Abstract
Thymosine β4 (Tß4) is a 43 amino acid long intrinsically disordered protein (IDP), which was initially identified as an actin-binding and sequestering molecule. Later it was described to have multiple other functions, such as regulation of endothelial cell differentiation, blood vessel formation, wound repair, cardiac cell migration, and survival.1 The various functions of Tβ4 are mediated by interactions with distinct and structurally unrelated partners, such as PINCH, ILK, and stabilin-2, besides the originally identified G-actin. Although the cellular readout of these interactions and the formation of these complexes have been thoroughly described, no attempt was made to study these interactions in detail, and to elucidate the thermodynamic, kinetic, and structural underpinning of this range of moonlighting functions. Because Tβ4 is mostly disordered, and its 4 described partners are structurally unrelated (the CTD of stabilin-2 is actually fully disordered), it occurred to us that this system might be ideal to characterize the structural adaptability and ensuing moonlighting functions of IDPs. Unexpectedly, we found that Tβ4 engages in multiple weak, transient, and fuzzy interactions, i.e., it is capable of mediating distinct yet specific interactions without adapting stable folded structures.
Keywords: ILK, PINCH, Thymosin β4, fuzzy complex, intrinsically disordered protein, moonlighting, stabilin, uncoupled binding and folding, weak interaction
Introduction
Intrinsically disordered proteins (IDPs) lack well-defined 3D structures under native conditions, yet they fulfill important functions primarily linked with signaling and regulation in cell physiology.2-5 Structural disorder abounds in all organisms, particularly in higher eukaryotes,6,7 and is thought to function either directly due to the lack of a stable structure (entropic chains) or via molecular recognition accompanied by induced folding.8 In certain cases, specific recognition of partner molecule(s) has been described without a transition to a fully-folded state, termed fuzziness.9,10 Structural disorder appears to impart many functional advantages in protein-protein interactions, such as increased speed of interaction, specificity without excessive binding strength, and structural adaptability to different binding partners. A key functional manifestation of this latter is moonlighting, when the protein carries out many, apparently unrelated functions.11 As appears in a few concrete cases, such as p53,12 binding to alternative partners is thought to occur via the acquisition of alternative structures, attesting to the extreme structural adaptability of IDPs. The functional complexity of the proteome may critically depend on this ability and ensuing moonlighting, because such proteins can function as molecular switches that effectively regulate the direction of information flow in the interactome.
Thymosine β4 (Tß4) is a small IDP (43 residues), with diverse experimental evidence attesting to its moonlighting functions.13 Being functionally, but not structurally well-characterized, it is an excellent candidate for studying the conformational underpinning of moonlighting functions. Tß4 was first identified as an actin-sequestring molecule,14 playing a key role in regulating the formation and modulation of the actin cytoskeleton.15 As actin-binding is considered its main function, this interaction is well-studied and well characterized,16 and the structure of Tß4 in complex with G-actin was published in 2004.17 During the following years many different functions were linked to this small protein, such as endothelial cell differentiation,18 angiogenesis,19 and wound repair.13 Tß4 increases the concentration of metalloproteinases, which is important in cell migration in many different cell types.20 The precise mechanism underlying this effect is unclear, but multiple related and in some cases independent mechanisms have been envisioned.21 Tß4 emerged as a potentially useful therapeutic agent in epidermolysis bullosa, an inherited disease characterized by the presence of recurrent blisters, mechanical fragility of the skin and other epithelial structures, and scar formation.22
Tß4 can also promote cardiac repair through increasing cardiac cell migration and survival,1 probably via interacting with 2 proteins, PINCH (Particularly Interesting New Cys His containing protein) and ILK (Integrin Linked Kinase).1 These partners also interact directly with each other and indirectly with the actin cytoskeleton as part of a larger complex known as the focal adhesion complex. Both proteins are required for cell survival and motility.23 The interaction with Tß4 has been mapped to the fourth and fifth LIM domains of PINCH, and to the N-terminal ankyrin domain of ILK.1 These interactions were later confirmed in other works24,25 and it was also shown that Tß4 plays an active role in NF-KB signaling by inhibiting TNF-α-induced NF-ƙB activation.26 This effect was independent of PINCH and ILK and it was suggested that there might be a binding competition between Tß4/PINCH/ILK and the newly identified Tß4-RelA/p65 complex. This raises the possibility that Tß4 targets the proinflammatory signaling mediated by PINCH/ILK, but not PINCH/ILK molecules themselves, to suppress inflammation.
Another Tß4-interacting protein was found by Lee and coworkers.27 Using GST-pull down, co-immunoprecipitation assays and immunofluorescence imaging they confirmed that the cytoplasmic domain (CTD) of stabilin-2 (an endocytic receptor for hyaluronic acid) binds Tß4. This interaction renders Tß4 a role in the stabilin-2 mediated phagocytosis of apoptotic cells, but its exact mechanism remains to be elucidated. Although the cellular function of these interactions and the formation of these complexes have been thoroughly characterized, no attempt was made to study them in vitro to elucidate the thermodynamic, kinetic, and structural underpinning of their formation. Whereas the different functional terms of Tβ4 are not orthogonal (that is, they refer to function at different levels), the molecule fits with the moonlighting concept, because its binding to different partners does entail different types of molecular effects. Upon binding to actin, it directly sequesters this protein from polymerization, whereas by binding to PINCH and ILK it affects upstream cytoskeletal signaling. With regards to stabilin, it may have a completely different function in endycytotic signaling. In addition, because Tβ4 is mostly disordered, and its 4 described partners are structurally unrelated (the CTD of stabilin-2 is in fact disordered itself), this system might be ideal to characterize the structural adaptability and ensuing moonlighting functions of IDPs. Unexpectedly, and without precedence in the literature, we found that Tβ4 engages in multiple fuzzy interactions, i.e., it is capable of mediating its distinct yet specific interactions without adapting distinct folded structures in the different complexes.
Results
Weak interaction of Tβ4 with PINCH LIM4–5 and ILK-Ank-GST
PINCH LIM4–5 was expressed as His-tagged protein, while the Ankyrin repeat region of ILK was stable only while conjugated to GST. Enzymatic cleavage of the GST tag was unsuccessful, leading to immediate aggregation of the cleaved protein.
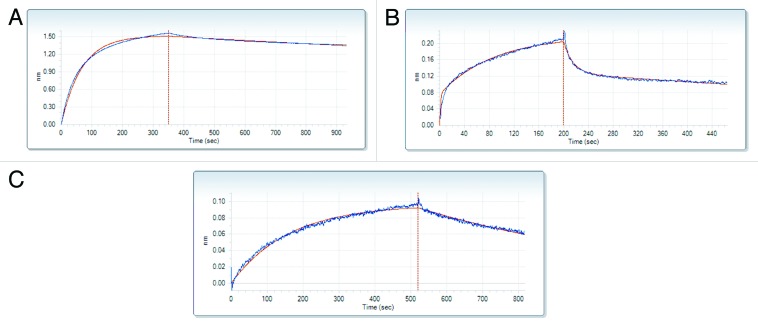
Both protein constructs associated with Tß4 very weakly, with a high KD, 0.6 mM for PINCH LIM4–5 and 0.4 mM for ILK-Ank-GST as confirmed by biolayer interferometry (BLI) (Fig. 1A and B; Table 1). An interesting feature of the reaction between Tß4 and PINCH LIM4–5 is that the dissociation is a much slower process (kd = 1.18x10−5 s−1) than the association (ka = 7.25 M−1s−1). The interaction with ILK-Ank-GST was somewhat stronger, but also in the millimolar range (Table 1). The negative control p27Kip1 did not bind to any of the partners. Due to the nature of the experiment there was no possibility to measure Tß4 binding to both of the partners together.
Figure 1. Biolayer interferometry (BLI). Corrected binding curves of Tß4 and its different partners: PINCH LIM4-5 (A), ILK-Ank-GST (B) and stabilin (C). The concentration of Tß4 was 3 mM in each case. Red line represents the fitted curve used to calculate kinetic parameters.
Table 1. Kinetic parameters determined in the BLI measurements.
| ka (M−1s−1) | kd (s−1) | KD (M) | |
| PINCH LIM4–5 | 7.5 | 1.2 x 10−5 | 6.3 x 10−4 |
| ILK-Ank-GST | 2.65 x 10−4 | 9.2 x 10−4 | 3.8 x 10−4 |
| Stabilin | 22.1 | 9.8 x 10−4 | 2.8 x 10−3 |
Isothermal titration calorimetry measurements were unsuccessful due to the extremely low heat capacity changes and low KD of the binding. Even though the interaction was confirmed by the results, data were not consistent enough to fit a binding curve and calculate thermodynamic constants with either of the partners.
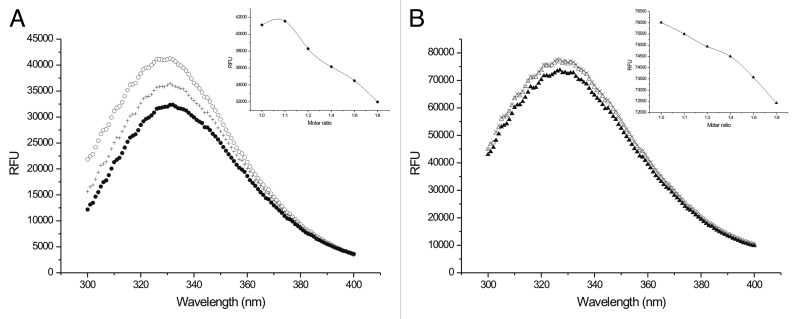
Steady-state fluorescence measurements confirmed the weak interactions with both partners (Fig. 2A and B). A concentration-dependent decrease in the intensity of the tryptophan fluorescence could be observed upon titration with Tß4. Since Tβ4 has no tryptophanes or tyrosines in its sequence, in this experiment the fluorescence of the partners were detected and Tβ4 was titrated to the solution of the partners. An important feature and a signal of weak binding is the fact that even at 1:8 (partner:Tβ4) molar ratio there was no saturation of the complex as could be seen from the constant decrease of the fluorescence intensity measured at 330 nm (insets of Fig. 2A and B). In order to exclude the unspecific effects of the changing composition of the solution control titrations were performed by adding 2 different tripeptides (GGG and GHG) to the partner molecules (PINCH LIM4-5 and ILK-Ank-GST). Since we could not detect any change in fluorescence throughout a broad concentration range, we concluded that the measured fluorescence intensity change originated from the specific interaction of Tβ4 and its partners. The higher fluorescence intensity and relative smaller changes with ILK-Ank-GST result from the high background fluorescence of GST. We performed control experiments with Tβ4 titrated to GST and there was no apparent change in the fluorescence signal (data not shown).
Figure 2. Steady-state fluorescence. (A) Fluorescence spectrum of PINCH LIM4-5 titrated with different concentrations of Tß4. ○: PINCH LIM-4-5, +:: 1:4 molar ratio of PINCH LIM4-5:Tß4; ●: 1:8 molar ratio of PINCH LIM4-5:Tß4. Inset: decrease of the maximum fluorescence measured at 330 nm at different molar ratios of PINCH LIM4-5 and Tß4. (B) Fluorescence spectrum of ILK-Ank-GST titrated with different concentrations of Tß4. △: ILK-Ank-GST, +: 1:4 molar ratio of ILK-Ank-GST:Tß4; ▲: 1:8 molar ratio of ILK-Ank-GST:Tß4. Inset: decrease of the maximum fluorescence measured at 330 nm at different molar ratios of ILK-Ank-GST and Tß4.
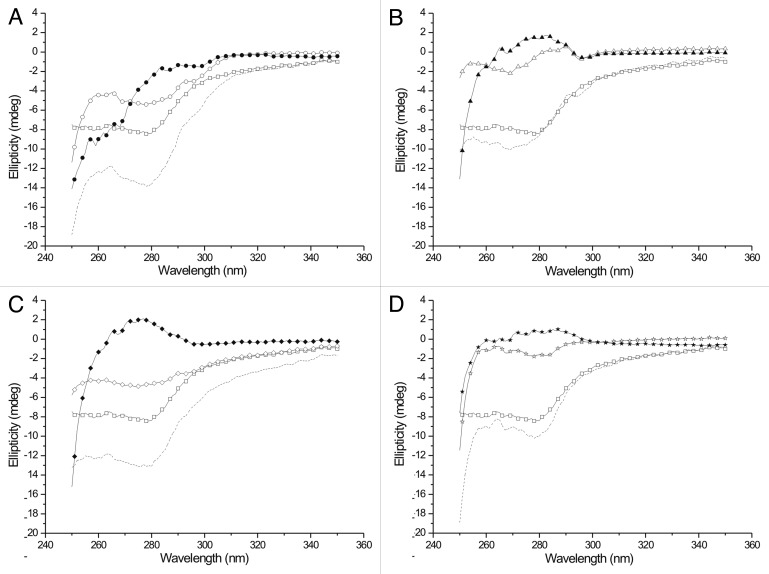
The near UV CD spectra of the complexes show that the binding involves 1 or several tryptophans of PINCH and ILK (Fig. 3.) As Tß4 has no tryptophan or tyrosine residues, the signal in the near UV CD spectrum arises mainly from the partner molecules. Although from these experiments no structural information can be subtracted, the fact of binding is clearly shown by the marked change of the spectra upon addition of Tß4. The measured spectra differ both from the spectra of the separated partners and the algebraic sum of their spectra. The spectrum of the 1:1 mixture of PINCH LIM4–5 and ILK-Ank-GST is a mixture of the separate molecules, which is reasonable, since these constructs lack the sites needed for their interaction. When Tß4 was added, the resulting spectrum was similar, but not identical to that of the ILK/Tß4 complex, suggesting that when both partners are present, the interaction of Tß4 with ILK-Ank-GST is preferable over the interaction with PINCH LIM4-5.
Figure 3. Near UV CD spectra of Tß4 and its partners. Tß4 is labeled with open squares and the algebraic sum of the spectra of the separated partners is labeled with dashed line on every panel. (A) interaction with PINCH LIM4-5. ○: PINCH LIM4-5 in the absence of Tß4; ●: complex of PINCH LIM4-5 and Tß4. (B) interaction with ILK-Ank-GST. △: ILK-Ank-GST in the absence of Tß4; ▲: complex of ILK-Ank-GST and Tß4. (C) interaction with PINCH LIM4-5 and ILK-Ank-GST. ◇: 1:1 mixture of PINCH LIM4-5 and ILK-Ank-GST; ◆: complex of PINCH LIM4-5, ILK-Ank-GST and Tß4. (D) interaction with Stabilin CTD. ☆: stabilin CTD in the absence of Tß4; ★: complex of stabilin CTD and Tß4.
Fuzziness in Tß4/PINCH LIM4-5 and Tß4/ILK-Ank-GST interactions
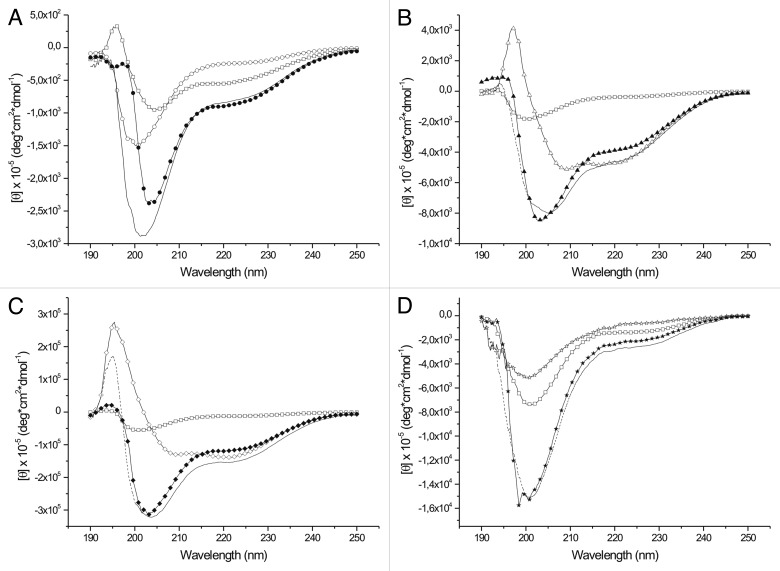
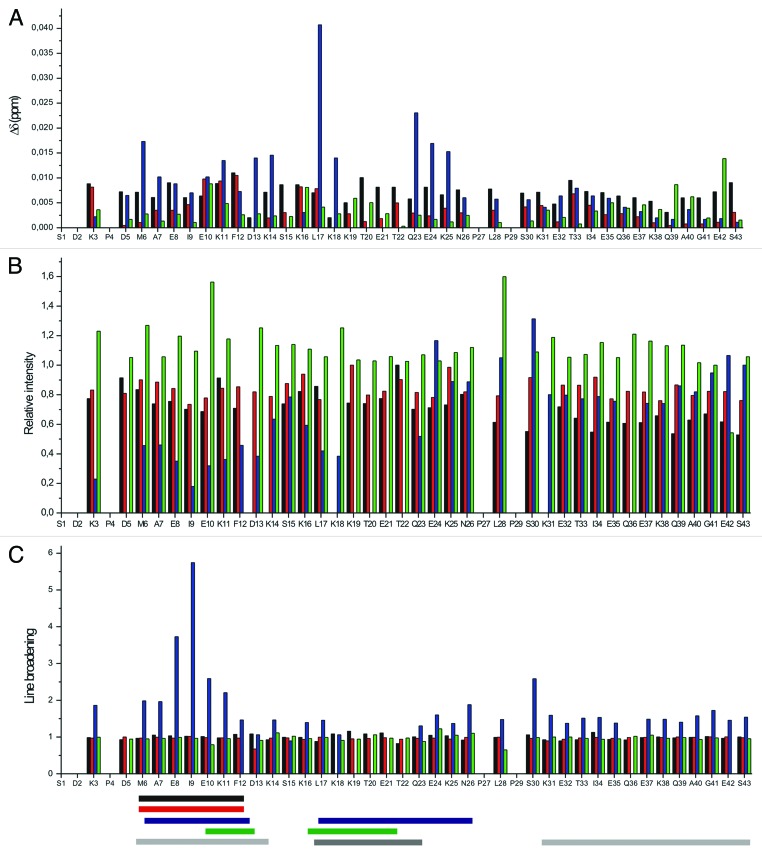
The results so far support the conclusion that Tß4 forms weak, but relatively stable complexes with PINCH LIM4-5 and ILK-Ank-GST. Surprisingly, formation of these complexes is not accompanied by significant changes in the far UV CD spectra of the proteins when mixed together (Fig. 4). As expected, Tß4 has a far UV CD spectrum typical of a disordered protein, characterized by a large negative peak around 200 nm. PINCH LIM4-5 and ILK-Ank-GST have CD spectra of globular proteins. PINCH LIM4–5 has a spectrum which is comprised from β sheet and α helix components (sheet and helix content 0.44 and 0.068 respectively), but there is a considerable amount of flexible regions present in the molecule (β turns: 0.19; unordered content: 0.3). ILK-Ank-GST had mostly α-helical spectrum which corresponds well to the structure of the ANK repeat region and the fusion partner GST (sheet and helix content 0.174 and 0.56, respectively). When Tβ4 was added to PINCH LIM4–5 the observable shift of the negative peak from 200 to 205 nm was slightly larger than in the algebraic sum of the spectra of the partners (Fig. 4A), but no significant differences could be observed when Tβ4 was added to ILK-GST (Fig. 4B) or to the mixture of the 2 partners (Fig. 4C). Ellipticity at 222 nm is proportional to the α-helix content of a protein and based on our far UV CD measurements there was no elevation in the helical content of the proteins when Tβ4 is added. In fact the measured ellipticity at 222 nm was lower than the sum of the ellipticity read of the 2 separate partners in each case, with the exception of PINCH LIM4–5, but the difference was only marginal and was not statistically significant. The results were quite similar when the titrations were performed in excess of the partners relative to Tβ4 (data not shown). If the interaction involved the folding of Tβ4 into a well-defined structure, the far UV CD spectrum of the protein should have changed significantly under these circumstances. This signifies that no stable secondary structure is formed upon the interaction with any of the partners, even when both are present in the reaction mixture. This notion was further strengthened by the results of the NMR experiments. 1H-15N HSQC spectra of Tß4 titrated with unlabeled partners confirm that the proteins interact but Tß4 undergoes very small structural changes (Fig. 5A–C). The amide proton resonances keep their narrow dispersion in all cases, indicating that the environment of the residues of Tß4 does not change dramatically (and remain characteristic of a disordered protein). The small perturbations of the indicated resonances outline the interaction sites of the protein, but the lack of a stable three dimensional structure is clear. It is interesting to note that the observed perturbations only appeared when Tβ4 was present in excess in the solution. From the 1H-15N HSQC spectra it is evident that the 2 stable α helices that are formed in Tß4 upon binding to G-actin17 do not arise in these complexes. The perturbations are more expressed when Tß4 is added to the 1:1 mixture of PINCH LIM4-5 and ILK-Ank-GST, but the signs of stable structures are still clearly missing. The major changes involve the N-terminal and central part of Tß4, including residues 17–23, the G-actin binding site.28 The peaks of residues 19–22 cannot be observed at all on the 1H-15N HSQC spectrum of Tß4 titrated with the mixture of PINCH LIM4-5 and ILK-Ank-GST, suggesting that these constitute the main binding site of Tß4. From the combined chemical shift perturbations (Fig. 6A) it is evident that the behavior of Tß4 is different when PINCH LIM4-5 and ILK-Ank-GST are present separately or together. By the analysis of the intensity changes and line broadening (Fig. 6B and C) 2 regions of Tβ4 seem to be involved in the interaction with PINCH LIM4–5 and ILK together. One region ranges from residues 6–11 and the other the above mentioned region between residues 17–23. Even though these stronger perturbations are still very small compared with those observed during Tß4 binding to G-actin,17 they are clear and evident based on our measurements.
Figure 4. Far UV CD spectra of Tß4 and its partners. Tß4 is labeled with open squares and the algebraic sum of the spectra of the separated partners is labeled with dashed line on every panel. (A) interaction with PINCH LIM4-5. ○: PINCH LIM4-5 in the absence of Tß4; ●: complex of PINCH LIM4-5 and Tß4. (B) interaction with ILK-Ank-GST. △: ILK-Ank-GST in the absence of Tß4; ▲: complex of ILK-Ank-GST and Tß4. (C) interaction with PINCH LIM4-5 and ILK-Ank-GST. ◇: 1:1 mixture of PINCH LIM4–5 and ILK-Ank-GST; ◆: complex of PINCH LIM4-5, ILK-Ank-GST and Tß4. (D) interaction with Stabilin CTD. ☆: stabilin CTD in the absence of Tß4; ★: complex of stabilin CTD and Tß4.
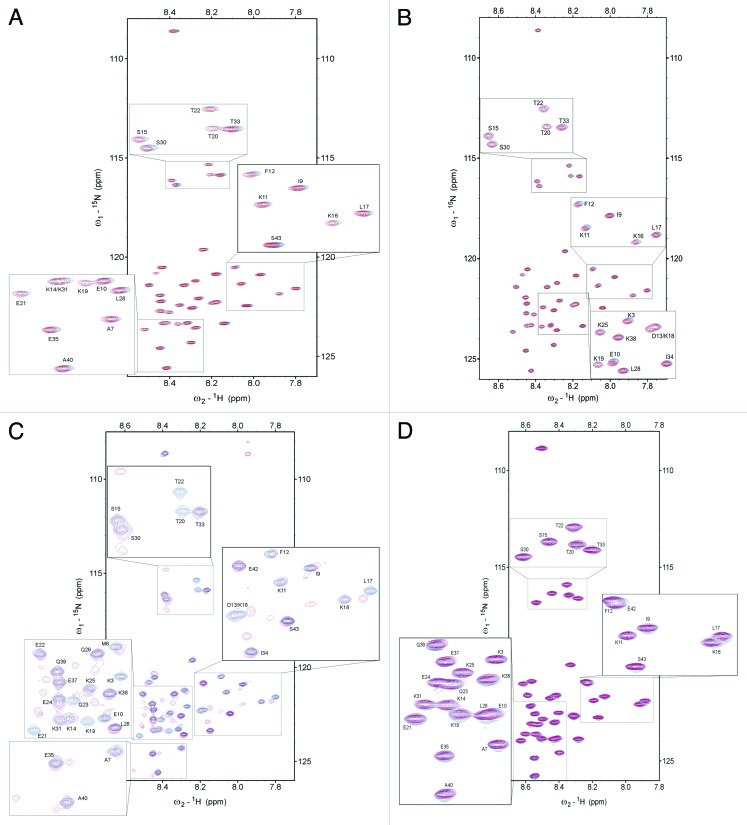
Figure 5.1H-15N HSQC spectra of Tß4 titrated with its partners. (A) 1H-15N HSQC overlaps of free (blue) 15N-Tß4 and with PINCH LIM4–5 (3:1 Tß4 excess, red). (B) 1H-15N HSQC overlaps of free (blue) 15N-tβ4 and with ILK-Ank-GST (3:1 Tß4 excess, blue). (C) 1H-15N HSQC overlaps of free (blue) 15N-Tß4 and with 1:1 mixture of PINCH LIM4–5 and ILK-Ank-GST (3:1 Tß4 excess, red). (D) 1H-15N HSQC overlaps of free (blue) 15N-Tß4 and stabilin CTD (2:1 Tß4 excess, red) in 50 mM sodium acetate and 50 mM NaCl pH 6.5 at 283 K.
Figure 6. Residue mapped perturbation based on amide combined chemical shift changes during interaction between tβ4 and its partners screened by 1H-15N-HSQC spectra seen on Figure 5 (A). (Note: D13/K18 and K14/K31 overlays.) (B) Relative intensity changes observed during the interaction between tβ4 and its partners. (C) Line broadening of the signals on the 1H-15N-HSQC spectra during the interaction between tβ4 and its partners. Black: interaction with PINCH LIM4-5; red: interaction with ILK-Ank-GST; blue: interaction with PINCH LIM4-5 and ILK-Ank-GST together; green: interaction with stabilin CTD. Horizontal bars represent the regions that primarily take part in the interaction with the specific partner. Black: PINCH LIM4-5; red: ILK-Ank-GST; blue: PINCH LIM4-5/ILK-Ank-GST; green: stabilin CTD; light gray: the 2 α helices that are formed upon G-actin binding; gray: the region important for matrix metalloproteinase activation.
Hydrodynamic and hydration properties of Tß4 in complexes with PINCH LIM4–5 and Tß4/ILK-Ank-GST
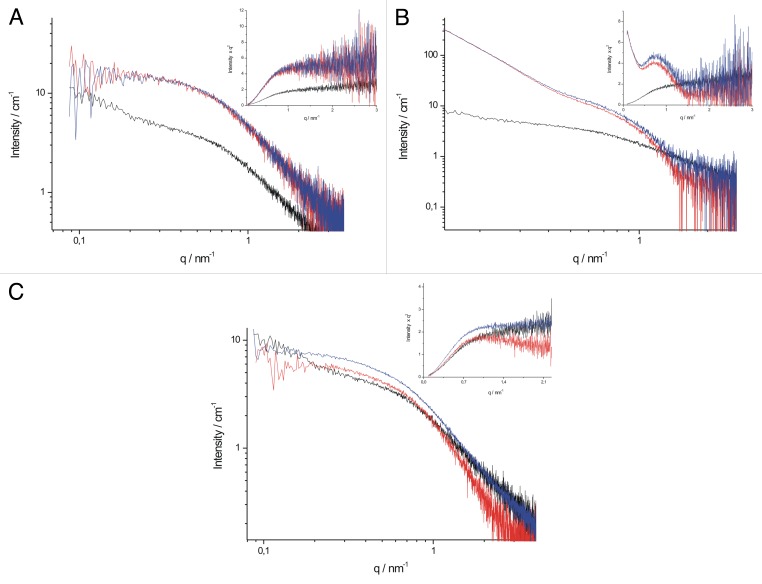
NMR experiments on residue-level disturbance suggest only limited local structural changes. To assess the overall hydrodynamic effects of the interactions, we performed small angle X-ray scattering (SAXS) measurements. SAXS gives low resolution picture of the shape and size of macromolecules and their complexes. The recorded scattering curves for the complexes of Tß4/PINCH LIM4-5 and Tß4/ILK-Ank-GST (Fig. 7A and B) differed only slightly from those of the globular partners, indicating that the overall shape of the partners does not change upon binding of Tß4. Unfortunately ILK-Ank-GST showed significant amount of aggregation due to the elongated storage, making the interpretation of the data unreliable. Kratky plots calculated from SAXS data give an indication of the structure of the studied macromolecule; disordered proteins lack a distinct peak on a Kratky plot, as demonstrated for Tß4 in the insets of Figure 7. The aggregation of ILK-Ank-GST is apparent on the Kratky plot and for this reason we concluded these results unreliable and they are not discussed in detail. It is evident from the curves of the complexes that there are no major structural changes and transition to a compact state upon complex formation with either of the partners. Unfortunately there was no possibility to record the SAXS curves of Tß4 with PINCH LIM4-5 and ILK-Ank-GST together.
Figure 7. SAXS scattering curves and Kratky plots of Tß4 and its partners. (A) scattering curves of PINCH LIM4–5 in the absence of Tß4 (red), Tß4 in the absence of PINCH LIM4-5 (black) and PINCH LIM4-5 in the presence of Tß4 (blue). Inset: Kratky plot of PINCH LIM4-5, Tß4 and their complexes. Labeling is the same as in (A). (B) scattering curves of ILK-Ank-GST in the absence of Tß4 (red), Tß4 in the absence of ILK-Ank-GST (black) and ILK-Ank-GST in the presence of Tß4 (blue). Inset: Kratky plot of ILK-Ank-GST, Tß4 and their complex. Labeling is the same as in (B). (C) scattering curves of stabilin CTD in the absence of Tß4 (red), Tß4 in the absence of stabilin CTD (black) and stabilin CTD in the presence of Tß4 (blue). Inset: Kratky plot of stabilin CTD, Tß4 and their complex. Labeling is the same as in (C).
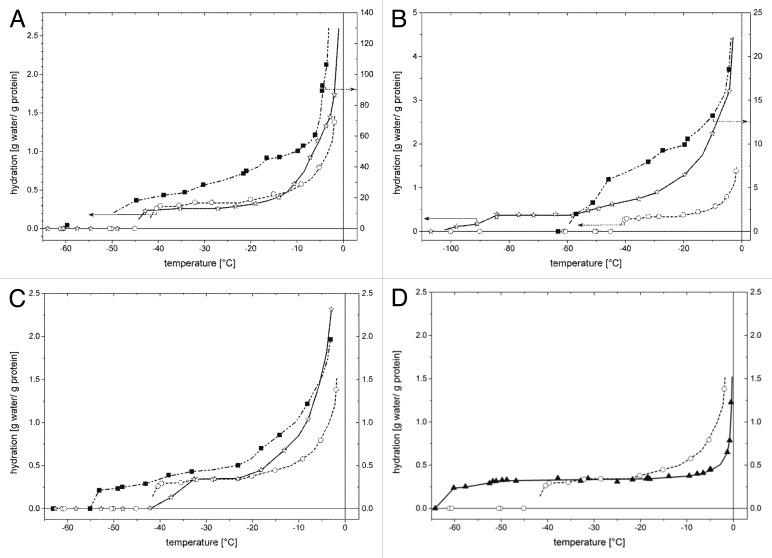
Wide-line 1H-NMR measurements can give information about the hydration of a protein or a protein complex which can be related to the level of order—or ordering upon complex formation—of a disordered protein. The wide-line 1H-NMR signal intensity is the measure of the number of mobile protons or equivalently, the number of mobile water molecules, which is expressed as hydration (h, gram water/gram protein). The slopes of hydration vs. temperature curves give a quantitative parameter characteristic of the heterogeneity of protein–water interactions.29,30 In our experience, the interactions of the IDPs with water molecules are characterized with broad energy distributions in contrast with the globular proteins for which these energies fall in a much narrower range.30 As a conclusive example, the hydration curves of the globular protein BSA and the IDP Tß4 are showed in Figure 8D. The low-temperature step in the hydration curve (i.e., the h vs. T data) appears when detectable molecular motion becomes active. The lower this ‘melting’ temperature, the weaker the interaction is between the water molecules and the protein compared with the water-water interaction in the bulk solvent. In this sense, PINCH LIM4-5 binds water less strongly than Tß4 (Fig. 8A). This is a consequence of the structure of the 2 proteins, since disordered proteins bind water molecules more strongly than structured molecules.30 The hydration curve of Tß4 dissolved in water shows an almost constant region below -20°C and a strongly changing part between -20 and 0°C (Fig. 8), which indicates that part of Tß4 has a tendency to gain structure known from previous studies.17
Figure 8. Hydration of Tß4 and its partners measured with wide-line 1H NMR. (A) Hydration of the proteins Tß4 (○, left ordinate), PINCH LIM4-5 (■, right ordinate), and Tß4/PINCH LIM4-5 complex (☆, left ordinate) measured for frozen solutions as a function of temperature. (B) Hydration of the proteins Tß4 (○, left ordinate), ILK-ANG-GST (■, right ordinate), and Tß4/ILK-Ank-GST complex (☆, left ordinate) measured for frozen solutions as a function of temperature. (C) Hydration of the proteins Tß4 (○), stabilin CTD (■), and Tß4/stabilin CTD complex (☆) measured for frozen solutions as a function of temperature. (D) Hydration of the proteins Tß4 (-) and bovine serum albumin (7) measured for frozen solutions as a function of temperature. The hydration values were calculated from the wide-line 1H-NMR signal intensity of the mobile water component. Lines are guides for the eye.
The hydration curve of PINCH LIM4-5 (Fig. 8A) dissolved in water increases continuously with increasing temperature. PINCH LIM4-5 has unusually higher hydration values than Tβ4 in the whole subzero temperature range. This rise of the hydration curve indicates that the mobile water molecules are involved in interactions which are energetically heterogeneous. This latter feature resembles the hydration curves of the IDPs.30,31 The hydration curves of globular proteins investigated earlier (ubiquitin29 and bovine serum albumin30,31) show a temperature independent, constant value below -10°C. The slope of a hydration curve does not depend on the precise concentration of the protein solution, and so it is a more error-proof parameter than the hydration values. The complex formed by Tß4 with PINCH LIM4-5 is hydrated similarly to the stand-alone Tß4 (Fig. 8A). The hydration vs. temperature curve measured for the Tß4/PINCH LIM4-5 solution cannot be reproduced as the sum of the independently hydrated Tß4 and PINCH LIM4-5 proteins. The Tß4/PINCH complex has somewhat lower hydration values below -10°C than Tß4 alone. Raising the temperature above -10°C causes the rapid melting of further amounts of water interacting with the complex. This part of the curve resembles more to that of PINCH LIM4-5 alone. The fact that the hydration values of the complex stay below the separate partners at all temperatures shows that certain surfaces of the partners get buried upon interaction, although Tß4 does not approach the state of folded proteins.
ILK-Ank-GST alone binds water less strongly (lower ‘melting’ temperature) than Tß4 but stronger than the Tß4/ILK-Ank-GST complex does (Fig. 8B). The hydration curve measured for the isolated ILK-Ank-GST dissolved in water (Fig. 8B) shows a lower ‘melting’ temperature than Tß4, increases very steeply with increasing temperature, and shows that the protein has the highest hydration values. These features suggest that ILK-Ank-GST is relatively unstable and binds water weaker than Tß4. The water molecules hydrating the Tß4/ILK-Ank-GST complex become mobile at a temperature even lower than for the isolated ILK-Ank-GST (Fig. 8B). The hydration values measured for the Tß4/ILK-Ank-GST solution are lower than the hydration values of ILK-Ank-GST (Fig. 8B), indicating the formation of the complex with a considerable loss of solvent accessible surface of ILK-Ank-GST. The h vs. temperature values for the complex show a thermal trend, which is characteristic to IDPs.
Tß4 binding to stabilin CTD
Due to disorder of both partners, we treated Tß4-stabilin CTD interaction separately. Whereas induced folding upon binding to a structured partner is now a common theme in the IDP literature,8 binding of 2 IDPs together is an exception rather than the rule.32,33
Stabilin CTD was expressed as a His-tagged protein and Tß4 was untagged, just like in the former experiments. The interaction of the 2 protein constructs was first confirmed by BLI measurements (Fig. 1C). The KD of Tß4 binding to stabilin CTD (2.8 mM) is even higher than the KD measured with the other 2 partners (Table 1). Although the binding is extremely weak, it is not unspecific, since Tß4 did not interact with the intrinsically disordered p27Kip1 in the same experiment. Isothermal titration calorimetry measurements gave similar results of weak binding. The binding was accompanied by a small, favorable enthalpy change (∆H = -1752 cal/mol) and a very small positive change in entropy (∆S = 19.9 cal/mol/deg). This latter suggests that the interaction of these 2 proteins is not accompanied by the formation of stable structures, in fact, the protein(s) become slightly more disordered in the complex.
The near UV CD measurements also confirmed the interaction of the 2 proteins, since the spectrum of the complex is clearly different from that of the sum of the 2 separated partners (Fig. 3D). Stabilin CTD contains 2 tyrosines and 1 phenylalanine residue, making the interpretation of the near UV CD spectrum more straightforward. The 2 tyrosines seem to be more buried than the phenylalanine in stabilin CTD when measured alone. Upon addition of Tß4 the local environment of the tyrosine residues seems to be affected most strongly while the phenylalanine in Tß4 or in stabilin CTD does not change dramatically. Far UV CD measurements confirmed the conjecture that there is no major structural reorganization upon binding of these 2 proteins. As shown on Figure 4D, both proteins are of disordered nature and this does not change after complex formation.
This type of fuzzy binding was further supported by the results of the NMR experiments. 1H-15N HSQC spectrum of Tß4 titrated with unlabeled stabilin CTD is very much like the one with the earlier described partners, i.e. the disordered nature of the protein is retained even in the complex (Fig. 5D). Observable changes in the chemical environment of the backbone cluster in the region of the 2 α helices at the N- and C-terminus of Tß4 which are formed upon binding to G-actin, but changes in chemical shifts fall way short of values characteristic of the formation of α-helices (Fig. 6A). Intensity changes and line broadening (Fig. 6B and C) do not show considerable change upon Tβ4 titration with stabilin CTD.
SAXS measurements provided yet another proof of Tß4 binding stabilin CTD while lacking stable tertiary structure. The scattering curve of the complex differs evidently from that of the separated partners (Fig. 7C), suggesting that there is interaction of the 2 proteins. The sharp increase of the scattering curve of Tß4 at small angles might be a result of a certain amount of protein association. It would be misleading to call it aggregation, since no aggregation of Tß4 could be detected with gel filtration or native PAGE and this could not be eliminated either with centrifugation or flitration. This upturn of the scattering curve is not present in the scattering curve of the complex, so it is reasonable to surmise that interaction with stabilin CTD disrupts these Tß4 associates. Kratky plots of the proteins reveal that stabilin CTD is less disordered than Tß4, even though there was no sign of stable secondary structures present in the far UV CD spectrum of the protein. This can be reconciled by a higher helical propensity in stabilin CTD which cannot be detected by CD, but is observable with SAXS. Interestingly, Kratky plot of the Tß4/stabilin CTD complex bears the characteristics of a more disordered protein, suggesting that instead of gaining structure, stabilin CTD loses some of its already present structural elements.
The wide-line 1H-NMR measurements also confirm that there is more order in stabilin CTD than in Tß4. As it can be deduced from the temperatures where detectable molecular motions become active, stabilin CTD alone binds water less strongly (lower ‘melting’ temperature) than Tß4 and the Tß4/stabilin CTD complex does (Fig. 8C). The temperature-trends of the hydration curves measured for the proteins (Fig. 8C) suggest that isolated Tß4 and stabilin CTD, as well as their complex, are IDPs. The hydration curves of Tß4 and the complex are indistinguishable between -35 and -20°C. The hydration of the complex approaches the hydration of stabilin CTD with increasing temperatures and these 2hydration curves become equal above -10°C. At this temperature several molecular layers of water become mobile and therefore the amount of the mobile water content cannot be considered as indicative of the size of the solvent accessible surface of the protein. The mobility and the amount of the unfrozen water are high enough even for translational diffusion of water in this temperature range. The formation of the complex results in a considerable loss of solvent accessible surface of stabilin CTD, again pointing to interaction with no structure.
Discussion
With increasingly deeper understanding of the structural and functional characteristics of IDPs, the notion of functional proteins not having a stable three dimensional structure becomes ever more accepted. Key observations published in the last few years outline a new feature of disordered proteins: binding without disorder-to-order transition (binding without induced folding).34 This type of binding is best characterized for the cytoplasmic domains of immune receptors,33,35-37 but there are other interactions that have been reported to form without clear disorder-to-order transition38,39 and “fuzziness,” i.e. structural disorder in the bound state of proteins might be a rather general phenomenon.9,10 Such fuzzy complexes may have major importance in moonlighting, when a protein fulfills more than one, often unrelated functions. IDPs are capable of structurally adapt to their partners, as shown for p21cip1 and p53, that can bind in many different conformations to many different partners.40,12 It has not been explored, however, if moonlighting can also be combined with fuzzy interactions, which would challenge our notion of specificity of interactions even further.
Tß4 is a typical moonlighting protein with many different functions.13 We chose 2 distinct functions and the related partners to study the structural background of the moonlighting ability of Tß4. Its involvement in cardiac cell repair is manifested through interactions with two focal adhesion proteins, ILK and PINCH. In the first relevant report,1 the ankyrin repeat region of ILK and the 4th and 5th LIM domains of PINCH were found to be involved in interaction with Tß4. We cloned and expressed these protein domains to characterize their interaction with Tß4 both from a kinetic and a structural point of view and we found that the recombinant proteins formed very weak complexes. The several independent methods we used to investigate these interactions and their brief results are listed in Table 2. All the methods that are capable of detecting the interaction itself gave indication of weak complexes, but no changes could be observed with methods that give information about the structure of the macromolecules. These weak interactions are extremely difficult to study from a structural point of view, but can be crucial, as shown for example for the Nck2/PINCH interaction.41 The quickly forming, transient complexes may play a key role in the rapid responses of the cells to the environmental changes.42 Tß4 is known to interact with the 4th and 5th LIM domains of PINCH and the ankyrin repeat region of ILK while the latter also forms a complex with the 1st LIM domain of PINCH. Taken into consideration that Tß4 is only 43 residues in length, its making contacts at so many different sites involves that the interacting surfaces should be small and the molecule should be mostly extended even in its bound state. Our results suggest that most of Tß4 remains in a disordered state when binding to PINCH or ILK separately and even upon binding to both partners only a few amino acids get completely buried. The structural changes that this type of binding induces in its partners remain to be elucidated in order to fully understand how the Tß4-induced upregulation of ILK occurs.24
Table 2. Summary of the methods used for the characterization of Tß4 binding to its partners.
| BLI | Steady-state fluorescence | Near UV CD | Far UV CD | ITC | NMR | SAXS | Wide-line NMR | |
| Tß4/PINCH LIM4–5 | KD = 630 µM | Weak interaction | Interaction with local changes | No structural changes | Too low heat of reaction | Interaction without gaining structure | No major structural changes | Interaction with some buried surfaces |
| Tß4/ILK-Ank-GST | KD = 380 µM | Weak interaction | Interaction with local changes | No structural changes | Too low heat of reaction | Interaction without gaining structure | No major structural changes | Interaction with some buried surfaces |
| Tß4/PINCH LIM4–5-ILK-Ank-GST | No information | No information | Interaction with local changes | No structural changes | Too low heat of reaction | Interaction without gaining structure | No information | No information |
| Tß4/Stabilin CTD | KD = 2.8 mM | No information | Interaction with local changes | No structural changes | Small enthalpy and entropy changes | Interaction without gaining structure | Interaction without gaining structure | Interaction with some buried surfaces |
Tß4 binding to G-actin is accompanied by the formation of 2 α helices at the N and C termini of the protein.17 Based on the results of our NMR experiments, the region 10EKFDKSKLKKTET takes part in the binding to PINCH LIM4-5, which overlaps with the N-terminal helix, but without stabilizing it. In G-actin binding, Asn26 plays a direct role,17 but it does not seem to take part in the interaction with PINCH LIM4–5. Binding to ILK-Ank-GST induced changes in the chemical environments of amino acids in the 9IEKFDKSKL region, suggesting that the binding regions of the 2 partners overlap significantly. This result is in accord with the predictions of the ANCHOR server43,44 that predicts a long binding region between residues 1–18 in Tß4. When both partners are present, the whole N-terminal part of Tß4 is affected, with the most pronounced changes of amino acids 6MAEIEK and 17LKKTETQLK (Fig. 6), the latter of which is the recognition site for G-actin.28 It is important to note that according to the observations of Domanski and coworkers,17 this region does not gain structure upon binding to G-actin and stays in and extended conformation. The fuzzy type of interaction was also confirmed by far UV CD and SAXS experiments. The fact that no significant change of the far UV CD spectra or of Kratky plots could be observed upon binding supports the hypothesis that a large population of Tβ4 remains in an unfolded state when binding PINCH LIM4-5 or ILK-ANK. Another important observation is that the only measurable changes in Tβ4 structure occurred when Tβ4 was present in excess in the reaction mixture. One possible explanation is that Tβ4 has to localize to the partners in large quantities for the association rate to overcome the dissociation rate. In these assemblies only a few Tβ4 molecules make direct contact with the partners at a given time point, making the detection of the bound structure difficult. An interesting study45 describes the capability of multivalent proteins of going through sol-gel phase transitions upon complex formation. One of the studied systems is the nephrin-Nck2-WASP complex which also takes part in the regulation of actin assembly. Since Tβ4 is the prototypical WH2 (WASP homology domain 2) protein, it would be intriguing to see whether such phase transition is involved in the interactions studied above. The presence of large Tβ4 associates measured by SAXS could be an indication that Tβ4 is capable of such transition.
The CTD of stabilin-2 also interacts with Tß4, and this interaction has a regulatory role in stabilin-mediated phagocytosis of apoptotic cells.46 This process has been characterized only in cellular systems and no structural or kinetic data are available on the complex of the two molecules. This domain of stabilin is predicted to be disordered throughout its whole sequence, thus we expected mutual induced folding (or co-folding) of the 2 IDPs to occur upon the interaction, in which both of the partners would become more structured in a functional, stable complex as observed in other cases.47 Instead, we observed that although the molecules interact with each-other (confirmed by several independent methods listed in Table 2) they do it without significant ordering of either partner. Titration of 15N labeled Tß4 with stabilin CTD or 15N labeled stabilin CTD with Tß4 (data not shown) resulted in very little shifts of the peaks of the 1H-15N HSQC spectra, with no sign of major structural reorganization in any of the experiments. The perturbations of the combined chemical shifts suggest that only a couple of amino acids of Tß4 make direct contact with stabilin CTD. Far UV CD and SAXS measurements confirmed the results of the NMR experiments as shown by the unchanged far UV CD spectra of the complex and Kratky plots of the complex and the free partners.
It is striking how different the modes of binding of Tß4 to G-actin and to the other proteins studied here, are: it seems we have encountered a new paradigm of multiple fuzzy interactions of a protein in its distinct functions. The first important difference is the strength of the binding. While the KD of the binding with G-actin is in the micromolar range,48 the KD of the binding with all 3 other partners is a couple of hundred micromolars. Since intracellular Tß4 concentrations can be as high as 600 µM,16 the conditions of the formation of these weak complexes may exist in cells that respond rapidly to external signals. Because actin is one of the most abundant proteins in the cell, the polymerization status of actin might be the primary factor regulating the availability of Tß4 for these other, weaker interactions. The structural differences of the binding most probably stem from the fundamental differences between the functions of Tß4 upon binding to G-actin and to the other partners. The aim of Tß4 binding to G-actin is to sequester it and keep it in monomeric form until it is necessary. For this, the complex must be more stable than a complex where Tß4 acts only as a transient activator, such as for ILK. This view is supported by the model of cell movement presented by Fan and coworkers in 200949 where they found that local regulated dissociation of Tß4/G-actin occurred during cell migration. G-actin availability promotes actin polymerization and Tß4 binding to ILK enhances Akt2-dependent matrix metalloproteinase 2 expression and extracellular matrix degradation. The authors reasoned that the sequestration of Tß4 prevents interaction with ILK in quiescent cells, until the stimulus mediated release of Tß4 at the cell front during migration. The fact that the region of Tβ4 that binds PINCH and ILK together is the same region that is important for matrix metalloproteinase activation21 underlines significance of our findings.
Tβ4 binding to stabilin CTD might be stabilized by polyelectrostatic interactions that make the interaction to form without stable structures possible. Both proteins are abundant of charged amino acids (20 in Tβ4 and 14 in stabilin CTD) and the charges are distributed more-or-less evenly throughout their sequence. Our results suggest a model where the interaction is formed through multiple specific contacts which contribute to the binding equally instead of a specific contact region between the 2 partners. This model was created to explain the interaction of the IDP Sic1 with Cdc450 but the generalization of the model is suggested. Mutational analysis could be useful to strengthen this hypothesis.
In all, the highly variable and adaptable nature of the functioning of IDPs is nicely demonstrated by the diverse modes Tβ4 can form complexes with its partners. Depending on the role it has to fulfill, Tß4 is capable of binding tightly through a disorder-to-order transition but it can act as part of very weak, transient complex(es) at the same time. It will be a challenge to find out if more such IDPs exist and, even more, to understand the fine structural details of this unique behavior of multiple fuzzy, yet specific interactions.
Materials and methods
Cloning and purification of the protein constructs
DNA coding for human Tß4 (UniProt: P62328) was synthesized by MWG Biotech AG and cloned into pET22b cloning vector. A stop codon was added to ensure that the protein is expressed without a His-tag. After overnight induction (28°C; 1 mM IPTG [Duchefa, I1401.0005]), cells were pelleted and lysed by sonication in lysis buffer (10 mM Na2HPO4 [Sigma, S0876], 150 mM NaCl [Sigma, S9888], pH7.0) with Complete Inhibitor Cocktail (Roche, 4693132001). Cell debris were removed by centrifugation (24 000 x g, 20 min, 4°C), the supernatant was boiled in a water bath for 5 min and centrifuged (24 000 x g, 20 min, 4°C). 3% perchloric acid (Sigma, 77230) was added to the supernatant and precipitated proteins were removed by centrifugation (24 000 x g, 20 min, 4°C). The pH of the supernatant was adjusted to 7.0 by adding 5 M KOH (Sigma, P1767). The final protein solution was filtered through a 0.2 µm nitrocellulose filter and purified on a reversed phase (RPC) column (GE Healthcare, 17-1182-01), on an Akta Explorer system (GE Healthcare). Eluted Tß4 was dialysed against distilled water and lyophylized. Lyophylized protein was dissolved in the appropriate buffer used in the different experiments: PBS (Sigma, P3813) for the BLI measurements, water for the wide-line NMR experiments and 10 mM Na2HPO4, 150 mM NaCl, pH 6.0 for all the other measurements.
The DNA sequence coding for the 4th and 5th LIM domains (amino acids 189–325) of human PINCH-1 (PINCH LIM4-5, UniProt: P48059) was amplified from Human Embryonic Kidney cell cDNA, using the following primers: forward: 5′ GTCCCCATCTGTGGTGCTTG, reverse: 5′ CGGAAATTTTTCATAGCATTTTTTGC. The resulting fragment was cloned into pET22b expression vector using NdeI/XhoI restriction sites. The protein expression was induced with 0.5 mM IPTG at 30°C for 3 h. After sonication in lysis buffer (50 mM Na2HPO4, 200 mM NaCl, 20 mM Imidazole [Sigma, 56750], pH7.4) with Complete Inhibitor Cocktail, the cell extract was centrifuged at 24 000 x g for 50 min at 4°C. The supernatant was filtered through a 0.2 µm nitrocellulose filter and loaded onto a HisTrap HP column (GE Healthcare, 17-5247-01). Proteins were eluted by step elution using different concentrations of imidazole. Imidazole was removed on a desalting column (GE Healthcare, 17-5087-01), using the buffer needed for further experiments: PBS (Sigma, P3813) for the BLI measurements, water for the wide-line NMR experiments and 10 mM Na2HPO4, 150 mM NaCl, pH 6.0 for all the other measurements.
The DNA coding for the N-terminal Ankyrin repeat region (amino acids 1–160) of human integrin linked kinase (ILK-Ank-GST, UniProt: Q13418) was generously provided by Professor Shoukat Dedhar (BCCRC). The region coding amino acids was amplified using the forward and reverse primers: 5′ CGGGATCCATATGGACGACATTTTCACTCAGTGC and 5′ CCGCTCGAGTTAGAGATTCTGGCCCATCTTCTC respectively. The amplified DNA was cloned into a pGEX-5X expression vector using BamHI/XhoI restriction sites. The expression of the protein was induced with 0.5 mM IPTG for 5 h at 16°C. Following induction, cells were pelleted and resuspended in PBS with Complete Inhibitor Cocktail, treated with lysozyme (Sigma, 62970) for 30 min at room temperature then sonicated and centrifuged (24 000 x g, 20 min, 4°C). Triton X-100 (Sigma, T8787) was added to the supernatant and left on ice for 30 min then the cell extract was centrifuged at 50 000 x g for 40 min at 4°C. The supernatant was filtered through a 0.2 µm nitrocellulose filter and loaded onto a GSTTrap FF column (GH Healthcare, 17-5130-02). GST-tagged protein was eluted from the column with 10 mM reduced glutathione (Sigma, G4251). Reduced glutathione was subsequently removed on a desalting column (GE Healthcare, 17-5087-01), using the buffer needed for further experiments.
The proper folding of the globular proteins was tested with far UV CD, NMR and SAXS. The far UV CD spectra and the secondary structure calculations (Table 2.) confirmed that the proteins adopted their natural folds, although they were not stable for more than a couple of days. 1H-15N HSQC spectra of 15N-labeled PINCH LIM4-5 and ILK-Ank-GST were characteristic of folded proteins. When an ab-initio modeling was applied to the SAXS scattering curve of the separate LIM4 domain of PINCH, the resulting model fitted acceptably to the crystal structure of the protein (pdb: 1NYP) so we assumed that PINCH LIM4-5 was probably properly folded as well. There is no available structure of LIM5 or the 2 domains together there was no chance to do the same with our full construct. The SAXS scattering curve of ILK-Ank-GST showed that the protein was aggregated which could be due to the elongated storage before the measurements, therefore, the SAXS experiments with this partner are not discussed in detail.
The DNA coding for the truncated intracellular region (amino acids 2501–2551) of human stabilin-2 (UniProt: Q8WWQ8) was synthesized by the MWG Biotech AG company and then cloned into pET16b cloning vector. Protein expression was induced overnight with 0.5 mM IPTG at 28°C. After sonication in lysis buffer (20 mM Na2HPO4, 500 mM NaCl, 20 mM imidazole, pH 7.4) with Complete Inhibitor Cocktail, the cell extract was centrifuged at 24 000 x g for 50 min at 4°C. The supernatant was filtered through a 0.2 µm nitrocellulose filter and loaded on a HisTrap HP column. Purified protein was dyalized against 20 mM ammonium acetate (Sigma, A7330) (pH 6.8) and lyophylized. Lyophylized protein was dissolved in the appropriate buffer used in the different experiments.
Affinity measurements by biolayer interferometry
For analyzing binding kinetics, biolayer interferometry (BLI) on an Octet Red384 system (ForteBio) was used. Biosensors (Ni-NTA for experiments with PINCH LIM4-5 and stabilin CTD and anti-GST for experiments with ILK-Ank-GST) were hydrated in sample diluent (ForteBio, 18-5028) for 30 min and then loaded onto the OctetRED instrument. Thirty-five micrograms per milliliter PINCH LIM4-5, ILK-Ank-GST or stabilin CTD was loaded to saturation after 60 s baseline in sample diluent. The baseline was re-established for 400 s. The sensors were moved to different concentrations of Tß4 or p27Kip1 as negative control. Association and dissociation were recorded for 350 and 600 s respectively in the case of PINCH LIM4-5, 1000 and 1000 s in the case of ILK-Ank-GST and 525 and 300 s in the case of stabilin CTD. All measurements were performed at 25°C. Equilibrium BLI data were analyzed using the Octet software provided by the manufacturer.
Steady-state fluorescence
Steady-state fluorescence measurements were performed on a Synergy (BioTek) microplate reader. Different molar ratios (ranging from 1:1 to 1:8) of Tß4 and its partners were mixed in a buffer of 10 mM Na2HPO4 and 50 mM NaCl, pH 6.0. In control experiments Tß4 was replaced with GGG or GHG (molar ratios 1:1 to 1:8). Tß4. Excitation wavelength was 270 nm and spectra were recorded between 400 and 300 nm at 25°C.
Circular dichroism
Near UV CD spectra were recorded in 1 cm pathlength cuvette on a Jasco J-720 spectropolarimeter in continuous mode with 1 nm bandwidth, 8 s response time and 20 nm/min scan speed. The temperature was maintained at 25°C with a Neslab RTE-111 circulating water bath. The experimental buffer was 10 mM Na2HPO4 and 50 mM NaCl, pH 6.0. Protein concentrations used were 22 µM PINCH LIM4-5, 6 µM ILK-Ank-GST, 2.5 mM stabilin CTD and a 10-fold concentration of Tß4 for each partner.
Far UV CD spectra were recorded in 10 mm pathlength cuvette, using the same instruments and experimental conditions as in the near UV CD measurements. Protein concentrations used were 5 µM PINCH LIM4-5, 3 µM ILK-Ank-GST, and 25 µM stabilin CTD and a 3-fold concentration of Tß4 for each partner.
Molar ellipticity was calculated according to the following equation: [θ] = 100xθ/(Cxl) Where θ is the ellipticity, C is the molar concentration and l is the pathlength in cm.
Data were analyzed with the CDPro program package using the SDP48 reference set that contains 43 soluble and 5 denatured proteins. The CDPro program package is available at the CDPro website: http://lamar.colostate.edu/~sreeram/CDPro/main.html.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) was done on a Microcal ITC 200 (GE Healthcare) instrument. Five mM Tß4 was titrated into 25 µM stabilin CTD in 25 steps. Protein samples were degassed prior to the experiments. The experimental buffer was 10 mM Na2HPO4 and 50 mM NaCl, pH 6.0 and the cell temperature was 25°C. The observed integrated reaction heat of interaction was corrected to the solvation by extracting the heat of titrating 5 mM Tß4 into buffer and fit to a single-site model of interaction and KD was determined using the Origin (MicroCal) software package.
Protein interaction assessed by NMR
For the NMR measurements all proteins were dissolved in a buffer containing 10 mM Na2HPO4 and 50 mM NaCl set to pH 6.0 with the exception of the stabilin CTD/15N -β4 titration where the buffer was 50 mM sodium acetate and 50 mM NaCl set to pH 6.5. All measurements were performed at 300 K (for stabilin CTD measurements were at 283 K).
To characterize the interaction between Tβ4 and and its partners, a series of 1H-15N HSQC spectra of 15N-labeled Tβ4 were acquired in the absence and presence of the partners The partners were added to the labeled Tß4 from stock solutions of 0.25 mM (PINCH LIM4–5), 60 µM (ILK-Ank-GST) and 4.5 mM (stabilin CTD) to reach 1:3, 1:1 and 3:1 molar ratios (Tß4:partner). The pH was measured and readjusted to pH 6.0 after each titration step. Time domain (TD) was set to 1024 in the direct dimension and incremented in 256 points and 10 ppm/20 ppm spectral width (SW) was applied (PINCH LIM4–5). For ILK-Ank-GST (both with and without PINCH LIM4–5) titrations with 2048 x 256 points for TD and 14 ppm/20 ppm for SW were used. As for stabilin CTD TD was set to 1024 in the direct dimension and incremented in 512 points and 12 ppm/20 ppm SW was applied. All measurements were acquired in 8 scans. 1H-15N HSQC spectra of free 15N-labeled Tβ4 were recorded in a pH range between pH 4–9 to be able to rule out the effect of buffer changes.
NMR spectra were acquired on 500 and 700 MHz Bruker instruments. The peak assignment of 15N-Tß4 was based on the chemical shift data of BRMB entry number 1065. NMR data were processed directly at the spectrometer with TopSpin software (provided by the manufacturer of the instrument) and analyzed with Cara51 and Sparky.52 Binding was followed by evaluating the perturbation effect on amides by means of chemical shift change of peaks on 1H-15N HSQC spectra. Residue specific chemical shift perturbations (CSP) were determined as follows:53
Small-angle X-ray scattering measurements
Synchrotron small-angle X-ray scattering (SAXS) data were collected at the X33 beamline of the EMBL (DESY, Hamburg).54 The wavelength of the incoming X-ray beam was 0.15 nm and the two dimensional scattering patterns were collected with a Pilatus 1M-W hybrid pixel detector. The sample to detector distance was 2.7 min, which corresponds to the range of momentum transfer of 0.08 < q < 5 nm−1. Prior to SAXS measurements 2 mM dithiothreitol (Sigma, 43815) was added to the samples in order to avoid radiation damage. The scattering curves were obtained by radial averaging of the 2D scattering patterns and were normalized to the primary beam intensity and corrected for transmission. Finally, the calibration of the curves to absolute units of macroscopic cross section (cm−1) was performed by using water as a reference. All measurements were performed at 298 K.
Proteins were mixed in molar ratios ranging from 1:3 to 3:1 Tß4:partner protein and protein concentrations ranged from 0.4 to 20 mg/ml.
The buffer used was 10 mM Na2HPO4, 50 mM NaCl, pH 6.0 and buffer correction was performed by measuring the scattering of the buffer in the same capillary as used for the sample measurements. The respective scattering curves of the sample and the buffer were normalized to the primary beam monitor counter and were subtracted.
Wide-line NMR measurements
1H NMR measurements and data acquisition were accomplished by a Bruker AVANCE III NMR pulse spectrometer at frequencies of 82.4 MHz with a stability of better than ± 10−6. The inhomogeneity of the magnetic field was 2 ppm. The data points in the figures are based on spectra recorded by averaging signals to reach a signal/noise ratio of 50. The extrapolation to zero time was done by fitting a stretched exponential. The principle and details of wide-line NMR spectroscopy are given in references 55,56 and in the references cited therein.
Proteins were dissolved in double-distilled water at a 25 mg/ml concentration, except bovine serum albumin (BSA, Sigma) which was dissolved in 50 mg/ml concentration. Free induction decays (FIDs) were measured in the temperature range between −200 and +45°C, following thermal equilibrium at every temperature that took typically 10 min in each case. The temperature was controlled by an open-cycle Janis cryostat with an uncertainty better than ± 1 K.
The phases of ice protons, protein protons, and mobile (water) protons are separated in the FID signal by virtue of large differences in the spin-spin relaxation rate. The portion of mobile proton (water) fraction is directly measured by the FID signal based on the comparison of the signal intensity extrapolated to t = 0 with the corresponding values measured at a temperature where the whole sample is in liquid state.56 We converted the measured signal intensities into a measure of hydration as h = x·mH2O/mprotein where mH2O is the mass of water, and mprotein is the mass of protein in the sample.
Glossary
Abbreviations:
- IDP
intrinsically disordered protein
- Tß4
Thymosin β 4
- PINCH LIM4-5
4th and 5th LIM domain of Particularly Interesting New Cys-His protein
- ILK-Ank-GST
Ankyrin repeat region of Integrin Linked Kinase fused to GST
- Stabilin CTD
C-terminal domain of stabilin
- ITC
isothermal calorimetric titration
- BLI
biolayer interferometry
- CD
circular dichroism
- NMR
nuclear magnetic resonance
- SAXS
small angle X-ray scattering
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the Korea-Hungarian Joint Laboratory grant from Korea Research Council of Fundamental Science and Technology, the Hungarian Research Fund (PD 105049) and the Janos Bolyai Research Scholarship of the Hungarian Academy of Sciences.
References
- 1.Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D. . Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature 2004; 432:466 - 72; http://dx.doi.org/ 10.1038/nature03000; PMID: 15565145 [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradović Z. . Intrinsic disorder and protein function. Biochemistry 2002; 41:6573 - 82; http://dx.doi.org/ 10.1021/bi012159+; PMID: 12022860 [DOI] [PubMed] [Google Scholar]
- 3.Dyson HJ, Wright PE. . Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005; 6:197 - 208; http://dx.doi.org/ 10.1038/nrm1589; PMID: 15738986 [DOI] [PubMed] [Google Scholar]
- 4.Smock RG, Gierasch LM. . Sending signals dynamically. Science 2009; 324:198 - 203; http://dx.doi.org/ 10.1126/science.1169377; PMID: 19359576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pancsa R, Tompa P. . Structural disorder in eukaryotes. PLoS One 2012; 7:e34687; http://dx.doi.org/ 10.1371/journal.pone.0034687; PMID: 22496841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. . Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol 2008; 4:728 - 37; http://dx.doi.org/ 10.1038/nchembio.127; PMID: 19008886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. . Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 2002; 323:573 - 84; http://dx.doi.org/ 10.1016/S0022-2836(02)00969-5; PMID: 12381310 [DOI] [PubMed] [Google Scholar]
- 8.Wright PE, Dyson HJ. . Linking folding and binding. Curr Opin Struct Biol 2009; 19:31 - 8; http://dx.doi.org/ 10.1016/j.sbi.2008.12.003; PMID: 19157855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hazy E, Tompa P. . Limitations of induced folding in molecular recognition by intrinsically disordered proteins. Chemphyschem 2009; 10:1415 - 9; http://dx.doi.org/ 10.1002/cphc.200900205; PMID: 19462392 [DOI] [PubMed] [Google Scholar]
- 10.Tompa P, Fuxreiter M. . Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci 2008; 33:2 - 8; http://dx.doi.org/ 10.1016/j.tibs.2007.10.003; PMID: 18054235 [DOI] [PubMed] [Google Scholar]
- 11.Tompa P, Szász C, Buday L. . Structural disorder throws new light on moonlighting. Trends Biochem Sci 2005; 30:484 - 9; http://dx.doi.org/ 10.1016/j.tibs.2005.07.008; PMID: 16054818 [DOI] [PubMed] [Google Scholar]
- 12.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. . Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics 2008; 9:Suppl 1 S1; http://dx.doi.org/ 10.1186/1471-2164-9-S1-S1; PMID: 18366598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein AL, Hannappel E, Kleinman HK. . Thymosin beta4: actin-sequestering protein moonlights to repair injured tissues. Trends Mol Med 2005; 11:421 - 9; http://dx.doi.org/ 10.1016/j.molmed.2005.07.004; PMID: 16099219 [DOI] [PubMed] [Google Scholar]
- 14.Safer D, Elzinga M, Nachmias VT. . Thymosin beta 4 and Fx, an actin-sequestering peptide, are indistinguishable. J Biol Chem 1991; 266:4029 - 32; PMID: 1999398 [PubMed] [Google Scholar]
- 15.Yarmola EG, Klimenko ES, Fujita G, Bubb MR. . Thymosin beta4: actin regulation and more. Ann N Y Acad Sci 2007; 1112:76 - 85; http://dx.doi.org/ 10.1196/annals.1415.008; PMID: 17947588 [DOI] [PubMed] [Google Scholar]
- 16.Mannherz HG, Hannappel E. . The beta-thymosins: intracellular and extracellular activities of a versatile actin binding protein family. Cell Motil Cytoskeleton 2009; 66:839 - 51; http://dx.doi.org/ 10.1002/cm.20371; PMID: 19405116 [DOI] [PubMed] [Google Scholar]
- 17.Domanski M, Hertzog M, Coutant J, Gutsche-Perelroizen I, Bontems F, Carlier MF, Guittet E, van Heijenoort C. . Coupling of folding and binding of thymosin beta4 upon interaction with monomeric actin monitored by nuclear magnetic resonance. J Biol Chem 2004; 279:23637 - 45; http://dx.doi.org/ 10.1074/jbc.M311413200; PMID: 15039431 [DOI] [PubMed] [Google Scholar]
- 18.Grant DS, Rose W, Yaen C, Goldstein A, Martinez J, Kleinman H. . Thymosin beta4 enhances endothelial cell differentiation and angiogenesis. Angiogenesis 1999; 3:125 - 35; http://dx.doi.org/ 10.1023/A:1009041911493; PMID: 14517430 [DOI] [PubMed] [Google Scholar]
- 19.Smart N, Rossdeutsch A, Riley PR. . Thymosin beta4 and angiogenesis: modes of action and therapeutic potential. Angiogenesis 2007; 10:229 - 41; http://dx.doi.org/ 10.1007/s10456-007-9077-x; PMID: 17632766 [DOI] [PubMed] [Google Scholar]
- 20.Qiu P, Kurpakus-Wheater M, Sosne G. . Matrix metalloproteinase activity is necessary for thymosin beta 4 promotion of epithelial cell migration. J Cell Physiol 2007; 212:165 - 73; http://dx.doi.org/ 10.1002/jcp.21012; PMID: 17348036 [DOI] [PubMed] [Google Scholar]
- 21.Philp D, Scheremeta B, Sibliss K, Zhou M, Fine EL, Nguyen M, Wahl L, Hoffman MP, Kleinman HK. . Thymosin beta4 promotes matrix metalloproteinase expression during wound repair. J Cell Physiol 2006; 208:195 - 200; http://dx.doi.org/ 10.1002/jcp.20650; PMID: 16607611 [DOI] [PubMed] [Google Scholar]
- 22.Fine JD. . Epidermolysis bullosa: a genetic disease of altered cell adhesion and wound healing, and the possible clinical utility of topically applied thymosin beta4. Ann N Y Acad Sci 2007; 1112:396 - 406; http://dx.doi.org/ 10.1196/annals.1415.017; PMID: 17468231 [DOI] [PubMed] [Google Scholar]
- 23.Rooney N, Streuli CH. . How integrins control mammary epithelial differentiation: a possible role for the ILK-PINCH-Parvin complex. FEBS Lett 2011; 585:1663 - 72; http://dx.doi.org/ 10.1016/j.febslet.2011.05.014; PMID: 21570968 [DOI] [PubMed] [Google Scholar]
- 24.Huang HC, Hu CH, Tang MC, Wang WS, Chen PM, Su Y. . Thymosin beta4 triggers an epithelial-mesenchymal transition in colorectal carcinoma by upregulating integrin-linked kinase. Oncogene 2007; 26:2781 - 90; http://dx.doi.org/ 10.1038/sj.onc.1210078; PMID: 17072345 [DOI] [PubMed] [Google Scholar]
- 25.Srivastava D, Saxena A, Michael Dimaio J, Bock-Marquette I. . Thymosin beta4 is cardioprotective after myocardial infarction. Ann N Y Acad Sci 2007; 1112:161 - 70; http://dx.doi.org/ 10.1196/annals.1415.048; PMID: 17600280 [DOI] [PubMed] [Google Scholar]
- 26.Zhao Y, Qiu F, Xu S, Yu L, Fu G. . Thymosin β4 activates integrin-linked kinase and decreases endothelial progenitor cells apoptosis under serum deprivation. J Cell Physiol 2011; 226:2798 - 806; http://dx.doi.org/ 10.1002/jcp.22624; PMID: 21935929 [DOI] [PubMed] [Google Scholar]
- 27.Ho JH, Tseng KC, Ma WH, Chen KH, Lee OK, Su Y. . Thymosin beta-4 upregulates anti-oxidative enzymes and protects human cornea epithelial cells against oxidative damage. Br J Ophthalmol 2008; 92:992 - 7; http://dx.doi.org/ 10.1136/bjo.2007.136747; PMID: 18480304 [DOI] [PubMed] [Google Scholar]
- 28.Sosne G, Qiu P, Goldstein AL, Wheater M. . Biological activities of thymosin beta4 defined by active sites in short peptide sequences. FASEB J 2010; 24:2144 - 51; http://dx.doi.org/ 10.1096/fj.09-142307; PMID: 20179146 [DOI] [PubMed] [Google Scholar]
- 29.Tompa K, Bánki P, Bokor M, Kamasa P, Lasanda G, Tompa P. . Interfacial water at protein surfaces: wide-line NMR and DSC characterization of hydration in ubiquitin solutions. Biophys J 2009; 96:2789 - 98; http://dx.doi.org/ 10.1016/j.bpj.2008.11.038; PMID: 19348762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hazy E, Bokor M, Kalmar L, Gelencser A, Kamasa P, Han KH, Tompa K, Tompa P. . Distinct hydration properties of wild-type and familial point mutant A53T of α-synuclein associated with Parkinson’s disease. Biophys J 2011; 101:2260 - 6; http://dx.doi.org/ 10.1016/j.bpj.2011.08.052; PMID: 22067166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tantos A, Szrnka K, Szabo B, Bokor M, Kamasa P, Matus P, Bekesi A, Tompa K, Han KH, Tompa P. . Structural disorder and local order of hNopp140. Biochim Biophys Acta 2013; 1834:342 - 50; http://dx.doi.org/ 10.1016/j.bbapap.2012.08.005; PMID: 22906532 [DOI] [PubMed] [Google Scholar]
- 32.Sigalov AB. . Unusual biophysics of immune signaling-related intrinsically disordered proteins. Self Nonself 2010; 1:271 - 81; http://dx.doi.org/ 10.4161/self.1.4.13641; PMID: 21487502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sigalov AB. . Interplay between protein order, disorder and oligomericity in receptor signaling. Adv Exp Med Biol 2012; 725:50 - 73; http://dx.doi.org/ 10.1007/978-1-4614-0659-4_4; PMID: 22399318 [DOI] [PubMed] [Google Scholar]
- 34.Sigalov AB. . Uncoupled binding and folding of immune signaling-related intrinsically disordered proteins. Prog Biophys Mol Biol 2011; 106:525 - 36; http://dx.doi.org/ 10.1016/j.pbiomolbio.2011.08.005; PMID: 21867726 [DOI] [PubMed] [Google Scholar]
- 35.Sigalov AB, Aivazian DA, Uversky VN, Stern LJ. . Lipid-binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry 2006; 45:15731 - 9; http://dx.doi.org/ 10.1021/bi061108f; PMID: 17176095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sigalov AB, Zhuravleva AV, Orekhov VY. . Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie 2007; 89:419 - 21; http://dx.doi.org/ 10.1016/j.biochi.2006.11.003; PMID: 17174464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sigalov AB. . Protein intrinsic disorder and oligomericity in cell signaling. Mol Biosyst 2010; 6:451 - 61; http://dx.doi.org/ 10.1039/b916030m; PMID: 20174674 [DOI] [PubMed] [Google Scholar]
- 38.Mittag T, Marsh J, Grishaev A, Orlicky S, Lin H, Sicheri F, Tyers M, Forman-Kay JD. . Structure/function implications in a dynamic complex of the intrinsically disordered Sic1 with the Cdc4 subunit of an SCF ubiquitin ligase. Structure 2010; 18:494 - 506; http://dx.doi.org/ 10.1016/j.str.2010.01.020; PMID: 20399186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goyal S, Gupta G, Qin H, Upadya MH, Tan YJ, Chow VT, Song J. . VAPC, an human endogenous inhibitor for hepatitis C virus (HCV) infection, is intrinsically unstructured but forms a “fuzzy complex” with HCV NS5B. PLoS One 2012; 7:e40341; http://dx.doi.org/ 10.1371/journal.pone.0040341; PMID: 22815741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. . Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A 1996; 93:11504 - 9; http://dx.doi.org/ 10.1073/pnas.93.21.11504; PMID: 8876165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaynberg J, Fukuda T, Chen K, Vinogradova O, Velyvis A, Tu Y, Ng L, Wu C, Qin J. . Structure of an ultraweak protein-protein complex and its crucial role in regulation of cell morphology and motility. Mol Cell 2005; 17:513 - 23; http://dx.doi.org/ 10.1016/j.molcel.2004.12.031; PMID: 15721255 [DOI] [PubMed] [Google Scholar]
- 42.Nooren IM, Thornton JM. . Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol 2003; 325:991 - 1018; http://dx.doi.org/ 10.1016/S0022-2836(02)01281-0; PMID: 12527304 [DOI] [PubMed] [Google Scholar]
- 43.Dosztányi Z, Mészáros B, Simon I. . ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics 2009; 25:2745 - 6; http://dx.doi.org/ 10.1093/bioinformatics/btp518; PMID: 19717576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mészáros B, Simon I, Dosztányi Z. . Prediction of protein binding regions in disordered proteins. PLoS Comput Biol 2009; 5:e1000376; http://dx.doi.org/ 10.1371/journal.pcbi.1000376; PMID: 19412530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, et al. . Phase transitions in the assembly of multivalent signalling proteins. Nature 2012; 483:336 - 40; http://dx.doi.org/ 10.1038/nature10879; PMID: 22398450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee SJ, So IS, Park SY, Kim IS. . Thymosin beta4 is involved in stabilin-2-mediated apoptotic cell engulfment. FEBS Lett 2008; 582:2161 - 6; http://dx.doi.org/ 10.1016/j.febslet.2008.03.058; PMID: 18519035 [DOI] [PubMed] [Google Scholar]
- 47.Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, Evans RM, Wright PE. . Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature 2002; 415:549 - 53; http://dx.doi.org/ 10.1038/415549a; PMID: 11823864 [DOI] [PubMed] [Google Scholar]
- 48.Didry D, Cantrelle FX, Husson C, Roblin P, Moorthy AM, Perez J, Le Clainche C, Hertzog M, Guittet E, Carlier MF, et al. . How a single residue in individual β-thymosin/WH2 domains controls their functions in actin assembly. EMBO J 2012; 31:1000 - 13; http://dx.doi.org/ 10.1038/emboj.2011.461; PMID: 22193718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan Y, Gong Y, Ghosh PK, Graham LM, Fox PL. . Spatial coordination of actin polymerization and ILK-Akt2 activity during endothelial cell migration. Dev Cell 2009; 16:661 - 74; http://dx.doi.org/ 10.1016/j.devcel.2009.03.009; PMID: 19460343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borg M, Mittag T, Pawson T, Tyers M, Forman-Kay JD, Chan HS. . Polyelectrostatic interactions of disordered ligands suggest a physical basis for ultrasensitivity. Proc Natl Acad Sci U S A 2007; 104:9650 - 5; http://dx.doi.org/ 10.1073/pnas.0702580104; PMID: 17522259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Keller R. The computer aided resonance assignment tutorial, (CANTINA Verlag, Goldau, Switzerland, 2004). [Google Scholar]
- 52.Goddard, T.D., Kneller, D.G. Sparky 3. (2009). [Google Scholar]
- 53.Kamphuis MB, Bonvin AM, Monti MC, Lemonnier M, Muñoz-Gómez A, van den Heuvel RH, Díaz-Orejas R, Boelens R. . Model for RNA binding and the catalytic site of the RNase Kid of the bacterial parD toxin-antitoxin system. J Mol Biol 2006; 357:115 - 26; http://dx.doi.org/ 10.1016/j.jmb.2005.12.033; PMID: 16413033 [DOI] [PubMed] [Google Scholar]
- 54.Koch MHJB. . J. X-ray diffraction and scattering on disordered systems using synchrotron radiation. Nucl Instrum Methods 1983; 208:461 - 9; http://dx.doi.org/ 10.1016/0167-5087(83)91169-9 [DOI] [Google Scholar]
- 55.Tompa K. B.M., Tompa P. PART II. Chapter 12. Hydration of intrinsically disordered proteins from wide-line NMR in: Instrumental Analysis of Intrinsically Disordered Proteins: Assessing Structure and Conformation. in Wiley Series in Protein and Peptide Science (ed. Vladimir Uversky, S.L.) (John Wiley & Sons, Incl., Hoboken, New Jersey, Hoboken, New Jersey, 2010). [Google Scholar]
- 56.Tompa K. B.M., Tompa P. Wide-line NMR and Protein Hydration in Intrinsically Disordered Protein Analysis. in Methods and Experimental Tools - Methods in Molecular Biology (ed. Vladimir N. Uversky, A.K.D.) (Humana Press Inc., 2012). [DOI] [PubMed] [Google Scholar]