Abstract
The correlation method from brain imaging has been used to estimate functional connectivity in the human brain. However, brain regions might show very high correlation even when the two regions are not directly connected due to the strong interaction of the two regions with common input from a third region. One previously proposed solution to this problem is to use a sparse regularized inverse covariance matrix or precision matrix (SRPM) assuming that the connectivity structure is sparse. This method yields partial correlations to measure strong direct interactions between pairs of regions while simultaneously removing the influence of the rest of the regions, thus identifying regions that are conditionally independent. To test our methods, we first demonstrated conditions under which the SRPM method could indeed find the true physical connection between a pair of nodes for a spring-mass example and an RC circuit example. The recovery of the connectivity structure using the SRPM method can be explained by energy models using the Boltzmann distribution. We then demonstrated the application of the SRPM method for estimating brain connectivity during stage 2 sleep spindles from human electrocorticography (ECoG) recordings using an 8 × 8 electrode array. The ECoG recordings that we analyzed were from a 32-year-old male patient with long-standing pharmaco-resistant left temporal lobe complex partial epilepsy. Sleep spindles were automatically detected using delay differential analysis and then analyzed with SRPM and the Louvain method for community detection. We found spatially localized brain networks within and between neighboring cortical areas during spindles, in contrast to the case when sleep spindles were not present.
1 Introduction
1.1 The Caveat in the Correlation Method
The correlation method is one of the most commonly used methods for estimating brain functional connectivity (Anand et al., 2005; Biswal, Yetkin, Haughton, & Hyde, 1995; Rubinov & Sporns, 2010; Siegle, Thompson, Carter, Steinhauer, & Thase, 2007; Smith et al., 2011; Uddin, Kelly, Biswal, Castellanos, & Milham, 2009; Vertes et al., 2012; Zhou, Thompson, & Siegle, 2009; Bullmore & Bassett, 2011; McIntosh, Rajah, & Lobaugh, 2003; Laufs et al., 2003). Biswal et al. (1995) analyzed the functional connectivity of the resting state human brain from fMRI data using the correlation method and reported that the regions of the primary sensory motor cortex that were activated secondary to hand movement were functionally connected. They also found that time courses of low-frequency (< 0.1 Hz) fluctuations in the resting brain had a high degree of correlation within these regions and also with time courses in several other regions associated with motor function. Other researchers (Cordes et al., 2000; Xiong, Parsons, Gao, and Fox, 1999) repeated this experiment by Biswal et al. (1995) and found similar results.
In another study of the resting state, Greicius, Krasnow, Reiss, and Menon (2003) found the posterior cingulate cortex (PCC) and ventral anterior cingulate cortex (vACC), via the correlation method, to be functionally connected within themselves and also with each other. The authors observed very high correlation in these regions under three specific conditions during a working memory task, a visual processing task, and at rest. They identified these regions with the default mode network of the brain. Fox, Corbetta, Snyder, Vincent, and Raichle (2006) and Fox et al. (2005) found similar results.
Uddin et al. (2009) also analyzed the functional connectivity of the default mode network with the correlation method on resting state data to find differences in functional connectivity between PCC and vACC and networks that are positively corrrelated and anticorrelated with these two regions. They observed that the positively correlated networks were the same for PCC and vACC; however, the anticorrelated networks were different. Activity in vACC negatively predicted activity in parietal visual spatial and temporal attention networks, whereas activity in PCC negatively predicted activity in prefrontal-based motor control circuits. Since the two major brain regions comprising the default mode network showed different behavior when correlated with other networks in the brain, the authors concluded that there is significant heterogeneity within the default mode network.
Anand et al. (2005) studied the effect of antidepressants on the functional connectivity of the human brain from fMRI data via the correlation method in depressed and healthy control subjects. They measured the connectivity between cortical and limbic regions during continuous exposure to neutral, positive, and negative pictures. Depressed patients showed decreased corticolimbic functional connectivity compared to healthy subjects during the resting state and on exposure to emotionally valenced pictures. At rest and on exposure to neutral and positive pictures, the functional connectivity between the anterior cingulate cortex and limbic regions was significantly increased in patients after treatment. However, on exposure to negative pictures, corticolimbic functional connectivity remained decreased in depressed patients. The authors concluded that antidepressant treatment increases corticolimbic connectivity in depressed patients.
Hampson, Peterson, Skudlarski, Gatenby, and Gore (2002) have found functional connectivity in low frequency using the correlation method. They found a functional connection between the Brocas and Wernickes areas in healthy subjects at rest. The functional connection increased when subjects started continuously listening to narrative text. Furthermore, significant correlation between the Brocas area and a region in the left premotor cortex was found at rest, and it increased during continuous listening.
Although the research on functional connectivity estimation using the correlation method is promising and exciting, the conclusions drawn from the experimental analysis can be misleading or wrong since the regions might show high correlation (i.e., they are functionally connected) due to a common input and not to strong physical connections between themselves (Wang, Kang, Kemmer, & Guo, 2016). Recently Glasser et al. (2016), in a study of multimodal magnetic resonance images from the Human Connectome Project (HCP), identified many new areas in the human cortex using a machine learning classifier. Pairs of these areas with a high degree of functional connectivity also received common input from other areas.
1.2 Solution: The Sparse Regularized Precision Matrix Method
Due to the shortcomings of the correlation method that we have outlined, some researchers have used partial correlations to measure strong direct interactions between pairs of regions while simultaneously removing the influence of the rest of the regions (Dempster, 1972; Lauritzen, 1996; Whittaker, 1990). Thus, partial correlations help identify pairwise brain regions that are conditionally independent given all other brain regions. When the output of the brain regions follows a multivariate gaussian distribution, the inverse of the covariance matrix (also known as the precision matrix, concentration matrix, or information matrix) can be used to calculate the pairwise partial correlations. A value of zero or very close to zero in the precision matrix indicates that the two brain regions are conditionally independent given the rest of the brain regions. In practice, the precision matrix can be estimated by simply inverting the sample covariance matrix provided a sufficiently large number of samples is available.
But when the number of samples is relatively small, the sample covariance matrix is a poor estimator of the eigenvalues of the covariance matrix, and thus the estimated precision matrix might produce a large number of false-positive and false-negative connections in a given brain network. If we assume that the number of connections between the regions in a given brain network is small (i.e., the precision matrix is sparse), then the sparse regularized precision matrix (SRPM) X can be estimated by solving the following Lq regularized optimization problem:
| (1.1) |
where λ is the regularization parameter balancing the error in the maximum likelihood estimate (MLE) of the precision matrix and the sparsity (The MLE of the precision matrix is the inverse of the sample covariance matrix according to the invariance principle), S is the sample covariance matrix and 0 ≤ q ≤ 1. Several algorithms (Banerjee, El Ghaoui, & d’Aspremont, 2008; Friedman, Hastie, & Tibshirani, 2008; Hsieh, Sustik, Dhillon, & Ravikumar, 2011; Hsieh, Sustik, Dhillon, Ravikumar, & Poldrack, 2013; Oztoprak, Nocedal, Rennie, & Olsen, 2012; Rothman, Bickel, Levina, & Zhu, 2008; Scheinberg, Ma, & Goldfarb, 2010; Yuan & Lin, 2007) have been proposed for the q = 1 case. For the q = 0 case, algorithms proposed in Marjanovic and Hero (2015) and Marjanovic and Solo (2014) can be used to estimate the SRPM. Marjanovic and Solo (2014) proposed an algorithm to solve the optimization problem in equation 1.1 for the general 0 ≤ q < 1 case.
Note that when q = 1, the optimization problem in equation 1.1 is a convex optimization problem. We use the QUIC algorithm (Hsieh et al., 2011, 2013) to estimate the SRPM for the simulations and experimental data analysis throughout this letter.
1.3 Prior Research for Human Brain Functional Connectivity Estimation Using the SRPM Method
The SRPM algorithm has been applied to obtain brain networks from voxels data in (Hsieh et al., 2011, 2013) using q = 1. Strong functionally connected regions were primarily found in gray matter regions in the human brain. Modularity-based clustering (Blondel, Guillaume, Lambiotte, & Lefebvre, 2008; Brandes et al., 2008; Newman & Girvan, 2004; Newman, 2006; Reichardt & Bornholdt, 2006; Ronhovde & Nussinov, 2009; Sporns, 2010; Sun, Danila, Josić, & Bassler, 2009) was then applied to the regularized precision matrix obtained from the algorithm. A number of resting state networks were identified, including default mode and sensorimotor networks. In addition, the method identified a number of structured coherent noise sources in the data set. The modules detected by the QUIC algorithm were similar to those identified using independent component analysis on the same data set without the need for the extensive dimensionality reduction (without statistical guarantees) inherent in such techniques.
Ryali, Chen, Supekar, and Menon (2012) applied the SRPM method with q = 1 to resting state fMRI data and found a modular architecture characterized by strong interhemispheric links, distinct ventral and dorsal stream pathways, and a major hub in the posterior medial cortex.
Varoquaux, Gramfort, Jean-Baptiste, and Thirion (2010) analyzed human brain functional connectivity using the SRPM method with q = 1 and after clustering found regions corresponding to important brain areas such as the primary visual system (medial visual areas), the dorsal visual pathway, the occipital pole, and the intraparietal areas comprising the default mode network, the fronto-parietal networks, the ventral visual pathway, the lateral visual areas, and the inferior temporal lobe. The default mode and the fronto-parietal networks appeared as hubs, connecting different networks with different functions, such as the visual streams, but also the motor areas, as well as the frontal regions.
Monti et al. (2014) analyzed the change in functional connectivity in the human brain with respect to time using the SRPM method with q = 1. Healthy patients were asked to perform a simple but attentionally demanding cognitive task. They observed that the activity of the right inferior frontal gyrus and the right inferior parietal lobe dynamically (with respect to time) changed with the task. The authors concluded that both regions play a key role in the attention and executive function during cognitively demanding tasks and may be fundamental in regulating the balance between other brain regions.
Rosa et al. (2015) applied the SRPM method with q = 1 for patient classification by analyzing brain functional connectivity from fMRI data. They were able to distinguish patients with major depressive disorder from healthy control subjects while the participants performed gender discrimination and emotional tasks during the visualization of emotionally valent faces.
Allen et al. (2012) estimated whole-brain functional connectivity dynamics using the SRPM method with q = 1 and clustering algorithms. They analyzed resting state data from a large sample of young adults and found connections between regions in the lateral parietal and cingulate cortex. This result was in contrast to other studies, which characterized such regions as separate entities. They also found that the dynamic functional connectivity of the human brain was markedly different from the stationary brain connectivity. The authors concluded that the study of time-varying connectivity patterns of the human brain will widen our understanding of cognitive and behavioral dynamics.
The SRPM method with q = 1 also has been used to estimate brain functional connectivity at a neuronal level in Yatsenko et al. (2015), where it was claimed that the SRPM method found more biologically plausible brain networks than the correlation method.
Wang et al. (2016) applied the SRPM method with q = 1 for human brain functional connectivity estimation from fMRI data and compared it with the functional connectivity estimated by the correlation method. They found that the SRPM method was able to remove considerable between-module connections, which were identified by the correlation method. When they applied the correlation method, the majority of the connections found were within-module connections. In addition, the authors found between-module connections, in particular between the three visual networks (Med Vis, Op Vis, Lat Vis) and between the auditory network and the sensori-motor network. But when they analyzed the partial correlations obtained from the SRPM method, they found relatively stronger within-module connections and very few between-module connections in comparison to the correlation method, indicating that the significant direct connections in the human brain are within-module connections. In particular, 34% of the significant connections found in the correlation method became insignificant after calculating the partial correlations from the SRPM method. This suggested that the between-module connections are mainly due to common inputs and not due to direct interactions between modules.
Although the SRPM method and the inverse covariance method have been previously applied in brain research to identify functionally connected networks in the human brain and conclusions have been drawn regarding how the brain processes information, more experimental analysis is necessary to verify these claims.
1.4 First Contribution: Demonstrating That SRPM Can Truly Find the Physical Connection in a Network
In this letter, we generate artificial networks by simulation and demonstrate that the SRPM method can indeed find a true physical connection between a pair of nodes in contrast to the correlation method and the inverse covariance method. This is the first contribution of the letter. To evaluate the performance of the SRPM method, we generate synthetic data from two physical models: (1) a mechanical model that considers a cascade connection of a number of springs and masses and (2) an electrical circuit model that consists of resistors (R) and capacitors (C). The purpose is to demonstrate that the connectivity pattern of the spring-mass model can be obtained from the SRPM associated with the displacements of the masses and the connectivity pattern of the RC circuit model can be obtained from the SRPM associated with the voltages measured at the nodes. The SRPM method is shown to give superior performance compared with the correlation method and the inverse covariance method and is also able to find the ground-truth connection. In other words, it is shown that the correlation method and the inverse covariance method may not be able to recover the true connectivity of the associated network in the simulated models in contrast to the SRPM method. Note that the inverse covariance matrix and the SRPM have to be normalized to obtain the partial correlations (Whittaker, 1990).
1.5 Second Contribution: Theoretical Analysis of the SRPM Method
Furthermore, we give interpretations for the recovery of the connectivity structure via the SRPM method by energy models using the Boltzmann distribution (MacKay, 2002). This is the second contribution of the letter. This shows that theoretically, if the inverse covariance matrix can estimate the connectivity structure between regions under consideration, then the SRPM method should be used to estimate the regularized version of the inverse covariance matrix to avoid false-positive and false-negative connections when a finite number of samples is available.
1.6 Third Contribution: Application of SRPM in Analyzing Brain Connectivity in the Presence and Absence of Sleep Spindles
During sleep, our brains are highly active. The low-amplitude, high-frequency activity in the neocortex characteristic of the awake state is replaced with a sequence of distinct phases with generally high-amplitude, low-frequency activity. Soon after the onset of sleep, brief episodes of 10 Hz to 14 Hz, synchronized spindling occur in the thalamus and cortex, producing large-scale spatiotemporal coherence throughout the forebrain. Throughout the night, the cortex alternates between periods of slow-wave sleep in the range of 2 Hz to 4 Hz and episodes of rapid eye movement sleep (REM), characterized by sharp waves of activity in the pons, the thalamus, and the occipital cortex, while also passing through intermediate non-REM sleep stages (Sejnowski, 1995; Sejnowski & Destexhe, 2000).
Activity in the sleeping brain is largely hidden from us because very little content of the brain activity that occurs during sleep directly enters consciousness. Hence, it becomes important to understand the patterns of electrical activity of neurons that occur in the brain during sleep. It has been suggested that the important functions of sleep are adaptive strategies, physical recovery, energy conservation, and information processing, among others. There is also evidence supporting the role of sleep in learning and memory consolidation, and neuronal plasticity (Sejnowski, 1995; Sejnowski & Destexhe, 2000; Stickgold & Walker, 2005, 2007; Martin et al., 2013).
Fogel, Nader, Cote, and Smith (2007) investigated the functional significance of the considerable interindividual differences in sleep spindles. The pattern of the sleep spindles within individuals is quite stable and varies little from night to night. Because of the remarkable intra-individual stability in sleep spindles from night to night, it was hypothesized that sleep spindles may serve as a “fingerprint” to account for interindividual differences.
Three separate studies were performed to broadly examine the relationship between spindles and learning potential as measured by an IQ test. In all three studies, it was found that the number of sleep spindles was positively correlated with performance IQ. Power in the 12–18 Hz frequency band, a more objective indicator of the level of stage 2 spindle activity, displayed even stronger correlations with performance IQ. These results indicated that performance IQ can be predicted simply by knowing the number of spindles and sigma power. This suggested that sleep spindles and sigma power may be biological markers for the specific abilities assessed by performance IQ. This relationship might reflect the efficiency of information processing that is dependent on thalamocortical communication. In other words, richer cortical representations would require more thalamocortical interconnectedness. Maintenance and encoding of new information in a more complex system may require more thalamocortical activity or a more efficient thalamocortical system. This efficiency or added complexity may be reflected in the higher number of sleep spindles in the cortical areas underlying perceptual or analytical abilities in individuals with a higher performance IQ.
Fogel and Smith (2006) examined the learning-dependent changes in sleep, including stage 2 sleep spindles, where subjects went through an intense period of simple motor procedural learning. Overall, the results from the study supported the hypothesis that sleep spindles are intimately involved with the consolidation of simple motor procedural memory and may be important for the offline reprocessing of recently acquired simple procedural tasks. They found an increase in the density of sleep spindles. Furthermore, they found that the overall improvement on motor tasks was positively correlated with the increase in sleep spindle density. In addition, there was an increase in the duration of stage 2 sleep following new learning. The magnitude of this change was very large: overall, there was a 15.8% increase in stage 2 sleep. The increase in the duration of stage 2 sleep would be expected to increase the total number of sleep spindles alone. In addition to the increased duration of stage 2 sleep, there was an increase in spindle density. This suggested that motor learning–dependent changes in sleep spindles are independent of the time spent in stage 2 sleep. To determine if the changes to sleep following simple procedural memory were limited to stage 2 sleep and sleep spindles, REM density was also considered in this experiment. The researchers found that neither the duration of REM sleep nor the density of REM changed following new learning, which suggested that the changes to sleep following new simple procedural learning affected only stage 2 sleep and is specific to sleep spindles.
Schabus et al. (2004) studied the functional significance of stage 2 sleep spindle activity for declarative memory consolidation. This study measured spindle activity during stage 2 sleep following a (declarative) word-pair association task as compared to a control task. Participants performed a cued recall in the evening after learning (160 word pairs), as well as in the subsequent morning after 8 hours of undisturbed sleep with full polysomnography. Overnight change in the number of recalled words correlated significantly with increased spindle activity during the experimental night. The results also suggested that the increase in spindle activity cannot simply be accounted for by changes in (stage 2) sleep architecture or subjects’ fatigue. They found that the relationship between memory performance and spindle activity was not an indirect effect of sleep-stage durations. Even when all sleep stages were controlled, the correlation between memory performance and spindle activity changes remained significant. Thus, their findings provided evidence for the involvement of sleep spindle activity in memory consolidation as measured by the declarative memory task performed before and after the experimental night. The fact that spindle activity was related only to changes in memory performance (increase or decrease over the night) was consistent with the hypothesis that spindle activity is specifically related to the consolidation of recently established memory traces. In other words, the findings of the study were in good agreement with the role of sleep spindles for memory consolidation. Several other researchers (Gais & Born, 2004; Gais, Mölle, Helms, & Born, 2002; Marshall & Born, 2007) also have reported similar results.
Walker, Brakefield, Morgan, Hobson, and Stickgold (2002) found that improvement in motor skill performance in humans was due to sleep. They found evidence that continued improvement on a motor skill task occurs only across a night of sleep, while an equivalent period of wake offers no significant benefit to performance. Furthermore, more than half the variance in overnight improvement was explained by the amount of stage 2 sleep obtained during night. The authors speculated that the enhancement in motor skill was due to the sleep spindles, which are thought to cause massive calcium entry into pyramidal cells of the cerebral cortex, triggering intracellular calcium-dependent mechanisms required for synaptic plasticity (Sejnowski & Destexhe, 2000) and have been shown to increase following training on a motor task (Fogel et al., 2007; Fogel & Smith, 2006) as described previously. These implications become most significant in the broader context of acquiring real-life skillful actions such as learning motor patterns required for movement-based sports, learning a musical instrument, or developing artistic movement control. All such learning of new actions may require sleep before the maximum benefit of practice is expressed.
Smith and Macneill (1994) found impaired motor memory for a pursuit rotor task following stage 2 sleep loss in college students. Among the subjects considered, one group was subjected to REM sleep deprivation and the other group to non-REM sleep deprivation following acquisition of a pure motor task, the pursuit rotor. Results showed that the REM sleep deprivation group had excellent memory for the task, whereas the non-REM sleep deprivation group had a deficit in memory for the task. It was concluded that stage 2 sleep (where sleep spindles occur) rather than REM sleep was the important stage of sleep for efficient memory processing of the pursuit motor task. In other words, the newly acquired pure motor skill was most efficient when posttraining stage 2 sleep was allowed and was impaired when this stage of sleep was reduced or interrupted in the sleep night following the training session.
Meier-Koll, Bussmann, Schmidt, and Neuschwander (1999) tried to link the storing of spatial information and episodic memory to sleep stages. Two city mazes, a simple and a complex one, were created by means of a computer program and were presented to the subjects on a TV screen. The task was to find various end points and the way back to the starting point with the help of a PC mouse. After the task, the subjects slept, and the sleep stages were measured polygraphically. The subjects exposed to this experiment had significantly enhanced sleep spindle activities in comparison to subjects who had experienced neither maze. The researchers concluded that there is a functional linkage between stage 2 sleep spindles and learning or information processing in cortical areas.
Furthermore, because of its well-organized and consistent structure, sleep can be a valuable instrument for investigating neurological disorders such as Alzheimer’s disease, progressive supranuclear palsy, REM sleep behavior disorder, Parkinson’s disease, dementia with Lewy bodies, multiple system atrophy (MSA), Huntington’s disease and Creutzfeldt-Jakob disease (Petit, Gagnon, Fantini, Ferini-Strambi, & Montplaisir, 2004). Ferrarelli et al. (2007) investigated whether sleep spindles differ between subjects with schizophrenia, healthy individuals, and a psychiatric control group with a history of depression. The authors found a decrease in sleep spindle number, amplitude, duration, and integrated spindle activity in patients with schizophrenia. Furthermore, integrated spindle activity had an effect size corresponding to 93.0% or 90.2% separation of the schizophrenia from the comparison or depression group. Since sleep spindles are generated by the thalamic reticular nucleus in conjunction with specific thalamic nuclei and are modulated by corticothalamic and thalamocortical connections, it was concluded that the deficit in sleep spindles in schizophrenia subjects may reflect dysfunction in thalamic-reticular and thalamocortical mechanisms and could represent a biological marker of illness. Hence, finding the functional pattern in the brain during sleep spindles can provide valuable information for understanding the pathophysiology and for assisting the diagnosis of neurodegenerative diseases.
From these studies, it is quite clear that sleep and sleep spindles play a vital role in memory and learning and in investigation of neurological disorders in the human brain. Thus, finding connectivity during sleep will surely give us insight into the strong activity regions in the brain areas and how they are organized, which may be responsible for information processing. Unfortunately, there has been very little research regarding the brain connectivity pattern during sleep. Moreover, researchers have termed the correlation of brain activity between brain regions as “functional connectivity” and have drawn conclusions based on that. These conclusions are flawed since the correlation method fails to identify the true connectivity pattern of a physical network, as we will show via extensive simulations using artificial networks.
Spoormaker et al. (2010) characterized the human functional brain network during non-REM sleep by using the correlation method on fMRI recordings. In this study, the authors used the correlation method to explore how physiological changes during sleep are reflected in functional connectivity and small-world network properties of a large-scale, low-frequency functional brain network. They observed that in the transition from wakefulness to light sleep, thalamocortical connectivity was sharply reduced, whereas corticocortical connectivity increased; corticocortical connectivity subsequently broke down in slow-wave sleep. Local clustering values were closest to random values in light sleep, whereas slow-wave sleep was characterized by the highest clustering ratio (gamma). The authors claimed that the changes in consciousness in the descent to sleep are subserved by reduced thalamocortical connectivity at sleep onset and a breakdown of general connectivity in slow-wave sleep, with both processes limiting the capacity of the brain to integrate information across functional modules.
Larson-Prior et al. (2009) studied cortical network functional connectivity during sleep using the correlation method. They examined functional connectivity using conventional seed-based analyses in three primary sensory and three association networks as normal young adults transitioned from wakefulness to light sleep. They found that functional connectivity in non-REM sleep was maintained in each network throughout all examined states of arousal. Further, these networks were consistent across subjects. The authors were surprised that they did not find any evidence of change in functional connectivity in the sensory (visual, auditory, and somatomotor) or cognitive (dorsal attention, default and executive control) networks examined.
Andrade et al. (2011) also investigated the functional connectivity between the hippocampal and neocortical regions of the human brain in non-REM sleep using the correlation method. They found increased connectivity between hippocampal and neocortical regions of the brain during stage 2 sleep spindles, suggesting increased capacity for global information transfer during sleep spindles.
Though the results of these studies are interesting, further experimental validation is necessary to support their claims and derive a more thorough physiological interpretation. A flaw in these analyses is the use of the correlation method to characterize the brain functional connectivity pattern during sleep since two brain regions might show very high correlation even if there is no strong physical connection between them; rather, the correlation could be due to a common input. Hence, this is the motivation for the application of the sparse regularized precision matrix (SRPM) method in connectivity estimation during sleep.
In our third contribution, we demonstrate the application of the SRPM method for estimating brain connectivity during sleep spindles from human electrocorticography (ECoG) data using an 8 × 8 electrode array. The ECoG recordings that we analyzed were from a 32-year-old male patient with longstanding pharmaco-resistant left temporal lobe complex partial epilepsy. Sleep spindles have a major role in learning and memory consolidation. Our purpose in this letter is to find and understand the functional organization of brain areas in the presence and absence of sleep spindles. We find that brain connectivity during the spindles is highly spatially localized in contrast to the case when sleep spindles are not present. We believe that this can give us further insight into the functioning of brain areas during spindles in stage 2 sleep. We can find the connectivity pattern in the presence and absence of sleep spindles by the SRPM method.
1.7 Fourth Contribution: Detection of Sleep Spindles Using Delay Differential Analysis (DDA)
For the detection of sleep spindles, we employ a novel method called DDA (Kremliovsky & Kadtke, 1997; Lainscsek, Hernandez, Weyhenmeyer, Sejnowski, & Poizner, 2013; Lainscsek & Sejnowski, 2015; Lainscsek, Weyhenmeyer, Hernandez, Poizner, & Sejnowski, 2013; Sampson, Lainscsek, Cash, Halgren, & Sejnowski, 2015). This is the fourth contribution of the letter. DDA is a time domain classification framework based on embedding theory in nonlinear dynamics. Given a recording (here, ECoG data) from some unknown dynamical system (here, the brain), an embedding will reveal the nonlinear invariant properties of the system, even from a single time series. The embedding in DDA provides a low-dimensional nonlinear functional basis onto which the data are mapped. Since this basis is built on the dynamical structure of the data, preprocessing of the data (such as filtering) is not necessary. DDA yields a low number of features (around four), as compared with traditional spectral techniques, which greatly reduces the risk of overfitting. Frequency-based approaches that have often been used for detecting spindles are sensitive to certain artifacts involving transient increases in spectral power in the band of interest; with DDA, we train on the real data to find the relevant dynamical features for spindle identification.
For clustering of similar brain regions, we use the Louvain method for community detection (LMCD) (Blondel et al., 2008; Brandes et al., 2008; Newman & Girvan, 2004; Newman, 2006; Reichardt & Bornholdt, 2006; Ronhovde & Nussinov, 2009; Sporns, 2010; Sun et al., 2009) on the SRPM. LMCD is a widely used method for clustering in human brain imaging data analysis (Bassett et al., 2010, 2011; Cole, Bassett, Power, Braver, & Petersen, 2014; Meunier, Lambiotte, & Bullmore, 2010; Rubinov & Sporns, 2010, 2011; Sporns, 2011; Zuo et al., 2012).
1.8 Organization of the Letter
The rest of the letter is organized as follows. In section 2, we describe the spring-mass model and show how the connectivity of the springs and masses can be recovered by the SRPM method. In section 3, we describe the RC circuit model and describe how to estimate the connectivity pattern of the nodes by using the SRPM method. In the RC circuit model, we consider two network topologies: the tree topology and the mesh topology. In section 4, we first describe the DDA method for automatically detecting sleep spindles from human ECoG data and then proceed to the application of SRPM to recover the connectivity among the brain regions during the sleep spindles. We draw conclusions in section 5.
2 The Spring-Mass Model
The spring-mass model that we use in our simulation is shown in Figure 1. We assume that we can measure the displacements of the masses in the model. There are 50 equal masses m connected in cascade via springs, each with a spring constant k. The left-most and the right-most springs are connected to a rigid wall. For our simulation (hereafter all units are in mks), we use m = 0.1 and k = 1. We assume that this spring-mass model is subject to thermal perturbation. In addition, we also assume that each spring is subjected to external force, and we model this external force as a white noise process with variance σ2. For our simulation, we use σ2 = 0.000025. We denote the displacements of the masses as x1, x2, …, x50 and the external forces associated with each of them as w1, w2, …, w50, respectively. Using Newton’s second law of motion and Hooke’s law, we can write the displacement equations of the masses as
| (2.1) |
In matrix-vector form, the above set of equations can be written as
| (2.2) |
Figure 1.

The spring-mass model. The black dots denote continuation of springs and masses.
In compact form, equation 2.2 can be represented as
| (2.3) |
where x is the displacement vector, w is the white noise vector, and C is the connectivity (or ground-truth) matrix and is shown in Figure 2. Using a second-order approximation of the double derivative on the left-hand side in equation 2.3, we have
| (2.4) |
where h is the step size and t denotes the time instant. We use a random initialization of x with variance . We then solve equation 2.4 repeatedly to generate N samples of the displacement vector x corresponding to N consecutive time points. For our simulation, we choose h = 0.007 and N = 50,000. We use these N samples to form the sample covariance matrix.
Figure 2.
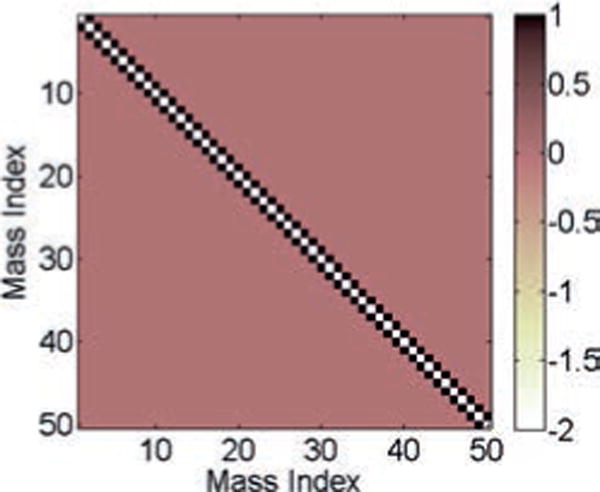
The connectivity matrix or the ground-truth matrix C for the spring-mass model.
We next apply the correlation method, the inverse covariance method, and the SRPM method to recover the connectivity pattern of the spring-mass system. The value of λ for the SRPM method is chosen to be 0.0009. The results are shown in Figure 3. Note that the ground-truth matrix in Figure 3 is binarized such that a one denotes either a diagonal element or a connectivity between two masses via a spring and a zero denotes no connectivity. For all three methods, we first estimate the connection matrix and choose the M largest elements (in absolute value) in the estimated connection matrix, where M is the number of nonzero elements in the ground-truth matrix. In Figure 3, for each method, we denote these M largest elements as ones and the rest as zeros. The binarization is done for clarity and better visualization. The error percentage in the figures for a particular method is calculated as the fraction of false-positive and false-negative connections between the binarized ground-truth matrix and the binarized estimated connection matrix for that method.
Figure 3.
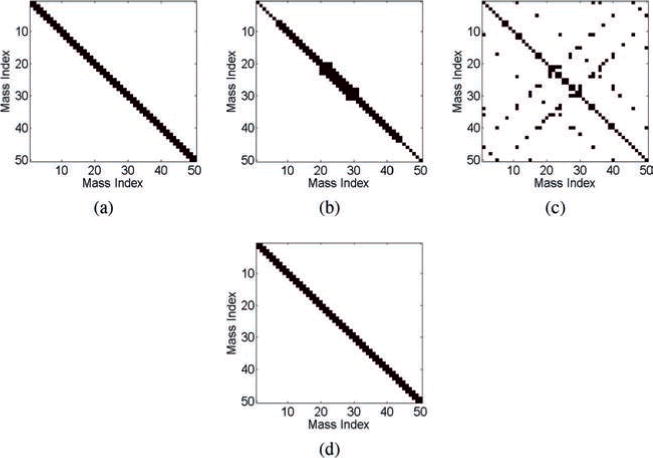
Results of the estimation methods for the spring-mass model. (a) The ground-truth matrix. (b) The estimated connection matrix from the correlation method (16% error). (c) The estimated connection matrix from the inverse covariance method (53% error). (d) The estimated connection matrix from the SRPM method (0% error). In panels a–d, black denotes either a diagonal element or a connectivity between two masses via a spring, and white denotes no connectivity.
From Figure 3b, we note that the correlation method is not able to fully recover the true connectivity structure of the spring-mass system. Since the sample covariance matrix is a poor estimator of the true covariance matrix, the inverse covariance method is even worse than the correlation method and results in a large number of false-positive and false-negative connections. The SRPM method is able to recover successfully the true connectivity pattern of the spring-mass model.
2.1 Explanation for the Recovery of the Connectivity Structure via the SRPM Method in the Spring-Mass Model
Assuming that the spring-mass model is subject only to thermal perturbation (i.e., if we assume w = 0 in equation 2.3), then the probability distribution of the displacement vector x can be given by the Boltzmann distribution (MacKay, 2002) as
| (2.5) |
where β = 1/(kT), k is the Boltzmann constant, T is the temperature, and Z is a normalization factor. In equation 2.5, E(x) is the energy of the spring-mass system. For the model in Figure 1, the energy E(x) can be written as
| (2.6) |
where xT denotes the transpose of x and . Hence, we have
| (2.7) |
We note that P(x) in equation 2.7 is a multivariate gaussian distribution in x whose inverse covariance matrix differs from only by a scalar multiple, implying that the inverse covariance matrix in equation 2.7 is just a scaled version of the connectivity matrix C in equation 2.3. Hence, the inverse covariance matrix can recover the connectivity structure of the cascade connection of springs and masses given in Figure 1.
Since the sample covariance matrix is a poor estimator of the eigenvalues of the covariance matrix, the inverse of the covariance matrix produces a large number of false-positive and false-negative connections as shown in Figure 3c. In contrast, since the connectivity structure is sparse, we can use the SRPM method to recover the connectivity pattern of the spring-mass system.
Remark 1
The performance (in terms of error percentage) of the correlation method, the inverse covariance method, and the SRPM method in the spring-mass model does not change under different types of noise distributions. We have tested the performance of these methods under gaussian, Poisson, and uniform distributions and have obtained similar performance to that given in Figure 3 for the three methods.
Remark 2
The performance (in terms of error percentage) of the three methods in the spring-mass model does not change for signal-to-noise ratio up to 30 dB, and the performance is very similar to that given in Figure 3 for the three methods.
These results are not surprising. Note that in the spring-mass model, we model the noise as an external force. So no matter how “forcefully” (noise variance) or in what way (noise distribution) we wobble the spring-mass system, the connectivity pattern will not change. Noise is not responsible for the connectivity pattern, and the connectivity patterns for different methods shown in Figure 3 are due to the methods themselves.
3 The RC Circuit Model
The RC circuit model is taken from Sojoudi and Doyle (2014), who applied an SRPM method with q = 1 (graphical LASSO). We consider two network topologies: the tree network and the mesh network. We assume that the voltages at the nodes are available for measurement and estimate the connectivity by the three methods from these measured voltages. An edge between a pair of nodes denotes a parallel resistor-capacitor (RC) circuit as shown in Figure 4a. We assume that the circuit is activated only by the thermal current (i.e., there is no supply of external current) and each node is subject to thermal current as shown in Figure 4b. We model this thermal current as white noise known as the Johnson-Nyquist noise. These stochastic currents produce stochastic voltages at the nodes, which in turn cause the charging and discharging of the capacitors in the network.
Figure 4.
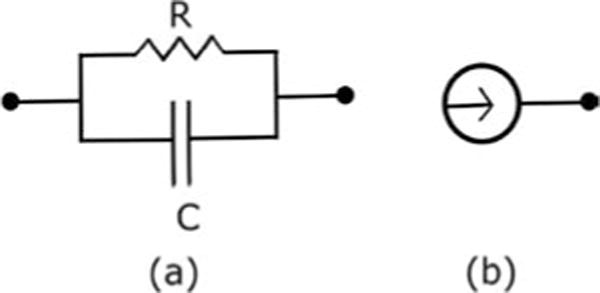
Elements of the RC circuit model. (a) Parallel RC circuit between a pair of nodes. (b) Thermal current associated with a node.
3.1 The Tree Network
This network is shown in Figure 5, where the nodes are denoted as numbers and each edge represents a parallel RC circuit as in Figure 4a. We consider 10 nodes in this example. Let v denote the vector of voltages at the nodes and i denote the vector of stochastic currents injected to the nodes by some external device. Using Kirchoff’s law, we have
| (3.1) |
where A is the capacitance matrix and G is the conductance matrix (see Sojoudi & Doyle, 2014, regarding how to construct the matrices A and G). For our simulation, we assume that each of the values of the capacitances and conductances denoted as edges in Figure 5 has a value of 1. Moreover, node 5 of the circuit is grounded through a parallel RC circuit, and for this, the values of both the capacitance and the conductance are taken to be 4. Hence, in this case, A ≡ G, and we call A or G the connectivity matrix or the ground-truth matrix of the RC circuit. Note that the values of the capacitances and conductances in the tree network are chosen such that the connectivity matrix A (≡ G) is positive definite. The connectivity matrix for the given tree network is shown in Figure 6. Assuming that the current vector i is due to the stochastic currents from the white thermal noise, we can write equation 3.1 as (Sojoudi & Doyle, 2014)
| (3.2) |
where is the square root (not the elementwise square root) of the matrix G. For our simulation, we choose variance σ2 = 4 for the white noise process w in equation 3.2. Using a first-order approximation of the derivative on the left-hand side in equation 3.2, we have
| (3.3) |
where h is the step size and t denotes the time instant. For our simulation, we choose h = 0.5. We use a random initialization of v with variance . As done in the spring-mass example, we solve equation 3.3 iteratively to generate N samples for the voltages. For our simulation, we choose N = 282.
Figure 5.
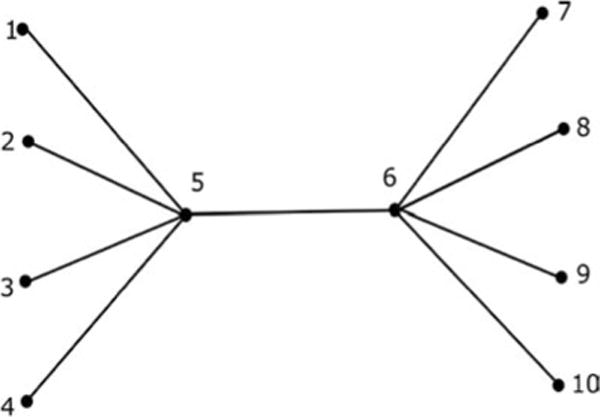
The tree network.
Figure 6.
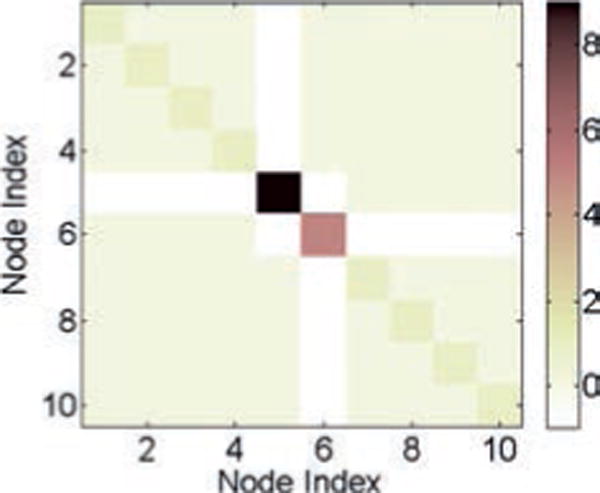
The connectivity matrix or the ground-truth matrix A (≡ G) for the tree network.
We next apply the correlation method, the inverse covariance method, and the SRPM method to recover the connectivity pattern of the tree network. The value of λ for the SRPM method is chosen to be 0.01. The results are shown in Figure 7. As done in the spring-mass example, we show only the binarized results for better visualization.
Figure 7.
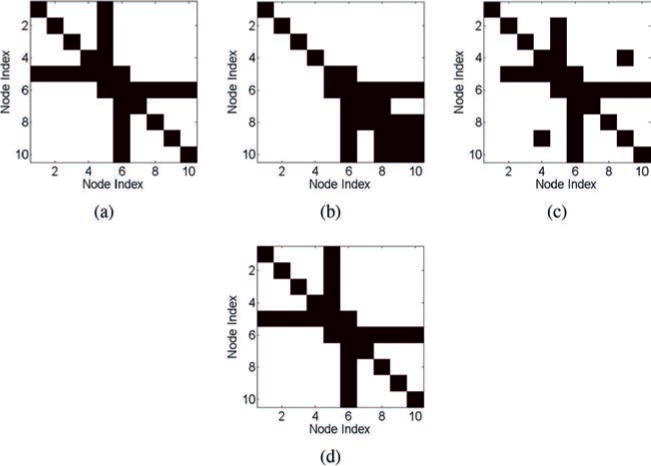
Results of the estimation methods for the tree network in the RC circuit model. (a) The ground-truth matrix. (b) The estimated connection matrix from the correlation method (29% error). (c) The estimated connection matrix from the inverse covariance method (7% error). (d) The estimated connection matrix from the SRPM method (0% error). In panels a–d, black denotes either a diagonal element or a connectivity between two nodes, and white denotes no connectivity.
We note that the correlation method and the inverse covariance method are not able to recover the true connectivity pattern of the tree network. The correlation method in particular has a large percentage of false-positive and false-negative connections. This is due to the fact that node 5 is connected to ground via a parallel RC circuit with relatively high conductance and capacitance. Thus, the grounded node and its neighbors will have low correlation and the ungrounded nodes and their neighbors will have high correlation even though they are not connected (Sojoudi & Doyle, 2014). In contrast, the SRPM method is able to recover successfully the ground truth of the tree network.
3.1.1 Explanation for the Recovery of the Connectivity Structure via the SRPM Method in the Tree Network
Since we have assumed that the tree network is only subject to thermal perturbation, the probability distribution of the voltage vector v can be given by the Boltzmann distribution as
| (3.4) |
where E(v) is the energy of the capacitors in the tree network and Z is the corresponding normalization factor. For the given tree network in Figure 5, the energy E(v) can be written as
| (3.5) |
where υi denotes the voltage at the ith node, i = 1, 2, …, 10 and A is the connectivity matrix of the tree network given in equation 3.2. Hence, we have
| (3.6) |
which is a multivariate gaussian distribution in v whose inverse covariance matrix differs from A only by a scalar multiple. Hence, the inverse covariance matrix can recover the connectivity pattern of the tree network given in Figure 5. But since the sample covariance matrix is a poor estimator of the eigenvalues of the covariance matrix, the inverse of the covariance matrix produces false-positive and false-negative connections, as shown in Figure 7c. In contrast, since the connectivity structure is sparse, the SRPM method is able to recover successfully the connectivity pattern of the tree network.
3.2 The Mesh Network
This network is shown in Figure 8, where the nodes are denoted as numbers and each edge represents a parallel RC circuit as in the tree network. We consider 24 nodes in this example. Equations 3.2 and 3.3 remain the same for this network with the corresponding A, G, and v. For our simulation, we assume that each of the values of the capacitances and conductances denoted as edges in Figure 8 has a value of 1. Moreover, all the nodes indexed from 1 to 18 of the circuit are grounded through parallel RC circuits, as is done for node 5 in the tree network, and the value of both the capacitance and the conductance are taken to be 5 for each parallel RC circuit connected to ground. Hence, in this case A ≡ G, and this is the connectivity matrix or the ground-truth matrix for the mesh network. Once again, note that the values of the capacitances and conductances in the mesh network are chosen such that the connectivity matrix A (≡ G) is positive definite. The connectivity matrix for the given mesh network is shown in Figure 9. We again choose σ2 = 4 for the white noise process w in equation 3.2. The value of step size is chosen to be h = 0.5. We also use a random initialization of v with variance . As done in the tree network, we solve equation 3.3 iteratively to generate N samples for the voltages, and in this case, we choose N = 2000.
Figure 8.
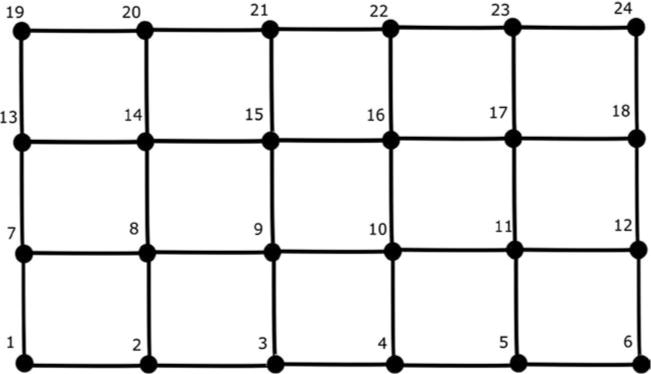
The mesh network.
Figure 9.
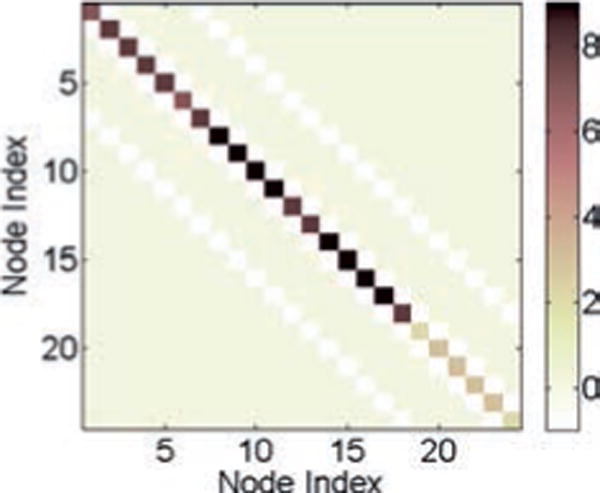
The connectivity matrix or the ground-truth matrix A (≡ G) for the mesh network.
We again apply the correlation method, the inverse covariance method, and the SRPM method to recover the connectivity pattern of the mesh network. The value of λ for the SRPM method is chosen to be 0.005. The results are shown in Figure 10. As done in the tree network, we show only the binarized results.
Figure 10.
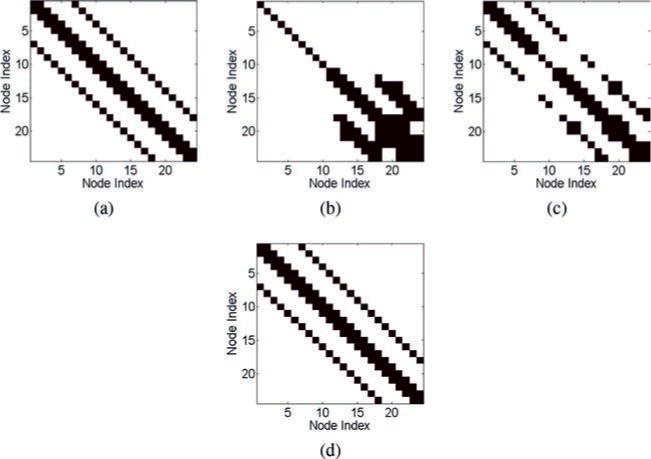
Results of the estimation methods for the mesh network in the RC circuit model. (a) The ground-truth matrix. (b) The estimated connection matrix from the correlation method (40% error). (c) The estimated connection matrix from the inverse covariance method (11% error). (d) The estimated connection matrix from the SRPM method (0% error). In panels a–d, black denotes either a diagonal element or a connectivity between two nodes, and white denotes no connectivity.
We note that the correlation method and the inverse covariance method are not able to recover the true connectivity pattern of the mesh network with the correlation method having a large percentage of false-positive and false-negative connections. We also observe that the ungrounded nodes and their neighbors have high correlation even though they are not connected. Once again, the SRPM method is able to successfully recover the ground truth of the mesh network.
3.2.1 Explanation for the Recovery of the Connectivity Structure via the SRPM Method in the Mesh Network
Following the procedure in the tree network, if we assume that the mesh network is subject only to thermal perturbation, it is straightforward to show that the voltage vector v in the mesh network follows a multivariate gaussian distribution whose inverse covariance matrix differs from the connectivity matrix A of the mesh network shown in Figure 8 only by a scalar multiple. Hence, the inverse covariance matrix can recover the connectivity pattern of the mesh network. Once again, the direct inverse of the covariance matrix produces false-positive and false-negative connections as shown in Figure 10c. In contrast, since the connectivity structure is sparse, the SRPM method is able to successfully recover the connectivity structure of the mesh network.
Also, note that for the voltage vector v to follow a multivariate normal distribution whose inverse covariance matrix differs from the connectivity matrix A only by a scalar multiple, the RC circuit network need not be one of the specific structures (tree and mesh) given in Figures 5 and 8. It is straightforward to show (following the steps as before via the Boltzmann distribution) that the inverse covariance matrix can recover the connectivity pattern of an arbitrarily connected RC circuit network in the limit as the number of samples increases to infinity.
Remark 3
The performance (in terms of error percentage) of the correlation method, the covariance method, and the SRPM method in the tree network and the mesh network does not change under different types of noise distributions. We have tested the performance of these methods under gaussian, Poisson, and uniform distributions and have obtained similar performance to that given in Figure 7 for the tree network and Figure 10 for the mesh network for the three methods.
Remark 4
The performance (in terms of error percentage) of the three methods in the tree network and the mesh network does not change for signal-to-noise ratio up to 30 dB, and the performance is very similar to that given in Figure 7 for the tree network and in Figure 10 for the mesh network for the three methods.
These results are not surprising. Note that in the RC networks, we have modeled the noise as stochastic current from white thermal noise. If the current goes up and down or changes direction, the connectivity pattern will not change. Noise is not responsible for the connectivity pattern, and the connectivity patterns for different methods shown in Figures 7 and 10 are due to the methods themselves.
4 Connectivity Estimation during Sleep Spindles from Human ECoG Recordings
We now proceed to the application of the SRPM method in estimating brain connectivity during sleep spindles from human ECoG data. We first describe the DDA method for sleep spindle detection. Sleep spindles are detected from single-channel time series data, and we repeat the DDA method on each channel. To collect the ECoG data, we use an 8 × 8 electrode array, hence 64 grid electrodes, or channels, in total. First, we describe the data acquisition protocol. Next, we briefly describe the DDA method. Then we apply the SRPM method for estimating the strongest connectivity between the brain regions during the sleep spindles. For connectivity estimation, we consider only time windows in which the spindles were present in a relatively large number of channels. After estimating the connectivity, we cluster similar brain regions together using the LMCD method and find spatially localized brain networks. These localized brain networks might suggest the flow of information in the brain areas during sleep spindles.
We clarify that the assumption of sparse connectivity is reasonable due to the following. During the non-REM sleep, the thalamic neurons excite the cortex with patterns of activity that are more spatially and temporally coherent than would normally be encountered in the awake state. Getting these neurons to fire together is a potent way of enhancing their impact on other neurons and the cortex, because the synaptic inputs arriving synchronously on a neuron produce greater output than the same number of inputs arriving asynchronously. Thus, if too many neurons fire together at the same time, this amplification may go awry and lead to an epileptic seizure (Steriade, McCormick, & Sejnowski, 1993). Therefore, the level of activity and degree of synchrony in the neural networks and cortex of the brain are strongly regulated through dynamic cellular mechanisms. Since at a given time very few neuronal cells in the brain are active, the assumption of sparse connectivity in the brain during sleep spindles is reasonable.
4.1 ECoG Data Acquisition and Protocol
ECoG recordings from a 32-year-old male patient with long-standing pharmaco-resistant left temporal lobe complex partial epilepsy were analyzed. Recordings were performed using a standard clinical recording system (XLTEK, Natus Medical, San Carlos, CA) with a 500 Hz sampling rate. The reference channel was a strip of electrodes placed outside the dura and facing the skull at a region remote from the other grid and strip electrodes. Subdural electrode arrays were placed to confirm the hypothesized seizure focus and locate epileptogenic tissue in relation to essential cortex, thus directing surgical treatment. The decision to implant, the electrode targets, and the duration of implantation were made entirely on clinical grounds with no input from this research study. All data acquisition was performed under protocols monitored by Institutional Review Board of the Massachusetts General Hospital according to National Institutes of Health guidelines. Data selected for use in this study were exclusively from stage 2 sleep, during time periods when no seizures were occurring. The medication information for the patient is given in Table 1. The recordings analyzed here are from day 10.
Table 1.
Patient Medication Information.
| Day | Medication |
|---|---|
| Home medications | Keppra 1000 bid |
| Day 1 | Levetiracetam 1000 bid |
| Day 2 | Levetiracetam 1000 bid |
| Day 3 | Levetiracetam 500 bid |
| Day 4 | Levetiracetam 500 bid |
| Day 5 | off |
| Day 9 | Levetiracetam 1500 qhs, lorazepam 1mg IV |
| Day 10 | Levetiracetam 1000 bid |
4.2 DDA Method for Sleep Spindle Detection in Human ECoG Data
In order to detect sleep spindles reliably in the ECoG data, we used DDA. In Figure 11a, DDA is introduced as a time domain classification framework based on embedding theory in nonlinear dynamics (Kremliovsky & Kadtke, 1997; Lainscsek, Hernandez et al., 2013; Lainscsek & Sejnowski, 2015; Lainscsek, Weyhenmeyer et al., 2013; Sampson et al., 2015). Given a recording (here, ECoG data) from some unknown dynamical system (here, the brain), an embedding will reveal the nonlinear invariant properties of the system, even from a single time series (Takens, 1981). The embedding in DDA provides a low-dimensional nonlinear functional basis onto which the data are mapped. Since this basis is built on the dynamical structure of the data, preprocessing of the data (such as filtering) is not necessary. DDA yields a low number of features (around four), as compared with traditional spectral techniques, which greatly reduces the risk of overfitting.
Figure 11.
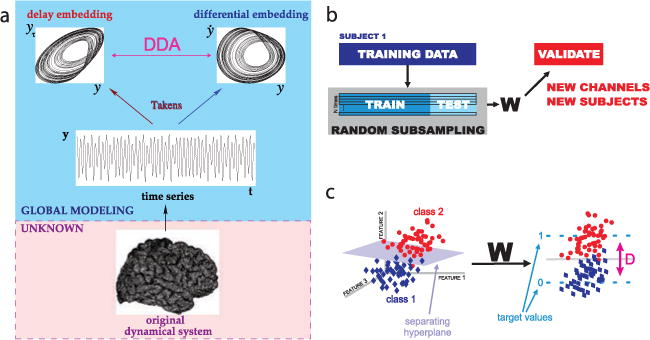
Delay differential analysis (DDA). (a) For an unknown dynamical system (such as the brain) from which we can record the value of a single variable over time (such as ECoG data), embedding theory states that we can recover the nonlinear invariant properties of the original system. DDA combines delay and differential embeddings in a functional form that allows time-domain classification of the data. (b) Performance of DDA model forms is evaluated with repeated random subsampling cross-validation. The data are repeatedly divided at random into training and testing sets. (c) Applying the weights (set by SVD) to the DDA features transforms from the feature space to a one-dimensional distance from the hyperplane of separation. This value is used as a measure of performance for classification.
Another way of viewing DDA models is as sparse Volterra series (Volterra, 1887, 1959). A general nonlinear real-valued function can be expressed as a Taylor series expansion of functionals of increasing complexity around a fixed point. When the function represents the behavior of a dynamical system, the expansion becomes a Volterra series. DDA restricts the complexity of the analysis by using a low-dimensional sparse delay differential equation model. In such a model, linear and nonlinear data components are analyzed in an interconnected manner. This reduces the computational load, and leaving some of the nonrelevant dynamics unmodeled highly reduces the effect of artifacts and other signals unrelated to those we aim to detect.
All of these properties make DDA well suited to the problem of spindle detection. Frequency-based approaches that have often been used for detecting spindles are sensitive to artifacts involving transient increases in spectral power in the band of interest, and parameters often have to be adjusted to fit individual subjects. In DDA, we train on real data from a single subject and a single electrode and find the relevant dynamical features for spindle identification that are highly generalizable to a wide class of subjects and recordings.
In practice, DDA (see Figure 11a) combines a differential embedding with a delay embedding by relating them in a polynomial function,
| (4.1) |
where I is the number of monomials in the model, N is the number of delays in each monomial, and mn,i is the order of the nth term in the ith monomial. The time derivative of the data, , is computed with a five-point center derivative (Miletics & Molnárka, 2005). The estimated coefficients ai for the model, as well as the least-squares error, form the low-dimensional feature space used for classifying the data (here, for detecting spindles). The least-squares error is defined as
| (4.2) |
where K denotes the number of time points, denotes , and denotes x(t − τn).
The particular form of the polynomial in equation 4.1 was chosen after an exhaustive search of all model forms subject to the following constraints: model forms were constrained to two delays (N ≤ 2), three terms (I ≤ 3), and third-order nonlinearity (Σn mn,i ≤ 3), and the delays were constrained to values up to 150 time points (τn ≤ 150). For model selection, we used a training data set with human expert–scored spindles in stereoelectroen-cephalogram (SEEG) data sampled at 500 Hz. As shown in Figure 11b, the best-performing model was selected using repeated random subsampling cross-validation (Kohavi, 1995). This method involves repeatedly dividing the data at random into training and testing sets. Each random split assigns 70% of the data to the training set and 30% to the testing set. The model form and values of the delays τ1 and τ2 in equation 4.3 were chosen to maximize the separation between spindle and nonspindle epochs. For spindle detection, we used the model
| (4.3) |
with τ1 = 19 δt and τ2 = 12 δt, where δt 1/fs = 2 ms. Different values of the delays would be selected for a different sampling rate. From the cross-validation procedure, we obtain weights using singular-value decomposition (SVD), which we apply to the four features from this model— a1, a2, a3, and ρ—to transform the four-dimensional feature space to a one-dimensional distance from a hyperplane of separation. This transformation is illustrated in Figure 11c. The small number of features is a general feature of DDA, which has the advantages of minimizing overfitting and reducing the influences of artifacts in the recordings. DDA is also more sensitive than traditional methods based on frequency analysis and thresholding, allowing spindles to be detected in most electrodes.
After selecting the model based on the SEEG training data, its performance was evaluated on a set of 20 recordings from SEEG, ECoG, and laminar electrodes. Across this data set, our DDA-based spindle detector agrees with the expert scoring with a mean area of 0.84 under the receiver operating characteristic (ROC) curve and a mean F1 score of 0.9. This is comparable to or better than typical interrater agreement between human experts. Warby et al. (2014) found a mean F1 score of 0.75 for marked spindles for a group of 24 experts as compared with the group consensus (designed to maximize mean individual F1 score). This finding is similar to those of other studies of human-expert sleep scoring, where typical inter rater agreement is in the range of 72% (Basner, Griefahn, & Penzel, 2008; Danker-Hopfe et al., 2009; Iber, 2007).
For detecting the spindles used in this analysis, we treat each channel separately and use sliding windows of 0.25 seconds, with a 0.05 second step size. In each window, we obtain a distance from the hyperplane as described above, which serves as an index indicating the presence of a spindle when it exceeds a set threshold. The threshold was set to maximize agreement with human scoring in the training data, as measured by the area under the ROC curve. Figure 12 shows, for two example spindles, the output detection index from DDA, along with the raw waveform and spectrogram for reference from one channel of ECoG data.
Figure 12.
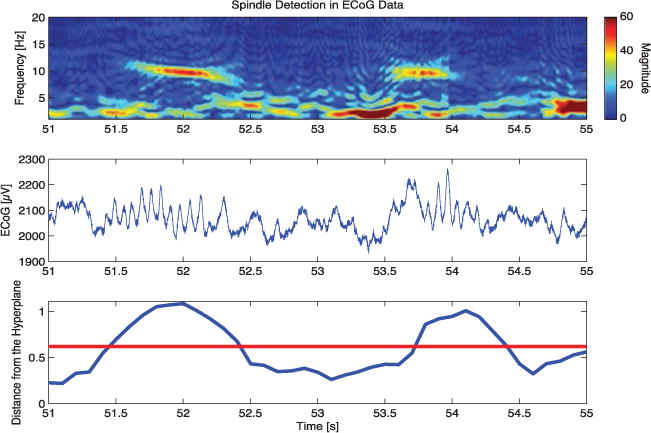
DDA spindle detection. The top panel shows a spectrogram of the ECoG data for a 4 second time period in which two sleep spindles are detected. The middle panel shows the same data in the time domain after the application of a 60 Hz notch filter for visualization. The bottom panel shows the spindle detection index from DDA; higher values above the set threshold, in red, correspond to the presence of spindles.
It is important to note that DDA is used here only to identify spindle epochs for study, and the outputs of DDA are not used in any of the subsequent analysis.
4.3 Sparse Connectivity Estimation by the SRPM Method during Sleep Spindles
After detecting the sleep spindles for each channel (see Figure 13) as described, we selected only time windows in which spindles were present in a relatively large number of channels. We selected 10 such time windows and estimated the SRPM for each of them. The average number of channels in which sleep spindles were present in the selected time windows was found to be 32.10 ± 6.78. We then applied the LMCD (Blondel et al., 2008; Brandes et al., 2008; Newman & Girvan, 2004; Newman, 2006; Reichardt & Bornholdt, 2006; Ronhovde & Nussinov, 2009; Sporns, 2010; Sun et al., 2009) on the SRPM to cluster similar electrodes together. The regularization parameter λ in the SRPM method was chosen to be large enough to minimize the intercluster connectivity after applying the LMCD, thus clustering the brain regions with the strongest connectivity. The range of values of λ used for the analysis was between 0.035 and 0.054. The SRPM method was found to be fairly robust to the regularization parameter, and small changes in the values of the regularization parameters did not significantly alter the results reported here. Figure 13 shows the clustered brain regions in the 10 time windows considered. Note that in all of the panels in Figure 13, the clusters are spatially localized, indicating spatially localized connectivity among brain regions. The average modularity across the 10 panels was found to be 0.41 defined as
| (4.4) |
where Bi j denotes the strength of connectivity (obtained from the SRPM) between brain regions i and j, si = Σj Bi j denotes the sum of connectivity strengths between brain region i and the rest of the brain regions, σi denotes the cluster to which brain region i belongs to, , and δ(σi, σj) is 1 if σi = σj and 0 otherwise (Blondel et al., 2008; Reichardt & Bornholdt, 2006). Also, the number of clusters is not the same in the panels. Observe that in different panels, different brain regions show the strongest connectivity.
Figure 13.
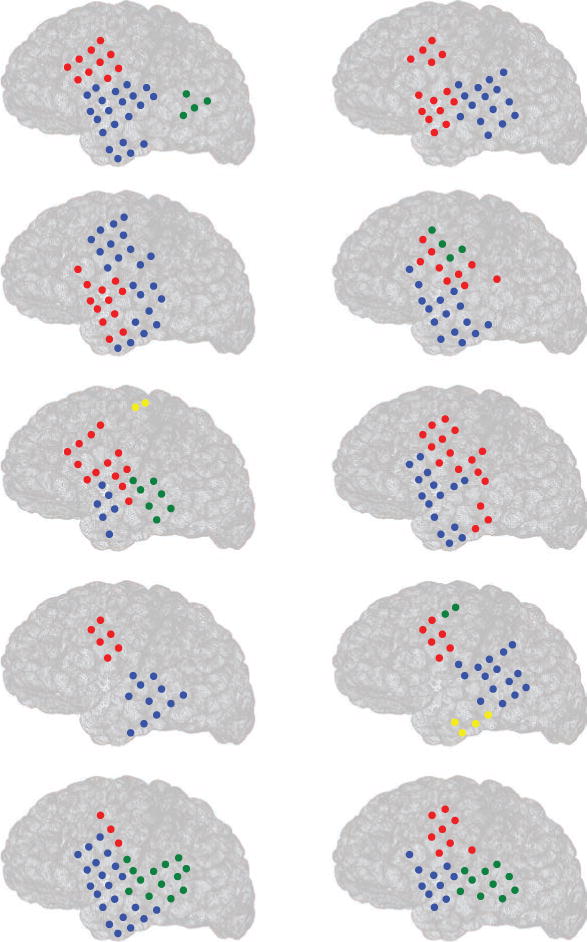
Estimating brain connectivity during sleep spindles from human ECoG data by the SRPM method in 10 epochs from a patient. Circles denote electrode locations, and clusters (of strongest activity) put together by the LMCD have the same color. For example, in the left top panel, there are three clusters of strongest activity denoted by red, blue, and green.
We next analyzed the time windows in which sleep spindles were not present in any of the 64 channels. We selected 21 such time windows in total and observed that the clustered brain regions were not as spatially localized as that of the time windows where sleep spindles were present. Figure 14 shows the clustered brain regions in four time windows in which there were no spindles. The average modularity across all the nonspindle time windows (including those not shown here) was found to be 0.49. The number of clusters is not the same in the panels, and in different panels, different brain regions show the strongest connectivity.
Figure 14.
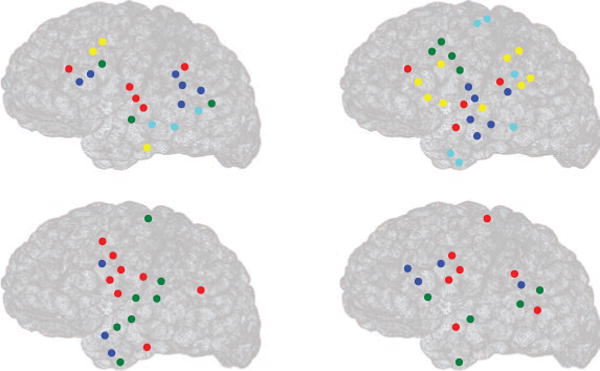
Estimating brain connectivity during absence of spindles from human ECoG data by the SRPM method in 4 epochs from a patient. Circles denote electrode locations, and clusters (of strongest activity) put together by the LMCD have the same color. For example, in the left top panel, there are five clusters of strongest activity, denoted by red, blue, green, yellow, and cyan.
In order to quantify the degree of spatial localization in spindle and nonspindle cases, we calculated the average relative surface area (ARSA) for both the cases. The ARSA for the spindle case was defined to be the average of the relative surface areas (RSAs) of all clusters across all time windows in which spindles were present and the ARSA for the nonspindle case was defined similarly. The RSA of a cluster was defined as
| (4.5) |
where SA denotes the the surface area spanned by the cluster, TA denotes the total surface area spanned by all the electrodes, and NA denotes the number of electrodes in that particular cluster. The ARSA for the spindle case was found to be 0.010 ± 0.006, and the ARSA for the nonspindle case was found to be 0.031 ± 0.009, which is significantly higher than that of the spindle case. This indicates that the brain networks during sleep spindles are highly spatially localized in comparison to during the absence of spindles.
Moreover, we applied the correlation method, and almost all brain regions showed very strong connectivity after applying the LMCD on the correlation matrix, indicating that the correlation method does not result in sparse connectivity between brain regions.
5 Discussion
The correlation method cannot find true physical connections in a network since two brain regions might show very high correlation even when the two regions are not directly connected; rather, the high correlation could be due to the strong interaction of the two regions with common input from a third region. Although research has primarily focused on the correlation method and conclusions have been drawn based on the results, those experiments need to be repeated using our proposed approach to validate the correctness of the findings. Researchers have proposed solutions to this problem and have suggested using a sparse regularized inverse covariance matrix or precision matrix (SRPM), assuming that the connectivity structure is sparse. This method yields partial correlations to measure strong direct interactions between pairs of regions while simultaneously removing the influence of the rest of the regions, thus identifying conditionally independent regions. Although the SRPM method and the inverse covariance method have been previously applied in brain research to identify functionally connected networks in the human brain and conclusions have been drawn regarding how the brain processes information, more experimental analyses are necessary to verify these claims.
Thus, we generated simple artificial networks via simulation and demonstrated through extensive analysis that the SRPM method can indeed find the true physical connection in a network. The spring-mass model and the two network topologies in the RC circuit model were used to evaluate the performance of the SRPM method. We showed successfully that as long as the connectivity structure is sparse, the SRPM method has the potential to outperform the correlation method and the inverse covariance method. For these two linear problems, SRPM recovered the exact network connectivity. This result is in contrast to the results in Sojoudi and Doyle (2014) and Sojoudi (2016), where the correlation method and the inverse covariance method gave equivalent or better performance than the SRPM method, although a different algorithm was used to solve the optimization problem in equation 1.1. Even though the algorithm in Sojoudi and Doyle (2014) and Sojoudi (2016) promoted sparsity, it did not successfully recover the connectivity pattern in a physical network.
The superior performance of the SRPM method on the artificial networks encouraged us to apply the same for analyzing the human brain. We applied the SRPM method for estimating brain connectivity during sleep spindles from human electrocorticography (ECoG) data using an 8 × 8 electrode array. For sleep spindle detection, we used DDA, a time domain classification framework based on embedding theory in nonlinear dynamics. After obtaining the SRPM during the sleep spindles, we clustered similar brain regions of strongest activity together using the LMCD and found spatially localized brain networks during spindles. Moreover, analyzing the time windows in which sleep spindles were not present, we found that the clusters were not as compact compared with that of the time windows where sleep spindles were present. These findings suggest that regional interactions in the cortex are stronger during sleep spindles. Moreover, this gives us insight into local information processing during spindle activity in brain. During sleep spindles, the connectivity pattern in the brain was transiently synchronized, thus providing evidence for globally organized coherence patterns. These findings on spatial localization during spindles provide us with new insights into how sleep spindles have a major role in learning, memory consolidation, and neuronal plasticity.
Furthermore, the clusters shown in Figure 13 obtained during sleep spindles were large, and in 7 of the 10 epochs, there was a cluster that included regions of both the temporal and prefrontal cortices. This suggests that some of the connectivity underlying the clustering is from association fiber bundles that project between the major cortical lobes. It is worth noting that the comparison we are making is between spindle epochs and all other epochs during this recording of stage 2 sleep. These nonspindle epochs, then, can be quite heterogeneous, with various other phenomena such as K-complexes or other slow oscillations occurring at different times throughout. As such, we expect the connectivity pattern to change accordingly throughout these periods. By including a number of different nonspindle epochs in our analysis, then, we establish a broad baseline for connectivity during stage 2 sleep that we can then compare to the specific connectivity patterns observed during spindles. Although the data were from a patient with epilepsy, the recordings we analyzed were during long seizure-free periods. Nonetheless, there is concern that the cortical sleep states may not be normal, and further work is needed to confirm our results in healthy control subjects. Another issue is that the ECoG recordings that we analyzed were from a single patient, and hence there is a need to test our methods in a large number of epileptic patients. It is important to note that the inputs from the thalamus also act as common inputs to the cortex and constitute latent (unobserved) variables in our analysis. In order to estimate the connectivity by modeling the latent inputs, we have applied the sparse-plus-latent regularized precision matrix (SLRPM) method (Chandrasekaran, Parrilo, & Willsky, 2012) on the same ECoG recordings shown here and found very similar results to the SRPM method, which is surprising. Hence, additional work is needed to test the SRPM and SLRPM methods on more subjects to find conditions on the applicability of these methods.
Recently, Brunton, Johnson, Ojemann, and Kutz (2016) used the dynamic mode decomposition algorithm and clustering methods to characterize brain networks during sleep spindles. Similar to our results, they found that the brain networks during sleep spindles are spatially localized. However, no results were reported in the absence of sleep spindles. Other researchers (Andrillon et al., 2011) also have found similar results. Recently Muller et al. (2016), by analyzing the phase coherence, found that sleep spindles are associated with traveling waves, which provides further support for our findings.
The analytical and experimental advances made in this letter on sleep, learning, and information processing in the brain suggest a possible resolution to one of the greatest mysteries in biology: the nature and function of sleep. The experimental results so far are incomplete and tentative, but they should lead us toward further advances that will widen our understanding of sleep. Additional analysis is needed to test these conclusions, and these are reserved for future research.
Finally, the same SRPM method we have used to analyze ECoG recordings could also be applied to single unit recordings, local field potentials, and fMRI data.
Acknowledgments
This research was supported by the Howard Hughes Medical Institute, the NIH under grant R01EB009282, and the Office of Naval Research under grant N000141310672. We also like to thank Dr. David J. C. MacKay for interesting discussions on the spring-mass model.
Contributor Information
Anup Das, Department of Electrical and Computer Engineering, University of California, San Diego, La Jolla, CA 92093, U.S.A.; Computational Neurobiology Laboratory, Salk Institute for Biological Studies, La Jolla, CA 92037, U.S.A.
Aaron L. Sampson, Computational Neurobiology Laboratory, Salk Institute for Biological Studies, La Jolla, CA 92037, U.S.A.
Claudia Lainscsek, Computational Neurobiology Laboratory, Salk Institute for Biological Studies, La Jolla, CA 92037, U.S.A..
Lyle Muller, Computational Neurobiology Laboratory, Salk Institute for Biological Studies, La Jolla, CA 92037, U.S.A..
Wutu Lin, Computational Neurobiology Laboratory, Salk Institute for Biological Studies, La Jolla, CA 92037, U.S.A..
John C. Doyle, Division of Engineering and Applied Science, California Institute of Technology, Pasadena, CA 91125, U.S.A.
Sydney S. Cash, Cortical Physiology Laboratory, Massachusetts General Hospital, Boston, MA 02114, U.S.A.
Eric Halgren, Department of Radiology, University of California, San Diego, La Jolla, CA 92093, U.S.A..
Terrence J. Sejnowski, Division of Biological Sciences and Institute of Neural Computation, University of California, San Diego, La Jolla, CA 92093, U.S.A. Howard Hughes Medical Institute, Salk Institute for Biological Studies, La Jolla, CA 92037, U.S.A.
References
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele E, Calhoun VD. Tracking whole-brain connectivity dynamics in the resting state. Cerebral Cortex. 2012;24:663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A, Li Y, Wang Y, Wu J, Gao S, Bukhari L, Lowe MJ. Antidepressant effect on connectivity of the mood-regulating circuit: An FMRI study. Neuropsychopharmacology. 2005;30(7):1334–1344. doi: 10.1038/sj.npp.1300725. [DOI] [PubMed] [Google Scholar]
- Andrade KC, Spoormaker VI, Dresler M, Wehrle R, Holsboer F, Samann PG, Czisch M. Sleep spindles and hippocampal functional connectivity in human NREM sleep. Journal of Neuroscience. 2011;31(28):10331–10339. doi: 10.1523/JNEUROSCI.5660-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrillon T, Nir Y, Staba RJ, Ferrarelli F, Cirelli C, Tononi G, Fried I. Sleep spindles in humans: Insights from intracranial EEG and unit recordings. Journal of Neuroscience. 2011;31(49):17821–17834. doi: 10.1523/JNEUROSCI.2604-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee O, El Ghaoui L, d’Aspremont A. Model selection through sparse maximum likelihood estimation for multivariate gaussian or binary data. Journal of Machine Learning Research. 2008;9:485–516. [Google Scholar]
- Basner M, Griefahn B, Penzel T. Inter-rater agreement in sleep stage classification between centers with different backgrounds. Somnologie-Schlafforschung und Schlafmedizin. 2008;12(1):75–84. [Google Scholar]
- Bassett DS, Greenfield DL, Lindenberg AM, Weinberger DR, Moore SW, Bullmore ET. Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Computational Biology. 2010;6(4):e1000748. doi: 10.1371/journal.pcbi.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. Dynamic reconfiguration of human brain networks during learning. Proceedings of the National Academy of Sciences USA. 2011;108(18):7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magnetic Resonance in Medicine. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Blondel VD, Guillaume JL, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment. 2008;2008(10):10008. [Google Scholar]
- Brandes U, Delling D, Gaertler M, Gorke R, Hoefer M, Nikoloski Z, Wagner D. On modularity clustering. IEEE Transactions on Knowledge and Data Engineering. 2008;20(2):172–188. [Google Scholar]
- Brunton BW, Johnson LA, Ojemann JG, Kutz JN. Extracting spatialtemporal coherent patterns in large-scale neural recordings using dynamic mode decomposition. Journal of Neuroscience Methods. 2016;258:1–15. doi: 10.1016/j.jneumeth.2015.10.010. [DOI] [PubMed] [Google Scholar]
- Bullmore ET, Bassett DS. Brain graphs: Graphical models of the human brain connectome. Annual Review of Clinical Psychology. 2011;7(1):113–140. doi: 10.1146/annurev-clinpsy-040510-143934. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran V, Parrilo PA, Willsky AS. Latent variable graphical model selection via convex optimization. Annals of Statistics. 2012;40(4):1935–1967. [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83(1):238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D, Haughton VM, Arfanakis K, Wendt GJ, Turski PA, Moritz CH, Meyerand ME. Mapping functionally related regions of brain with functional connectivity MR imaging. American Journal of Neuroradiology. 2000;21(9):1636–1644. [PMC free article] [PubMed] [Google Scholar]
- Danker-Hopfe H, Anderer P, Zeitlhofer J, Boeck M, Dorn H, Gruber G, Dorffner G. Interrater reliability for sleep scoring according to the Rechtschaffen & Kales and the new AASM standard. Journal of Sleep Research. 2009;18(1):74–84. doi: 10.1111/j.1365-2869.2008.00700.x. [DOI] [PubMed] [Google Scholar]
- Dempster AP. Covariance selection. Biometrics. 1972;28(1):157–175. [Google Scholar]
- Ferrarelli F, Huber R, Peterson MJ, Massimini M, Murphy M, Riedner BA, Tononi G. Reduced sleep spindle activity in schizophrenia patients. American Journal of Psychiatry. 2007;164(3):483–492. doi: 10.1176/ajp.2007.164.3.483. [DOI] [PubMed] [Google Scholar]
- Fogel SM, Nader R, Cote KA, Smith CT. Sleep spindles and learning potential. Behavioral Neuroscience. 2007;121(1):1–10. doi: 10.1037/0735-7044.121.1.1. [DOI] [PubMed] [Google Scholar]
- Fogel SM, Smith CT. Learning-dependent changes in sleep spindles and stage 2 sleep. Journal of Sleep Research. 2006;15(3):250–255. doi: 10.1111/j.1365-2869.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Spontaneous neuronal activity distinguishes human dorsal and ventral attention systems. Proceedings of the National Academy of Sciences USA. 2006;26(103):10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proceedings of the National Academy of Sciences USA. 2005;102(27):9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gais S, Born J. Declarative memory consolidation: Mechanisms acting during human sleep. Learning and Memory. 2004;11(6):679–685. doi: 10.1101/lm.80504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gais S, Mölle M, Helms K, Born J. Learning-dependent increases in sleep spindle density. Journal of Neuroscience. 2002;22(15):6830–6834. doi: 10.1523/JNEUROSCI.22-15-06830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Van Essen DC. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V. Functional connectivity in the resting brain: A network analysis of the default mode hypothesis. Proceedings of the National Academy of Sciences USA. 2003;1(100):253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M, Peterson BS, Skudlarski P, Gatenby JC, Gore JC. Detection of functional connectivity using temporal correlations in MR images. Human Brain Mapping. 2002;15(4):247–262. doi: 10.1002/hbm.10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CJ, Sustik MA, Dhillon IS, Ravikumar P. Sparse inverse covariance matrix estimation using quadratic approximation. In: Shawe-Taylor J, Zemel RS, Bartlett PL, Pereira F, Weinberger KQ, editors. Advances in neural information processing systems. Red Hook, NY: Curran; 2011. pp. 2330–2338. [Google Scholar]
- Hsieh CJ, Sustik MA, Dhillon IS, Ravikumar P, Poldrack R. BIG & QUIC: Sparse inverse covariance estimation for a million variables. In: Burges CJC, Bottou L, Welling M, Ghahramani Z, Weinberger KO, editors. Advances in neural information processing systems. Red Hook, NY: Curran; 2013. pp. 3165–3173. [Google Scholar]
- Iber C. The AASM manual for the scoring of sleep and associated events: rules, terminology and technical specifications. Darien: American Academy of Sleep Medicine; 2007. [Google Scholar]
- Kohavi R. Proceedings of the 14th International Joint Conference on Artificial Intelligence. Vol. 2. San Francisco: Morgan Kaufmann; 1995. A study of cross-validation and bootstrap for accuracy estimation and model selection; pp. 1137–1143. [Google Scholar]
- Kremliovsky MN, Kadtke JB. Proceedings: Applied nonlinear dynamics and stochastic systems near the millennium. Vol. 411. College Par, MD: AIP Publishing; 1997. Using delay differential equations as dynamical classifiers; pp. 57–62. [Google Scholar]
- Lainscsek C, Hernandez ME, Weyhenmeyer J, Sejnowski TJ, Poizner H. Non-linear dynamical analysis of EEG time series distinguishes patients with Parkinsons disease from healthy individuals. Frontiers in Neurology. 2013:4. doi: 10.3389/fneur.2013.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainscsek C, Sejnowski TJ. Delay differential analysis of time series. Neural Computation. 2015;27:615–627. doi: 10.1162/NECO_a_00706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lainscsek C, Weyhenmeyer J, Hernandez ME, Poizner H, Sejnowski TJ. Non-linear dynamical classification of short time series of the Rössler system in high noise regimes. Frontiers in Neurology. 2013;4:182. doi: 10.3389/fneur.2013.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Prior LJ, Zempel JM, Nolan TS, Prior FW, Snyder AZ, Raichle ME. Cortical network functional connectivity in the descent to sleep. Proceedings of the National Academy of Sciences USA. 2009;1(106):4489–4494. doi: 10.1073/pnas.0900924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufs H, Krakow K, Sterzer P, Eger E, Beyerle A, Salek-Haddadi A, Kleinschmidt A. Electroencephalographic signatures of attentional and cognitive default modes in spontaneous brain activity fluctuations at rest. Proceedings of the National Academy of Sciences USA. 2003;100(19):11053–11058. doi: 10.1073/pnas.1831638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauritzen SL. Graphical models. New York: Oxford University Press; 1996. [Google Scholar]
- MacKay DJC. Information theory, inference and learning algorithms. Cambridge: Cambridge University Press; 2002. [Google Scholar]
- Marjanovic G, Hero AO. ℓ0 sparse inverse covariance estimation. IEEE Transactions on Signal Processing. 2015;63(12):3218–3231. [Google Scholar]
- Marjanovic G, Solo V. On ℓq optimization and sparse inverse covariance selection. IEEE Transactions on Signal Processing. 2014;62(7):1644–1654. [Google Scholar]
- Marshall L, Born J. The contribution of sleep to hippocampus-dependent memory consolidation. Trends in Cognitive Sciences. 2007;11(10):442–450. doi: 10.1016/j.tics.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Martin N, Lafortune M, Godbout J, Barakat M, Robillard R, Poirier G, Carrier J. Topography of age-related changes in sleep spindles. Neurobiology of Aging. 2013;34(2):468–476. doi: 10.1016/j.neurobiolaging.2012.05.020. [DOI] [PubMed] [Google Scholar]
- McIntosh AR, Rajah MN, Lobaugh NJ. Functional connectivity of the medial temporal lobe relates to learning and awareness. Journal of Neuroscience. 2003;23(16):6520–6528. doi: 10.1523/JNEUROSCI.23-16-06520.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Koll A, Bussmann B, Schmidt C, Neuschwander D. Walking through a maze alters the architecture of sleep. Perceptual and Motor Skills. 1999;88(3):1141–1159. doi: 10.2466/pms.1999.88.3c.1141. [DOI] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Bullmore ET. Modular and hierarchically modular organization of brain networks. Frontiers in Neuroscience. 2010;4:200. doi: 10.3389/fnins.2010.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletics E, Molnárka G. Implicit extension of Taylor series method with numerical derivatives for initial value problems. Computers and Mathematics with Applications. 2005;50(7):1167–1177. [Google Scholar]
- Monti RP, Hellyer P, Sharp D, Leech R, Anagnostopoulos C, Montana G. Estimating time-varying brain connectivity networks from functional MRI time series. NeuroImage. 2014;103:427–443. doi: 10.1016/j.neuroimage.2014.07.033. [DOI] [PubMed] [Google Scholar]
- Muller L, Piantoni S, Koller D, Cash SS, Halgren E, Sejnowski TJ. Rotating waves during human sleep spindles organize global patterns of activity that repeat precisely through the night. eLife. 2016;5:e17267. doi: 10.7554/eLife.17267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Modularity and community structure in networks. Proceedings of the National Academy of Sciences USA. 2006;103(23):8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Physical Review E. 2004;69(2):026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- Oztoprak F, Nocedal J, Rennie S, Olsen PA. Newton-like methods for sparse inverse covariance estimation. In: Pereira F, Burges CJC, Bottou L, Weinberger KQ, editors. Advances in neural information processing systems. Red Hook, NY: Curran; 2012. pp. 764–772. [Google Scholar]
- Petit D, Gagnon JF, Fantini ML, Ferini-Strambi L, Montplaisir J. Sleep and quantitative EEG in neurodegenerative disorders. Journal of Psychosomatic Research. 2004;56(5):487–496. doi: 10.1016/j.jpsychores.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Reichardt J, Bornholdt S. Statistical mechanics of community detection. Physical Review E. 2006;74(1):016110. doi: 10.1103/PhysRevE.74.016110. [DOI] [PubMed] [Google Scholar]
- Ronhovde P, Nussinov Z. Multiresolution community detection for megascale networks by information-based replica correlations. Physical Review E. 2009;80(1):016109. doi: 10.1103/PhysRevE.80.016109. [DOI] [PubMed] [Google Scholar]
- Rosa MJ, Portugal L, Hahn T, Fallgatter AJ, Garrido MI, Taylor JS, Mourao-Miranda J. Sparse network-based models for patient classification using fMRI. NeuroImage. 2015;105:493–506. doi: 10.1016/j.neuroimage.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman AJ, Bickel PJ, Levina E, Zhu J. Sparse permutation invariant covariance estimation. Electronic Journal of Statistics. 2008;2:494–515. [Google Scholar]
- Rubinov M, Sporns O. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage. 2010;52(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. NeuroImage. 2011;56(4):2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]
- Ryali S, Chen T, Supekar K, Menon V. Estimation of functional connectivity in fMRI data using stability selection-based sparse partial correlation with elastic net penalty. NeuroImage. 2012;59(4):3852–3861. doi: 10.1016/j.neuroimage.2011.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson AL, Lainscsek C, Cash SS, Halgren E, Sejnowski TJ. Society for Neuroscience. 2015. Nonlinear dynamical sleep spindle detection using delay differential analysis. abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schabus M, Gruber G, Parapatics S, Sauter C, Klösch G, Anderer P, Zeitlhofer J. Sleep spindles and their significance for declarative memory consolidation. Sleep. 2004;27(8):1479–1485. doi: 10.1093/sleep/27.7.1479. [DOI] [PubMed] [Google Scholar]
- Scheinberg K, Ma S, Goldfarb D. Sparse inverse covariance selection via alternating linearization methods. In: Lafferty JD, Williams CKI, Shawe-Taylor J, Zemel RS, Culotta A, editors. Advances in neural information processing systems. Red Hook, NY: Curran; 2010. pp. 2101–2109. [Google Scholar]
- Sejnowski TJ. Neural networks: Sleep and memory. Current Biology. 1995;5(8):832–834. doi: 10.1016/s0960-9822(95)00165-5. [DOI] [PubMed] [Google Scholar]
- Sejnowski TJ, Destexhe A. Why do we sleep? Brain Research. 2000;886(1–2):208–223. doi: 10.1016/s0006-8993(00)03007-9. [DOI] [PubMed] [Google Scholar]
- Siegle GJ, Thompson W, Carter CS, Steinhauer SR, Thase ME. Increased amygdala and decreased dorsolateral prefrontal BOLD responses in unipolar depression: Related and independent features. Biological Psychiatry. 2007;61(2):198–209. doi: 10.1016/j.biopsych.2006.05.048. [DOI] [PubMed] [Google Scholar]
- Smith C, Macneill C. Impaired motor memory for a pursuit rotor task following stage 2 sleep loss in college students. Journal of Sleep Research. 1994;3(4):206–213. doi: 10.1111/j.1365-2869.1994.tb00133.x. [DOI] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, Woolrich MW. Network modelling methods for FMRI. NeuroImage. 2011;54(2):875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Sojoudi S. Equivalence of graphical lasso and thresholding for sparse graphs. Journal of Machine Learning Research. 2016;17(115):1–21. [Google Scholar]
- Sojoudi S, Doyle J. Proceedings of the 52nd Annual Allerton Conference on Communication, Control, and Computing. Piscataway, NJ: IEEE; 2014. Study of the brain functional network using synthetic data; pp. 350–357. [Google Scholar]
- Spoormaker VI, Schröter MS, Gleiser PM, Andrade KC, Dresler M, Wehrle R, Czisch M. Development of a large-scale functional brain network during human non-rapid eye movement sleep. Journal of Neuroscience. 2010;30(34):11379–11387. doi: 10.1523/JNEUROSCI.2015-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O. Brain connectivity toolbox. 2010 https://sites.google.com/site/bctnet/visualization.
- Sporns O. The nonrandom brain: Efficiency, economy, and complex dynamics. Frontiers in Computational Neuroscience. 2011;5(5) doi: 10.3389/fncom.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steriade M, McCormick DA, Sejnowski TJ. Thalamocortical oscillations in the sleeping and aroused brain. Science. 1993;262(5134):679–685. doi: 10.1126/science.8235588. [DOI] [PubMed] [Google Scholar]
- Stickgold R, Walker MP. Memory consolidation and reconsolidation: What is the role of sleep? Trends in Neurosciences. 2005;28(8):408–415. doi: 10.1016/j.tins.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Stickgold R, Walker MP. Sleep-dependent memory consolidation and reconsolidation. Sleep Medicine. 2007;8(4):331–343. doi: 10.1016/j.sleep.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Danila B, Josić K, Bassler KE. Improved community structure detection using a modified fine-tuning strategy. Europhysics Letters. 2009;86(2):28004. [Google Scholar]
- Takens F. Detecting strange attractors in turbulence. Lecture Notes in Mathematics. 1981;898:366–381. [Google Scholar]
- Uddin LQ, Kelly AC, Biswal BB, Castellanos FX, Milham MP. Functional connectivity of default mode network components: Correlation, anti-correlation, and causality. Human Brain Mapping. 2009;30(2):625–637. doi: 10.1002/hbm.20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux G, Gramfort A, Jean-Baptiste P, Thirion B. Brain covariance selection: Better individual functional connectivity models using population prior. In: Lafferty JD, Williams CKI, Shawe-Taylor J, Zemel RS, Weinberger KQ, editors. Advances in neural information processing systems. Red Hook, NJ: Curran; 2010. pp. 2334–2342. [Google Scholar]
- Vertes PE, Alexander-Bloch AF, Gogtay N, Giedd JN, Rapoport JL, Bullmore ET. Simple models of human brain functional networks. Proceedings of the National Academy of Sciences. 2012;109(15):5868–5873. doi: 10.1073/pnas.1111738109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volterra V. Sopra le funzioni che dipendono da altre funzioni. Atti della Reale Accademia dei Lincei. 1887;3:97–105. [Google Scholar]
- Volterra V. Theory of functionals and of integral and integro-differential equations. Mineola, NY: Dover Publications; 1959. [Google Scholar]
- Walker MP, Brakefield T, Morgan A, Hobson JA, Stickgold R. Practice with sleep makes perfect: Sleep-dependent motor skill learning. Neuron. 2002;35(1):205–211. doi: 10.1016/s0896-6273(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Wang Y, Kang J, Kemmer PB, Guo Y. An efficient and reliable statistical method for estimating functional connectivity in large scale brain networks using partial correlation. Frontiers in Neuroscience. 2016;10:123. doi: 10.3389/fnins.2016.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Wendt SL, Welinder P, Munk EGS, Carrillo O, Sorensen HBD, Mignot E. Sleep-spindle detection: Crowdsourcing and evaluating performance of experts, non-experts and automated methods. Nature Methods. 2014;11:385–392. doi: 10.1038/nmeth.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker J. Graphical models in applied multivariate statistics. Hoboken, NJ: Wiley; 1990. [Google Scholar]
- Xiong J, Parsons LM, Gao JH, Fox PT. Interregional connectivity to primary motor cortex revealed using MRI resting state images. Human Brain Mapping. 1999;8(2–3):151–156. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<151::AID-HBM13>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko D, Josić K, Ecker AS, Froudarakis E, Cotton RJ, Tolias AS. Improved estimation and interpretation of correlations in neural circuits. PLoS Computational Biology. 2015;11(3):e1004083. doi: 10.1371/journal.pcbi.1004083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Lin Y. Model selection and estimation in the gaussian graphical model. Biometrika. 2007;94(1):19–35. [Google Scholar]
- Zhou D, Thompson WK, Siegle G. MATLAB toolbox for functional connectivity. NeuroImage. 2009;47(4):1590–1607. doi: 10.1016/j.neuroimage.2009.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, Milham MP. Network centrality in the human functional connectome. Cerebral Cortex. 2012;22(8):1862–1875. doi: 10.1093/cercor/bhr269. [DOI] [PubMed] [Google Scholar]


