Abstract
Objective
Oral lichen planus (OLP) is a disease of the oral mucosa of unknown cause producing lesions with an intense band-like inflammatory infiltrate of T cells to the subepithelium and keratinocyte cell death. We performed gene expression analysis of the oral epithelium of lesions in subjects with OLP and its sister disease, oral lichenoid reaction (OLR), in order to better understand the role of the keratinocytes in these diseases.
Design
Fourteen patients with OLP or OLR were included in the study, along with a control group of 23 subjects with a variety of oral diseases and a normal group of 17 subjects with no clinically visible mucosal abnormalities. Various proteins have been associated with OLP, based on detection of secreted proteins or changes in RNA levels in tissue samples consisting of epithelium, stroma, and immune cells. The mRNA level of twelve of these genes expressed in the epithelium was tested in the three groups.
Results
Four genes showed increased expression in the epithelium of OLP patients: CD14, CXCL1, IL8, and TLR1, and at least two of these proteins, TLR1 and CXCL1, were expressed at substantial levels in oral keratinocytes.
Conclusions
Because of the large accumulation of T cells in lesions of OLP it has long been thought to be an adaptive immunity malfunction. We provide evidence that there is increased expression of innate immune genes in the epithelium with this illness, suggesting a role for this process in the disease and a possible target for treatment.
Keywords: lichen planus, innate immunity, epithelium, RNA, gene expression
1. Introduction
Oral lichen planus (OLP) is a disease of the oral mucosa characterized by a dysfunctional basal epithelium.1–3 The keratinization cycle is abnormal with an elevated rate of apoptosis of the basal keratinocytes and increased cell proliferation that may be compensatory. This is accompanied by vacular degeneration of the basement membrane. The cause is believed to be an abnormal chronic inflammation marked by an intense band-like infiltration including cytotoxic T cells below the basement membrane which trigger the death of the keratinocytes. While the disease is thought to be autoimmune in nature, it is not clear what causes the infiltration and adherence of the CD8+ cells to the disease sites. Some studies of tissue from lichen planus lesions have noted higher rates of Hepatitis C Virus (HCV) infection and even Candida in lesions, which suggested an abnormal immune response to infected keratinocytes may initiate the disease.2,4 The fact that oral lichenoid reactions (OLR), which are histopathologically indistinguishable from OLP lesions, are known to be due to an allergic reaction to drugs or dental amalgam further supports the role of adaptive immunity in OLP.3,5 The treatment of OLP consists of topical application of corticosteroid, which shows some reduction of symptoms possibly by dampening lymphocyte migration and activation.2
It has long been thought that the key to determining the etiology of lichen planus, be it in the skin or in the oral cavity, is to uncover what causes the buildup of T cells. Because the differential expression of chemokines, chemokine receptors, and adhesion molecules plays an important role in the migration and buildup of immune cells in the periphery, much effort has been made to characterize the expression of these protein types in the lymphocytes of diseased versus normal patients. In addition, abnormalities of dendritic cells, mast cells and other immune cells, endothelial cells, and keratinocytes have all been suggested as the source of the factors that draw the T cells to the site just below the basement membrane. For example, while specialized cells like Langerhans cells play a major role in antigen processing and T cell recruitment, it is known that keratinocytes of the skin and mucosa play a fundamental immunological role as a first barrier to infection.6 Abnormal antigen presentation by either of these cell types has been considered as a first step to this disease, which eventually results in the autoimmune-like features of OLP.4,7
As the first barrier to infection, the epithelium of the skin and mucosa play a role in the innate immune response that works to recognize and eliminate various microbial components.6,8 In recent years there has been a renewed focus on the innate immune system, including its ability to affect adaptive immunity and possible roles for innate immunity in chronic inflammatory and autoimmune diseases, such as psoriasis, asthma, diabetes, and others.9–12 A primary part of the innate immune system are the toll-like receptors (TLRs), transmembrane factors that are pattern-recognition proteins that recognize ligands that are part of microbes.6,13,14 There are more than 11 TLRs with specificities for different organisms that, when engaged, activate cellular pathways resulting in changes in gene expression. Additional receptor proteins, such as CD14, can act as co-receptors and aid in recognizing pathogen-associated molecular patterns and also play key roles in TLR1, TLR2, and TLR4 recognition of triacylated lipoproteins15 and the initiation of the innate immune response. Activation of the TLRs typically results in activation of NFK-B and other regulatory complexes that signal the production of inflammatory chemokines, such as CXCLs, which can recruit neutrophils for acute inflammation. Increases in certain cytokines and receptors in immune cells via TLR activation can have direct effects on T cell response in autoimmune diseases.12,16 Some years ago Xao et al. published the only global gene expression analysis of tissue from this disease 17 and we have tried to use this as a starting point to discern genes differentially expressed in the epithelium of patients with OLP or OLR. Various factors have been shown to be associated with OLP based on changes in protein or RNA levels in OLP mucosal tissue or even protein levels in saliva. 18–23 Focusing on these and related genes we performed gene expression analysis on highly expressed genes of the oral epithelium and were able to identify a set of genes linked to OLP.
2. Material and methods
2.1 Clinical sampling
Brush cytology samples were collected from 14 patients with OLP based on the clinical features of a bilateral, near symmetrical pattern of white, lacy lines on the mucosa, called Wickham’s striae, which is the reticular form of OLP. 24 Several cases were of the erosive type, also referred to as the atrophic or ulcerative type, which presents as erythematous, ulcerative lesions. Care was taken to sample oral epithelium only. Most cases (12) and all ambiguous cases were verified by surgical biopsy and histopathology to show the World Health Organization criteria of a dense subepithelial lymph-histiocytic infiltrate, overlying keratinization and degeneration of basal keratinocytes.7 While OLP in the classic presentation can be recognized without biopsy based on the lesion appearance, the lesions are typically biopsied to rule out the presence of malignancy which may be associated with the disease. 2,3,24
Two cases were consistent with OLR while the remainder were OLP based on the case history. Diagnosis was done by one of two specialists in oral medicine. Patients were seen in the Oral and Maxillofacial Surgery Clinic and the Multidisciplinary Head and Neck Cancer Clinic in the University of Illinois Medical Center.25 The healthy normal group samples were from mucosa that appeared to be clinically healthy in patients there for tooth extraction. The alternative disease group consisted of 19 brush cytology samples sites of apparent mucosal abnormalities, all diagnosed after tissue histopathology. All subjects provided consent to participate in accordance with guidelines of the Institutional Review Board of the University of Illinois at Chicago.
2.2 Brush cytology
Brush cytology was performed on patients as they presented in the clinic. Samples were immediately placed in Trizol (Invitrogen, Carlsbad, CA, USA), mixed, and frozen. We used a cervical cytology brush with RNA purification as described using the RNAeasy Mini Kit with DNAse digestion (Qiagen, Germantown MD). 26 Typically 400 to 2000 nanograms were harvested per sample.
2.3 Gene target selection
A number of studies have compared mRNAs in surgically obtained mucosa of OLP patients versus control mucosa in order to define genes differentially expressed with this disease. Much of this work has focused on the T cells that invade the stroma. Primers were designed for q-RT-PCR analysis of CD1418, TLR1, TLR2, TLR619, IL1020, HIF127, hTR 28, and TRPV129. RNA for IL6 and IL8, two proteins that have been shown in multiple reports to be increased with OLP and to be synthesized by keratinocytes were also examined.21–23,30 In a small study Ichimura et al. studied differential gene expression in epithelial samples from 3 OLP patients and saw 20 to 40x increases in expression of CXCL9, 10 and 11, so we designed primers to test these despite low basal expression in normal epithelium. 31 We did not test VEGFa due to the difficulty in designing specific primers probes for this mRNA.27 Brush oral cytology supplies limited amounts of sample so that the focus was on mRNAs with more than average expression levels in the epithelium, as tested by global gene expression analysis of 6 epithelial samples of a variety of oral illnesses using hybridization to Affymetrix Human Genome U133 Plus 2.0 microarrays32 (data not shown). In 2009 Tao et al. published the first, and perhaps only, global gene expression analysis study of OLP gene expression17. Using whole mucosa (epithelium, stroma, immune cells, blood) as a source of lichen planus and normal tissue they showed 22 genes to be overexpressed at least 8x with this disease. We designed qRT-PCR primers for those that showed expression levels above background in our epithelial samples: CCR7, CXCL1, MMP12, GBP5, and MMP9. We also examined the genes shown to be downregulated at least 8x in the Tao study, focusing on the highest expressers in normal epithelia samples we tested: SEC61B, FOSB, ALOX12, ANXA1, and ADH7.
2.4 RT-PCR
One hundred nanograms of each RNA from brush cytology was converted to cDNA and quantitative RT-PCR was carried out using the iCycler iQ (Bio-Rad, Hercules, CA, USA) and SYBR Green fluorescence as described.25 Approximately equal amounts of each cDNA were amplified using Brilliant II SYBR Green QPCR Master Mix (Agilent Life Technologies Santa Clara, CA, USA) at 95 degrees C for 10 minutes followed by 40 cycles at 95 degrees C for 30 seconds, 60 or 57 degrees C for 60 seconds, followed by 95 degrees C for 60 seconds and 55 to 95 C degrees in 0.5 degree steps for melt curve analysis. Hybridization temperatures were optimized empirically. Standard curves were created for each primer pair. Values were normalized to the geometric mean of the controls: GAPD, RPLPO, and RPL4. Primers for these mRNAs, and those to detect the target mRNAs, were designed using Primer Express to give products of approximately 100 bases; sequences were previously published and/or included in the supplemental data section (supplemental figure).25 The qRT-PCR for FOSB and ADH7 did not produce a single product and IL6, MMP1, MMP9, and MMP12 were undetectable in most samples and were not pursued (data not shown).
2.5 Immunohistochemical staining
Thin sections from paraffin blocks of tissue obtained by biopsy were stained immunohisto-chemically for CXCL1 (GRO alpha) and TLR1 proteins. After mounting, slides were deparaffinized using xylene washes, rehydrated in graded ethanol washes, microwave heated for 12 minutes in 10 mM Sodium Citrate, exposed 15 minutes to 3% hydrogen peroxide in methanol, followed by 2 PBS washes, 40 minutes incubation with 2.5% horse serum and then overnight with rabbit anti-CXCL1(GRO alpha) (NBP1-40227)(1:600) or rabbit anti-TLR1 (NBP1-02993) (1:600) (both from Abcam Inc., Cambridge, MA, USA). To complete the staining, the ImmPRESS reagent kit and ImmPACT DAB peroxidase substrate (Vector Laboratories, Burlingame, CA, USA) were used. Counterstaining used Mayers Hematoxalin (Thermo Fisher Scientific, Pittsburgh, PA, USA). Positive controls for CXCL1 were squamous cell carcinoma tissue and for TLR1, lymphocytes (data not shown and Figure 1).
Figure 1.
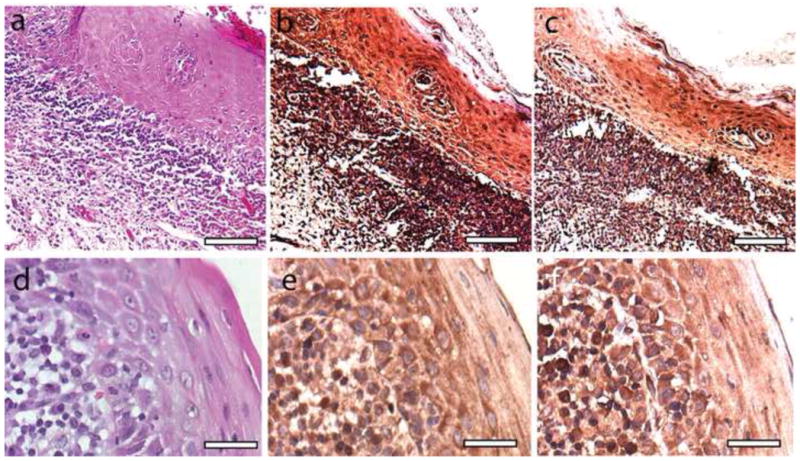
Histopathology and immunohistochemical staining of OLP mucosa. (A) Hematoxalin and eosin staining reveals characteristic subepithelial lymphocytic infiltration and hyperkertatosis in representative OLP mucosa. Scale bar represents 60 microns (B) Immunohistochemical detection of CXCL1 reveals positive staining in both stromal lymphocytes and epithelial keratinocytes. Scale bar represents 60 microns (C) Immunohistochemical detection of TLR1 reveals positive staining in both stromal lymphocytes and epithelial keratinocytes.Sc ale bar represents 60 microns (D) Hematoxalin and eosin staining of second sample of lichen planus mucosa viewed at high power. Scale bar represents 25 microns. (E) High power histochemical detection of CXCL1 in OLP mucosa. Scale bar represents 25 microns. (F) High power histochemical detection of CXCL1 in OLP mucosa. Scale bar represents 25 microns.
2.6 Statistical analysis, class comparison and class prediction using RNA from brush cytology
The class comparison function of BRB-Array Tools 3.9 (two-sample t-test with random variance model) allowed the determination of mRNA of genes that show differential expression in oral epithelium in patients with OLP versus those with visually normal or “healthy” mucosa with a maximum allowed proportion of false positive genes of 0.1 (R. Simon and A. Pang Lam (http://linus.nci.nih.gov/BRB-ArrayTools.html). A gene expression classifier for OLP and for OLR was developed using leave-one-out cross-validation (LOO CV) and BRB-Array Tools as described earlier.25,33 Briefly, normalized mRNA levels for the 9 targets were log2 transformed and entered for each sample of the training set. We allowed the program to choose the significance level for gene inclusion for class prediction using an imbedded optimization scheme to prevent overfitting. Seven separate algorithms were used to generate optimized predictors while simultaneously performing LOO CV of the generated classifiers.
3. Results
Brush oral cytology samples were obtained from 14 patients with OLP or OLR. Table 1 provides a description of these subjects. In a bid to focus on genes differentially expressed uniquely with OLP and OLR and not in other oral diseases, we compared gene expression to a second group with samples from a variety of oral lesions (Table 2). This alternative disease set included OSCCs and hyperkeratotic lesions from 23 patients with apparent oral mucosal lesions that required a biopsy, but were not OLP or OLR based on clinical examination and histopathology. RT-PCR analysis of 21 different potential mRNA markers of OLP was attempted but only 10 revealed detectable expression in at least half of one group. Of these, 4 were likely to be differentially expressed between the two groups (Table 3).
Table 1.
OLP Patient Data
| Sample | Site* | Sex | Age | Tobacco/Betel† | Diagnosis |
|---|---|---|---|---|---|
| BL142255 | Bu | F | 53 | F-Tob | OLP |
| BL225 | Bu | F | 56 | Tob | OLP |
| BL259 | Bu | F | 71 | Tob | OLP |
| BL263 | T | M | 60 | F-Tob | OLP |
| BL298 | UG | F | 67 | NU | OLP |
| BL374 | Bu | M | 36 | NU | OLP |
| BL377 | Bu | M | 30 | Tob | OLR |
| BL381 | T | M | 62 | Tob | OLR |
| BL394 | Bu | F | 57 | NU | OLP |
| BL397 | Bu | F | 72 | NU | OLP |
| BL12010 | G | F | 53 | NU | OLP |
| BL400 | Bu | F | 76 | NU | OLP |
| BL403 | HP | M | 60 | Tob | OLP |
| BL419 | Bu | M | 18 | NU | OLP |
T, tongue; SP, soft palate; HP, hard palate; LG/UG, lower/upper gingival; FOM, floor of mouth; Bu, buccal mucosa, LM, lip mucosa.
Tob, Tobacco user; Bet, betel nut user, if former user than F-Tob or F-Bet; NU, Nonuser
Table 2.
Non-OLP Patient Data
| Sample | Site* | Sex | Age | Tobacco/ Betel† | Diagnosis |
|---|---|---|---|---|---|
| BL100109 | Bu | F | 18 | Tob | odontoma |
| BL 21410 | UG | M | 31 | Mar | maxillary dentigerous cyst |
| BL 31110-3 | L | M | 78 | F-Tob | herpetic viral mucositis |
| BL 31110-5 | UG | M | 48 | NU | periapical granuloma |
| BL 278 | Bu | M | 56 | Tob | mild dysplasia |
| OSCC 301 | FOM | M | 60 | Tob | OSCC |
| BL45 | T | M | 23 | NU | candidiasis, hyperkeratosis |
| BL147 | UG | F | 21 | NU | periapical granuloma with focal abscess |
| BL153 | Bu | M | 52 | T | tobacco related hyperkeratosis |
| BL165 | Bu | F | 56 | NU | hypkeratosis |
| BL197 | UG | F | 66 | Tob | leukoplakia# |
| BL200 | T | M | 74 | NU | hyperplastic foliate papilla |
| BL206 | T | M | 53 | Tob | atypia, candidiasis |
| OSCC 231 | T | M | 49 | NU | OSCC |
| BL 235 | LM | M | 52 | Tob | canalicular adenoma |
| BL 272 | FOM | M | 34 | F-Tob | condyloma acuminatum |
| OSCC 305K | T | M | 79 | NU | OSCC |
| BL 345 | G | F | 82 | F-Tob | keratinizing dysplasia |
| BL366 | Bu | M | 44 | Tob | hyperkeratosis |
| BL 368 | T | M | 71 | NU | epithelial hyperplasia with dysplasia |
| OSCC 383 | T | M | 45 | Bet | OSCC |
| OSCC 393 | LG | M | 68 | F-Tob | OSCC |
| OSCC 42810 | Bu | F | 59 | NU | OSCC |
T, tongue; SP, soft palate; HP, hard palate; LG/UG, lower/upper gingival; FOM, floor of mouth; Bu, buccal mucosa, LM, lip mucosa.
Tob, tobacco user; if former user than F-Tob.; Mar, marijuana user; Bet, betel use; NU, never used
BL 197 was not biopsied but indicated no clinical signs of OLP on multiple visits
Table 3.
RT-PCR analysis of gene expression using RNA from brush cytology of OLP versus other oral disease or condition
| Parametric p-value | FDR | Fold- change | mRNA | |
|---|---|---|---|---|
| 1 | 3.15E-05 | 0.000284 | 13.1 | CD14 |
| 2 | 0.00191 | 0.0062 | 12.5 | IL8 |
| 3 | 0.00207 | 0.0062 | 11.2 | CXCL1 |
| 4 | 0.00612 | 0.0138 | 5.0 | TLR1 |
| 5 | 0.0331 | 0.0596 | 0.55 | ANXA1 |
| 6 | 0.0893 | 0.134 | 1.72 | SEC61B |
| 7 | 0.192 | 0.229 | 3.0 | GBP5 |
| 8 | 0.204 | 0.229 | 2.0 | CCR7 |
| 9 | 0.299 | 0.332 | 2.4 | IL6 |
| 10 | 0.771 | 0.771 | 1.1 | ALOX12 |
Sorted by p-value of the univariate test
FDR is the false discovery rate, a measure of the probability that the data point is truly differentially expressed.
Due to the overwhelming association of the top 4 genes differentially expressed with OLP and the innate immune response we then compared gene expression in the 14 lichen planus samples with expression in normal healthy tissue in subjects lacking oral soft tissue disease. If genes were truly differentially expressed with OLP/OLR, then the expression changes might be marked in a comparison to normal tissue. RNA from normal or “healthy mucosa” was from 17 subjects without any oral soft tissue lesions based on visual inspection. The mean subject age was 42.2; 12 were females, 5 males; and 9 samples were from buccal mucosa, 7 from the gingiva, and 1 from tongue. There was one smoker and 2 former smokers. RT-PCR analysis of 19 different mRNAs was done but only 9 mRNAs revealed detectable expression in at least half of one group. Six of the nine showed differential expression between the two groups at a statistically significant level, taking into account multiple testing. These are shown in Table 4 as mRNAs with a change in level of at least 2x and a False Discovery Rate of less than 0.05.
Table 4.
RT-PCR analysis of gene expression using RNA from brush cytology of OLP versus normal mucosa
| Parametric p-value | FDR | Fold- change | mRNA | |
|---|---|---|---|---|
| 1 | 1.15E-05 | 5.31E-05 | 39.5 | IL8 |
| 2 | 1.18E-05 | 5.31E-05 | 15.9 | CXCL1 |
| 3 | 8.26E-05 | 0.000248 | 14.8 | CD14 |
| 4 | 0.000611 | 0.00137 | 17.5 | TLR1 |
| 5 | 0.00485 | 0.00873 | 0.34 | ANXA1 |
| 6 | 0.0333 | 0.0499 | 0.25 | ALOX12 |
| 7 | 0.0531 | 0.0682 | 13.7 | GBP5 |
| 8 | 0.114 | 0.129 | 1.67 | SEC61B |
| 9 | 0.418 | 0.418 | 1.98 | CCR7 |
Sorted by p-value of the univariate test
FDR is the false discovery rate, a measure of the probability that the data point is truly differentially expressed.
Increased levels of specific mRNAs in cells of the epithelium that occur with OLP could be due to increased levels of expression across the tissue or due to very high levels in one particular cell type. Immunohistochemistry analysis of CXCL1 and TLR1 proteins showed substantial accumulation of these proteins in the cytoplasm of keratinocytes of the epithelium of OLP tissue and little evidence for extremely high protein levels in professional immune cells, such as lymphocytes, Langerhans, and dendritic cells which are present in the epithelium but at much lower numbers (Figure 1).
A classifier for OLP and OLR would be of interest because although these sister diseases are typically diagnosed with a visual exam, only specialists have this expertise. Thus a gene expression classifier may ease detection of these treatable diseases. To perform class prediction for OSCC using the RNA from cytology samples, BRB-Array Tools was used to do LOO CV, simultaneously testing 7 classifiers for their ability to differentiate OLP and OLR from healthy mucosa samples based on the levels of the 9 genes in table 4. We found that among the 6 methods (compound covariate predictor, diagonal linear discriminant analysis, 1-nearest neighbor, 3-nearest neighbor, nearest centroid, and support vector machines) there was on average 79% accuracy in identifying these samples.
A more valuable class predictor is one that can differentiate OLP/OLR not from healthy tissue but from other oral soft tissue pathologies. BRB-Array Tools was used again, this time to develop a classifier to differentiate OLP/OLR from alternative pathologies based on epithelia gene expression. Note all non-OLP/OLR diagnoses were confirmed by biopsy and histopathology with one exception. Using LOO CV this new classifier showed on average 78% accuracy in classifying OLP/OLR versus other pathologies.
4. Discussion
The original goal of this study was to identify genes highly expressed in oral epithelium at or near the site of OLP or OLR lesions that showed differential expression compared to control oral epithelium without the disease.18–23 Some of the genes tested showed differential expression of much greater magnitude than those recorded in earlier studies. For example Ohno et al. saw a less than 2-fold increase of TLR1 in oral tissue with OLP, and did not focus on this gene, while we saw a 7x change.19 This is likely due to the fact that our screen focuses on cells of the epithelium, which consists mainly of keratinocytes, with small numbers of immune cells including Langerhans cells, mast cells, and minimal numbers of lymphocytes. One earlier study of the OLP epithelial layer used laser micro-dissection but focused on lymphocytes and their gene expression and used minimal numbers of samples.31 In the end the small group of genes we tested did not produce a highly accurate classifier for OLP, but by focusing on the epithelium the study did highlight a possible role for innate immunity in this disease. Of the top 4 genes shown to be differentially expressed in our brush oral cytology samples of OLP/OLR and non-OLP/OLR mucosa, all 4 are part of the innate immune system. A fifth gene that was mildly downregulated on the RNA level versus all non-OLP/OLR epithelium tested was ANXA1, a widely expressed protein that promotes resolution of inflammation through various mechanisms.34,35 All are known to be expressed by cells that include keratinocytes and professional immune cells, Langerhans, and mast cells in the mucosa, all of which have the ability to recognize microbial invaders and signal an innate immune response. CD14 has already been shown to be elevated on the protein and RNA levels in oral keratinocytes with OLP.18 We showed here that two of these proteins, CXCL1 and TLR1, were at significant levels in the cytoplasm of mucosal keratinocytes of OLP lesions (see Figure 1). This allows us to conclude that the changes in expression seen are likely to represent changes in the keratinocytes, at least for CXCL1, TLR1, and CD14.
TLRs are expressed in cells of the mucosal and dermal epithelium with the role of recognizing microbial invasions. Their engagement results in activation of these cells and their expression can be increased with activation of an innate immune response.6,14 TLR1 protein shows changes in distribution in the epithelium with the cutaneous version of lichen planus, though the overall mRNA level does not change.36. Functionally TLR1 can contribute to the TLR4 directed recognition of HCV proteins in human macrophage and in a kidney cell line.37 It is curious in that HCV has been suggested to play a role in OLP induction.4 Recent work showed TLR4 and TLR9 increases in oral keratinocytes in OLP lesions supporting the role for innate immunity component in this disease.38 CD14 functions as a co-receptor along with other pattern-recognition receptors of the innate immune system such as TLR2.39 We note that CD14 can augment the uptake of bacteria by macrophage and keratinocytes and facilitate the generation of an innate immune response by these cells.40 CXCL1 is produced in response to engagement of multiple TLRs in different tissues.41–44 For example, aperigillus fumigatos when recognized by TLR1 in mice macrophages and human cell lines induces the production of CXCL1.45 IL8 protein, which can be produced by monocytes, macrophage, neutrophils, fibroblasts, and keratinocytes, has been shown to be elevated in levels in the saliva and serum with OLP.22,23 IL8 can be induced, for example, by TLR3 activation in human esophagus keratinocytes in cell culture.46. Secreted IL8 plays a role in the neutrophil migration and activation and T lymphocyte migration. CXCL1 which functions in response to microbial invasion and tissue injury also is a potent neutrophil recruiter. Both CXCL1 and IL8 work to create inflammation.47,48
It was shown some time ago that the innate immune response can lead to the activation of adaptive immunity. Initially cytokines and chemokines released can attract various leukocytes including immature neutrophils and Langerhans cells that can perform antigen processing with the activation of Th1 and B cells.14 It has been shown for multiple hypersensitivities or diseases with auto-immune components such as asthma, diabetes, and psoriasis, that the presence of microbial products that activate the innate immune system or the artificial activation of the TLRs along with antigen exposure results in induction of these diseases in experimental models.9,10,12,13,23,49–51 The present study showed changes in innate immunity response gene expression in keratinocytes with OLP but did not rule out changes in other cells as being involved in the disease. Indeed there is evidence that mast cells, cells that can stimulate and inhibit the innate immune response by regulating acute inflammation and can stimulate T cell migration to a site, show increased activation with OLP. Mast cells also have a role in immune-related diseases such as rheumatoid arthritis, asthma, psorisaisis, etc.52,53
A key question is what is the microbe, or foreign entity, that induces the innate immune response by engaging the TLRs that trigger OLP. While HCV has received some attention as a possible initiator in the chain of events that lead to OLP, other sources are possible 54. A similar question is what cells change to initiate the major defects in OLP of T cell invasion and keratinocyte death. While our results show changes in gene expression that occur in keratinocytes, and others have indicated changes in mast cell activation with this disease, it is unclear what initiates the illness.52,53 Another major question is why there is recruitment of T cells but the general lack of acute inflammatory cells like neutrophils with OLP. If there is increased IL8 and CXCL1 protein, and not just RNA, at the OLP site then we would expect to see more acute inflammatory cells there, unless some other regulatory mechanism is preventing it. Finally, it is also important to determine if each OLP episode requires a new activation of the innate immune system in the mucosa or if a once-in-a-lifetime exposure is sufficient to induce the disease episodically. If the former is the explanation then drugs that block the innate immune response may show efficacy in treating this disease. Curiously, it has been shown that plaque control, one method of reducing infection associated with periodontal disease, simultaneously reduces symptoms of oral lichen planus 55,56
Supplementary Material
Acknowledgments
This work was supported by R21CA139137 to JS and RO3CA150076 to JS and GA. We are grateful to Drs. Douglas Damm, Dean White, and Craig Fowler of the Oral Pathology Service at the University of Kentucky for supplying cut and mounted tissue sections from biopsies of OLP mucosa.
Abbreviations
- OLP
oral lichen planus
- OLR
oral lichenoid reaction
- LO OCV
leave-one-out cross validation
- FDR
false discovery rate
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roopashree MR, Gondhalekar RV, Shashikanth MC, George J, Thippeswamy SH, Shukla A. Pathogenesis of oral lichen planus--a review. J Oral Pathol Med. 2010;39(10):729–734. doi: 10.1111/j.1600-0714.2010.00946.x. [DOI] [PubMed] [Google Scholar]
- 2.Scully C, Carrozzo M. Oral mucosal disease: Lichen planus. Br J Oral Maxillofac Surg. 2008;46(1):15–21. doi: 10.1016/j.bjoms.2007.07.199. [DOI] [PubMed] [Google Scholar]
- 3.Payeras MR, Cherubini K, Figueiredo MA, Salum FG. Oral lichen planus: Focus on etiopathogenesis. Arch Oral Biol. 2013;58(9):1057–1069. doi: 10.1016/j.archoralbio.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Lodi G, Scully C, Carrozzo M, Griffiths M, Sugerman PB, Thongprasom K. Current controversies in oral lichen planus: report of an international consensus meeting. Part 1. Viral infections and etiopathogenesis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2005;100(1):40–51. doi: 10.1016/j.tripleo.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 5.van der Waal I. Oral lichen planus and oral lichenoid lesions; a critical appraisal with emphasis on the diagnostic aspects. Med Oral Patol Oral Cir Bucal. 2009;14(7):E310–314. [PubMed] [Google Scholar]
- 6.Dwivedy A, Aich P. Importance of innate mucosal immunity and the promises it holds. Int J Gen Med. 2011;4:299–311. doi: 10.2147/IJGM.S17525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugerman PB, Savage NW, Walsh LJ, Zhao ZZ, Zhou XJ, Khan A, et al. The pathogenesis of oral lichen planus. Crit Rev Oral Biol Med. 2002;13(4):350–365. doi: 10.1177/154411130201300405. [DOI] [PubMed] [Google Scholar]
- 8.Kollisch G, Kalali BN, Voelcker V, Wallich R, Behrendt H, Ring J, et al. Various members of the Toll-like receptor family contribute to the innate immune response of human epidermal keratinocytes. Immunology. 2005;114(4):531–541. doi: 10.1111/j.1365-2567.2005.02122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirota JA, Knight DA. Human airway epithelial cell innate immunity: relevance to asthma. Curr Opin Immunol. 2012;24(6):740–746. doi: 10.1016/j.coi.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 10.McInturff JE, Modlin RL, Kim J. The role of toll-like receptors in the pathogenesis and treatment of dermatological disease. J Invest Dermatol. 2005;125(1):1–8. doi: 10.1111/j.0022-202X.2004.23459.x. [DOI] [PubMed] [Google Scholar]
- 11.Sigsgaard T, Hoffmann HJ, Thorne PS. The role of innate immunity in occupational allergy: recent findings. Curr Opin Allergy Clin Immunol. 2008;8(2):120–125. doi: 10.1097/ACI.0b013e3282f82492. [DOI] [PubMed] [Google Scholar]
- 12.Lang KS, Recher M, Junt T, Navarini AA, Harris NL, Freigang S, et al. Toll-like receptor engagement converts T-cell autoreactivity into overt autoimmune disease. Nat Med. 2005;11(2):138–145. doi: 10.1038/nm1176. [DOI] [PubMed] [Google Scholar]
- 13.Hammad H, Lambrecht BN. Dendritic cells and epithelial cells: linking innate and adaptive immunity in asthma. Nat Rev Immunol. 2008;8(3):193–204. doi: 10.1038/nri2275. [DOI] [PubMed] [Google Scholar]
- 14.Olson TS, Ley K. Chemokines and chemokine receptors in leukocyte trafficking. Am J Physiol Regul Integr Comp Physiol. 2002;283(1):R7–28. doi: 10.1152/ajpregu.00738.2001. [DOI] [PubMed] [Google Scholar]
- 15.Ranoa DR, Kelley SL, Tapping RI. Human LBP and CD14 independently deliver triacylated lipoproteins to TLR1 and TLR2 and enhance formation of the ternary signaling complex. J Biol Chem. 2013;288(14):9729–9741. doi: 10.1074/jbc.M113.453266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2(8):675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 17.Tao XA, Li CY, Xia J, Yang X, Chen XH, Jian YT, et al. Differential gene expression profiles of whole lesions from patients with oral lichen planus. J Oral Pathol Med. 2009;38(5):427–433. doi: 10.1111/j.1600-0714.2009.00764.x. [DOI] [PubMed] [Google Scholar]
- 18.Srinivasan M, Kodumudi KN, Zunt SL. Soluble CD14 and toll-like receptor-2 are potential salivary biomarkers for oral lichen planus and burning mouth syndrome. Clin Immunol. 2008;126(1):31–37. doi: 10.1016/j.clim.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Ohno S, Tateishi Y, Tatemoto Y, Morishita K, Sasabe E, Yamamoto T. Enhanced expression of Toll-like receptor 2 in lesional tissues and peripheral blood monocytes of patients with oral lichen planus. J Dermatol. 2011;38(4):335–344. doi: 10.1111/j.1346-8138.2010.00956.x. [DOI] [PubMed] [Google Scholar]
- 20.Simark-Mattsson C, Bergenholtz G, Jontell M, Eklund C, Seymour GJ, Sugerman PB, et al. Distribution of interleukin-2, -4, -10, tumour necrosis factor-alpha and transforming growth factor-beta mRNAs in oral lichen planus. Arch Oral Biol. 1999;44(6):499–507. doi: 10.1016/s0003-9969(99)00013-8. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto T, Osaki T. Characteristic cytokines generated by keratinocytes and mononuclear infiltrates in oral lichen planus. J Invest Dermatol. 1995;104(5):784–788. doi: 10.1111/1523-1747.ep12606990. [DOI] [PubMed] [Google Scholar]
- 22.Rhodus NL, Cheng B, Myers S, Bowles W, Ho V, Ondrey F. A comparison of the pro-inflammatory, NF-kappaB-dependent cytokines: TNF-alpha, IL-1-alpha, IL-6, and IL-8 in different oral fluids from oral lichen planus patients. Clin Immunol. 2005;114(3):278–283. doi: 10.1016/j.clim.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Sun A, Wang JT, Chia JS, Chiang CP. Serum interleukin-8 level is a more sensitive marker than serum interleukin-6 level in monitoring the disease activity of oral lichen planus. Br J Dermatol. 2005;152(6):1187–1192. doi: 10.1111/j.1365-2133.2005.06497.x. [DOI] [PubMed] [Google Scholar]
- 24.Al-Hashimi I, Schifter M, Lockhart PB, Wray D, Brennan M, Migliorati CA, et al. Oral lichen planus and oral lichenoid lesions: diagnostic and therapeutic considerations. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2007;103 (Suppl):S25, e21–12. doi: 10.1016/j.tripleo.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 25.Kolokythas A, Schwartz JL, Pytynia KB, Panda S, Yao M, Homann B, et al. Analysis of RNA from brush cytology detects changes in B2M, CYP1B1 and KRT17 levels with OSCC in tobacco users. Oral Oncol. 2011;47(6):532–536. doi: 10.1016/j.oraloncology.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 26.Schwartz JL, Panda S, Beam C, Bach LE, Adami GR. RNA from brush oral cytology to measure squamous cell carcinoma gene expression. J Oral Pathol Med. 2008;37(2):70–77. doi: 10.1111/j.1600-0714.2007.00596.x. [DOI] [PubMed] [Google Scholar]
- 27.Ding M, Xu JY, Fan Y. Altered expression of mRNA for HIF-1alpha and its target genes RTP801 and VEGF in patients with oral lichen planus. Oral Dis. 2010;16(3):299–304. doi: 10.1111/j.1601-0825.2009.01645.x. [DOI] [PubMed] [Google Scholar]
- 28.O'Flatharta C, Leader M, Kay E, Flint SR, Toner M, Robertson W, et al. Telomerase activity detected in oral lichen planus by RNA in situ hybridisation: not a marker for malignant transformation. J Clin Pathol. 2002;55(8):602–607. doi: 10.1136/jcp.55.8.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ban A, Marincsak R, Biro T, Perkecz A, Gomori E, Sandor K, et al. Upregulation of transient receptor potential vanilloid type-1 receptor expression in oral lichen planus. Neuroimmunomodulation. 2010;17(2):103–108. doi: 10.1159/000258693. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto T, Osaki T, Yoneda K, Ueta E. Cytokine production by keratinocytes and mononuclear infiltrates in oral lichen planus. J Oral Pathol Med. 1994;23(7):309–315. doi: 10.1111/j.1600-0714.1994.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 31.Ichimura M, Hiratsuka K, Ogura N, Utsunomiya T, Sakamaki H, Kondoh T, et al. Expression profile of chemokines and chemokine receptors in epithelial cell layers of oral lichen planus. J Oral Pathol Med. 2006;35(3):167–174. doi: 10.1111/j.1600-0714.2006.00402.x. [DOI] [PubMed] [Google Scholar]
- 32.Kolokythas A, Bosman MJ, Pytynia KB, Panda S, Sroussi HY, Dai Y, et al. A prototype tobacco-associated oral squamous cell carcinoma classifier using RNA from brush cytology. J Oral Pathol Med. 2013 doi: 10.1111/jop.12068. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Informatics. 2007;2:11–17. [PMC free article] [PubMed] [Google Scholar]
- 34.Kamal AM, Flower RJ, Perretti M. An overview of the effects of annexin 1 on cells involved in the inflammatory process. Mem Inst Oswaldo Cruz. 2005;100 (Suppl 1):39–47. doi: 10.1590/s0074-02762005000900008. [DOI] [PubMed] [Google Scholar]
- 35.Oliani SM, Paul-Clark MJ, Christian HC, Flower RJ, Perretti M. Neutrophil interaction with inflamed postcapillary venule endothelium alters annexin 1 expression. Am J Pathol. 2001;158(2):603–615. doi: 10.1016/S0002-9440(10)64002-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salem SA, Abu-Zeid RM, Nada OH. Immunohistochemical study of toll-like receptors 1 and 2 expression in cutaneous lichen planus lesions. Arch Dermatol Res. 2013;305(2):125–131. doi: 10.1007/s00403-012-1267-8. [DOI] [PubMed] [Google Scholar]
- 37.Chang S, Dolganiuc A, Szabo G. Toll-like receptors 1 and 6 are involved in TLR2-mediated macrophage activation by hepatitis C virus core and NS3 proteins. J Leukoc Biol. 2007;82(3):479–487. doi: 10.1189/jlb.0207128. [DOI] [PubMed] [Google Scholar]
- 38.Siponen M, Kauppila JH, Soini Y, Salo T. TLR4 and TLR9 are induced in oral lichen planus. J Oral Pathol Med. 2012;41(10):741–747. doi: 10.1111/j.1600-0714.2012.01169.x. [DOI] [PubMed] [Google Scholar]
- 39.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, Barton GM, et al. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147(4):868–880. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takayama A, Satoh A, Ngai T, Nishimura T, Ikawa K, Matsuyama T, et al. Augmentation of Actinobacillus actinomycetemcomitans invasion of human oral epithelial cells and up-regulation of interleukin-8 production by saliva CD14. Infect Immun. 2003;71(10):5598–5604. doi: 10.1128/IAI.71.10.5598-5604.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao J, Ren G, Gong Y, Dong S, Yin Y, Zhang L. Bronchial epithelial cells release IL-6, CXCL1 and CXCL8 upon mast cell interaction. Cytokine. 2011;56(3):823–831. doi: 10.1016/j.cyto.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 42.Kimura G, Ueda K, Eto S, Watanabe Y, Masuko T, Kusama T, et al. Toll-like receptor 3 stimulation causes corticosteroid-refractory airway neutrophilia and hyperresponsiveness in mice. Chest. 2013;144(1):99–105. doi: 10.1378/chest.12-2610. [DOI] [PubMed] [Google Scholar]
- 43.Leow-Dyke S, Allen C, Denes A, Nilsson O, Maysami S, Bowie AG, et al. Neuronal toll-like receptor 4 signaling induces brain endothelial activation and neutrophil transmigration in vitro. J Neuroinflammation. 2012;9:230. doi: 10.1186/1742-2094-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nischalke HD, Berger C, Luda C, Muller T, Berg T, Coenen M, et al. The CXCL1 rs4074 A allele is associated with enhanced CXCL1 responses to TLR2 ligands and predisposes to cirrhosis in HCV genotype 1-infected Caucasian patients. J Hepatol. 2012;56(4):758–764. doi: 10.1016/j.jhep.2011.10.019. [DOI] [PubMed] [Google Scholar]
- 45.Rubino I, Coste A, Le Roy D, Roger T, Jaton K, Boeckh M, et al. Species-specific recognition of Aspergillus fumigatus by Toll-like receptor 1 and Toll-like receptor 6. J Infect Dis. 2012;205(6):944–954. doi: 10.1093/infdis/jir882. [DOI] [PubMed] [Google Scholar]
- 46.Lim DM, Narasimhan S, Michaylira CZ, Wang ML. TLR3-mediated NF-{kappa}B signaling in human esophageal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2009;297(6):G1172–1180. doi: 10.1152/ajpgi.00065.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koelink PJ, Overbeek SA, Braber S, de Kruijf P, Folkerts G, Smit MJ, et al. Targeting chemokine receptors in chronic inflammatory diseases: an extensive review. Pharmacol Ther. 2012;133(1):1–18. doi: 10.1016/j.pharmthera.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Blanchet X, Langer M, Weber C, Koenen RR, von Hundelshausen P. Touch of chemokines. Front Immunol. 2012;3:175. doi: 10.3389/fimmu.2012.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker BS, Ovigne JM, Powles AV, Corcoran S, Fry L. Normal keratinocytes express Toll-like receptors (TLRs) 1, 2 and 5: modulation of TLR expression in chronic plaque psoriasis. Br J Dermatol. 2003;148(4):670–679. doi: 10.1046/j.1365-2133.2003.05287.x. [DOI] [PubMed] [Google Scholar]
- 50.Hari A, Flach TL, Shi Y, Mydlarski PR. Toll-like receptors: role in dermatological disease. Mediators Inflamm. 2010;2010:437246. doi: 10.1155/2010/437246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khoo JJ, Forster S, Mansell A. Toll-like receptors as interferon-regulated genes and their role in disease. J Interferon Cytokine Res. 2011;31(1):13–25. doi: 10.1089/jir.2010.0095. [DOI] [PubMed] [Google Scholar]
- 52.Ghalayani P, Jahanshahi G, Saberi Z. Degranulated mast cells and TNF-alpha in oral lichen planus and oral lichenoid reactions diseases. Adv Biomed Res. 2012;1:52. doi: 10.4103/2277-9175.100161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharma R, Sircar K, Singh S, Rastogi V. Role of mast cells in pathogenesis of oral lichen planus. J Oral Maxillofac Pathol. 2011;15(3):267–271. doi: 10.4103/0973-029X.86674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holmstrup P, Schiotz AW, Westergaard J. Effect of dental plaque control on gingival lichen planus. Oral Surg Oral Med Oral Pathol. 1990;69(5):585–590. doi: 10.1016/0030-4220(90)90241-j. [DOI] [PubMed] [Google Scholar]
- 56.Salgado DS, Jeremias F, Capela MV, Onofre MA, Massucato EM, Orrico SR. Plaque control improves the painful symptoms of oral lichen planus gingival lesions. A short-term study. J Oral Pathol Med. 2013 May; doi: 10.1111/jop.12093. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


