ABSTRACT
More than 5 decades of work support the idea that cell envelope synthesis, including the inward growth of cell division, is tightly coordinated with DNA replication and protein synthesis through central metabolism. Remarkably, no unifying model exists to account for how these fundamentally disparate processes are functionally coupled. Recent studies demonstrate that proteins involved in carbohydrate and nitrogen metabolism can moonlight as direct regulators of cell division, coordinate cell division and DNA replication, and even suppress defects in DNA replication. In this minireview, we focus on studies illustrating the intimate link between metabolism and regulation of peptidoglycan (PG) synthesis during growth and division, and we identify the following three recurring themes. (i) Nutrient availability, not growth rate, is the primary determinant of cell size. (ii) The degree of gluconeogenic flux is likely to have a profound impact on the metabolites available for cell envelope synthesis, so growth medium selection is a critical consideration when designing and interpreting experiments related to morphogenesis. (iii) Perturbations in pathways relying on commonly shared and limiting metabolites, like undecaprenyl phosphate (Und-P), can lead to pleotropic phenotypes in unrelated pathways.
KEYWORDS: FtsZ, MreB, UDP-glucose, cell division, gluconeogenesis, metabolism, morphogenesis, peptidoglycan, phosphoenolpyruvate, undecaprenyl phosphate
INTRODUCTION
To accurately partition chromosomes and other cell contents during reproduction, cells must possess mechanisms to organize repeated cycles of cell growth, chromosome replication, and division. Eukaryotes orchestrate this coordination using the cell cycle and separate growth, DNA synthesis, and cytokinesis into distinct, temporally sequestered phases. Bacteria, by contrast, simultaneously increase in cell size and replicate DNA before (or concurrently with) cell division. Elucidating the molecular mechanisms prokaryotes employ to achieve spatiotemporal organization of these intertwined yet functionally disparate processes is of considerable interest to scientists seeking to understand bacterial reproduction, and many outstanding questions remain to be answered. For example, how is DNA replication kept in sync with changing growth and division rates? How are cell dimensions maintained or actively rearranged in response to environmental or developmental cues? What signals do cells sense to switch between increasing in cell size and dividing during the cell cycle? Relatedly, how are these signals transduced to activate/deactivate the distinct machineries required for each process?
Perhaps one of the biggest mysteries remaining in bacterial cell biology relates to understanding the regulatory cross talk that must occur to integrate central metabolism with macromolecular biosynthesis. Nutrients are converted into stored energy and precursors used to synthesize macromolecules like DNA and peptidoglycan (PG), so it is no surprise that nutrient availability has a profound impact on growth capacity. However, a growing body of evidence also suggests that metabolites and metabolic enzymes play a more direct role in regulating critical aspects of cell growth and division than previously appreciated. These findings raise the intriguing possibility that metabolism itself is the major determinant in shaping the underlying organization of the bacterial cell.
Actively growing bacteria respond rapidly to changing conditions by adjusting their overall shape and size. When nutrients are unrestricted, bacteria often capitalize on the available resources by increasing in cell size and reproducing more often. For rod-shaped bacteria, including Escherichia coli and Bacillus subtilis, cell size is determined by the length and width of the cell envelope. During steady-state growth, rapidly growing cells are generally longer and sometimes wider than their slower growing counterparts (1–4), at least when nutrients are unrestricted (3). The positive correlation between cell size and growth rate is likely due to nutrient availability rather than the growth rate itself because the relationship can be broken under conditions where growth rate is controlled by restricting nutrients. For example, in minimal media with different tryptophan concentrations, E. coli cells growing at steady state are largest (by volume) at concentrations of tryptophan that result in approximately one-half of the maximal growth rate achieved with nonlimiting tryptophan (5).
During balanced growth, cell size is remarkably homogenous across a population, suggesting that the signals cuing growth and division cycles are regulated and not random. Single-cell experiments performed on E. coli and Caulobacter crescentus show that cells achieve cell size homeostasis not by triggering cell division when a specific cell volume is achieved but rather by elongating a constant amount (and thus adding a constant volume) before dividing (6–9). Precise division at midcell allows for a homogenous cell size population to be maintained over time (7). A more recent study demonstrates that when bacteria grow, surface area and cell volume scale together across a variety of bacteria (10). The authors of this study also provide data implicating the levels of a limiting PG precursor as the likely signal for cuing cell division, thus providing a possible mechanistic basis to describe how bacteria integrate central metabolism with growth and division (10).
In bacteria, growth and division cycles are largely coordinated by the actin-like protein MreB (and its paralogs) and/or the tubulin-like protein FtsZ. In many bacteria, MreB is associated primarily with cell elongation and FtsZ is associated with cell division. However, these functional generalizations are probably oversimplified for bacteria as a whole, as the roles of MreB and FtsZ can overlap (11–14) and even be completely reversed (15–17). MreB and FtsZ are hypothesized to act as scaffolds, directing sites of new PG synthesis and old PG turnover (18). MreB and FtsZ are part of larger, multiprotein complexes (the elongasome and divisome) (19, 20) that ultimately define cellular dimensions by imposing spatial constraints on elongation and cell division (21, 22). The dependency of cell size on nutrient availability indicates that metabolism itself plays a critical role in controlling MreB and FtsZ activity, although possible mechanisms for this regulation are only beginning to emerge. One of the emerging themes discussed in more detail in this minireview is that the type of medium in which bacteria are cultured (particularly whether the medium supports primarily glycolytic or gluconeogenic growth regimes) can have a profound impact on cell shape and cell shape mutants and, thus, is an important consideration when designing and interpreting studies.
METABOLISM AND CELL ELONGATION
Amdinocillin resistance.
The close relationship between metabolism and cell elongation is illustrated by studies with mutants that are resistant to amdinocillin (mecillinam), a β-lactam antibiotic that selectively targets the cell elongation transpeptidase PBP 2 (23–25). In E. coli (as well as many other Gram-negative bacteria), treatment with amdinocillin results in rounded cells that lyse (25, 26). Amdinocillin resistance can be conferred by mutations in genes in the mrd and mre loci that encode components of the elongasome, such as PBP 2 and MreB (24), as well as in several dozen other targets, many of which have been shown to result in increased levels of the growth-rate-regulating molecule (p)ppGpp (synthesized by RelA and SpoT in E. coli) (27). Although amdinocillin resistance has been studied for more than 3 decades, the mechanism by which enhanced (p)ppGpp levels lead to amdinocillin resistance has not been elucidated. In E. coli, overexpression of the cell division proteins FtsQ, FtsA, and FtsZ (but not FtsZ alone) or artificially induced synthesis of (p)ppGpp bypasses the lethality associated with amdinocillin treatment, so it has been proposed that increased (p)ppGpp levels result in an enhanced cell division capacity that allows the wider cells to divide (28, 29). However, subsequent studies demonstrated that increased FtsZ levels are not responsible for amdinocillin resistance, at least for the mutants that have been examined (30, 31), indicating that another mechanism confers resistance.
One way that the effects of (p)ppGpp may confer resistance is through the modulation of carbon metabolism. In E. coli, (p)ppGpp has been shown in combination with the RpoS regulator DksA to activate the expression of csrB and csrC, genes encoding antagonists of the global carbon storage regulator CsrA (32). Downregulation of CsrA represses the expression of glycolytic genes (pgi, pfkA, pfkB, tpi, eno, pykF, and pykA), promotes the expression of genes important for flux through gluconeogenesis (pck, fbp, pps, and pgm) (33, 34), and leads to elevated levels of several intermediates in cell envelope synthesis, including phosphoenolpyruvate (PEP) (35, 36). Since gluconeogenesis is the primary pathway for generating precursors for cell envelope biogenesis, the higher levels of (p)ppGpp may result in a gain of function in PG synthesis by enhancing the available PG precursor pool. A similar relationship to enhanced gluconeogenesis and PG precursors may also explain why crp and cya mutants are resistant to amdinocillin (37–39), since in E. coli the cyclic AMP receptor protein-cyclic AMP complex (CRP-cAMP) represses the expression of genes involved in shifting flux through gluconeogenesis, including pck (40, 41).
One amdinocillin-resistant E. coli mutant that depends on SpoT but not RelA for resistance possesses a mutation in aroK. aroK encodes a shikimate kinase that catalyzes the ATP-dependent conversion of shikimate to shikimate-3-phosphate in the aromatic amino acid synthesis pathway (31). The mutant also shows partial rescue of the filamentation and lethality effects associated with ftsZ84(Ts), a temperature-sensitive allele of ftsZ, indicating that aroK inactivation acts as a general suppressor of defects in cell envelope synthesis. Based on the fact that FtsZ levels were not detectably different in the aroK ftsZ84 double mutant strain and that the amdinocillin resistance was not dependent on AroK's characterized enzymatic activity or relA, the authors concluded that AroK likely possesses a second, moonlighting function related to cell division regulation (31). Another possibility is that aroK inactivation causes a change in carbon flux. We can envision two ways that this may occur, both of which involve enhancing the availability of PG precursor pools to the elongasome and divisome machineries. First, the next step in the shikimate pathway utilizes PEP so the aroK mutant can possess elevated levels of PEP. PEP is a critical intermediate in gluconeogenesis and is required for the first dedicated step of PG synthesis, the MurA-dependent conversion of UDP-N-acetylglucosamine (UDP-GlcNAc) to UDP-GlcNAc-enolpyruvate (42, 43). In addition, the levels of (p)ppGpp, although doubled in the aroK mutant, were lower than those of the wild type in the amdinocillin-resistant relA aroK double mutant, leading the authors to conclude that enhanced (p)ppGpp could not account for the resistance. However, the quantification of (p)ppGpp was performed in a minimal medium supplemented with glucose, amino acids, and shikimate, whereas the amdinocillin resistance was assayed on LB plates (31). Therefore, the possibility that the resistance was conferred by (p)ppGpp accumulation on LB was not directly ruled out. If there were elevated (p)ppGpp levels, this could lead to enhanced PG precursor synthesis through the mechanism discussed above.
Gluconeogenic factor YvcK.
Another example of a connection between carbon metabolism and cell elongation is demonstrated by studies on YvcK. Homologs exist in a variety of bacteria (44–46), but YvcK is most extensively studied in B. subtilis (44, 47, 48). Although yvcK is not essential during growth in LB, cells with a yvcK deletion are round and lyse under gluconeogenic growth regimes, indicating a deficiency in elongasome function (44). Consistent with this hypothesis, yvcK's conditional essentiality is bypassed by overexpressing MreB (47) or by providing a growth condition that suppresses problems with elongasome function (extra magnesium) (44, 49–52). YvcK is phosphorylated by the highly conserved PrkC Ser/Thr kinase (53–55), which also regulates the essential WalRK two-component system involved in regulating PG metabolism (56). PrkC also phosphorylates several proteins involved in metabolic regulation (Hpr, Icd, and GlnA) (55) and protein synthesis and cell shape determination (EF-Tu, EF-G, and CpgA) (57–59) and one protein implicated in mediating the switch between cell elongation and cell division, GpsB (60–62). Although phosphorylation of YvcK is not required for its role in gluconeogenesis, only the phosphorylated form can bypass mreB essentiality (48) and restore the septal localization of the major transglycosylase/transpeptidase PBP 1 (47, 48).
YvcK's role in promoting gluconeogenic growth in B. subtilis is unknown, but its essentiality is bypassed by transposon insertions in several genes related to carbon and nitrogen metabolism (zwf, cggR, and mfd) and cell envelope synthesis (yfnI, dgkA, and yqfF) (44). For example, the yvcK mutant may be rescued by inactivation of Zwf, which feeds glucose-6-phosphate into the pentose phosphate pathway, or CggR, which inhibits synthesis of glucose-6-phosphate by repressing genes in the bottom half of gluconeogenesis (44). The transposon insertion in mfd, which results in decreased synthesis of an extracellular polymer of glutamate called poly-γ-glutamate (PGA), would be expected to result in enhanced flux through other glutamate-utilizing pathways (63). The insertions in yfnI and the first gene in the yqfF operon likely perturb different processes in cell envelope synthesis. YfnI is important for generation of the polyglycerolphosphate and GroP-Glc2-diacylglycerol (DAG) glycolipid components of lipoteichoic acid during cell stress (64), whereas the last gene in the yqfF operon, dgkA, may be involved in phosphorylating undecaprenol to generate undecaprenyl phosphate (Und-P) (65). yqfF itself encodes a protein involved in turnover of cyclic di-AMP, an essential molecule involved in cell envelope homeostasis in Gram-positive organisms (66). Intriguingly, the deletion of whiA (a cotranscribed gene downstream of yvcK) (67) in combination with the deletion of zapA (68) causes a synthetic cell division defect. The cell division defect is also rescued by mutations in genes involved in carbon transformations, specifically gtaB and pgcA (68), the latter of which is discussed in more detail below. These results hint that YvcK and WhiA take part in related processes (68).
Phosphomannose isomerase (ManA) (B. subtilis).
ManA is another protein in B. subtilis that is implicated in both carbon transformations and cell shape. In B. subtilis, ManA is required for rod shape and viability in LB but not in minimal media containing glucose as a carbon source (69). Characterized ManA homologs catalyze the reversible conversion between mannose-6-phosphate and fructose-6-phosphate. In some archaea and bacteria, the enzyme responsible for phosphomannose isomerase activity is also a phosphoglucose isomerase, catalyzing the isomerization between glucose-6-phosphate and fructose-6-phosphate (70). In most bacteria, including B. subtilis, the characterized phosphoglucose isomerase is encoded by pgi. However, B. subtilis ManA can carry out isomerization of several sugars in vitro, suggesting that there may be substrate promiscuity in the active site (71); therefore, one speculation is that it can also act as a phosphoglucose isomerase.
In B. subtilis, a ΔmanA mutant becomes rounded and stops growing shortly after shifting from a minimal medium to LB. The requirement for ManA in cell shape is likely attributable to its enzymatic activity, as several predicted catalytic mutants are unable to complement the ΔmanA mutant (69). Since LB does not contain mannose, these results are somewhat perplexing. The cell envelope of the manA mutant shows reduced levels of galactose and GalNAc (N-acetylglucosamine, a component of teichoic acid), so it was proposed that the defect in the manA mutant was due to problems with teichoic acid synthesis (71). However, since the UDP-GalNAc used to synthesize teichoic acids is primarily generated from epimerization of UDP-GlcNAc (72), the result hints that the manA mutant is also likely compromised in its ability to synthesize UDP-GlcNAc. Consistent with this possibility, the manA mutant phenocopied cells inhibited for synthesis of lipid I by tunicamycin (71). No difference in GlcNAc levels was observed between the manA mutant and the wild type (71); however, the mutant also stopped growing shortly after the shift to LB, and neither intracellular GlcNAc levels nor PG synthesis were measured directly; thus, the possibility that PG synthesis was essentially paused in the manA mutant growing in LB was not ruled out.
The widened cells and altered chromosome structures of the manA mutant are remarkably similar to those observed when the pools of pyrimidine precursors are limited in E. coli (73). Since DNA and PG synthesis can share a precursor, UTP (used to generate UDP-GlcNAc and dCTP/dTTP, respectively), these results suggest that cell growth is particularly sensitive to perturbations in UTP and possibly provide a link for the correlation observed between cell growth and DNA replication during steady state (74). One possibility that may explain all of the observed results is that in LB not supplemented with glucose, ManA contributes significantly to the conversion of fructose-6-phosphate to glucose-6-phosphate to feed pathways for PG, teichoic acid, and DNA synthesis. In principle, this would be an easy hypothesis to test since the addition of glucose to the LB medium would be predicted to alleviate the cell shape defect. Alternatively, ManA may possess a moonlighting role, regulating some aspect of cell envelope biogenesis directly, although its diffuse localization provides no clues as to how such a mechanism would occur (69).
METABOLISM AND CELL DIVISION
Evidence for genetic interactions between carbon and nitrogen metabolism and cell division are also abundant, and several examples are summarized in more depth in recent reviews (7, 75, 76). In most cases, a metabolic enzyme is implicated in inhibiting FtsZ activity either directly or indirectly. In some cases, the loss of the corresponding gene leads to more active FtsZ (pgm, pgcA, ugtP, opgH, and pykA in an FtsZ temperature-sensitive background), causing cells to divide more often, while in other cases (gdhZ, kidO, and pycK in a wild-type background), the loss of the gene inhibits cell division.
UDP-glucose and UgtP (B. subtilis).
The first study to demonstrate a direct link between FtsZ regulation and a metabolic enzyme was published in 2007 (77). The authors identified a gene disruption in pgcA (encoding phosphoglucomutase) that resulted in cells that were approximately one-third shorter than the wild type yet maintained a wild-type growth rate in LB. PgcA catalyzes the reversible conversion between glucose-6-phosphate and glucose-1-phosphate in the production of UDP-glucose (78). These investigations revealed that UgtP, an enzyme that can transfer glucose from UDP-glucose to diacylglycerol-containing sugar acceptors in vitro (79), also contributed to a short cell phenotype. UgtP interacts directly with FtsZ and inhibits its polymerization in a manner that depends upon UDP-glucose both in vivo and in vitro (77). According to their model, nutrient-rich conditions result in excess glucose-6-phosphate, resulting in an increased flux toward UDP-glucose synthesis. In the presence of its substrate, UgtP shows preference for interaction with FtsZ over itself, resulting in the inhibition of FtsZ and longer cells (80).
UDP-glucose and OpgH (E. coli).
Regulation of FtsZ assembly by another enzyme utilizing UDP-glucose has also been observed in E. coli. In this study, the authors observed that a knockout of pgm, the functional equivalent of pgcA, also produced small cells (81). The authors eventually implicated OpgH (formerly MdoA) in the small cell size. OpgH is a glucosyltransferase involved in the synthesis of osmoregulated periplasmic glucans. OpgH is not homologous to UgtP and is involved in the synthesis of a distinct macromolecule, yet both proteins appear to inhibit FtsZ polymerization in a UDP-glucose-dependent manner (77, 80, 81). Deletion of opgH results in more frequent divisions, while overexpression results in filamentation. OpgH also colocalizes with FtsZ in an FtsZ-dependent manner during fast growth. A truncation of OpgH consisting of amino acids 1 to 138 (and devoid of the membrane spanning domains) strongly inhibits FtsZ assembly in vitro, indicating that OpgH interacts directly with FtsZ to inhibit cell division (81). In vitro assays suggest that OpgH inhibits FtsZ polymerization by binding FtsZ monomers, thereby increasing the apparent concentration of FtsZ required for GTP hydrolysis. How this mechanism would suffice in vivo given that the native copy number of OpgH is likely to be low (82) is less clear. Unlike UgtP in B. subtilis, OpgH appears to localize to FtsZ in a UDP-glucose-independent manner. Curiously, OpgH still localizes to FtsZ during fast growth but becomes more diffuse in the membrane as growth slows (81).
The data presented above strongly indicate that UgtP and OpgH interact directly with FtsZ, although it is less clear if these enzymes possess an express role in regulating FtsZ dynamics in vivo. As with any data, it is worth considering alternative models. Another possibility is that UgtP and/or OpgH utilizes interactions with FtsZ to localize to regions in the cell where UDP-glucose is initially produced and metabolized. There are numerous protein-protein interaction and substrate channeling studies that certainly argue that bacteria are too clever to depend on diffusion alone to control metabolite flow (83–90). Although the in vitro data argue for possible FtsZ-regulating mechanisms, especially for UgtP, any protein that interacts with an FtsZ surface may conceivably act as an inhibitor of FtsZ activity in vitro. How then can one account for the UDP-glucose levels having such a profound impact on two entirely different organisms? Assuming that the pathways outlined in Fig. 1 are accurate, mutations in several of the genes discussed above would likely result in increased availability of glucose-6-phosphate to other pathways. Such a shift may be expected to increase the availability of lipid II to support FtsZ-dependent cell division and MreB-dependent cell elongation. The net result that one may expect, assuming FtsZ is more sensitive to substrate availability than MreB, would be more active FtsZ and shorter cells. Some of the mutations (in opgH and ugtP) may also result in enhanced Und-P availability (discussed more below). Synthesis of teichoic acids requires Und-PP-sugar intermediates (91, 92), so defects in the pathway prior to Und-P commitment would be expected to increase overall Und-P pools. Similarly, the glycosyltransferase activity of OpgH uses polyprenyl phosphates (including decaprenyl-phosphate) as substrates (93). Although Und-P has not been formally tested as a substrate, an opgH mutant exhibits increased resistance to bacitracin (which inhibits recycling of Und-PP to Und-P) (94) and enhanced exopolysaccharide (EPS) production (95, 96); each of these phenotypes is consistent with the enhanced availability of Und-P. The idea that elevated pools of cell envelope precursors (especially PG precursors) can lead to enhanced cell division is also supported by additional studies discussed below.
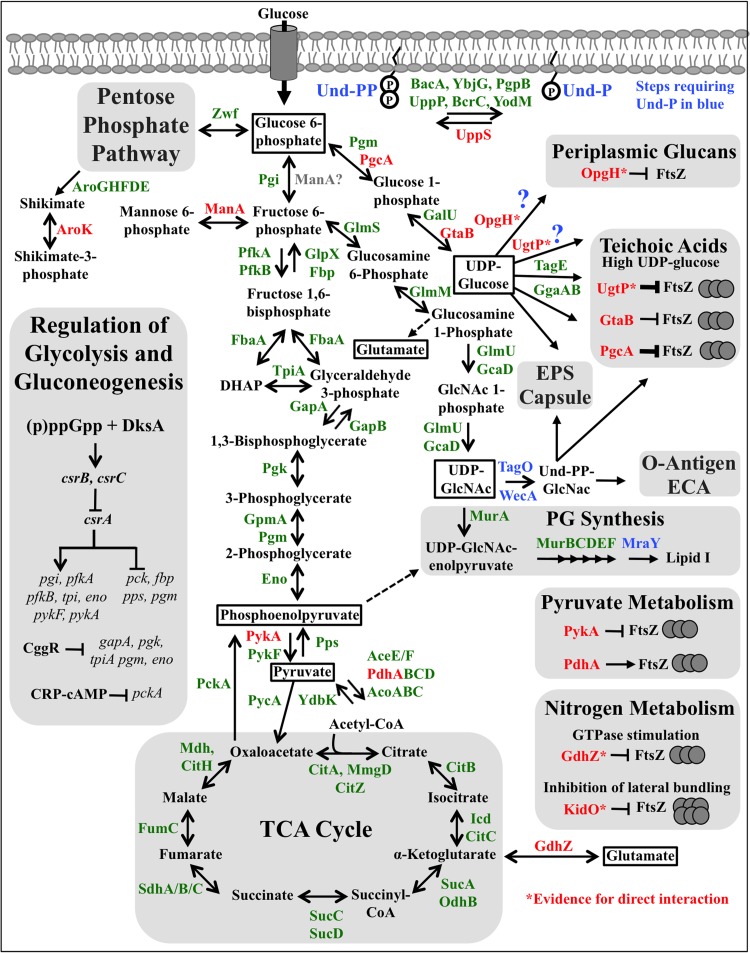
FIG 1.
Metabolic pathways implicated in the regulation of cell shape and size. Enzymes are indicated by green, red, or blue text. Enzymes discussed in the text are indicated in red or blue. Enzymes shown in blue denote steps dedicating Und-P (also in blue) to one or more pathways. Enzymes predicted to coincide with or precede Und-P dedication to one or more pathways are followed by blue question marks. In most cases, the relevant enzyme(s) for both E. coli and B. subtilis is given; however, not all organisms possess every enzyme shown. Enzymes that are less studied or have not been tested experimentally are generally excluded. Enzymes shown to interact directly with FtsZ are denoted with an asterisk. Only regulators of glycolysis and gluconeogenesis discussed in the text are shown in the left-hand block.
Pyruvate, PykA, and PdhA (B. subtilis).
In B. subtilis, deletion of pykA (encoding pyruvate kinase), which catalyzes the conversion of PEP to pyruvate, rescues a temperature-sensitive allele of ftsZ (97), possibly suggesting that enhanced PEP levels (or enhanced gluconeogenic flux) rescue defects in cell division. However, in the wild type, deletion of pykA also disrupts normal FtsZ regulation, resulting in ∼40% of cells possessing either subpolar or multiple FtsZ rings. Although no direct interaction between PykA and FtsZ could be detected, subsequent experiments demonstrated that overexpression of an enzyme that feeds pyruvate into the tricarboxylic acid (TCA) cycle, PdhA, promotes additional polar FtsZ ring formation in the pykA mutant (97). PdhA also shows an unusual localization pattern that is reminiscent of nucleoid staining in the wild type, which undergoes redistribution to nucleoid-free regions in the pykA mutant (97). Notably, the latter pattern is very similar to what has been observed for several components of the B. subtilis RNA degradosome (98). Colocalization with the nucleoid is restored in the pykA mutant by the addition of exogenous pyruvate, suggesting that PdhA depends on the product of PykA (pyruvate) and not PykA itself for wild-type localization and normal FtsZ assembly. The authors propose that in the presence of pyruvate, PdhA acts as a positive regulator of FtsZ assembly and that PdhA's subpolar positioning in the pykA mutant promotes subpolar FtsZ ring formation. More studies will be required to assess if PdhA's role in controlling FtsZ dynamics is direct or indirect.
Nitrogen metabolism, GdhZ, and KidO (Caulobacter crescentus).
Glutamate is a key central metabolite, connecting pathways for cellular anabolism and catabolism; glutamate is also the central donor for nearly all anaplerotic metabolites containing nitrogen and is maintained at high intracellular concentrations under all growth conditions (99). In E. coli, mutations in several genes important for nitrogen uptake and metabolism (glnD, glnG, and glnL) restore growth of an ftsZ84 mutant at nonpermissive temperatures by promoting (p)ppGpp accumulation (100). Studies in C. crescentus have also implicated nitrogen metabolism, and in particular glutamate conversions, in the direct regulation of FtsZ assembly. The enzyme responsible for the catalytic conversion of glutamate and NAD+ to α-ketoglutarate, ammonia, and NADH in C. crescentus is called GdhZ (101). GdhZ was identified in a screen for proteins interacting with FtsZ and comes down as an FtsZ partner in coimmunoprecipitation experiments. Deletion of gdhZ results in abnormal cell divisions, leading to a pleiomorphic mixture of short, normal, and filamented cells in mixed population cultures. GdhZ colocalizes with FtsZ in vivo and stimulates FtsZ's GTPase activity in a glutamate-dependent manner in vitro. GdhZ's enzymatic activity is required for its effect on FtsZ in vivo, as a catalytically dead mutant phenocopies the gdhZ deletion strain; the same variant does not stimulate GTPase activity in vitro (101), suggesting that a conformational cycling of GdhZ may be required to stimulate hydrolysis of GTP. Alternatively, the effects of pH on FtsZ's GTPase activity resulting from the conversion of glutamate to α-ketoglutarate and ammonia in the reaction were not ruled out.
KidO is another protein shown to promote FtsZ disassembly in C. crescentus. KidO is believed to act in conjunction with GdhZ to regulate FtsZ disassembly during the cell cycle and under conditions of nitrogen limitation (101). The phenotypes associated with a kidO deletion are not as pronounced as those associated with a gdhZ deletion; however, the proteins follow similar cell cycle regulation, colocalize with FtsZ rings, and copurify (101, 102). In vitro, KidO inhibits lateral bundling of FtsZ filaments in an NADH-dependent manner. KidO is proposed to act in cooperation with GdhZ, utilizing the NADH produced from GdhZ's enzymatic activity to inhibit FtsZ bundling (101).
Although much of the data point to the direct regulation of FtsZ by GdhZ and KidO, as with UgtP and OpgH, other (metabolically based) models can be invoked. In a subsequent study by the same group, the authors found that when intracellular pools of glutamine drop, the (p)ppGpp hydrolysis function of SpoT becomes inhibited by the phosphorylated form of enzyme IIA (EIIA) (part of the phosphotransferase system responsible for nitrogen uptake [PTSNtr]). This leads to (p)ppGpp accumulation, slowed growth, and an extended G1 phase in a proportion of synchronized cells (103). In the gdhZ mutant, the glutamate that is unable to feed into the TCA cycle may conceivably be converted to glutamine by GlnA (glutamine synthetase), also generating the preferred carbon source of C. crescentus, ammonium, as a by-product (103). However, the glnA gene is also repressed by high levels of glutamine (103), so the balance between glutamate and glutamine may be disrupted in the gdhZ mutant, leading to elevated (p)ppGpp levels. This scenario would also be consistent with the authors' observation that exogenous addition of glutamine did not rescue the observed cell division defects in the gdhZ mutant. This alternative possibility may presumably be tested by determining if the gdhZ phenotype depends on SpoT.
UND-P AVAILABILITY AS A REGULATOR OF GROWTH AND DIVISION
The lipid carrier Und-P is critical for inner membrane export of a highly diverse array of sugar-containing molecules, including various exopolysaccharides, capsule, PG, teichoic acids, periplasmic glucans, O antigen, and even glycosylated proteins, such as those associated with the flagella of several pathogens (91); as such, Und-P is an excellent target candidate for the development of novel antimicrobials. A growing body of research also indicates that Und-P is a limited resource that each of the aforementioned pathways must compete for, and disruptions in cell division and cell elongation are associated with Und-P perturbations. How the distribution of Und-P among these pathways is regulated remains an open question, but one can assume that the most essential processes (e.g., PG synthesis) are preferentially fed before others. Und-P is generated from Und-PP de novo by UppS, and Und-P can subsequently act as an acceptor of sugars/glycans linked to a nucleotide diphosphate donor. Once the sugars/glycans are flipped across the membrane, they can be acted upon by additional enzymes, released, or transferred to another acceptor, such as PG or lipoteichoic acid. Following the termination reaction, Und-PP is recycled to Und-P and reutilized (91). Mutations that block steps following the commitment of Und-P to a pathway (transfer of the first sugar/glycan to the lipid carrier) trap the Und-PP-conjugated intermediates in a dead-end pathway that effectively depletes the cellular pool of Und-P, resulting in cell division and cell shape defects (104–107). The phenotypic consequences of Und-P depletion likely depend on the drain imposed by the broken pathway pulling from the remaining Und-P pool, the metabolic demand for Und-P in other active and competing pathways, and the level of coordination required for these pathways (which may become disrupted as Und-P becomes limiting).
Many Gram-positive organisms utilize Und-P for synthesis of wall teichoic acids (WTAs) and PG, and synthesis in the two pathways is likely coordinated (108). Consistent with this idea, B. subtilis WTAs are essential for normal rod cell shape. However, WTAs are not required for viability, and lethality only occurs in mutants defective in the stages following commitment of Und-P (via TagO) to the pathway (109). Similarly, Staphylococcus aureus WTA mutations are lethal in most cases except when the gene encoding the Und-P dedicating enzyme (tarO) is deleted (110). Moreover, epistasis experiments demonstrate that the lethality associated with deletions in latter stages of the WTA synthesis pathway can be rescued by the tarO deletion (110). These results suggest that the sequestration of Und-P, not the essentiality of the pathway products, leads to lethality. Similar results were found in pathways for Streptococcus pneumoniae capsule production (111) and E. coli O-antigen and enterobacterial common antigen (ECA) synthesis (104, 105), implying that any mutation that significantly affects Und-P levels has the potential to have adverse consequences for any other pathway utilizing Und-P.
In the case of ECA or O-antigen synthesis, the deleterious effects of Und-P sequestration (manifested as cell division and cell shape defects) can be overcome by increasing the expression of UppS or MurA, the latter of which makes the PG synthesis pathway more competitive for the available Und-P (104, 105). These results strongly argue that the phenotypes observed in at least some mutants disrupted at intermediate steps in Und-P-utilizing pathways do not result from toxic build-up of intermediates but rather dysregulation of Und-P-utilizing reactions. In contrast, UppS overexpression does not rescue mutants halted at intermediate steps in colonic acid biosynthesis, leaving open the possibility that at least some intermediate products are toxic (112). Although little is understood about how Und-P availability is regulated, Und-P recycling is a likely candidate. Given the large number of Und-P recycling enzymes already discovered, it is even possible that certain pathways possess their own dedicated recycler. Of course, this would suggest that Und-P production or utilization is controlled spatially, as multiple enzymes are still competing for an identical substrate. There is at least some evidence for this possibility in B. subtilis, where MreB localization was shown to depend on the synthesis of Und-P-linked precursors (113). This extraordinary finding suggests that the availability of precursors at specific sites in the cell envelope is likely to act upstream of MreB (and likely FtsZ) in determining where and when cells grow or divide.
CONCLUSIONS
It is likely that many readers have either carried out or are at least familiar with results from an impeccably designed screen related to their favorite cellular process that implicated one or more seemingly boring, if not completely incomprehensible, metabolic genes. For many of us, metabolism is a white elephant in the room. We know it is lurking there, but ignoring it is easier than facing Fig. 1. With metabolism, everything is connected and one can quickly become overwhelmed. However, even though cells are rather complex amalgamations of nucleic acid, protein, lipid, and carbohydrate, a surprisingly small number of key metabolites weave these macromolecules neatly together.
One of the themes we encountered while assembling this minireview is that growth is profoundly impacted by changes in carbon flux between glucose-6-phosphate and pyruvate. In particular, PG synthesis seems to be finely tuned to the relative levels of PEP, pyruvate, and glucose-6-phosphate present within cells. Often mutations that enhance glucose-6-phosphate availability or promote carbon flow through gluconeogenesis rescue mutants with elongasome and/or divisome defects. The activity of both the divisome and elongasome is highest when there is an abundance of glucose-6-phosphate available and the PEP/pyruvate ratios are high. When cells are grown in minimal medium utilizing TCA intermediates or non-PTS sugars, sugars such as glucose-6-phosphate must be synthesized. Not surprisingly, this slows down PG synthesis and cell growth in general. It is striking that many of the mutations that we highlighted either enhance production of glucose-6-phosphate or result in higher PEP/pyruvate ratios. Either of these conditions would be expected to increase the pool of Und-P and UDP-GlcNAc to the PG synthesis machinery. Conversely, decreasing the PEP/pyruvate ratio artificially (by depleting PdhA) delays cell division (97). Notably, knocking down gapA in LB (which should decrease the PEP/pyruvate ratio and the precursors available for synthesis of Und-P and UDP-GlcNAc) results in cell bulging in B. subtilis (114). Could these results hint that the final arbitrator of cell size (the decision of whether to grow or divide) is not a protein but the availability of the metabolites themselves?
Many factors outlined in this review are proposed to have moonlighting functions, acting as both enzymes and regulators of FtsZ. In the space allotted, we were barely able to scratch the surface of proteins predicted to act in similar ways to coordinate metabolism with regulation of cytoskeletal function or DNA replication (115–119). Based on the growing number of factors that appear to interact with MreB and/or FtsZ and have a seemingly unrelated enzymatic function, we would like to end with the following speculation: that at least some of the proteins proposed to have moonlighting functions may actually be utilizing interactions with FtsZ and MreB to localize their enzymatic activities to sites in the cell where their substrate is actively metabolized. For example, at least some evidence suggests that the lipid I and II generated for cell elongation are synthesized at nonrandom, punctate-helical locations in the cell where they then attract components of the elongasome, including MreB (113). Since many of the components of the cell envelope share precursors like Und-P, UDP-glucose, and UDP-GlcNAc, it is conceivable that at least some of the enzymes involved in making these components utilize interactions with the elongasome and divisome to coordinate synthesis and/or to compete for substrate. From this perspective, MreB and FtsZ may not be “cytoskeletal” factors so much as they are middlemen, mediating interactions between metabolites and enzymes. Regardless of the ultimate mechanism, understanding the regulatory cross talk that must occur to coordinate cell growth, chromosome replication, and cell division requires delving into metabolism, and there is likely considerable insight to be gained by analyzing the cell biology from this additional perspective. Perhaps it is time we all rummaged through our freezers and gave those perplexing mutants another chance.
ACKNOWLEDGMENT
Thanks go to members of the Herman lab for helpful comments on this review.
Biographies

Anthony M. Sperber obtained his B.S. in biochemistry at the University of Missouri, where he worked on the E. coli secretory system as an undergraduate researcher in the laboratory of Linda Randall. He is currently a Ph.D. student in the Department of Biochemistry and Biophysics at Texas A&M University, where he works on projects related to both bacteriophage biology and characterizing genes of unknown function in Bacillus subtilis in the laboratories of Jennifer Herman and Ryland Young. He became interested in bacterial physiology when one of his Ph.D. projects collided head-on with carbon metabolism.

Jennifer K. Herman is an assistant professor in the Department of Biochemistry and Biophysics at Texas A&M University. She completed her Ph.D. at Indiana University in 2005, working on Caulobacter crescentus stalk synthesis in the laboratory of Yves Brun. This was followed by postdoctoral training at Harvard Medical School, where she studied Bacillus subtilis sporulation under the mentorship of David Rudner. Her interest in bacterial metabolism dates back to creating her first Winogradsky column in an undergraduate bacterial physiology course. Current projects in her laboratory focus on understanding the mechanistic relationship between bacterial morphogenesis and gluconeogenic carbon flux.
REFERENCES
- 1.Pierucci O. 1978. Dimensions of Escherichia coli at various growth rates: model for envelope growth. J Bacteriol 135:559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sargent MG. 1975. Control of cell length in Bacillus subtilis. J Bacteriol 123:7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaechter M, Maaloe O, Kjeldgaard NO. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella Typhimurium. J Gen Microbiol 19:592–606. doi: 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 4.Sharpe ME, Hauser PM, Sharpe RG, Errington J. 1998. Bacillus subtilis cell cycle as studied by fluorescence microscopy: constancy of cell length at initiation of DNA replication and evidence for active nucleoid partitioning. J Bacteriol 180:547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shehata TE, Marr AG. 1971. Effect of nutrient concentration on the growth of Escherichia coli. J Bacteriol 107:210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campos M, Surovtsev IV, Kato S, Paintdakhi A, Beltran B, Ebmeier SE, Jacobs-Wagner C. 2014. A constant size extension drives bacterial cell size homeostasis. Cell 159:1433–1446. doi: 10.1016/j.cell.2014.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S. 2015. Cell-size control and homeostasis in bacteria. Curr Biol 25:385–391. doi: 10.1016/j.cub.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deforet M, van Ditmarsch D, Xavier JB. 2015. Cell-size homeostasis and the incremental rule in a bacterial pathogen. Biophys J 109:521–528. doi: 10.1016/j.bpj.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanouchi Y, Pai A, Park H, Huang S, Stamatov R, Buchler NE, You L. 2015. A noisy linear map underlies oscillations in cell size and gene expression in bacteria. Nature 523:357–360. doi: 10.1038/nature14562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris LK, Theriot JA. 2016. Relative rates of surface and volume synthesis set bacterial cell size. Cell 165:1479–1492. doi: 10.1016/j.cell.2016.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slovak PM, Wadhams GH, Armitage JP. 2005. Localization of MreB in Rhodobacter sphaeroides under conditions causing changes in cell shape and membrane structure. J Bacteriol 187:54–64. doi: 10.1128/JB.187.1.54-64.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenton AK, Gerdes K. 2013. Direct interaction of FtsZ and MreB is required for septum synthesis and cell division in Escherichia coli. EMBO J 32:1953–1965. doi: 10.1038/emboj.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szwedziak P, Lowe J. 2013. Do the divisome and elongasome share a common evolutionary past? Curr Opin Microbiol 16:745–751. doi: 10.1016/j.mib.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 14.van der Ploeg R, Verheul J, Vischer NO, Alexeeva S, Hoogendoorn E, Postma M, Banzhaf M, Vollmer W, den Blaauwen T. 2013. Colocalization and interaction between elongasome and divisome during a preparative cell division phase in Escherichia coli. Mol Microbiol 87:1074–1087. doi: 10.1111/mmi.12150. [DOI] [PubMed] [Google Scholar]
- 15.Liechti G, Kuru E, Packiam M, Hsu YP, Tekkam S, Hall E, Rittichier JT, VanNieuwenhze M, Brun YV, Maurelli AT. 2016. Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog 12:e1005590. doi: 10.1371/journal.ppat.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouellette SP, Karimova G, Subtil A, Ladant D. 2012. Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol Microbiol 85:164–178. doi: 10.1111/j.1365-2958.2012.08100.x. [DOI] [PubMed] [Google Scholar]
- 17.Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, Brun YV, VanNieuwenhze MS, Vollmer W, Horn M, Jensen GJ. 2013. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun 4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egan AJF, Cleverley RM, Peters K, Lewis RJ, Vollmer W. 23 November 2016 Regulation of bacterial cell wall growth. FEBS J doi: 10.1111/febs.13959. [DOI] [PubMed] [Google Scholar]
- 19.den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. 2008. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev 32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- 20.Nanninga N. 1991. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol 5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 21.Perez-Nunez D, Briandet R, David B, Gautier C, Renault P, Hallet B, Hols P, Carballido-Lopez R, Guedon E. 2011. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol Microbiol 79:759–771. doi: 10.1111/j.1365-2958.2010.07483.x. [DOI] [PubMed] [Google Scholar]
- 22.Elhadi D, Lv L, Jiang XR, Wu H, Chen GQ. 2016. CRISPRi engineering E. coli for morphology diversification. Metab Eng 38:358–369. doi: 10.1016/j.ymben.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Tamaki S, Matsuzawa H, Matsuhashi M. 1980. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J Bacteriol 141:52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Boer PA. 2016. Classic spotlight: staying in shape and discovery of the mrdAB and mreBCD operons. J Bacteriol 198:1479. doi: 10.1128/JB.00180-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spratt BG. 1975. Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc Natl Acad Sci U S A 72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spratt BG. 1977. The mechanism of action of mecillinam. J Antimicrob Chemother 3Suppl):13–19. doi: 10.1093/jac/3.suppl_B.13. [DOI] [PubMed] [Google Scholar]
- 27.Thulin E, Sundqvist M, Andersson DI. 2015. Amdinocillin (mecillinam) resistance mutations in clinical isolates and laboratory-selected mutants of Escherichia coli. Antimicrob Agents Chemother 59:1718–1727. doi: 10.1128/AAC.04819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joseleau-Petit D, Thevenet D, D'Ari R. 1994. ppGpp concentration, growth without PBP2 activity, and growth-rate control in Escherichia coli. Mol Microbiol 13:911–917. doi: 10.1111/j.1365-2958.1994.tb00482.x. [DOI] [PubMed] [Google Scholar]
- 29.Vinella D, Joseleau-Petit D, Thevenet D, Bouloc P, D'Ari R. 1993. Penicillin-binding protein 2 inactivation in Escherichia coli results in cell division inhibition, which is relieved by FtsZ overexpression. J Bacteriol 175:6704–6710. doi: 10.1128/jb.175.20.6704-6710.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navarro F, Robin A, D'Ari R, Joseleau-Petit D. 1998. Analysis of the effect of ppGpp on the ftsQAZ operon in Escherichia coli. Mol Microbiol 29:815–823. doi: 10.1046/j.1365-2958.1998.00974.x. [DOI] [PubMed] [Google Scholar]
- 31.Vinella D, Gagny B, Joseleau-Petit D, D'Ari R, Cashel M. 1996. Mecillinam resistance in Escherichia coli is conferred by loss of a second activity of the AroK protein. J Bacteriol 178:3818–3828. doi: 10.1128/jb.178.13.3818-3828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards AN, Patterson-Fortin LM, Vakulskas CA, Mercante JW, Potrykus K, Vinella D, Camacho MI, Fields JA, Thompson SA, Georgellis D, Cashel M, Babitzke P, Romeo T. 2011. Circuitry linking the Csr and stringent response global regulatory systems. Mol Microbiol 80:1561–1580. doi: 10.1111/j.1365-2958.2011.07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romeo T, Gong M, Liu MY, Brun-Zinkernagel AM. 1993. Identification and molecular characterization of csrA, a pleiotropic gene from Escherichia coli that affects glycogen biosynthesis, gluconeogenesis, cell size, and surface properties. J Bacteriol 175:4744–4755. doi: 10.1128/jb.175.15.4744-4755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabnis NA, Yang H, Romeo T. 1995. Pleiotropic regulation of central carbohydrate metabolism in Escherichia coli via the gene csrA. J Biol Chem 270:29096–29104. doi: 10.1074/jbc.270.49.29096. [DOI] [PubMed] [Google Scholar]
- 35.McKee AE, Rutherford BJ, Chivian DC, Baidoo EK, Juminaga D, Kuo D, Benke PI, Dietrich JA, Ma SM, Arkin AP, Petzold CJ, Adams PD, Keasling JD, Chhabra SR. 2012. Manipulation of the carbon storage regulator system for metabolite remodeling and biofuel production in Escherichia coli. Microb Cell Fact 11:79. doi: 10.1186/1475-2859-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu K. 2013. Regulation systems of bacteria such as Escherichia coli in response to nutrient limitation and environmental stresses. Metabolites 4:1–35. doi: 10.3390/metabo4010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aono R, Yamasaki M, Tamura G. 1979. High and selective resistance to mecillinam in adenylate cyclase-deficient or cyclic adenosine 3′,5′-monophosphate receptor protein-deficient mutants of Escherichia coli. J Bacteriol 137:839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Ari R, Jaffe A, Bouloc P, Robin A. 1988. Cyclic AMP and cell division in Escherichia coli. J Bacteriol 170:65–70. doi: 10.1128/jb.170.1.65-70.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogura T, Bouloc P, Niki H, D'Ari R, Hiraga S, Jaffe A. 1989. Penicillin-binding protein 2 is essential in wild-type Escherichia coli but not in lov or cya mutants. J Bacteriol 171:3025–3030. doi: 10.1128/jb.171.6.3025-3030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gosset G, Zhang Z, Nayyar S, Cuevas WA, Saier MH Jr. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J Bacteriol 186:3516–3524. doi: 10.1128/JB.186.11.3516-3524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sauer U, Eikmanns BJ. 2005. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev 29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Marquardt JL, Siegele DA, Kolter R, Walsh CT. 1992. Cloning and sequencing of Escherichia coli murZ and purification of its product, a UDP-N-acetylglucosamine enolpyruvyl transferase. J Bacteriol 174:5748–5752. doi: 10.1128/jb.174.17.5748-5752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown ED, Vivas EI, Walsh CT, Kolter R. 1995. MurA (MurZ), the enzyme that catalyzes the first committed step in peptidoglycan biosynthesis, is essential in Escherichia coli. J Bacteriol 177:4194–4197. doi: 10.1128/jb.177.14.4194-4197.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorke B, Foulquier E, Galinier A. 2005. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 151:3777–3791. doi: 10.1099/mic.0.28172-0. [DOI] [PubMed] [Google Scholar]
- 45.Mir M, Prisic S, Kang CM, Lun S, Guo H, Murry JP, Rubin EJ, Husson RN. 2014. Mycobacterial gene cuvA is required for optimal nutrient utilization and virulence. Infect Immun 82:4104–4117. doi: 10.1128/IAI.02207-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pensinger DA, Boldon KM, Chen GY, Vincent WJ, Sherman K, Xiong M, Schaenzer AJ, Forster ER, Coers J, Striker R, Sauer JD. 2016. The Listeria monocytogenes PASTA kinase PrkA and its substrate YvcK are required for cell wall homeostasis, metabolism, and virulence. PLoS Pathog 12:e1006001. doi: 10.1371/journal.ppat.1006001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foulquier E, Pompeo F, Bernadac A, Espinosa L, Galinier A. 2011. The YvcK protein is required for morphogenesis via localization of PBP1 under gluconeogenic growth conditions in Bacillus subtilis. Mol Microbiol 80:309–318. doi: 10.1111/j.1365-2958.2011.07587.x. [DOI] [PubMed] [Google Scholar]
- 48.Foulquier E, Pompeo F, Freton C, Cordier B, Grangeasse C, Galinier A. 2014. PrkC-mediated phosphorylation of overexpressed YvcK protein regulates PBP1 protein localization in Bacillus subtilis mreB mutant cells. J Biol Chem 289:23662–23669. doi: 10.1074/jbc.M114.562496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Formstone A, Errington J. 2005. A magnesium-dependent mreB null mutant: implications for the role of mreB in Bacillus subtilis. Mol Microbiol 55:1646–1657. doi: 10.1111/j.1365-2958.2005.04506.x. [DOI] [PubMed] [Google Scholar]
- 50.Kruse T, Bork-Jensen J, Gerdes K. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol 55:78–89. doi: 10.1111/j.1365-2958.2004.04367.x. [DOI] [PubMed] [Google Scholar]
- 51.Kawai Y, Asai K, Errington J. 2009. Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis. Mol Microbiol 73:719–731. doi: 10.1111/j.1365-2958.2009.06805.x. [DOI] [PubMed] [Google Scholar]
- 52.Leaver M, Errington J. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol 57:1196–1209. doi: 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- 53.Ruggiero A, De Simone P, Smaldone G, Squeglia F, Berisio R. 2012. Bacterial cell division regulation by Ser/Thr kinases: a structural perspective. Curr Protein Pept Sci 13:756–766. doi: 10.2174/138920312804871201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pereira SF, Goss L, Dworkin J. 2011. Eukaryote-like serine/threonine kinases and phosphatases in bacteria. Microbiol Mol Biol Rev 75:192–212. doi: 10.1128/MMBR.00042-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pietack N, Becher D, Schmidl SR, Saier MH, Hecker M, Commichau FM, Stulke J. 2010. In vitro phosphorylation of key metabolic enzymes from Bacillus subtilis: PrkC phosphorylates enzymes from different branches of basic metabolism. J Mol Microbiol Biotechnol 18:129–140. doi: 10.1159/000308512. [DOI] [PubMed] [Google Scholar]
- 56.Dubrac S, Bisicchia P, Devine KM, Msadek T. 2008. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol 70:1307–1322. doi: 10.1111/j.1365-2958.2008.06483.x. [DOI] [PubMed] [Google Scholar]
- 57.Absalon C, Obuchowski M, Madec E, Delattre D, Holland IB, Seror SJ. 2009. CpgA, EF-Tu and the stressosome protein YezB are substrates of the Ser/Thr kinase/phosphatase couple, PrkC/PrpC, in Bacillus subtilis. Microbiology 155:932–943. doi: 10.1099/mic.0.022475-0. [DOI] [PubMed] [Google Scholar]
- 58.Libby EA, Goss LA, Dworkin J. 2015. The eukaryotic-like Ser/Thr kinase PrkC regulates the essential WalRK two-component system in Bacillus subtilis. PLoS Genet 11:e1005275. doi: 10.1371/journal.pgen.1005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaidenko TA, Kim TJ, Price CW. 2002. The PrpC serine-threonine phosphatase and PrkC kinase have opposing physiological roles in stationary-phase Bacillus subtilis cells. J Bacteriol 184:6109–6114. doi: 10.1128/JB.184.22.6109-6114.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rued BE, Zheng JJ, Mura A, Tsui HT, Boersma MJ, Mazny JL, Corona F, Perez AJ, Fadda D, Doubravova L, Buriankova K, Branny P, Massidda O, Winkler ME. 2017. Suppression and synthetic-lethal genetic relationships of ΔgpsB mutations indicate that GpsB mediates protein phosphorylation and penicillin-binding protein interactions in Streptococcus pneumoniae D39. Mol Microbiol 103:931–957. doi: 10.1111/mmi.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claessen D, Emmins R, Hamoen LW, Daniel RA, Errington J, Edwards DH. 2008. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol Microbiol 68:1029–1046. doi: 10.1111/j.1365-2958.2008.06210.x. [DOI] [PubMed] [Google Scholar]
- 62.Rismondo J, Cleverley RM, Lane HV, Grosshennig S, Steglich A, Moller L, Mannala GK, Hain T, Lewis RJ, Halbedel S. 2016. Structure of the bacterial cell division determinant GpsB and its interaction with penicillin-binding proteins. Mol Microbiol 99:978–998. doi: 10.1111/mmi.13279. [DOI] [PubMed] [Google Scholar]
- 63.Chan JM, Guttenplan SB, Kearns DB. 2014. Defects in the flagellar motor increase synthesis of poly-γ-glutamate in Bacillus subtilis. J Bacteriol 196:740–753. doi: 10.1128/JB.01217-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wormann ME, Corrigan RM, Simpson PJ, Matthews SJ, Grundling A. 2011. Enzymatic activities and functional interdependencies of Bacillus subtilis lipoteichoic acid synthesis enzymes. Mol Microbiol 79:566–583. doi: 10.1111/j.1365-2958.2010.07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jerga A, Lu YJ, Schujman GE, de Mendoza D, Rock CO. 2007. Identification of a soluble diacylglycerol kinase required for lipoteichoic acid production in Bacillus subtilis. J Biol Chem 282:21738–21745. doi: 10.1074/jbc.M703536200. [DOI] [PubMed] [Google Scholar]
- 66.Gundlach J, Mehne FM, Herzberg C, Kampf J, Valerius O, Kaever V, Stulke J. 2015. An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J Bacteriol 197:3265–3274. doi: 10.1128/JB.00564-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Hartig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rugheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stulke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Volker U, Bessieres P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 68.Surdova K, Gamba P, Claessen D, Siersma T, Jonker MJ, Errington J, Hamoen LW. 2013. The conserved DNA-binding protein WhiA is involved in cell division in Bacillus subtilis. J Bacteriol 195:5450–5460. doi: 10.1128/JB.00507-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elbaz M, Ben-Yehuda S. 2010. The metabolic enzyme ManA reveals a link between cell wall integrity and chromosome morphology. PLoS Genet 6:e1001119. doi: 10.1371/journal.pgen.1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hansen T, Wendorff D, Schonheit P. 2004. Bifunctional phosphoglucose/phosphomannose isomerases from the Archaea Aeropyrum pernix and Thermoplasma acidophilum constitute a novel enzyme family within the phosphoglucose isomerase superfamily. J Biol Chem 279:2262–2272. doi: 10.1074/jbc.M309849200. [DOI] [PubMed] [Google Scholar]
- 71.Yeom SJ, Ji JH, Kim NH, Park CS, Oh DK. 2009. Substrate specificity of a mannose-6-phosphate isomerase from Bacillus subtilis and its application in the production of l-ribose. Appl Environ Microbiol 75:4705–4710. doi: 10.1128/AEM.00310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Young FE, Arias L. 1967. Biosynthesis of the N-acyl-galactosamine in cell walls of Bacillus subtilis. J Bacteriol 94:1783–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zaritsky A, Woldringh CL, Einav M, Alexeeva S. 2006. Use of thymine limitation and thymine starvation to study bacterial physiology and cytology. J Bacteriol 188:1667–1679. doi: 10.1128/JB.188.5.1667-1679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zaritsky A. 2015. Cell-shape homeostasis in Escherichia coli is driven by growth, division, and nucleoid complexity. Biophys J 109:178–181. doi: 10.1016/j.bpj.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Monahan LG, Harry EJ. 2016. You are what you eat: metabolic control of bacterial division. Trends Microbiol 24:181–189. doi: 10.1016/j.tim.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 76.Vadia S, Levin PA. 2015. Growth rate and cell size: a re-examination of the growth law. Curr Opin Microbiol 24:96–103. doi: 10.1016/j.mib.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weart RB, Lee AH, Chien AC, Haeusser DP, Hill NS, Levin PA. 2007. A metabolic sensor governing cell size in bacteria. Cell 130:335–347. doi: 10.1016/j.cell.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lazarevic V, Soldo B, Medico N, Pooley H, Bron S, Karamata D. 2005. Bacillus subtilis α-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl Environ Microbiol 71:39–45. doi: 10.1128/AEM.71.1.39-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jorasch P, Wolter FP, Zahringer U, Heinz E. 1998. A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol Microbiol 29:419–430. doi: 10.1046/j.1365-2958.1998.00930.x. [DOI] [PubMed] [Google Scholar]
- 80.Chien AC, Zareh SK, Wang YM, Levin PA. 2012. Changes in the oligomerization potential of the division inhibitor UgtP co-ordinate Bacillus subtilis cell size with nutrient availability. Mol Microbiol 86:594–610. doi: 10.1111/mmi.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hill NS, Buske PJ, Shi Y, Levin PA. 2013. A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet 9:e1003663. doi: 10.1371/journal.pgen.1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Debarbieux L, Bohin A, Bohin JP. 1997. Topological analysis of the membrane-bound glucosyltransferase, MdoH, required for osmoregulated periplasmic glucan synthesis in Escherichia coli. J Bacteriol 179:6692–6698. doi: 10.1128/jb.179.21.6692-6698.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang X, Holden HM, Raushel FM. 2001. Channeling of substrates and intermediates in enzyme-catalyzed reactions. Annu Rev Biochem 70:149–180. doi: 10.1146/annurev.biochem.70.1.149. [DOI] [PubMed] [Google Scholar]
- 84.Bulutoglu B, Garcia KE, Wu F, Minteer SD, Banta S. 2016. Direct evidence for metabolon formation and substrate channeling in recombinant TCA cycle enzymes. ACS Chem Biol 11:2847–2853. doi: 10.1021/acschembio.6b00523. [DOI] [PubMed] [Google Scholar]
- 85.Hollinshead WD, Rodriguez S, Martin HG, Wang G, Baidoo EE, Sale KL, Keasling JD, Mukhopadhyay A, Tang YJ. 2016. Examining Escherichia coli glycolytic pathways, catabolite repression, and metabolite channeling using Δpfk mutants. Biotechnol Biofuels 9:212. doi: 10.1186/s13068-016-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, Davey M, Parkinson J, Greenblatt J, Emili A. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- 87.Marchadier E, Carballido-Lopez R, Brinster S, Fabret C, Mervelet P, Bessieres P, Noirot-Gros MF, Fromion V, Noirot P. 2011. An expanded protein-protein interaction network in Bacillus subtilis reveals a group of hubs: exploration by an integrative approach. Proteomics 11:2981–2991. doi: 10.1002/pmic.201000791. [DOI] [PubMed] [Google Scholar]
- 88.Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J. 2009. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics 8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yamamoto H, Kurosawa S, Sekiguchi J. 2003. Localization of the vegetative cell wall hydrolases LytC, LytE, and LytF on the Bacillus subtilis cell surface and stability of these enzymes to cell wall-bound or extracellular proteases. J Bacteriol 185:6666–6677. doi: 10.1128/JB.185.22.6666-6677.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Meyer FM, Gerwig J, Hammer E, Herzberg C, Commichau FM, Volker U, Stulke J. 2011. Physical interactions between tricarboxylic acid cycle enzymes in Bacillus subtilis: evidence for a metabolon. Metab Eng 13:18–27. doi: 10.1016/j.ymben.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 91.Manat G, Roure S, Auger R, Bouhss A, Barreteau H, Mengin-Lecreulx D, Touze T. 2014. Deciphering the metabolism of undecaprenyl-phosphate: the bacterial cell-wall unit carrier at the membrane frontier. Microb Drug Resist 20:199–214. doi: 10.1089/mdr.2014.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Brown S, Santa Maria JP Jr, Walker S. 2013. Wall teichoic acids of gram-positive bacteria. Annu Rev Microbiol 67:313–336. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weissborn AC, Rumley MK, Kennedy EP. 1991. Biosynthesis of membrane-derived oligosaccharides. Membrane-bound glucosyltransferase system from Escherichia coli requires polyprenyl phosphate. J Biol Chem 266:8062–8067. [PubMed] [Google Scholar]
- 94.Fiedler W, Rotering H. 1988. Properties of Escherichia coli mutants lacking membrane-derived oligosaccharides. J Biol Chem 263:14684–14689. [PubMed] [Google Scholar]
- 95.Bontemps-Gallo S, Cogez V, Robbe-Masselot C, Quintard K, Dondeyne J, Madec E, Lacroix JM. 2013. Biosynthesis of osmoregulated periplasmic glucans in Escherichia coli: the phosphoethanolamine transferase is encoded by opgE. Biomed Res Int 2013:371429. doi: 10.1155/2013/371429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ebel W, Vaughn GJ, Peters HK III, Trempy JE. 1997. Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J Bacteriol 179:6858–6861. doi: 10.1128/jb.179.21.6858-6861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Monahan LG, Hajduk IV, Blaber SP, Charles IG, Harry EJ. 2014. Coordinating bacterial cell division with nutrient availability: a role for glycolysis. mBio 5:e00935-14. doi: 10.1128/mBio.00935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Cascante-Estepa N, Gunka K, Stulke J. 2016. Localization of components of the RNA-degrading machine in Bacillus subtilis. Front Microbiol 7:1492. doi: 10.3389/fmicb.2016.01492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Commichau FM, Gunka K, Landmann JJ, Stulke J. 2008. Glutamate metabolism in Bacillus subtilis: gene expression and enzyme activities evolved to avoid futile cycles and to allow rapid responses to perturbations of the system. J Bacteriol 190:3557–3564. doi: 10.1128/JB.00099-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Powell BS, Court DL. 1998. Control of ftsZ expression, cell division, and glutamine metabolism in Luria-Bertani medium by the alarmone ppGpp in Escherichia coli. J Bacteriol 180:1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beaufay F, Coppine J, Mayard A, Laloux G, De Bolle X, Hallez R. 2015. A NAD-dependent glutamate dehydrogenase coordinates metabolism with cell division in Caulobacter crescentus. EMBO J 34:1786–1800. doi: 10.15252/embj.201490730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Radhakrishnan SK, Pritchard S, Viollier PH. 2010. Coupling prokaryotic cell fate and division control with a bifunctional and oscillating oxidoreductase homolog. Dev Cell 18:90–101. doi: 10.1016/j.devcel.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 103.Ronneau S, Petit K, De Bolle X, Hallez R. 2016. Phosphotransferase-dependent accumulation of (p)ppGpp in response to glutamine deprivation in Caulobacter crescentus. Nat Commun 7:11423. doi: 10.1038/ncomms11423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jorgenson MA, Young KD. 2016. Interrupting biosynthesis of O antigen or the lipopolysaccharide core produces morphological defects in Escherichia coli by sequestering undecaprenyl phosphate. J Bacteriol 198:3070–3079. doi: 10.1128/JB.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jorgenson MA, Kannan S, Laubacher ME, Young KD. 2016. Dead-end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate. Mol Microbiol 100:1–14. doi: 10.1111/mmi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Farha MA, Czarny TL, Myers CL, Worrall LJ, French S, Conrady DG, Wang Y, Oldfield E, Strynadka NC, Brown ED. 2015. Antagonism screen for inhibitors of bacterial cell wall biogenesis uncovers an inhibitor of undecaprenyl diphosphate synthase. Proc Natl Acad Sci U S A 112:11048–11053. doi: 10.1073/pnas.1511751112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Grover S, Alderwick LJ, Mishra AK, Krumbach K, Marienhagen J, Eggeling L, Bhatt A, Besra GS. 2014. Benzothiazinones mediate killing of Corynebacterineae by blocking decaprenyl phosphate recycling involved in cell wall biosynthesis. J Biol Chem 289:6177–6187. doi: 10.1074/jbc.M113.522623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Atilano ML, Pereira PM, Yates J, Reed P, Veiga H, Pinho MG, Filipe SR. 2010. Teichoic acids are temporal and spatial regulators of peptidoglycan cross-linking in Staphylococcus aureus. Proc Natl Acad Sci U S A 107:18991–18996. doi: 10.1073/pnas.1004304107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.D'Elia MA, Millar KE, Beveridge TJ, Brown ED. 2006. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol 188:8313–8316. doi: 10.1128/JB.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.D'Elia MA, Pereira MP, Chung YS, Zhao W, Chau A, Kenney TJ, Sulavik MC, Black TA, Brown ED. 2006. Lesions in teichoic acid biosynthesis in Staphylococcus aureus lead to a lethal gain of function in the otherwise dispensable pathway. J Bacteriol 188:4183–4189. doi: 10.1128/JB.00197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xayarath B, Yother J. 2007. Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J Bacteriol 189:3369–3381. doi: 10.1128/JB.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ranjit DK, Young KD. 2016. Colanic acid intermediates prevent de novo shape recovery of Escherichia coli spheroplasts, calling into question biological roles previously attributed to colanic acid. J Bacteriol 198:1230–1240. doi: 10.1128/JB.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schirner K, Eun YJ, Dion M, Luo Y, Helmann JD, Garner EC, Walker S. 2015. Lipid-linked cell wall precursors regulate membrane association of bacterial actin MreB. Nat Chem Biol 11:38–45. doi: 10.1038/nchembio.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Peters JM, Colavin A, Shi H, Czarny TL, Larson MH, Wong S, Hawkins JS, Lu CH, Koo BM, Marta E, Shiver AL, Whitehead EH, Weissman JS, Brown ED, Qi LS, Huang KC, Gross CA. 2016. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165:1493–1506. doi: 10.1016/j.cell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. 2010. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol 12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Janniere L, Canceill D, Suski C, Kanga S, Dalmais B, Lestini R, Monnier AF, Chapuis J, Bolotin A, Titok M, Le Chatelier E, Ehrlich SD. 2007. Genetic evidence for a link between glycolysis and DNA replication. PLoS One 2:e447. doi: 10.1371/journal.pone.0000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu F, Qimuge, Hao J, Yan H, Bach T, Fan L, Morigen . 2014. AspC-mediated aspartate metabolism coordinates the Escherichia coli cell cycle. PLoS One 9:e92229. doi: 10.1371/journal.pone.0092229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Murray H, Koh A. 2014. Multiple regulatory systems coordinate DNA replication with cell growth in Bacillus subtilis. PLoS Genet 10:e1004731. doi: 10.1371/journal.pgen.1004731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yadavalli SS, Carey JN, Leibman RS, Chen AI, Stern AM, Roggiani M, Lippa AM, Goulian M. 2016. Antimicrobial peptides trigger a division block in Escherichia coli through stimulation of a signalling system. Nat Commun 7:12340. doi: 10.1038/ncomms12340. [DOI] [PMC free article] [PubMed] [Google Scholar]