Abstract.
Computational modeling of visual attention is an active area of research. These models have been successfully employed in applications such as robotics. However, most computational models of visual attention are developed in the context of natural scenes, and their role with medical images is not well investigated. As radiologists interpret a large number of clinical images in a limited time, an efficient strategy to deploy their visual attention is necessary. Visual saliency maps, highlighting image regions that differ dramatically from their surroundings, are expected to be predictive of where radiologists fixate their gaze. We compared 16 state-of-art saliency models over three medical imaging modalities. The estimated saliency maps were evaluated against radiologists’ eye movements. The results show that the models achieved competitive accuracy using three metrics, but the rank order of the models varied significantly across the three modalities. Moreover, the model ranks on the medical images were all considerably different from the model ranks on the benchmark MIT300 dataset of natural images. Thus, modality-specific tuning of saliency models is necessary to make them valuable for applications in fields such as medical image compression and radiology education.
Keywords: visual attention, saliency, eye tracking, bottom-up
1. Introduction
Visual attention is the cognitive process of selectively attending to a region or object while ignoring the surrounding stimuli.1 This mechanism guides human visual systems to important parts of the scene to gather more detailed information, and there are two components for the guidance: (1) “bottom-up,” image-driven saliency and (2) “top-down,” task-driven saliency.2 An item or location that differs significantly from its surroundings is “salient,” and more salient regions tend to attract more attention. Visual saliency maps are topographic maps that represent the conspicuity of objects and locations.3 If the regions of an image identified as salient in a saliency map coincide with the regions on which an observer’s eyes fixate, then the saliency map is considered to be an accurate prediction of the observer’s deployment of attention.
Several computational models of human’s visual attention have been developed and applied in fields such as computer vision,4–6 robotics,7,8 and computer graphics.9–11 Borji and Itti2 suggested that state-of-art saliency models, mainly those capturing bottom-up attention, can be grouped into eight categories: (1) cognitive models (CM) that are based on psychological and/or neurophysiological concepts; (2) Bayesian models (BM) that combine sensory information (e.g., object features) with prior knowledge (e.g., context) using Bayes’ rule; (3) decision theoretic models (DM) that describe the states of the surrounding environment with respect to the decision task; (4) information theoretic models (IM) that compute localized saliency to maximize information sampled from the selected parts; (5) graphical models (GM) that model the conditional independence structure of hidden variables influencing the generation of eye movements as a time series; (6) spectral analysis models (SM) that estimate saliency in the frequency domain; (7) pattern classification models (PM) that learn saliency models using training eye-movement data; (8) other models (OM) that incorporate different sources of information than the other seven.
Computational models of visual attention are typically evaluated in terms of their ability to predict human fixations. There are several publicly available image datasets (e.g., Bruce and Tsotsos,12 Kootstra and Schomaker,13 and Judd et al.14), but the most comprehensive and objective source of saliency model performances is the MIT saliency benchmark.15 It contains the MIT300 dataset of 300 natural images with eye-tracking data from 39 observers,16 and the CAT2000 dataset of 2000 images from 20 different categories with eye-tracking data from 24 observers.17 Using testing datasets where human eye movements are not public prevents models specifically training on, and fitting to, the particular testing datasets for good performance. An up-to-date ranked list of models, measured using multiple figures of merit (FOM), is maintained on the benchmark website.
Most saliency models are developed and validated in the context of natural scenes. Only a very limited number of studies have investigated the role of saliency models on medical images. Matsumoto et al.18 used the graph-based visual saliency (GBVS) model19 to predict where neurologists look when they view brain CT images depicting cerebrovascular accidents. Both neurologists and controls tended to gaze at high-salience areas of the saliency maps, but neurologists gazed more often at inconspicuous but clinically important areas outside the salient areas. Jampani et al.20 reported that GBVS, among three tested saliency models (GBVS, Itti-Koch,21 and SR22), performed the best for chest x-ray images with diffuse lesions, while SR performed the best for retinal images with hard exudates. Alzubaidi et al.23 used 13 different types of features that may predict which regions catch the eye of experienced radiologists when diagnosing chest x-ray images. The results showed that four features (localized edge orientation histograms, Haar wavelets, Gabor filters, and steerable filters) were particularly useful. In a prior study,24 we focused on volumetric medical imaging data (stack of chest CT slices with lung nodules), and used two three-dimensional (3-D) saliency models [3-D dynamic saliency, a stack of two-dimensional (2-D)-based GBVS] to assess radiologists’ different search strategies. One of the key insights from these studies is that bottom-up saliency plays an important role in examining medical images, although incorporating some top-down factors such as lung segmentation on chest CT images would likely improve the accuracy of saliency models in predicting radiologists’ gaze positions. However, no study has attempted to comprehensively evaluate the performance of saliency models on medical images or developed a mechanism for choosing appropriate models for this application domain. Saliency models that are informative in natural scenes may not be equally informative in the context of medical images. For example, as many conventional medical images (e.g., mammograms and radiographs) are present as graylevel images, would lack of color information significantly degrade the performance of saliency models? Given that different medical imaging techniques produce images with distinct characteristics (e.g., an MRI image has significantly higher resolution than an ultrasound image25), would certain saliency models perform consistently well across the modalities? These questions remain to be answered.
The purpose of medical imaging is to enable human observers (e.g., radiologists) to locate and evaluate medically relevant abnormalities (e.g., nodules in a chest radiograph) amidst normal anatomy and physiology (e.g., normal lung tissues). It is reasonable to suppose that a modality will be more effective if it makes medically relevant abnormalities more salient. One of the challenges in reading medical images is that significant image findings are sometimes not salient—they may be too subtle for the human eye or infiltrative with ill-defined margins. Hence, having a saliency model that will accurately predict the intrinsic saliency of targets as displayed by the modality would be beneficial for evaluating new technologies.24 For example, better medical image compression algorithms could decrease file sizes while maintaining image quality.26–28 Similarly, radiologists must interpret a large number of clinical images in a limited time, thus an efficient strategy to deploy their visual attention is necessary. A wide range of eye-tracking studies have been conducted to examine radiologists’ attention strategies (e.g., lung cancer in radiographs,29,30 breast masses in mammograms,31,32 and lesions in brain CT18). In addition to qualitative descriptions of the strategies reported in these studies, it would also be valuable to examine saliency maps of medical images to assess the degree to which they capture radiologists’ visual attention. This is especially interesting considering that radiologists are likely to adjust their visual search strategies according to specific clinical tasks being performed (e.g., Refs. 18, 33, and 34). For example, Matsumoto et al.18 suggested that radiologists tend to use a “look-detect-scan” strategy when reading mammograms for breast lesions, but use a “scan-look-detect” strategy when reading brain CT images. Even with the same chest radiograph, radiologists’ eye movement patterns are significantly different when they are asked to search for pulmonary nodules versus to detect bone fractures.33 Hence, our study of saliency models would help identify efficient strategies of attention deployment that may improve diagnostic accuracy and training of early-career radiologists.
The objective of this comparative study was to evaluate saliency models in medical imaging relative to natural scenes. Sixteen representative saliency models, covering the eight model categories, were ranked by how well the saliency maps agreed with radiologists’ eye positions. Limited top-down factors (i.e., organ segmentation) were added into the models to incorporate task-relevant effects on saliency estimation. An eye-tracking study, approved by the Institutional Review Board (IRB) at the University of Texas at MD Anderson Cancer Center, was conducted to collect radiologists’ eye movements when reading 2-D medical images. The ranking of the models on the MIT300 benchmark served as the reference.
2. Materials and Methods
2.1. Experimental Data
The experimental data utilized in this study were acquired from an existing dataset35 that we collected for a prior study on radiology reporting. In that study, we monitored the eye position of radiologists as they read 2-D medical images. The study protocol received IRB approval from MD Anderson. Written informed consent was obtained from all the participating radiologists for performing the image interpretation. The radiological images used in this study were deidentified in accordance with the Health Insurance Portability and Accountability Act.
Ten board-certified radiologists (seven radiology faculty, three radiology fellows; four females, six males; mean years of experience = 6.2 years, range = 1 to 13 years) were recruited, of which four radiologists (three faculty, one fellow; two females, two males) were excluded from the present analysis due to incomplete eye-tracking data. As a result, eye-movement data from six radiologists (four faculty and two fellows; two females, four males) were included in this analysis. All the participants had normal or corrected-to-normal vision. The image dataset consisted of four imaging modalities: chest computed tomography (CT), chest x-ray images (CXR), whole-body positron emission tomography (PET), and mammograms. The modalities were chosen as representative of clinical images that radiologists encounter in daily practice. However, data with mammograms were only collected with three radiologists in order to validate the experimental design, and thus excluded from the present analysis. As a result, there were a total of three imaging modalities (i.e., CT, CXR, and PET) and 30 experimental images, 10 images per modality. For each imaging modality, the radiologists were instructed to perform a particular set of two tasks (Table 1): (1) for PET images, they were instructed to detect the most significant metabolic abnormality and the degree of distention of the bladder; (2) for CT images, they were instructed to detect the largest liver metastasis and comment on the abdominal aorta; (3) For CXR images, they were instructed to detect the largest lung lesion, and comment on the heart size. The radiologists were allowed unlimited time to read the images, and they could choose when to move on to a new case. The user interface of the experiment was implemented in C#, using Tobii software development kit (Tobii AB, Stockholm, Sweden). There were three reading sessions, one session per imaging modality. The reading order of the three sets of images was randomized across the participants. At the beginning of the first reading session, the participants read 10 training images (mix of the three imaging modalities, different from the 30 experiment images) to familiarize themselves with the user-interface and the tasks to be performed.
Table 1.
Summary of three imaging modalities, and radiologists’ corresponding tasks with the modalities.
| Modality | Resolution | Radiologists’ tasks |
|---|---|---|
| CT | Identify and comment on the largest liver metastasis and then comment on the abdominal aorta | |
| CXR | Identify and comment on the largest lung lesion, and then comment on the heart size | |
| PET | Identify and comment on the most significant metabolic abnormality, and then comment on the degree of bladder distention |
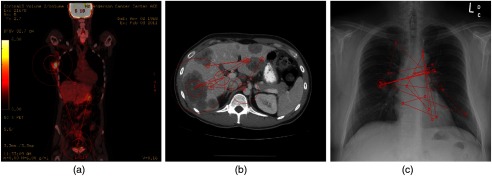
A Tobii T60 XL eye-tracker was used to sample the and positions of the eye(s) at 60 Hz. The eye tracker was integrated into a 24-in. widescreen monitor with a resolution of for displaying the images. To avoid additional factors that could affect performance in the study, the same fixed display parameters were used for all radiologists, and radiologists were not allowed to change the settings. The radiologists were seated away from the monitor, and they were able to move freely and naturally, within a limited range, in front of the monitor. No chin rest was used to immobilize subjects’ heads, as the eye tracker had large head movement tolerance, and head movement compensation algorithms were applied when calculating position of observers’ gaze. All the experiments were conducted in a darkened room with consistent ambient lighting roughly 15 lux ( lumen per square meter). Conventional methods as used in many prior eye-tracking studies on medical images36–39 were employed to analyze the eye-tracking data, i.e., fixations were formed by grouping - and -coordinates of the raw eye positions using an area-based, spatial distance filter that corresponded to a radius threshold of 1-deg visual angle.40 Example gaze maps generated from one of the radiologist faculty’s eye-tracking data are shown in Fig. 1. We have observed that there was some interobserver variability in the six participants’ search strategies, especially when comparing the four faculty’s eye movements with those of the two fellows. However, given the limited number of radiologists in each group, we could not observe any consistent trends across modalities or tasks. In Sec. 3.5, we briefly explored if considering the difference in the level of expertise and/or experience would alter any of the general trends of the study.
Fig. 1.
Example gaze maps generated from one of the radiologist’ eye-position recorded when reading (a) PET, (b) CT, and (c) CXR images. The red circles are fixations, with the radius representing the corresponding dwell time of the fixation. Connecting red lines are saccades between two consecutive fixations.
2.2. Saliency Maps
2.2.1. Bottom-up saliency models
We considered 16 state-of-art saliency models that cover the eight categories of models.2 This section lists the model abbreviations used in the rest of the paper, and briefly summarizes the main idea of each model. Each bullet point of the section describes one model and is organized according to the following: full model name, model abbreviation in parenthesis, acronym of the model category in braces, literature reference in which the model was originally proposed in square brackets, and brief summary of the model. The models are introduced in alphabetical order of their abbreviations. We acknowledge that one limitation of this study is that there are many OMs that we did not test which may also perform well. Since different models output saliency maps at different resolutions, we resized the computed saliency maps, using nearest neighbor interpolation, to the size of the original images.
-
•
Attention based on information maximization (AIM) {IM}:12 This model calculates saliency using Shannon’s self-information measure.41 Saliency of a local image region is the information that region conveys relative to its surroundings, where the information of a visual feature is inversely proportional to the log likelihood of observing the feature.
-
•
Adaptive whitening saliency model {OM}:42 This model adopts the variability in local energy for predicting saliency. Given an image in Lab color space (i.e., lightness channel plus two-color channels), a bank of multioriented multiresolution Gabor filters is used to represent the luminance channel, and principal component analysis over a set of multiscale low-level features is applied to decorrelate the color components. In this way, a local measure of variability, using Hoteling’s T2 statistic, is extracted.
-
•
Context-aware saliency (ContAware) {OM}:43 This model follows four principles of human visual attention to detect salient image regions: (1) local low-level features (e.g., color and contrast), (2) global considerations (i.e., suppress regularly occurring features but preserve features that differ from the norm), (3) visual organization rules (i.e., visual forms may possess multiple centers of gravity about which the form is organized), and (4) high-level factors (e.g., human faces).
-
•
Covariance saliency (CovSal) {PM}:44 This model formulates saliency estimation as a supervised learning problem and uses multiple kernel learning to integrate information from different feature dimensions at an intermediate level. It also uses an object-bank, a large filter bank of object detectors, to extract additional semantic high-level features.
-
•
Fast and efficient saliency (FES) {BM}:45 This center-surround model estimates saliency of local feature contrast in a Bayesian framework. Sparse sampling and kernel density estimation are used to estimate the needed probability distributions. The method also implicitly addresses the center bias.
-
•
GBVS {GM}:19 This model first extracts features (e.g., intensity and orientation) at multiple spatial scales and constructs a fully connected graph over all grid locations for each feature map. With weights between two nodes proportional to the similarity of feature values and their spatial distance, the resulting graphs are treated as Markov chains, and their equilibrium distribution is adopted as the saliency maps.
-
•
Image signature (ImgSig) {SM}:46 This model uses the image signature, the sign function of the discrete cosine transform (DCT) of an image, to spatially approximate the foreground of the images. Specially, the saliency map is built on the idea that inverse DCT of the image signature concentrates the image energy at the locations of a spatially sparse foreground, relative to a spectrally sparse background.
-
•
Itti’s saliency model {CM}:21 This basic model uses three feature channels, color, intensity, and orientation, to construct “center-surround” feature maps. Then these maps are linearly combined and normalized to yield the saliency map.
-
•
Phase spectrum of quaternion fourier transform {SM}:47 This model incorporates the phase spectrum of an image’s Fourier transform, and uses a quaternion representation of an image combining intensity, color, and motion features. The use of multiresolution wavelets allows the model to compute the saliency map of an image under various resolutions from coarse to fine.
-
•
RARE {DM}:48 This model selects information worthy of attention based on multiscale spatial rarity, which detects both locally contrasted and globally rare regions in the image. After extracting low-level color and medium-level orientation features, a multiscale rarity mechanism is applied to compute the cross-scale occurrence probability of each pixel, and represent the attention score using self-information. The rarity maps are then fused into a single final saliency map.
-
•
Salicon {PM}:49 This model takes advantage of the representational power of high-level semantics encoded in deep neural networks (DNNs) for object recognition. Two key components are fine-tuning the DNNs with an objective function based on the saliency evaluation metrics and integrating information at different scales.
-
•
Self-resemblance saliency (SelfResem) {IM}:50 This model uses local regression kernels to measure how similar a pixel is to its surroundings. Specifically, the self-resemblance measure slides a window within the surrounding window and computes the matrix cosine similarity between the center window and the cropped window. The saliency map is an ensemble of resemblance scores indicating the rarity of the center features.
-
•
Saliency by induction mechanisms (SIM) {CM}:51 Based on a low-level vision system, the model computes the saliency map in three steps: (1) process visual stimuli according to the early human visual pathway (e.g., color-opponent, luminance channels, a multiscale decomposition), (2) simulate the inhibition mechanisms in cells of the visual cortex, and (3) integrate information at multiple scales by an inverse wavelet transform directly on weights.
-
•
Saliency using natural statistics (SUN) {BM}:52 This model considers what objective human visual system is optimizing when deploying attention and constructs a Bayesian framework using natural statistics. When visually searching for a target, bottom-up saliency represents the self-information of visual features, and overall saliency, after adding top-down factors, emerges as the pointwise mutual information between local image features and the target’s features.2
-
•
Spatially weighted dissimilarity {OM}:53 This model measures the saliency by integrating three elements: (1) the dissimilarities between image patches, (2) the spatial distance between image patches, and (3) the central bias. Principal components of image patches are extracted by sampling the patches from the current image. The dissimilarities are inversely proportional to the spatial distance, and a bias for human fixations toward the center of the image is reflected in a weighting mechanism.
-
•
Torralbas saliency model (Torralba) {BM}:54 This model uses a Bayesian framework to compute the probability of the target object being present in the scene. A context-based prior on the target location, as top-down knowledge of global features of the scene, is integrated with a pure bottom-up prior that is independent from the target.55
2.2.2. Top-down factors
In addition to the image-driven features, human visual attention is also influenced by goal-driven top-down features.2 In the context of medical images, top-down features correspond to factors such as radiologists’ knowledge, their a priori expectations of a task (e.g., identifying size and shape of a lesion). For example, for PET images, the participants had a high number of fixations on the bladder region [Fig. 1(a)], as they were asked to comment on the degree of bladder distention. Hence, for all the saliency models, in order to incorporate some of the role of top-down factors, we performed manual segmentation before applying the saliency models to exclude some image regions from being salient. Specifically, as radiologists were asked to perform a given set of tasks for each imaging modality, we segmented the relevant anatomical regions (i.e., liver and aorta in CT images, lungs and heart in CXR images) so that the other regions (e.g., kidney in CT images, shoulder in CXR images) were not allowed to be salient. For each image, a binary mask, with 1’s indicating the potential salient regions, was produced. However, manual segmentation can result in unsmooth, “spikelike” edges, which may be mistakenly considered as salient by some of the saliency models. To avoid such potential errors, morphological operations, with disk-shaped structure elements, were applied to the binary masks to smooth the edges. Elementwise multiplications between the final binary masks and the original images were done to create the segmented images for saliency map estimation.
2.2.3. Example saliency images
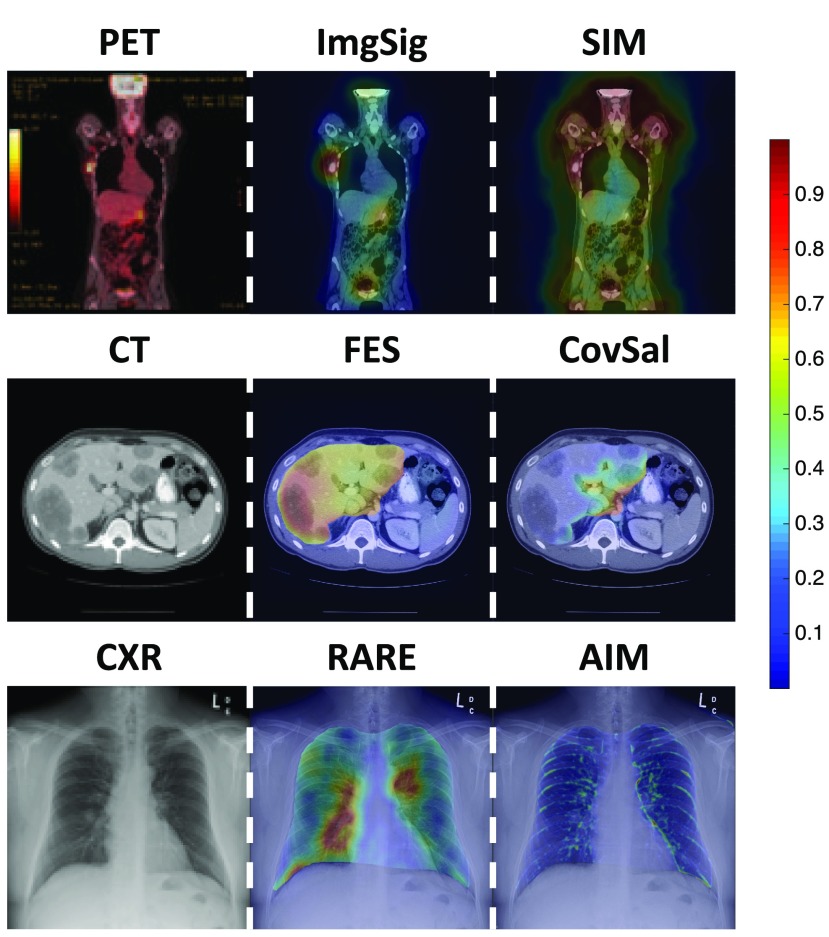
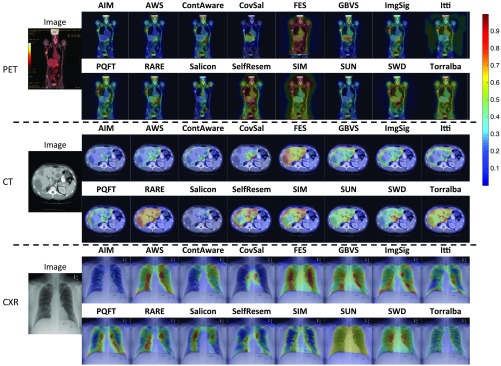
The computed saliency maps highlight different image regions that a human observer might fixate on. Figure 2 shows example saliency maps overlapped upon a stimulus image. One example image from each of the three medical imaging modalities is shown (left column), together with an example of a saliency map (middle column) that highlights the regions of interest that radiologists did look at when performing the given tasks, and an example of a saliency map (right column) that highlights some task-irrelevant regions. For example, for the particular CXR image, a saliency map by RARE had highest saliency located around the largest lung lesion in the image. In contrast, a saliency map by AIM only focused on the ribs. Detailed quantitative evaluations of the model performance are discussed in Sec. 3. Generally, it is observed that the 16 saliency maps on the medical images were visually different from each other, i.e., they make different predictions about where the radiologists will fixate. Segmentation of task-relevant anatomical regions (e.g., liver in CT images) before applying the saliency models did help to prevent irrelevant regions (e.g., kidney in CT images) from being salient. Visual inspections of the saliency maps suggest that most of the models are able to produce saliency maps that indicate the conspicuity of image regions and their relevance to radiologists’ tasks. However, it is also apparent that some of the models may be ineffective for medical images. A comprehensive figure showing example saliency maps from all 16 of the saliency models on the medical images is available in Appendix A (Fig. 7).
Fig. 2.
Example medical images and saliency maps overlapped upon them. Saliency maps, with normalized saliency value in the range of [0,1] are represented as heat maps, and the color indicates the saliency at that location: red is more salient than blue. The left column shows one example image for each of the three imaging modalities. The middle column shows examples of saliency maps that accurately highlighted the regions of interest. The right column shows examples of saliency maps that only highlighted task-irrelevant regions of the image.
Fig. 7.
Example images from the three imaging modalities (from top to bottom: PET, CT, CXR), and the computed saliency maps, overlapped upon the image. Saliency maps with normalized saliency value in the range of [0,1] are represented as heat maps, and the color indicates the saliency at that location: red is more salient than blue. Generally, the 16 saliency maps were visually different from each other. Segmentation of task-relevant anatomical regions before applying the saliency models did help reduce the number of irrelevant regions identified as salient. Some of the models are able to produce saliency maps that indicate the conspicuity of image regions and their relevance to radiologists’ tasks, but some of the models may be ineffective on medical images.
2.3. Evaluation Metrics
To compare the performance of the models in predicting radiologists’ fixations, we computed three commonly used evaluation metrics: (1) area under the receiver operating characteristic (ROC) curve (AUC), (2) linear correlation coefficient (CC), and (3) normalized saliency scanpath (NSS).2 The motivation for using multiple metrics was to ensure that any qualitative conclusion of the study was independent of the choice of metric.56,57 For each experimental image, the output of a saliency model is a saliency map, denoted by . The ground-truth fixation map, denoted by , is constructed by assigning 1’s at the fixation locations of all participants, and smoothing the binary image with a Gaussian blob.2 For each fixation, the spread of the Gaussian blob reflected the radius of the fixated area (i.e., 1 deg of visual angle around the fixation58). For each of the three medical imaging modalities, the mean and standard deviation of the three metrics across the 10 images in the set were reported, respectively. Counterpart results with the MIT300 dataset of natural images were extracted from the MIT saliency benchmark for comparison purposes.
-
•
AUC: The saliency map , as a binary classifier, classified a given percent of image pixels as fixated and the rest as not fixated.2 Radiologists’ fixations were considered as the ground truth. A pixel location is a “hit” if classified the pixel as fixated, and the pixel was actually fixated by the radiologist. A pixel location is a “false-alarm” if classified the pixel as fixated but it was never fixated upon by the radiologist. Varying thresholds of the saliency value resulted in different classifiers, which yielded different pairs of sensitivity and specificity. In this way, an ROC curve was constructed, and AUC in the range of [0, 1] was calculated as the FOM. A score of 1 indicates perfect prediction while a score of 0.5 indicates chance level. Specifically, Borji’s shuffled AUC59 was computed to avoid the effect of centered fixations.56
-
•CC: CC measures the strength of a linear relationship between two random variables (i.e., and ).2 It is computed as
where and are the mean and variance of the values in and , respectively. CC provides a single scalar score in the range of , where larger scores (absolute value) indicate more accurate predictions. -
•
NSS: The saliency map was considered as a random variable. After normalizing to have zero mean and unit standard deviation, NSS was computed as the average of the response values in at radiologists’ eye positions .60 indicates that the eye positions fall in a region whose predicted saliency is one standard deviation above average. Thus, means that the fixated locations have significantly higher saliency values in than other locations. Meanwhile, indicates that the model performs at chance level.2
3. Results
3.1. Correlations Among the FOMs
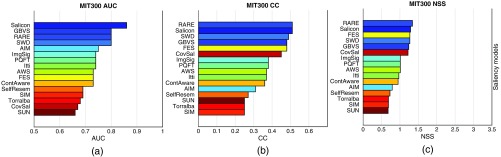
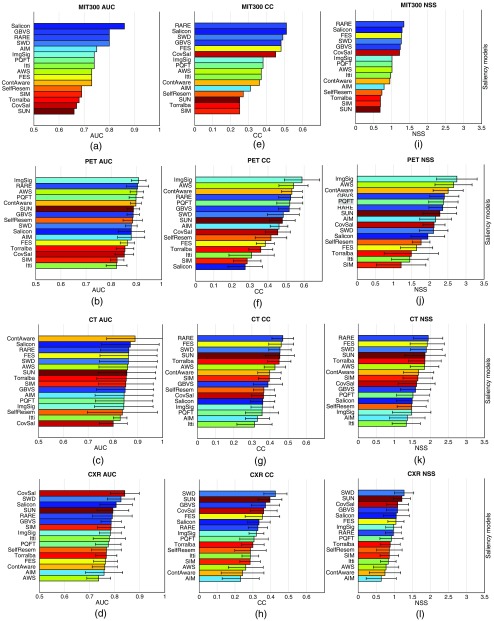
Three FOMs (i.e., AUC, CC, and NSS) were used in the study to measure the performance of the saliency models, thus there were three corresponding rank orders of the models. We observe that the rank orders over the benchmark MIT300 image dataset, indicated by the color patterns in the three bar graphs of Fig. 3, are visually highly similar. This suggests that the rank order of the saliency models is relatively independent of the choice of FOM. We conducted the same analysis for the three medical image datasets, and similar trends as in Fig. 3 were observed. For example, for PET images, ImgSig ranks at the top half of all three lists, while SIM ranks in the bottom half. Detailed figures of the three FOMs on medical images are shown in Appendix B (Fig. 8).
Fig. 3.
Bar graphs of the scores achieved by the saliency models over the benchmark images. The models are shown in the descending order of the mean score. Each model is indicated by a unique color, and the color patterns are similar across the three subfigures: (a) the AUC scores, (b) the CC scores, and (c) the NSS scores. This suggests that the rank order of the saliency models is relatively independent of the choice of the figure of merit.
Fig. 8.
Bar graphs of the FOM scores achieved by the saliency models. The models are shown in descending order of the mean scores. Each model is indicated by a unique color, and the color patterns are the same across the 12 subfigures. The ranking by AUC in (a), as on the MIT saliency benchmark, serves as the reference of the colors of the models. (a)–(d) AUC scores with the MIT300 dataset, the PET images, the CT images, and the CXR images. (e)–(h) CC scores with the four image sets. (i)–(l) NSS scores with the four image sets. Visually, the rank orders across the columns of the same row are similar to each other, while the rank orders across the rows of the same column are significantly different.
To further determine if the three rank orders for medical images were statistically consistent, we calculated the Spearman’s rank correlation coefficients ()61 among the three rankings obtained using the three FOMs. The analysis was repeated for all three imaging modalities (i.e., PET, CT, and CXR). There were strong correlations among the three rank orders as all the correlation coefficients were above 0.5, and the corresponding -values for testing against the null hypothesis of zero correlation were significant (Table 2). The correlation between AUC and CC, and the correlation between AUC and NSS, was lower than the correlation between CC and NSS. This was expected because both CC and NSS consider the saliency map as a random variable, while AUC considers the saliency map as a binary classifier.
Table 2.
Spearman’s rank correlation coefficients () among the three ranking orders. Each row of the table shows the correlation coefficients for one imaging modality. Each column of the table shows the correlation coefficients between the two FOMs being compared, as indicated in the header row of the same column. The -value for testing against a null hypothesis of zero correlation is also reported.
| AUC and CC | AUC and NSS | CC and NSS | |
|---|---|---|---|
| PET | () | () | () |
| CT | () | () | () |
| CXR | () | () | () |
In conclusion, although the rank orders were not exactly the same over the FOMs, the choice of FOM does not dramatically alter the qualitative conclusions that we draw from this study. Hence, for the rest of the paper, we focus on the AUC results since it is the mostly commonly used FOM for evaluating saliency models,2,55,59 and it is most familiar to the medical imaging community.
3.2. Comparisons between Medical Images and Natural Images
Most saliency models, including the 16 models investigated in this study, were validated on images of natural scenes. The MIT300 dataset of 300 natural images with eye-tracking data from 39 observers provides an up-to-date benchmark model performance and ranking of these models. We compared this benchmark rank order, labeled as MIT, to the rank orders from the three medical image datasets (labeled as PET, CT, and CXR, respectively). Similarly, correlation coefficients (Spearman) and associated -values for testing against a null hypothesis of zero correlation are reported in Table 3. It can be observed that the correlation coefficients all fell in the range of [0.1, 0.3], which indicates weak correlation between the two rank orders being compared. Even for colored PET images, which one might suppose to be more similar to natural images than grayscale medical images would be, no strong correlation was observed with the model rank order on the benchmark set of natural scene images. We failed to reject the null hypothesis that the correlation coefficient was not zero, but this may be due in part to the limited number of saliency models tested, e.g., the post-hoc power analysis using G*Power62,63 on the correlation between MIT and PET shows a relatively low power of 0.137 at the significance level 0.05. However, we emphasize that the magnitude of observed correlations was weak [0.1, 0.3] even if they were actually not zero.
Table 3.
Correlation coefficients (Spearman) between the benchmark MIT rank order and the three rank orders over medical images. Each column of the table shows the correlations for one imaging modality. There is only weak correlation between the two rank orders, which was not statistically distinguishable from zero.
| MIT and PET | MIT and CT | MIT and CXR |
|---|---|---|
| () | () | () |
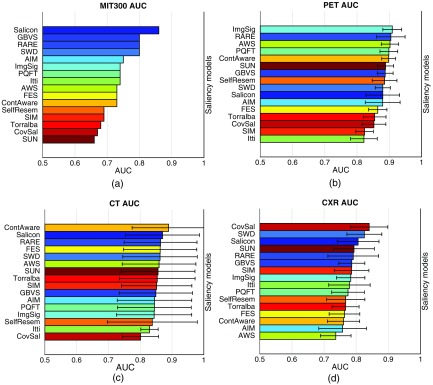
Figure 4 shows the bar graphs of the AUCs achieved by the saliency models over the four image datasets. From visual inspections of the color patterns in the four subfigures, it is obvious that the rank orders for the three medical image sets [Figs. 4(b)–4(d)] are significantly different from the rank order for the benchmark MIT300 dataset [Fig. 4(a)]. For example, according to the benchmark MIT ranking, AIM ranks in the top five high-performing models, but for the medical images, AIM ranks in the bottom five low-performing models. This could be explained by the fact that AIM defines the saliency based on the self-information of each local image patch, but the basis independent components for estimating the probability distribution are drawn from natural images. Hence, if saliency models such as AIM contain modules that are trained specifically for natural scenes, they may fail to capture the characteristics of medical images when directly applied. On the other hand, despite the huge differences among the four rank orders, some general patterns can be observed. RARE, ranking among the top five in all four rankings, appears to work satisfactorily for both the natural scene images and the three medical imaging sets. Recall the main idea of RARE is to detect locally contrasted and globally rare regions in the image. Multiscale rarity appears to be generally applicable for fixation prediction over 2-D still images. SelfResem, using only local regression kernels as features to measure how much a pixel stands out from its local surroundings, did not perform well in any of the four rankings; hence, accounting for certain global factors may be broadly beneficial. Similar results are observed with CC scores and NSS scores, and the details are available in Appendix B (Fig. 8).
Fig. 4.
Bar graphs of the AUC scores achieved by the saliency models. The models are shown in the descending order of the mean AUC score. Each model is indicated by a unique color to facilitate color pattern assessment across the four subfigures. The ranking listed on the MIT saliency benchmark serves as the reference of the colors of the models. (a) The AUC scores with the MIT300 dataset. No standard deviation of AUC is reported on the benchmark. (b) The AUC scores with the PET images. (c) The AUC scores with CT images, (d) The AUC scores with the CXR images. Visually, it is obvious that the rank orders of the models are significantly different across the four subfigures.
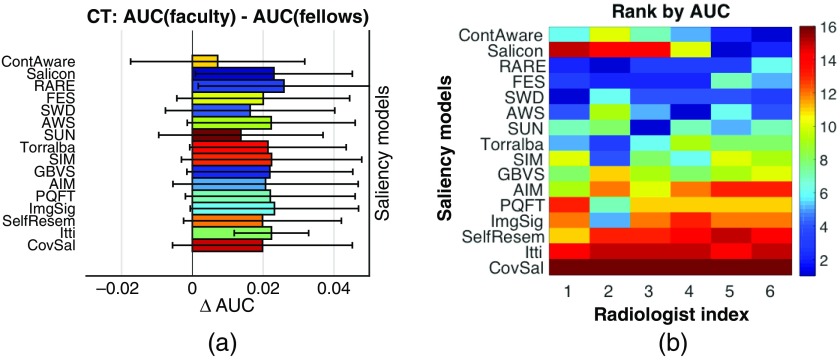
Fig. 6.
(a) Bar graphs of the difference in AUC scores achieved by the saliency models for faculty and for fellows. Each of the saliency models achieved a positive , suggesting that the saliency models may be more effectively in predicting faculty’ eye-movements than fellows’. (b) Rank orders of the saliency models with respect to each of the six radiologists. Certain saliency models may perform well for some radiologists than others.
3.3. Comparisons Among Different Medical Imaging Modalities
We compared the performance of the saliency models with the three medical image datasets [Figs. 4(b)–4(d)]. First, we can observe that the overall scores for PET [Fig. 4(b)] are higher, and with a smaller variance than the scores for CT [Fig. 4(c)] and CXR [Fig. 4(d)]. Such difference may be because PET images are presented as color images, while CT and CXR images are grayscale images. As most of natural scene images and videos contain three channels (e.g., red, green, and blue), color-relevant features are commonly included in the design of saliency models, and these features may remain valuable for medical images presented in color. Another potential contributing factor to the larger variations for CT and CXR images is that the tasks for CT and CXR images required a higher coverage of the image, while the task for PET images only need analysis of a few specific regions of the image. Hence, differences in radiologists’ visual search strategies may have resulted in larger interreader and intercase variations in their fixations. Second, the overall mean scores for CXR [Fig. 4(d)] are lower than the mean scores for CT [Fig. 4(c)]. This underperformance may be because a CXR image contains a number of high-intensity, high-contrast anatomical structures (e.g., long curved ribs). Though they are completely irrelevant to the task being performed, they often partially overlap with the target lung lesions being searched for. The saliency models for CXR images may have been mistakenly attracted to these image regions. Third, the scores for CT [Fig. 4(c)] have larger variations (i.e., longer error bars) than the scores for PET [Fig. 4(b)] and CXR [Fig. 4(d)]. Recall that the radiologists’ tasks with CT images were to find the largest liver metastasis and comment on abdominal aorta. The sizes and shapes of the metastases varied substantially across the 10 experimental images. For example, several cases contain a metastasis that was even larger than the defined fixated areas. When one radiologist fixated at the bottom-left corner of the metastasis while another radiologist fixated at the top-right corner of the metastasis, they both detected the metastasis, but obviously, their eye positions and movements would be notably different. Another factor that may contribute to the larger variations is that some models (e.g., SelfResem and ContAware) compute the saliency map at a lower resolution for efficiency, so the resized computed saliency maps may not be accurate enough when the fixated regions to be predicted were large. Correlation coefficients (Spearman) and the associated -values for testing against a null hypothesis of zero correlation are reported in Table 4. It can be observed that: (1) the correlations between PET and CT, and between PET and CT are moderate for the two rank orders being compared, but the correlation between CT and CXR is very weak. (2) None of the observed correlations was statistically distinguishable from zero, albeit this assessment is limited by sample size as previously discussed. There are three conflicting remarks to highlight here: (1) ImgSig was the best-performing model on PET, but it only ranked 8th for CXR and 13th for CT. As the model is based on the amplitude spectrum of Fourier transform, the appearances of anatomical structures in CXR and CT, as high-frequency components, seem to degrade its performance dramatically. (2) FES ranked 2nd for CT, 6th for CXR, but only 13th for PET. Unlike the liver and aorta regions in a CT image, and lung regions in a CXR image, the bladder in a PET image is not near the center of the image, and hence the “center-surround” feature of the model may have caused the performance decline. (3) CovSal, a pretrained model with natural scene images, ranked at 1st for CXR, but only 14th for PET and 16th for CT. Though the model worked excellently with CXR images, we suspect that it may be simply a coincidence that the trained model was a good descriptor for this particular set of CXR images. Hence, modality-specific tuning of a saliency model may be essential for medical images. In addition, we considered the possibility that the performance of saliency models might vary by model category, i.e., that performance trends could be explained by commonalities in the models’ underlying mathematical formulations. However, due to the limited number of models in some categories investigated in this work, e.g., only GBVS in the GM category, we could not draw definitive conclusions from the observed performance patterns. Further studies on more models would be helpful to address this question.
Table 4.
Correlation coefficients (Spearman) across the three rank orders over three sets of medical images. The correlations between PET and CT and between PET and CXR are moderate, but the correlation between CT and CXR was very weak. None of the observed correlations was statistically distinguishable from zero.
| PET and CT | PET and CXR | CT and CXR |
|---|---|---|
| () | () | () |
3.4. Comparisons Across Saliency Models
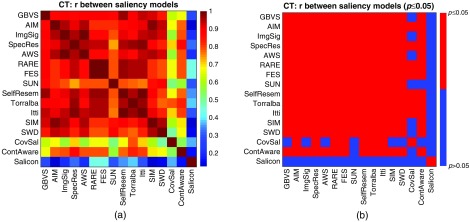
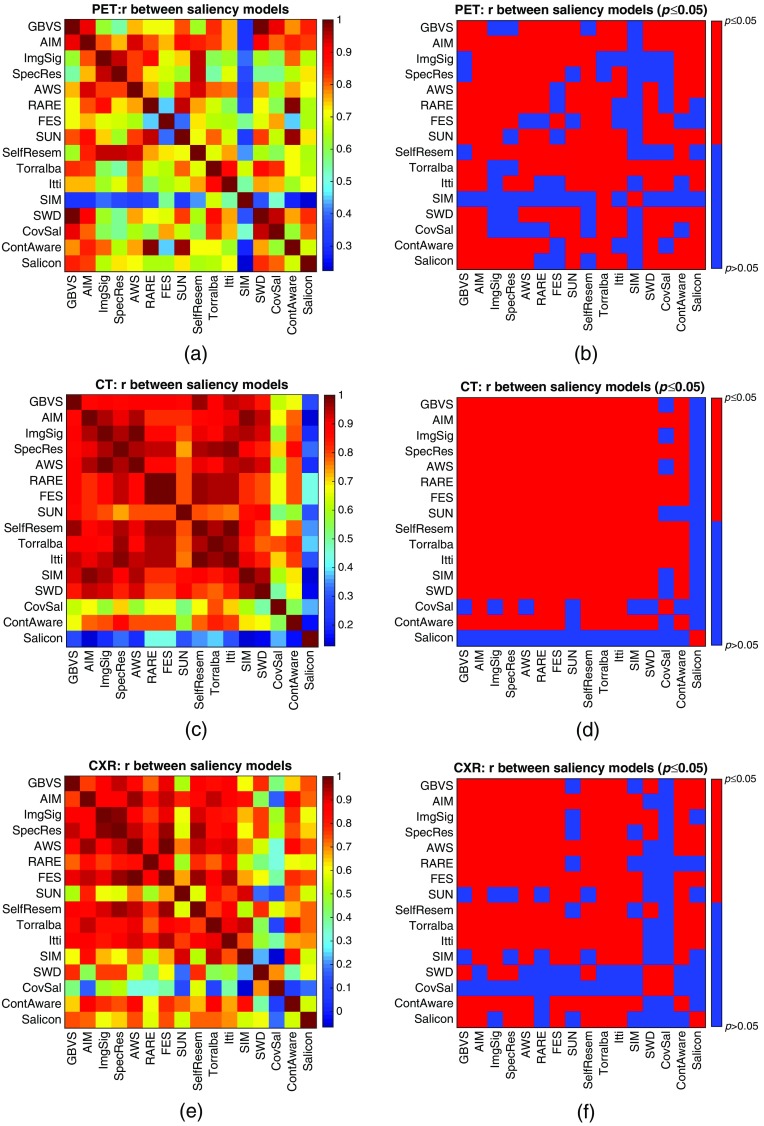
On natural scenes where saliency models are extensively studied, it has been noted that some categories of images are more difficult than others in the sense that models are less accurate in predicting where humans will fixate on them.59 Even within a given category of images, saliency models are not equally effective across all images at predicting where a human will fixate. For example, a red bottle lying on lawn is more salient than a green bottle lying on the lawn so it is easier for saliency models to highlight the red bottle than the green bottle. In order to investigate possible image-specific effects in medical imaging, we compared the performance of one particular saliency model over 10 images from the same imaging modality. Hence, for the 10 images in each imaging modality, 16 rank orders of the accuracy of the 10 corresponding saliency maps were created, one per saliency model. Correlation coefficients (Spearman) and the associated -values for testing against a null hypothesis of zero correlation for each pair of the rank orders were computed. Figure 5 visualizes the results with CT images in the matrix form, where each element of the matrix, represented as a colored square patch, shows the correlation between the two indexed saliency models [Fig. 5(a)], and whether the -value is less than 0.05 [Fig. 5(b)]. Most pairs of the rank orders were moderately correlated, and the corresponding -values are all less than 0.05. This agrees with the studies in nature scenes that existing saliency models may have common difficulty over some particular images. One exception from this general trend is Salicon, whose correlations with the other 15 models were all weak or close to none. However, interestingly, Salicon is one of the top-performing saliency models for CT images. This may also suggest some hints for future research on integrating deep learning techniques into saliency models to provide alternative features, and different mechanisms of selecting salient image regions. Additional results with PET and CXR images are available in Appendix C (Fig. 9).
Fig. 5.
Visualizations of the correlations among the performances of the saliency models over the CT images. The rank orders of the 10 images represent how accurately the model predicts the radiologists’ fixations on each image. (a) Matrix of correlations: each element of the matrix, represented as a colored square patch, shows the correlation coefficient (Spearman) between the two indexed saliency models. (b) Matrix of : each element of the matrix indicates whether the -value is smaller than 0.05 for testing against a null hypothesis of zero correlation. Most of the correlations are moderate to strong, and the corresponding -values are all smaller than 0.05. This suggests that an image for which one saliency model has difficulty predicting where a human will fixate will likewise be difficult for another saliency model.
Fig. 9.
Visualizations of the correlations among the performances of the saliency models over the medical images. The rank orders represent how difficult the 10 images in the same imaging modality are for saliency prediction. (a) Matrix of for PET images, (b) matrix of for PET images, (c) matrix of correlation coefficients (Spearman) for CT images, (d) matrix of for CT images, (e) matrix of correlation coefficients (Spearman) for CXR images, and (f) matrix of for CXR images. Most of the correlations are moderate to strong, and are statistically distinguishable from zero (). This suggests that the saliency models may have common difficulty over some sets of images.
3.5. Effect of Radiologists’ Experience Level
There are numerous eye-movement studies on the role of expertise in medical image perception (e.g., Refs. 18, 31, 34, 38, 39, and 64–69). Experts have shown superior perceptual ability in their specific domain of expertise.65 For example, in comparison to nonexperts or novices, experienced radiologists typically interpret images with fewer fixations,38,68 longer saccades,38,68 and less coverage of the image.34,66 They were able to achieve an accuracy of 70% in detecting lesions in chest radiographs when images were viewed for only 200 ms.69 Hence, it is worthwhile to investigate whether the radiologists’ level of experience would potentially alter any of the general conclusions of the study. We first computed two sets of aggregated fixation maps, one for the group of four faculties, and the other for the group of two fellows. Then same evaluation procedures were conducted to compare the performance of the saliency models in predicting faculty’s fixations with the performance for fellows’ fixations. For illustration, Fig. 6(a) shows the differences in the AUC scores that the saliency models achieved for faculty and for fellows over CT images. Similar results are observed with the other two imaging modalities. It can be observed that for all the saliency models, the mean AUC for faculty was higher than the mean AUC for fellows (i.e., ). This suggests that the visual attention of the faculty participants was better captured by the saliency maps of an image. As those visually salient objects/areas were often clinically significant, faculty, with more expertise and experience, are generally more efficient than the fellows in deploying their visual attention. However, due to the small number of images and participants in each group, not all the differences were statistically significant. Future studies are needed to draw a definitive conclusion on this issue.
Previous eye-movement studies with medical images have also demonstrated the interobserver variability in radiologists’ visual attention deployment.70 We explored this issue by evaluating the saliency models with respect to each of the radiologists. The objective was to study whether certain saliency models may perform better for some radiologists than for others. We compared the rank orders of the models across the six radiologists. Similarly, we reported the AUC scores for CT images for illustration [Fig. 6(b)], and similar trends are observed with PET and CXR images. It is observed that: (1) CovSal was the worst performing model for all six radiologists; (2) RARE was among the top three for all six radiologists; (3) Salicon ranked in the top two for radiologists five and six (one faculty and one fellow), but ranked below 14th for the other four radiologists. Having some individual radiologists better represented by certain saliency models implies that different image information may be utilized in their visual searches. This could potentially help identify efficient strategies of attention deployment that may improve diagnostic accuracy and radiologist training. Follow up studies are needed to draw a definitive conclusion.
4. Conclusions and Future Work
We conducted a comparative study of computational visual attention models, mainly bottom-up saliency models, in the context of 2-D medical images. We investigated 16 models, by comparing saliency maps over three image sets (PET, CT, and CXR) with radiologists’ gaze positions. We conclude that (1) all the saliency models performed reasonably well over medical images. (2) The rank orders of the models over medical images were different from the benchmark rank-order over natural images. (3) The overall performance and the rank orders of the models were also different across different medical imaging modalities. (4) Some medical images are more difficult than others in the sense of the accuracy of saliency models predicting where a human will fixate upon them. Thus, we have provided some insight to the choice of the appropriate saliency model. Models trained in the context of natural scenes may not be directly applicable to medical images, and modality-specific tuning of the models is necessary. Models that capture both local and global image characteristics may be more accurate in predicting radiologists’ fixations. We acknowledge two main limitations of the study: (1) we incorporated limited task-driven factors by segmenting the relevant anatomical regions. Other top-down attention factors such as prior knowledge of the typical appearance of lung lesions were not considered. (2) The sample size, 30 images (10 images per modality) and 6 radiologists, restricted the power for statistically distinguishing the observed correlations from zero correlation. Additional research with more types of medical images, more cases, and more radiologists is needed in order to identify more subtle differences and optimize parameters used in saliency models (e.g., a priori basis functions of SUN) without overfitting.
In the future, other saliency models that may improve the predictions of radiologists’ fixations need be investigated. Beyond simple segmentation, a more complete account of top-down factors used by expert radiologists to guide their attention is necessary. Experiments could be conducted to investigate the influence of the tasks being conducted. For example, one could study how radiologists’ eye positions and movements differ when asked to read the same abdominal CT image for different tasks (e.g., detect liver lesions versus detect renal masses). Another example would be to study the impact of certain imaging parameters (e.g., if intravenous contrast is used in a CT scan) on the effectiveness of saliency models. Moreover, rather than simply assessing 2-D images, it would be interesting to study the value of saliency models with 3-D medical imaging data (e.g., stacks of CT slices), especially considering the increasing usage of volumetric medical imaging in clinical practice. Radiologists have to develop efficient strategies of attention deployment for reading volumetric images and storage of data. Finally, additional assessments with larger sets of images and more radiologists would be valuable to achieve a more definitive assessment of saliency in medical imaging. Large, publicly available datasets of eye-tracking studies would be beneficial to the whole medical imaging community, especially to researchers who are interested in image perception, observer performance, and technology evaluations.
Acknowledgments
We would like to thank Ms. Tejaswani Ganapathi for her work on the database that was used to answer research questions that are addressed in this paper. Ms. Ganapathi described her data collection efforts in a document35 that she submitted to arXiv, an e-print service. We also recognize the following current and former radiologists at The University of Texas MD Anderson Cancer Center for their participation in data collection: Dr. Dhakshinamoorthy Ganeshan, Dr. Thomas Yang, Dr. Ott Le, Dr. Gregory W. Gladish, Dr. Piyaporn Boonsirikamchai, Dr. Catherine E. Devine, Dr. Madhavi Patnana, Dr. Nicolaus A. Wagner-Bartak, Dr. Priya Bhosale, and Dr. Raghunandan Vikram.
Biographies
Gezheng Wen is a PhD candidate in electrical and computer engineering at the University of Texas at Austin. He received his BS degree from the University of Hong Kong in 2011 and his MS degree from the University of Texas at Austin in 2014. He is a research trainee in diagnostic radiology at the University of Texas MD Anderson Cancer Center. His research interests include medical image quality assessment, model observer, and visual search of medical images.
Brenda Rodriguez-Niño is an undergraduate student of mechanical engineering with an emphasis in biomechanics at the University of Texas at Austin. She is expected to obtain her BS degree in December 2017.
Furkan Y. Pecen is an undergraduate student majoring in biomedical engineering with an emphasis in biomedical imaging and instrumentation at the University of Texas at Austin. He is expected to obtain his BS degree in May 2017.
David J. Vining is a professor of diagnostic radiology at the University of Texas MD Anderson Cancer Center located in Houston, Texas, USA, and the founder of VisionSR, a medical informatics company developing a multimedia structured reporting solution for diagnostic imaging. His professional interests include colorectal cancer screening and big data analytics for the determination of medical outcomes.
Naveen Garg is an assistant professor of radiology at the University of Texas MD Anderson Cancer Center. His clinical specialty is body imaging. His research interest is in computing applications in radiology and medical informatics. He is also interested in user interfaces, natural language processing, image processing, vision, and deep learning.
Mia K. Markey is a professor of biomedical engineering and engineering foundation endowed faculty fellow in engineering at the University of Texas at Austin as well as adjunct professor of imaging physics at the University of Texas MD Anderson Cancer Center. She is a fellow of both the American Association for the Advancement of Science and the American Institute for Medical and Biological Engineering, and a senior member of both the IEEE and SPIE.
Appendix A.
Figure 7 shows the example saliency maps by the 16 saliency models over the three imaging modalities. Generally, the saliency maps were visually different from each other, indicating that the predicted fixations from these models are different. Most of the models are able to produce saliency maps that indicate the conspicuity of image regions and their relevance to radiologists’ tasks, but some of the models only highlight task-irrelevant image regions, and thus may not perform well in fixation predictions.
Appendix B.
Figure 8 shows the FOM scores of the saliency models. Visually, the rank orders across the columns of the same row (i.e., comparisons across the FOMs for the same imaging modality) are similar to each other, while the rank orders across the rows of the same column (i.e., comparison between the benchmark and the three medical modalities) are significantly different.
Appendix C.
Figure 9 visualizes the correlations across the saliency models in the matrix form, where each element of the matrix, represented as a colored square patch, shows one correlation coefficient between the two saliency models [Figs. 9(a), 9(c), and 9(e)], and whether the -value is smaller than or equal to 0.05 [Figs. 9(b), 9(d), and 9(f)]. For all three imaging modalities, most pairs of the rank orders have moderate to strong correlation, which are statistically different from zero. This suggests that saliency models may have common difficulty over some medical images.
Disclosures
No conflicts of interest, financial or otherwise, are declared by the authors.
References
- 1.Carrasco M., “Visual attention: the past 25 years,” Vision Res. 51(13), 1484–1525 (2011). 10.1016/j.visres.2011.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borji A., Itti L., “State-of-the-art in visual attention modeling,” IEEE Trans. Pattern Anal. Mach. Intell. 35(1), 185–207 (2013). 10.1109/TPAMI.2012.89 [DOI] [PubMed] [Google Scholar]
- 3.Itti L., Koch C., “Computational modelling of visual attention,” Nat. Rev. Neurosci. 2(3), 194–203 (2001). 10.1038/35058500 [DOI] [PubMed] [Google Scholar]
- 4.Itti L., “Automatic foveation for video compression using a neurobiological model of visual attention,” IEEE Trans. Image Process. 13(10), 1304–1318 (2004). 10.1109/TIP.2004.834657 [DOI] [PubMed] [Google Scholar]
- 5.Frintrop S., VOCUS: A Visual Attention System for Object Detection and Goal-Directed Search, Vol. 3899, Springer-Berlin Heidelberg, Germany: (2006). [Google Scholar]
- 6.Salah A. A., Alpaydin E., Akarun L., “A selective attention-based method for visual pattern recognition with application to handwritten digit recognition and face recognition,” IEEE Trans. Pattern Anal. Mach. Intell. 24(3), 420–425 (2002). 10.1109/34.990146 [DOI] [Google Scholar]
- 7.Siagian C., Itti L., “Biologically inspired mobile robot vision localization,” IEEE Trans. Rob. 25(4), 861–873 (2009). 10.1109/TRO.2009.2022424 [DOI] [Google Scholar]
- 8.Borji A., et al. , “Online learning of task-driven object-based visual attention control,” Image Vision Comput. 28(7), 1130–1145 (2010). 10.1016/j.imavis.2009.10.006 [DOI] [Google Scholar]
- 9.DeCarlo D., Santella A., “Stylization and abstraction of photographs,” ACM Trans. Graph. 21(3), 769–776 (2002). 10.1145/566654.566650 [DOI] [Google Scholar]
- 10.Marchesotti L., Cifarelli C., Csurka G., “A framework for visual saliency detection with applications to image thumbnailing,” in IEEE 12th Int. Conf. on Computer Vision, pp. 2232–2239, IEEE; (2009). 10.1109/ICCV.2009.5459467 [DOI] [Google Scholar]
- 11.El-Nasr M. S., et al. , “Dynamic intelligent lighting for directing visual attention in interactive 3-D scenes,” IEEE Trans. Comput. Intell. AI Games 1(2), 145–153 (2009). 10.1109/TCIAIG.2009.2024532 [DOI] [Google Scholar]
- 12.Bruce N., Tsotsos J., “Saliency based on information maximization,” Adv. Neural Inf. Process. Syst. 18, 155 (2006). [Google Scholar]
- 13.Kootstra G., Schomaker L. R., “Prediction of human eye fixations using symmetry,” in The 31st Annual Conf. of the Cognitive Science Society (CogSci ’09), pp. 56–61, Cognitive Science Society; (2009). [Google Scholar]
- 14.Judd T., et al. , “Learning to predict where humans look,” in IEEE 12th Int. Conf. on Computer Vision, pp. 2106–2113, IEEE; (2009). 10.1109/ICCV.2009.5459462 [DOI] [Google Scholar]
- 15.http://saliency.mit.edu.
- 16.Judd T., Durand F., Torralba A., “A benchmark of computational models of saliency to predict human fixations,” MIT CSAIL Technical Reports, TR–2012–001 (2012).
- 17.Borji A., Itti L., “Cat2000: a large scale fixation dataset for boosting saliency research,” arXiv preprint arXiv:1505.03581 (2015).
- 18.Matsumoto H., et al. , “Where do neurologists look when viewing brain ct images? an eye-tracking study involving stroke cases,” PloS One 6(12), e28928 (2011). 10.1371/journal.pone.0028928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harel J., et al. , “Graph-based visual saliency,” in Neural Information Processing Systems (NIPS), Vol. 1, p. 5 (2006). [Google Scholar]
- 20.Jampani V., et al. , “Assessment of computational visual attention models on medical images,” in Proc. of the Eighth Indian Conf. on Computer Vision, Graphics and Image Processing, p. 80, ACM; (2012). [Google Scholar]
- 21.Itti L., Koch C., Niebur E., “A model of saliency-based visual attention for rapid scene analysis,” IEEE Trans. Pattern Anal. Mach. Intell. 20(11), 1254–1259 (1998). 10.1109/34.730558 [DOI] [Google Scholar]
- 22.Hou X., Zhang L., “Saliency detection: a spectral residual approach,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR ’07), pp. 1–8, IEEE; (2007). 10.1109/CVPR.2007.383267 [DOI] [Google Scholar]
- 23.Alzubaidi M., et al. , “What catches a radiologist’s eye? A comprehensive comparison of feature types for saliency prediction,” Proc. SPIE 7624, 76240W (2010). 10.1117/12.844508 [DOI] [Google Scholar]
- 24.Wen G., et al. , “Computational assessment of visual search strategies in volumetric medical images,” J. Med. Imaging 3(1), 015501 (2016). 10.1117/1.JMI.3.1.015501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Webb S., The Physics of Medical Imaging, CRC Press; (1988). [Google Scholar]
- 26.Rabbani M., Jones P. W., “Image compression techniques for medical diagnostic imaging systems,” J. Digital Imaging 4(2), 65–78 (1991). 10.1007/BF03170414 [DOI] [PubMed] [Google Scholar]
- 27.Smutek D., “Quality measurement of lossy compression in medical imaging,” Prague Med. Rep. 106(1), 5–26 (2005). [PubMed] [Google Scholar]
- 28.Flint A. C., “Determining optimal medical image compression: psychometric and image distortion analysis,” BMC Med. Imaging 12(1), 24 (2012). 10.1186/1471-2342-12-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manning D., Ethell S., Donovan T., “Detection or decision errors? Missed lung cancer from the posteroanterior chest radiograph,” Br. J. Radiol. 77, 231–235 (2014). 10.1259/bjr/28883951 [DOI] [PubMed] [Google Scholar]
- 30.Kundel H. L., Nodine C. F., Toto L., “Searching for lung nodules: the guidance of visual scanning,” Invest. Radiol. 26(9), 777–781 (1991). 10.1097/00004424-199109000-00001 [DOI] [PubMed] [Google Scholar]
- 31.Kundel H. L., et al. , “Holistic component of image perception in mammogram interpretation: gaze-tracking study 1,” Radiology 242(2), 396–402 (2007). 10.1148/radiol.2422051997 [DOI] [PubMed] [Google Scholar]
- 32.Mello-Thoms C., “How much agreement is there in the visual search strategy of experts reading mammograms?” Proc. SPIE 6917, 691704 (2008). 10.1117/12.768835 [DOI] [Google Scholar]
- 33.Hu C. H., et al. , “Searching for bone fractures: a comparison with pulmonary nodule search,” Acad. Radiol. 1(1), 25–32 (1994). 10.1016/S1076-6332(05)80780-9 [DOI] [PubMed] [Google Scholar]
- 34.Krupinski E. A., “Visual scanning patterns of radiologists searching mammograms,” Acad. Radiol. 3(2), 137–144 (1996). 10.1016/S1076-6332(05)80381-2 [DOI] [PubMed] [Google Scholar]
- 35.Ganapathi T., et al. , “A human computer interaction solution for radiology reporting: evaluation of the factors of variation,” arXiv preprint arXiv:1607.06878 (2016).
- 36.Kundel H. L., Nodine C. F., Carmody D., “Visual scanning, pattern recognition and decision-making in pulmonary nodule detection,” Invest. Radiol. 13(3), 175–181 (1978). 10.1097/00004424-197805000-00001 [DOI] [PubMed] [Google Scholar]
- 37.Nodine C., et al. , “Recording and analyzing eye-position data using a microcomputer workstation,” Behav. Res. Methods 24(3), 475–485 (1992). 10.3758/BF03203584 [DOI] [Google Scholar]
- 38.Krupinski E. A., et al. , “Eye-movement study and human performance using telepathology virtual slides. Implications for medical education and differences with experience,” Hum. Pathol. 37(12), 1543–1556 (2006). 10.1016/j.humpath.2006.08.024 [DOI] [PubMed] [Google Scholar]
- 39.Krupinski E. A., Graham A. R., Weinstein R. S., “Characterizing the development of visual search expertise in pathology residents viewing whole slide images,” Hum. Pathol. 44(3), 357–364 (2013). 10.1016/j.humpath.2012.05.024 [DOI] [PubMed] [Google Scholar]
- 40.Salvucci D. D., Goldberg J. H., “Identifying fixations and saccades in eye-tracking protocols,” in Proc. of the 2000 Symp. on Eye Tracking Research and Applications, pp. 71–78, ACM; (2000). [Google Scholar]
- 41.Shannon C. E., “A mathematical theory of communication,” ACM Sigmobile Mob. Comput. Commun. Rev. 5(1), 3–55 (2001). 10.1145/584091 [DOI] [Google Scholar]
- 42.Garcia-Diaz A., et al. , “On the relationship between optical variability, visual saliency, and eye fixations: a computational approach,” J. Vision 12(6), 17–17 (2012). 10.1167/12.6.17 [DOI] [PubMed] [Google Scholar]
- 43.Goferman S., Zelnik-Manor L., Tal A., “Context-aware saliency detection,” IEEE Trans. Pattern Anal. Mach. Intell. 34(10), 1915–1926 (2012). 10.1109/TPAMI.2011.272 [DOI] [PubMed] [Google Scholar]
- 44.Kavak Y., Erdem E., Erdem A., “Visual saliency estimation by integrating features using multiple kernel learning,” arXiv preprint arXiv:1307.5693 (2013).
- 45.Tavakoli H. R., Rahtu E., Heikkilä J., “Fast and efficient saliency detection using sparse sampling and kernel density estimation,” in Scandinavian Conf. on Image Analysis, Heyden A., Kahl F., Eds., pp. 666–675, Springer; (2011). [Google Scholar]
- 46.Hou X., Harel J., Koch C., “Image signature: highlighting sparse salient regions,” IEEE Trans. Pattern Anal. Mach. Intell. 34(1), 194–201 (2012). 10.1109/TPAMI.2011.146 [DOI] [PubMed] [Google Scholar]
- 47.Guo C., Zhang L., “A novel multiresolution spatiotemporal saliency detection model and its applications in image and video compression,” IEEE Trans. Image Process. 19(1), 185–198 (2010). 10.1109/TIP.2009.2030969 [DOI] [PubMed] [Google Scholar]
- 48.Riche N., et al. , “Rare2012: a multi-scale rarity-based saliency detection with its comparative statistical analysis,” Signal Process. Image Commun. 28(6), 642–658 (2013). 10.1016/j.image.2013.03.009 [DOI] [Google Scholar]
- 49.Huang X., et al. , “Salicon: reducing the semantic gap in saliency prediction by adapting deep neural networks,” in Proc. of the IEEE Int. Conf. on Computer Vision, pp. 262–270 (2015). 10.1109/ICCV.2015.38 [DOI] [Google Scholar]
- 50.Seo H. J., Milanfar P., “Static and space-time visual saliency detection by self-resemblance,” J. Vision 9(12), 15–15 (2009). 10.1167/9.12.15 [DOI] [PubMed] [Google Scholar]
- 51.Murray N., et al. , “Saliency estimation using a non-parametric low-level vision model,” in IEEE conference on Computer Vision and Pattern Recognition (CVPR ’11), pp. 433–440, IEEE; (2011). 10.1109/CVPR.2011.5995506 [DOI] [Google Scholar]
- 52.Zhang L., et al. , “Sun: a Bayesian framework for saliency using natural statistics,” J. Vision 8(7), 32–32 (2008). 10.1167/8.7.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duan L., et al. , “Visual saliency detection by spatially weighted dissimilarity,” in IEEE Conf. on Computer Vision and Pattern Recognition (CVPR ’11), pp. 473–480, IEEE; (2011). 10.1109/CVPR.2011.5995676 [DOI] [Google Scholar]
- 54.Torralba A., et al. , “Contextual guidance of eye movements and attention in real-world scenes: the role of global features in object search,” Psychol. Rev. 113(4), 766–786 (2006). 10.1037/0033-295X.113.4.766 [DOI] [PubMed] [Google Scholar]
- 55.Kimura A., Yonetani R., Hirayama T., “Computational models of human visual attention and their implementations: a survey,” IEICE Trans. Inf. Syst. E96.D(3), 562–578 (2013). 10.1587/transinf.E96.D.562 [DOI] [Google Scholar]
- 56.Riche N., et al. , “Saliency and human fixations: state-of-the-art and study of comparison metrics,” in Proc. of the IEEE Int. Conf. on Computer Vision, pp. 1153–1160 (2013). 10.1109/ICCV.2013.147 [DOI] [Google Scholar]
- 57.Bylinskii Z., et al. , “What do different evaluation metrics tell us about saliency models?” arXiv preprint arXiv:1604.03605 (2016). [DOI] [PubMed]
- 58.Le Meur O., Baccino T., “Methods for comparing scan paths and saliency maps: strengths and weaknesses,” Behav. Res. Methods 45(1), 251–266 (2013). 10.3758/s13428-012-0226-9 [DOI] [PubMed] [Google Scholar]
- 59.Borji A., Sihite D. N., Itti L., “Quantitative analysis of human-model agreement in visual saliency modeling: a comparative study,” IEEE Trans. Image Process. 22(1), 55–69 (2013). 10.1109/TIP.2012.2210727 [DOI] [PubMed] [Google Scholar]
- 60.Peters R. J., et al. , “Components of bottom-up gaze allocation in natural images,” Vision Res. 45(18), 2397–2416 (2005). 10.1016/j.visres.2005.03.019 [DOI] [PubMed] [Google Scholar]
- 61.Spearman C., “The proof and measurement of association between two things,” Am. J. Psychol. 15(1), 72–101 (1904). 10.2307/1412159 [DOI] [PubMed] [Google Scholar]
- 62.Faul F., et al. , “G* power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences,” Behav. Res. Methods 39(2), 175–191 (2007). 10.3758/BF03193146 [DOI] [PubMed] [Google Scholar]
- 63.Faul F., et al. , “Statistical power analyses using g* power 3.1: tests for correlation and regression analyses,” Behav. Res. Methods 41(4), 1149–1160 (2009). 10.3758/BRM.41.4.1149 [DOI] [PubMed] [Google Scholar]
- 64.Reingold E. M., Sheridan H., “Eye movements and visual expertise in chess and medicine,” in Oxford Handbook on Eye Movements, pp. 528–550, Oxford University Press Oxford, England: (2011). [Google Scholar]
- 65.Bertram R., et al. , “The effect of expertise on eye movement behaviour in medical image perception,” PloS One 8(6), e66169 (2013). 10.1371/journal.pone.0066169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manning D., et al. , “How do radiologists do it? The influence of experience and training on searching for chest nodules,” Radiography 12(2), 134–142 (2006). 10.1016/j.radi.2005.02.003 [DOI] [Google Scholar]
- 67.Drew T., et al. , “Scanners and drillers: characterizing expert visual search through volumetric images,” J. Vision 13(10), 3–3 (2013). 10.1167/13.10.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kocak E., et al. , “Eye motion parameters correlate with level of experience in video-assisted surgery: objective testing of three tasks,” J. Laparoendoscopic Adv. Surg. Techn. 15(6), 575–580 (2005). 10.1089/lap.2005.15.575 [DOI] [PubMed] [Google Scholar]
- 69.Kundel H. L., Nodine C. F., “Interpreting chest radiographs without visual search 1,” Radiology 116(3), 527–532 (1975). 10.1148/116.3.527 [DOI] [PubMed] [Google Scholar]
- 70.Krupinski E. A., “Current perspectives in medical image perception,” Attention Percept. Psychophys. 72(5), 1205–1217 (2010). 10.3758/APP.72.5.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]