Abstract
Background
Hippocampal formation (HF) volume and episodic memory performance are substantially heritable, but HF subregion heritability estimates and their possible shared genetic variance with episodic memory performance remain to be determined.
Methods and findings
This study provides heritability estimates for hippocampal subregions (e.g, Cornu Amonis, Subiculum, Parasubiculum, Molecular and Granule Cell Layers of the Dentate Gryus) and Total HF volumes obtained using FreeSurfer 6.0. In addition, this study assesses the heritability of object sequence and verbal episodic memory performance, and the amount of shared genetic variance between HF subregions and Total HF volume and episodic memory performance. HF volumes were obtained from high-resolution brain scans from a sample of 499 siblings (mean age±SD=30.0±3.1, 203 men), including 51 monozygotic and 46 dizygotic twin pairs and 305 non-twin siblings, collected by the Human Connectome Project (www.humanconnectome.org). Heritability estimates for HF subregions ranged from 0.42–0.87 and shared genetic variance of HF subregions with hippocampal volume was substantial (mean=0.79, range=0.50–0.98). HF subregion volumes residualized for Total HF and percent HF subregion volumes were also found to be substantially heritable (range=0.04–0.86 and 0.07–0.84, respectively). Verbal (h2=0.47) but not object sequence episodic memory was found to be significantly heritable; though the amount of shared genetic variance between HF subregions and verbal episodic memory was low (mean=0.10, range=0.01–0.20).
Conclusions
These findings suggest that HF subregion volumes are heritable and can be used as quantitative phenotypes in genetic association studies. The low shared genetic variance between HF subregions and verbal episodic memory suggests that quantitative trait analyses may not benefit from including both HF volume and episodic memory as bivariate traits in healthy individuals. The extent to which HF subregion volumes share genetic variance with neuropsychiatric disorders, and as such add value to our ability to identify genetic risk loci for these disorders, remains to be determined.
Keywords: hippocampal formation, subregion, subfield, heritability, Human Connectome Project, twin
Introduction
Overall hippocampal formation volume, either measured by manual tracing or by automated methods, is significantly heritable with estimates in the range of 0.40 to 0.80 (1, 2). Episodic memory, a cognitive function known to depend on the hippocampal formation, is also significantly heritable with estimates in the range of 0.30 to 0.60 (3). However, heritability estimates of hippocampal formation subregions and possible shared genetic variance between subregions and episodic memory measures have not yet been reported on. Genome wide association studies (GWAS) have already identified loci significantly associated with hippocampal volume (4, 5) and episodic memory performance (6). The determination of heritability of hippocampal formation subregion volumes is important because subregions may provide additional quantitative traits (7, 8) that can be used in genetic association analysis studies, in particular in the search for genes that convey vulnerability to neuropsychiatric disorders. Moreover, evidence of significant shared genetic variance between hippocampal formation and episodic memory measures could allow for bivariate quantitative trait loci (qtl) association and linkage analyses, which can be more powerful than univariate qtl analyses (9).
The hippocampal formation is comprised of Cornu Amonis regions 1–4 (CA1–CA4), the dentate gyrus, and the subiculum. A recent review has hypothesized differential subfield involvement in several neuropsychiatric disorders (10). Overall hippocampal and hippocampal subregion volume abnormalities have been reported in many neuropsychiatric disorders, including schizophrenia (11–13), bipolar disorder (13, 14), depression (15), post-traumatic stress disorder (16), Alzheimer’s disease (17), and mild cognitive impairment (18).
This study estimates the heritability of hippocampus subfield volumes based on the Human Connectome Project’s [HCP] extended twin sample (19). The study assessed hippocampal subfield volumes using Freesurfer 6.0 (20, 21), which provides volumes for Cornu Amonis regions 1, 2 and 3 combined, and 4 (CA1, CA2/3, and CA4), Fimbria, Hippocampal Fissure, Presubiculum, Subiculum, Hippocampal Tail, Parasubiculum, GCMLDG [Molecular (ML) and Granule Cell Layers (GC) of the Dentate Gryus (DG)], Molecular Layer, and the Hippocampal Amygdala Transition Area (HATA). In addition, this study estimates the heritability of HF subregions controlling for Total HF volume and the shared additive genetic variance between subregions and Total HF volume and episodic memory performance. To control for Total HF volume we performed heritability analyses of subregion volumes residualized for total HF volume as well as analyses of percent (of Total HF) subregion volumes. We hypothesized that 1) HF subregion volumes are significantly heritable, and 2) that they share genetic variance with Total HF volume and episodic memory measures.
Materials and Methods
Participants
Prior to study commencement, the use of de-identified Human Connectome Project (HCP) data was determined non-human subjects research by the University of California, Irvine Institutional Review Board. In addition, all authors obtained permission from the HCP to work with both the unrestricted and restricted HCP data. Five-hundred-and-eleven bias-field corrected, high-resolution, twin and sibling, T1-weighted brain scans (T1w/T1w_acpc_dc_restore.nii) were downloaded from the HCP database (www.humanconnectome.org). We excluded: a) one scan that showed poor HF segmentation based on visual inspection, b) 8 singletons, which are not informative in heritability analyses, and c) 3 subjects who were missing episodic memory performance [NIH Toolbox Picture Sequence Memory Test (PSMT) or Penn Word Memory Test (PWMT)] data. The final study sample comprised 51 monozygotic (MZ) and 46 dizygotic (DZ) twin pairs and 305 non-twin siblings (n=499; see Table 1).
Table 1.
Sample Demographics
| Human Connectome Project Sample (n=499) | |
|---|---|
| Age at Scan in Years (SD) | 29.2 (3.5) |
| Sex (Men / Women) | 203 / 296 |
| Ethnicity (Not Hispanic / Hispanic / Unknown) | 446 / 52 / 1 |
| MZ Twin Pairs / DZ Twin Pairs / Siblings | 51 / 46 / 305 |
| Race | |
| White | 364 |
| Black or African American | 111 |
| Asian / Native Hawaiian | 9 |
| Mixed | 5 |
| Unknown | 10 |
| Education in Years (SD)a | 14.8 (1.9) |
| Handedness (Right / Left / Ambidextrous) | 428 / 38 / 33 |
Estimated from 489 subjects.
Image Acquisition Parameters
The HCP high-resolution, sagittal, T1-weighted (MP-RAGE) scans (TR/TE/TI = 2400/2.14/1000ms, flip angle=8°, FOV=224mm2, in-plane resolution=320×320, 256 slices, 0.7mm3 isotropic voxels, GRAPPA Acceleration factor=2) were acquired with a 3 Tesla Siemens Skyra scanner equipped with a 32-channel head coil at the University of Minnesota (for details see HCP_S500_Release_Appendix_I.pdf on the HCP website).
Image Processing
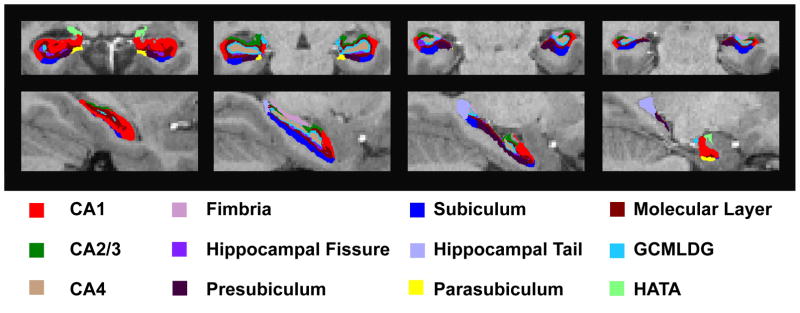
Left and right hippocampal formation (HF) subregion, total HF, and total intracranial volumes were extracted using FreeSurfer 6.0 (20, 21), which is currently available to the public as part of the FreeSurfer development release (ftp://surfer.nmr.mgh.harvard.edu/pub/dist/freesurfer/dev). FreeSurfer 6.0 HF subregions (21) are based on an ultra-high resolution (0.13 mm3 isotropic voxels) ex-vivo atlas and include Cornu Amonis regions 1, 2 and 3 combined, and 4 (CA1, CA2/3, and CA4), Fimbria, Hippocampal Fissure, Presubiculum, Subiculum, Hippocampal Tail, Parasubiculum, GCMLDG [Molecular (ML) and Granule Cell Layers (GC) of the Dentate Gryus (DG)], Molecular Layer, HATA (Hippocampal Amygdala Transition Area), and Total HF (see Figure 1). Given no known differences in left and right HF laterality, for each subject mean of left and right HF subregion volumes were computed for heritability analyses (see Table 2, Volume). In addition to HF subregion volumes, we also computed the percent subregion volume of the Total HF [( HF subregion volume / Total HF volume ) * 100%], a phenotype that should be largely independent from overall HF volume (see Table 2, Percent).
Figure 1.
FreeSurfer 6.0 Hippocampal Subregion Segmentations
Table 2.
Absolute Means
| Volume | Percent | |
|---|---|---|
| Hippocampal Formation Measures | ||
| CA1 | 659 (72) | 18.6 (0.8) |
| CA2/3 | 211 (26) | 6.0 (0.5) |
| CA4 | 261 (26) | 7.4 (0.3) |
| Fimbria | 94 (16) | 2.7 (0.4) |
| Hippocampal Fissure | 137 (19) | 3.9 (0.5) |
| Presubiculum | 322 (36) | 9.1 (0.6) |
| Subiculum | 441 (47) | 12.5 (0.6) |
| Hippocampal Tail | 521 (66) | 14.7 (1.4) |
| Parasubiculum | 68 (10) | 1.9 (0.2) |
| Molecular Layer | 587 (56) | 16.6 (0.3) |
| GCMLDG | 305 (30) | 8.6 (0.3) |
| HATA | 65 (8) | 1.8 (0.2) |
| Total HF | 3534 (331) | |
| Episodic Memory Measures | ||
| PSMT | 111.1 (13.5) | |
| PWMT | 35.4 (3.0) | |
Mean absolute volumes are in mm3 (SD). Percent = (HF subregion / Total HF ) * 100%. PSMT = NIH Toolbox Picture Sequence Memory. PWMT = Penn Word Memory Test. The mean Intracranial Volume (SD) for the sample was 1556 (185) cm3.
Quality Assurance
Quality assurance (QA) procedures included 1) visual inspection of sagittal snapshots of HF subregion segmentations overlaid on corresponding T1-weighted anatomical images, created using Freesurfer’s Freeview called by custom in-house scripts, and 2) inspection of normal distributions of all HF subregion volumes. No significant motion artifacts were detected on any of the scans. One twin subject’s HF data was eliminated based on poor quality of the HF segmentation.
Episodic Memory Measures
The Human Connectome Project assessed object episodic memory with the NIH ToolBox Picture Sequence Memory Test (PSMT) and verbal episodic memory with the Penn Word Memory Test (PWMT). In the PSMT subjects have to recall the order of series of 6 to 18 images. The number of images presented depends on the age of the subjects. Points are given for each correctly recalled adjacent set of images. The total test score is the sum of the points converted to a theta score based on item response theory (IRT; see http://www.nihtoolbox.org/HowDoI/Pages/ScoringAndInterpretation.aspx). The PWMT provides subjects with a list of 20 words to remember. Subjects are then given a second list of 40 words, comprised of 20 targets and 20 foils, and are asked whether each word was on the first list. Options are “definitely yes,” “probably yes,” “probably no,” and “definitely no.” The total accuracy score is the number of correctly identified targets (with “definitely yes,” “probably yes,” responses) and foils (with “probably no,” and “definitely no” responses). Given that age is covariate in the heritability analyses, the unadjusted (for age) scores for each of the tests were used in the heritability analyses.
Statistical Analysis
Prior to the heritability analyses, HF subregion, hippocampal, and ICV volumes were examined for normality via visual inspection of normal distribution plots (Proc Univariate, SAS v9.2, SAS Institute Inc.). Univariate and bivariate heritability analyses were performed using SOLAR version 6.6.2 (22, 23), which fits a mixed model that estimates additive polygenetic (g), household or common environment (c), and unique environment or error variance (e). The significance of heritability is determined by comparing the housepoly (ACE) with the poly (AE), and sporadic (g=0) models and the poly (AE) model with a sporadic (g=0) model based a chi-square tests with df=1 (the difference in the number of terms in the model). The univariate models computed heritability estimates for the HF subregion, Total HF, and ICV volumes as well as PSMT and PWMT episodic memory performance measures. The bivariate models estimated the amount of shared genetic variance (RhoG) between the HF subregions and Total HF (FS 6.0 Whole_hippocampus label) volume and HF subregions and episodic memory performance. All models included sex and age as covariates (Model 1). The univariate heritability analyses were also run including Total HF volume as an additional covariate (Model 2, HF subregions residualized for Total HF volume), and with the percent HF subregion (of total HF volume) as traits, which may provide useful quantitative phenotypes that are independent from Total HF volume. False discovery rate (FDR) was used to control for multiple comparison correction (24).
Results
Heritability Estimates
The heritability estimate for Total HF volume was 0.87. Univariate heritability estimates for HF subregions ranged from 0.20 to 0.83 and were significant (FDR corrected) for all regions with exception of the fimbria (see Table 3, Model 1). Additionally, heritability estimates of HF subfield volumes residualized for Total HF volume (Table 3, Model 2) ranged from 0.04 to 0.86 and the heritability estimates for percent HF subfield volumes ranged from 0.07 to 0.84 (see Table 3, Percent), and were significant (FDR corrected) for all regions except for the fimbria, hippocampal fissure, and HATA. Verbal episodic memory, as measured with the PWMT was significantly heritable (h2=0.47), while object sequence episodic memory as measured with the NIH Toolbox PSMT was not (h2=0.29; Table 3).
Table 3.
Heritability of Hippocampal Formation and Episodic Memory Measures
| Model 1 | Model 2 | Percent | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| h2 | p-value | c2 | p-value | h2 | p-value | c2 | p-value | h2 | p-value | c2 | p-value | |
| Hippocampal Formation | ||||||||||||
| CA1 | 0.83 | 1.07E-18 | 0.70 | 4.69E-15 | 0.69 | 6.19E-15 | ||||||
| CA2/3 | 0.81 | 1.52E-13 | 0.86 | 1.27E-18 | 0.84 | 5.46E-18 | ||||||
| CA4 | 0.79 | 1.09E-14 | 0.65 | 4.19E-10 | 0.63 | 4.98E-10 | ||||||
| Fimbria | 0.20 | 2.12E-01 | 0.30 | 1.26E-02 | 0.04 | 4.47E-01 | 0.30 | 3.17E-02 | 0.07 | 4.00E-01 | 0.26 | 4.92E-02 |
| Hip. Fissure | 0.53 | 9.43E-09 | 0.47 | 1.88E-02 | 0.08 | 2.89E-01 | 0.43 | 1.84E-02 | 0.14 | 1.40E-01 | ||
| Presubiculum | 0.64 | 1.06E-04 | 0.13 | 1.20E-01 | 0.66 | 3.18E-13 | 0.62 | 1.34E-11 | ||||
| Subiculum | 0.83 | 1.81E-23 | 0.69 | 3.96E-17 | 0.69 | 5.17E-17 | ||||||
| Hip. Tail | 0.82 | 1.89E-20 | 0.75 | 6.64E-19 | 0.73 | 2.39E-05 | 0.02 | 4.21E-01 | ||||
| Parasubiculum | 0.65 | 2.18E-04 | 0.06 | 3.05E-01 | 0.56 | 1.82E-08 | 0.49 | 6.00E-07 | ||||
| Molecular Layer | 0.81 | 6.00E-19 | 0.63 | 5.57E-14 | 0.63 | 3.00E-14 | ||||||
| GCMLDG | 0.81 | 2.21E-16 | 0.67 | 2.14E-10 | 0.65 | 5.72E-10 | ||||||
| HATA | 0.62 | 1.69E-04 | 0.12 | 1.39E-01 | 0.45 | 2.20E-02 | 0.13 | 1.59E-01 | 0.38 | 5.29E-02 | 0.15 | 1.29E-01 |
| Total HF | 0.87 | 1.71E-23 | ||||||||||
| Episodic Memory | ||||||||||||
| PSMT | 0.29 | 1.00E-01 | ||||||||||
| PWMT | 0.47 | 1.00E-07 | ||||||||||
h2 = hertiabilty estimate, c2 = common environment variance component estimate. HF subregion heritability estimates that survive the false discovery rate (FDR) multiple comparison correction are underlined. For comparison purposes, the heritability estimate for intracranial volume is 0.88 (p=1.21E-31).
Shared Genetic Variance
Bivariate heritability analyses showed that the mean shared genetic variance (RhoG) between HF subregions and Total HF volume was 0.79 (range = 0.50 – 0.98). Phenotypic correlations (RhoP) between HF subregion volumes and the hippocampus volumes ranged from 0.46 to 0.97 (Table 4, Total HF). The mean shared genetic variance (RhoG) between HF subregions and verbal episodic memory performance as measured by the PWMT was small (mean=0.10, range = 0.01–0.20) as were the phenotypic correlations (Table 4, PWMT).
Table 4.
Shared Genotypic and Phenotypic Variance of Hippocampal Formation Subregions with Hippocampal Volume and PWMT
| Total HF | PWMT | |||
|---|---|---|---|---|
|
| ||||
| RhoG (SE) | RhoP | RhoG (SE) | RhoP | |
| CA1 | 0.91 (0.01) | 0.90 | 0.14 (0.11) | 0.08 |
| CA2/3 | 0.69 (0.04) | 0.70 | 0.04 (0.12) | 0.03 |
| CA4 | 0.91 (0.02) | 0.89 | 0.09 (0.12) | 0.07 |
| Fimbria | 0.67 (0.06) | 0.50 | 0.04 (0.12) | 0.07 |
| Hippocampal Fissure | 0.50 (0.07) | 0.46 | 0.20 (0.14) | −0.01 |
| Presubiculum | 0.78 (0.04) | 0.73 | 0.07 (0.10) | 0.08 |
| Subiculum | 0.90 (0.02) | 0.88 | 0.10 (0.10) | 0.10 |
| Hippocampal Tail | 0.69 (0.04) | 0.65 | 0.20 (0.11) | 0.07 |
| Parasubiculum | 0.72 (0.09) | 0.52 | 0.01 (0.11) | 0.10 |
| Molecular Layer | 0.98 (0.003) | 0.97 | 0.11 (0.11) | 0.09 |
| GCMLDG | 0.93 (0.01) | 0.92 | 0.06 (0.11) | 0.06 |
| HATA | 0.76 (0.04) | 0.68 | 0.08 (0.10) | 0.14 |
| Total HF | 0.12 (0.10) | 0.09 | ||
RhoG = shared genotypic variance. RhoP = shared phentotypic variance. HF = Hippocampal Formation.
Discussion
The main findings of this study are 1) that HF subregion volumes obtained using FreeSurfer 6.0 and verbal memory performance as measured by the Penn Word Memory Test are significantly heritable, and 2) that HF subregions volumes, in predominantly healthy individuals, share a significant amount of genetic variance with overall hippocampal volumes but only a limited amount of genetic variance with verbal episodic memory as measured by the Penn Word Memory Test.
The heritability estimates for HF subregions observed in this study, based on Human Connectome Project (HPC), closely resemble those recently reported based on the Queensland Twins Imaging Study (QTIM): heritability for all regions, except the fimbria differed by less than 0.10 (25). One interpretation of our findings is that almost all hippocampal subfields are substantially heritable. Only the heritability estimate for the fimbria was not significant in this study. It is possible that this is due to lower reliability of smaller brain regions, though the parasubiculum and HATA regions are of similar size and did show significant heritability.
An alternative interpretation, predominantly based on the finding of a large amount of shared genetic variance between the hippocampal subregion and whole hippocampal volumes, is that the 0.7 mm3 isotropic voxels do not provide sufficient resolution to discriminate at least some individual HF subregions. When the resolution of the scans is insufficient, the volumes of the HF subregions may predominantly be determined by Total HF volume, and the HF subregion heritability estimates may simply reflect Total HF volume heritability. While, we cannot fully distinguish between these two alternative interpretations, the fact that 9 out of 12 HF subregion volumes residualized for Total HF volumes were significantly heritable, and b) the same 9 out of 12 percent HF volumes were also significantly heritable suggests that the FreeSurfer 6.0 method is able to reliably measure variation in HF subregions at the 0.7mm3 isotropic voxel resolution. Family studies using data with higher resolution scans will be needed to firmly address these distinct possibilities.
Strengths of the study include 1) the extraction of hippocampal subregion volumes from a publically available extended twin sample with high quality, high-resolution (0.7 mm3 isotropic) structural imaging data collected by the Human Connectome Project (19); 2) the use of publically available FreeSurfer version 6.0, which allows for the estimation of HF subregion volumes based on a high-resolution post-mortem template that addresses a number of issues raised about prior versions of the software (26); 3) the visual inspection of hippocampal segmentations performed on the segmentations for each individual subject; 4) the use of the ACE (SOLAR’s housepoly) model for heritability estimation, a model that includes the common environment term to help avoid overestimation of heritability effects; 5) the use of FDR multiple comparison correction, and 6) the estimation of HF subregion heritability controlling for Total HF volume (both subregions residualized for Total HF as well as percent HF subregion).
A shortcoming must also be noted. While the HF subregion heritability analyses were arguably performed on the highest resolution extended twin structural imaging data set, it is not fully clear whether the 0.7mm3 isotropic voxel size of images provides sufficient resolution to adequately dissociate HF subregions.
There are numerous manual (27) and automated (21, 28–31) methods to assess hippocampal subregions. The opportunities provided by these tools drive the need for the collection of new high-quality, high-resolution brain scans to improve our understanding of the role of hippocampal circuitry in health and disease.
In conclusion, hippocampal formation subregion volumes are heritable and therefore can be used as quantitative traits in genetic association and linkage studies. In healthy individuals, HF subregions share substantial additive genetic variance with overall hippocampal volumes (between 50–98%). The field of high-resolution hippocampal imaging needs more data to provide guidance on the minimum resolution required to properly dissociate HF subregions. Finally, the extent to which individual HF subregions differ in terms of their shared genetic variance with neuropsychiatric disorders and the extent to which they add value to our ability to identify genetic risk loci for these disorders remain to be determined.
Acknowledgments
Data were provided [in part] by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. The project described was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1 TR000153. Dr. Van Erp is supported in part by NIMH grant number MH97196 and the NIH BD2K award, U54EB020403. The authors would like to thank Dr. Mangalam, Mr. Fahran, and Mr. Brenner for their support of the University of California, Irvine High Performance Computing cluster. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflict of Interest
Dr. Van Erp has consulted for Roche Pharmaceuticals and has contract with Otsuka Pharmaceutical Co., Ltd. (OPCJ). The remaining authors declare no potential conflict of interest.
References
- 1.Blokland GA, de Zubicaray GI, McMahon KL, Wright MJ. Genetic and environmental influences on neuroimaging phenotypes: a meta-analytical perspective on twin imaging studies. Twin Res Hum Genet. 2012;15(3):351–71. doi: 10.1017/thg.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Swagerman SC, Brouwer RM, de Geus EJ, Hulshoff Pol HE, Boomsma DI. Development and heritability of subcortical brain volumes at ages 9 and 12. Genes Brain Behav. 2014;13(8):733–42. doi: 10.1111/gbb.12182. [DOI] [PubMed] [Google Scholar]
- 3.Papassotiropoulos A, de Quervain DJ. Genetics of human episodic memory: dealing with complexity. Trends Cogn Sci. 2013;15(9):381–7. doi: 10.1016/j.tics.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, et al. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44(5):552–61. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, Fornage M, et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 2012;44(5):545–51. doi: 10.1038/ng.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Milnik A, Heck A, Vogler C, Heinze HJ, de Quervain DJ, Papassotiropoulos A. Association of KIBRA with episodic and working memory: a meta-analysis. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(8):958–69. doi: 10.1002/ajmg.b.32101. [DOI] [PubMed] [Google Scholar]
- 7.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160(4):636–45. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 8.Martin N, Boomsma D, Machin G. A twin-pronged attack on complex traits. Nat Genet. 1997;17(4):387–92. doi: 10.1038/ng1297-387. [DOI] [PubMed] [Google Scholar]
- 9.Visscher PM, Hemani G, Vinkhuyzen AA, Chen GB, Lee SH, Wray NR, et al. Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genet. 2014;10(4):e1004269. doi: 10.1371/journal.pgen.1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small SA, Schobel SA, Buxton RB, Witter MP, Barnes CA. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat Rev Neurosci. 2011;12(10):585–601. doi: 10.1038/nrn3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narr KL, Thompson PM, Szeszko P, Robinson D, Jang S, Woods RP, et al. Regional specificity of hippocampal volume reductions in first-episode schizophrenia. Neuroimage. 2004;21(4):1563–75. doi: 10.1016/j.neuroimage.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Haukvik UK, Westlye LT, Morch-Johnsen L, Jorgensen KN, Lange EH, Dale AM, et al. In Vivo Hippocampal Subfield Volumes in Schizophrenia and Bipolar Disorder. Biol Psychiatry. 2014 doi: 10.1016/j.biopsych.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 13.Mathew I, Gardin TM, Tandon N, Eack S, Francis AN, Seidman LJ, et al. Medial temporal lobe structures and hippocampal subfields in psychotic disorders: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) study. JAMA Psychiatry. 2014;71(7):769–77. doi: 10.1001/jamapsychiatry.2014.453. [DOI] [PubMed] [Google Scholar]
- 14.Elvsashagen T, Westlye LT, Boen E, Hol PK, Andersson S, Andreassen OA, et al. Evidence for reduced dentate gyrus and fimbria volume in bipolar II disorder. Bipolar Disord. 2013;15(2):167–76. doi: 10.1111/bdi.12046. [DOI] [PubMed] [Google Scholar]
- 15.Ballmaier M, Narr KL, Toga AW, Elderkin-Thompson V, Thompson PM, Hamilton L, et al. Hippocampal morphology and distinguishing late-onset from early-onset elderly depression. Am J Psychiatry. 2008;165(2):229–37. doi: 10.1176/appi.ajp.2007.07030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Neylan TC, Mueller SG, Lenoci M, Truran D, Marmar CR, et al. Magnetic resonance imaging of hippocampal subfields in posttraumatic stress disorder. Arch Gen Psychiatry. 2010;67(3):296–303. doi: 10.1001/archgenpsychiatry.2009.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mueller SG, Weiner MW. Selective effect of age, Apo e4, and Alzheimer’s disease on hippocampal subfields. Hippocampus. 2009;19(6):558–64. doi: 10.1002/hipo.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yassa MA, Stark SM, Bakker A, Albert MS, Gallagher M, Stark CE. High-resolution structural and functional MRI of hippocampal CA3 and dentate gyrus in patients with amnestic Mild Cognitive Impairment. Neuroimage. 2010;51(3):1242–52. doi: 10.1016/j.neuroimage.2010.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K. The WU-Minn Human Connectome Project: an overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischl B. FreeSurfer. Neuroimage. 2012;62(2):774–81. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, et al. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage. 2015;115:117–37. doi: 10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almasy L, Dyer TD, Blangero J. Bivariate quantitative trait linkage analysis: pleiotropy versus co-incident linkages. Genet Epidemiol. 1997;14(6):953–8. doi: 10.1002/(SICI)1098-2272(1997)14:6<953::AID-GEPI65>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling for False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. JR statist Soc. 1995;57(1):289–300. [Google Scholar]
- 25.Whelan CD, Hibar DP, van Velzen LS, Zannas AS, Carrillo-Roa T, McMahon K, et al. Heritability and reliability of automatically segmented human hippocampal formation subregions. Neuroimage. 2016;128:125–37. doi: 10.1016/j.neuroimage.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wisse LE, Biessels GJ, Geerlings MI. A Critical Appraisal of the Hippocampal Subfield Segmentation Package in FreeSurfer. Front Aging Neurosci. 2014;6:261. doi: 10.3389/fnagi.2014.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yushkevich PA, Amaral RS, Augustinack JC, Bender AR, Bernstein JD, Boccardi M, et al. Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol. Neuroimage. 2015 doi: 10.1016/j.neuroimage.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yushkevich PA, Wang H, Pluta J, Das SR, Craige C, Avants BB, et al. Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI. Neuroimage. 2010;53(4):1208–24. doi: 10.1016/j.neuroimage.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yushkevich PA, Avants BB, Pluta J, Minkoff D, Detre JA, Grossman M, et al. Shape-based alignment of hippocampal subfields: evaluation in postmortem MRI. Med Image Comput Comput Assist Interv. 2008;11(Pt 1):510–7. doi: 10.1007/978-3-540-85988-8_61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, et al. Automated segmentation of hippocampal subfields from ultra-high resolution in vivo MRI. Hippocampus. 2009;19(6):549–57. doi: 10.1002/hipo.20615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Leemput K, Bakkour A, Benner T, Wiggins G, Wald LL, Augustinack J, et al. Model-based segmentation of hippocampal subfields in ultra-high resolution in vivo MRI. Med Image Comput Comput Assist Interv. 2008;11(Pt 1):235–43. doi: 10.1007/978-3-540-85988-8_29. [DOI] [PMC free article] [PubMed] [Google Scholar]