Abstract
A significant fraction of every proteome is occupied by biologically active proteins that do not form unique three-dimensional structures. These intrinsically disordered proteins (IDPs) and IDP regions (IDPRs) have essential biological functions and are characterized by extensive structural plasticity. Such structural and functional behavior is encoded in the amino acid sequences of IDPs/IDPRs, which are enriched in disorder-promoting residues and depleted in order-promoting residues. In fact, amino acid residues can be arranged according to their disorder-promoting tendency to form an alphabet of intrinsic disorder that defines the structural complexity and diversity of IDPs/IDPRs. This review is the first in a series of publications dedicated to the roles that different amino acid residues play in defining the phenomenon of protein intrinsic disorder. We start with proline because data suggests that of the 20 common amino acid residues, this one is the most disorder-promoting.
Keywords: cis-trans isomerization, conformational restriction, intrinsically disordered protein, post-translational modification, protein solubility, protein surfaces
Introduction
Intrinsically disordered proteins (IDPs) and intrinsically disordered protein regions (IDPRs) have recently become a hot topic in molecular and structural biology.1,2 Computational analyses show that about 10–20% of full-length eukaryotic proteins are IDPs and that 25–40% of all protein residues are classified as IDPRs.3-7 Furthermore, more than half of IDPs experimentally characterized by NMR are in fact IDPRs.8 Despite the fact that IDPs/IDPRs do not form regular, three dimensional structures on their own,9 they are nevertheless associated with various important cellular roles10-24 and implicated in a number of prominent human diseases.14,25-33 The unique structural properties of IDPs/IDPRs require new methods for their analyses34 and new concepts for understanding their functions.10,11,15
Structural and functional properties of a protein are encoded by the alphabet of the 20 naturally occurring amino acids. Therefore, to understand the unique structural and functional properties of IDPs/IDPRs it is necessary to determine how their amino acid sequences differ from ordered proteins. A number of research groups, including ours, have interrogated this problem using computational methods and determined that the amino acid compositions of IDPs and IDPRs are biased in relation to ordered proteins.5,9,11,35-37 (add ref. 0) Based on these studies, the concept of “order-promoting” (cysteine, tryptophan, tyrosine, isoleucine, phenylalanine, valine, leucine, histidine, threonine, asparagine) and “disorder-promoting” residues (aspartic acid, methionine, lysine, arginine, serine, glutamine, proline, glutamic acid) has been proposed.38 From a physico-chemical point of view, the majority of order-promoting residues are non-polar and commonly found within the hydrophobic cores of ordered proteins, whereas the majority of disorder-promoting residues are polar, often charged, and commonly found on the surfaces of ordered proteins. This notion is consistent with our current understanding of the highly dynamic structures of IDPs/IDPRs that do not form stable hydrophobic cores and probably expose most of their amino acids to the solvent.5,11 Important exceptions to the just stated polar or charged tendencies are prolines, which are the most disorder-promoting residues39 despite the non-polar nature of their side chains.
The differences in composition between ordered and disordered proteins are coupled to distinct evolutionary patterns, with IDPs and IDPRs typically displaying higher global mutation rates than ordered proteins.40 Despite this, some IDP residues, such as aromatic amino acids (tryptophans, tyrosines, and phenylalanines), leucines and prolines are well-conserved.41 With the exception of prolines, all other conserved residues are generally less abundant in IDPs than in ordered proteins. Conserved aromatic and hydrophobic IDP residues are frequently found in protein segments with molecular recognition features (MoRFs)42,43 and in the pre-structured motifs (PreSMos).8 MoRFs are short IDPRs that of 10-fold upon binding to other proteins, as well as to DNA. MoRFs determine the functions of many IDPs because they define specific protein-protein interaction surfaces, which likely explain their higher degree of evolutionary conservation.
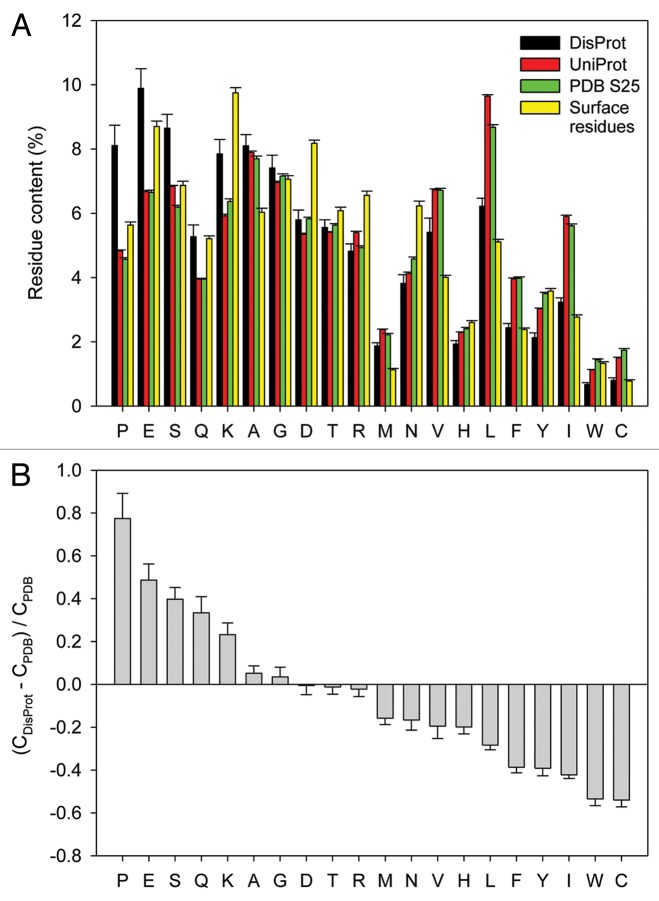
Figure 1 and Table 1 show the statistics of amino acid compositions of proteins in four standard data sets, Swiss-Prot,45 PDB Select 25,46 surface residues37 and DisProt,44 where Figure 1A recapitulates Table 1 in a graphical form, and Figure 1B shows the compositional differences between the structured and disordered data sets. The Swiss-Prot database (UniProtKB/Swiss-Prot) was chosen because it contains sequence and functional information on ~550,000 proteins from all kingdoms of life and therefore represents the unbiased distribution of amino acids throughout nature.37 PDB Select 2546 contains a representative set of PDB entries with less than 25% of sequence identity. This database was chosen because of its bias toward “structural” proteins that are likely to crystalize.37 Surface residues were determined with the Molecular Surface Package and a number of PDB structures of monomeric proteins that were found suitable for studying biological activities associated with protein surface properties, such as protein binding, for example.37 Finally, the DisProt44 database comprises entries of proteins and protein regions that had been experimentally verified to be intrinsically disordered.37Figure 1A and Table 1 show that average proline contents in these four data sets are 4.83 ± 0.03%, 4.57 ± 0.05%, 5.6 ± 0.1% and 8.1 ± 0.6%, respectively (cprofiler.org/help.html).37 Hence, IDPs contain, on average, 1.7- to 1.8-times more prolines than proteins in UniProt, or PDB Select 25, respectively. Furthermore, the overall proline content in IDPs is 1.4-times higher than on surfaces of folded proteins.
Figure 1. Amino acid determinants defining structural and functional differences between the ordered and intrinsically disordered proteins. (A) Amino acid compositions of several data sets discussed in the text (DisProt,44 UniProt,45 PDB Select 2546 and surface residues37). (B) Fractional difference in the amino acid composition (compositional profile) between the typical IDPs from the DisProt database44 and a set of completely ordered proteins46 calculated for each amino acid residue. The fractional difference was evaluated as (CDisProt-CPDB)/CPDB, where CDisProt is the content of a given amino acid in a DisProt databse, and CPDB is the corresponding content in the data set of fully ordered proteins. Positive bars correspond to residues found more abundantly in IDPs, whereas negative bars show residues, in which IDPs are depleted. Amino acid types were ranked according to their decreasing disorder-promoting potential.36
Table 1. Amino acid compositions of the standard data sets (modified from ref. 37).
| Residuea | Disorder propensityb | SwissProtc | PDB S25d | Surface residuese | DisProtf |
|---|---|---|---|---|---|
| Cys (C) | 0.000 | 1.50 ± 0.02 | 1.74 ± 0.05 | 0.78 ± 0.04 | 0.80 ± 0.08 |
| Trp (W) | 0.004 | 1.13 ± 0.01 | 1.44 ± 0.03 | 1.33 ± 0.05 | 0.67 ± 0.06 |
| Ile (I) | 0.090 | 5.90 ± 0.04 | 5.61 ± 0.06 | 2.77 ± 0.07 | 3.24 ± 0.13 |
| Tyr (Y) | 0.113 | 3.03 ± 0.02 | 3.50 ± 0.04 | 3.58 ± 0.08 | 2.13 ± 0.15 |
| Phe (F) | 0.117 | 3.96 ± 0.03 | 3.98 ± 0.04 | 2.38 ± 0.05 | 2.44 ± 0.13 |
| Leu (L) | 0.195 | 9.65 ± 0.04 | 8.68 ± 0.08 | 5.11 ± 0.08 | 6.22 ± 0.25 |
| His (H) | 0.259 | 2.29 ± 0.02 | 2.41 ± 0.04 | 2.60 ± 0.06 | 1.93 ± 0.11 |
| Val (V) | 0.263 | 6.73 ± 0.03 | 6.72 ± 0.06 | 4.01 ± 0.06 | 5.41 ± 0.44 |
| Asn (N) | 0.285 | 4.13 ± 0.04 | 4.58 ± 0.06 | 6.23 ± 0.15 | 3.82 ± 0.27 |
| Met (M) | 0.291 | 2.38 ± 0.02 | 2.22 ± 0.04 | 1.13 ± 0.04 | 1.87 ± 0.10 |
| Arg (R) | 0.394 | 5.40 ± 0.04 | 4.93 ± 0.06 | 6.56 ± 0.13 | 4.82 ± 0.23 |
| Thr (T) | 0.401 | 5.41 ± 0.02 | 5.63 ± 0.05 | 6.08 ± 0.11 | 5.56 ± 0.24 |
| Asp (D) | 0.407 | 5.35 ± 0.03 | 5.83 ± 0.05 | 8.18 ± 0.10 | 5.80 ± 0.30 |
| Gly (G) | 0.437 | 6.96 ± 0.04 | 7.16 ± 0.07 | 7.06 ± 0.11 | 7.41 ± 0.40 |
| Ala (A) | 0.450 | 7.89 ± 0.05 | 7.70 ± 0.08 | 6.03 ± 0.13 | 8.10 ± 0.35 |
| Lys (K) | 0.588 | 5.92 ± 0.05 | 6.37 ± 0.08 | 9.75 ± 0.16 | 7.85 ± 0.45 |
| Gln (Q) | 0.665 | 3.95 ± 0.03 | 3.95 ± 0.05 | 5.21 ± 0.09 | 5.27 ± 0.37 |
| Ser (S) | 0.713 | 6.83 ± 0.04 | 6.19 ± 0.06 | 6.87 ± 0.13 | 8.65 ± 0.43 |
| Glu (E) | 0.781 | 6.67 ± 0.04 | 6.65 ± 0.07 | 8.70 ± 0.17 | 9.89 ± 0.61 |
| Pro (P) | 1.000 | 4.83 ± 0.03 | 4.57 ± 0.05 | 5.63 ± 0.10 | 8.11 ± 0.63 |
a Residues are arranged according to their decreasing intrinsic disorder propensity; bDisorder propensity is calculated based on the fractional difference in the amino acid compositions between the disordered and ordered proteins obtained by renormalizing these values to lie between 0 and 1; cSwissProt 51 is closest to the distribution of amino acids in nature among the four data sets;45dPDB Select 25 is a subset of proteins from the Protein Data Bank with less than 25% sequence identity, biased toward the composition of proteins amenable to crystallization studies;46eSurface residues determined by the Molecular Surface Package over a sample of PDB structures of monomeric proteins suitable for protein surface analysis; fDisProt 3.4 comprised of a set of experimentally determined disordered regions.44
Figure 1B shows that proline exhibits the largest fractional change between structured and disordered proteins, and the fractional changes for the various residues provide the basis for estimating the disorder propensities given in Table 1 (see Table 1, footnote b). Indeed, the disorder propensities here yield the same P, E and S ranking for the most disorder-promoting residues as obtained in a previous study,39 while the remaining amino acids show some alterations in the ranking compared with the previous study, especially for amino acids with similar disorder propensity values. Of course such estimates depend on both the methods used and the sets of proteins in the databases, which were both significantly different in the previous study39 as compared with this one. Overall, the disorder propensity ranking between the two studies differ in detail but these differences are not significant.
This article starts a series of publications on the alphabet of intrinsic disorder, which is dedicated to exploring the amino acid determinants of intrinsic protein disorder. Here, we review the functions of prolines in IDPs/IDPRs and provide compelling evidence for proline-specific biological activities that may provide explanations for their high levels of abundance and conservation in disordered proteins and protein regions.
Structural Properties of Prolines
Chemical structure of prolines
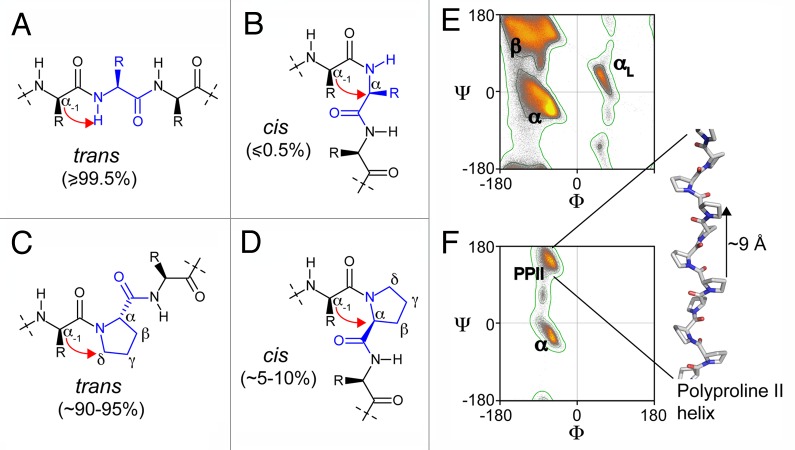
Among the 20 natural amino acids, proline is unique in that it is the only imino acid; that is, the proline backbone nitrogen is bound to two alkyl carbons and lacks the usual proton (see Fig. 2). Proline’s distinctive cyclic structure renders the backbone conformation more rigid than in any other amino acid. Hence, proline peptide bonds exhibit structural features that differ substantially from other residues, also because they do not contain backbone amide hydrogen atoms at physiological pH and therefore do not form stabilizing hydrogen bonds in α-helices, or β-sheets. In consequence, prolines are rarely found as integral parts of secondary structure elements,47,48 but rather at the ends of a-helices, or in protein loop regions.49 Their characteristic backbone angle properties and unique structural properties in proteins and polypeptides (see below) also give rise to atypical Ramachandran plot features.50-53 Prolines sample restricted areas of the Ramachandran space, which are primarily defined by their backbone pyrrolidine constraints.54 They also exert pronounced effects on the backbone geometries of residues preceding them, i.e., pre-prolines.55
Figure 2. Chemical structure of peptide fragments in trans (A) and (C) and cis conformation (B) and (D); (C) and (D) show a proline-containing fragment. The red arrows point out the steric hindrances between the Cα of the residue (−1) with the Hamide (A) or the Cα of the residue (0) (B) for the non-proline-containing peptides, and between the Cα of the residue (-1) with the Cδ (C) or the Cα of the proline (D). Ramachandran plots of non-proline, non-glycine, non-isoleucine, non-valine residues (E) and proline residues (F) result from the analysis of 1.5 million residues in 8,000 protein chains with resolution < 2 Å and backbone B-factors < 30.The contours separate the “outlier,” “allowed” and “favored” regions of the Ramachandran plots. The Ramachandran plots were adapted from commons.wikimedia.org/wiki/User:Dcrjsr. The β-strand (β), α-helix (α), α-L-helix (αL), poly-proline II (PPII) regions of the Ramachandran plots are indicated and we show a representation of a model poly-proline II helix.
Cis-trans isomerization
Although most amino acids form peptide bonds that are in their trans-isomer conformations (> 99.5%),56,57 Xaa-Pro peptide bonds populate both cis- and trans-states. Xaa-Pro trans isomers are indeed less favored because of relatively high steric conflicts between Xaa-Cα atoms and Pro-Cδ’s (see Fig. 2). The energy differences between proline cis/trans conformers are less pronounced than in other amino acids, which, in connection with a high energy barrier between the two isomers (~20 kcal/mol)58,59 results in slow cis/trans interconversion rates (10−3 s−1).56 Hence, on average, ordered proteins contain 5–10% cis-conformers of the Xaa-Pro peptide bonds, whereas the occurrence of cis-isoforms of usual amide bonds in proteins is typically below 0.5%.56,57 The cis-isomer content is influenced by the nature of the surrounding residues and by the types of surrounding secondary structure.60-62 Despite these similar energy levels in disordered peptides, prolines in natively folded proteins tend to display exclusive cis-, or trans-conformations, which are primarily established via the protein fold and the resulting specific interactions with residues close in space.63,64
Within protein Xaa-Pro motifs, Cα(Xaa)/Cα(Pro) distances of trans-proline conformations are on average 1.5 Å larger than for cis proline isomers;65,66 however, these effects are not systematic and strongly influenced by the nature of Xaa. In most folded proteins, isomer-specific structural changes are local, and vanish at a distance of 2–3 residues from the proline of interest. More extended conformational rearrangements have only been observed for a few cases.67 From a local point of view the effects that proline cis/trans isomers induce in polypeptide chains are important. Cis-isoforms result in turn-like structures, whereas trans-isoforms favor locally extended conformations (see Fig. 2). In protein folding cis/trans isomerization plays an important role and often functions as the rate limiting step in the overall folding process.64 Important cellular enzymes such as peptidyl-prolyl isomerases (PPIases) accelerate proline isomerization processes and thereby enhance the kinetic rates with which thermodynamic equilibrium states are reached. The relationships between PPIases and IDPs will be discussed in more detail, later in the article. One aspect that we want to stress is that proline cis-trans characteristics and behaviors of IDPs are similar to those of peptides. IDPs display cis population averages of ~5–10% and, therefore, IDPs with 10 or more prolines have high probabilities for multiple cis conformations. This creates substantial diversity in population conformers that sample a vast conformational space.
On the hydrophobicity of the proline residue
In the initial hydrophobicity scale development, the backbone was considered to be constant for all of the amino acids, and thus only the side chain was considered to be contributing to the values of the scale.68 However, with regard to residue hydrophobicity, the proline imine brings the backbone into play. That is, upon burying a typical amino acid residue, the backbone has both hydrogen bond donors and acceptors, leading to helices, sheets, turns or other structures in which the backbone hydrogen bonding potential is self-satisfied. For proline, on the other hand, the backbone has hydrogen bond acceptors but no donors, and for this reason it is costly from an energetic point of view to sequester the proline backbone from the solvent. The consequences of this donor / acceptor imbalance in the backbone are that, compared with valine, the other amino acid with a side chain containing 3 aliphatic carbons, proline is less frequently buried and more frequently on protein surfaces (Table 1; Fig. 1B). In this regard, the solubility of the individual amino acids is generally inversely correlated with hydrophobicity, yet proline is by far the most soluble of the amino acids at neutral pH,69 and furthermore, polyproline is much more water soluble than polyglycine, polyalanine and polyleucine due to polyproline’s lack of an NH group.70 Thus, despite its hydrophobic side chain, the proline residue is very hydrophilic.
Prolines in IDPs/IDPRs: Structural and Functional Roles
The polyproline type II helix as a unique binding interface
The unusual chemistry of prolines imposes several constraints on neighboring residues and proline-rich motifs (PRMs) have high propensities for adopting non-classical conformations such as the polyproline type II (PPII) helix.71-73 PPII helices are left-handed, extended structures that contain three residues per turn and no internal hydrogen bonding. They are surprisingly abundant structural scaffolds in virtually every proteome. Even ordered proteins contain short PPII stretches, and PPII backbone dihedral angles (−75°, 150°) are frequently observed in amino acids other than prolines.74,75 In PPII helices, side-chain and backbone carbonyls are solvent-exposed and often engage in intermolecular hydrogen bonds, thereby mediating generic intermolecular recognition events of rather low ligand specificities. In turn, a great number of proline-recognition domains (PRDs) interact with PRMs and PPII helices, among which SH3 and WW domains are probably the most well-known examples. The giant human protein titin, with a total of 34,000 amino acids, contains ~550 SH3 binding motifs, of which ~100 are found in PRMs.76-79
PPII-mediated interactions regulate diverse sets of particular cellular functions.72,80,81 A statistical analysis on 74 scaffolding proteins for example, has revealed that this class of proteins contained predicted degrees of disorder (i.e., 49.7% by IUPred, 63.36% by VSL2 and 47.82% by FoldIndex82) that were comparable to highly disordered classes of proteins, such as transcription factors14 and RNA chaperones.83 Furthermore, 26 of the most disordered scaffolding proteins contained average proline contents of 11.2 ± 0.4%, which appears to predispose PRM-proteins to function as hubs in protein-protein interaction networks.84-91 PRMs, or polyproline regions (PPRs) are also found in the proteomes of several viruses, such as hepatitis E (HEV), rubivirus and cutthroat virus (CTV).92 Although the functional significance of PPRs in viruses remains poorly understood, they appear to mediate interactions of viral proteins with cellular host factors to modulate viral replication efficiencies.93 A recent study further demonstrated that sequence variabilities in viral PPRs play important roles in adaptation and in specifying the range of host cells.92 PPRs of HEV genotypes 3 and 4, for example, indicating viral variants of zoonotic origins that can infect humans and animals, are twice as heterogeneous than PPRs in the HEV genotype 1 variant, which is purely anthropotropic and can infect humans only.92
Also, in these PRM-containing binding regions, proline not only is involved in maintaining an open conformational state compatible with binding, it is also the most important residue that contacts the partner protein. An analysis of short linear motifs (SLiMs, also termed Eukaryotic Linear Motifs, ELMs) showed that Pro is the residue most significantly enriched in sites that determine binding specificity of the motif (restricted sites, RSs).94
PRMs and IDP conformations
Based on the high levels of PPII sequence conservations in folded proteins, it has been suggested that these structural elements constituted a separate class of secondary structure elements,75 with two major functions: To promote super-secondary structures, such as PPII/α-helical interactions, and to form inter-domain linkers.75 In IDPs, the unique propensities of PPII structures in rigidifying polypeptide backbone conformations is thought to spatially separate functionally important protein regions.13 An example for such a separation function is provided by the human oncoprotein and transcription factor p53 that contains two PRMs in PPII-type conformations. One separating the intrinsically disordered N-terminal transactivation domain (NTAD) of p53 from its folded DNA-binding domain (DBD), the other one within the NTAD separating a helical pre-structured segment and two pre-structured turns32 that mediate distinct protein-protein interactions.33,95-97 Similarly, two transactivation domains within the C-terminus of herpes simplex virus protein 16 (VP16) are separated by a conserved PRM (452PGPGFTPHDSAP464).98,99 In both cases, spatial positioning via PRMs likely regulates independent transcription activation processes that rely on different interactions with the RNA polymerase II machinery.98,100 By analogy, two helical segments within the C-terminal portion of human securin, potentially mediating the interactions with separase,101 are separated by a PRM (162PPSPVKMPSPP173), whereas a PRM in the human transcription factor FoxA3 (250PPQPPPPAPEP260) separates its DNA- from its histone-binding domain.
Whereas PRMs often induce extended conformations, many IDPs are usually more compact than chemically denatured proteins of comparable lengths,16,33 whose conformational behaviors still cannot be described as random coils.102 Because most IDPs are not restricted to stable three-dimensional architectures, to seamlessly vary their degrees of global compactions is thought to constitute an important functional IDP feature.103,104 Therefore, the ability of PRMs to elongate and stiffen polypeptide chains has to be discussed in this context. For example, proline-rich salivary proteins possess significantly higher radii of gyration than are expected for unfolded polypeptides of similar lengths.105 It has been proposed that organized PPII helices in these proteins result in larger collisional cross sections that facilitate their interactions with tannins,106 which form the basis of the sensory perception of astringency.107
Extending IDP structures via PRM-mediated effects may not necessarily be restricted to long proline sequences alone. In fact, a strong correlation between the number of prolines in an IDP and its radius of gyration has been established.108 Such expansions have been attributed to the unique properties of Xaa-Pro peptide bonds to adopt backbone dihedral angles that correspond to extended conformations. However, prolines can also promote β-turn conformations, which elicit various degrees of polypeptide chain compactions.109,110 The degree of compaction can moreover be tuned by cis/trans equilibria.111 In line with these observations, mutating proline residues in a short, disordered elastin-like peptide has been shown to induce a stepwise expansion.112 In contrast, the overall stiffness of four disordered peptides were reported to be more correlated with their PPII contents than their proline counts, whereas the intrinsic capacities for hairpin structures strongly correlated with the numbers of glycines and prolines.113 Therefore, the possible role(s) of prolines in compacting, or expanding IDPs conformations would depend on the context. While increasing the number of prolines in PPII conformations appears to rigidify IDPs, a high-abundance of prolines in combination with favorable glycine contents, or with selective positioning of charged and/or hydrophobic residues, gives rise to preferred hairpin conformations that result in more collapsed structures.114
Prolines as secondary structure-breakers
Because of their unique chemical and structural properties, and because of their negative influence on classical secondary structure, it is tantalizing to speculate that proline positions in folded, but also in intrinsically disordered proteins, had been evolutionary selected, as well as conserved, for their unique capacities to modulate the structural propensities of neighboring protein residues. In folded proteins, a preference for prolines at helix-capping positions had been recognized very early on.115 Depending on the data set, or the methods for defining secondary-structures, prolines in N- or C-cap positions preferentially occur between Ncap-1 and Ncap+2 and between Ccap and Ccap+3, respectively.116-120 In these instances, high proline frequencies do not relate to helix stabilization effects, but more likely function as border elements that confine existing secondary structures to certain lengths.121,122 In IDPs, proline positions may have been evolutionarily conserved to ensure that protein regions with residual structural propensities, such as MoRFs for example, retain their partially folded states in a balanced manner. Recent findings support this notion by showing that prolines at positions that flank partially folded IDP segments (PreSMos) occur more frequently33 and display higher levels of positional conservation, than elsewhere in these proteins.94 In essence, this notion represents an extension of the “proline bracket” concept,123,124 according to which prolines in segments flanking protein interaction sites negatively modulate the propagation of α-helices and β-strands. Such effects may preserve various degrees of conformational IDP plasticity, which may eventually steer different binding behaviors in protein-protein interactions.
Prolines and prevention of amyloid-like aggregation
As mentioned earlier, positional proline effects in IDPs may preserve levels of disorder in regions with residual structural propensities. This, in turn, may also reduce the likelihood for spontaneous IDP aggregation, which is often cytotoxic, results in cell death and produces several devastating disease phenotypes.125 In fact, many different IDP aggregation processes proceed via intermediate conformations that harbor folded aggregation cores, which progressively expand into highly ordered macromolecular assemblies such as amyloids fibrils, for example. In folded proteins, uncontrolled association events via existing secondary structure elements are often prevented by combinations of dedicated structural features that “protect” aggregation-prone entities such as peripheral β-strands. These include “covering” interactions with loop- or helical-segments, β-strand distortions via inward-pointing, charged residues, incorporation of prolines, β-bulges, or glycine-promoted bends and twists, or via formations of continuous β-sheets to yield β-barrels.126 Therefore, prolines at the domain boundaries are often highly conserved and mutating them usually promotes aggregation.125,127 In depth analyses of various protein segments that display high propensities for β-aggregation have shown that β-breaking prolines, together with charged amino acids such as lysines, arginines, glutamates and aspartates, are specifically enriched at these positions and thought to serve as anti-aggregation “gatekeepers.”128
Elastomeric proteins
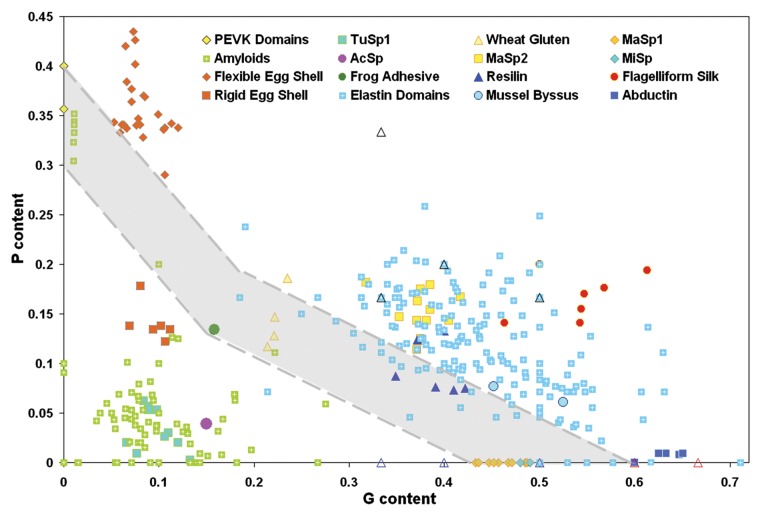
Elastomeric proteins exemplify another important aspect of the “usage” of prolines for specific biological functions. These proteins display remarkable propensities for elastic recoiling behaviors and undergo innumerous reversible deformations in the course of their lifetimes, which are directly related to their specific biological functions in tissues and other biomaterials.129 In all vertebrates, elastomeric proteins constitute the building blocks of blood vessels; in insects, they give rise to specialized structures such as a spider’s silk; in arthropods they make up the intrinsic energy storage apparatus that enables jumping. Some of these proteins are IDPs that have evolved to aggregate in a controlled manner to form dedicated, rubber-like structures that are able to be stretched under extreme physical circumstances and to recoil by itself later.129 Although these elastomeric proteins can spontaneously organize themselves into elastomeric protein complexes, they are surprisingly resistant to forming β-rich amyloid structures.125 Despite their sequence and functional diversities, all elastomeric proteins and IDPs contain unusually high proline and glycine contents,130 which clearly separates elastomeric proteins from amyloidogenic proteins and peptides (Fig. 3).130 Prolines in these structures, together with glycines, prevent the formation of long, stable amyloid structures, whereas their relatively high hydrophobicities promote aggregation-like behaviors such as recoiling. Thus, amino-acid compositions of elastomeric proteins depend on a fine balance between polypeptide hydrophobicity and high proline and glycine contents.125,130
Figure 3. A two-dimensional plot correlating proline and glycine content for a wide variety of elastomeric and amyloidogenic peptides. Elastomeric proteins are characterized by high GP content and are located in the upper-right part of this plot. Contrarily, amyloidogenic peptides are characterized by low PG content and therefore are located in the left bottom corner of the plot. The coexistence region (shaded in gray) contains P and G compositions consistent with both amyloidogenic and elastomeric properties. Elastomeric proteins, including the domains of elastin, major ampullate spindroin (MaSp) 2, flagelliform silk, the elastic domains of mussel byssus thread, and abductin, appear above a composition threshold (upper dashed line). Amyloidogenic sequences are primarily found below the PG-threshold, along with rigid lizard egg shells, tubulliform silk (TuSp1), a protective silk for spider eggs, and aciniform silk (AcSp), used for wrapping prey. The coexistence region contains amyloid-like peptides as well as the elastomeric adhesive produced by the frog Notaden bennetti, the PEVK domains of titin, wheat glutenin protein, and the strongest spider silks, namely MaSp1 and minor ampullate spindroin (MiSp). Figure reproduced from ref. 130 Abbreviations: AcSp, aciniform silk; MaSp, major ampullate spindroin; MiSp, minor ampullate spindroin; TuSp1, tubulliform silk.
Proline-Directed Post-translational Modifications
Post-translational protein modifications (PTMs) range from enzymatic cleavage reactions of peptide bonds to covalent additions of particular chemical groups, lipids, carbohydrates or even entire proteins onto selected subsets of amino acid side chains. PTMs extend the range of amino acid structures and properties and greatly diversify the functional space of virtually every proteome.131 With regard to our subject, strong correlations between predicted, and experimentally verified protein disorder and the occurrence of PTMs exist,26 the most common among which are phosphorylation,132,133 ubiquitination,134 acetylation,135 methylation136,137 and glycosylation138 reactions. These PTMs are typically involved in the regulation and control of various signaling and recognition processes (for example see ref. 139). Although direct post-translational modifications of proline residues only have a limited range of functions, prolines play important roles in the regulation of the occurrences of other PTMs.
Proline PTMs
Annotated lists of experimentally verified PTMs, in Swiss-Prot and other databases, clearly indicate that prolines are primarily subject to post-translational hydroxylation (selene.princeton.edu/PTMCuration/),140 which can occur on Cβ ((2S,3S)-3-hydroxyproline) or Cγ ((2S,4R)-4-hydroxyproline) positions. These nonreversible conversions of prolines to (2S,4R)-4-hydroxyprolines (Hyps) are catalyzed by prolyl 4-hydroxylase enzymes and surprisingly, represent the most common PTM in humans.141 In fact, Hyps are more abundant in animals than seven of the most “common” amino-acid types: Cys, Gln, His, Met, Phe, Trp and Tyr.142 The best known roles for Hyp’s are in stabilizing collagen triple helices.141 Proline hydroxylation enhances the stability of trans-isoforms of Xaa-Pro peptide bonds relative to cis-isoforms.141 Since proline trans-isoforms already constitute the major conformations in IDPs (~90%), hydroxylation is not thought to play additional important roles in their conformational behaviors. Apart from their roles in collagen-like coiled-coil structures, Hyp’s are also found in many other connective tissue proteins, in proteins with collagen-like domains, as well as in the (partially) disordered proteins elastin, conotoxin and argonaute 2.141
The best example for Pro-hydroxylation generating a signal for regulation is hypoxia-inducible transcription factor 1α (HIF-1α). At low oxygen conditions (hypoxia), HIF-1α activates transcription by recruiting the general coactivator CBP/p300 via interaction with its TAZ1 domain. Upon elevation of oxygen level, Pro564 of HIF-1α becomes hydroxylated, it binds to the ubiquitin ligase von Hippel-Lindau factor and undergoes ubiquitination that targets the protein for degradation.143
Proline-directed limited proteolysis
Structural disorder and the extended structure ensured by Pro residue(s) are also involved in directing the action of proteases in limited proteolysis. Due to being an irreversible modification, limited proteolysis is a serious and tightly regulated signaling decision by the cell. For example, calpain, the intracellular protease only cleaves specific substrates if activated by calcium and released by its tight inhibitor, calpastatin, and shows a strong preference for regions of local structural disorder dominated by Pro residues (Tompa et al. J Biol Chem 2004; 279: 20775-85).144 Actually, Pro is depleted around the scissile bond (positions P2, P1 and P1’), but is highly significantly enriched in flanking regions (positions P4, P3 and P2’ to P6’).144
Roles of prolines in protein phosphorylation
Many serine/threonine kinases modify substrate sites that constitute integral or distal parts of kinase consensus motifs.145,146 Within these consensus motifs, proline residues often define substrate site specificities. Examples include many proline-directed protein kinases, such as cyclin-dependent kinases (CDKs),147,148 the mitogen-activated family of protein kinases (MAPKs),149,150 extracellular signal-regulated kinases (ERKs), stress-activated protein kinases/c-Jun-N-terminal kinases (SAPKs/JNKs), p38 kinases, glycogen synthase kinase-3 (GSK3) and Polo-like kinases (PLKs),151 all of which require prolines at positions +1 with respect to the sites of modification. These kinases play important roles in diverse cellular processes, such as cell-cycle progression, sensing of metabolic states, regulating of cellular growth, mediating of intracellular signaling, as well as executing deterministic cell response behaviors. In turn, mutations of proline-directed kinase consensus- and phosphorylation-sites are often involved in different forms of cancer and in neurodegenerative disorders.152-157 A second, less stringent proline −2 position has recently been identified as a supplementary specificity determinant for some proline +1-directed kinases.158,159
Whereas many prolines positively regulate kinase activities, by targeting them to their phosphorylation sites, proline residues within kinase consensus motifs can also weaken kinase activities, especially when they occur at positions −1 and −2, relative to the PTM sites,159 or even at positions +1.160-163 In other phosphorylation reactions, prolines play important roles in serving as specific kinase docking sites that are distal from actual phosphorylation sites but key to recruiting kinases to substrate proteins.164-166 In addition to kinases, the enzymatic properties of phosphatases are also modulated by prolines, either in the vicinities of phospho-sites167 or at distal docking sites.168,169 Finally, prolines that are close to modified substrate residues may critically influence PTM-mediated protein-protein interactions. It has been shown that phosphorylated serines, or threonines, followed by a proline, are more specifically recognized by subsets of 14–3-3 proteins170 or by Group IV WW domains.171,172
Roles of prolines in protein glycosylation
Glycosyltransferases are classes of enzymes that transfer sugar moieties onto proteins and they are strongly influenced by the presence of prolines in their substrate proteins. N-glycosylation of asparagines within the Asn-Xaa-Ser/Thr motif has been found to have a very low penetrance when the Xaa residue is a proline or when prolines are present at the +1 positions. In contrast, N-glycosylation is greatly enhanced when prolines are present at the −2 positions.173,174
O-glycosylation preferentially occurs in protein regions with high proline contents138,175 and particularly high proline frequencies have been reported for positions −1 and +3 relative to O-glycosylation sites.176 Both phosphorylation and glycosylation do not affect proline cis-conformer contents of phospho-Ser/Thr/Tyr-Pro motifs177-179 and of glyco-Ser-Pro motifs,180 respectively.
Roles of proline isomerases in PTM establishments
As mentioned previously, proline cis/trans isomerization reactions play important roles in protein folding and refolding processes, via the establishment of rather long-lived kinetic intermediates. Therefore, classes of cellular enzymes, so-called peptidyl-prolyl isomerases (PPIases), specifically enhance proline cis/trans isomerization without affecting their thermodynamic equilibrium states.181 PPIases are evolutionarily conserved and often characterized as foldases, or annotated as catalytic structural chaperones.182 Due to their inherent differences in stereochemistry, proline cis/trans isomers can also define different functional states of proteins.183 In these cases, PPIase activity drastically impacts protein function, as has been shown for the folded SH2 domain of the interleukin-2 inducible T-cell kinase (Itk)184-188 and the PHD-BRD tandem domain of the MLL1 protein.189-204 In both cases, proline cis/trans isomerization leads to large inter-domain conformational changes that subsequently affect protein-protein interaction behaviors.
Enhanced proline cis/trans isomerization in the presence of PPIases, leads to rapid sequestration of binding-competent protein states, which shifts the global population equilibrium toward the structure with which the more abundant binding partner interacts.184,189,191 Therefore, without changing protein free energies of cis/trans isomers, PPIases are capable of promoting new cis/trans distributions via additional factors that form complexes with, and thereby stabilize, individual isomer states. Because many IDPs are PPIase substrates,192-194 enzyme-controlled proline cis/trans isomerization processes provide intricate extensions to the long list of possible proline functions in IDPs. For example, proline isomerization controls switching of the adaptor protein Crk between two conformations: an auto-inhibitory state is stabilized by intramolecular association of two, tandem SH3 domains via a flexible linker IDPR containing a cis-proline isomer and a non-inhibited, activated conformation results from the promoted interconversion of this proline into its trans form. In turn, this particular cis/trans isomerization is targeted by the PPIase cyclophilin A.191
Among other PPIase enzymes, the phospho-dependent Pin1 [protein interacting with NIMA (never in mitosis A)-1] enzyme is of special interest. Pin1 functions in phospho-dependent signaling by catalyzing cis/trans interconversions of pSer/pThr-Pro peptide bonds in their phosphorylated states.151 Structurally, Pin1 consists of an N-terminal phospho-recognition WW domain and a C-terminal, catalytic PPIase domain.195 Whereas cis/trans population ratios in these Ser/Thr-Pro motifs are not affected by phosphorylation in a peptide/IDP context, cis/trans isomerization rates are severely reduced when the motif is modified.177-179 In folded proteins, the protein fold and amino acids that surround these Ser/Thr-Pro sites often stabilize, or de-stabilize one of the isomers. Enzymes such as Pin1 establish faster inter-conversion rates upon phosphorylation, which enables a 2-way control over the protein’s function:151,196-198 One way is regulation via phosphorylation, processed by a kinase or removed by a phosphatase, and a second way is control via isomerization, accelerated by a non-phospho-dependent PPIase or by the phospho-dependent PPIase Pin1.
Could similar 2-way controls be utilized by IDPs? A limitation is that Ser/Thr-Pro cis/trans thermodynamic equilibrium is not greatly affected by protein phosphorylation but is substantially affected in folded proteins. A supplementary IDP protein partner is thus required for the emergence of a function of the phospho-dependent cis/trans isomerization. For example, 2-way control like that discussed above has been observed for the pSer7-Pro8 motif within the intrinsically disordered, C-terminal domain (CTD) of RNA polymerase II, whose phosphorylation status correlates with transcriptional activity. Only the cis-isomer of the modified peptide motif serves as a substrate for the Ssu72 phosphatase.199,200 Hence, Ssu72-mediated dephosphorylation of the CTD pSer7-Pro8 sequence occurred much faster when Pin1 was present and proline cis/trans isomerization has been identified as the rate-limiting step in Ser7 dephosphorylation.
Another interesting example is afforded by pSer62 of the c-Myc oncoprotein, a key regulator of cell growth that is stabilized by Ser62 phosphorylation. Dephosphorylation by PP2A only occurs when Thr58-Pro59 is phosphorylated and Pin1 is present. Therefore, pSer62 dephosphorylation may similarly require Pro59 to be in the cis isomer state.201 Analogous relations between the Alzheimer disease-associated protein Tau, Pin1 and PP2 have been observed.202 Based on these examples, it is evident that PPIase activities represent important supplementary levels of regulatory controls in many cellular processes, although, in some cases, it remains unclear whether Pin1 binding, or catalysis, constitutes is the mechanism of action.203,204
Conclusions
Examples presented in this review show that there are multiple, distinct mechanisms by which proline regulates IDP and IDPR structure and function. The unique chemical properties of proline define its role as a modulator of secondary structural elements, but also its propensity to promote specific structural motifs such as the polyproline type II helix. In turn, these features appear to be especially important in regulating a multitude of functional IDP and IDPR properties that include their aggregation propensities. In addition, nature seems to have taken full advantage of the slow proline cis/trans isomerization characteristics in a number of biological processes that, altogether, extend the impressive functional range of this unique imino acid.
Glossary
Abbreviations:
- intrinsically disordered proteins
IDPs
- intrinsically disordered protein regions
IDPRs
- proline-rich motifs
PRMs
- proline-recognition domains
PRDs
- polyproline type II
PPII
- hydroxyproline
Hyp
- protein interacting with NIMA
Pin1
- never in mitosis A
NIMA
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Uversky VN. . Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol 2011; 43:1090 - 103; http://dx.doi.org/ 10.1016/j.biocel.2011.04.001; PMID: 21501695 [DOI] [PubMed] [Google Scholar]
- 2.Tompa P. . Unstructural biology coming of age. Curr Opin Struct Biol 2011; 21:419 - 25; http://dx.doi.org/ 10.1016/j.sbi.2011.03.012; PMID: 21514142 [DOI] [PubMed] [Google Scholar]
- 3.Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Garner E, Guilliot S, et al. . Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput 1998; •••:437 - 48; PMID: 9697202 [PubMed] [Google Scholar]
- 4.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. . Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform 2000; 11:161 - 71; PMID: 11700597 [PubMed] [Google Scholar]
- 5.Uversky VN, Gillespie JR, Fink AL. . Why are “natively unfolded” proteins unstructured under physiologic conditions?. Proteins 2000; 41:415 - 27; http://dx.doi.org/; PMID: 11025552 [DOI] [PubMed] [Google Scholar]
- 6.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. . Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 2004; 337:635 - 45; http://dx.doi.org/ 10.1016/j.jmb.2004.02.002; PMID: 15019783 [DOI] [PubMed] [Google Scholar]
- 7.Xue B, Dunker AK, Uversky VN. . Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn 2012; 30:137 - 49; http://dx.doi.org/ 10.1080/07391102.2012.675145; PMID: 22702725 [DOI] [PubMed] [Google Scholar]
- 8.Lee SH, Kim DH, Han JJ, Cha EJ, Lim JE, Cho YJ, et al. . Understanding pre-structured motifs (PreSMos) in intrinsically unfolded proteins. Curr Protein Pept Sci 2012; 13:34 - 54; http://dx.doi.org/ 10.2174/138920312799277974; PMID: 22044148 [DOI] [PubMed] [Google Scholar]
- 9.Uversky VN, Dunker AK. . Understanding protein non-folding. Biochim Biophys Acta 2010; 1804:1231 - 64; http://dx.doi.org/ 10.1016/j.bbapap.2010.01.017; PMID: 20117254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright PE, Dyson HJ. . Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 1999; 293:321 - 31; http://dx.doi.org/ 10.1006/jmbi.1999.3110; PMID: 10550212 [DOI] [PubMed] [Google Scholar]
- 11.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, et al. . Intrinsically disordered protein. J Mol Graph Model 2001; 19:26 - 59; http://dx.doi.org/ 10.1016/S1093-3263(00)00138-8; PMID: 11381529 [DOI] [PubMed] [Google Scholar]
- 12.Tompa P. . Intrinsically unstructured proteins. Trends Biochem Sci 2002; 27:527 - 33; http://dx.doi.org/ 10.1016/S0968-0004(02)02169-2; PMID: 12368089 [DOI] [PubMed] [Google Scholar]
- 13.Dyson HJ, Wright PE. . Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005; 6:197 - 208; http://dx.doi.org/ 10.1038/nrm1589; PMID: 15738986 [DOI] [PubMed] [Google Scholar]
- 14.Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. . Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 2002; 323:573 - 84; http://dx.doi.org/ 10.1016/S0022-2836(02)00969-5; PMID: 12381310 [DOI] [PubMed] [Google Scholar]
- 15.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradović Z. . Intrinsic disorder and protein function. Biochemistry 2002; 41:6573 - 82; http://dx.doi.org/ 10.1021/bi012159+; PMID: 12022860 [DOI] [PubMed] [Google Scholar]
- 16.Uversky VN. . Natively unfolded proteins: a point where biology waits for physics. Protein Sci 2002; 11:739 - 56; http://dx.doi.org/ 10.1110/ps.4210102; PMID: 11910019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uversky VN, Oldfield CJ, Dunker AK. . Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit 2005; 18:343 - 84; http://dx.doi.org/ 10.1002/jmr.747; PMID: 16094605 [DOI] [PubMed] [Google Scholar]
- 18.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. . Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J 2005; 272:5129 - 48; http://dx.doi.org/ 10.1111/j.1742-4658.2005.04948.x; PMID: 16218947 [DOI] [PubMed] [Google Scholar]
- 19.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, et al. . Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res 2007; 6:1882 - 98; http://dx.doi.org/ 10.1021/pr060392u; PMID: 17391014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, et al. . Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res 2007; 6:1899 - 916; http://dx.doi.org/ 10.1021/pr060393m; PMID: 17391015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J, Faeder JR, Camacho CJ. . Toward a quantitative theory of intrinsically disordered proteins and their function. Proc Natl Acad Sci U S A 2009; 106:19819 - 23; PMID: 19903882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim PM, Sboner A, Xia Y, Gerstein M. . The role of disorder in interaction networks: a structural analysis. Mol Syst Biol 2008; 4:179; http://dx.doi.org/ 10.1038/msb.2008.16; PMID: 18364713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. . Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 2008; 9:Suppl 1 S1; http://dx.doi.org/ 10.1186/1471-2164-9-S1-S1; PMID: 18366598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright PE, Dyson HJ. . Linking folding and binding. Curr Opin Struct Biol 2009; 19:31 - 8; http://dx.doi.org/ 10.1016/j.sbi.2008.12.003; PMID: 19157855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y, LeGall T, Oldfield CJ, Dunker AK, Uversky VN. . Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry 2006; 45:10448 - 60; http://dx.doi.org/ 10.1021/bi060981d; PMID: 16939197 [DOI] [PubMed] [Google Scholar]
- 26.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, et al. . Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res 2007; 6:1917 - 32; http://dx.doi.org/ 10.1021/pr060394e; PMID: 17391016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uversky VN, Oldfield CJ, Dunker AK. . Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys 2008; 37:215 - 46; http://dx.doi.org/ 10.1146/annurev.biophys.37.032807.125924; PMID: 18573080 [DOI] [PubMed] [Google Scholar]
- 28.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. . Unfoldomics of human genetic diseases: illustrative examples of ordered and intrinsically disordered members of the human diseasome. Protein Pept Lett 2009; 16:1533 - 47; http://dx.doi.org/ 10.2174/092986609789839377; PMID: 20001916 [DOI] [PubMed] [Google Scholar]
- 29.Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, et al. . Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics 2009; 10:Suppl 1 S7; http://dx.doi.org/ 10.1186/1471-2164-10-S1-S7; PMID: 19594884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. . Protein disorder in the human diseasome: unfoldomics of human genetic diseases. BMC Genomics 2009; 10:Suppl 1 S12; http://dx.doi.org/ 10.1186/1471-2164-10-S1-S12; PMID: 19594871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.James TL, Liu H, Ulyanov NB, Farr-Jones S, Zhang H, Donne DG, et al. . Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc Natl Acad Sci U S A 1997; 94:10086 - 91; http://dx.doi.org/ 10.1073/pnas.94.19.10086; PMID: 9294167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, et al. . Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem 2000; 275:29426 - 32; http://dx.doi.org/ 10.1074/jbc.M003107200; PMID: 10884388 [DOI] [PubMed] [Google Scholar]
- 33.Chi SW, Lee SH, Kim DH, Ahn MJ, Kim JS, Woo JY, et al. . Structural details on mdm2-p53 interaction. J Biol Chem 2005; 280:38795 - 802; http://dx.doi.org/ 10.1074/jbc.M508578200; PMID: 16159876 [DOI] [PubMed] [Google Scholar]
- 34.Uversky VN, Dunker AK. . Multiparametric analysis of intrinsically disordered proteins: looking at intrinsic disorder through compound eyes. Anal Chem 2012; 84:2096 - 104; http://dx.doi.org/ 10.1021/ac203096k; PMID: 22242801 [DOI] [PubMed] [Google Scholar]
- 35.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, et al. . Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput 1998; •••:473 - 84; PMID: 9697205 [PubMed] [Google Scholar]
- 36.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. . Intrinsic disorder and functional proteomics. Biophys J 2007; 92:1439 - 56; http://dx.doi.org/ 10.1529/biophysj.106.094045; PMID: 17158572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vacic V, Uversky VN, Dunker AK, Lonardi S. . Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics 2007; 8:211; http://dx.doi.org/ 10.1186/1471-2105-8-211; PMID: 17578581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams RM, Obradovi Z, Mathura V, Braun W, Garner EC, Young J, et al. . The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac Symp Biocomput 2001; •••:89 - 100; PMID: 11262981 [DOI] [PubMed] [Google Scholar]
- 39.Campen A, Williams RM, Brown CJ, Meng J, Uversky VN, Dunker AK. . TOP-IDP-scale: a new amino acid scale measuring propensity for intrinsic disorder. Protein Pept Lett 2008; 15:956 - 63; http://dx.doi.org/ 10.2174/092986608785849164; PMID: 18991772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown CJ, Johnson AK, Dunker AK, Daughdrill GW. . Evolution and disorder. Curr Opin Struct Biol 2011; 21:441 - 6; http://dx.doi.org/ 10.1016/j.sbi.2011.02.005; PMID: 21482101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown CJ, Johnson AK, Daughdrill GW. . Comparing models of evolution for ordered and disordered proteins. Mol Biol Evol 2010; 27:609 - 21; http://dx.doi.org/ 10.1093/molbev/msp277; PMID: 19923193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. . Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 2005; 44:12454 - 70; http://dx.doi.org/ 10.1021/bi050736e; PMID: 16156658 [DOI] [PubMed] [Google Scholar]
- 43.Vacic V, Oldfield CJ, Mohan A, Radivojac P, Cortese MS, Uversky VN, et al. . Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res 2007; 6:2351 - 66; http://dx.doi.org/ 10.1021/pr0701411; PMID: 17488107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, et al. . DisProt: the Database of Disordered Proteins. Nucleic Acids Res 2007; 35:Database issue D786 - 93; http://dx.doi.org/ 10.1093/nar/gkl893; PMID: 17145717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, et al. . The Universal Protein Resource (UniProt). Nucleic Acids Res 2005; 33:Database issue D154 - 9; http://dx.doi.org/ 10.1093/nar/gki070; PMID: 15608167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. . The Protein Data Bank. Nucleic Acids Res 2000; 28:235 - 42; http://dx.doi.org/ 10.1093/nar/28.1.235; PMID: 10592235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li SC, Goto NK, Williams KA, Deber CM. . α-helical, but not beta-sheet, propensity of proline is determined by peptide environment. Proc Natl Acad Sci U S A 1996; 93:6676 - 81; http://dx.doi.org/ 10.1073/pnas.93.13.6676; PMID: 8692877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chou PY, Fasman GD. . Empirical predictions of protein conformation. Annu Rev Biochem 1978; 47:251 - 76; http://dx.doi.org/ 10.1146/annurev.bi.47.070178.001343; PMID: 354496 [DOI] [PubMed] [Google Scholar]
- 49.Deber CM, Brodsky B, Rath A. Proline residues in proteins. In: Encyclopedia of Life Sciences (ELS). Chichester: John Wiley & Sons Ltd, 2010. [Google Scholar]
- 50.Ramachandran GN, Ramakrishnan C, Sasisekharan V. . Stereochemistry of polypeptide chain configurations. J Mol Biol 1963; 7:95 - 9; http://dx.doi.org/ 10.1016/S0022-2836(63)80023-6; PMID: 13990617 [DOI] [PubMed] [Google Scholar]
- 51.Maccallum PH, Poet R, Milner-White EJ. . Coulombic interactions between partially charged main-chain atoms not hydrogen-bonded to each other influence the conformations of alpha-helices and antiparallel beta-sheet. A new method for analysing the forces between hydrogen bonding groups in proteins includes all the Coulombic interactions. J Mol Biol 1995; 248:361 - 73; http://dx.doi.org/ 10.1016/S0022-2836(95)80056-5; PMID: 7739046 [DOI] [PubMed] [Google Scholar]
- 52.Ho BK, Thomas A, Brasseur R. . Revisiting the Ramachandran plot: hard-sphere repulsion, electrostatics, and H-bonding in the alpha-helix. Protein Sci 2003; 12:2508 - 22; http://dx.doi.org/ 10.1110/ps.03235203; PMID: 14573863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho BK, Brasseur R. . The Ramachandran plots of glycine and pre-proline. BMC Struct Biol 2005; 5:14; http://dx.doi.org/ 10.1186/1472-6807-5-14; PMID: 16105172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho BK, Coutsias EA, Seok C, Dill KA. . The flexibility in the proline ring couples to the protein backbone. Protein Sci 2005; 14:1011 - 8; http://dx.doi.org/ 10.1110/ps.041156905; PMID: 15772308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.MacArthur MW, Thornton JM. . Influence of proline residues on protein conformation. J Mol Biol 1991; 218:397 - 412; http://dx.doi.org/ 10.1016/0022-2836(91)90721-H; PMID: 2010917 [DOI] [PubMed] [Google Scholar]
- 56.Dugave C, Demange L. . Cis-trans isomerization of organic molecules and biomolecules: implications and applications. Chem Rev 2003; 103:2475 - 532; http://dx.doi.org/ 10.1021/cr0104375; PMID: 12848578 [DOI] [PubMed] [Google Scholar]
- 57.Pal D, Chakrabarti P. . Cis peptide bonds in proteins: residues involved, their conformations, interactions and locations. J Mol Biol 1999; 294:271 - 88; http://dx.doi.org/ 10.1006/jmbi.1999.3217; PMID: 10556045 [DOI] [PubMed] [Google Scholar]
- 58.Steinberg IZ, Berger A, Katchalski E. . Reverse mutarotation of poly-L-proline. Biochim Biophys Acta 1958; 28:647 - 8; http://dx.doi.org/ 10.1016/0006-3002(58)90537-7; PMID: 13560425 [DOI] [PubMed] [Google Scholar]
- 59.Steinberg IZ, Harrington WF, Berger A, Sela M, Katchalski E. . The configurational changes of poly-L-proline in solution. J Am Chem Soc 1960; 82:5263 - 79; http://dx.doi.org/ 10.1021/ja01505a001 [DOI] [Google Scholar]
- 60.Pahlke D, Freund C, Leitner D, Labudde D. . Statistically significant dependence of the Xaa-Pro peptide bond conformation on secondary structure and amino acid sequence. BMC Struct Biol 2005; 5:8; http://dx.doi.org/ 10.1186/1472-6807-5-8; PMID: 15804350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Exarchos KP, Papaloukas C, Exarchos TP, Troganis AN, Fotiadis DI. . Prediction of cis/trans isomerization using feature selection and support vector machines. J Biomed Inform 2009; 42:140 - 9; http://dx.doi.org/ 10.1016/j.jbi.2008.05.006; PMID: 18586558 [DOI] [PubMed] [Google Scholar]
- 62.Exarchos KP, Exarchos TP, Papaloukas C, Troganis AN, Fotiadis DI. . Detection of discriminative sequence patterns in the neighborhood of proline cis peptide bonds and their functional annotation. BMC Bioinformatics 2009; 10:113; http://dx.doi.org/ 10.1186/1471-2105-10-113; PMID: 19379512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zimmerman SS, Scheraga HA. . Stability of cis, trans, and nonplanar peptide groups. Macromolecules 1976; 9:408 - 16; http://dx.doi.org/ 10.1021/ma60051a005; PMID: 940354 [DOI] [PubMed] [Google Scholar]
- 64.Wedemeyer WJ, Welker E, Scheraga HA. . Proline cis-trans isomerization and protein folding. Biochemistry 2002; 41:14637 - 44; http://dx.doi.org/ 10.1021/bi020574b; PMID: 12475212 [DOI] [PubMed] [Google Scholar]
- 65.Valiaev A, Lim DW, Oas TG, Chilkoti A, Zauscher S. . Force-induced prolyl cis-trans isomerization in elastin-like polypeptides. J Am Chem Soc 2007; 129:6491 - 7; http://dx.doi.org/ 10.1021/ja070147r; PMID: 17469821 [DOI] [PubMed] [Google Scholar]
- 66.Anderson RJ, Weng Z, Campbell RK, Jiang X. . Main-chain conformational tendencies of amino acids. Proteins 2005; 60:679 - 89; http://dx.doi.org/ 10.1002/prot.20530; PMID: 16021632 [DOI] [PubMed] [Google Scholar]
- 67.Reimer U, Fischer G. . Local structural changes caused by peptidyl-prolyl cis/trans isomerization in the native state of proteins. Biophys Chem 2002; 96:203 - 12; http://dx.doi.org/ 10.1016/S0301-4622(02)00013-3; PMID: 12034441 [DOI] [PubMed] [Google Scholar]
- 68.Nozaki Y, Tanford C. . The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J Biol Chem 1971; 246:2211 - 7; PMID: 5555568 [PubMed] [Google Scholar]
- 69.Amend JP, Helgeson HC. . Solubilities of the common L-alpha-amino acids as a function of temperature and pH. Pure Appl Chem 1997; 59:935 - 42; http://dx.doi.org/ 10.1351/pac199769050935 [DOI] [Google Scholar]
- 70.Berger A, Kurtz J, Katchalski E. . Poly-L-proline. J Am Chem Soc 1954; 76:5552 - 4; http://dx.doi.org/ 10.1021/ja01650a082 [DOI] [Google Scholar]
- 71.Rath A, Davidson AR, Deber CM. . The structure of “unstructured” regions in peptides and proteins: role of the polyproline II helix in protein folding and recognition. Biopolymers 2005; 80:179 - 85; http://dx.doi.org/ 10.1002/bip.20227; PMID: 15700296 [DOI] [PubMed] [Google Scholar]
- 72.Cubellis MV, Caillez F, Blundell TL, Lovell SC. . Properties of polyproline II, a secondary structure element implicated in protein-protein interactions. Proteins 2005; 58:880 - 92; http://dx.doi.org/ 10.1002/prot.20327; PMID: 15657931 [DOI] [PubMed] [Google Scholar]
- 73.Moradi M, Babin V, Sagui C, Roland C. . PPII propensity of multiple-guest amino acids in a proline-rich environment. J Phys Chem B 2011; 115:8645 - 56; http://dx.doi.org/ 10.1021/jp203874f; PMID: 21630640 [DOI] [PubMed] [Google Scholar]
- 74.Adzhubei AA, Sternberg MJ. . Left-handed polyproline II helices commonly occur in globular proteins. J Mol Biol 1993; 229:472 - 93; http://dx.doi.org/ 10.1006/jmbi.1993.1047; PMID: 8429558 [DOI] [PubMed] [Google Scholar]
- 75.Adzhubei AA, Sternberg MJ. . Conservation of polyproline II helices in homologous proteins: implications for structure prediction by model building. Protein Sci 1994; 3:2395 - 410; http://dx.doi.org/ 10.1002/pro.5560031223; PMID: 7756993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma K, Kan L, Wang K. . Polyproline II helix is a key structural motif of the elastic PEVK segment of titin. Biochemistry 2001; 40:3427 - 38; http://dx.doi.org/ 10.1021/bi0022792; PMID: 11297408 [DOI] [PubMed] [Google Scholar]
- 77.Gutierrez-Cruz G, Van Heerden AH, Wang K. . Modular motif, structural folds and affinity profiles of the PEVK segment of human fetal skeletal muscle titin. J Biol Chem 2001; 276:7442 - 9; http://dx.doi.org/ 10.1074/jbc.M008851200; PMID: 11084039 [DOI] [PubMed] [Google Scholar]
- 78.Ma K, Wang K. . Malleable conformation of the elastic PEVK segment of titin: non-co-operative interconversion of polyproline II helix, beta-turn and unordered structures. Biochem J 2003; 374:687 - 95; http://dx.doi.org/ 10.1042/BJ20030702; PMID: 12816538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ma K, Forbes JG, Gutierrez-Cruz G, Wang K. . Titin as a giant scaffold for integrating stress and Src homology domain 3-mediated signaling pathways: the clustering of novel overlap ligand motifs in the elastic PEVK segment. J Biol Chem 2006; 281:27539 - 56; http://dx.doi.org/ 10.1074/jbc.M604525200; PMID: 16766517 [DOI] [PubMed] [Google Scholar]
- 80.Li SS. . Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J 2005; 390:641 - 53; http://dx.doi.org/ 10.1042/BJ20050411; PMID: 16134966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ball LJ, Kühne R, Schneider-Mergener J, Oschkinat H. . Recognition of proline-rich motifs by protein-protein-interaction domains. Angew Chem Int Ed Engl 2005; 44:2852 - 69; http://dx.doi.org/ 10.1002/anie.200400618; PMID: 15880548 [DOI] [PubMed] [Google Scholar]
- 82.Balázs A, Csizmok V, Buday L, Rakács M, Kiss R, Bokor M, et al. . High levels of structural disorder in scaffold proteins as exemplified by a novel neuronal protein, CASK-interactive protein1. FEBS J 2009; 276:3744 - 56; http://dx.doi.org/ 10.1111/j.1742-4658.2009.07090.x; PMID: 19523119 [DOI] [PubMed] [Google Scholar]
- 83.Tompa P, Csermely P. . The role of structural disorder in the function of RNA and protein chaperones. FASEB J 2004; 18:1169 - 75; http://dx.doi.org/ 10.1096/fj.04-1584rev; PMID: 15284216 [DOI] [PubMed] [Google Scholar]
- 84.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. . Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J 2005; 272:5129 - 48; http://dx.doi.org/ 10.1111/j.1742-4658.2005.04948.x; PMID: 16218947 [DOI] [PubMed] [Google Scholar]
- 85.Patil A, Nakamura H. . Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett 2006; 580:2041 - 5; http://dx.doi.org/ 10.1016/j.febslet.2006.03.003; PMID: 16542654 [DOI] [PubMed] [Google Scholar]
- 86.Ekman D, Light S, Björklund AK, Elofsson A. . What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae?. Genome Biol 2006; 7:R45; http://dx.doi.org/ 10.1186/gb-2006-7-6-r45; PMID: 16780599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, et al. . Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol 2006; 2:e100; http://dx.doi.org/ 10.1371/journal.pcbi.0020100; PMID: 16884331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dosztányi Z, Chen J, Dunker AK, Simon I, Tompa P. . Disorder and sequence repeats in hub proteins and their implications for network evolution. J Proteome Res 2006; 5:2985 - 95; http://dx.doi.org/ 10.1021/pr060171o; PMID: 17081050 [DOI] [PubMed] [Google Scholar]
- 89.Singh GP, Dash D. . Intrinsic disorder in yeast transcriptional regulatory network. Proteins 2007; 68:602 - 5; http://dx.doi.org/ 10.1002/prot.21497; PMID: 17510967 [DOI] [PubMed] [Google Scholar]
- 90.Singh GP, Ganapathi M, Dash D. . Role of intrinsic disorder in transient interactions of hub proteins. Proteins 2007; 66:761 - 5; http://dx.doi.org/ 10.1002/prot.21281; PMID: 17154416 [DOI] [PubMed] [Google Scholar]
- 91.Cortese MS, Uversky VN, Dunker AK. . Intrinsic disorder in scaffold proteins: getting more from less. Prog Biophys Mol Biol 2008; 98:85 - 106; http://dx.doi.org/ 10.1016/j.pbiomolbio.2008.05.007; PMID: 18619997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Purdy MA, Lara J, Khudyakov YE. . The hepatitis E virus polyproline region is involved in viral adaptation. PLoS One 2012; 7:e35974; http://dx.doi.org/ 10.1371/journal.pone.0035974; PMID: 22545153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pudupakam RS, Kenney SP, Córdoba L, Huang YW, Dryman BA, Leroith T, et al. . Mutational analysis of the hypervariable region of hepatitis e virus reveals its involvement in the efficiency of viral RNA replication. J Virol 2011; 85:10031 - 40; http://dx.doi.org/ 10.1128/JVI.00763-11; PMID: 21775444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fuxreiter M, Tompa P, Simon I. . Local structural disorder imparts plasticity on linear motifs. Bioinformatics 2007; 23:950 - 6; http://dx.doi.org/ 10.1093/bioinformatics/btm035; PMID: 17387114 [DOI] [PubMed] [Google Scholar]
- 95.Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, et al. . Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci U S A 2008; 105:5762 - 7; http://dx.doi.org/ 10.1073/pnas.0801353105; PMID: 18391200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, et al. . Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci U S A 2005; 102:15412 - 7; http://dx.doi.org/ 10.1073/pnas.0504614102; PMID: 16234232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Di Lello P, Jenkins LM, Jones TN, Nguyen BD, Hara T, Yamaguchi H, et al. . Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell 2006; 22:731 - 40; http://dx.doi.org/ 10.1016/j.molcel.2006.05.007; PMID: 16793543 [DOI] [PubMed] [Google Scholar]
- 98.Jonker HR, Wechselberger RW, Boelens R, Folkers GE, Kaptein R. . Structural properties of the promiscuous VP16 activation domain. Biochemistry 2005; 44:827 - 39; http://dx.doi.org/ 10.1021/bi0482912; PMID: 15654739 [DOI] [PubMed] [Google Scholar]
- 99.Kim DH, Lee SH, Nam KH, Chi SW, Chang I, Han KH. . Multiple hTAF(II)31-binding motifs in the intrinsically unfolded transcriptional activation domain of VP16. BMB Rep 2009; 42:411 - 7; http://dx.doi.org/ 10.5483/BMBRep.2009.42.7.411; PMID: 19643037 [DOI] [PubMed] [Google Scholar]
- 100.Ikeda K, Stuehler T, Meisterernst M. . The H1 and H2 regions of the activation domain of herpes simplex virion protein 16 stimulate transcription through distinct molecular mechanisms. Genes Cells 2002; 7:49 - 58; http://dx.doi.org/ 10.1046/j.1356-9597.2001.00492.x; PMID: 11856373 [DOI] [PubMed] [Google Scholar]
- 101.Csizmok V, Felli IC, Tompa P, Banci L, Bertini I. . Structural and dynamic characterization of intrinsically disordered human securin by NMR spectroscopy. J Am Chem Soc 2008; 130:16873 - 9; http://dx.doi.org/ 10.1021/ja805510b; PMID: 19053469 [DOI] [PubMed] [Google Scholar]
- 102.Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, et al. . Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci U S A 2004; 101:12491 - 6; http://dx.doi.org/ 10.1073/pnas.0403643101; PMID: 15314214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Müller-Späth S, Soranno A, Hirschfeld V, Hofmann H, Rüegger S, Reymond L, et al. . From the Cover: Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc Natl Acad Sci U S A 2010; 107:14609 - 14; http://dx.doi.org/ 10.1073/pnas.1001743107; PMID: 20639465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yamada J, Phillips JL, Patel S, Goldfien G, Calestagne-Morelli A, Huang H, et al. . A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol Cell Proteomics 2010; 9:2205 - 24; http://dx.doi.org/ 10.1074/mcp.M000035-MCP201; PMID: 20368288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Boze H, Marlin T, Durand D, Pérez J, Vernhet A, Canon F, et al. . Proline-rich salivary proteins have extended conformations. Biophys J 2010; 99:656 - 65; http://dx.doi.org/ 10.1016/j.bpj.2010.04.050; PMID: 20643086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Canon F, Ballivian R, Chirot F, Antoine R, Sarni-Manchado P, Lemoine J, et al. . Folding of a salivary intrinsically disordered protein upon binding to tannins. J Am Chem Soc 2011; 133:7847 - 52; http://dx.doi.org/ 10.1021/ja200534f; PMID: 21524106 [DOI] [PubMed] [Google Scholar]
- 107.Jöbstl E, O’Connell J, Fairclough JP, Williamson MP. . Molecular model for astringency produced by polyphenol/protein interactions. Biomacromolecules 2004; 5:942 - 9; http://dx.doi.org/ 10.1021/bm0345110; PMID: 15132685 [DOI] [PubMed] [Google Scholar]
- 108.Marsh JA, Forman-Kay JD. . Sequence determinants of compaction in intrinsically disordered proteins. Biophys J 2010; 98:2383 - 90; http://dx.doi.org/ 10.1016/j.bpj.2010.02.006; PMID: 20483348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilmot CM, Thornton JM. . Analysis and prediction of the different types of beta-turn in proteins. J Mol Biol 1988; 203:221 - 32; http://dx.doi.org/ 10.1016/0022-2836(88)90103-9; PMID: 3184187 [DOI] [PubMed] [Google Scholar]
- 110.Wilmot CM, Thornton JM. . Beta-turns and their distortions: a proposed new nomenclature. Protein Eng 1990; 3:479 - 93; http://dx.doi.org/ 10.1093/protein/3.6.479; PMID: 2371257 [DOI] [PubMed] [Google Scholar]
- 111.Pierson NA, Chen L, Russell DH, Clemmer DE. . Cis-trans isomerizations of proline residues are key to bradykinin conformations. J Am Chem Soc 2013; 135:3186 - 92; http://dx.doi.org/ 10.1021/ja3114505; PMID: 23373819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Glaves R, Baer M, Schreiner E, Stoll R, Marx D. . Conformational dynamics of minimal elastin-like polypeptides: the role of proline revealed by molecular dynamics and nuclear magnetic resonance. Chemphyschem 2008; 9:2759 - 65; http://dx.doi.org/ 10.1002/cphc.200800474; PMID: 18972488 [DOI] [PubMed] [Google Scholar]
- 113.Cheng S, Cetinkaya M, Gräter F. . How sequence determines elasticity of disordered proteins. Biophys J 2010; 99:3863 - 9; http://dx.doi.org/ 10.1016/j.bpj.2010.10.011; PMID: 21156127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Choi UB, McCann JJ, Weninger KR, Bowen ME. . Beyond the random coil: stochastic conformational switching in intrinsically disordered proteins. Structure 2011; 19:566 - 76; http://dx.doi.org/ 10.1016/j.str.2011.01.011; PMID: 21481779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Richardson JS, Richardson DC. . Amino acid preferences for specific locations at the ends of alpha helices. Science 1988; 240:1648 - 52; http://dx.doi.org/ 10.1126/science.3381086; PMID: 3381086 [DOI] [PubMed] [Google Scholar]
- 116.Kruus E, Thumfort P, Tang C, Wingreen NS. . Gibbs sampling and helix-cap motifs. Nucleic Acids Res 2005; 33:5343 - 53; http://dx.doi.org/ 10.1093/nar/gki842; PMID: 16174845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fonseca NA, Camacho R, Magalhães AL. . Amino acid pairing at the N- and C-termini of helical segments in proteins. Proteins 2008; 70:188 - 96; http://dx.doi.org/ 10.1002/prot.21525; PMID: 17654550 [DOI] [PubMed] [Google Scholar]
- 118.Gunasekaran K, Nagarajaram HA, Ramakrishnan C, Balaram P. . Stereochemical punctuation marks in protein structures: glycine and proline containing helix stop signals. J Mol Biol 1998; 275:917 - 32; http://dx.doi.org/ 10.1006/jmbi.1997.1505; PMID: 9480777 [DOI] [PubMed] [Google Scholar]
- 119.Engel DE, DeGrado WF. . Amino acid propensities are position-dependent throughout the length of alpha-helices. J Mol Biol 2004; 337:1195 - 205; http://dx.doi.org/ 10.1016/j.jmb.2004.02.004; PMID: 15046987 [DOI] [PubMed] [Google Scholar]
- 120.Kim MK, Kang YK. . Positional preference of proline in alpha-helices. Protein Sci 1999; 8:1492 - 9; http://dx.doi.org/ 10.1110/ps.8.7.1492; PMID: 10422838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cochran DA, Penel S, Doig AJ. . Effect of the N1 residue on the stability of the alpha-helix for all 20 amino acids. Protein Sci 2001; 10:463 - 70; http://dx.doi.org/ 10.1110/ps.31001; PMID: 11344315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cochran DA, Doig AJ. . Effect of the N2 residue on the stability of the alpha-helix for all 20 amino acids. Protein Sci 2001; 10:1305 - 11; http://dx.doi.org/ 10.1110/ps.50701; PMID: 11420432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kini RM. . Proline brackets and identification of potential functional sites in proteins: toxins to therapeutics. Toxicon 1998; 36:1659 - 70; http://dx.doi.org/ 10.1016/S0041-0101(98)00159-7; PMID: 9792183 [DOI] [PubMed] [Google Scholar]
- 124.Kini RM, Evans HJ. . A hypothetical structural role for proline residues in the flanking segments of protein-protein interaction sites. Biochem Biophys Res Commun 1995; 212:1115 - 24; http://dx.doi.org/ 10.1006/bbrc.1995.2084; PMID: 7626100 [DOI] [PubMed] [Google Scholar]
- 125.Monsellier E, Chiti F. . Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep 2007; 8:737 - 42; http://dx.doi.org/ 10.1038/sj.embor.7401034; PMID: 17668004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Richardson JS, Richardson DC. . Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci U S A 2002; 99:2754 - 9; http://dx.doi.org/ 10.1073/pnas.052706099; PMID: 11880627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Steward A, Adhya S, Clarke J. . Sequence conservation in Ig-like domains: the role of highly conserved proline residues in the fibronectin type III superfamily. J Mol Biol 2002; 318:935 - 40; http://dx.doi.org/ 10.1016/S0022-2836(02)00184-5; PMID: 12054791 [DOI] [PubMed] [Google Scholar]
- 128.Rousseau F, Serrano L, Schymkowitz JW. . How evolutionary pressure against protein aggregation shaped chaperone specificity. J Mol Biol 2006; 355:1037 - 47; http://dx.doi.org/ 10.1016/j.jmb.2005.11.035; PMID: 16359707 [DOI] [PubMed] [Google Scholar]
- 129.Rauscher S, Pomès R. . Structural disorder and protein elasticity. Adv Exp Med Biol 2012; 725:159 - 83; http://dx.doi.org/ 10.1007/978-1-4614-0659-4_10; PMID: 22399324 [DOI] [PubMed] [Google Scholar]
- 130.Rauscher S, Baud S, Miao M, Keeley FW, Pomès R. . Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure 2006; 14:1667 - 76; http://dx.doi.org/ 10.1016/j.str.2006.09.008; PMID: 17098192 [DOI] [PubMed] [Google Scholar]
- 131.Walsh CT, Garneau-Tsodikova S, Gatto GJ Jr.. . Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl 2005; 44:7342 - 72; http://dx.doi.org/ 10.1002/anie.200501023; PMID: 16267872 [DOI] [PubMed] [Google Scholar]
- 132.Gsponer J, Futschik ME, Teichmann SA, Babu MM. . Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 2008; 322:1365 - 8; http://dx.doi.org/ 10.1126/science.1163581; PMID: 19039133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, et al. . The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 2004; 32:1037 - 49; http://dx.doi.org/ 10.1093/nar/gkh253; PMID: 14960716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, et al. . Identification, analysis, and prediction of protein ubiquitination sites. Proteins 2010; 78:365 - 80; http://dx.doi.org/ 10.1002/prot.22555; PMID: 19722269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Xue B, Jeffers V, Sullivan WJ, Uversky VN. . Protein intrinsic disorder in the acetylome of intracellular and extracellular Toxoplasma gondii. Mol Biosyst 2013; 9:645 - 57; http://dx.doi.org/ 10.1039/c3mb25517d; PMID: 23403842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Daily KM, Radivojac P, Dunker AK. Intrinsic disorder and protein modifications: building an SVM predictor for methylation. IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology, CIBCB 2005:475-81.
- 137.Shao J, Xu D, Tsai SN, Wang Y, Ngai SM. . Computational identification of protein methylation sites through bi-profile Bayes feature extraction. PLoS One 2009; 4:e4920; http://dx.doi.org/ 10.1371/journal.pone.0004920; PMID: 19290060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nishikawa I, Nakajima Y, Ito M, Fukuchi S, Homma K, Nishikawa K. . Computational Prediction of O-linked Glycosylation Sites That Preferentially Map on Intrinsically Disordered Regions of Extracellular Proteins. Int J Mol Sci 2010; 11:4991 - 5008; http://dx.doi.org/ 10.3390/ijms11124991; PMID: 21614187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Deribe YL, Pawson T, Dikic I. . Post-translational modifications in signal integration. Nat Struct Mol Biol 2010; 17:666 - 72; http://dx.doi.org/ 10.1038/nsmb.1842; PMID: 20495563 [DOI] [PubMed] [Google Scholar]
- 140.Khoury GA, Baliban RC, Floudas CA. . Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci Rep 2011; 1; http://dx.doi.org/ 10.1038/srep00090; PMID: 22034591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gorres KL, Raines RT. . Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol 2010; 45:106 - 24; http://dx.doi.org/ 10.3109/10409231003627991; PMID: 20199358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.McCaldon P, Argos P. . Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide sequences. Proteins 1988; 4:99 - 122; http://dx.doi.org/ 10.1002/prot.340040204; PMID: 3227018 [DOI] [PubMed] [Google Scholar]
- 143.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. . Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 2001; 292:468 - 72; http://dx.doi.org/ 10.1126/science.1059796; PMID: 11292861 [DOI] [PubMed] [Google Scholar]
- 144.Tompa P, Buzder-Lantos P, Tantos A, Farkas A, Szilágyi A, Bánóczi Z, et al. . On the sequential determinants of calpain cleavage. J Biol Chem 2004; 279:20775 - 85; http://dx.doi.org/ 10.1074/jbc.M313873200; PMID: 14988399 [DOI] [PubMed] [Google Scholar]
- 145.Ubersax JA, Ferrell JE Jr.. . Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 2007; 8:530 - 41; http://dx.doi.org/ 10.1038/nrm2203; PMID: 17585314 [DOI] [PubMed] [Google Scholar]
- 146.Goldsmith EJ, Akella R, Min X, Zhou T, Humphreys JM. . Substrate and docking interactions in serine/threonine protein kinases. Chem Rev 2007; 107:5065 - 81; http://dx.doi.org/ 10.1021/cr068221w; PMID: 17949044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Brown NR, Noble ME, Endicott JA, Johnson LN. . The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat Cell Biol 1999; 1:438 - 43; http://dx.doi.org/ 10.1038/15674; PMID: 10559988 [DOI] [PubMed] [Google Scholar]
- 148.Harper JW, Adams PD. . Cyclin-dependent kinases. Chem Rev 2001; 101:2511 - 26; http://dx.doi.org/ 10.1021/cr0001030; PMID: 11749386 [DOI] [PubMed] [Google Scholar]
- 149.Avruch J. . MAP kinase pathways: the first twenty years. Biochim Biophys Acta 2007; 1773:1150 - 60; http://dx.doi.org/ 10.1016/j.bbamcr.2006.11.006; PMID: 17229475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Songyang Z, Lu KP, Kwon YT, Tsai LH, Filhol O, Cochet C, et al. . A structural basis for substrate specificities of protein Ser/Thr kinases: primary sequence preference of casein kinases I and II, NIMA, phosphorylase kinase, calmodulin-dependent kinase II, CDK5, and Erk1. Mol Cell Biol 1996; 16:6486 - 93; PMID: 8887677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lu KP, Zhou XZ. . The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol 2007; 8:904 - 16; http://dx.doi.org/ 10.1038/nrm2261; PMID: 17878917 [DOI] [PubMed] [Google Scholar]
- 152.Blume-Jensen P, Hunter T. . Oncogenic kinase signalling. Nature 2001; 411:355 - 65; http://dx.doi.org/ 10.1038/35077225; PMID: 11357143 [DOI] [PubMed] [Google Scholar]
- 153.Nigg EA. . Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol 2001; 2:21 - 32; http://dx.doi.org/ 10.1038/35048096; PMID: 11413462 [DOI] [PubMed] [Google Scholar]
- 154.Lee MS, Tsai LH. . Cdk5: one of the links between senile plaques and neurofibrillary tangles?. J Alzheimers Dis 2003; 5:127 - 37; PMID: 12719630 [DOI] [PubMed] [Google Scholar]
- 155.Lu KP, Liou YC, Vincent I. . Proline-directed phosphorylation and isomerization in mitotic regulation and in Alzheimer’s Disease. Bioessays 2003; 25:174 - 81; http://dx.doi.org/ 10.1002/bies.10223; PMID: 12539244 [DOI] [PubMed] [Google Scholar]
- 156.Lim J, Lu KP. . Pinning down phosphorylated tau and tauopathies. Biochim Biophys Acta 2005; 1739:311 - 22; http://dx.doi.org/ 10.1016/j.bbadis.2004.10.003; PMID: 15615648 [DOI] [PubMed] [Google Scholar]
- 157.Lu KP. . Pinning down cell signaling, cancer and Alzheimer’s disease. Trends Biochem Sci 2004; 29:200 - 9; http://dx.doi.org/ 10.1016/j.tibs.2004.02.002; PMID: 15082314 [DOI] [PubMed] [Google Scholar]
- 158.Mok J, Kim PM, Lam HY, Piccirillo S, Zhou X, Jeschke GR, et al. . Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci Signal 2010; 3:ra12; http://dx.doi.org/ 10.1126/scisignal.2000482; PMID: 20159853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Alexander J, Lim D, Joughin BA, Hegemann B, Hutchins JR, Ehrenberger T, et al. . Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci Signal 2011; 4:ra42; http://dx.doi.org/ 10.1126/scisignal.2001796; PMID: 21712545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zhu G, Fujii K, Belkina N, Liu Y, James M, Herrero J, et al. . Exceptional disfavor for proline at the P + 1 position among AGC and CAMK kinases establishes reciprocal specificity between them and the proline-directed kinases. J Biol Chem 2005; 280:10743 - 8; http://dx.doi.org/ 10.1074/jbc.M413159200; PMID: 15647260 [DOI] [PubMed] [Google Scholar]
- 161.Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. . Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr Biol 1994; 4:973 - 82; http://dx.doi.org/ 10.1016/S0960-9822(00)00221-9; PMID: 7874496 [DOI] [PubMed] [Google Scholar]
- 162.Tuazon PT, Spanos WC, Gump EL, Monnig CA, Traugh JA. . Determinants for substrate phosphorylation by p21-activated protein kinase (gamma-PAK). Biochemistry 1997; 36:16059 - 64; http://dx.doi.org/ 10.1021/bi9717845; PMID: 9405039 [DOI] [PubMed] [Google Scholar]
- 163.Marin O, Meggio F, Draetta G, Pinna LA. . The consensus sequences for cdc2 kinase and for casein kinase-2 are mutually incompatible. A study with peptides derived from the beta-subunit of casein kinase-2. FEBS Lett 1992; 301:111 - 4; http://dx.doi.org/ 10.1016/0014-5793(92)80221-2; PMID: 1451779 [DOI] [PubMed] [Google Scholar]
- 164.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. . Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev 1999; 13:163 - 75; http://dx.doi.org/ 10.1101/gad.13.2.163; PMID: 9925641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Murphy LO, Blenis J. . MAPK signal specificity: the right place at the right time. Trends Biochem Sci 2006; 31:268 - 75; http://dx.doi.org/ 10.1016/j.tibs.2006.03.009; PMID: 16603362 [DOI] [PubMed] [Google Scholar]
- 166.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, et al. . The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell 2003; 115:83 - 95; http://dx.doi.org/ 10.1016/S0092-8674(03)00725-6; PMID: 14532005 [DOI] [PubMed] [Google Scholar]
- 167.Gray CH, Good VM, Tonks NK, Barford D. . The structure of the cell cycle protein Cdc14 reveals a proline-directed protein phosphatase. EMBO J 2003; 22:3524 - 35; http://dx.doi.org/ 10.1093/emboj/cdg348; PMID: 12853468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shi Y. . Serine/threonine phosphatases: mechanism through structure. Cell 2009; 139:468 - 84; http://dx.doi.org/ 10.1016/j.cell.2009.10.006; PMID: 19879837 [DOI] [PubMed] [Google Scholar]
- 169.Roy J, Cyert MS. . Cracking the phosphatase code: docking interactions determine substrate specificity. Sci Signal 2009; 2:re9; http://dx.doi.org/ 10.1126/scisignal.2100re9; PMID: 19996458 [DOI] [PubMed] [Google Scholar]
- 170.Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. . Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J 2010; 427:69 - 78; http://dx.doi.org/ 10.1042/BJ20091834; PMID: 20141511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. . Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol 2000; 7:639 - 43; http://dx.doi.org/ 10.1038/77929; PMID: 10932246 [DOI] [PubMed] [Google Scholar]
- 172.Kato Y, Ito M, Kawai K, Nagata K, Tanokura M. . Determinants of ligand specificity in groups I and IV WW domains as studied by surface plasmon resonance and model building. J Biol Chem 2002; 277:10173 - 7; http://dx.doi.org/ 10.1074/jbc.M110490200; PMID: 11751914 [DOI] [PubMed] [Google Scholar]
- 173.Ben-Dor S, Esterman N, Rubin E, Sharon N. . Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology 2004; 14:95 - 101; http://dx.doi.org/ 10.1093/glycob/cwh004; PMID: 14514714 [DOI] [PubMed] [Google Scholar]
- 174.Igura M, Kohda D. . Quantitative assessment of the preferences for the amino acid residues flanking archaeal N-linked glycosylation sites. Glycobiology 2011; 21:575 - 83; http://dx.doi.org/ 10.1093/glycob/cwq196; PMID: 21115605 [DOI] [PubMed] [Google Scholar]
- 175.Thanka Christlet TH, Veluraja K. . Database analysis of O-glycosylation sites in proteins. Biophys J 2001; 80:952 - 60; http://dx.doi.org/ 10.1016/S0006-3495(01)76074-2; PMID: 11159462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wilson IB, Gavel Y, von Heijne G. . Amino acid distributions around O-linked glycosylation sites. Biochem J 1991; 275:529 - 34; PMID: 2025231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schutkowski M, Bernhardt A, Zhou XZ, Shen M, Reimer U, Rahfeld JU, et al. . Role of phosphorylation in determining the backbone dynamics of the serine/threonine-proline motif and Pin1 substrate recognition. Biochemistry 1998; 37:5566 - 75; http://dx.doi.org/ 10.1021/bi973060z; PMID: 9548941 [DOI] [PubMed] [Google Scholar]
- 178.Guo YT, Li YM, Zhu ZT, Zhao YF. . Effect of the phosphate group with different negative charges on the conformation of phosphorylated Ser/Thr-Pro motif. Int J Pept Res Ther 2005; 11:159 - 65; http://dx.doi.org/ 10.1007/s10989-005-4710-2 [DOI] [Google Scholar]
- 179.Byun BJ, Kang YK. . Conformational preferences and prolyl cis-trans isomerization of phosphorylated Ser/Thr-Pro motifs. Biopolymers 2010; 93:330 - 9; PMID: 19885922 [DOI] [PubMed] [Google Scholar]
- 180.Pao YL, Wormarld MR, Dwek RA, Lellouch AC. . Effect of serine O-glycosylation on cis-trans proline isomerization. Biochem Biophys Res Commun 1996; 219:157 - 62; http://dx.doi.org/ 10.1006/bbrc.1996.0198; PMID: 8619800 [DOI] [PubMed] [Google Scholar]
- 181.Schmid FX. . Prolyl isomerases. Adv Protein Chem 2001; 59:243 - 82; http://dx.doi.org/ 10.1016/S0065-3233(01)59008-7; PMID: 11868274 [DOI] [PubMed] [Google Scholar]
- 182.Wang CC, Tsou CL. . Enzymes as chaperones and chaperones as enzymes. FEBS Lett 1998; 425:382 - 4; http://dx.doi.org/ 10.1016/S0014-5793(98)00272-5; PMID: 9563498 [DOI] [PubMed] [Google Scholar]
- 183.Andreotti AH. . Native state proline isomerization: an intrinsic molecular switch. Biochemistry 2003; 42:9515 - 24; http://dx.doi.org/ 10.1021/bi0350710; PMID: 12911293 [DOI] [PubMed] [Google Scholar]
- 184.Min L, Fulton DB, Andreotti AH. . A case study of proline isomerization in cell signaling. Front Biosci 2005; 10:385 - 97; http://dx.doi.org/ 10.2741/1536; PMID: 15574377 [DOI] [PubMed] [Google Scholar]
- 185.Brazin KN, Fulton DB, Andreotti AH. . A specific intermolecular association between the regulatory domains of a Tec family kinase. J Mol Biol 2000; 302:607 - 23; http://dx.doi.org/ 10.1006/jmbi.2000.4091; PMID: 10986122 [DOI] [PubMed] [Google Scholar]
- 186.Mallis RJ, Brazin KN, Fulton DB, Andreotti AH. . Structural characterization of a proline-driven conformational switch within the Itk SH2 domain. Nat Struct Biol 2002; 9:900 - 5; http://dx.doi.org/ 10.1038/nsb864; PMID: 12402030 [DOI] [PubMed] [Google Scholar]
- 187.Brazin KN, Mallis RJ, Fulton DB, Andreotti AH. . Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proc Natl Acad Sci U S A 2002; 99:1899 - 904; http://dx.doi.org/ 10.1073/pnas.042529199; PMID: 11830645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Colgan J, Asmal M, Neagu M, Yu B, Schneidkraut J, Lee Y, et al. . Cyclophilin A regulates TCR signal strength in CD4+ T cells via a proline-directed conformational switch in Itk. Immunity 2004; 21:189 - 201; http://dx.doi.org/ 10.1016/j.immuni.2004.07.005; PMID: 15308100 [DOI] [PubMed] [Google Scholar]
- 189.Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, et al. . Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell 2010; 141:1183 - 94; http://dx.doi.org/ 10.1016/j.cell.2010.05.016; PMID: 20541251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ayton PM, Cleary ML. . Molecular mechanisms of leukemogenesis mediated by MLL fusion proteins. Oncogene 2001; 20:5695 - 707; http://dx.doi.org/ 10.1038/sj.onc.1204639; PMID: 11607819 [DOI] [PubMed] [Google Scholar]
- 191.Sarkar P, Saleh T, Tzeng SR, Birge RB, Kalodimos CG. . Structural basis for regulation of the Crk signaling protein by a proline switch. Nat Chem Biol 2011; 7:51 - 7; http://dx.doi.org/ 10.1038/nchembio.494; PMID: 21131971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Takahashi K, Uchida C, Shin RW, Shimazaki K, Uchida T. . Prolyl isomerase, Pin1: new findings of post-translational modifications and physiological substrates in cancer, asthma and Alzheimer’s disease. Cell Mol Life Sci 2008; 65:359 - 75; http://dx.doi.org/ 10.1007/s00018-007-7270-0; PMID: 17965833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Meuvis J, Gerard M, Desender L, Baekelandt V, Engelborghs Y. . The conformation and the aggregation kinetics of α-synuclein depend on the proline residues in its C-terminal region. Biochemistry 2010; 49:9345 - 52; http://dx.doi.org/ 10.1021/bi1010927; PMID: 20828147 [DOI] [PubMed] [Google Scholar]
- 194.Nelson CJ, Santos-Rosa H, Kouzarides T. . Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 2006; 126:905 - 16; http://dx.doi.org/ 10.1016/j.cell.2006.07.026; PMID: 16959570 [DOI] [PubMed] [Google Scholar]
- 195.Wintjens R, Wieruszeski JM, Drobecq H, Rousselot-Pailley P, Buée L, Lippens G, et al. . 1H NMR study on the binding of Pin1 Trp-Trp domain with phosphothreonine peptides. J Biol Chem 2001; 276:25150 - 6; http://dx.doi.org/ 10.1074/jbc.M010327200; PMID: 11313338 [DOI] [PubMed] [Google Scholar]
- 196.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, et al. . Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 1997; 278:1957 - 60; http://dx.doi.org/ 10.1126/science.278.5345.1957; PMID: 9395400 [DOI] [PubMed] [Google Scholar]
- 197.Ranganathan R, Lu KP, Hunter T, Noel JP. . Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent. Cell 1997; 89:875 - 86; http://dx.doi.org/ 10.1016/S0092-8674(00)80273-1; PMID: 9200606 [DOI] [PubMed] [Google Scholar]
- 198.Lu PJ, Zhou XZ, Shen M, Lu KP. . Function of WW domains as phosphoserine- or phosphothreonine-binding modules. Science 1999; 283:1325 - 8; http://dx.doi.org/ 10.1126/science.283.5406.1325; PMID: 10037602 [DOI] [PubMed] [Google Scholar]
- 199.Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, et al. . cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J Biol Chem 2011; 286:5717 - 26; http://dx.doi.org/ 10.1074/jbc.M110.197129; PMID: 21159777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Bataille AR, Jeronimo C, Jacques PE, Laramée L, Fortin ME, Forest A, et al. . A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 2012; 45:158 - 70; http://dx.doi.org/ 10.1016/j.molcel.2011.11.024; PMID: 22284676 [DOI] [PubMed] [Google Scholar]
- 201.Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, et al. . A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol 2004; 6:308 - 18; http://dx.doi.org/ 10.1038/ncb1110; PMID: 15048125 [DOI] [PubMed] [Google Scholar]
- 202.Landrieu I, Smet-Nocca C, Amniai L, Louis JV, Wieruszeski JM, Goris J, et al. . Molecular implication of PP2A and Pin1 in the Alzheimer’s disease specific hyperphosphorylation of Tau. PLoS One 2011; 6:e21521; http://dx.doi.org/ 10.1371/journal.pone.0021521; PMID: 21731772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lu KP, Finn G, Lee TH, Nicholson LK. . Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol 2007; 3:619 - 29; http://dx.doi.org/ 10.1038/nchembio.2007.35; PMID: 17876319 [DOI] [PubMed] [Google Scholar]
- 204.Lippens G, Landrieu I, Smet C. . Molecular mechanisms of the phospho-dependent prolyl cis/trans isomerase Pin1. FEBS J 2007; 274:5211 - 22; http://dx.doi.org/ 10.1111/j.1742-4658.2007.06057.x; PMID: 17892493 [DOI] [PubMed] [Google Scholar]