Abstract
The transcriptional activator AphB has been implicated in acid resistance and pathogenesis in the food borne pathogens Vibrio vulnificus and Vibrio cholerae. To date, the full-length AphB crystal structure of V. cholerae has been determined and characterized by a tetrameric assembly of AphB consisting of a DNA binding domain and a regulatory domain (RD). Although acidic pH and low oxygen tension might be involved in the activation of AphB, it remains unknown which ligand or stimulus activates AphB at the molecular level. In this study, we determine the crystal structure of the AphB RD from V. vulnificus under aerobic conditions without modification at the conserved cysteine residue of the RD, even in the presence of the oxidizing agent cumene hydroperoxide. A cysteine to serine amino acid residue mutant RD protein further confirmed that the cysteine residue is not involved in sensing oxidative stress in vitro. Interestingly, an unidentified small molecule was observed in the inter-subdomain cavity in the RD when the crystal was incubated with cumene hydroperoxide molecules, suggesting a new ligand-binding site. In addition, we confirmed the role of AphB in acid tolerance by observing an aphB-dependent increase in cadC transcript level when V. vulnificus was exposed to acidic pH. Our study contributes to the understanding of the AphB molecular mechanism in the process of recognizing the host environment.
Keywords: crystal structure, low pH, transcriptional regulator AphB, vibrio vulnificus
INTRODUCTION
Many pathogenic bacteria increase the expression of virulence factors by recognizing and responding to the host environment. Vibrio vulnificus is a facultative aerobic gram-negative species that lives in marine environments and in the human body (Horseman and Surani, 2011; Lee et al., 2014). Oral ingestion of food contaminated with V. vulnificus or direct administration of the bacteria to injured skin can cause acute gastroenteritis or invasive septicemia, respectively. Once in the human host, the bacteria can rapidly expand by sensing the human environment. V. vulnificus produces toxins during the pathogenic response, whose gene expression is governed by global regulators that recognize the host environments (Lee et al., 2014).
Vibrio cholerae, closely related to V. vulnificus, activates the ToxR virulence cascade, which is initiated by two transcriptional regulators: the winged-helix AphA and the LysR family transcriptional regulator (LTTR) AphB. The active form of AphB binds to the tcpPH promoter and induces its transcription by cooperating with AphA. TcpPH functions cooperatively with ToxR in the virulence signaling pathway by expression of ToxT, a major regulator of virulence factor transcription in V. cholerae (Krukonis et al., 2000).
LTTRs comprise one of the largest families of transcriptional regulators in prokaryotes that are involved in diverse biological processes. They are composed of an N-terminal DNA binding domain (DBD) and a C-terminal regulatory domain (RD). Crystal structures of full-length AphB from V. cholerae has been determined (Taylor et al., 2012) and comprise a homotetrameric structure that can be described as a dimer of dimers that assembles via two distinct dimerization interfaces, similar to other LTTR proteins.
In V. cholerae, AphB was characterized as responsive to intracellular pH and a lack of oxygen (Taylor et al., 2012). AphB takes on an active conformation at low pH or under low oxygen tension, but it remains inactive at high pH (or pH 8.5). Based on AphB crystal structure, the putative ligand binding pocket region was identified, suggesting that the binding of a ligand triggers conformational changes that alter DNA binding (Taylor et al., 2012). An alternative mechanism for AphB was proposed as a thiol-based switch protein, where the thiol is oxidized depending on the presence of oxygen (Liu et al., 2011).
AphB of V. vulnificus is not involved with tcpPH expression because this species lacks tcpPH genes, indicating a different role for AphB in V. vulnificus compared to AphB from V. cholerae (Jeong and Choi, 2008; Rhee et al., 2006). AphB is known to directly induce cadC at the transcriptional level in V. cholerae and V. vulnificus, which facilitates pathogen survival under acid stress (Kovacikova and Skorupski, 1999; Rhee et al., 2006). Microarray analysis revealed that AphB affected the expression of virulence-involved genes in V. vulnificus ATCC 29307, indicating that AphB might be a global regulator contributing to the pathogenesis of V. vulnificus (Jeong and Choi, 2008). AphA of V. vulnificus does not seem to cooperate with AphB; instead, it is known to upregulate gene expression of the Fe-S cluster regulator iscR (Lim et al., 2014). In this study, we determined the crystal structure of the RD of AphB from V. vulnificus, followed by functional analyses.
MATERIALS AND METHODS
Construction of the expression vector and protein purification
DNA encoding the AphB RD (residues 88-291) was amplified by PCR using the V. vulnificus MO6-24/O genome (accession number CP002469) as a template and was inserted into the expression vector pProEx-HTa (Invitrogen) using NcoI/XbaI restriction enzyme sites. The resulting plasmid (pProEx-HTa-VvAphB-RD) was used to transform Escherichia coli strain C43 (DE3) (Miroux and Walker, 1996) for protein production. The E. coli strain was cultured in 2.0 L of LB medium including appropriate antibiotics until an OD600 of 0.6 and protein production was induced with 0.5 mM IPTG at 30°C. Cells were harvested 5 h after induction, and the cell pellet was resuspended with 50 ml lysis buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 2 mM 2-mercaptoethanol. After homogenization by French press, the cell lysate was acquired by centrifugation at 13,000 rpm for 30 min. The protein was subsequently purified using a Ni-NTA column and anion-exchange chromatography (HiTrap Q, GE Healthcare, USA). When necessary, the purified protein was incubated with several oxidants at this step. Then, the protein was further purified by size exclusion chromatography (HiLoad Superdex 200 26/600; GE Healthcare) pre-equilibrated with lysis buffer. The final purified proteins were concentrated to 16 mg/ml using a centrifugal filter concentration device (Millipore, USA; 10 kDa cutoff) and stored frozen at −80°C until use.
Site-directed mutagenesis
The cysteine codon at position 227 (C227) was changed to a serine codon (C227S) using the overlapping PCR method with Pfu polymerase based on pProEx-HTa-VvAphB-RD (Patel et al., 1993).
Crystallization, data collection, and structural determination
Crystallization of the wild type AphB RD and RD treated with the oxidant cumene hydroperoxide (CHP) was performed using the vapor-diffusion hanging drop method at 14°C under a mother liquor containing 0.1 M HEPES (pH 7.5), 15% (wt/vol) PEG 8K, and 10% (vol/vol) ethylene glycol. The crystals were flash-frozen using 25% (vol/vol) glycerol as a cryoprotectant in a nitrogen stream at −173°C prior to collecting the X-ray diffraction dataset with the Pohang Accelerator Laboratory beamline 5C (Park et al., 2017) and were processed with the HKL2000 package (Otwinowski and Minor, 1997). Both RD crystals belonged to the spacegroup P212121, with unit cell dimensions of a = 125.0 Å, b = 188.0 Å, and c = 57.4 Å for the wild type RD and a = 124.8 Å, b = 189.4 Å and c = 57.4 Å for the CHP-treated RD (Table 1). The structure was determined using the MOLREP program in the CCP4 package by the molecular replacement method and a search model taken from the full-length AphB from V. cholerae (PDB code: 3SZP) (Winn et al., 2011). The final structure of wild type AphB RD was refined at a 1.9 Å resolution with an R factor of 21.9% and an Rfree of 26.3% using the PHENIX program (Adams et al., 2010) and the CHP-treated RD at a 2.4 Å resolution. Further details on the structure determination and refinement are given in Table 1.
Table 1.
Statistics for data collection and refinement
| Native | CHP-incubated | C227S | |
|---|---|---|---|
| Data collection | |||
| Beam line | PAL 5C | PAL 5C | PAL 5C |
| Wavelength | 0.97960 | 0.97960 | 0.97930 |
| Space group | P212121 | P212121 | C2 |
| Cell dimensions | |||
| a,b,c (Å) | 125.0, 188.0, 57.4 | 124.8, 189.4, 57.4 | 230.3,72.4, 112.2 |
| α,β,γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.0–1.90 (1.93–1.90) | 50.0–2.40 (2.44–2.40) | 50.0–3.0 (3.05–3.00) |
| Rmerge | 0.111 (0.639) | 0.122 (0.433) | 0.169 (0.445) |
| I/σI | 14.1 (3.7) | 13.4 (3.0) | 7.2 (2.1) |
| Completeness (%) | 98.1 (85.4) | 99.7 (99.4) | 97.2 (90.6) |
| Redundancy | 8.7 (6.2) | 8.8 (5.7) | 4.2 (2.6) |
| Refinement | |||
| Resolution (Å) | 42.26–1.90 | 45.70–2.40 | 38.93 – 3.00 |
| No. reflections | 90980 | 51444 | 28983 |
| Rwork/Rfree | 0.219/0.263 | 0.21/0.26 | 0.24/0.29 |
| No. of total atoms | 9655 | 9382 | 9186 |
| Wilson B-factor (Å) | 19.40 | 30.79 | 47.55 |
| R.M.S deviations | |||
| Bond lengths (Å) | 0.005 | 0.003 | 0.003 |
| Bond angles (°) | 1.11 | 0.56 | 0.60 |
| Ramachandran plot | |||
| Favored (%) | 98.3 | 97.1 | 94.9 |
| Allowed (%) | 1.8 | 2.9 | 5.1 |
| Outliers (%) | 0.00 | 0.00 | 0.00 |
| PDB ID | 5FHK | 5X0O | 5X0N |
Values in parentheses are for the highest resolution shell.
Values in parentheses are for the highest-resolution shell.
R merge = ∑ hkl ∑ i |I i (hkl) – [I(hkl)]|/∑ hkl ∑ i I i (hkl), where I i (hkl) is the intensity of the ith observation of reflection hkl and [I(hkl)] is the average intensity of the i observations.
R free calculated for a random set of 10% of reflections not used in the refinement
Crystallization of the AphB C227S mutant RD was performed using the same method as described above with a mother liquor containing 0.35 M potassium thiocyanate (pH 7.0) and 17% (wt/vol) PEG 3350, and the dataset was collected using Paratone-N as a cryoprotectant. The crystal belongs to the spacegroup C2, with unit cell dimensions of a = 230.3 Å, b = 72.4 Å, and c = 112.2 Å. The final structure was refined at a 3.0 Å resolution, and further details on the structure determination and refinement are given in Table 1.
Construction of aphB mutant strain
pJR0325, which was constructed previously to carry a mutant allele of V. vulnificus aphB on pDM4 (Table 2) (Rhee et al., 2006), was used to generate the aphB mutant of V. vulnificus. The E. coli S17-1 λpir, tra strain (Simon et al., 1983) containing pJR0325 was used as a conjugal donor in conjugation with the V. vulnificus MO6-24/O as a recipient. The resulting aphB mutant was named KK1419 (Table 2). The conjugation and isolation of the transconjugants were conducted using the method described previously (Jang et al., 2016).
Table 2.
Plasmids and bacterial strains used in this study
| Strain or plasmid | Relevant characteristics a | Reference or source |
|---|---|---|
| Bacterial strains | ||
| V. vulnificus | ||
| MO6-24/O | Clinical isolate; virulent | Wright et al., 1990 |
| KK1419 | MO6-24/O with ΔaphB | This study |
| E. coli | ||
| S17-1λpir | λ-pir lysogen; thi pro hsdR hsdM+ recA RP4-2 Tc::Mu-Km::Tn7;Tpr Smr; host for π-requiring plasmids; conjugal donor | Simon et al., 1983 |
| C43 (DE3) | F− ompT hsdSB (rB− mB−) gal dcm (DE3) with uncharacterized mutations | Miroux and Walker, 1996 |
| Plasmids | ||
| pProEx-HTa-VvAphB-RD | His6–tag fusion protein expression vector; Apr | Invitrogen |
| pDM4 | R6K γ ori sacB; suicide vector; oriT of RP4; Cmr | Milton et al., 1996 |
| pJR0325 | pDM4 with ΔaphB; Cmr | Rhee et al., 2006 |
Tpr, trimethoprim resistance; Smr, streptomycin resistance; Apr, ampicillin resistance; Cmr, chloramphenicol resistance
Growth kinetics under acid stress
The wild type and aphB mutant strains were grown with LB medium supplemented with 2% (wt/vol) NaCl (LBS, pH 6.8) or LBS buffered at pH 5.2 with HCl (DUKSAN, Korea) in 24-well culture plates (SPL, Korea). The cultures were further incubated at 30°C with shaking for 10 h, and their growth was monitored at OD600 with a spectrophotometer (Tecan Infinite M200 reader, Männedorf, Switzerland).
RNA purification and transcript analysis
The wild type and the aphB mutant grown to an OD600 of 0.5 were exposed to LBS or LBS adjusted to pH 5.2, 6.0 with HCl (DUKSAN) and 7.5 with NaOH (Sigma, USA) for 30 min and harvested to isolate total RNA using the RNeasy® mini kit (Qiagen, USA). For quantitative real-time PCR (qRT-PCR), the concentration of total RNA from the strains was measured using a NanoVue Plus spectrophotometer (GE Healthcare). cDNA was synthesized from 1 μg of total RNA using the iScript™ cDNA synthesis kit (Bio-Rad, USA), and real-time PCR amplification of the cDNA was performed using the Chromo 4 real time PCR detection system (Bio-Rad) with a pair of specific primers (Supplementary Table S1), as described previously (Park et al., 2015). Relative cadC and aphB mRNA expression levels in the same amount of total RNA were calculated using the 16S rRNA expression level as the internal reference for normalization.
RESULTS
Structural determination of the wild-type AphB regulatory domain from V. vulnificus
We initially attempted to obtain the full-length AphB from V. vulnificus (VvAphB), but the expression level was not suitable for the ensuing structural study. The RD region (residues 88-291) of the wild type AphB from V. vulnificus was successfully produced in the E. coli expression system. Rod-shaped crystals were grown at pH 7.5, and a native dataset was collected at a 1.9 Å resolution. The crystals belong to the spacegroup P212121, and the structure was determined using a molecular replacement model taken from the full-length AphB structure of V. cholerae, as previously reported (Taylor et al., 2012). The asymmetric unit of the crystal contains three dimers, resulting in a Matthew’s coefficient of 2.11 Å3/Da (Kantardjieff and Rupp, 2003). Similar to the RD structure of AphB from V. cholerae (VcAphB), the VvAphB RD forms a stable dimer in the crystal structure (Fig. 1A). Interestingly, we observed that the VvAphB RD had conformational variants among the three dimers in the asymmetric units when superposed (Fig. 1B), with one dimer relatively distinguished from the other two.
Fig. 1.
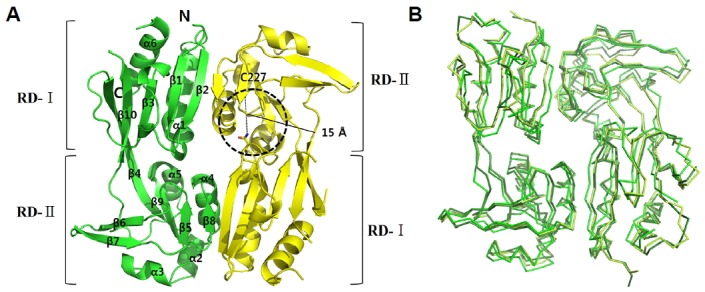
Overall structure of wild type VvAphB RD.
(A) The structure of the wild type VvAphB RD. The two protomers are colored green and yellow. The secondary structures are displayed in the ribbon representations, and the two subdomains (RD-I and RD-II) are labeled. C227 residues are in the RD-II in the second protomer (yellow). A broken-line circle indicates the putative ligand-binding site between the two subdomains. The distance indicates C227 and N100 in the ligand-binding site. (B) Superposition of three dimers from the wild type VvAphB in the asymmetric unit. Each protomer is differently colored.
Like other LTTR RD structures, the VvAphB RD consists of two subdomains named RD-I and RD-II. The two subdomains are connected through an extended β strand, similar to the VcAphB structure. The N-terminus of RD-I is connected to the DNA binding domain. VvAphB RD monomers are composed of 10 β-strands and 6 α-helices connected by an α/β fold (Fig. 1A).
Structural comparison to AphB from V. cholerae
VvAphB and VcAphB share high sequence homology (80% amino acid sequence identity). We found that a VvAphB RD dimer showed a somewhat different conformation from the wild type VcAphB RD (rmsd 0.908 Å between 347 atoms; Fig. 2A), while the other VvAphB RD dimers were very similar to VcAphB (rmsd 0.270 Å between 380 atoms; Fig. 2B).
Fig. 2.
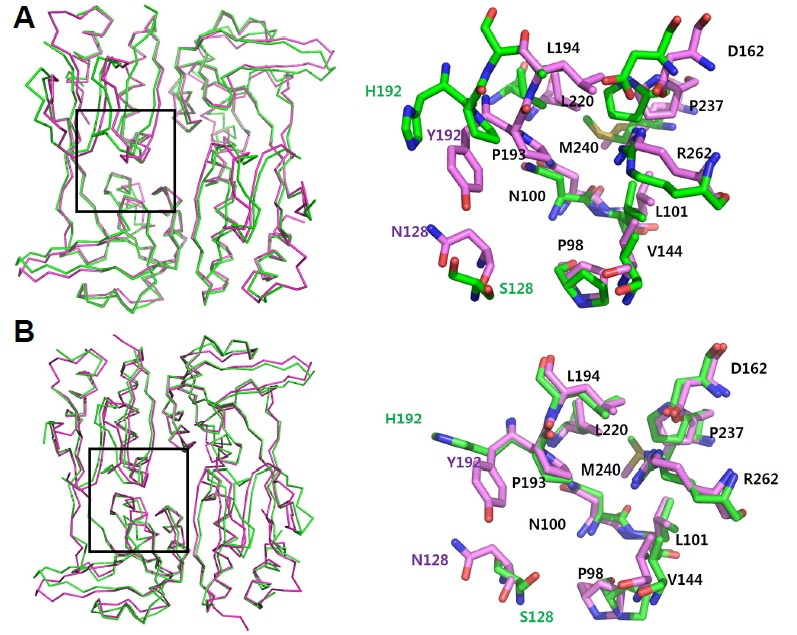
Structural superposition of wild type VvAphB RD with VcAphB RD.
(A) VvAphB RD dimer (green) is superposed onto the wild type VcAphB RD (magenta). (B) The other VvAphB RD dimer (green) is superposed onto the wild type VcAphB RD (magenta). The boxed region in the left panel is enlarged in the right panel that is drawn with stick representations. Each residue is labeled (the common residues in black, with the other residues following the same color scheme) in the right panel.
Importantly, a small cavity was found at the interface between RD-I and RD-II, which was also observed in the VcAphB structure (Taylor et al., 2012). In VvAphB, the small cavity was lined with 12 residues, P98, N100, L101, S128, V144, D162, H192, P193, L220, P237, M240, and R262. In the VcAphB, the corresponding cavity was proposed as the putative ligand-binding site despite the fact that the specific co-inducing ligand is not yet known (Taylor et al., 2012). When comparing the residues lining the cavity with VcAphB, the S128 and H192 from the VvAphB RD were substituted in VcAphB with N128 and Y192, respectively (Fig. 2).
C227 in RD-II
We noted the local chemical environment around C227 in RD-II, which is the only cysteine residue in the VvAphB RD (Fig. 1). The cysteine residue is ~15 Å from the small inter-subdomain cavity (Fig. 1). Liu et al. proposed that the cysteine residue senses oxygen level by changing the oxidation state of the thiol moiety (Liu et al., 2011). The role of this cysteine residue is under debate because cysteine residue mutants did not show consistent results (Liu et al., 2011; 2016; Taylor et al., 2012).
To assess the structural impact of the C227 sulfur atom in VvAphB, we determined the crystal structure of a VvAphB C227S mutant variant that was not subjected to oxidation. The crystal belongs to spacegroup C2, which is different from the wild type structure, and three dimers are present in the asymmetric unit. The crystal structure was solved at a 3.0 Å resolution by the molecular replacement method using the wild type VvAphB structure. When the three dimers of the mutant protein were superposed, no substantial differences were observed (rmsd 0.496 Å between 328 atoms, rmsd 0.473 Å between 337 atoms (Fig. 3A)). By structural comparison with the wild type VvAphB structure, we found that the two structures were very similar, with a slight difference of the subdomains in the dimeric assembly (Fig. 3B) at the region near the cysteine residue, indicating that the cysteine residue or the sulfur atom might not be important for the function of AphB (Fig. 3B, inset).
Fig. 3.
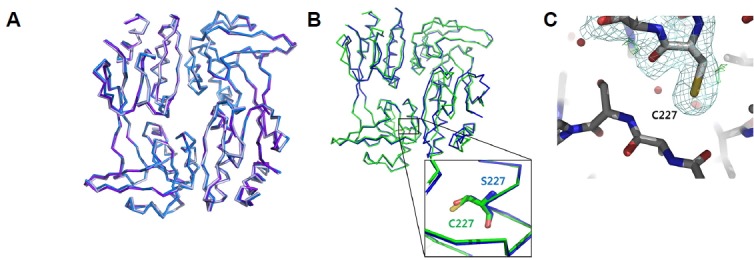
Structural analysis of the C227 residue in VvAphB RD.
Superposition of three dimers from the VvAphB C227S variant in the asymmetric unit. (B) Structural superposition of the wild type VvAphB (green) and the C227S variant (blue) in C-alpha tracing representations. The magnified box shows C227 (or C227S in the variant structure). (C) Electron density maps around C227 in the CHP-treated structure. No modification at C227 was observed. 2FoFc (pale blue mesh) and FoFc (green mesh) maps were contoured at 1.0σ and 3.0σ, respectively.
A new cavity on the back side of AphB RD
In order to determine reactivity of the cysteine residue, we incubated the crystals with various peroxides such as hydrogen peroxide and alkyl hydroperoxides and determined the structures. No oxidation modification at the cysteine residue was found in the structures under any of the tested conditions (Fig. 3C). In the structure of VvAphB RD treated with cumene hydroperoxide (CHP), we unexpectedly found an unidentified residual electron density map in a different small cavity located at the interface between two monomers on the opposite side of the dimer to the previously known ligand binding site (Fig. 4A). Unfortunately, we failed to identify the compound of the residual density maps since the density maps are rather heterogeneous in the different CHP-treated VvAphB RDs in the asymmetric unit. K103, R104, N221, R224, M240, and Y244 seem to participate in ligand binding (Fig. 4B). suggesting that the cavity might be functionally related. However, further study is required to elucidate the role of the new cavity.
Fig. 4.
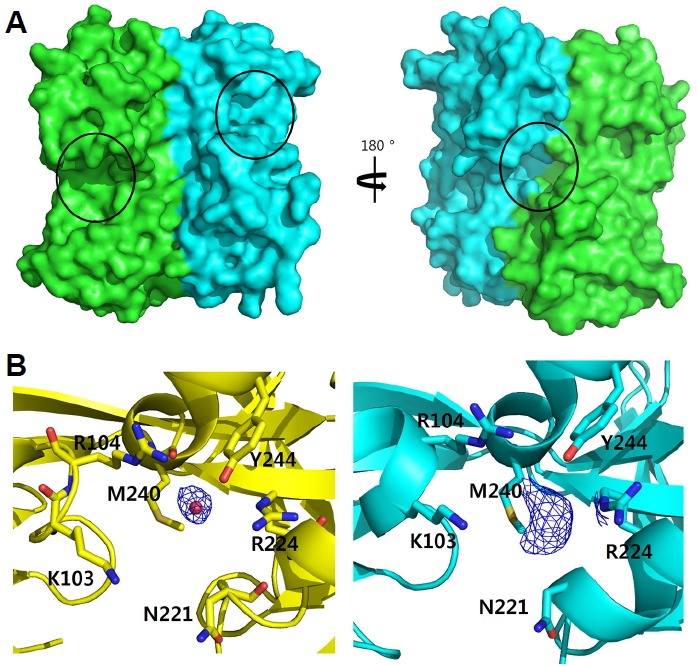
New ligand binding cavity in the dimeric interface of AphB.
(A) The front and back sides of the surface representations of the VvAphB RD dimer. The putative ligand binding sites previously characterized at the interface between RD-I and RD-II are shown in the same orientation of Fig. 1A (left). The newly found cavity is at the interface of the two protomers on the back side of the dimer (right). Each protomer is colored differently (green and cyan), and the putative ligand binding sites or cavity are indicated by black circles. (B) Electron density maps around the newly found cavity between the protomers in the back side of the dimer. While the non-treated structure (yellow) contains a water molecule (red ball, left), the CHP-treated structure (cyan) contains an unidentified molecule displayed by the density map (right). Each of FoFc electron density maps (blue mesh) were contoured at 1.0σ. Residues in the cavity are displayed in the stick representations.
AphB from V. vulnificus is activated at low pH and is important for acid tolerance in vivo
Both VcAphB and VvAphB contribute to bacterial acid resistance because the known target gene of AphB regulators is cadC, which encodes the master positive regulator of the acid tolerance genes cadA and cadB encoding lysine decarboxylase and cadaverine antiporter, respectively (Merrell and Camilli, 2000; Rhee et al., 2006). In this study, we compared growth kinetics of the wild type V. vulnificus MO6-24/O strain and an aphB-deleted strain at acidic and neutral pH (pH 5.2 and pH 6.8, respectively). As shown in Fig. 5A, the aphB-deleted strain showed a delayed lag phase at acidic pH compared to the wild type strain. At neutral pH, deletion of the aphB gene did not affect the growth kinetics. These results are consistent with previous results using V. vulnificus ATCC 29307 (Rhee et al., 2006).
Fig. 5.
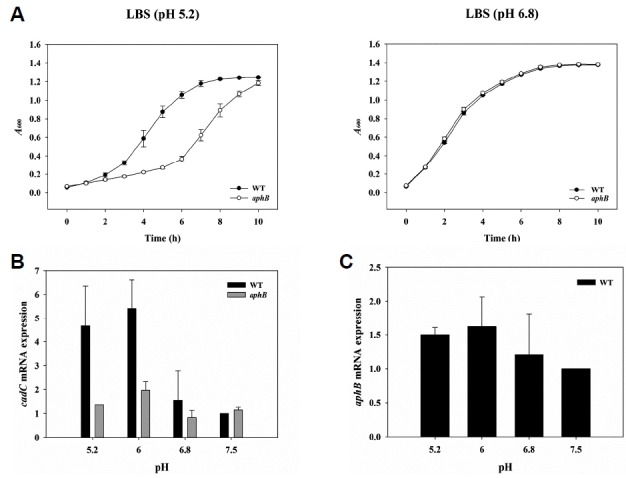
Effect of AphB on V. vulnificus growth and expression of cadC and aphB under acid stress.
(A) V. vulnificus strains were compared for their ability to grow in LBS (right) or LBS adjusted to pH 5.2 (left). The result is representative of at least three independent experiments, and the standard deviations (SD) are displayed as the error bars. (B, C) Total RNAs were isolated from V. vulnificus cultures grown in LBS or LBS adjusted to pH 5.2, 6.0, or 7.5. The cadC and aphB mRNA levels were determined by qRT-PCR analyses, and the cadC (B) and aphB (C) mRNA levels in the wild type grown in LBS adjusted to pH 7.5 were set as 1. Three independent experiments were carried out, and the SDs are displayed as error bars. WT, wild type; aphB, aphB mutant.
We next carried out qRT-PCR to examine pH-dependent AphB transcriptional activity. When we measured cadC transcript level in bacteria exposed to pH shock for 30 min, the wild type strain exhibited much higher levels at pH 5.2 and 6.0 compared to pH 7.5 and 6.8 (Fig. 5B). In contrast, the aphB-deleted strain was not influenced by external pH; however, the level of aphB mRNA was not significantly affected by low pH shock (Fig. 5C). These results suggest that AphB might sense acid shock for V. vulnificus and induces the expression of cadC and further suggest that the transcriptional activity of AphB protein is changed; however, we note that our findings do not mean that VvAphB directly recognizes acid pH because the external pH variation did not significantly change cytosolic pH, and we cannot exclude the possibility that an unknown pH-sensitive element might affect the activity of AphB.
DISCUSSION
In this study, we determined the crystal structures of VvAphB RD under crystallization conditions at pH 7.5. The structure revealed two different conformations similar to those of the wild-type and the constitutive active N100E variant VcAphB, indicating an intrinsic structural flexibility of VvAphB between two states. The putative ligand-binding site in the RD-I and RD-II interface of the front side of the dimer, previously proposed in VcAphB structure (Taylor et al., 2012), was also well defined. Even though it is not clear whether VvAphB operates with the same mechanism as VcAphB, structural resemblance between the two proteins strongly suggests that the same mechanism for sensing those stressful environments are shared.
Since AphB senses anoxic environments, the cysteine residue (C227) was investigated based on the C227S mutant structure and the reaction by various peroxide molecules. The mutation of C227S did not give substantial structural alteration to AphB. Furthermore, incubation of peroxides did not result in any oxidation on the cysteine residue. Instead, the CHP-treated structure showed a new cavity on the opposite side of the RD dimer with an unidentified ligand molecule in it. Taken together, our findings suggest that the cysteine in AphB RD might not directly participate in sensing oxygen.
AphB was responsive to low pH as well as low oxygen tension in V. cholerae (Kovacikova et al., 2010; Merrell and Camilli, 2000). Another group performed a mutational analysis of the VcAphB RD by changing amino acid residues lining the cavity to residues with negative charge (P98D, N100E, L101E, P193D, L220E) and revealed that the mutations completely restored the ability of AphB to activate the tcpPH promoter at the non-permissive pH of 8.5 in V. cholerae (Taylor et al., 2012). The authors proposed that certain residues within the ligand-binding pocket of AphB might be involved in the activity of the protein in response to acid pH. Moreover, our study, together with a previous report (Rhee et al., 2006), strongly suggests that the transcriptional activity of AphB is increased at acidic pH in V. vulnificus.
Then, how are the anoxic conditions recognized by AphB in V. cholerae and V. vulnificus? We noted that the metabolism would be shifted to the anaerobic fermentation producing organic acids, leading to lowering pH in the cytosol when the bacteria are placed under the anoxic condition. Thus it is likely that the anoxic conditions may give the acidic stress to the bacteria. We hypothesized that the ligand for AphB would be a small compound that is accumulated under external acidic or an anoxic stress-induced acidic environment. We don’t support that AphB directly sense the cytosolic pH because the cytosolic pH of the bacteria would not substantially change due to the cellular buffering systems. Instead we believe that the ligand might bind to the ligand binding site(s) of AphB, resulting in the conformational change of AphB. We first paid attention to the CadBA system which antiports cadaverine and lysine across the cell membrane to counteract acidic pH in V. vulnificus (Rhee et al., 2005). Given the size of the putative ligand-binding site, an amino acid or its derivative might fit the AphB site. We chose lysine and cadaverine as possible ligands, which are components of the acid resistance system in bacteria. Unfortunately, we failed to determine if lysine and cadaverine bind to the purified VvAphB protein, as judged by the results using isothermal titration calorimetry (data not shown). Although the ligand for AphB was not identified in this study, we suggest molecules in the cellular buffering system as ligand candidates.
In this study, we presented the crystal structures of AphB RD of V. vulnificus with the conformational flexibility of AphB. Our findings might exclude the possible mechanism mediated by the cysteine residue in RD, and instead suggests the acidic pH might be more important in activation of AphB. Further studies are still needed to clarify the role and action mechanism of AphB, which help elucidate the virulence mechanism of bacteria at the molecular level.
Supplementary data
ACKNOWLEDGMENTS
This research was supported by the R&D Convergence Center Support Program (to SHC and NCH) funded by the Ministry for Food, Agriculture, Forestry, and Fisheries, Republic of Korea.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horseman M.A., Surani S. A comprehensive review of Vibrio vulnificus: an important cause of severe sepsis and skin and soft-tissue infection. Int J Infect Dis. 2011;15: e157–166. doi: 10.1016/j.ijid.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Jang K.K., Gil S.Y., Lim J.G., Choi S.H. Regulatory Characteristics of Vibrio vulnificus gbpA Gene Encoding a Mucin-binding Protein Essential for Pathogenesis. J Biol Chem. 2016;291:5774–5787. doi: 10.1074/jbc.M115.685321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H.G., Choi S.H. Evidence that AphB, essential for the virulence of Vibrio vulnificus, is a global regulator. J Bacteriol. 2008;190:3768–3773. doi: 10.1128/JB.00058-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantardjieff K.A., Rupp B. Matthews coefficient probabilities: Improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003;12:1865–1871. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova G., Lin W., Skorupski K. The LysR-type virulence activator AphB regulates the expression of genes in Vibrio cholerae in response to low pH and anaerobiosis. J Bacteriol. 2010;192:4181–4191. doi: 10.1128/JB.00193-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacikova G., Skorupski K. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol. 1999;181:4250–4256. doi: 10.1128/jb.181.14.4250-4256.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukonis E.S., Yu R.R., Dirita V.J. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol. 2000;38:67–84. doi: 10.1046/j.1365-2958.2000.02111.x. [DOI] [PubMed] [Google Scholar]
- Lee M.A., Kim J.A., Yang Y.J., Shin M.Y., Park S.J., Lee K.H. VvpM, an extracellular metalloprotease of Vibrio vulnificus, induces apoptotic death of human cells. J Microbiol. 2014;52:1036–1043. doi: 10.1007/s12275-014-4531-0. [DOI] [PubMed] [Google Scholar]
- Lim J.G., Park J.H., Choi S.H. Low cell density regulator AphA upregulates the expression of Vibrio vulnificus iscR gene encoding the Fe-S cluster regulator IscR. J Microbiol. 2014;52:413–421. doi: 10.1007/s12275-014-3592-4. [DOI] [PubMed] [Google Scholar]
- Liu Z., Yang M., Peterfreund G.L., Tsou A.M., Selamoglu N., Daldal F., Zhong Z., Kan B., Zhu J. Vibrio cholerae anaerobic induction of virulence gene expression is controlled by thiol-based switches of virulence regulator AphB. Proc Natl Acad Sci USA. 2011;108:810–815. doi: 10.1073/pnas.1014640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Wang H., Zhou Z., Naseer N., Xiang F., Kan B., Goulian M., Zhu J. Differential Thiol-Based Switches Jump-Start Vibrio cholerae Pathogenesis. Cell Rep. 2016;14:347–354. doi: 10.1016/j.celrep.2015.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell D.S., Camilli A. Regulation of vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J Bacteriol. 2000;182:5342–5350. doi: 10.1128/jb.182.19.5342-5350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton D.L., O’Toole R., Horstedt P., Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroux B., Walker J.E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Park J.H., Jo Y., Jang S.Y., Kwon H., Irie Y., Parsek M.R., Kim M.H., Choi S.H. The cabABC operon essential for biofilm and rugose colony development in vibrio vulnificus. PLoS Pathogens. 2015;11:e1005252. doi: 10.1371/journal.ppat.1005192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Ha S., Kim Y. The protein crystallography beamlines at the pohang light source ll. BIODESIGN. 2017;5:30–34. [Google Scholar]
- Patel V.P., Rojas M.R., Paplomatas E.J., Gilbertson R.L. Cloning biologically active geminivirus DNA using PCR and overlapping primers. Nucleic Acids Res. 1993;21:1325–1326. doi: 10.1093/nar/21.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J.E., Kim K.S., Choi S.H. CadC activates pH-dependent expression of the Vibrio vulnificus cadBA operon at a distance through direct binding to an upstream region. J Bacteriol. 2005;187:7870–7875. doi: 10.1128/JB.187.22.7870-7875.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee J.E., Jeong H.G., Lee J.H., Choi S.H. AphB influences acid tolerance of Vibrio vulnificus by activating expression of the positive regulator CadC. J Bacteriol. 2006;188:6490–6497. doi: 10.1128/JB.00533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R., Priefer U., Phuler A. A broad host range mobilization system for invivo genetic-engineering - transposon mutagenesis in gram-negative bacteria. Bio-Technol. 1983;1:784–791. [Google Scholar]
- Taylor J.L., De Silva R.S., Kovacikova G., Lin W., Taylor R.K., Skorupski K., Kull F.J. The crystal structure of AphB, a virulence gene activator from Vibrio cholerae, reveals residues that influence its response to oxygen and pH. Mol Microbiol. 2012;83:457–470. doi: 10.1111/j.1365-2958.2011.07919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A.C., Simpson L.M., Oliver J.D., Morris J.G. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect Immun. 1990;58:1769–1773. doi: 10.1128/iai.58.6.1769-1773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


