Abstract
Ran-binding protein family member, RanBP9 has been reported in various basic cellular mechanisms and neuropathological conditions including schizophrenia. Previous studies have reported that RanBP9 is highly expressed in the mammalian brain and retina; however, the role of RanBP9 in retinal development is largely unknown. Here, we present the novel and regulatory roles of RanBP9 in retinal development of a vertebrate animal model, zebrafish. Zebrafish embryos exhibited abundant expression of ranbp9 in developing brain tissues as well as in the developing retina. Yeast two-hybrid screening demonstrated the interaction of RanBP9 with Mind bomb, a component of Notch signaling involved in both neurogenesis and neural disease autism. The interaction is further substantiated by co-localization studies in cultured cells. Knockdown of ranbp9 resulted in retinal dysplasia with defective proliferation of retinal cells, downregulation of neuronal differentiation marker huC, elevation of neural proliferation marker her4, and alteration of cell cycle marker p57kip2. Expression of the Müller glial cell marker glutamine synthase was also affected in knockdown morphants. Our results suggest that Mind bomb-binding partner RanBP9 plays a role during retinal cell development of zebrafish embryogenesis.
Keywords: mind bomb, notch, RanBP9, retinogenesis, zebrafish
INTRODUCTION
Notch signaling is an evolutionarily conserved mechanism which controls a broad spectrum of cell fates and developmental processes through cell–cell interaction and lateral inhibition. Notch signaling is initiated when the extracellular domain of the ligand interacts with the Notch receptor, resulting in proteolytic cleavage of the Notch intracellular domain, NICD. Subsequent nuclear translocation of NICD regulates the transcription of various target genes. Defects in Notch signaling have been implicated in a wide variety of developmental defects and diseases (Selkoe and Kopan, 2003).
We previously identified Mind bomb, a key component of the Notch pathway, via positional cloning of zebrafish neurogenic mutants (Itoh et al., 2003). Mutations in mind bomb have also been shown to cause defects in a variety of tissues such as somites, pancreas, intestine, hematopoietic stem cells, and vasculature where Notch signaling is thought to play an important role (Lawson et al., 2001). mind bomb mutants generate neurons in appropriate spatial regions but in excessive quantity. The gene encodes for a highly conserved E3 ubiquitin ligase which ubiquitinates and promotes endocytosis of Notch ligands. The endocytic process is crucial for efficient activation of Notch receptor signaling and is regulated spatiotemporally by several distinct mechanisms (Gwak et al., 2010; Koo et al., 2005). Post-translational modifications involving ubiquitination play a critical role in the regulation of Delta-mediated Notch signaling activity (Le Borgne et al., 2005). Delta is a major ligand in the Notch pathway that is regulated by the E3 ubiquitin ligase, Mind bomb.
In this study we identified a Mind bomb-binding protein, Ran-binding protein 9 (RanBP9), using the yeast two-hybrid screening method. RanBP9 is a member of the Ran-binding protein family and is encoded by the gene, ranbp9. RanBP9 has five conserved functional domains, including the N-terminal proline-rich domain (PRD), a SPRY domain, a lissencephaly type-I-like homology (LisH) motif, a C-terminal to LisH (CTLH) motif, and a C-terminal CRA motif (Nishitani et al., 2001). The RanBP9 protein binds Ran, a RAS family GTP binding protein that plays a significant role in translocation of RNA and protein through the nuclear pore complex.
In our study, the binding of RanBP9 with Mind bomb was confirmed by co-immunoprecipitation in vitro and subcellular colocalization assays in cultured cells. The spatiotemporal expression pattern of ranbp9 from whole-mount in situ hybridization analysis implicates the necessity of RanBP9 in early brain and retina development in zebrafish. Also, knockdown experiments using antisense morpholino oligo-nucleotide were performed to examine the functional role of ranbp9 in zebrafish retinal cell development.
MATERIALS AND METHODS
Yeast two-hybrid screening
For the construction of bait, the mid-region of the Ankyrin repeat domain (from amino acids 410–774) of the MIB cDNA (Genbank Accession No. AY147849) was amplified by PCR using the primers, 5′-CAGAATTCGTGGTGGAAGGGGT TGGCGCTCGGGT-3′ (Forward Primer) and 5′-GATGCATTGCTAATGGCTGATCCTG-3′ (Reverse Primer). The amplified PCR fragment was then cloned into the EcoRI site of the pGBT9 vector (encoding the GAL4 DNA-binding domain), pGBT9/Ank-MIB. The cloned vectors were transformed into the yeast host strain, AH109, which was then mated with the Y187 yeast cells. The cells were pre-transformed using the Human Matchmaker cDNA Library (Clontech). The pre-transformed cDNA library in the yeast Y187 strain was screened for MIB-interaction partners. The toxicity of baits on the AH109 yeast cells and their transcriptional activation were tested and screened. Positive colonies were allowed to grow on Trp-, Leu-, His-, and Ade-depleted, minimum synthetic dropout medium (SD) for yeast, and were selected by β-Galactosidase quantitative assay. Plasmid DNAs isolated from the yeast cells were used as templates for further PCR amplification and sequencing. From DNA sequencing analysis, we identified six clones for RanBP9, one clone for SNX5 (Yoo et al., 2006), one clone for FIH-1 (So et al., 2014), and clones for other genes.
Isolation of RanBP9 full-length cDNA and sequence analysis
The zebrafish RanBP9 was isolated from its genome by performing the RT-PCR, using specific primers, 5′-TAGCTGGCAAGCTGTTGGTATCGAC-3′ as the forward primer and 5′-GGCCTCTGTCACGTGGTCGTTTAA-3′ as the reverse primer, with the total RNA isolated from different developmental stages of the zebrafish. The amplified full-length RanBP9 fragment was then cloned into the pGEM-T Easy vector and later confirmed by sequencing. The full-length RanBP9 was then cloned into the pCS2+ expression vector. To make capped mRNA, vectors were linearized using respective digestive enzyme and transcribed with SP6 RNA polymerase using the mMESSAGE mMACHINE SP6 in vitro transcription kit (Ambion), following manufacturer instructions.
Also, to study the evolutionary relationship between the RanBP9 from the zebrafish and other organisms, the translated amino acid sequences were aligned with the ‘ClustalOmega’ Protein Alignment Tool, (https://www.ebi.ac.uk/Tools/msa/clustalo/)
RNA and antisense morpholino oligonucleotides injection
Synthetic capped RNA for RanBP9 was transcribed in vitro using linearized plasmid DNA as the template. Later, the RNA was dissolved in 0.2 M KCl with 0.2% Phenol Red (tracking dye), and injected into the cytoplasm of the two-cell stage embryos using a microinjector (PV820 Pneumatic PicoPump, WPI). An antisense morpholino oligonucleotide having the sequence 5′-CAAATGTTTCCTACCTAGACTGTGG-3′ (Gene Tools) was designed to target the exon 4 splicing donor site of ranbp9. The oligonucleotide was resuspended in distilled nuclease-free water and over 0.5 nl of 0.75 mM MO (4 ng/embryo) was injected into the yolk of one to two-cell stage embryos.
Whole-mount in situ hybridization, cryostat and paraffin sections
DIG-labelled antisense RNA probe for RanBP9 and MIB were synthesized from corresponding linearized plasmids using SP6 RNA polymerase following manufacturer instructions (Ambion). Fluorescein labeled probes for gata1 and flk-1 were synthesized as previously described. Whole mount in situ hybridization was done as previously described with little modifications. For sectioning, the embryos were soaked in 30% sucrose at 4°C overnight, and later embedded in 1.5% agarose and 5% sucrose. The 8 μm thick sections were cut using a cryostat microtome. For paraffin sectioning, the embryos were dehydrated in ethanol and washed in isopropanol for 10 min. The embryos were, later equilibrated in xylene for 15 min and transferred to fresh solution of xylene. Embryos were transferred to Xylene: Paraffin (1:1), at 60°C for 20 min, replaced with paraffin, 3 times at 60°C after every 20 min. After complete equilibration of the embryos with paraffin, the embryos were oriented and molten paraffin was poured and solidified. Trimmed paraffin blocks were sectioned at 8 μm thickness, using a histostat microtome. Later, the dry sections were soaked in xylene to remove the paraffin, loaded with the Canada balsam mounting medium and covered with cover slips.
Hematoxylin and eosin staining
Paraffin from the paraffin section slides were removed using xylene, rehydrated in a graded ethanol series (100%, 95%, 85%, 70%) for 3 min, and washed in double distilled water. Later the slides were stained with Harris’ Alum Hematoxylin (Harleco) with 4% acetic acid at room temperature for 5 min. The slides were again washed with double distilled water for 2 min, followed by dipping 8 times in acid-alcohol (1% HCl in 70% ethanol) for 30 s. Subsequently, the sections were rinsed in double distilled water for 2 min and dipped 20 times in 0.5% ammonium sulphate (w/v) for 1 min, and again rinsed with double distilled water for 2 min. Later, the sections were stained with eosin Y (1 mg/ml eosin Y in 95% ethanol + 2.8 μl/ml acetic acid) at room temperature for 3 min. The sections were, then, rehydrated in a graded ethanol series (70%, 85%, 95%, 100%) for 3 min. Sections were soaked in xylene and left at room temperature to slightly dry before loading on with Canada balsam. Slides were viewed using a Leica DM5000B microscope under white light.
Cell cultures and transient transfection
Transfectable COS7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% heat inactivated fetal bovine serum and antibiotics. The cDNA of the zebrafish RanBP9 was sub-cloned into pCS2+GFP via EcoRI and XhoI. Appropriate amounts of RanBP9-pCS2+GFP were co-transfected with mouse HA-MIB1 pcDNA into the cultured cells using Lipofectamine Plus (Invitrogen).
Immunocytochemistry
At 24–48 h post-transfection of plasmids into the COS7 cells, the cells were washed in Phosphate Buffered Saline (PBS) and fixed in 4% paraformaldehyde with 3% sucrose at 4°C for 30 min. The fixed cells were then incubated in blocking solution (3% skim milk and 0.1% TritonX-100 in PBS) at 4°C overnight, and later stained with respective primary antibody in 3% skim milk in PBS at room temperature for 1 h. Eventually, the cells were incubated with a mouse antibody conjugated with Alex549 at room temperature for 30 min. For DNA staining, the cells were stained with Hoechst 33342 (10 μg/ml) for 2 min. After thorough rinsing with 0.1% TritonX-100 in PBS for three times, the cells were mounted on glass slides and analyzed with a fluorescent microscope (Zeiss).
RESULTS
Identification of RanBP9 as a Mind bomb-binding protein
Yeast two-hybrid screening was performed to identify putative interacting members or partner proteins of Mind bomb (Mib)-mediated Notch signaling pathways, using the Ankyrin (Ank) mid-region of the Mib as bait (Fig. 1A). We previously isolated the mib gene from positional cloning of zebrafish mind bomb neurogenic mutant (Itoh et al., 2003). Mib is an ubiquitin ligase that is essential for efficient activation of Notch signaling. The Notch signaling pathway is essential for neuronal and glial cell specification during CNS development. Mib is seen as highly conserved throughout the evolutionary process as observed in Drosophila and humans. On screening of 1.44 × 106 colonies of a cDNA library, several candidate putative genes were identified using the mid-region containing Ank repeats; included among them the prey gene, ranbp9 which is the cDNA encoding RanBP9. Further screening of zebrafish EST and genomic database led to the identification of several other ESTs and fragments of RanBP9. Zebrafish ranbp9 coding regions were amplified by PCR with their respective primers using cDNA of 24 hpf zebrafish embryos. Zebrafish RanBP9 showed significant amino acid identity with RanBP9 of other organisms (Fig. 1B), with conserved domains of SPRY, LiSH, CTLH, and CRA (Fig. 1C). However, zebrafish RanBP9 did not share the long stretch of proline and glutamine within the N-terminal region, which is shown in mammals.
Fig. 1.
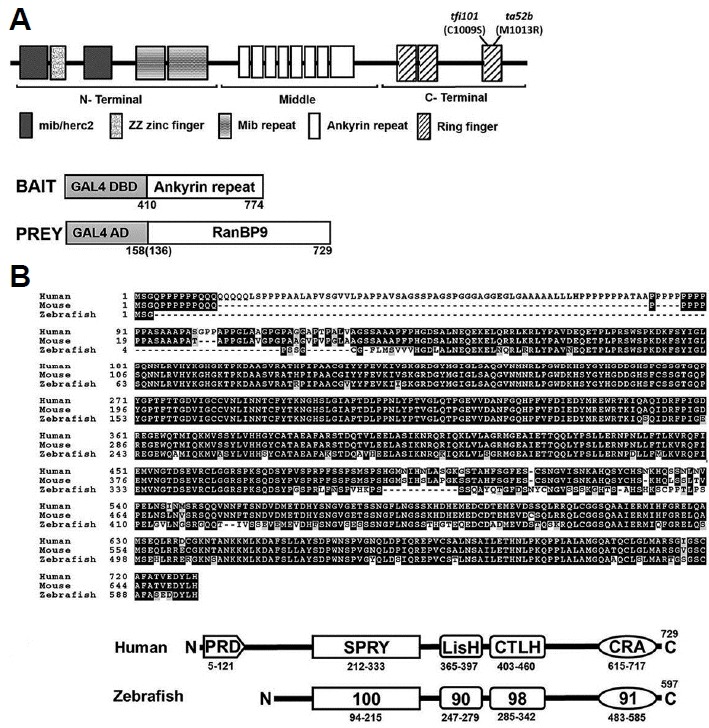
Isolation of RanBP9 as a Mib-binding protein from yeast two-hybrid screening.
(A) Schematic representation of Mib bait and RanBP9 prey proteins. The Mib bait contains the middle region of ankyrin repeats from amino acids 410 to 744. Multiple clones of RanBP9 prey were identified. Here, RanBP9 polypeptides starting from 136th or 158th amino acid were represented. (B) Protein sequence alignment between human, mouse and zebrafish RanBP9 homologues. (C) Comparison of domain similarity (%) between human and zebrafish RanBP9. PRD, proline-rich domain; SPRY, domain in sp1A and Ryanodine receptor; LisH, Lissencephaly type-1-like homology motif; CTLH, C-Terminal to the LisH; and a CRA motif.
Spatiotemporal distribution of ranbp9 mRNA in embryogenesis of zebrafish
To better understand the roles of RanBP9 in developing zebrafish embryos, we first tested spatiotemporal distribution of ranbp9 mRNA in zebrafish embryos using whole mount in situ hybridization analysis (Fig. 2). During early development, ranbp9 expression can be observed as early as the 2-cell stage (Fig. 2A), which strongly suggests that ranbp9 mRNA is present in the embryo maternally. Expression was also detected ubiquitously throughout early developmental stages, as in the 4 hpf (Fig. 2B). However, it was hard to detect the expression at the shield stage (Fig. 2C). At the early segmentation stage the ranbp9 showed a broad expression pattern (Fig. 2D). At 48 hpf the distribution of ranbp9 transcripts was found to be confined to the developing brain and the retina, while it disappeared in the trunk region (Fig. 2E). At 72 hpf, zebrafish larvae were cryosectioned and mounted on slides (Fig. 2F). The distribution of ranbp9 transcripts was found to be confined to the developing brain. In the retina, ranbp9 transcripts is highly detected in the developing retinal layers, particularly in the ganglion cell layer and inner nuclear layer (Fig. 2F).
Fig. 2.
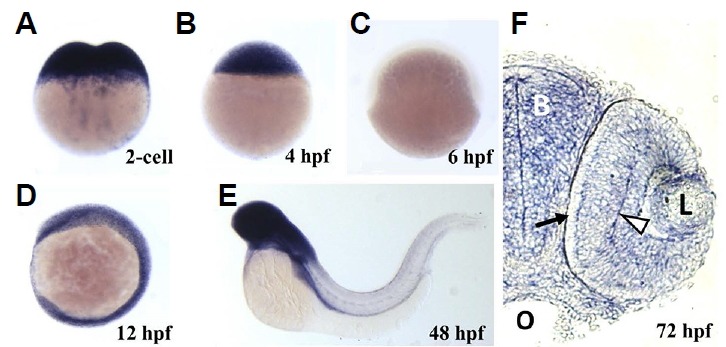
Spatiotemporal expression of ranbp9 mRNA during embryonic development.
Whole-mount in situ hybridization was performed with DIG-labeled antisense RNA probe (hpf, hours after fertilization). (A) At 2-cell stage, maternal transcripts were detected in high density. (B) At dome stage (4 hpf), high level of transcription persisted. (C) At the beginning of gastrulation, shield stage (6 hpf), ranbp9 mRNA was hardly detected. (D) At 6-somite stage (12 hpf), ranbp9 was seen expressed ubiquitously. (E) At 48 hpf, ranbp9 expression was mainly observed in the head region. (F) Section of head region at 72 hpf, showing ranbp9 expression in the brain (B) and eye (L, lens). Its expression is more prominent in forming layers (arrow head) of the retina, compared to the periphery (arrow). O, oral cavity.
Physical and subcellular co-localization of RanBP9 and Mib
The physical interaction between RanBP9 and Mib was tested by co-immunoprecipitation of the proteins expressed in culture cells (Fig. 3). RanBP9 was co-transfected with Mib into the COS7 cells. Interestingly, RanBP9 tagged at the C- terminal (RanBP9-Flag) did not show any interaction with Mib (Fig. 3B, lane 2). The CRA motif at the C-terminus of RanBP9 protein, which is comprised of approximately 100 amino acids, was found to be important for the interaction of RanBP9 with fragile X mental retardation protein (FMRP), but its functional significance has yet to be determined (Menon et al., 2004). However, RanBP9 tagged at the N-terminal (Myc-RanBP9) was co-immunoprecipitated with Mib (Fig. 3B, lane 5). Its interaction was higher with wild type Mib than a mutant form of Mib (ta52b), point mutation at the ring finger domain (M1013R), although the expression level of ta52b was higher than wild type Mib in transfected cells (Fig. 3B). As a positive control we used Moe/Epb41l5 which is known to bind to the N-terminal fragment of Mib, but not to the Ankyrin mid-region and the C-terminal domain (Matsuda et al., 2016). Moe showed higher expression level than that of Mib proteins. The interaction between RanBP9 and Mib was further tested by examining their subcellular localization in cultured cells (Fig. 4). The RanBP9 was localized both in the nucleus and the punctate structures of the cytoplasm (Fig. 4B). Interestingly, in the cytoplasmic punctate structures, Mib was co-localized with the RanBP9 (Figs. 4A and 4C), suggesting that RanBP9 associates with some roles in the Notch signaling pathway. Previously, we showed that Mib is localized in the early endosomes and involved in Notch ligand Delta processing (Yoo et. al., 2006).
Fig. 3.
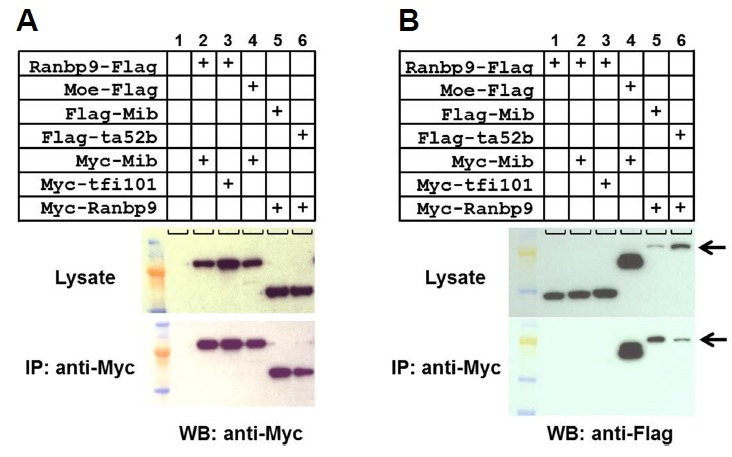
Physical interaction between RanBP9 and Mib.
(A) Each cell lysate transfected with a combination of expression vectors was pulled down with anti-Myc antibody and the western blot was stained with anti-Myc antibody. Top panels show lysates without co-immunoprecipitation. (B) Cell lysates were pulled down with anti-Myc antibody and the western blot was stained with anti-Frag antibody. ta52b (M1013R) and tfi101 (C1009S) are mutant forms of Mib, having point mutation in the 3rd ring finger. Moe was used as a positive control for Mib binding. Arrows indicate the size for Frag-Mib and Frag-Mib/ta52b.
Fig. 4.
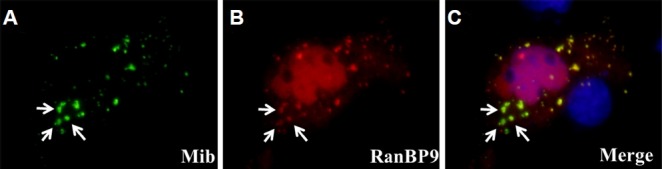
Subcellular co-localization of RanBP9 and Mib in COS7 cells.
COS7 cells were transiently transfected with expression vectors encoding Mib-GFP and HA-RanBP9. (A) Mib-GFP was detected in the cytoplasm in a punctuate pattern. (B) HA-RanBP9 was localized in a similar punctuate pattern in the cytoplasm, in addition to its localization in the nucleus. Transfected cells were fixed and stained with anti-HA antibody. The yellow color and arrows in the merged image in (C) indicate co-localization of Mib and RanBP9 in the cytoplasmic punctuate structures. Hoechst dye staining (blue) shows the nucleus.
RanBP9 functions in brain and retinal development
To test the roles of RanBP9 in embryonic development, gene knockdown experiments were performed using morpholino antisense oligonucleotides (Fig. 5). A morpholino oligonucleotide was designed to target the exon splice donor site of RanBP9 and produce premature translation termination, which was confirmed by RT-PCR (Fig. 5D). Morpholino injected embryos exhibited defects in brain development and swim-bladder inflation (Fig. 6A). By acridine orange staining method, we identified specific cell death in the brain, especially in the optic tectum, in 54 hpf morphants (Fig. 6B). Because RanBP9 is also expressed in the developing retina, we concentrated on examining retinal development in the ranbp9 morphant larvae. Furthermore, we tested the effects of RanBP9 knockdown on embryos through histological examinations. Hematoxylin and eosin stained transverse sections of zebrafish eyes at 73 hpf showed severe defects in retinal lamination. Particularly, the cell layers GCL, INL and ONL were morphologically different in contrast to wild type (Fig. 6C). This finding points to a possible reason for the failure of retinal cell differentiation.
Fig. 5.
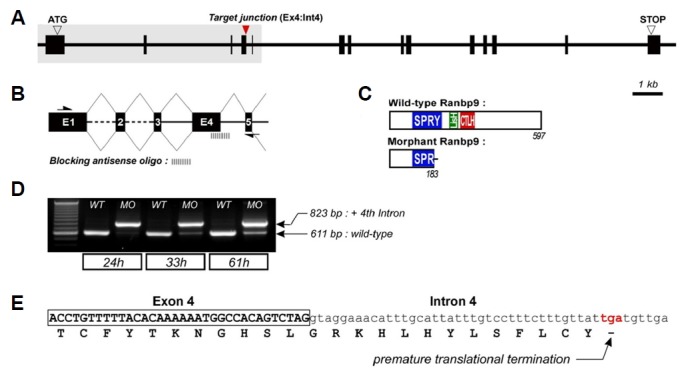
Genomic structure of ranbp9 and design of antisense morpholino to block the pre-mRNA splicing.
(A) 14 exons of ranbp9 gene was stretched for 20 kb on the genome. The target junction is the splicing donor site at Ex4:Int4. (B) Diagram illustrating the process of interference with premature mRNA splicing. Enlargement of the grey boxed area in (A). Arrows in (B) indicate the locations of primers used for the PCR analysis. (C) Predicted proteins encoded by the normal and morphant transcripts. Truncated RanBP9 was produced by an induced stop codon. (D) RT-PCR analysis for the efficacy of antisense morpholino. Abnormal transcripts in the morphant embryos were detected (823 bp, compared to 611 bp wild-type). The efficacy gradually decreased at later stages. (E) The truncated RanBP9 was confirmed by sequencing of morphant transcripts.
Fig. 6.
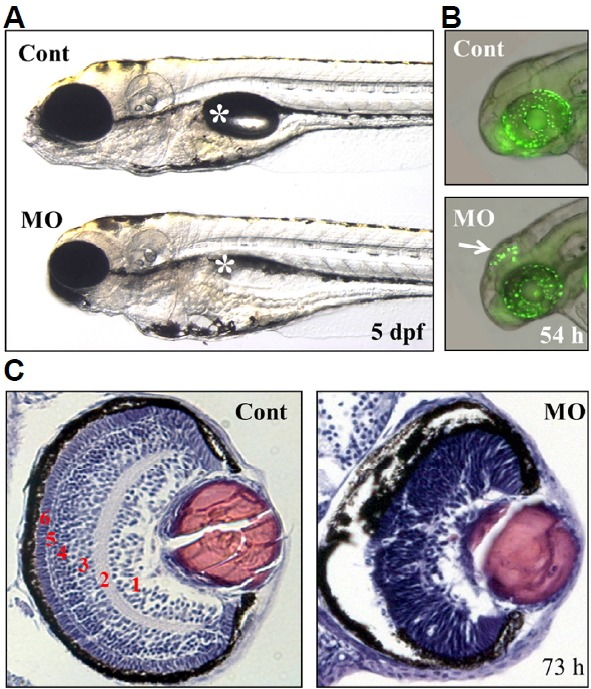
Defects in the brain and retina of ranbp9-MO injected zebrafish embryos.
(A) Control (Cont) and ranbp9-MO injected (MO) zebrafish larvae at 5 dpf. Compared to normal development of body trunk region, reduced size of brain and eye was observed in the MO injected larva. Asterisk indicates swim-bladder. (B) By acridine orange staining, a specific cell death was detected in the brain tectum of 54 hpf morphants. (C) Disruption of retinal development in ranbp9 morphant. Section of HE-stained retina in control and ranbp9 morphant at 73 hpf. In wild-type, the retina was established into several layers; 1, ganglion cell layer: 2, inner plexiform layer: 3, inner nuclear layer: 4, outer plexiform layer: 5, outer nuclear layer: and 6, photoreceptor cell layer, respectively. In morphants, however, these layer formations were not well established.
Knockdown of RanBP9 delayed proliferation of retinal cell types
To understand the molecular mechanisms of RanBP9 in retinal cell development, we tested the expression of several molecular markers in the retina of ranbp9 knockdown morphants. At about 48 hpf, the retina mostly consisted of pigmented epithelium and retinal neuro-epithelium; however, distinct layers were not yet distinguishable at this stage. At 72 hpf, the major nuclear and plexiform layers, the optic nerves, and the pigmented epithelium were well differentiated in the retina (Avanesov and Malicki, 2004). Interestingly, ranbp9 knockdown morphant larvae exhibited a reduced number of retinal cells and lacked properly established nuclear and plexiform layers. To examine the role of RanBP9 in retinal glial cell differentiation, we tested the expression domain of huC, a molecular marker for the differentiated neurons (Kim et al., 1996). huC expression domains were greatly reduced in ranbp9 knockdown morphants in contrast with control embryos at 48 hpf, indicating that elimination of RanBP9 may inhibit neuronal differentiation. Next, we examined the expression of her4 in the retina at 48 hpf. her4 is a molecular marker for undifferentiated neuronal precursors (Jung et al., 2012) that could survey the role of RanBP9 in retinal cell proliferation. Surprisingly, the expression domain for her4 was found to be expanded in the retinal layers (Fig. 7A) which would suggest that RanBP9 is required in retinal cell proliferation during retinogenesis. Previous studies have also shown that Notch signaling activates her4 to inhibit neurogenesis during retinogenesis (Yamaguchi et al., 2005). Based on the above findings we propose that RanBP9 is expressed in the brain and retina and regulates the proliferation and differentiation of neuronal cells during retinogenesis.
Fig. 7.
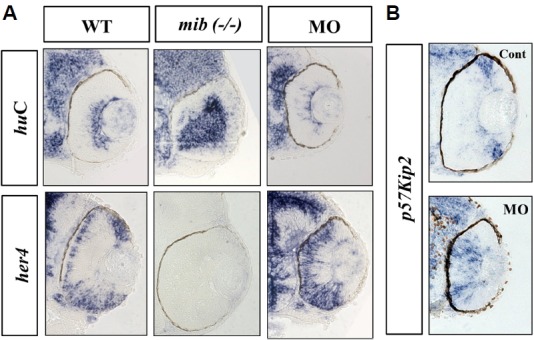
Molecular characterization of retinal development in ranbp9 morphants.
(A) Whole-mount in situ hybridization analysis for the expression of neural markers; huC for neuronal differentiation marker and her4 for neural progenitor marker, respectively. The neurogenic mutant mind bomb (mib, −/−) was used for comparison. In the WT larvae at 48 hpf, huC was expressed in the apical cells of the retina, but her4 was expressed in the basal cells. In the mib mutant larvae, huC was seen stained in most of the retinal cells near the apico-basal axis, but her4-positive cells were not observed. In the ranbp9 morphant larvae, huC expression domain was significantly reduced, however, the her4 expression domain was expanded. (B) Altered-regulation of p57kip2, a molecular marker for cell cycle exit, in ranbp9 morphants. p57kip2 transcripts were observed in the peripheral marginal regions of wild-type retina, while its expression was expanded to the central region of retina in ranbp9 morphants.
Furthermore, the expression of a cell cycle modulator, p57kip2, was examined in ranbp9 knockdown morphants. Expression domains of p57kip2 were shifted from marginal regions to the center in the retina of morphants (Fig. 7B), which contrasts with control embryos where p57kip2 expression was confined to the peripheral and marginal region. This data suggests the possible role of RanBP9 in regulating cell cycle exit and thus delay in neuronal differentiation. It has been reported that several hours after the formation of initial neurons in zebrafish retina, p57kip2 are expressed in a domain surrounding the region of differentiated neurons in the central retina, and, as the differentiation proceeds, transcription is observed to progress from the central region to the peripheral retina (Shkumatava and Neumann, 2005). To further elucidate the failure of retinal cell differentiation in ranbp9 morphants, we also tested the expression of glutamine synthetase (GS), a Müller glia-specific marker. Interestingly, severe blockage of Müller glia cell differentiation was observed in morphants in contrast with control MO injected embryos (Fig. 8). This data clearly demonstrated that elimination of RanBP9 induced defects both in neuronal cells and glia cells during retinogenesis of zebrafish.
Fig. 8.
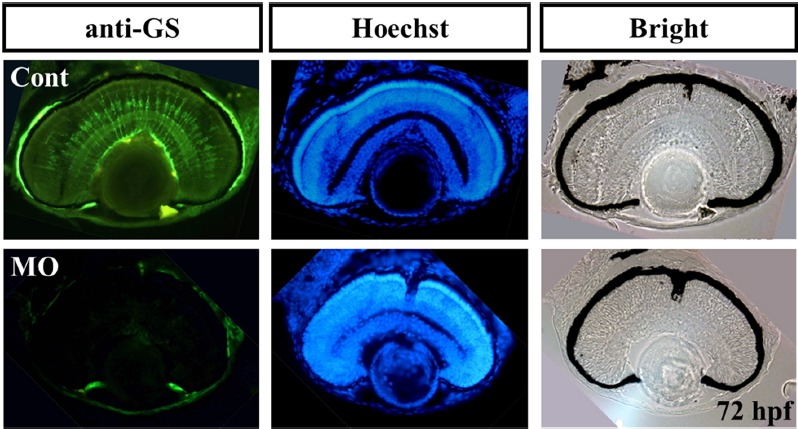
Characterization of glial cells in ranbp9 morphants.
Retinal sections were stained with an antibody against Glutamine synthetase, which is used as a molecular marker for the Müller glia cells. Glial cell differentiation is also affected in ranbp9 morphants (MO), compared to that of control retina. Hoechst staining and bright field image were shown for the comparison of retinal structures.
DISCUSSION
In this study, we report of the molecular interaction between RanBP9 and Mind bomb. Mind bomb is a key component of the Notch signaling pathway, which plays a critical role in neural development. RanBP9 is known to interact with multiple proteins and modulate various processes including synaptic plasticity, actin cytoskeleton remodeling and mitogen-activated protein kinase cascade (Haase et al., 2008). It has also been shown to simultaneously inhibit cell-adhesive processes and enhance amyloid β generation by accelerating APP, LRP, and β1-integrin endocytosis (Woo et al., 2012); and inhibit agonist-induced Mu opioid receptor internalization (Talbot et al., 2009). Reports have demonstrated through genetic epistasis experiments the interaction of RanBP9 with fragile-X mental retardation protein (Menon et al., 2004), as well as disrupted in schizophrenia 1 (DISC1) which is associated with autism, schizophrenia and bipolar disorder (Bae et al., 2015). However, previous study with neo-natal RanBP9 null mice showed suckling problems caused by defects in brain development and subsequent premature death (Palavicini et al., 2013).
The retinal phenotype of RanBP9 knock-down zebrafish was different from that of the Mib knock-out zebrafish. As RanBP9 interacts with multiple proteins, Mib also binds to several other proteins, such as Snx5, Fih-1, Usp1, Usp9, Trabid, and Moe (Matsuda et al., 2016; So et al., 2014; Tseng et al., 2014; Yoo et al., 2006). Notch signaling controls a broad spectrum of developmental processes in various different kinds of tissues. There are multiple homologues of Notch receptors and Notch ligands in zebrafish, which are differentially regulated in a temporal and spatial expression pattern. To our knowledge, no knock-out animal of these Notch signaling components have shown morphological phenotypes similar to that of the Mib knock-out; this being in spite of defects in Notch signaling in specific tissue types of the KO animal. We also identified another Mib member, Mib2, which showed normal development and was completely viable in its homozygous mutant (Koo et al., 2007). Mib-like E3 ubiquitin ligases, Neuralized and Neuralized-like 1, are additionally involved in the Notch signaling (Koutelou et al., 2008). There was also no detectable phenotype in the double knock-out of Neuralized1 and Neuralized2 animals (Koo et al., 2007). Thus we can speculate all these multiple components may have different combination of complementary functions in different cell types and tissues.
The loss-of-function phenotype of ranbp9 in zebrafish resulted in disorientation of the cellular laminae in the retina. The retina in ranbp9 knockdown morphants showed collapsed phenotype both in layer formation and cellular differentiation, suggesting that RanBP9 is actively involved in a pathway which regulates cell cycle control and differentiation. In mind bomb mutant zebrafish embryos, her4-expressing retinal progenitor cells were found to be reduced in the retina instead of the expected overproduction of huC-expressing differentiated neuronal cells (Fig. 7A). This finding indicates an essential role of Mind bomb in the maintenance of progenitor cell identity in the early steps of retinal neurogenesis. In contrast, ranbp9 morphants showed an excess of her4-positive cells with a reduction in huC-positive cells. A possible mechanism to support this phenomenon is the failure of proper cellular division during the appropriate time points in retinal development that result in aberrant cell division and proliferation (Bernardos et al., 2005). The loss of aberrant division in response to RanBP9 disruption suggests the necessity of ranbp9 for proper cell division in retinal neurogenesis. To examine this aspect further, we used the cell cycle regulator marker, p57kip2 (Fig. 7B) which is a Kip family member regulating cell cycle exit in progenitor cells, especially in the mouse retina (Dyer and Cepko, 2000). Thus, these results suggest an antagonistic role of RanBP9 against Mib in the Notch signaling pathway during neural development.
Proliferation of retinoblast cells to glial cell differentiation is a complex process comprising various events such as cell cycle exit (Dyer and Cepko, 2001). Cyclin kinase inhibitors (CKIs) have been shown to initiate cell cycle exit in various contexts. In the present study, inhibition of huC-expressing cells was consistent with the expansion of her4-expressing cells. We found that p57kip2 expression is up-regulated in the retina, especially in the central region (Fig. 7B). Recently, it has been shown that microRNA-216a is expressed in the retina during early development and regulates snx5 to precisely regulate Notch signaling (Olena et al, 2015). We previously identified SNX5 as a Mind bomb binding partner and demonstrated the localization of SNX5 to a distinct domain of the early endosome where it could potentially participate in multiple roles in cellular trafficking (Yoo et al., 2006). Another Mind bomb-binding protein, Moe/Epb41l5, is reported to be involved in delamination and differentiation of neural progenitor cells in the zebrafish hindbrain (Matsuda et al., 2016). Thus, an investigation into the interrelationship of these molecules (Mib, Snx5, Moe, and RanBP9) along with the Notch signaling pathway during neural development is needed in further studies.
In conclusion, our data suggest that the novel Mib-interacting protein, RanBP9, is critically required for neural development during embryogenesis of zebrafish. Based on the knockdown experiments and expression analysis, it can additionally be inferred that RanBP9 plays a significant role in the generation of various retinal cell types. RanBP9 may be particularly important for the modulation of Notch target gene expression and the proliferation and differentiation of neural cells by delaying cell cycle exit. In future experiments, it would be of interest to study these novel roles further in ranbp9 knockout zebrafish embryos.
ACKNOWLEDGEMENTS
This work was supported by the National R&D Program for Cancer Control (1020090) and National Research Foundation of Korea (NRF) grants funded by the Korean government Ministry of Science, ICT and Future Planning (MSIP) (2014R1A2A1A11053562, 2015M3A9A8029261).
REFERENCES
- Avanesov A., Malicki J. Approaches to study neurogenesis in the zebrafish retina. Methods Cell Biol. 2004;76:333–384. doi: 10.1016/s0091-679x(04)76016-1. [DOI] [PubMed] [Google Scholar]
- Bae J.S., Kim J.Y., Park B., Cheong H.S., Kim J., Namgoong S., Kim J., Park C.S., Kim B., Lee C.S., et al. Investigating the potential genetic association between RANBP9 polymorphisms and the risk of schizophrenia. Mol Med Rep. 2015;11:2975–2980. doi: 10.3892/mmr.2014.3045. [DOI] [PubMed] [Google Scholar]
- Bernardos R.L., Lentz S.I., Wolfe M.S., Raymond P.A. Notch-Delta signaling is required for spatial patterning and Müller glia differentiation in the zebrafish retina. Dev Biol. 2005;278:381–395. doi: 10.1016/j.ydbio.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Dyer M.A., Cepko C.L. p57(Kip2) regulates progenitor cell proliferation and amacrine interneuron development in the mouse retina. Development. 2000;127:3593–3605. doi: 10.1242/dev.127.16.3593. [DOI] [PubMed] [Google Scholar]
- Dyer M.A., Cepko C.L. Regulating proliferation during retinal development. Nat Rev Neurosci. 2001;2:333–342. doi: 10.1038/35072555. [DOI] [PubMed] [Google Scholar]
- Gwak J.W., Kong H.J., Bae Y.K., Kim M.J., Lee J., Park J.H., Yeo S.Y. Proliferating neural progenitors in the developing CNS of zebrafish require Jagged2 and Jagged1b. Mol Cells. 2010;30:155–159. doi: 10.1007/s10059-010-0101-4. [DOI] [PubMed] [Google Scholar]
- Haase A., Nordmann C., Sedehizade F., Borrmann C., Reiser G. RanBPM, a novel interaction partner of the brain-specific protein p42(IP4)/centaurin alpha-1. J Neurochem. 2008;105:2237–2248. doi: 10.1111/j.1471-4159.2008.05308.x. [DOI] [PubMed] [Google Scholar]
- Itoh M., Kim C.H., Palardy G., Oda T., Jiang Y.J., Maust D., Yeo S.Y., Lorick K., Wright G.J., Ariza-McNaughton L., et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of notch signaling by delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jung S.H., Kim H.S., Ryu J.H., Gwak J.W., Bae Y.K., Kim C.H., Yeo S.Y. Her4-positive population in the tectum opticum is proliferating neural precursors in the adult zebrafish brain. Mol Cells. 2012;33:627–632. doi: 10.1007/s10059-012-0091-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.H., Ueshima E., Muraoka O., Tanaka H., Yeo S.Y., Huh T.L., Miki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett. 1996;216:109–112. doi: 10.1016/0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- Koo B.K., Lim H.S., Song R., Yoon M.J., Moon J.S., Kim Y.W., Kwon M.C., Yoo K.W., Kong M.P., Lee J., et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- Koo B.K., Yoon M.J., Yoon K.J., Im S.K., Kim Y.Y., Kim C.H., Suh P.G., Jan Y.N., Kong Y.Y. An obligatory role of mind bomb-1 in notch signaling of mammalian development. PLoS One. 2007;2:e1221. doi: 10.1371/journal.pone.0001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E., Sato S., Tomomori-Sato C., Florens L., Swanson S.K., et al. Neuralized-like 1 (Neurl1) targeted to the plasma membrane by N-myristoylation regulates the Notch ligand Jagged1. J Biol Chem. 2008;283:3846–3853. doi: 10.1074/jbc.M706974200. [DOI] [PubMed] [Google Scholar]
- Lawson N.D., Scheer N., Pham V.N., Kim C.H., Chitnis A.B., Campos-Ortega J.A., Weinstein B.M. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- Le Borgne R., Bardin A., Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132:1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Rand K., Palardy G., Shimizu N., Ikeda H., Dalle Nogare D., Itoh M., Chitnis A.B. Epb41l5 competes with Delta as a substrate for Mib1 to coordinate specification and differentiation of neurons. Development. 2016;143:3085–3096. doi: 10.1242/dev.138743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon R.P., Gibson T.J., Pastore A. The C terminus of fragile X mental retardation protein interacts with the multi-domain Ran-binding protein in the microtubule-organising centre. J Mol Biol. 2004;343:43–53. doi: 10.1016/j.jmb.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Nishitani H., Hirose E., Uchimura Y., Nakamura M., Umeda M., Nishii K., Mori N., Nishimoto T. Full-sized RanBPM cDNA encodes a protein possessing a long stretch of proline and glutamine within the N-terminal region, comprising a large protein complex. Gene. 2001;272:25–33. doi: 10.1016/s0378-1119(01)00553-4. [DOI] [PubMed] [Google Scholar]
- Olena A.F., Rao M.B., Thatcher E.J., Wu S.Y., Patton J.G. miR-216a regulates snx5, a novel notch signaling pathway component, during zebrafish retinal development. Dev Biol. 2015;400:72–81. doi: 10.1016/j.ydbio.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palavicini J.P., Lloyd B.N., Hayes C.D., Bianchi E., Kang D.E., Dawson-Scully K., Lakshmana M.K. RanBP9 plays a critical role in neonatal brain development in mice. PLoS One. 2013;8:e66908. doi: 10.1371/journal.pone.0066908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe D., Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- Shkumatava A., Neumann C.J. Shh directs cell-cycle exit by activating p57Kip2 in the zebrafish retina. EMBO Rep. 2005;6:563–569. doi: 10.1038/sj.embor.7400416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So J.H., Kim J.D., Yoo K.W., Kim H.T., Jung S.H., Choi J.H., Lee M.S., Jin S.W., Kim C.H. FIH-1, a novel interactor of mindbomb, functions as an essential anti-angiogenic factor during zebrafish vascular development. PLoS One. 2014;9:e109517. doi: 10.1371/journal.pone.0109517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbot J.N., Skifter D.A., Bianchi E., Monaghan D.T., Toews M.L., Murrin L.C. Regulation of mu opioid receptor internalization by the scaffold protein RanBPM. Neurosci Lett. 2009;466:154–158. doi: 10.1016/j.neulet.2009.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng L.C., Zhang C., Cheng C.M., Xu H., Hsu C.H., Jiang Y.J. New classes of mind bomb-interacting proteins identified from yeast two-hybrid screens. PLoS One. 2014;9:e93394. doi: 10.1371/journal.pone.0093394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J.A., Jung A.R., Lakshmana M.K., Bedrossian A., Lim Y., Bu J.H., Park S.A., Koo E.H., Mook-Jung I., Kang D.E. Pivotal role of the RanBP9-cofilin pathway in A beta-induced apoptosis and neurodegeneration. Cell Death Differ. 2012;19:1413–1423. doi: 10.1038/cdd.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Tonou-Fujimori N., Komori A., Maeda R., Nojima Y., Li H., Okamoto H., Masai I. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development. 2005;132:3027–3043. doi: 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- Yoo K.W., Kim E.H., Jung S.H., Rhee M., Koo B.K., Yoon K.J., Kong Y.Y., Kim C.H. Snx5, as a Mind bomb-binding protein, is expressed in hematopoietic and endothelial precursor cells in zebrafish. FEBS Lett. 2006;580:4409–4416. doi: 10.1016/j.febslet.2006.07.009. [DOI] [PubMed] [Google Scholar]


