Abstract
The genus Porphyrobacter belongs to aerobic anoxygenic phototrophic bacteria cluster. Porphyrobacter neustonensis DSM 9434 was isolated from a eutrophic freshwater pond in Australia, and is able to synthesize Bacteriochlorophyll a as well as grow under aerobic conditions. It is the type species of the genus Porphyrobacter. Here we describe the characteristics of the strain DSM 9434, including the genome sequence and annotation, synthesis of BChl a, and metabolic pathways of the organism. The genome of strain DSM 9434 comprises 3,090,363 bp and contains 2,902 protein-coding genes, 47 tRNA genes and 6 rRNA genes. Strain DSM 9434 encodes 46 genes which participate in BChl a synthesis and this investigation shed light on the evolution and functional implications regarding bacteriochlorophyll synthesis.
Electronic supplementary material
The online version of this article (doi:10.1186/s40793-017-0243-5) contains supplementary material, which is available to authorized users.
Keywords: Porphyrobacter neustonensis DSM 9434, Aerobic anoxygenic phototrophic bacteria, Bacteriochlorophyll synthesis, Genome sequence, Alphaproteobacteria
Introduction
Aerobic anoxygenic phototrophic bacteria probably evolved after the accumulation of oxygen in the earth’s biosphere [1]. They are widely distributed in the euphotic zone of the ocean as well as terrestrial water, and play an ecologically and biogeochemical important role in aquatic systems, especially marine carbon cycling [2–4]. AAP bacteria harvest light by Bacteriochlorophyll a and possess various carotenoids as auxiliary pigments [5]. They derive a significant portion of their energy requirements from light but perform photoheterotrophic metabolism based on an obligatory supply of organic substrates for growth [6]. Until now, all the AAP bacteria that have been discovered belong to the Proteobacteria, and the majority of cultured AAP strains are members of the Alphaproteobacteria [5].
Porphyrobacter has been proposed as a genus along with four Porphyrobacter strains being isolated from a eutrophic freshwater pond in Australia [7]. They are obligate aerobes in the AAP bacteria cluster. Porphyrobacter neustonensis strain DSM 9434 is the type strain of the genus Porphyrobacter [7]. To get insight into the capability of Porphyrobacter in adapt to harvest energy photosynthetically, recently, we obtained the complete genome of P. neustonensis strain DSM 9434 and detected key genes for synthesizing BChl a and mediating aerobic anoxygenic phototrophic metabolism. We also describe the genomic sequencing related to its annotation for understanding their physiological, metabolic and ecological functions in the environments.
Organism information
Classification and features
P. neustonensis DSM 9434 was purified from a peptone-yeast extract alga plate after being isolated from the euphotic freshwater pond in Australia [7]. The strain grew with temperature between 10 and 37 °C [7]. The cell is rod-shaped, and occasionally coccoid and ovoid (Fig. 1). The strain produced BChl a and carotenoid, analyzed by extracting cells with ethanol (Additional file 1: Figure S1). It grew aerobically in the dark and used a series of organic carbon, such as galactose, glucose, maltose, mannose, sucrose, xylose, arginine, as sole sources of carbon and energy [7]. Analysis of cell wall materials isolated from strain DSM 9434 detected muramic acid and diaminopimelic acid, the major components of peptidoglycan cell wall layer [7]. A high proportion of fatty acids identified as octadecenoic acids (18:1, 84%) is present in the cell with minor components of fatty acids, such as octadecadienoic acid (18:2, 6.1%), 2-hydroxytetradecanoic acid (2OH14:0, 2.7%) and hexadecanoic acid (16:0, 2.6%) [7]. Based on phylogenetic analysis of 16S rRNA gene sequence, the strain belongs to the Alphaproteobacteria class and falls into the cluster comprising the Porphyrobacter species (Fig. 2). The classification and features of P. neustonensis DSM 9434 are summarized in Table 1.
Fig. 1.
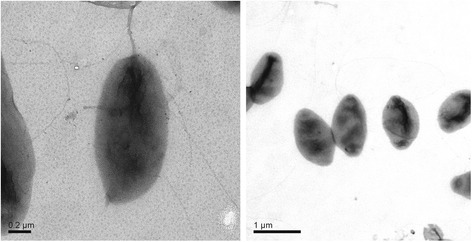
Transmission electron microscopy of cells of Porphyrobacter neustonensis DSM 9434. The peritrichous flagella are present. Bars represent scales of 0.2 μm (a) and 1 μm (b), respectively
Fig. 2.
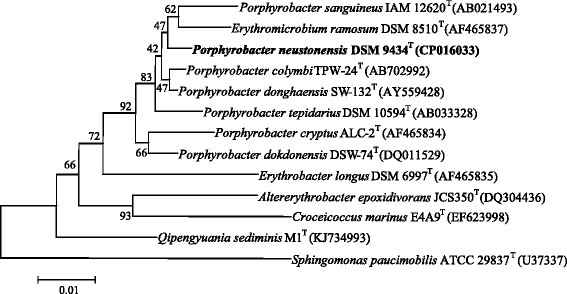
Phylogenetic tree based on 16S rRNA gene sequences was constructed by neighbor-joining algorithms. Related sequences were aligned with Clustal W [21]. Evolutionary distances were calculated according to the algorithm of the Kimura two-parameter model with bootstraps analysis set to 1000 replicates. Bar, 0.01 substitutions per nucleotide position
Table 1.
Classification and general features of Porphyrobacter neustonensis DSM 9434 according to the MIGS recommendations [22]
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Classification | Domain Bacteria | TAS [23] | |
| Phylum Proteobacteria | TAS [24] | ||
| Class Alphaproteobacteria | TAS [25, 26] | ||
| Order Sphingomonadales | TAS [25, 27] | ||
| Family Erythrobacteraceae | TAS [28] | ||
| Genus Porphyrobacter | TAS [7] | ||
| Species Porphyrobacter neustonensis | TAS [7] | ||
| Type strain DSM 9434 | |||
| Gram stain | Negative | IDA | |
| Cell shape | Rod or cocci | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Non-sporulation | IDA | |
| Temperature range | 10–37 °C | TAS [7] | |
| Optimum temperature | Not reported | ||
| pH range; Optimum | Not reported | ||
| Carbon source | Organic carbon | TAS [7] | |
| MIGS-6 | Habitat | Freshwater | TAS [7] |
| MIGS-6.3 | Salinity | Not reported | |
| MIGS-22 | Oxygen requirement | Strictly aerobic | TAS [7] |
| MIGS-15 | Biotic relationship | free-living | TAS [7] |
| MIGS-14 | Pathogenicity | Non-pathogen | NAS |
| MIGS-4 | Geographic location | University of Queensland, Australia | TAS [7] |
| MIGS-5 | Sample collection | Not reported | |
| MIGS-4.1 | Latitude | Not reported | |
| MIGS-4.2 | Longitude | Not reported | |
| MIGS-4.4 | Altitude | Sea level | NAS |
aEvidence codes - IDA Inferred from Direct Assay, TAS Traceable Author Statement (i.e., a direct report exists in the literature), NAS Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project
Genome sequencing information
Genome project history
P. neustonensis DSM 9434 was selected for sequencing in the project of Porphyrobacter Genome Sequencing and Assembly because it is relevant to genomic sequencing of the whole family of Erythrobacteraceae and BChl a synthesis. The complete genome sequence was finished on May 31, 2016 and presented for public access on June 22, 2016. This whole genome has been deposited at DDBJ/EMBL/GenBank under the accession number CP016033. The main genome sequence information is present in Table 2.
Table 2.
Genome sequencing project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS 31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | 10 kb |
| MIGS 29 | Sequencing platforms | A PacBio RS II platform |
| MIGS 31.2 | Fold coverage | 203-fold |
| MIGS 30 | Assemblers | HGAP Assembly version 2, Pacific Biosciences |
| MIGS 32 | Gene calling method | RAST |
| Locus Tag | A9D12 | |
| Genbank ID | CP016033 | |
| GenBank Date of Release | June 22, 2016 | |
| GOLD ID | Go0029942 | |
| BIOPROJECT | PRJNA322640 | |
| MIGS 13 | Source Material Identifier | DSM (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH) |
| Project relevance | Bacteriochlorophyll a synthesis |
Growth conditions and genomic DNA preparation
P. neustonensis DSM 9434 was aerobically cultivated in Luria-Bertani medium at 28 °C. High-quality genomic DNA was extracted using Qiagen DNA extraction kit based on its protocol. DNA sequencing of P. neustonensis DSM 9434 was performed using SMRT technology. One Library with insert size of 10 kb was constructed according to the large SMRTbell gDNA protocol (Pacific Biosciences, USA).
Genome sequencing and assembly
Genomic DNA was sequenced with a PacBio RS II platform yielding 48,527 reads with an average length of 12,972 nt (600 Mb, 203-fold genome coverage; Pacific Biosciences). These reads were assembled using HGAP Assembly version 2 (Pacific Biosciences, USA). The final contigs were checked for circularization and the overlapping ends were trimmed.
Genome annotation
The tRNA genes were identified using tRNAscan-SE 1.21 [8] with bacterial model, and rRNA genes were found via RNAmmer 1.2 Server [9]. The open reading frames (ORFs) and the functional annotation of translated ORFs were predicted and achieved by using the RAST server online [10]. Classification of some predicted genes and pathways were analyzed using COG database [11] and KEGG database [12, 13].
Genome properties
The genome of strain DSM 9434 contains a single circular chromosome (Fig. 3). The complete genome of strain DSM 9434 comprises 3,090,363 bp with an average G + C content of 65.3%. The contig contains 2,902 coding sequences of total 2955 genes, 47 tRNAs and 2 operons of 16S-23S-5S rRNA gene. The summary of features and statistics of the genome is shown in Table 3 and genes belonging to COG functional categories are listed in Table 4.
Fig. 3.
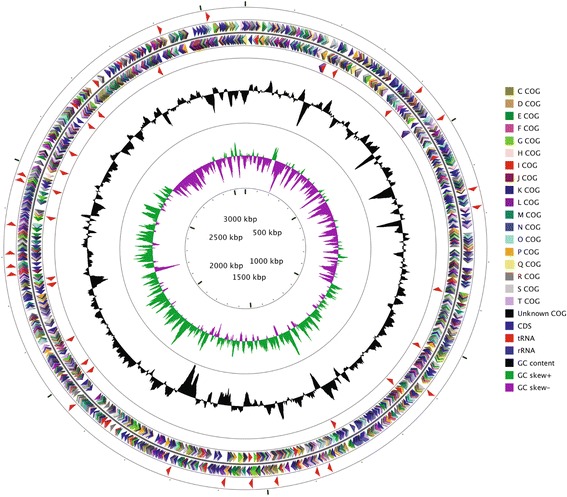
Circular map of the chromosome of Porphyrobacter neustonensis DSM 9434. From outside to the center: RNA genes on the forward strand (tRNAs red, rRNAs blue), genes on the forward strand (colored by COG categories), genes on the reverse strand (colored by COG categories), RNA genes on the reverse strand (tRNAs red, rRNAs blue), G + C content (peaks out/inside the circle indicate values higher or lower than the average G + C content, respectively), GC skew (calculated as (G-C)/(G + C), green/purple peaks out/inside the circle indicate values higher or lower than 1, respectively), genome size (3,090,363 bp)
Table 3.
Genome statistics
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 3,090,363 | 100 |
| DNA coding (bp) | 2,809,376 | 90.91 |
| DNA G + C (bp) | 2,016,518 | 65.25 |
| DNA scaffolds | 1 | - |
| Total genes | 2955 | 100 |
| Protein coding genes | 2902 | 98.21 |
| RNA genes | 53 | 1.79 |
| Pseudo genes | - | - |
| Genes in internal clusters | 350 | 11.84 |
| Genes with function prediction | 2189 | 74.08 |
| Genes assigned to COGs | 2326 | 78.71 |
| Genes with Pfam domains | 2373 | 80.30 |
| Genes with signal peptides | 400 | 13.54 |
| Genes with transmembrane helices | 674 | 22.81 |
| CRISPR repeats | 2 | - |
Table 4.
Number of genes associated with general COG functional categories
| Code | Value | % age | Description |
|---|---|---|---|
| J | 158 | 6.13 | Translation, ribosomal structure and biogenesis |
| A | 2 | 0.08 | RNA processing and modification |
| K | 129 | 5.00 | Transcription |
| L | 110 | 4.27 | Replication, recombination and repair |
| B | 4 | 0.16 | Chromatin structure and dynamics |
| D | 25 | 0.97 | Cell cycle control, Cell division, chromosome partitioning |
| V | 43 | 1.67 | Defense mechanisms |
| T | 165 | 6.40 | Signal transduction mechanisms |
| M | 167 | 6.48 | Cell wall/membrane biogenesis |
| N | 61 | 2.37 | Cell motility |
| U | 87 | 3.37 | Intracellular trafficking and secretion |
| O | 117 | 4.54 | Posttranslational modification, protein turnover, chaperones |
| C | 169 | 6.56 | Energy production and conversion |
| G | 97 | 3.76 | Carbohydrate transport and metabolism |
| E | 172 | 6.67 | Amino acid transport and metabolism |
| F | 63 | 2.44 | Nucleotide transport and metabolism |
| H | 123 | 4.77 | Coenzyme transport and metabolism |
| I | 152 | 5.90 | Lipid transport and metabolism |
| P | 127 | 4.93 | Inorganic ion transport and metabolism |
| Q | 79 | 3.06 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 286 | 11.09 | General function prediction only |
| S | 242 | 9.39 | Function unknown |
| - | 546 | 19.01 | Not in COGs |
Insights from the genome sequence
Bacteriochlorophyll a synthesis and phototropic activity
The genome of P. neustonensis DSM 9434 harbors 46 genes which participate in BChl a synthesis (Additional file 2: Table S1). A complete photosynthesis gene cluster structures was observed. The PGC is 38 kb and includes 5 main sets of genes: bch genes encoding enzymes involved in the BChl a biosynthetic pathway, puf operons encoding proteins forming the reaction center, puh operons involved in the RC assembly, crt genes responsible for biosynthesis of carotenoids and a variety of regulatory genes. The complete PGC in the genome of P. neustonensis DSM 9434 genome consists of bchIDO-crtCDF-bchCXYZ-pufALM-tspO-bchP-bchG-ppsR-ppaA-bchFNBHLM-lhaA-puhABC-ascF-puhE-hemA-cycA (Additional file 2: Table S1).
The heart of aerobic anoxygenic phototrophy is the RC encoded by the puf and puh operons. The puf operon encodes the subunits of the light-harvesting (LH1) (pufA, ANK11803) and RC complex (pufL and pufM, ANK11804 -11805). The puh operons encoding RC assembly indirectly effect on LH1 assembly (puhABC, ANK11818-11820, puhE, ANK11823). Gene lhaA (ANK11817) encodes a possible LH1 assembly protein [14]. Genes bchBCDFGHILMNOPXYZ (ANK11793-11795, 11800–11802, 13992, 11806, 11808, 11811–11816) and ascF (ANK11822), with exception of 8-vinyl reductase (ANK12775), represent the complete biosynthetic pathway from protoporphyrin XI to BChl a. The cluster of three carotenoid biosynthesis genes, crtC (ANK11797), crtD (ANK11798) and crtF (ANK11799) may participate in the formation of acyclic xanthophylls from lycopene [15]. Other carotenoid biosynthesis genes are located outside the cluster (crtE, ANK13491; crtB, ANK12836; crtI, ANK14187; crtY, ANK14188; crtZ, ANK11768; crtW, ANK13982, 14112 and 13340). Three regulatory genes (ppsR, ppaA and tspO) were found in the genome of strain DSM 9434. Regulatory genes ppsR (DNA-binding repressor, ANK11809) and ppaA (oxygen sensor, ANK11810) are sensitive to light intensity and oxygen concentration [16], and the gene tspO (tryptophan-rich sensory protein precursor, ANK13994) negatively affects the transcriptional expression of several photosynthesis genes [17].
Metabolism of P. neustonensisDSM 9434
The complete genome of P. neustonensis DSM 9434 was annotated for understanding the major metabolic pathways of carbon, nitrogen, sulfur and phosphorus based on the key genes it processes. As we mentioned, although it has bacteriochlorophyll-synthesis genes and acquires energy from light, the absence of carbon fixation and CO-oxidizing genes indicates that strain DSM 9434 is not able to grow autotrophically. They can only use organic carbon sources. It does not have a complete glycolysis pathway but processes key genes for the Entener-Doudoroff, the pentose phosphate pathway, and the tricarboxylic acid cycle. The genome of P. neustonensis DSM 9434 harbors a variety of transporter genes for ammonium (amtB) and other organic nitrogen substrates (e.g. amino acids, polyamines). It is lack of genes involved in nitrate/nitrite reduction, nitrogen fixation or anaerobic ammonium oxidation, thus strain DSM 9434 only relies on reduced nitrogen sources. The genes encoding urea transporter and urease (ureABC) are absent in the genome of DSM 9434, suggesting its incapability of utilizing urea as a C or N source in the environment. The lack of urea uptake and degradation may reflect the environmental adaption of strain DSM 9434 from a eutrophic pond, where ammonium and algae-derived organic N (e.g. amino acids and polyamines) are usually enriched [18, 19]. P. neustonensis DSM 9434 processes genes involved in assimilatory SO4 reduction (e.g. sulP encoding sulfate permease). Sulfate can be reduced to sulfide (cys), subsequently being incorporated into amino acids. The strain DSM 9434 is also able to utilize organic sulfur compounds (e.g. amino acids, alkanesulfonates); however, it missed the transporter genes (ssuACB) for uptake of extracellular alkanesulfonates. Strain DSM 9434 possesses the high-affinity phosphate transporter (pstSCAB) and regulatory genes (phoUBR), and genes for inorganic P storage as polyphosphate (ppk), a signal of using an alternative strategy for maintaining a phosphate supply [20]. The presence of genes encoding alkaline phosphatase in the genome of strain DSM 9434 indicates that it is capable of using both inorganic and organic forms of phosphorus.
Conclusion
The complete genome sequence of the BChl a synthesizing bacteria P. neustonensis DSM 9434 provide an insight into the genomic basis of its metabolic characteristics and bacteriochlorophyll-synthesis pathway. This investigation sheds light on the evolution of PGCs of aerobic anoxygenic phototrophs and provides the possibility for comparative genomics of AAP bacteria isolated from marine, freshwater and terrestrial environments.
Additional files
Porphyrobacter neustonensis DSM 9434. In vitro absorption spectrum of ethanol extract of showing peaks of carotenoid (452–484 nm) and Bacteriochlorophyll a (765 nm). (EPS 1267 kb)
Genes related to bacteriochlorophyll-synthesis in the genome of Porphyrobacter neustonensis DSM 9434. (DOCX 35 kb)
Acknowledgments
This work was supported by grants from the National Key Basic Research Program of China (2014CB441503), the Natural Science Foundation of Zhejiang Province (LR17D060001 and LY14D060006), the Regional Demonstration of Marine Economy Innovative Development Project (NO.12PYY001SF08) and the Top-Notch Young Talents Program of China.
Authors’ contributions
XX and CW organized the study. YW, LX and QL performed laboratory experiments. QL and HC analyzed the data and drafted the manuscript. XX edited the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- AAP
Aerobic anoxygenic phototrophic
- BChl a
Bacteriochlorophyll a
- COG
Clusters of Orthologous Groups
- LH1
Light-harvesting
- ORF
Open reading frame
- PGC
Photosynthesis gene cluster
- RC
Reaction center
References
- 1.Beatty JT. On the natural selection and evolution of the aerobic phototrophic bacteria. Photosynth Res. 2002;73:109–114. doi: 10.1023/A:1020493518379. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Chaves MC, Cottrell MT, Kirchman DL, Ruiz-González C, del Giorgio P A. Single-cell activity of freshwater aerobic anoxygenic phototrophic bacteria and their contribution to biomass production. ISME J. 2016; doi:10.1038/ismej.2015.242. [DOI] [PMC free article] [PubMed]
- 3.Jiao N, Zhang Y, Zeng Y, Hong N, Liu R, Chen F, Wang P. Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean. Environ Microbiol. 2007;9:3091–3099. doi: 10.1111/j.1462-2920.2007.01419.x. [DOI] [PubMed] [Google Scholar]
- 4.Kolber ZS, Gerald F, Lang AS, Beatty JT, Blankenship RE, VanDover CL, Vetriani C, Koblizek M, Rathgeber C, Falkowski PG. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science. 2001;292:2492–2495. doi: 10.1126/science.1059707. [DOI] [PubMed] [Google Scholar]
- 5.Zheng Q, Zhang R, Fogg PC, Beatty JT, Wang Y, Jiao N. Gain and loss of phototrophic genes revealed by comparison of two Citromicrobium bacterial genomes. PLoS One. 2012;7(4):2010–2026. doi: 10.1371/journal.pone.0035790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yurkov V, Csotonyi JT. New light on aerobic anoxygenic phototrophs. In: Hunter CN, Daldal F, Thurnauer MC, Beatty JT, editors. The purple phototrophic bacteria. Dordrecht: Springer; 2009. pp. 31–55. [Google Scholar]
- 7.Fuerst JA, Hawkins JA, Holmes A, Sly LI, Moore CJ, Stackebrandt E. Porphyrobacter neustonensis gen. nov., sp. nov., an aerobic bacteriochlorophyll-synthesizing budding bacterium from fresh water. Int J Syst Evol Microbiol. 1993;43:125–134. doi: 10.1099/00207713-43-1-125. [DOI] [PubMed] [Google Scholar]
- 8.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005;33:686–689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mothersole DJ, Jackson PJ, Vasilev C, Tucker JD, Brindley AA, Dickman MJ, Hunter CN. PucC and LhaA direct efficient assembly of the light-harvesting complexes in Rhodobacter sphaeroides. Mol Microbiol. 2016;99:307–327. doi: 10.1111/mmi.13235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Armstrong GA. Genetics of eubacterial carotenoid biosynthesis: a colorful tale. Annu Rev Microbiol. 1997;51:629–659. doi: 10.1146/annurev.micro.51.1.629. [DOI] [PubMed] [Google Scholar]
- 16.Komiya H, Yeates T, Rees D, Allen JP, Feher G. Structure of the reaction center from Rhodobacter sphaeroides R-26 and 2.4. 1: symmetry relations and sequence comparisons between different species. Proc Natl Acad Sci U S A. 1988;85:9012–9016. doi: 10.1073/pnas.85.23.9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeliseev AA, Kaplan S. A sensory transducer homologous to the mammalian peripheral-type benzodianzepine receptor regulates photosynthetic membrane complex formation in Rhodobacter sphaeroides 2.4.1. J Biol Chem. 1995;270:21167–21175. doi: 10.1074/jbc.270.36.21167. [DOI] [PubMed] [Google Scholar]
- 18.Lee C, Jørgensen NOG. Seasonal cycling of putrescine and amino acids in relation to biological production in a stratified coastal salt pond. Biogeochemistry. 1995;29:131–157. doi: 10.1007/BF00000229. [DOI] [Google Scholar]
- 19.Park MG, Shim JH, Cho BC. Urea decomposition activities in an ammonium enriched freshwater pond. Aquat Microb Ecol. 1997;13:303–311. doi: 10.3354/ame013303. [DOI] [Google Scholar]
- 20.Moran MA, Belas R, Schell MA, González JM, Sun F, et al. Ecological genomics of marine roseobacters. Appl Environ Microbiol. 2007;73:4559–4569. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proc Natl Acad Sci U S A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrity GM, Bell JA, Lilburn TG. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. Second. New York: Springer; 2005. p. 1. [Google Scholar]
- 25.Validation List No. 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1–6. [DOI] [PubMed]
- 26.Garrity GM, Bell JA, Lilburn TG. Class I. Alphaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. Second. New York: Springer; 2005. p. 1. [Google Scholar]
- 27.Yabuuchi E, Kosako Y. Order IV. Sphingomonadales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT, editors. Bergey’s Manual of Systematic Bacteriology. Second. New York: Springer; 2005. p. 230. [Google Scholar]
- 28.Lee KB, Liu CT, Anzai Y, Kim H, Aono T, Oyaizu H. The hierarchical system of the ‘Alphaproteobacteria’: description of Hyphomonadaceae fam. nov., Xanthobacteraceae fam. nov. and Erythrobacteraceae fam. nov. Int J Syst Evol Microbiol. 2005;55:1907–1919. doi: 10.1099/ijs.0.63663-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Porphyrobacter neustonensis DSM 9434. In vitro absorption spectrum of ethanol extract of showing peaks of carotenoid (452–484 nm) and Bacteriochlorophyll a (765 nm). (EPS 1267 kb)
Genes related to bacteriochlorophyll-synthesis in the genome of Porphyrobacter neustonensis DSM 9434. (DOCX 35 kb)


