Abstract
Huntington’s disease is a devastating, incurable neurodegenerative disease affecting up to 12 per 100,000 patients worldwide. The disease is caused by a mutation in the Huntingtin (Htt) gene. There is interest in reducing mutant Huntingtin by targeting it at the mRNA level, but the maximum tolerable dose and long-term effects of such a treatment are unknown. Using a self-complementary AAV9 vector, we delivered a mir-155-based artificial miRNA under the control of the chicken β-actin or human U6 promoter. In mouse brain, the artificial miRNA reduced the human huntingtin mRNA by 50%. The U6, but not the CβA promoter, produced the artificial miRNA at supraphysiologic levels. Embedding the antisense strand in a U6-mir-30 scaffold reduced expression of the antisense strand but increased the sense strand. In mice treated with scAAV9-U6-mir-155-HTT or scAAV9-CβA-mir-155-HTT, activated microglia were present around the injection site 1 month post-injection. Six months post-injection, mice treated with scAAV9-CβA-mir-155-HTT were indistinguishable from controls. Those that received scAAV9-U6-mir-155-HTT showed behavioral abnormalities and striatal damage. In conclusion, miRNA backbone and promoter can be used together to modulate expression levels and strand selection of artificial miRNAs, and in brain, the CβA promoter can provide an effective and safe dose of a human huntingtin miRNA.
Keywords: Huntington’s disease, gene therapy, RNAi, AAV, miRNA, shRNA, huntingtin, HTT
Introduction
Huntington’s disease (HD) is a devastating inherited neurodegenerative disease caused by an expansion of the CAG repeat region in exon 1 of the huntingtin gene.1 Although huntingtin is expressed throughout the body,2 the polyglutamine expanded protein is especially toxic to medium spiny neurons in the striatum and their cortical connections.3 Patients struggle with emotional symptoms, including depression and anxiety, and with characteristic movement disturbances and chorea. Whereas the mutant protein exerts its toxic effects through myriad cellular pathways, elimination of huntingtin in the striatum has the potential to improve the lives of patients by treating some of the severe effects of the disease. Our goal is to reduce huntingtin in the brain using an artificial microRNA (miRNA) targeting human huntingtin mRNA, which can be delivered using a recombinant adeno-associated virus (AAV) vector and which will be safe and effective for long-term use.
RNAi-based therapy depends on successful delivery to striatal medium spiny neurons and to neurons in layers 5 and 6 of the cortex. Unfortunately, the blood-brain barrier limits the distribution of systemically delivered oligonucleotide therapeutics to the CNS. Following a single injection, antisense oligonucleotides targeting the human huntingtin mRNA can provide a sustained reduction in human huntingtin mRNA lasting up to 3 months.4 Nevertheless, therapeutics based on chemically synthesized oligonucleotides would necessitate repeated administration to maintain silencing. Recombinant AAV vectors can deliver an RNAi effector in the form of a short hairpin RNA (shRNA) or artificial miRNA and, in the non-dividing cells of the brain, a single dose is expected to last indefinitely.5 AAV vector-mediated RNAi has enormous potential for chronic, severe diseases such as HD.6, 7
Initial studies using AAV-mediated RNAi focused on shRNAs as the effector molecules. shRNAs are transcribed from polymerase III promoters (usually the U6 or H1 promoter) and are designed to bypass Drosha/DGCR8 cleavage. After export from the nucleus by exportin 5, shRNAs are cleaved by Dicer to form a duplex that can be loaded into Argonaute-RISC complexes. shRNAs are simple and effective and, with care, off-target effects due to improper strand loading or imprecise Dicer cleavage can be minimized.8, 9 However, shRNAs are often produced at extremely high levels, and toxicity due to oversaturation of the RNAi machinery has been reported in the liver, heart, and CNS.10, 11, 12, 13, 14, 15 In contrast to shRNAs, artificial miRNAs are designed to undergo cleavage both by Drosha/DGCR8 and Dicer. They can be transcribed from their own promoters, embedded in an intron, or located in the 3′-UTR of a protein-coding gene. Although in theory artificial miRNAs could also saturate the endogenous RNAi machinery11, in practice the incorporation of endogenous miRNA flanking arms reduces expression of the mature small RNA and improves the safety of vector delivered small RNAs.8
Results
Design and Selection of Huntingtin Targeting Artificial miRNAs
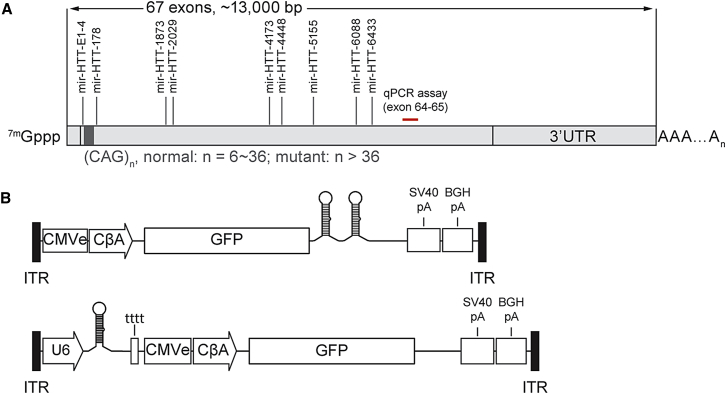
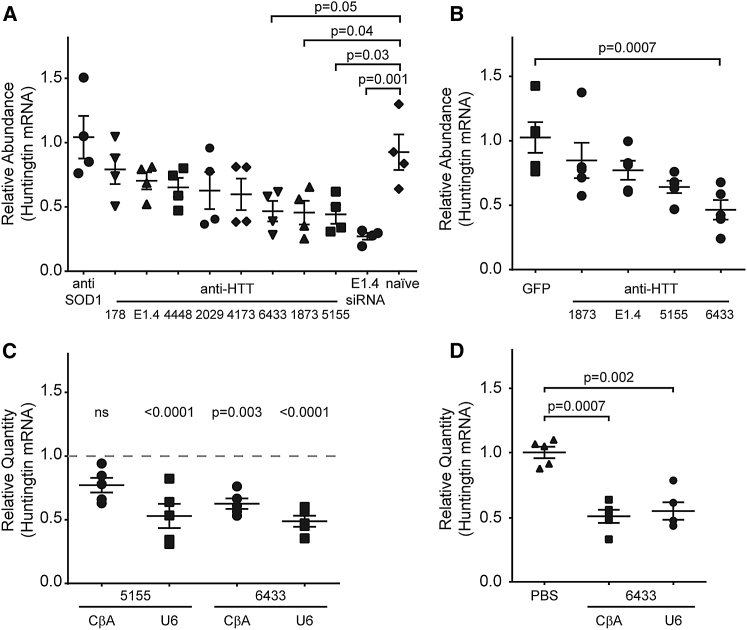
We selected nine sequences targeting the human huntingtin mRNA (Table 1; Figure 1A). These sequences were selected on the basis of known targeting rules.16 We cloned two copies of the artificial miRNA in tandem into a backbone on the basis of the endogenous miRNA 155 and placed the entire artificial miRNA into the 3′-UTR of EGFP (Figure 1B, top), which was expressed under the control of a chicken beta-actin promoter. The resulting plasmids were transfected into HeLa cells, and 48 hr later, we harvested the cells and measured the levels of endogenous huntingtin mRNA using a qRT-PCR probe targeting the boundary between exons 64 and 65 (Figure 1A). We have previously shown that probes upstream and downstream of this target site report consistent levels of silencing of Yac128 mRNA using assays upstream and downstream of the artificial miRNA cleavage site.17 Three of the nine artificial miRNAs reduced huntingtin by >50% (Figure 2A).
Table 1.
Predicted Antisense Sequences Targeting Human Huntingtin
| Name | Predicted Antisense Sequence |
|---|---|
| miR-E14-anti-HTT | 5′-UUCAUCAGCUUUUCCAGGGUC-3′ |
| miR-178-anti-HTT | 5′-UUGAGGCAGCAGCGGCUGUGC-3′ |
| miR-1873-anti-HTT | 5′-UAAAUGUGCCUGUUGAAGGGC-3′ |
| miR-2029-anti-HTT | 5′-AAGAGGUGCAGAGUCAUCAUC-3′ |
| miR-4173-anti-HTT | 5′-UUCUGGAGGACAUCAAACCAU-3′ |
| miR-4448-anti-HTT | 5′-UGAACUGGCCCACUUCAAUGU-3′ |
| miR-5155-anti-HTT | 5′-UAGCGUUGAAGUACUGUCCCC-3′ |
| miR-6088-anti-HTT | 5′-UUCCAUUGGCAACUGGGCCAU-3′ |
| miR-6433-anti-HTT | 5′-UAAGCAUGGAGCUAGCAGGCU-3′ |
Figure 1.
Design of Artificial miRNAs Targeting Human Huntingtin
(A) Position of target sites on the human huntingtin mRNA. Target sites are distributed throughout the coding region. The location of the qPCR assay, which spans the junction between exons 64 and 65, is also depicted. (B) AAV9 constructs expressing a miRNA from the CβA (polII) and U6 promoters. The CβA promoter-driven miRNA is located in the 3′-UTR of the GFP gene (top), whereas the construct containing the U6 promoter-driven artificial miRNA co-expresses GFP from a separate promoter (bottom).
Figure 2.
Artificial miRNAs Targeting Human Huntingtin Reduce the Huntingtin mRNA in Cell Culture and In Vivo
Expressing an artificial miRNA from the U6 promoter does not improve silencing of huntingtin in the mouse striatum. (A) HeLa cells were transfected with plasmids expressing artificial miRNAs targeting human huntingtin, and the huntingtin mRNA levels were measured after 48 hr by qPCR. Huntingtin expression was normalized to HPRT to account for well-to-well variation in cell number and is expressed relative to the naive cells. Error bars represent SE of three independent experiments. The negative control is an anti-SOD1 miRNA, and the positive control is an siRNA (E1-4) targeting exon 1 of the huntingtin mRNA. Data were analyzed by one-way ANOVA with Bonferroni comparison. Three artificial miRNAs, 5155 (p = 0.03), 1873 (p = 0.04), and 6433 (p = 0.05), significantly reduce the native huntingtin mRNA relative to untreated HeLa cells. (B) Top candidates were selected for in vivo testing on the basis of the results in cell culture. Mice (n = 5/group) were injected unilaterally in the striatum. One month post-injection, the striatum was harvested, and GFP expression was verified. Data are normalized to HPRT and expressed relative to the GFP-only control. Only scAAV9-CβA-anti-6433 reduced human huntingtin significantly (p = 0.0007, one-way ANOVA) relative to the GFP control. (C) Relative quantity of huntingtin mRNA in the injected striatum 1 month after injection of the U6 and CβA promoter-driven artificial miRNAs targeting sites 5155 (left) and 6433 (right). Data are expressed relative to the non-injected side (n = 5 mice/group). scAAV9-U6-anti-5155, scAAV9-U6-anti-6433, and scAAV9-CβA-anti-6433 reduce human huntingtin significantly in the striatum (p < 0.0001, p = 0.003, and p < 0.0001 respectively, two-way ANOVA). (D) Relative quantity of huntingtin mRNA in mice 1 month after injection with the U6 and CβA promoter-driven miRNA targeting site 6433. In this experiment, a fourth group of mice were injected with PBS (vehicle) only, and data are expressed relative to this group (n = 5 mice/group). Again, both scAAV9-CβA-anti-6433 and scAAV9-U6-anti-6433 reduce human huntingtin significantly relative to PBS injection (p = 0.0007 and p = 0.002, respectively), but there is no significant difference between the two promoters. Error bars in all panels represent SE of independent replicates.
We selected the three best sequences from our initial screen for in vivo experiments. In addition, we included an artificial miRNA on the basis of a previously published small interfering RNA (siRNA) (E1.4).18 We packaged these candidates into a self-complementary AAV9 vector and injected it directly into the striatum of transgenic mice expressing human huntingtin with a stretch of approximately 128 polyglutamine encoding repeats (Yac128 mice).19 At a vector dose of 3.0 × 109 vg/striatum, GFP staining was present throughout the striatum, and human huntingtin mRNA was significantly reduced in mice treated with either scAAV9-CβA-anti-HTT-6433 (p = 0.0007) (Figure 2B) compared with mice treated with an scAAV9-GFP.
Expressing the Artificial miRNA from the CβA Promoter Is Sufficient for Maximal Silencing of Huntingtin mRNA
To investigate the possibility that increasing the expression further would improve silencing, we cloned a single copy of the most potent miRNA into an AAV9 vector under the control of the U6 promoter (Figure 1B, bottom). Mice were injected unilaterally with the original two-copy scAAV9-CβA-anti-HTT-6433, scAAV9-CβA-anti-HTT-5155, scAAV9-U6-anti-HTT-6433, or scAAV9-U6-anti-HTT-5155. One month later, we harvested the striatum and confirmed GFP expression. We measured the levels of huntingtin mRNA by qRT-PCR. There was a significant reduction in human huntingtin in mice treated with scAAV9-U6-anti-HTT-5155, scAAV9-CβA-anti-HTT-6433, or scAAV9-U6-anti-HTT-6433 but not in those treated with scAAV9-CβA-anti-HTT-5155 compared with the contralateral (non-injected) side (Figure 2C). We compared the relative quantity of the mature artificial miRNA guide strand by qPCR. Expression from scAAV9-U6-anti-HTT-6433 was about 150 times higher than from scAAV9-CβA-anti-HTT-6433 (Figure S1).
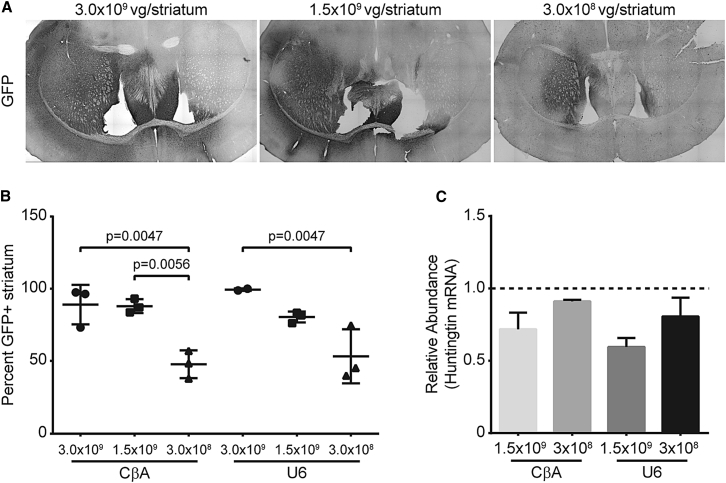
Using the contralateral side as a control for each animal reduces the inter-animal variability by controlling for animal to animal variation in huntingtin expression. This approach assumes that there is no spread of AAV or artificial miRNA from the injected to the non-injected, contralateral side. In mice injected with scAAV9-GFP unilaterally, we often see a small number of GFP-positive neurons on the contralateral side. Therefore, using the contralateral side as the control may underestimate silencing. To eliminate this potential confounding effect, we repeated the experiment using a group of animals injected with PBS as the control. We confirmed that both scAAV9-CβA-anti-HTT-6433 and scAAV9-U6-anti-HTT-6433 reduced huntingtin mRNA by approximately 50% in the striatum (Figure 2D). We did not observe a difference in silencing between the two studies, suggesting that spread to the contralateral side is insufficient to produce silencing. To determine if we could achieve huntingtin silencing with a lower vector dose, we injected mice (n = 3/group) with vector diluted by 0.5 log (final dose 1.5 × 109 vg/striatum) and 1 full log (final dose 3.0 × 108 vg/striatum). GFP is present in 89% of the striatum with the highest dose of the vector, but reducing the dose of the vector results in reduced spread (Figures 3A and 3B) and decreased silencing of human huntingtin mRNA (Figure 3C).
Figure 3.
Reducing the Vector Dose Results in Reduced Spread and Reduced mRNA Silencing in the Mouse Striatum
(A) Representative images of GFP spread in mice injected with a vector encoding both the huntingtin-targeting artificial miRNA and EGFP at three different doses. (B) Quantification of GFP spread in the mouse striatum 1 month after injection (n = 3/group). ImageJ was used to outline the GFP-positive area and the striatum and to calculate the percentage of the striatum that contained GFP. (C) qRT-PCR to measure human huntingtin mRNA in the striatum of Yac128 mice (n = 3/group) 1 month post-injection. Data for 3.0 × 109 are shown in Figure 2B. Error bars in (B) and (C) represent SE of independent replicates.
Expression of the Artificial miRNA Targeting Huntingtin from a U6 Promoter Results in Expression of Multiple Small RNA Species at Levels Comparable with Those of Endogenous miRNAs
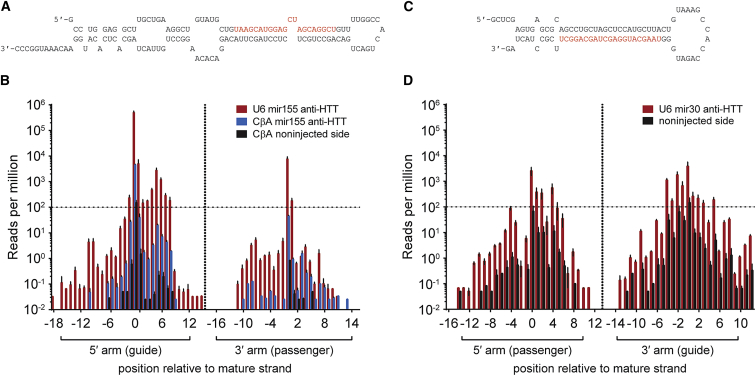
To examine whether the same small RNA species were produced from processing of both the U6 and CβA promoter-driven artificial miRNA, we injected groups of mice unilaterally with either scAAV9-CβA-anti-HTT-6433 or scAAV9-U6-anti-HTT-6433. We cloned and sequenced the total 18- to 30-nucleotide small RNAs at 2 weeks post-injection. We mapped the sequences back onto the predicted hairpin structure of the artificial miRNA (Figure 4A). In both scAAV9-CβA-anti-HTT-6433- and scAAV9-U6-anti-HTT-6433-injected groups, 96% of the sequences mapping to the AAV genome were the expected small RNA product. Imprecise Dicer or Drosha cleavage of miRNA precursors can result in small RNAs with heterogeneous 5′ ends. These noncanonical small RNA isoforms (isomirs) can have a different target profile than the canonical isoform. Therefore, we looked at the distribution of 5′ ends along the sequence of the pre-miRNA. Fewer than 4% of the small RNAs produced from scAAV9-CβA-anti-HTT-6433 and scAAV9-U6-anti-HTT-6433 were noncanonical isomirs (Figure 4B; Table 2). However, because of the high levels of expression produced by the U6 promoter, it should be noted that these represent a much higher proportion of the total small RNA pool in the scAAV9-U6-anti-HTT-6433 group than in those mice injected with scAAV9-CβA-anti-HTT-6433. In the group injected with the U6 promoter-driven artificial miRNA, however, the huntingtin-targeting sequence dominated the sequencing results, accounting for half (50%) of the combined genome matching and AAV vector matching sequences, whereas in the mice injected with the CβA vector, only 5% of the sequences matched the vector-encoded small RNA (Figure 4B). This finding means that small RNAs with alternative seed sequences, including the sense strand, +1 and −1 products, could be present at levels comparable with functional endogenous miRNAs.
Figure 4.
Expression of Artificial mir-155-Based miRNA from a U6 Promoter Results in Overexpression of the Huntingtin-Targeting Guide Strand, whereas the mir-30-Based Artificial miRNA Produces Both the Guide and Other Sequences
Predicted hairpin structures of the mir-155-6433 (A) and mir-30-6433 (C) hairpins. (B) Start positions of reads mapping to the huntingtin-targeting mir-155-based artificial miRNA hairpin under control of the U6 (red) and CβA (blue) promoters or non-injected side of the CβA mice (black). Positions are reported relative to the mature strand, and reads are normalized to the total number of endogenous miRNA mapped in each sample. The horizontal line represents the background levels of the artificial miRNA found in control samples. Data were analyzed using n = 3 mice/group. (D) Start positions of reads mapping to the huntingtin-targeting artificial miRNA embedded in a mir-30 backbone and expressed from a U6 promoter (injected side, red; non-injected side, black). Error bars in all panels represent SE of independent replicates.
Table 2.
Percentage of Total Hairpin Mapping Reads for Each Seed Sequence Represented in the Small RNA Sequencing Results
| Seed Sequence (2–8) | U6-Anti-HTT-6433 | CβA-Anti-HTT-6433 |
|---|---|---|
| GUAAGCA (−2) | 0.01 | 0.01 |
| UAAGCAU (−1) | 0.05 | 0.57 |
| AAGCAUG (antisense) | 96.43 | 96.68 |
| AGCAUGG (+1) | 0.97 | 0.85 |
| GCAUGGA (+2) | 0.03 | 0.04 |
| CAUGGAG (+3) | 0.03 | 0.02 |
| AUGGAGC (+4) | 0.09 | 0.07 |
| UGGAGCU (+5) | 0.53 | 0.42 |
| GGAGCUA (+6) | 0.23 | 0.15 |
| GAGCUAG (−2) | 0.06 | 0.09 |
| AGCUAGC (−1) | 0.04 | 0.04 |
| AGCCUGC (sense) | 1.49 | 0.97 |
| GCCUGCU (+1) | 0.03 | 0.02 |
| CUGCUCU (+2) | 0.00 | 0.03 |
| UGCUCUC (+3) | 0.00 | 0.01 |
| Total | 99.99 | 99.97 |
Endogenous miRNA 30 sequences are commonly used as a scaffold for artificial miRNA.8, 20 To determine if the isomir profiles derived from this scaffold were more favorable, we embedded the anti-HTT-6433 sequence in a miR-30 backbone and injected into 10-week-old Yac128 mice (Figure 4C). The mir-30 scaffold produces levels of the mature artificial miRNA comparable with those produced by the CβA promoter (Figure 4D) and also reduces human huntingtin by close to 50% (Figure S2). Unlike the mir-155 scaffold, the mir-30 scaffold produces the mature sense (passenger) strand at levels comparable with the antisense (guide) strand. When designing artificial miRNA, target selection can be critical to introducing the correct asymmetry. Here, miRNA backbone can be used as an additional method to control the asymmetry of artificial miRNA. For this huntingtin-targeting sequence, the combination of CβA promoter with mir-155 backbone is unique, as it is the only combination that produces only the intended antisense strand above background (Figure 4B).
Long-Term Striatal Expression of mir-HTT-6433 from a U6 Promoter Is Toxic
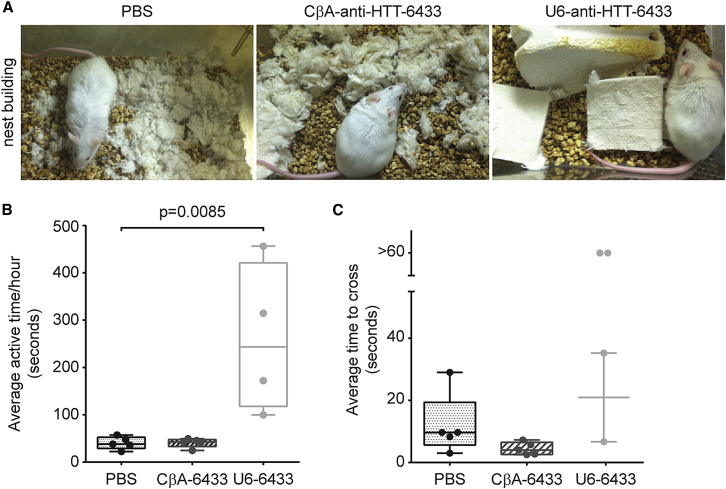
Having established that both scAAV9-U6-anti-HTT-6433 and scAAV9-CbA-anti-HTT-6433 silence human huntingtin in the short term, we wished to evaluate the duration of effect and long-term consequences of expression and overexpression of the huntingtin-targeting artificial miRNA. We injected scAAV9-U6-anti-HTT-6433 or scAAV9-CβA-anti-HTT-6433 unilaterally into the striatum of Yac128 mice. Six months after injection, we noticed that the mice injected with scAAV9-U6-anti-HTT-6433 appeared behaviorally abnormal. When a new nestlet is placed in the cage, normal mice will shred the material, producing a nest. The mice injected with scAAV9-U6-anti-HTT-6433 appeared to leave the bedding material untouched. To document this, we replaced the nestlets in each cage. Twenty-four hours later, the mice treated with scAAV9-U6-anti-HTT-6433 had not used the new nestlets, whereas the bedding of PBS and scAAV9-CβA-anti-HTT-6433 mice looked as expected (Figure 5A). Using an automated home-cage monitoring system, we recorded the mice for 24 hr. This system produces unbiased tracking of the movements of the mice over the course 24 hr, without interference from the experimenter. Individual mice are placed in the cage, and the computer records the amount of time they spend moving versus remaining stationary. The mice treated with scAAV9-U6-anti-HTT-6433 spent significantly more time moving around their home cage than mice treated with PBS or with scAAV9-CβA-anti-HTT-6433 (Figure 5B). Finally, we measured the average time it took for the mice to cross an elevated beam. For this test, we require that the mice complete the beam crossing three times. Whereas both scAAV9-CβA-anti-HTT-6433-injected and PBS-injected mice crossed the beam easily, two of the four remaining mice in the scAAV9-U6-anti-HTT-6433 group were unable to successfully cross, either jumping or falling off the beam (Figure 5C). We repeated this experiment with a larger group of animals, testing them on the beam at regular intervals. Mice injected with scAAV-U6-anti-HTT-6433 at 6 weeks of age were unable to cross the beam by 4 months post-injection (Figure S3A). Older Yac128 mice (injected at 7 months of age) exhibited an age-related increase in time to cross the beam. This increase was present in both naive mice and in mice treated with AAV9-CβA-anti-HTT-6433. Like the younger animals, older mice treated with scAAV9-U6-anti-HTT-6433 showed a deterioration of beam crossing 4 months post-injection (Figure S3B). By 6 months post-injection, three of five animals injected with AAV9-U6-anti-HTT-6433 failed to cross the beam in under 1 min, whereas all of the mice in the AAV9-CβA-anti-HTT-6433 and naive groups crossed within that time.
Figure 5.
Long-Term Striatal Expression of mir-HTT-6433 from a U6 Promoter Causes Behavioral Abnormalities
(A) Six months post-injection, mice injected with the U6 promoter-driven mir-HTT-6433 failed to make nests. When new nestlets are placed in their cages, mice injected intrastriatally with PBS (left) or with CβA-mir-HTT-6433 (center) shred the bedding, making a nest. In contrast, mice injected with U6-mir-HTT-6433 do not. Pictures were taken 24 hr after placing new nestlets in the cage. (B) Home cage monitoring of Yac128 mice treated with PBS, CBA-mir-HTT-6433, or U6-mir-HTT-6433. Animals were individually placed in an automated home cage monitoring system. The amount of time spent moving around the cage was recorded for 24–27 hr. Average time per hour was calculated by dividing the total amount of time by the number of hours of recording (n = 4 U6-mir-HTT-6433, n = 5 PBS and CβA-mir-HTT-6433). (C) Animals were trained to cross a round beam and enter a darkened box. For each mouse, the average time to cross from one end was calculated for three recorded trials. If a mouse failed to cross within 60 s, we recorded the time as >60 s. Error bars in (B) and (C) represent SE of independent replicates.
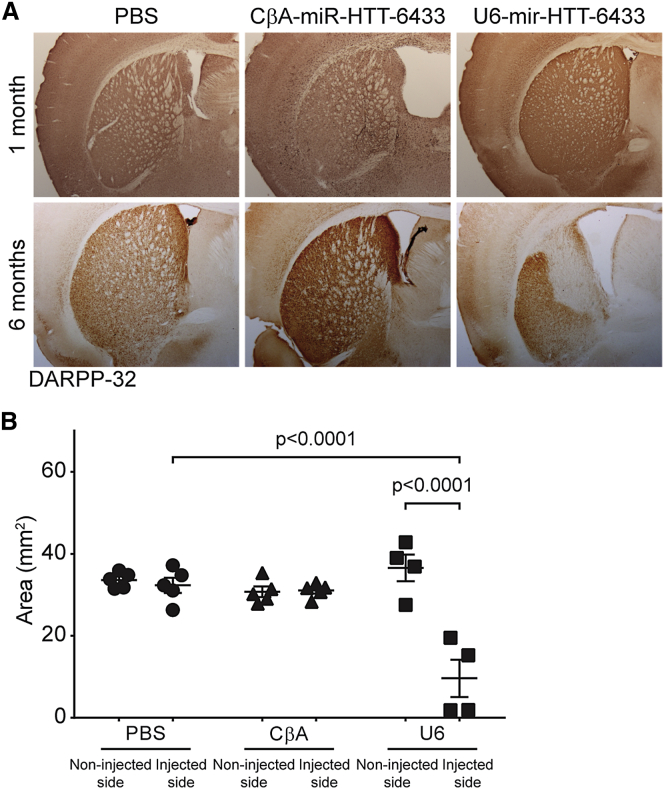
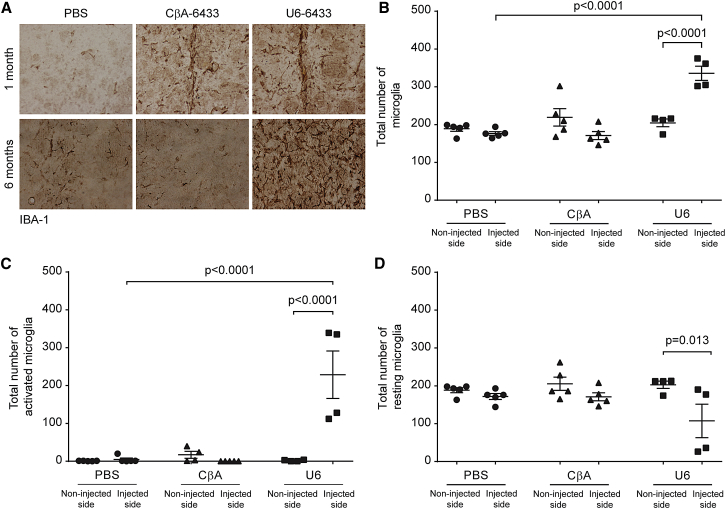
Neuropathological findings explained the behavioral outcomes. On the injected side, the scAAV9-U6-anti-HTT-6433 mice showed enlargement of the ventricle, loss of DARPP-32-positive neurons, and striatal shrinkage (Figure 6). It has previously been shown that Yac128 mice display increased numbers of activated microglia compared with wild-type mice.21 We expected, on the basis of the loss of DARPP-32 neurons, that the mice injected with scAAV9-U6-anti-HTT-6433 would exhibit increased microglial activation, but we were curious whether the scAAV9-CβA-anti-HTT-6433 mice would also exhibit signs of immune activation. We classified Iba-1 positive cells as resting or active on the basis of morphology.22 One month after injection, we were able to locate the needle track in mice injected with both scAAV9-U6-anti-HTT-6433 and scAAV9-CβA-anti-HTT-6433 (Figure 7A, top). Five months later, this was no longer true in the CβA-treated mice. Meanwhile, in the surviving striatum, the scAAV9-U6-anti-HTT-6433 mice exhibited increased Iba1 staining (Figure 7A, bottom), an increase in total and activated microglia, and a decrease in the number of resting microglia (Figures 7B–7D). This suggests an ongoing innate immune response, which could be a result of or participate in striatal cell death. In contrast, by 6 months post-injection, the mice treated with scAAV9-CβA-anti-HTT-6433 do not show an increase in microglial activation. We considered the possibility that Yac128 mice, which may have a “primed” immune system, may be more susceptible to toxicity by miRNA overexpression than wild-type mice. To determine if toxicity was dependent on the presence of mutant huntingtin, we injected wild-type C57BL/6 mice and FVB mice with the same vectors and assessed the consequences of the U6-driven miRNA in that context. In FVB mice, the effect was similar to that in Yac128 mice with rapid degeneration on the beam and severely enlarged ventricles (data not shown). In C57BL6 mice, the effect was present but less pronounced (Figure S3). Although there was an initial increase in time to cross the beam in the U6 cohort, at the study endpoint, there was no significant difference between groups (Figure S4A). Striatal shrinkage was also less severe in the C57BL6 mice (Figure S4B).
Figure 6.
Long-Term Expression of Anti-HTT-6433 from a U6 Promoter Causes Striatal Shrinkage
(A) Representative images of DARPP-32 staining on the injected side in Yac128 mice at 1 (top) and 6 (bottom) months post-injection. (B) Quantification of DARPP-32 positive area 6 months post-injection. Error bars in (B) represent SE of independent replicates.
Figure 7.
Long-Term Expression of mir-HTT-6433 from a U6 Promoter Causes Persistent Activation of Microglia
(A) Representative images of Iba1 staining on the injected side in Yac128 mice at 1 (top) and 6 (bottom) months post-injection. When possible, images were taken at the site of injection. Quantification of total (B), activated (C), and resting (D) microglia at 6 months post-injection. Error bars in (B)–(D) represent SE of independent replicates.
We examined the small RNA sequencing for clues to explain the toxicity. At 1 week post-injection, the mice treated with CβA-anti-HTT-6433 produced 32 endogenous miRNAs whose expression was changed greater than 2-fold compared with PBS-injected mice (Table S1). Of these, 18 (56%) were downregulated, whereas 14 (44%) were upregulated. In the mice treated with U6-anti-HTT-6433, 58 miRNAs were significantly altered, with 33 being downregulated (57%) and 25 (43%) being upregulated. We also performed RNA sequencing (RNA-seq) on total RNA. In the mice injected with AAV9-U6-anti-HTT-6433, 44 transcripts were significantly downregulated, while 30 were significantly upregulated compared with the naive control. In the AAV-CβA-anti-HTT-6433 group, 12 transcripts were downregulated and 4 were upregulated. Overall, transcripts containing a seed sequence target are downregulated in the scAAV-U6-anti-HTT-6433 group but not the scAAV9-CβA-anti-HTT-6433 group (Figures S5B and S5F). Those originating from the +1 and −1 positions and the sense strand (Figures S5A, S5C, S5D, S5E, S5G, and S5H) were not altered. This result suggests that off-target effects are exacerbated by overexpression and are likely to be due to overexpression of the intended huntingtin-targeting artificial guide strand rather than to the presence of other small RNAs with alternative seed sequences.
Discussion
We have achieved a higher level of expression of the vector-encoded small RNA than has been previously described for artificial miRNAs. We show that the mir-155 backbone produces mostly the intended small RNA species, reducing the chance of off-target effects due to improper strand loading or imprecise processing. We also show that the mir-155-based artificial miRNA using a CβA promoter reduces human huntingtin by 50% at 1 month and causes no overt toxicity up to 6 months. Using a U6 promoter increases the levels of this artificial miRNA guide strand but does not result in additional huntingtin lowering. Expression of the anti-HTT artificial miRNA from a CβA promoter in the context of the scAAV9 and the mir-155 backbone is sufficient to achieve maximum silencing from a direct injection. We have previously reported that the same artificial miRNA reduces human huntingtin by up to 80% in liver.17 Unlike the liver, the striatum contains a highly heterogeneous population of cell types, including medium spiny neurons, cholinergic and GABAergic interneurons, astrocytes, oligodendrocytes, and microglia. In homozygous Q140 knockin mice, individual medium spiny neurons show variable levels of huntingtin mRNA expression.23 Treatment with an artificial miRNA targeting mouse huntingtin shifts the distribution, but does not eliminate huntingtin mRNA from DARPP32-positive medium spiny neurons.23 This suggests that a relatively small amount of huntingtin mRNA is present in non-neuronal cell types and hints at the presence of a miRNA-inaccessible pool of huntingtin mRNA in medium spiny neurons. Additional delivery routes, AAV optimization, improved formulation, and additional therapeutic moieties could improve silencing by improving delivery to other cell types, increasing distribution and targeting inaccessible huntingtin mRNA.
There are several possible explanations for the observed toxicity of the AAV9-U6-anti-HTT-6433. First, overexpression of the miR-155-based hairpin results not only in overexpression of the intended mature artificial miRNA strand, but it also reveals additional processing products. These additional products are expressed at levels comparable with functional endogenous and artificial miRNAs and increase the potential for sequence-specific off-target effects. However, our results indicate that only the artificial miRNA guide strand is expressed at a high enough level to produce a detectable global pattern of seed-mediated silencing. So, although we have been unable to find a single off-target mRNA that would explain the observed toxicity, overexpression of the guide strand may in fact be revealing an otherwise hidden off-target effect. However, species-specific toxicities due to seed-mediated off-target effects have been reported for other huntingtin-targeting artificial miRNAs.24 Second, overexpression of an exogenous miRNA might disrupt the balance of endogenous miRNAs. In liver, expression of an shRNA causes a reduction in the predominant liver miRNA, mir-122.15 In brain, there is no corresponding dominant miRNA; nonetheless, it is conceivable that overexpression of an artificial miRNA causes a disruption in endogenous miRNA in brain. Additional explanations for the observed toxicity include saturation of various components of the cellular RNAi machinery (Argonaute2, exportin-5) and an innate immune response. We do see evidence for a long-lived immune response in the form of activated microglia that persist up to 1 month post-injection, even in mice treated with AAV9-CβA-anti-HTT-6433. These toxicities may be exacerbated by ongoing disease processes or the presence of a toxic disease-related transgene. The fact that some strains of mice exhibit accelerated pathology should encourage us to consider factors that might influence susceptibility to such toxicity and caution when transitioning vectors from one animal species or model to another.
In non-human primates, partial silencing of huntingtin with a U6 promoter-driven, mir-30-based artificial miRNA appears safe up to 6 weeks,25 and intrathecal delivery of a U6 promoter-driven artificial miRNA targeting SOD1 is safe in non-human primates.26. It is possible that the toxicity of the vector carrying the U6 promoter is exacerbated by the method of delivery, particularly in the small brain of a mouse. When we deliver an AAV vector by direct injection to the striatum, there is a limited transport of vector away from the injection site, and the local concentration is high. In some cases, a vector with a weaker promoter may be sufficient to achieve maximum silencing. Likewise, the mir-30 backbone, which seems to produce lower levels of the mature artificial miRNA, may also be preferable. As we move toward other modes of delivery in which the vector distributes to a wider area, individual cells will receive fewer copies of the AAV, and high levels expression of the artificial miRNA produced from a strong promoter and optimized backbone may be necessary. In our hands, the mir-155 backbone produces higher levels of the mature artificial miRNA than the mir-30 backbone. The level and expression pattern of the mature artificial miRNA can be further tuned by promoter choice. Such tuning may be necessary as we explore additional AAV capsids and delivery methods. Promoter, backbone, AAV serotype, and injection method should be considered together when attempting to deliver vector-mediated RNAi to the brain.
Finally, we show using the same anti-HTT-6433 sequence that the miRNA backbone can influence the strand biasing of artificial miRNA processing. When we placed our targeting sequence in the mir-30 backbone, the sense and antisense strands were produced at very similar levels. In contrast, in the context of the mir-155 backbone, there was strong asymmetry, and the antisense strand predominated. Asymmetric strand selection of miRNA is due primarily to thermodynamic instability at the 5′ end of the guide/antisense strand. When a different, luciferase-targeting sequence is embedded in the mir-155 backbone, both strands were produced in roughly equal proportions.27 However, components of the pri-miRNA may also influence asymmetry. Slight differences in the construction of the miRNA backbone are common. These differences likely affect processing efficiency and fidelity of artificial miRNAs, while interactions between the targeting sequence and backbone determine strand selection. A comprehensive analysis of pri-miRNA features28 was used to design from scratch a miRNA scaffold that is processed as efficiently as the most commonly used endogenously derived miRNA scaffolds. This should allow further refinement and improvement of artificial miRNA.
Materials and Methods
Cell Culture and Screening Assays
HeLa cells were maintained in DMEM, high glucose with 10% heat-inactivated fetal bovine serum (FBS), and 1% penicillin streptomycin (ThermoFisher). Twenty-four hours before transfection, cells were seeded onto six-well plates at 0.8 × 106 to 1.0 × 106 cells/well. On the day of transfection, we first replaced the growth medium with 1.6 mL of Opti-MEM (ThermoFisher). Plasmids were transfected in triplicate using 2 μL/well of DharmaFECT Duo (Dharmacon). Each well received 0.5 μg of plasmid DNA. Forty-eight hours after transfection, the cells were harvested, and total RNA was extracted using the MirVana RNA isolation kit. We made cDNA using 1 μg of RNA per reaction using oligo-dT and Superscript III (Invitrogen). Htt mRNA was measured using TaqMan assay # (ThermoFisher). Relative levels of Htt mRNA were calculated using the ΔΔC(T) method,29 with human hypoxanthine-guanine phosphoribosyltransferase (Hprt) as the housekeeping gene. Each experiment was performed three times using three independent cell passages.
Mouse Housing, Injections, and Maintenance
YAC128 and wild-type FVB mice were purchased from The Jackson Laboratory. They were bred on the FVB background by mating wild-type male mice with YAC128 females. The resulting heterozygous YAC128 and wild-type mice were maintained on a 12:12 light schedule and were given access to food and water ad libitum. All animals were maintained and used according to the Institutional Animal Care and Use Committee guidelines of University of Massachusetts Medical School (docket A978-12 or A978-15). Genotypes were verified by PCR of DNA extracted from tail snips or ear punches. Mice were injected with selected AAV directly into the striatum by means of a small animal stereotax SAS-4100 (ASI Instruments) aided by UMPC3 or UMPC4 microinjectors (World Precision Instruments). Mice were anesthetized with 284 mg/kg of tribromoethanol and placed in the stereotax. Surgery was performed using the bregma as the zero point, measuring anterior 1.0 mm, lateral 2.0 mm, and lowering a 33 gauge needle 3.0 mm into the striatum. The pumps were set to deliver 3.0 μL at a rate of 125 nL/min. After the injections the mice were allowed to recover on a warming pad and then placed back in their cages in the housing area. Females were housed together in groups of three and males were housed separately to prevent fighting.
Tissue Extraction
At the appropriate time point, mice were sacrificed and tissue was extracted for RNA analysis or immunohistochemistry. For RNA extraction, mice were anesthetized and killed by cervical dislocation. Brains were removed, and the striatum was dissected out manually, removing as much white matter as possible. To ensure that only animals in which the vector was delivered to the striatum were analyzed, GFP expression was visualized by placing the entire tissue chunk under the fluorescent scope. Tissue was then placed in RNALater (Ambion) for 24 hr, at which point the RNAlater was removed and the tissue was stored at −80°C until use. Separate mice were used for immunocytochemistry. These mice were deeply anesthetized and perfused intracardially first with saline and then with 4% paraformaldehyde. They were postfixed overnight in cold 2% paraformaldehyde and then stored in PBS at 4°C. Coronal sections were made by slicing 40 μm sections on the Leica VT1000s vibratome.
Mouse Behaviors
Beam Walking
Mice were trained to cross a beam 2 cm in diameter. After training, we recorded the mice as they crossed from one end of the beam to the other. We recorded three trials per mouse. From the recording we measured the amount of time it took for the mice to cross from mark on one end of the beam to the other. The investigator was not blinded to the treatment group of each mouse.
Home Cage Activity
Mice were placed singly in an automated home cage phenotyping scanning system (Clever Sys) for 26 hr. This system records and automatically calculates the activity of the mouse during both the active and inactive period without the presence or interference of the investigator. To calculate the average active time per hour, we first removed the first hour of data, during which the mouse acclimates to the new environment. We then divided the total time spent walking by the total time.
Immunohistochemistry and Quantification
Fixed tissue slices were blocked with 3% hydrogen peroxide for 3 min and then incubated with 0.5% triton x for 20 min. Immunocytochemistry was performed using Vector Laboratories Elite ABC kit reagents and protocols for rabbit- or mouse-derived antibodies against DARPP32 (Abcam ab40801, 1:10,000 dilution), Iba1 (Wako 019-19741, 1:1,000 dilution), GFP (Life Technologies G10362, 1:1,000 dilution), and NeuN (EMD Millipore MAB377, 1:1,000 dilution). Sections were stained for 2 min with diaminobenzidine using the Metal Enhanced DAB Substrate Kit (Pierce). For quantification of GFP in the striatum, eight coronal sections spanning from anterior to posterior of mouse striatum were stained for GFP and mounted onto microscope slides. Pictures were taken at 2× magnification on Nikon Eclipse E600 microscope of entire striatum using Nikon DS-Qi1MC camera with NIS-Elements. From these pictures, area of striatum on either side of each mouse brain and GFP within striatum were outlined using the polygon selection tool in ImageJ and calculated with the analyze:measure function. The percentage of striatal area saturated with GFP was calculated using the areas measured by ImageJ to show the percentage distribution of GFP-tagged AAV within each coronal section. The total percentage area of GFP within striatum was calculated by adding the area of GFP among all coronal sections within each brain, beside the total striatal area of all sections.
Cloning of Artificial miRNA and AAV Packaging
Artificial miRNAs were designed and cloned according to the protocol detailed by Toro Cabrera and Mueller.16 AAVs were packaged by the University of Massachusetts vector core, according to published protocols.30
Small RNA Library Cloning and Analysis
Total RNA was extracted using the MirVana RNA isolation kit, according to the manufacturer’s protocol. Size selection of the 18- to 30-nucleotide RNAs was performed using 5 μg of total RNA on a 15% denaturing polyacrylamide gel. Following size selection, the small RNAs were ethanol precipitated and ligated to a pre-adenylated 3′-adaptor (5′-rAppTGGAATTCTCGGGTGCCAAGG/ddC/-3′). The ligated products were annealed to the RT primer (5′-CCTTGGCACCCGAGAATTCCA-3′) and ligated to a 5′-adaptor (RNA: 5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′). Reverse transcription was performed using AMV Reverse transcriptase mix (NEB) and PCR-amplified using AccuPrime Pfx DNA Polymerase (Invitrogen) with one universal primer (5′-AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGA-3′) and one barcoded primer (5′-CAAGCAGAAGACGGCATACGAGATNNNNNNGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′). Libraries were sequenced on the Illumina HiSeq at the University of Massachusetts Deep Sequencing Core. Libraries were mapped to the mm9 genome and to the AAV genome using bowtie.31 We classified miRNA species on the basis of the position of the 5′ end mapping on the miRNA hairpin; therefore each species consists of all the small RNAs with shared seed sequences. The 3′ end was not considered in species assignment. Differential expression of endogenous miRNAs was analyzed using the edgeR package,32 which is not sensitive to changes in distribution caused by overexpression of the AAV-derived artificial miRNA in the U6 group. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus33 (GEO: GSE97353).
mRNA Library Cloning and Analysis
RNA was extracted as described above. Libraries were constructed by standard methods.34 Reads were mapped using TopHat2,35 and differential expression was calculated using the DESeq2 package.36
Author Contributions
Conceptualization, E.L.P., N.A., and C.M.; Methodology, E.L.P. and C.M.; Investigation, K.O.C., L.A.K., R.M., F.C., E.J., F.B.; Formal Analysis, H.S.; Writing—Original Draft, E.L.P.; Writing—Review and Editing, N.A. and C.M.; Project Management, E.L.P.; Funding Acquisition, N.A. and C.M.; Supervision, N.A. and C.M.
Acknowledgments
We wish to thank Lina Song (University of Massachusetts Medical School) for help in cloning of the AAV plasmids and the University of Massachusetts Gene Therapy Center Viral Vector Core for production of the viral vectors. This work was supported by the NIH (NS038194 to N.A.); the CHDI Foundation (A-5038); and University of Massachusetts start-up funds to C.M. Funding for open access charge was provided by University of Massachusetts RTF funds to N.A. and start-up funds to C.M.
Footnotes
Supplemental Information includes five figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.omtn.2017.04.011.
Contributor Information
Neil Aronin, Email: neil.aronin@umassmed.edu.
Christian Mueller, Email: chris.mueller@umassmed.edu.
Supplemental Information
References
- 1.Huntington T., Macdonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Sharp A.H., Loev S.J., Schilling G., Li S.H., Li X.J., Bao J., Wagster M.V., Kotzuk J.A., Steiner J.P., Lo A. Widespread expression of Huntington’s disease gene (IT15) protein product. Neuron. 1995;14:1065–1074. doi: 10.1016/0896-6273(95)90345-3. [DOI] [PubMed] [Google Scholar]
- 3.Graveland G.A., Williams R.S., DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington’s disease. Science. 1985;227:770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- 4.Kordasiewicz H.B., Stanek L.M., Wancewicz E.V., Mazur C., McAlonis M.M., Pytel K.A., Artates J.W., Weiss A., Cheng S.H., Shihabuddin L.S. Sustained therapeutic reversal of Huntington’s disease by transient repression of huntingtin synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borel F., Kay M.A., Mueller C. Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol. Ther. 2014;22:692–701. doi: 10.1038/mt.2013.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stanek L.M., Sardi S.P., Mastis B., Richards A.R., Treleaven C.M., Taksir T., Misra K., Cheng S.H., Shihabuddin L.S. Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington’s disease. Hum. Gene Ther. 2014;25:461–474. doi: 10.1089/hum.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. U S A. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu S., Jin L., Zhang Y., Huang Y., Zhang F., Valdmanis P.N., Kay M.A. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell. 2012;151:900–911. doi: 10.1016/j.cell.2012.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 11.Grimm D., Wang L., Lee J.S., Schürmann N., Gu S., Börner K., Storm T.A., Kay M.A. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J. Clin. Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bish L.T., Sleeper M.M., Reynolds C., Gazzara J., Withnall E., Singletary G.E., Buchlis G., Hui D., High K.A., Gao G. Cardiac gene transfer of short hairpin RNA directed against phospholamban effectively knocks down gene expression but causes cellular toxicity in canines. Hum. Gene Ther. 2011;22:969–977. doi: 10.1089/hum.2011.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin J.N., Wolken N., Brown T., Dauer W.T., Ehrlich M.E., Gonzalez-Alegre P. Lethal toxicity caused by expression of shRNA in the mouse striatum: implications for therapeutic design. Gene Ther. 2011;18:666–673. doi: 10.1038/gt.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Gestel M.A., van Erp S., Sanders L.E., Brans M.A.D., Luijendijk M.C.M., Merkestein M., Pasterkamp R.J., Adan R.A. shRNA-induced saturation of the microRNA pathway in the rat brain. Gene Ther. 2014;21:205–211. doi: 10.1038/gt.2013.76. [DOI] [PubMed] [Google Scholar]
- 15.Valdmanis P.N., Gu S., Chu K., Jin L., Zhang F., Munding E.M., Zhang Y., Huang Y., Kutay H., Ghoshal K. RNA interference-induced hepatotoxicity results from loss of the first synthesized isoform of microRNA-122 in mice. Nat. Med. 2016;22:557–562. doi: 10.1038/nm.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toro Cabrera G., Mueller C. Design of shRNA and miRNA for delivery to the CNS. Methods Mol. Biol. 2016;1382:67–80. doi: 10.1007/978-1-4939-3271-9_5. [DOI] [PubMed] [Google Scholar]
- 17.Liu W., Pfister E.L., Kennington L.A., Chase K.O., Mueller C., DiFiglia M., Aronin N. Does the mutant CAG expansion in huntingtin mRNA interfere with exonucleolytic cleavage of its first exon? J. Huntingtons Dis. 2016;5:33–38. doi: 10.3233/JHD-150183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R.K., Rajeev K.G. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. U S A. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slow E.J., van Raamsdonk J., Rogers D., Coleman S.H., Graham R.K., Deng Y., Oh R., Bissada N., Hossain S.M., Yang Y.Z. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum. Mol. Genet. 2003;12:1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- 20.Silva J.M., Li M.Z., Chang K., Ge W., Golding M.C., Rickles R.J., Siolas D., Hu G., Paddison P.J., Schlabach M.R. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 21.Franciosi S., Ryu J.K., Shim Y., Hill A., Connolly C., Hayden M.R., McLarnon J.G., Leavitt B.R. Age-dependent neurovascular abnormalities and altered microglial morphology in the YAC128 mouse model of Huntington disease. Neurobiol. Dis. 2012;45:438–449. doi: 10.1016/j.nbd.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Johnson E., Chase K., McGowan S., Mondo E., Pfister E., Mick E., Friedline R.H., Kim J.K., Sapp E., DiFiglia M., Aronin N. Safety of striatal infusion of siRNA in a transgenic Huntington’s disease mouse model. J. Huntingtons Dis. 2015;4:219–229. doi: 10.3233/JHD-150163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeler A.M., Sapp E., Chase K., Sottosanti E., Danielson E., Pfister E., Stoica L., DiFiglia M., Aronin N., Sena-Esteves M. Cellular analysis of silencing the Huntington’s disease gene using AAV9 mediated delivery of artificial micro RNA into the striatum of Q140/Q140 mice. J. Huntingtons Dis. 2016;5:239–248. doi: 10.3233/JHD-160215. [DOI] [PubMed] [Google Scholar]
- 24.Monteys A.M., Spengler R.M., Dufour B.D., Wilson M.S., Oakley C.K., Sowada M.J., McBride J.L., Davidson B.L. Single nucleotide seed modification restores in vivo tolerability of a toxic artificial miRNA sequence in the mouse brain. Nucleic Acids Res. 2014;42:13315–13327. doi: 10.1093/nar/gku979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBride J.L., Pitzer M.R., Boudreau R.L., Dufour B., Hobbs T., Ojeda S.R., Davidson B.L. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol. Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borel F., Gernoux G., Cardozo B., Metterville J.P., Toro Cabreja G.C., Song L., Su Q., Gao G.P., Elmallah M.K., Brown R.H., Jr., Mueller C. Therapeutic rAAVrh10 Mediated SOD1 silencing in adult SOD1(G93A) mice and nonhuman primates. Hum. Gene Ther. 2016;27:19–31. doi: 10.1089/hum.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galka-Marciniak P., Olejniczak M., Starega-Roslan J., Szczesniak M.W., Makalowska I., Krzyzosiak W.J. siRNA release from pri-miRNA scaffolds is controlled by the sequence and structure of RNA. Biochim. Biophys. Acta. 2016;1859:639–649. doi: 10.1016/j.bbagrm.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 28.Fang W., Bartel D.P. The menu of features that define primary microRNAs and enable de novo design of microRNA genes. Mol. Cell. 2015;60:131–145. doi: 10.1016/j.molcel.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Mueller C., Ratner D., Zhong L., Esteves-Sena M., Gao G. Production and discovery of novel recombinant adeno-associated viral vectors. Curr. Protoc. Microbiol. 2012;Chapter 14 doi: 10.1002/9780471729259.mc14d01s26. Unit14D.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z., Theurkauf W.E., Weng Z., Zamore P.D. Strand-specific libraries for high throughput RNA sequencing (RNA-Seq) prepared without poly(A) selection. Silence. 2012;3:9. doi: 10.1186/1758-907X-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.