (A) Modeling of the N37-recognizing motif of SepRS. The crystal structure of A. fulgidus SepRS⋅tRNACys (PDB accession no. 2du3) was used for the modeling. Although G37 in the crystal structure is unmodified, N1-methylation may create a van der Waals interaction between the methyl group and the side chain of Ile444, as indicated with spheres. In bacterial SepRS species, Ile444 is replaced by hydrophilic Asp in most cases, and the following helix is totally missing. (B) Modeling of the dimer structure of SepCysS. The crystal structure of A. fulgidus SepCysS1 (PDB accession no. 2e7j) was used for the modeling. In bacterial SepCysS species, a loop (amino acids 144 to 147) is missing, and there is no insertion between amino acids 234 and 235. (C) Bacterial SepRS activity in vitro. Time course for plateau aminoacylation obtained by monitoring the accumulation of phosphoseryl-[32P]tRNACys using thin-layer chromatograms in Fig. S5A. tRNACys substrates from the “Ca. Parcubacteria” DG_74_2 bin are indicated (G37 containing tRNACys, [tRNACysarch], as a circle, its variant [tRNACysarchG37A] as a triangle, and “Ca. Parcubacteria”-type tRNACys as rectangles [tRNACysbac]). (D) Thin-layer chromatograms showing SepCysS-dependent O-phosphoseryl- to cysteinyl-[32P]tRNACys conversion. Reaction mixtures contained either “Ca. Parcubacteria” or Chloroflexi SepCysS (marked above the chromatogram). As a control, reaction mixtures containing only SepRS were also inspected (denoted “No SepCysS”). tRNA substrates are indicated below the chromatograms. Reaction products (O-phosphoseryl- and cysteinyl-[32P]tRNACys) were monitored as O-phosphoseryl- and cysteinyl-adenylates (Sep-AMP and Cys-AMP, respectively) after P1 nuclease digestion.
Bacterial SepCysS has the same structure as the archaeal clade I SepCysSs (e.g., A. fulgidus SepCysS1 [PDB accession no. 2e7j] [44]). Bacterial SepCysS lacks a fragment corresponding to a loop of A. fulgidus SepCysS1 (residues 144 to 147) (Fig. 2B). Only the CG (“Ca. Parcubacteria”) bacterium SepCysS retains this loop. Like the other clade I SepCysS genes (except Methanopyrus kandleri), bacterial SepCysS has an 8-amino-acid deletion between residues 234 and 235 (Fig. 2B). Because clade II-to-VII SepCysS species (with two exceptions) and M. kandleri SepCysS have an 8-amino-acid insertion here, the bacterial SepRS-SepCysS operons form a new lineage of SepCysS.
(i) Bacterial SepRSs aminoacylate bacterial type (A37) tRNACys species.
We established the function of two bacterial SepRS proteins (from “Ca. Parcubacteria” and Chloroflexi) with two tRNACys species (A37 or G37) found in the “Ca. Parcubacteria” DG_74_2 bin (Fig. S2). The tRNACys with A37 is similar to other “Ca. Parcubacteria” tRNAsCys and is designated tRNACysbac, whereas the tRNACys with G37, which is similar to some archaeal tRNAsCys species, is designated tRNACysarch. We also examined the artificial G37A variant of tRNACysarch. Recombinant “Ca. Parcubacteria” SepRS aminoacylated both tRNACysbac and tRNACysarch, albeit at a low level (10%) (Fig. 2C; Fig. S5A). The aminoacylation plateau level of the variant tRNACysarchG37A was about twice that of tRNACysarch, suggesting that G37 is not a determinant for “Ca. Parcubacteria” SepRS (55). Recombinant Chloroflexi SepRS acylated tRNACysarch and tRNACysarchG37A to 50 to 60%, while tRNACysbac was charged less well (to 35%) (Fig. 2C; Fig. S5A). Interestingly, an archaeal SepRS from Methanococcus maripaludis was also able to aminoacylate all three tRNACys types (Fig. S5B), indicating a more relaxed tRNA specificity than expected from previous studies (3, 53–55, 59).
In vitro and in vivo assays of bacterial SepRS, SepCysS, and tRNACys. (A) Bacterial SepRS activity in vitro. This is the original data for Fig. 2C. Thin-layer chromatograms showing phosphoseryl-[32P]tRNACys accumulation during a reaction time course in the case of “Ca. Parcubacteria” (left) and Chloroflexi SepRS (right). tRNA substrates are indicated below and time points are indicated above the chromatograms. A reaction mixture without the enzyme was loaded prior (c) and after (d) nuclease P1 digestion. (B) Bacterial tRNACys variants are substrates for Methanococcus maripaludis SepRS. Thin-layer chromatograms showing phosphoseryl-[32P]tRNACys (Sep-AMP) accumulation after 20 min (marked as “No SepCysS”). The produced phosphoseryl-[32P]tRNACys is a substrate for Chloroflexi SepCysS (denoted as “Chloroflexi” above the chromatogram). Only phosphoseryl-(Sep-AMP) and cysteinyl-[32P]adenylate (Cys-AMP) are monitored after nuclease P1 digestion of phosphoseryl- and cysteinyl-[32P]tRNACys, respectively. tRNA substrates are indicated below and SepCysS presence is indicated above the chromatograms. (C) Bacterial SepCysS activity in E. coli. E. coli JS1 (ΔselA) was transformed with pACYC-MjPSTK-EcselC, pBAD-CDF-fdhF, and either of the SepCysS-expressing plasmids and an empty vector. In JS1 cells, conversion of Sep-tRNASec to Cys-tRNASec by SepCysS leads to the expression of full-length formate dehydrogenase H (FDHH), which converts benzyl viologen into a purple dye. Only the edges of the cell spots on agar plates were stained, because Cys-containing FDHH is far less active than the wild-type-Sec-containing FDHH. Supplementation of the growth medium with sodium selenite (final concentration, 1 μM) enhanced the FDHH activity of the cells expressing the “Ca. Parcubacteria” SepCysS, possibly due to the formation of Sec-tRNASec. Download FIG S5, PDF file, 1.1 MB (1.2MB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(ii) Bacterial SepCysSs catalyze the tRNA-dependent Sep-to-Cys conversion.
Although the exact mechanism of the SepCysS-catalyzed reaction has not yet been fully elucidated, the Sep-to-Cys conversion most likely proceeds through a PLP-dependent generation of a dehydroalanyl-tRNACys intermediate, which is subsequently attacked by a persulfide group to form Cys-tRNACys (60, 61). Both “Ca. Parcubacteria” and Chloroflexi SepCysS possess residues involved in PLP binding (59), while only Chloroflexi SepCysS harbors conserved Cys residues implicated in persulfide (60, 61) and Fe−S cluster formation (6). “Ca. Parcubacteria” SepCysS is one of the two exceptional SepCysSs that lacks 2 out of 3 conserved cysteines (see above).
Both bacterial SepCysS proteins were expressed in Escherichia coli and purified anaerobically. Consistently with the presence of an Fe−S cluster, samples containing purified Chloroflexi SepCysS displayed a brown color, while “Ca. Parcubacteria” SepCysS samples were colorless, in agreement with the lack of cysteines needed for an Fe−S cluster formation (6). To demonstrate SepCysS activity, reaction mixtures containing 5 µM Chloroflexi SepRS and either 40 µM Chloroflexi or 20 µM “Ca. Parcubacteria” SepCysS were performed. Like SepRS, both SepCysSs were shown to be functional in the cases of all three tRNACys variants (Fig. 2D). While conversion was complete in the case of “Ca. Parcubacteria” SepCysS (95 to 99% of the total Sep-tRNACys intermediate), Chloroflexi SepCysS converted ~30 to 60% of Sep-tRNACys to Cys-tRNACys (Fig. 2D). We also found using a previously reported method (45) that “Ca. Parcubacteria” and Chloroflexi SepCysS proteins are active in E. coli (Fig. S5C).
Cooccurrence of PylRS and SepRS in some archaea and bacteria.
Diverse archaea and a group of Chloroflexi bacteria possess both SepRS and Pyl-encoding systems. Methane-producing Methermicoccus shengliensis strains AmaM and ZC-1 (62, 63) contain a Methanomassiliicoccales-type PylSc and tRNAPyl (Fig. 3). M. shengliensis uses trimethylamine for methanogenesis (62, 63), consistent with the presence of Pyl-containing mono-, di-, and trimethylamine methyltransferases (MtmB, MtbB, MttB) (Fig. 3) (23, 63). Two closely related MSBL1 archaea, SCGC-AAA382A20 and SCGC-AAA382A03 (37), have an incomplete or defective Pyl-encoding system and MtmB/MtbB/MttB genes. Very similar and complete Pyl-encoding operons of another halophilic archaeon have been reported (BioSample accession no. SAMN05770050). The V1 strain of “Candidatus Verstraetearchaeota,” or “Ca. Methanomethylicus” sp. strain V1, a newly proposed methanogen within the Terrestrial Miscellaneous Crenarchaeota group (TMCG), was reported to have PylSc, PylSn, and MtmB/MtbB (21), although tRNAPyl was not identified. We found the V1 to V5 strains of “Ca. Verstraetearchaeota” (21) to have SepRS of the pJP 41 SepRS group (Fig. 1A). Surprisingly, the PylRS species of the MSBL1 and “Ca. Verstraetearchaeota” archaea are not grouped within the three known clades of PylRS (Fig. 3).
FIG 3 .
Distribution of PylRS in the prokaryotic domains of life. The bootstrap values (percentages) are shown for the unrooted maximum likelihood tree made with 100 replicates using MEGA 7. Bacterial origin is indicated with brown letters, while archaeal origin is indicated with black letters. Purple letters indicate a pending phylogenetic inference. Metagenomic origins are described in black parentheses, whereas “(pylSn)” indicates the existence of a pylSn gene. The occurrence of SepRS and Pyl-containing methyltransferases are shown next to the tree. For some of the archaeal bins in the hot spring metagenomes, the presence and absence of SepRS is pending, which is designated with “?.”
In analyzing the metagenomic data sets, we focused on particular metagenomes because a whole Pyl-encoding gene cluster is rarely contained in a single metagenomic contig, which hampers the phylogenetic inference of PylRS genes. We chose data sets from deep marine and hot spring environments because of the abundance of archaeal species in these niches, the presence of SepRS genes, and high-quality data that are provided by Microbial Dark Matter, phase II.
Invaluable information was obtained from the metagenome of a deep-oceanic, basalt-hosted subsurface ecosystem from Juan de Fuca Ridge flank, Pacific Ocean (CORK borehole 1362A_J2.573). Three dominant archaeal species, Methermicoccaceae, marine benthic group E (MBG-E), and “Ca. Bathyarchaeota” (64), possess both SepRS and PylRS genes as well as the genes for Pyl-utilizing MtmB/MtbB/MttB enzymes (Fig. 3). The PylRS species of the Methermicoccaceae and MBG-E archaea divide the bacterial PylRS clade into two (Acetohalobium and others) (Fig. 3), indicating the occurrence of horizontal gene transfer of a Pyl-encoding system between bacteria and archaea (65). The “Ca. Bathyarchaeota” PylRS (PylSc) forms a new PylRS clade (Fig. 3) together with the V1 PylSc and some PylSc species found in the Deep Marine Sediments White Oak River (WOR) estuary metagenomes (data not shown). Pyl-encoding systems are also present in the hot spring metagenomes, although their metagenomic bins are less reliable due to the complex composition of the prokaryotic communities (Fig. 3). In the sulfidic Washburn Spring metagenome, one Archaeoglobus-type and several Crenarchaeota-type SepRS genes were also found. Thus, it is tempting to assume that a few subgroups of Archaeoglobus and TMCG possess both SepRS and PylRS.
The metagenome data revealed many Chloroflexi-type pylSc and pylSn genes (Fig. 3; Fig. S6A). Interestingly, a lineage of Dehalococcoides was found to have both SepRS and PylRS (Fig. 3). In one case, a SepRS-SepCysS operon and a pylSn gene exist on the same metagenomic contig (Fig. S6B).
Cooccurrence of SepRS and PylRS in bacteria. (A) Lineage of Dehalococcoides carrying SepRS, SepCysS, and PylRS; (B) binning of metagenomic contigs of the three Dehalococcoides strains with SepRS, SepCysS, and PylRS. Download FIG S6, PDF file, 0.01 MB (198.5KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
In this study, we searched all the genomic and metagenomic protein sequence data in the public databases for the RNA-dependent cysteine biosynthesis pathway. Previous studies used only genome sequences and a particular metagenome sequence datum to search for a particular aminoacyl-tRNA synthesis system, in part due to the low reliability and accessibility of metagenomic sequence data. We encountered a similar problem with the “Ca. Parcubacteria” DG_74_2 bin, which is apparently composed of a few different genomes, including two “Ca. Parcubacteria” species. Our contig binning was greatly facilitated by the fact that minor genetic code systems rely on multiple components, which are, in turn, frequently dispersed on different metagenomic contigs. This approach eventually led us to the detection of rRNA and protein genes useful for phylogenetic inference. This work and other recent studies (66–68) will lead future studies of gene evolution in uncultured microbes.
Our phylogenetic analyses demonstrate that (i) the well-investigated class I/II/III methanogen and Archaeoglobi SepRSs constitute only a terminal branch of one of the clades, (ii) the TRAX-SepRS genes, SepCysE, the Sec-encoding system, and the four selenoproteins are shared by Euryarchaeota and TACK/Asgard, (iii) a few groups of proteins accompany SepCysS genes within the genetic loci, (iv) new PylRS types occur in nature and represent a missing link between the three known clades of PylRS, and (v) modern archaea may have fused the adapter peptides PylSn and SepCysSn to PylSc and SepCysS, respectively. In addition, our biochemical analyses confirm that bacterial SepRS and SepCysS species from uncultured “Ca. Parcubacteria” and Chloroflexi bacteria possess canonical activity. It is still not clear whether the SepRS system was present in the last common archaeal ancestor (3, 4), because the SepRS system was rarely found in the DPANN group, which was predicted (29) to have diverged first among archaea. It is also unclear why the SepRS system is absent or sparsely distributed in many branches of archaea. Was it gradually replaced by CysRS in each branch or horizontally transferred from another branch?
There may be diverse mechanisms for RNA-dependent cysteine biosynthesis in nature. The composite genome of a bacterium, ADurb.Bin236 (BioSample accession no. SAMN05004151), encodes a noncanonical SepRS homolog (GenBank accession no. OQA87054.1) and a SepCysSn-SepCysS operon (GenBank accession no. OQA83877.1 and OQA83876.1). Their protein sequences are highly diverged and may have archaeal origins. Surprisingly, this SepRS homolog has an additional N-terminal domain corresponding to the serine-editing domain of archaeal ThrRS (ThrRS-R). This is consistent with the genetic coupling of some clade VII SepCysS and tRNA-Thr-ED genes in Euryarchaeota. Although further study and validation are required, one may hypothesize that some SepRS species might possess a serine-editing activity in cis or in trans, because dephosphorylation of Sep-tRNACys produces Ser-tRNACys, which may translate cysteine codons as serine.
The presence of the SepRS and SepCysS system may correlate with a high demand for iron-sulfur proteins, one of which is SepCysS (6), in obligate anaerobes for methanogenesis and for other metabolisms (14, 69, 70). For example, organohalide respiration in Dehalococcoides relies on iron-sulfur proteins (71). The coexisting tendency of PylRS and SepRS in archaea and bacteria may be partially explained by the facts that methylamine metabolism by Pyl-utilizing enzymes requires an iron-sulfur protein, RamA (8, 72), and that methylornithine synthase (PylB) is an iron-sulfur enzyme (73). Apart from assisting iron-sulfur proteins, the SepRS system may be useful for extreme thermophiles, because free phosphoserine is stable even at an extremely high temperature (74).
It has been shown that genes characteristic of methanogen-type sulfur assimilation and mobilization exist in some deltaproteobacteria and Chloroflexi (75). These genes encode proteins involved in methanogen-idiosyncratic homocysteine synthesis and facilitate growth when sulfide is provided as the sole sulfur source (5, 61, 75). As predicted, these genes cooccur with SepCysS and are present in SepRS-carrying Chloroflexi Dehalococcoidia bacterium CG2_30_46_9, Dehalococcoidia bacterium CG2_30_46_19, and Chloroflexi bacterium RBG_13_51_36 (see Fig. S1 in the supplemental material). Because of the vast abundance of Chloroflexi in deep sediments, their metabolic traits have a direct impact on sulfur cycling within the marine subsurface.
MATERIALS AND METHODS
Bioinformatics.
A BLAST search was performed by using three public Web servers, JGI IMG/MER (35), NCBI BLAST, and NCBI SRA BLAST. Some of the SepCysSn and tRNA sequences were manually identified. The SepRS sequences of “Ca. Verstraetearchaeota” were obtained by using tBLASTn from accession no. PRJNA321438. Multiple-alignment analyses of protein sequences were performed using Clustal X 2.1 (40), followed by manual curation based on the reported structure-based alignment analyses of SepRS (59), SepCysS (44), SepCysE (4), the ThrRS editing domain (76), and PylRS (77) using SeaView (78). The phylogeny reconstruction analyses of the alignment files were performed by using MEGA 7 (79) with the default settings (maximum likelihood, Jones-Taylor-Thornton [JTT] model, uniform rates, use all gaps/missing sites). Protein structure models were made with PyMol 1.7.6.0 (Schrödinger, LLC). The sequence and alignment data used in this study are provided in the supplemental material (see Data Set S1).
Sequence and alignment data. Download DATA SET S1, RTF file, 0.4 MB (473.5KB, rtf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Binning of metagenomic contigs was performed based on GC contents and read depths. Some of the WOR metagenomic contigs lack the read depth information. For the binning of metagenomic contigs of the AK8/W8A-19 group archaea, contaminating “Ca. Bathyarchaeota” contigs were removed. For the binning of metagenomic contigs of the BOG (Asgard) archaeon, each contig was confirmed to harbor Asgard-like protein genes, and contaminating Methanocella contigs were removed. The automatic annotation pipelines of the NCBI and JGI databases and our manual annotation/curation identified or predicted the host archaea and bacteria of these metagenomic contigs (Table S1). Our 16S rRNA phylogeny revealed that unclassified “LHC4-2-B” archaea JGI MDM2 LHC4sed-1-M8 and N8 belong to the pSL50 group and that unclassified “LHC4-2-B” archaeon JGI MDM2 LHC4sed-1-M18 belongs to the pJP 33 group. It was revealed that W8A-19 archaea, which were annotated to belong to the Korarchaeota (46), have ribosomal protein operons very similar to those of AK8 archaea (Fig. S3).
In vitro and in vivo assays of bacterial SepRS and SepCysS.
Assays were performed using traditional methods (45, 80, 81). Detailed materials and methods are provided in Text S2 in the supplemental material.
Supplemental materials and methods. Download TEXT S2, DOCX file, 0.2 MB (26.4KB, docx) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Jessica Jarett, Eric Becraft, Ramunas Stepanauskas, and Brian P. Hedlund for permission to use the Microbial Dark Matter (phase II) data produced through the DOE JGI’s community sequencing program. We also thank many others for permission to use unpublished sequence data through the program. We are grateful to Yuchen Liu, Patrick O’Donoghue, Noah M. Reynolds, Tateki Suzuki, and Oscar Vargas-Rodriguez for enlightened discussions.
T.M. is a Japan Society for the Promotion of Science postdoctoral fellow for research abroad. This work was supported by grants from the National Institute for General Medical Sciences (R01GM22854 and R35GM122560 to D.S.) and from the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the Department of Energy (DE-FG02-98ER20311 to D.S. [for funding the genetic experiments]). The work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported under contract no. DE-AC02-05CH11231.
Footnotes
Citation Mukai T, Crnković A, Umehara T, Ivanova NN, Kyrpides NC, Söll D. 2017. RNA-dependent cysteine biosynthesis in bacteria and archaea. mBio 8:e00561-17. https://doi.org/10.1128/mBio.00561-17.
REFERENCES
- 1.Srinivasan G, James CM, Krzycki JA. 2002. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 2.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. 2002. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 3.Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR III, Ibba M, Söll D. 2005. RNA-dependent cysteine biosynthesis in archaea. Science 307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 4.Liu Y, Nakamura A, Nakazawa Y, Asano N, Ford KA, Hohn MJ, Tanaka I, Yao M, Söll D. 2014. Ancient translation factor is essential for tRNA-dependent cysteine biosynthesis in methanogenic archaea. Proc Natl Acad Sci U S A 111:10520–10525. doi: 10.1073/pnas.1411267111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rauch BJ, Perona JJ. 2016. Efficient sulfide assimilation in Methanosarcina acetivorans is mediated by the MA1715 protein. J Bacteriol 198:1974–1983. doi: 10.1128/JB.00141-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Vinyard DJ, Reesbeck ME, Suzuki T, Manakongtreecheep K, Holland PL, Brudvig GW, Söll D. 2016. A [3Fe 4S] cluster is required for tRNA thiolation in archaea and eukaryotes. Proc Natl Acad Sci U S A 113:12703–12708. doi: 10.1073/pnas.1615732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Nakazawa Y, Kubo Y, Asano N, Kato K, Tanaka I, Yao M. 2016. Crystallographic analysis of a subcomplex of the transsulfursome with tRNACys-tRNACys synthesis. Acta Crystallogr F Struct Biol Commun 72:569–572. doi: 10.1107/S2053230X16009559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rother M, Krzycki JA. 2010. Selenocysteine, pyrrolysine, and the unique energy metabolism of methanogenic archaea. Archaea 2010:453642. doi: 10.1155/2010/453642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mariotti M, Lobanov AV, Manta B, Santesmasses D, Bofill A, Guigó R, Gabaldón T, Gladyshev VN. 2016. Lokiarchaeota marks the transition between the archaeal and eukaryotic selenocysteine encoding systems. Mol Biol Evol 33:2441–2453. doi: 10.1093/molbev/msw122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Donoghue P, Sethi A, Woese CR, Luthey-Schulten ZA. 2005. The evolutionary history of Cys-tRNACys formation. Proc Natl Acad Sci U S A 102:19003–19008. doi: 10.1073/pnas.0509617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li T, Graham DE, Stathopoulos C, Haney PJ, Kim HS, Vothknecht U, Kitabatake M, Hong KW, Eggertsson G, Curnow AW, Lin W, Celic I, Whitman W, Söll D. 1999. Cysteinyl-tRNA formation: the last puzzle of aminoacyl-tRNA synthesis. FEBS Lett 462:302–306. doi: 10.1016/S0014-5793(99)01550-1. [DOI] [PubMed] [Google Scholar]
- 12.Newberry KJ, Hou YM, Perona JJ. 2002. Structural origins of amino acid selection without editing by cysteinyl-tRNA synthetase. EMBO J 21:2778–2787. doi: 10.1093/emboj/21.11.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang CM, Christian T, Newberry KJ, Perona JJ, Hou YM. 2003. Zinc-mediated amino acid discrimination in cysteinyl-tRNA synthetase. J Mol Biol 327:911–917. doi: 10.1016/S0022-2836(03)00241-9. [DOI] [PubMed] [Google Scholar]
- 14.Klipcan L, Frenkel-Morgenstern M, Safro MG. 2008. Presence of tRNA-dependent pathways correlates with high cysteine content in methanogenic Archaea. Trends Genet 24:59–63. doi: 10.1016/j.tig.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 15.Meyerdierks A, Kube M, Kostadinov I, Teeling H, Glöckner FO, Reinhardt R, Amann R. 2010. Metagenome and mRNA expression analyses of anaerobic methanotrophic archaea of the ANME-1 group. Environ Microbiol 12:422–439. doi: 10.1111/j.1462-2920.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 16.Sethi A, O’Donoghue P, Luthey-Schulten Z. 2005. Evolutionary profiles from the QR factorization of multiple sequence alignments. Proc Natl Acad Sci U S A 102:4045–4050. doi: 10.1073/pnas.0409715102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, Krzycki JA. 2004. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature 431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 18.Polycarpo C, Ambrogelly A, Bérubé A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Söll D. 2004. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc Natl Acad Sci U S A 101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fournier GP, Andam CP, Gogarten JP. 2015. Ancient horizontal gene transfer and the last common ancestors. BMC Evol Biol 15:70. doi: 10.1186/s12862-015-0350-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borrel G, Gaci N, Peyret P, O’Toole PW, Gribaldo S, Brugère JF. 2014. Unique characteristics of the pyrrolysine system in the 7th order of methanogens: implications for the evolution of a genetic code expansion cassette. Archaea 2014:374146. doi: 10.1155/2014/374146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanwonterghem I, Evans PN, Parks DH, Jensen PD, Woodcroft BJ, Hugenholtz P, Tyson GW. 2016. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat Microbiol 1:16170. doi: 10.1038/nmicrobiol.2016.170. [DOI] [PubMed] [Google Scholar]
- 22.Longstaff DG, Larue RC, Faust JE, Mahapatra A, Zhang L, Green-Church KB, Krzycki JA. 2007. A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine. Proc Natl Acad Sci U S A 104:1021–1026. doi: 10.1073/pnas.0610294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ticak T, Kountz DJ, Girosky KE, Krzycki JA, Ferguson DJ Jr.. 2014. A nonpyrrolysine member of the widely distributed trimethylamine methyltransferase family is a glycine betaine methyltransferase. Proc Natl Acad Sci U S A 111:E4668–E4676. doi: 10.1073/pnas.1409642111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, Tyson GW. 2015. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350:434–438. doi: 10.1126/science.aac7745. [DOI] [PubMed] [Google Scholar]
- 25.Borrel G, Adam PS, Gribaldo S. 2016. Methanogenesis and the Wood-Ljungdahl pathway: an ancient, versatile, and fragile association. Genome Biol Evol 8:1706–1711. doi: 10.1093/gbe/evw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mondav R, Woodcroft BJ, Kim EH, McCalley CK, Hodgkins SB, Crill PM, Chanton J, Hurst GB, VerBerkmoes NC, Saleska SR, Hugenholtz P, Rich VI, Tyson GW. 2014. Discovery of a novel methanogen prevalent in thawing permafrost. Nat Commun 5:3212. doi: 10.1038/ncomms4212. [DOI] [PubMed] [Google Scholar]
- 27.Nobu MK, Narihiro T, Kuroda K, Mei R, Liu WT. 2016. Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J 10:2478–2487. doi: 10.1038/ismej.2016.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcy Y, Ouverney C, Bik EM, Lösekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, Quake SR. 2007. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A 104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 30.Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 31.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJ. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 32.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 33.Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJG. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anantharaman K, Brown CT, Hug LA, Sharon I, Castelle CJ, Probst AJ, Thomas BC, Singh A, Wilkins MJ, Karaoz U, Brodie EL, Williams KH, Hubbard SS, Banfield JF. 2016. Thousands of microbial genomes shed light on interconnected biogeochemical processes in an aquifer system. Nat Commun 7:13219. doi: 10.1038/ncomms13219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen IA, Markowitz VM, Chu K, Palaniappan K, Szeto E, Pillay M, Ratner A, Huang J, Andersen E, Huntemann M, Varghese N, Hadjithomas M, Tennessen K, Nielsen T, Ivanova NN, Kyrpides NC. 2017. IMG/M: integrated genome and metagenome comparative data analysis system. Nucleic Acids Res 45:D507–D516. doi: 10.1093/nar/gkw929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker BJ, Saw JH, Lind AE, Lazar CS, Hinrichs KU, Teske AP, Ettema TJ. 2016. Genomic inference of the metabolism of cosmopolitan subsurface Archaea, Hadesarchaea. Nat Microbiol 1:16002. doi: 10.1038/nmicrobiol.2016.2. [DOI] [PubMed] [Google Scholar]
- 37.Mwirichia R, Alam I, Rashid M, Vinu M, Ba-Alawi W, Anthony Kamau A, Kamanda Ngugi D, Göker M, Klenk HP, Bajic V, Stingl U. 2016. Metabolic traits of an uncultured archaeal lineage—MSBL1—from brine pools of the Red Sea. Sci Rep 6:19181. doi: 10.1038/srep19181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He Y, Li M, Perumal V, Feng X, Fang J, Xie J, Sievert SM, Wang F. 2016. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol 1:16035. doi: 10.1038/nmicrobiol.2016.35. [DOI] [PubMed] [Google Scholar]
- 39.Baker BJ, Lazar CS, Teske AP, Dick GJ. 2015. Genomic resolution of linkages in carbon, nitrogen, and sulfur cycling among widespread estuary sediment bacteria. Microbiome 3:14. doi: 10.1186/s40168-015-0077-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 41.Mukai T, Englert M, Tripp HJ, Miller C, Ivanova NN, Rubin EM, Kyrpides NC, Söll D. 2016. Facile recoding of selenocysteine in nature. Angew Chem Int Ed Engl 55:5337–5341. doi: 10.1002/anie.201511657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta GD, Kale A, Kumar V. 2012. Molecular evolution of Translin Superfamily proteins within the genomes of eubacteria, Archaea and eukaryotes. J Mol Evol 75:155–167. doi: 10.1007/s00239-012-9534-z. [DOI] [PubMed] [Google Scholar]
- 43.Dowell F, Cardman Z, Dasarathy S, Kellermann MY, Lipp JS, Ruff SE, Biddle JF, McKay LJ, MacGregor BJ, Lloyd KG, Albert DB, Mendlovitz H, Hinrichs KU, Teske A. 2016. Microbial communities in methane- and short chain alkane-rich hydrothermal sediments of Guaymas Basin. Front Microbiol 7:17. doi: 10.3389/fmicb.2016.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fukunaga R, Yokoyama S. 2007. Structural insights into the second step of RNA-dependent cysteine biosynthesis in archaea: crystal structure of Sep-tRNA:Cys-tRNA synthase from Archaeoglobus fulgidus. J Mol Biol 370:128–141. doi: 10.1016/j.jmb.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 45.Yuan J, Hohn MJ, Sherrer RL, Palioura S, Su D, Söll D. 2010. A tRNA-dependent cysteine biosynthesis enzyme recognizes the selenocysteine-specific tRNA in Escherichia coli. FEBS Lett 584:2857–2861. doi: 10.1016/j.febslet.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li JW, Peng XT, Zhang LX, Jiang L, Chen S. 2016. Linking microbial community structure to S, N and Fe biogeochemical cycling in the hot springs at the Tengchong geothermal fields, Southwest China. Geomicrobiol J 33:135–150. doi: 10.1080/01490451.2015.1043165. [DOI] [Google Scholar]
- 47.McKay L, Klokman VW, Mendlovitz HP, LaRowe DE, Hoer DR, Albert D, Amend JP, Teske A. 2016. Thermal and geochemical influences on microbial biogeography in the hydrothermal sediments of Guaymas Basin, Gulf of California. Environ Microbiol Rep 8:150–161. doi: 10.1111/1758-2229.12365. [DOI] [PubMed] [Google Scholar]
- 48.Kanokratana P, Chanapan S, Pootanakit K, Eurwilaichitr L. 2004. Diversity and abundance of Bacteria and Archaea in the Bor Khlueng Hot Spring in Thailand. J Basic Microbiol 44:430–444. doi: 10.1002/jobm.200410388. [DOI] [PubMed] [Google Scholar]
- 49.Saw JH, Spang A, Zaremba-Niedzwiedzka K, Juzokaite L, Dodsworth JA, Murugapiran SK, Colman DR, Takacs-Vesbach C, Hedlund BP, Guy L, Ettema TJ. 2015. Exploring microbial dark matter to resolve the deep archaeal ancestry of eukaryotes. Philos Trans R Soc Lond B Biol Sci 370:20140328. doi: 10.1098/rstb.2014.0328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seitz KW, Lazar CS, Hinrichs KU, Teske AP, Baker BJ. 2016. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J 10:1696–1705. doi: 10.1038/ismej.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andam CP, Gogarten JP. 2011. Biased gene transfer and its implications for the concept of lineage. Biol Direct 6:47. doi: 10.1186/1745-6150-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Korenčić D, Ahel I, Schelert J, Sacher M, Ruan B, Stathopoulos C, Blum P, Ibba M, Söll D. 2004. A freestanding proofreading domain is required for protein synthesis quality control in Archaea. Proc Natl Acad Sci U S A 101:10260–10265. doi: 10.1073/pnas.0403926101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hauenstein SI, Perona JJ. 2008. Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei. J Biol Chem 283:22007–22017. doi: 10.1074/jbc.M801839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang CM, Liu C, Slater S, Hou YM. 2008. Aminoacylation of tRNA with phosphoserine for synthesis of cysteinyl-tRNACys. Nat Struct Mol Biol 15:507–514. doi: 10.1038/nsmb.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hohn MJ, Park HS, O’Donoghue P, Schnitzbauer M, Söll D. 2006. Emergence of the universal genetic code imprinted in an RNA record. Proc Natl Acad Sci U S A 103:18095–18100. doi: 10.1073/pnas.0608762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ito T, Masuda I, Yoshida K, Goto-Ito S, Sekine S, Suh SW, Hou YM, Yokoyama S. 2015. Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc Natl Acad Sci U S A 112:E4197–E4205. doi: 10.1073/pnas.1422981112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christian T, Lahoud G, Liu C, Hoffmann K, Perona JJ, Hou YM. 2010. Mechanism of N-methylation by the tRNA m1G37 methyltransferase Trm5. RNA 16:2484–2492. doi: 10.1261/rna.2376210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goto-Ito S, Ito T, Ishii R, Muto Y, Bessho Y, Yokoyama S. 2008. Crystal structure of archaeal tRNA(m1G37)methyltransferase aTrm5. Proteins 72:1274–1289. doi: 10.1002/prot.22019. [DOI] [PubMed] [Google Scholar]
- 59.Fukunaga R, Yokoyama S. 2007. Structural insights into the first step of RNA-dependent cysteine biosynthesis in archaea. Nat Struct Mol Biol 14:272–279. doi: 10.1038/nsmb1219. [DOI] [PubMed] [Google Scholar]
- 60.Liu Y, Dos Santos PC, Zhu X, Orlando R, Dean DR, Söll D, Yuan J. 2012. Catalytic mechanism of Sep-tRNA:Cys-tRNA synthase: sulfur transfer is mediated by disulfide and persulfide. J Biol Chem 287:5426–5433. doi: 10.1074/jbc.M111.313700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rauch BJ, Klimek J, David L, Perona JJ. 2017. Persulfide formation mediates cysteine and homocysteine biosynthesis in Methanosarcina acetivorans. Biochemistry 56:1051–1061. doi: 10.1021/acs.biochem.6b00931. [DOI] [PubMed] [Google Scholar]
- 62.Cheng L, Qiu TL, Yin XB, Wu XL, Hu GQ, Deng Y, Zhang H. 2007. Methermicoccus shengliensis gen. nov., sp. nov., a thermophilic, methylotrophic methanogen isolated from oil-production water, and proposal of Methermicoccaceae fam. nov. Int J Syst Evol Microbiol 57:2964–2969. doi: 10.1099/ijs.0.65049-0. [DOI] [PubMed] [Google Scholar]
- 63.Mayumi D, Mochimaru H, Tamaki H, Yamamoto K, Yoshioka H, Suzuki Y, Kamagata Y, Sakata S. 2016. Methane production from coal by a single methanogen. Science 354:222–225. doi: 10.1126/science.aaf8821. [DOI] [PubMed] [Google Scholar]
- 64.Jungbluth SP, Bowers RM, Lin HT, Cowen JP, Rappé MS. 2016. Novel microbial assemblages inhabiting crustal fluids within mid-ocean ridge flank subsurface basalt. ISME J 10:2033–2047. doi: 10.1038/ismej.2015.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prat L, Heinemann IU, Aerni HR, Rinehart J, O’Donoghue P, Söll D. 2012. Carbon source-dependent expansion of the genetic code in bacteria. Proc Natl Acad Sci U S A 109:21070–21075. doi: 10.1073/pnas.1218613110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukai T, Reynolds NM, Crnković A, Söll D. 2017. Bioinformatic analysis reveals archaeal tRNATyr and tRNATrp identities in bacteria. Life (Basel) 7:e8. doi: 10.3390/life7010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Burstein D, Harrington LB, Strutt SC, Probst AJ, Anantharaman K, Thomas BC, Doudna JA, Banfield JF. 2017. New CRISPR-Cas systems from uncultivated microbes. Nature 542:237–241. doi: 10.1038/nature21059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wrighton KC, Castelle CJ, Varaljay VA, Satagopan S, Brown CT, Wilkins MJ, Thomas BC, Sharon I, Williams KH, Tabita FR, Banfield JF. 2016. RuBisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J 10:2702–2714. doi: 10.1038/ismej.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin WF. 2016. The physiology and habitat of the last universal common ancestor. Nat Microbiol 1:16116. doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- 70.Rothman DH, Fournier GP, French KL, Alm EJ, Boyle EA, Cao C, Summons RE. 2014. Methanogenic burst in the end-Permian carbon cycle. Proc Natl Acad Sci U S A 111:5462–5467. doi: 10.1073/pnas.1318106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kublik A, Deobald D, Hartwig S, Schiffmann CL, Andrades A, von Bergen M, Sawers RG, Adrian L. 2016. Identification of a multi-protein reductive dehalogenase complex in Dehalococcoides mccartyi strain CBDB1 suggests a protein-dependent respiratory electron transport chain obviating quinone involvement. Environ Microbiol 18:3044–3056. doi: 10.1111/1462-2920.13200. [DOI] [PubMed] [Google Scholar]
- 72.Ferguson T, Soares JA, Lienard T, Gottschalk G, Krzycki JA. 2009. RamA, a protein required for reductive activation of corrinoid-dependent methylamine methyltransferase reactions in methanogenic archaea. J Biol Chem 284:2285–2295. doi: 10.1074/jbc.M807392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quitterer F, List A, Eisenreich W, Bacher A, Groll M. 2012. Crystal structure of methylornithine synthase (PylB): insights into the pyrrolysine biosynthesis. Angew Chem Int Ed Engl 51:1339–1342. doi: 10.1002/anie.201106765. [DOI] [PubMed] [Google Scholar]
- 74.Makino Y, Sato T, Kawamura H, Hachisuka SI, Takeno R, Imanaka T, Atomi H. 2016. An archaeal ADP-dependent serine kinase involved in cysteine biosynthesis and serine metabolism. Nat Commun 7:13446. doi: 10.1038/ncomms13446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rauch BJ, Gustafson A, Perona JJ. 2014. Novel proteins for homocysteine biosynthesis in anaerobic microorganisms. Mol Microbiol 94:1330–1342. doi: 10.1111/mmi.12832. [DOI] [PubMed] [Google Scholar]
- 76.Hussain T, Kruparani SP, Pal B, Dock-Bregeon AC, Dwivedi S, Shekar MR, Sureshbabu K, Sankaranarayanan R. 2006. Post-transfer editing mechanism of a d-aminoacyl-tRNA deacylase-like domain in threonyl-tRNA synthetase from archaea. EMBO J 25:4152–4162. doi: 10.1038/sj.emboj.7601278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nozawa K, O’Donoghue P, Gundllapalli S, Araiso Y, Ishitani R, Umehara T, Söll D, Nureki O. 2009. Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature 457:1163–1167. doi: 10.1038/nature07611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 79.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ledoux S, Uhlenbeck OC. 2008. [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44:74–80. doi: 10.1016/j.ymeth.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolfson AD, Pleiss JA, Uhlenbeck OC. 1998. A new assay for tRNA aminoacylation kinetics. RNA 4:1019–1023. doi: 10.1017/S1355838298980700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kubo K, Lloyd KG, F Biddle J, Amann R, Teske A, Knittel K. 2012. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J 6:1949–1965. doi: 10.1038/ismej.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barns SM, Delwiche CF, Palmer JD, Pace NR. 1996. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci U S A 93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robertson CE, Spear JR, Harris JK, Pace NR. 2009. Diversity and stratification of archaea in a hypersaline microbial mat. Appl Environ Microbiol 75:1801–1810. doi: 10.1128/AEM.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ, Frischkorn KR, Tringe SG, Singh A, Markillie LM, Taylor RC, Williams KH, Banfield JF. 2015. Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol 25:690–701. doi: 10.1016/j.cub.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 86.Durbin AM, Teske A. 2012. Archaea in organic-lean and organic-rich marine subsurface sediments: an environmental gradient reflected in distinct phylogenetic lineages. Front Microbiol 3:168. doi: 10.3389/fmicb.2012.00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Restrepo-Ortiz CX, Casamayor EO. 2013. Environmental distribution of two widespread uncultured freshwater Euryarchaeota clades unveiled by specific primers and quantitative PCR. Environ Microbiol Rep 5:861–867. doi: 10.1111/1758-2229.12088. [DOI] [PubMed] [Google Scholar]
- 88.Probst AJ, Weinmaier T, Raymann K, Perras A, Emerson JB, Rattei T, Wanner G, Klingl A, Berg IA, Yoshinaga M, Viehweger B, Hinrichs KU, Thomas BC, Meck S, Auerbach AK, Heise M, Schintlmeister A, Schmid M, Wagner M, Gribaldo S, Banfield JF, Moissl-Eichinger C. 2014. Biology of a widespread uncultivated archaeon that contributes to carbon fixation in the subsurface. Nat Commun 5:5497. doi: 10.1038/ncomms6497. [DOI] [PubMed] [Google Scholar]
- 89.Bird JT, Baker BJ, Probst AJ, Podar M, Lloyd KG. 2016. Culture independent genomic comparisons reveal environmental adaptations for altiarchaeales. Front Microbiol 7:1221. doi: 10.3389/fmicb.2016.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Probst AJ, Moissl-Eichinger C. 2015. ‘Altiarchaeales’: uncultivated archaea from the subsurface. Life (Basel) 5:1381–1395. doi: 10.3390/life5021381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Probst AJ, Castelle CJ, Singh A, Brown CT, Anantharaman K, Sharon I, Hug LA, Burstein D, Emerson JB, Thomas BC, Banfield JF. 2017. Genomic resolution of a cold subsurface aquifer community provides metabolic insights for novel microbes adapted to high CO2 concentrations. Environ Microbiol 19:459–474. doi: 10.1111/1462-2920.13362. [DOI] [PubMed] [Google Scholar]
- 92.Liu R, Zhang Y, Ding R, Li D, Gao Y, Yang M. 2009. Comparison of archaeal and bacterial community structures in heavily oil-contaminated and pristine soils. J Biosci Bioeng 108:400–407. doi: 10.1016/j.jbiosc.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 93.Lazar CS, Baker BJ, Seitz KW, Teske AP. 2017. Genomic reconstruction of multiple lineages of uncultured benthic archaea suggests distinct biogeochemical roles and ecological niches. ISME J 11:1058. doi: 10.1038/ismej.2017.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Full trees for SepRS and SepCysS. Download FIG S1, PDF file, 0.1 MB (149KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genetic loci of SepRS and SepCysS genes and bacterial tRNACys genes. Bacterial tRNACys species are shown with residue 37 marked with a red circle. Download FIG S2, PDF file, 0.5 MB (606.5KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cooccurrence of SepRS-SepCysS-SepCysE and Sec-encoding systems in the clone AK8 W8A-19 group archaea and the BOG (Asgard) archaeon. See Table S1 for other members. Download FIG S3, PDF file, 0.4 MB (507KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Binning and naming of genomic and metagenomic contigs. Download TABLE S1, DOCX file, 0.03 MB (39.2KB, docx) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental text about tRNA-Thr-ED proteins. Download TEXT S1, DOCX file, 0.01 MB (18.2KB, docx) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification and analysis of homologs of the editing domain of archaeal ThrRS. (A) Crystal structure of the editing domain of ThrRS from M. jannaschii with l-Ser3AA (a substrate analog) (PDB accession no. 4RRF). (B) Phylogeny of tRNA-Thr-ED homologs and the editing domains of archaeal ThrRS (ThrRS-R) and ThrRS-ED. The bootstrap values (percentages) are shown for the unrooted maximum likelihood tree made with 100 replicates using MEGA 7. The tRNA-Thr-ED proteins associating with clade VII SepCysS (shown in the red box) are very similar to the archaeal ThrRS (ThrRS-R) and ThrRS-ED proteins. d-Tyrosyl-tRNATyr deacylase (DTD), used as an outgroup, was previously known as the closest homolog of the editing domain of ThrRS-R/ThrRS-ED. The full tree is shown below the compressed tree. (C) The SepCysS-associating tRNA-Thr-ED proteins from class II/III methanogens and rice cluster II methanogens have an additional C-terminal cysteine-rich motif, while Hadesarchaea MSBL1 proteins have no extension. Download FIG S4, PDF file, 0.3 MB (398.7KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
In vitro and in vivo assays of bacterial SepRS, SepCysS, and tRNACys. (A) Bacterial SepRS activity in vitro. This is the original data for Fig. 2C. Thin-layer chromatograms showing phosphoseryl-[32P]tRNACys accumulation during a reaction time course in the case of “Ca. Parcubacteria” (left) and Chloroflexi SepRS (right). tRNA substrates are indicated below and time points are indicated above the chromatograms. A reaction mixture without the enzyme was loaded prior (c) and after (d) nuclease P1 digestion. (B) Bacterial tRNACys variants are substrates for Methanococcus maripaludis SepRS. Thin-layer chromatograms showing phosphoseryl-[32P]tRNACys (Sep-AMP) accumulation after 20 min (marked as “No SepCysS”). The produced phosphoseryl-[32P]tRNACys is a substrate for Chloroflexi SepCysS (denoted as “Chloroflexi” above the chromatogram). Only phosphoseryl-(Sep-AMP) and cysteinyl-[32P]adenylate (Cys-AMP) are monitored after nuclease P1 digestion of phosphoseryl- and cysteinyl-[32P]tRNACys, respectively. tRNA substrates are indicated below and SepCysS presence is indicated above the chromatograms. (C) Bacterial SepCysS activity in E. coli. E. coli JS1 (ΔselA) was transformed with pACYC-MjPSTK-EcselC, pBAD-CDF-fdhF, and either of the SepCysS-expressing plasmids and an empty vector. In JS1 cells, conversion of Sep-tRNASec to Cys-tRNASec by SepCysS leads to the expression of full-length formate dehydrogenase H (FDHH), which converts benzyl viologen into a purple dye. Only the edges of the cell spots on agar plates were stained, because Cys-containing FDHH is far less active than the wild-type-Sec-containing FDHH. Supplementation of the growth medium with sodium selenite (final concentration, 1 μM) enhanced the FDHH activity of the cells expressing the “Ca. Parcubacteria” SepCysS, possibly due to the formation of Sec-tRNASec. Download FIG S5, PDF file, 1.1 MB (1.2MB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Cooccurrence of SepRS and PylRS in bacteria. (A) Lineage of Dehalococcoides carrying SepRS, SepCysS, and PylRS; (B) binning of metagenomic contigs of the three Dehalococcoides strains with SepRS, SepCysS, and PylRS. Download FIG S6, PDF file, 0.01 MB (198.5KB, pdf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequence and alignment data. Download DATA SET S1, RTF file, 0.4 MB (473.5KB, rtf) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental materials and methods. Download TEXT S2, DOCX file, 0.2 MB (26.4KB, docx) .
Copyright © 2017 Mukai et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.