Abstract
The endoplasmic reticulum (ER) serves as a warehouse for factors that augment and control the biogenesis of nascent proteins entering the secretory pathway. In turn, this compartment also harbors the machinery that responds to the presence of misfolded proteins by targeting them for proteolysis via a process known as ER-associated degradation (ERAD). During ERAD, substrates are selected, modified with ubiquitin, removed from the ER, and then degraded by the cytoplasmic 26S proteasome. While integral membrane proteins can directly access the ubiquitination machinery that resides in the cytoplasm or on the cytoplasmic face of the ER membrane, soluble ERAD substrates within the lumen must be retrotranslocated from this compartment. In either case, nearly all ERAD substrates are tagged with a polyubiquitin chain, a modification that represents a commitment step to degrade aberrant proteins. However, increasing evidence indicates that the polyubiquitin chain on ERAD substrates can be further modified, serves to recruit ERAD-requiring factors, and may regulate the ERAD machinery. Amino acid side chains other than lysine on ERAD substrates can also be modified with ubiquitin, and post-translational modifications that affect substrate ubiquitination have been observed. Here, we summarize these data and provide an overview of questions driving this field of research.
Introduction
Proteins exhibit a wide variety of structural and chemical features, which are essential for their function. To attain these features, cotranslational and post-translational modifications occur, along with protein folding. Protein heterogeneity in the cell is made even more complex because some proteins are transported into intracellular organelles. The unique chemical environments within these organelles are often encountered concomitant with cotranslational and post-translational events. For example, the endoplasmic reticulum (ER) receives approximately one-third of all newly synthesized proteins in eukaryotes [1]. Not only do these substrates traverse or become embedded within a lipid bilayer, but also they encounter a more oxidizing and calcium-rich environment compared to the cytoplasm [2–4]. Moreover, the machinery to catalyze specific post-translational modifications resides in the ER. Therefore, proteins translocated into the ER may acquire N-linked glycans, lipid appendages, and/or disulfide bonds [5–9]. In the absence of these modifications, protein folding in the ER is abrogated.
The protein-folding ‘problem’ is made even worse by the fact that nascent cotranslationally inserted polypeptide chains enter the ER in an N- to C-terminal fashion [10, 11]. Should folding require more C-terminal residues, the folding of N-terminal portions is delayed [12, 13]. In addition, membrane proteins must fold in three unique environments: the ER lumen, the lipid bilayer, and the cytoplasm. Owing to the fact that the native and unfolded states of many proteins are differentiated by only a few kCals per mole [14, 15], and that genetic mutations or stochastic errors in amino acid incorporation might take place, protein folding is quite error-prone. If uncorrected, proteins may aggregate in the ER, leading to compromised cellular and organelle homeostasis [16–20].
Fortunately, the ER is replete with molecular chaperones and enzymes that directly facilitate protein folding [21, 22]. Molecular chaperones capture unfolded polypeptides by virtue of their ability to bind amino acid patches containing exposed hydrophobic side chains [23–26]. Moreover, eukaryotes have evolved two systems to temper the potentially toxic effects of misfolded proteins. First, the unfolded protein response (UPR) may be induced, which leads to: (1) the induction of factors that increase the protein-folding capacity of the ER; (2) expanded ER volume; and (3) the transport of unfolded proteins to other compartments, such as the vacuole/ lysosome in which they may be degraded [27–29]. Second, an ER-resident protein ensemble directly selects, exports, and degrades misfolded proteins in the ER. This second pathway is known as ER-associated degradation (ERAD), and components of the ERAD machinery are also induced by the UPR [30–34]. Together, the UPR and ERAD constitute two complementary legs of the ER quality control apparatus. However, growing evidence indicates that ERAD also targets properly folded proteins in order to regulate: (1) metabolically controlled enzymes, (2) transcription factor activity, and (3) the amount and thus activity of a plasma membrane metal transporter [35–40].
ERAD substrates are selected by molecular chaperones and by chaperone-like lectins. Once selected, soluble substrates that completely reside within the ER must be retrotranslocated, so that at least a portion of the protein becomes exposed to the cytoplasm. Here, the substrate is modified with the 76 amino acid peptide, ubiquitin. Ubiquitin modification of proteins can be important for protein trafficking decisions (for review, see ref [41]) and as shown recently for the folding and ER exit of a membrane protein [42]. During ERAD, the subsequent acquisition of a polyubiquitin chain helps recruit an ATP-dependent ‘engine’, known as p97 in mammalian cells or Cdc48 in yeast, which extracts ERAD substrates from the ER [43–45]. The cytoplasmic portion(s) of misfolded integral membrane proteins are also modified with ubiquitin. After or concordant with the complete retrotranslocation of the selected and modified soluble or integral membrane proteins, ERAD substrates are degraded by the 26S proteasome [46–49]. The proteasome is a multicatalytic protease that recognizes polyubiquitinated proteins, which leads to the unfolding and spooling of captured substrates into a chamber in which three proteolytic activities (tryptic, chymotryptic, and caspase-like) reside [50–52]. Because a subpopulation of proteasomes associates with the ER membrane [53], retrotranslocated ERAD substrates are efficiently degraded.
In this review, we will discuss the step in the ERAD pathway that represents an important decision-making point: the acquisition of a polyubiquitin tag. We will focus on the link between ubiquitination and ERAD by describing many of the initial findings that established which key factors perform this crucial function. We will also discuss how the field has expanded since those initial discoveries, along with some of the variations on the protein ubiquitination theme that have been identified. Finally, we will briefly introduce what we believe are critical questions in future research on the ubiquitination of ERAD substrates.
The ubiquitination pathway
The ubiquitination of a misfolded protein can represent a rate-limiting step during ERAD. But, before the substrate can be ubiquitinated, cells utilize an E1 ubiquitin-activating enzyme that hydrolyzes ATP to create a thiol-ester bond with the C-terminal carboxylate in ubiquitin (Figure 1) [54–56]. After activation, an E2 ubiquitin-conjugating enzyme is utilized to transfer ubiquitin to a substrate that may be bound to an E3-ubiquitin ligase. In other cases, the substrate may be linked to the E3 by a molecular chaperone (see below). Currently, only 1 E1 ubiquitin-activating enzyme and 11 E2 ubiquitin-conjugating enzymes have been identified in yeast, whereas there are 2 E1 ubiquitin-activating enzymes and 35 E2 ubiquitin-conjugating enzymes that have been identified in humans [57, 58]. In contrast, there are roughly 80 and 600 putative E3s in yeast and humans [59].
Figure 1. The ubiquitination pathway.
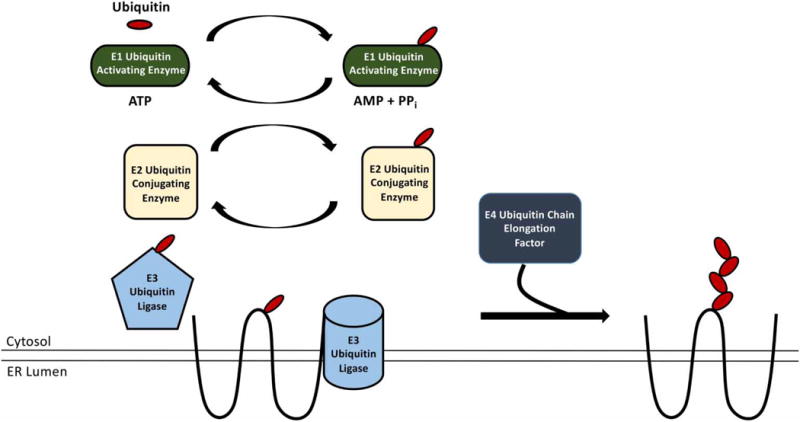
The cytosolic E1 ubiquitin-activating enzyme hydrolyzes ATP to activate the ubiquitin molecule. ATP hydrolysis and the formation of a transient AMP-derivative lead to the formation of a thioester bond between the E1 and the C-terminus of ubiquitin. The E1 then transfers ubiquitin to one of the ~11 yeast or the ~35 mammalian E2 ubiquitin-conjugating enzymes. The covalently bound ubiquitin—E2 adduct then binds one of the ~80 yeast or the ~300 mammalian E3 ubiquitin ligases. The E3 enzymes may also be bound to the ERAD substrate and facilitate transfer of ubiquitin to the substrate, or a chaperone intermediate (not shown) may facilitate transfer. Some E3 ubiquitin ligases (e.g. HECT domain E3s) become covalently modified with ubiquitin during ERAD substrate modification, while other E3s (e.g. RING and U-box domain proteins) facilitate the transfer of ubiquitin from the E2 ubiquitin-conjugating enzyme to the substrate. Importantly, select E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases are cytoplasmic, while others reside at the ER membrane. Once a substrate is ubiquitinated by an E3 ubiquitin ligase, other enzymes, such as E4s, may further extend the ubiquitin chain on the ERAD substrate.
Among these many ligases, three classes of E3s function in ERAD: (1) RING domain, (2) HECT domain, and (3) U-box domain E3s. RING domain and U-box domain E3s facilitate the transfer of ubiquitin from the E2 ubiquitin-conjugating enzyme to the substrate, whereas HECT domain E3s are directly ubiquitinated before the ubiquitin is transferred [60–64]. Once the substrate becomes mono-ubiquitinated, a ubiquitin chain can be synthesized and elongated. The most prominent Lys in ubiquitin that is elongated in this manner and used to target substrates for degradation is Lys-48 [56, 65–67]. However, substrate ubiquitination via Lys-11-derived isopeptide linkages are also recognized by the proteasome [68]. A more recently discovered class of components of the ubiquitination machinery are the ubiquitin chain elongation factors, called E4s [69]. As the name implies, the activity of the E4 elongates the polyubiquitin chain in order to more effectively recruit factors that facilitate substrate degradation and expedite proteasomal degradation [70–72].
After an ERAD substrate has been adequately ubiquitinated (Figures 2 and 3), the p97/Cdc48 complex is recruited to the substrate by its cofactors, Ufd1, Npl4, and in some cases Ubx2 (UBXD8 in mammalian cells) [73, 74]. Interestingly, the viral protein, US11, which promotes the ERAD of specific substrates (see below), interacts with p97 in a ubiquitin-independent manner in mammalian cells [75]. However, most p97/Cdc48 substrates are ubiquitinated. The recruitment of the p97/Cdc48 complex leads to substrate ‘retrotranslocation’ (or ‘dislocation’ for membrane proteins) from the ER and into the cytosol, where it is bound and shuttled to the 26S proteasome by delivery factors, such as Rad23 and Dsk2, in yeast [76]. Interaction of Rad23 with the Cdc48–Ufd1–Npl4 complex is mediated by another Cdc48 cofactor, the E4 Ufd2 [72]. Once Rad23 and Dsk2 bind the Cdc48 complex, they link the complex to the proteasome through a component that resides on the proteasome ‘cap’ (also known as the 19S particle or PA700), Rpn1 [77]. In addition to binding by Rad23–Dsk2–Rpn1, another component of the 19S cap, called Rpn10, can bind ubiquitin in an Rad23-independent manner [78]. After substrate binding to the 19S particle of the proteasome, the proteasome-associated deubiquitinating enzymes (DUBs), Ubp6 and Rpn11, cleave the ubiquitin chain from the substrate, and the substrate is subsequently threaded into the 20S core particle and degraded [79–83]. Entry into the 20S particle requires the activity of six AAA-ATPases that drive substrate entry into the core and facilitate the opening of an aperture that otherwise retains the 20S particle in a closed state [84, 85].
Figure 2. Function of the Hrd1 complex during ERAD in yeast.
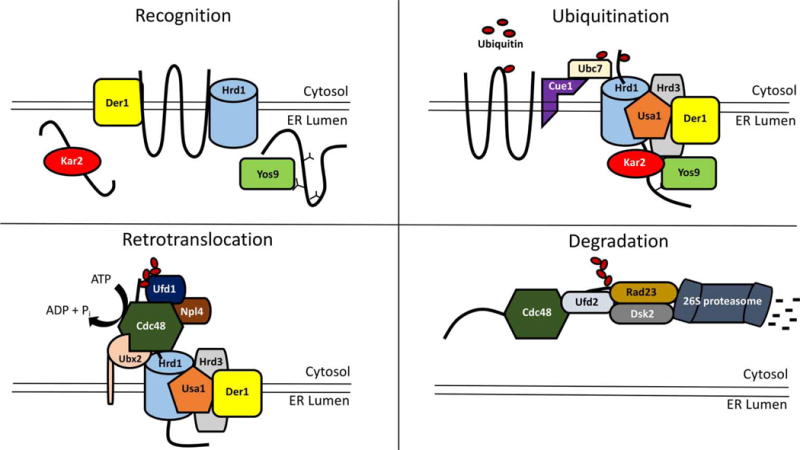
In the first step during ERAD (‘Recognition’), a misfolded substrate is recognized by a subset of factors, namely Kar2 (ER luminal Hsp70 chaperone), Yos9 (ER luminal lectin), Der1 (transmembrane core Hrd1 complex member), or directly by the E3, Hrd1. Once the substrate has been recognized, the substrate is transferred to the Hrd1 complex for polyubiquitination. Kar2 and Yos9 bind to the Hrd1 core complex member, Hrd3, and the substrate is transferred to Hrd1 (‘Ubiquitination’). Der1 is bound by Usa1, which helps link Der1 to Hrd1. Cue1 is an ER membrane protein that recruits the E2 ubiquitin-conjugating enzyme, Ubc7, to the Hrd1 complex. After the substrate is polyubiquitinated, the dislocation machinery is linked to the complex. This dislocation complex consists of the membrane protein, Ubx2, which helps recruit the AAA+ ATPase, Cdc48. Cdc48 is stabilized at the Hrd1 complex through an interaction with Hrd1 and through an interaction with the Cdc48 cofactors, Ufd1 and Npl4, with the polyubiquitin chain. Assembly is believed to be due to Ufd1 binding to the polyubiquitin chain, as yeast Npl4 lacks a zinc finger domain. Another class of cofactors that bind Cdc48 and affect retrotranslocation includes DUBS, such as Otu1 (not shown here). Once bound to the substrate, Cdc48 hydrolyzes ATP and liberates the substrate from the ER (‘Retrotranslocation’). The Cdc48 cofactor, Ufd2, then extends the polyubiquitin chain and interacts with the ubiquitinated protein shuttles, Rad23 and Dsk2 (‘Degradation’). Although not shown in this figure, ubiquitin chains may be trimmed by DUBS prior to substrate passage through Cdc48 and then extended again by Ufd2. Rad23 and Dsk2 can also interact with the 19S cap of the cytosolic 26S proteasome, which leads to substrate degradation. Another DUB associated with the 19S cap of the proteasome, called Rpn11 (not shown), removes the polyubiquitin chain attached to the ERAD substrate, so it efficiently threads into the core of the 26S proteasome for degradation.
Figure 3. Function of the Hrd1 complex during ERAD in mammalian cells.
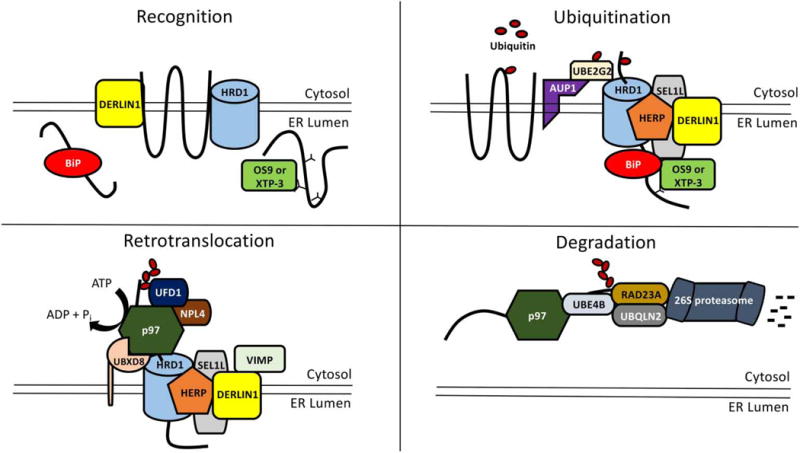
In the first step during ERAD (‘Recognition’), a substrate is recognized by a group of luminal and membrane-associated factors, namely BiP (Hsp70 chaperone), OS9 and XTP-3 (lectins), Derlin1 (transmembrane core Hrd1 complex member), and/or HRD1. While OS9 and XTP-3 recognize misfolded ER luminal glycosylated substrates, they may also recognize nonglycosylated substrates. The substrate is then transferred to the HRD1 complex and polyubiquitinated. BiP, OS9, and XTP-3 bind the HRD1 core complex member, SEL1L, and the substrate is next transferred to HRD1 (‘Ubiquitination’). DERLIN1 is bound by HERP1, which helps link Derlin1 to HRD1. The mammalian HRD1 complex can utilize DERLIN2 and DERLIN3 as well. AUP1 is a Cue domain-containing ER membrane protein that recruits the E2 ubiquitin-conjugating enzyme, UBE2G2, to the HRD1 complex. Once polyubiquitinated, the dislocation machinery is recruited to the HRD1 complex. This complex includes the membrane protein, UBXD8, which augments p97 recruitment to the HRD1 complex. Along with UBXD8, the HRD1 complex can utilize another protein, called UBXD2, to recruit p97. p97 is further stabilized at the HRD1 complex through interaction with HRD1 along with the p97 cofactors, UFD1 and NPL4, via the polyubiquitin chain. In an alternative mechanism of p97 recruitment, the cytoplasmic protein, VIMP, binds p97 (not shown) at the HRD1 complex through a VIMP interaction domain in DERLIN1. Another class of cofactors that bind p97 and act during ERAD include DUBS, such as YOD1 (not shown). Once bound to the substrate, p97 hydrolyzes ATP and removes the substrate from the ER (‘Retrotranslocation’). After retrotranslocation, the p97 cofactor and a mammalian homolog of Ufd2, UBE4A/B, may extend the polyubiquitin chain and associate with the protein shuttles, RAD23A, and a specific UBIQUILIN, UBQLN2 (‘Degradation’). Ubiquitin chains may be trimmed by DUBS prior to substrate transit through p97 and could then be extended again by UBE4A/B. RAD23A and UBIQUILIN also interact with the 19S cap of the cytosolic 26S proteasome, which facilitates substrate degradation. In addition, there is a DUB associated with the 19S cap of the proteasome, RPN11, which removes the polyubiquitin chain attached to the ERAD substrate, which aids efficient entry of the substrate into the 26S proteasome core.
The first link between the ubiquitin pathway and ERAD
Ubiquitination was first identified as an ATP-dependent process important for protein degradation in the late 1970s/early 1980s [86, 87]. However, it was not until the 1990s that a component of the ubiquitination machinery was discovered at the ER [88]. This factor is an ER membrane-resident E2 ubiquitin-conjugating enzyme, Ubc6. It was shown that deleting UBC6 leads to the rescue of a sec61 mutant allele that is defective for protein translocation, or entry into the ER. In the absence of Ubc6, the mutant Sec61 channel has a longer half-life, which allows for partial rescue of protein translocation.
Other early discoveries led to the identification of proteins in the ER that were ubiquitinated. At least initially these were substrates that trafficked through the secretory pathway, but were retained in this compartment due to errors in folding or maturation. One of the most notable substrates identified was the cystic fibrosis transmembrane conductance regulator (CFTR), whose failure to properly fold and mature due to rapid degradation is the cause of cystic fibrosis (CF) [47, 49]. Degradation of both immature forms of the wild-type and the ΔF508 mutant form of CFTR, which accounts for the majority of CF cases, was shown to be dependent on both the E1 ubiquitin-activating enzyme and the 26S proteasome for degradation. CFTR and ΔF508 were also directly shown to be ubiquitinated.
Substrates used to identify additional components of the ubiquitination machinery in the yeast ER included a mutated form of a vacuole-targeted protein, carboxypeptidase yscY, which was subsequently termed CPY* [46]. Analyses of CPY* degradation uncovered the importance of an E2 ubiquitin-conjugating enzyme, Ubc7, as well as the proteasome in removing this trapped protein from the ER [46, 89, 90]. In parallel, an enzyme that catalyzes the rate-limiting step in cholesterol synthesis, known as hydroxymethylglutaryl coenzyme A reductase, was known to be metabolically regulated and degraded in the ER [91, 92]. By examining the genetic requirements for the degradation of the yeast homolog, Hmg2, three HRD genes that are important for the degradation of Hmg2 at the ER were identified [38]. Two of these HRD genes, HRD1 and HRD3, are vital for the ubiquitination and degradation of many other ERAD substrates (Table 1) [93–95]. Hrd1 is one of the major ER membrane-resident E3s involved in ERAD in yeast and is the central component of the Hrd1 complex, which is conserved between yeast and mammals (see below). Hrd3 is an ER membrane-resident protein that interacts with Hrd1. When HRD3 is deleted, Hrd1 degradation increases, leading to a loss of function of the Hrd1 complex [94, 96, 97]. Finally, the major histocompatability complex class I (MHCI) protein was utilized to identify a novel mechanism of immune evasion by human cytomegalovirus (HCMV). A virally encoded, ER-localized protein, US11, triggered nascent MHCI heavy chain transport from the ER into the cytosol, where it is acted upon by a cytosolic N-glycanase, ubiquitinated, and subsequently, degraded by the 26S proteasome [98, 99]. Together, these substrates have proved essential for continued analysis of the factors important for ubiquitination and degradation during ERAD.
Table 1.
List of cellular factors shown to affect the ubiquitination of ERAD substrates.
| Factors | Select substrates | Notes | References | |
|---|---|---|---|---|
| E1 ubiquitin-activating enzymes1 | ||||
| Yeast | ||||
| Uba1 | [262] | |||
| Mammalian | ||||
| E1a, E1b | Isoforms resulting from alternative splicing | [263–265] | ||
| E2 ubiquitin-conjugating enzymes2 | ||||
| Yeast | ||||
| Ubc1 | CPY*, Hmg2 | Cytosolic E2 that is recruited to the ER membrane by Hrd1 | [93, 113] | |
| Ubc6 | Sss1, sec61, Deg1–Ura3, Pma1–D378N, Ste6* | Integral ER membrane E2 | [89, 148, 239, 266, 267] | |
| Ubc7 | Hmg2, CPY*, SSS1, sec61, Deg1–Ura3, Pma1–D378N, Ste6* | Cytosolic E2 that is recruited to the ER membrane by Cue1 | [46, 89, 90, 93, 95, 148, 239, 266–268] | |
| Ubc2 | Ste6* | Cytosolic E2 only shown to be important with Ubr1 | [171] | |
| Mammalian | ||||
| UBE2D1 | CFTR, p53, APP | Homolog of yeast Ubc4/5 | [161, 265, 269] | |
| UBE2J1 | MHCI, CFTR, TCRα, OS-9, EDEM, SEL1L | Homolog of yeast Ubc6 | [135, 138, 170, 270] | |
| UBE2K | MHCI | Homolog of yeast Ubc1, used by viral protein US11 | [271] | |
| UBE2G2 | HERP, gp78, CD3-δ, TCRα, InsP3R | Homolog of yeast Ubc7 | [137, 272–274] | |
| E3 ubiquitin ligases | ||||
| Yeast | ||||
| Hrd1 | CPY*, sec61-2, Hmg2, pdr5–C1427Y, Huntingtin, unglycosylated PrP | Integral ER membrane E3 | [38, 93, 100, 275–278] | |
| Doa10 | Deg1-Ura3, Ubc6, Ste6*, Erg1, α, β, γ subunits of ENaC, Pca1, Zrt1, Sbh2 | Integral ER membrane E3 | [147, 148, 152, 156, 239, 279, 280] | |
| Rsp5 | CPY* | Cytosolic E3 required only when substrates are significantly overexpressed and cells are under oxidative stress | [173, 174] | |
| Ubr1 | Ste6*, CFTR | Cytoplasmic E3 | [171] | |
| Asi1/Asi1/ Asi3 | Erg11 | Integral nuclear membrane-localized E3 complex | [175, 176] | |
| Mammalian | ||||
| HRD13 | HMGR, TCR-α, CD3-δ, MHCI, NHK, APP, α1AT, gp78, NS1 LC, CD95/Fas, p53, CD147 | Homolog of yeast Hrd1 | [36, 121, 123, 138, 219, 265, 269, 281–285] | |
| TEB4 | Type 2 iodothyronine deiodinase (D2), squalene monooxygenase (SM), HMGR | Homolog of yeast Doa10 | [154, 156–158] | |
| gp78 | CD3-δ, apoB100, CFTR, HMGR, α1AT, Kai1, Cytochrome P450 3A | Homolog of yeast Hrd1 | [137, 164, 165, 286–290] | |
| CHIP | CFTR, NCC, Pael-R, cytochrome P450 3A | Cytoplasmic E3 | [161, 168, 169, 290–292] | |
| RMA1/ RNF5 | CFTR | Integral ER membrane-resident E3 | [163, 165, 170] | |
| TRC8 | MHCI, SREBP-1, SREBP-2, Heme oxygenase-1, XBP1u, α1 integrin, α2 integrin, α4 integrin, β1 integrin, thrombomodulin, PTPRJ and IL-12 receptor β1 | Integral ER membrane-resident E3 | [37, 293–296] | |
| SCFFbx2 | Pre-integrin β1, TCR-α, CFTR | Cytoplasmic/ER membrane-associated E3 | [180, 221] | |
| SCFFbx6 | TCR-α | Cytoplasmic/ER membrane-associated E3 | [181] | |
| SCFβ-TrCP | CD4, Tetherin | Cytoplasmic E3 utilized by HIV protein, Vpu | [177, 178] | |
| RNF4 | CFTR | Cytoplasmic/nuclear E3 that recognizes poly-SUMOylated substrates | [227] | |
| RNF103 | Autoubiquitination | Integral ER membrane-resident E3 | [184] | |
| RNF170 | Inositol 1,4,5-trisphosphate (IP3) | Integral ER membrane-resident E3 | [297] | |
| RNF185 | CFTR | Integral ER membrane-resident E3 that is homologous to RMA1/RNF5 | [298] | |
| PARKIN | Pael-R, mutant glucocerebrosidase | Cytoplasmic E3 | [168, 299] | |
| TRIM13 | CD3-δ, Cav1.2 | Integral ER membrane-resident E3 that also plays a role in autophagy | [300, 301] | |
| SMURF1 | WFS1 | Cytoplasmic/nuclear-localized E3 | [302] | |
| TMEM129 | MHCI | Integral ER membrane-resident E3 utilized by HCMV | [179] | |
| NIXIN/ ZNRF4 | Calnexin | Integral ER membrane-resident E3 | [303] | |
| NRDP1 | ErbB3 | Cytoplasmic E3 | [304] | |
| SYVN1 | CFTR | ER/cytoplasmic E3 | [221] | |
| Ubiquitination modifiers | ||||
| Yeast | ||||
| Cue1 | CPY*, Hmg2, KWW | Required for Ubc7 activation and integral ER membrane protein | [101, 111, 112, 305] | |
| Hrd3 | Hmg2, CPY*, sec61-2, Pdr5*, unglycosylated PrP | Integral membrane component of Hrd1 complex | [38, 94, 105, 106, 277] | |
| Ssa1 | Ste6*, Pma1–D378S, CFTR | Cytosolic Hsp70 | [71, 150, 152, 153] | |
| Ydj1 | Ste6*, Pma1–D378S | Cytosolic Hsp40 that can be farnesylated | [71, 152, 153] | |
| Hlj1 | Ste6*, CFTR | Homolog of Ydj1 | [71, 151] | |
| Otu14,5 | CPY*, Spt23 | Cytosolic deubiquitinating enzyme | [119, 192, 306] | |
| Ufd34 | Spt23 | Cytosolic ubiquitin chain length regulator | [192] | |
| Ufd24 | Ole1, HMG2, DEG1Sec62 | Cytosolic ubiquitin extension enzyme (E4) | [69, 72] | |
| Mammalian | ||||
| HDJ2 | CFTR, Pael-R | Homolog of yeast Ydj1 | [159, 161, 168] | |
| SEL1L | NHK, TTRD18G, RI332, tyrosinase | Homolog of yeast Hrd3 | [123, 134, 135] | |
| FAM8A1 | NHK, TTRD18G | Hrd1-binding partner believed to regulate Hrd1 function | [134] | |
| HSC70 | CFTR, Pael-R, ApoB100, ENaC, NCC | Homolog of yeast Ssa1 | [160, 161, 168, 240, 241, 307] | |
| DNAJB12 | CFTR | Integral ER membrane-resident Hsp40 | [162, 163] | |
| USP134 | Ubl4A | Deubiquitinating enzyme | [204, 205] | |
| USP19 | CFTR, TCRα | Integral ER membrane-resident deubiquitinating enzyme | [206] | |
| USP25 | CD3-δ, APP, CFTR | Integral ER membrane-resident deubiquitinating enzyme | [208] | |
| AUP1 | MHCI, NHK, HMG CoA | Integral ER-membrane Cue domain-containing protein that recruits Ube2g2 to the ER membrane and lipid droplets | [128, 308] | |
| YOD14,5,6 | RI332, NHK, TCRα | Homolog of yeast Otu1 | [191] | |
| ATAXIN-34 | TCRα, CD3-δ, BACE457Δ | Deubiquitinating enzyme | [201, 202, 204] | |
| UBE4B4 | Homolog of yeast Ufd2 | [69, 72] | ||
| NEDD8 | CFTR | Cytoplasmic ubiquitin-like protein | [221] | |
Substrates for the E1 are the E2s.
Substrates for the E2s are the E3s.
These components can recognize and bind to the Cdc48/p97 complex in the cytosol.
May trim ubiquitin chains prior to entry into p97/Cdc48.
May also act on ERAD machinery [310].
Expanding the links between ubiquitination and ERAD
Identification of contributing E3s and their associated complexes
While these initial breakthroughs were crucial to better understand the importance of protein ubiquitination in ERAD, the years that followed led to the identification of additional factors, complexes, and substrates associated with this process (Table 1). In yeast, there are two primary E3s, Hrd1 and Doa10, that are required for ubiquitination and degradation; however, there are currently eight E3s that play some role during the ubiquitination of ERAD substrates in yeast. In mammals, there are four E3s, Hrd1, TEB4, gp78, and CHIP, that are associated with the degradation of several notable ERAD substrates, but ~19 E3s have been linked more generally to ERAD (Table 1). The expanded number of E3s involved in ERAD in mammals when compared with yeast is most probably due to the greater number of potential substrates (e.g. transmembrane proteins) in mammalian cells.
While Hrd1 was the first E3 linked to the ERAD pathway, it was appreciated later that the Hrd1 complex (Figure 2) ubiquitinates substrates that possess misfolded regions in their ER luminal (ERAD-L) or membrane-spanning domains (ERAD-M) [38, 93, 95, 100, 101]. In yeast, the Hrd1 complex consists of Hrd3, Usa1, Der1, Dfm1, Yos9, Kar2, Ubc7, and Cue1 [97, 102]. While these factors are all known to be a part of the Hrd1 complex, not all of them are considered components of the core Hrd1 complex. When the complex was purified, Hrd3, Usa1, Der1, and Yos9 copurified, thus defining components of the core complex [102]. As noted previously, Hrd3 stabilizes the Hrd1 oligomer, but also binds glycosylated misfolded luminal substrates by interacting with a luminal lectin, Yos9, as well as nonglycosylated misfolded substrates via an Hsp70 molecular chaperone, Kar2 [94, 103–105]. Until recently, it was difficult to study the role of Hrd3 in ERAD due to the increased turnover of Hrd1 when HRD3 is deleted. However, when the ubiquitin-like domain of Usa1 is removed, Hrd1 remains stable and a direct role for Hrd3 in ERAD was established [106]. Der1 also binds soluble luminal substrates and interacts with Hrd1 through Usa1 [107, 108]. In addition, a Der1 homolog, Dfm1, interacts with Hrd1 and the E3 ligase, Doa10, and helps degrade a Doa10 substrate, Ste6* [73, 109]. In a mechanism believed to be independent of ERAD, Dfm1 also interacts with Cdc48 in the absence of Ubx2 [109, 110]. As the Hrd1 complex engages substrates, Ubc7 is recruited to the ER membrane by a transmembrane protein, Cue1 [111, 112]. In addition to Ubc7, another E2, Ubc1, ubiquitinates select ERAD substrates [93, 113]. Hrd1 also recruits the ER membrane protein, Ubx2, to the complex, which anchors Cdc48 to the membrane [73, 74]. Ubx2 anchoring facilitates efficient substrate retrotranslocation. Some evidence suggests that Der1 and the mammalian homolog (also see below) may act as the retrotranslocation channel for substrates, or at least are intimately associated with the retrotranslocation process [114–118], while other evidence suggests that Hrd1 could serve as the channel [119, 120]. In either case, this would position the ubiquitination machinery near the site of retrotranslocation.
Mammalian homologs for many of the components discussed above were subsequently identified (Figure 3). The mammalian homolog of Hrd1, also called HRD1 or synoviolin [121, 122], is important to ubiquitinate several ERAD substrates (Table 1). Other mammalian homologs of yeast Hrd1 complex members have been identified as well: SEL1L (Hrd3), HERP (Usa1), DERLIN-1, DERLIN-2, DERLIN-3 (Der1), OS-9 and XTP3-B (Yos9), BiP (Kar2), UBE2G2 (Ubc7), and Aup1 (Cue1) [102, 116, 123–129].
Perhaps not surprisingly, mammalian HRD1 complex members have also been implicated in the ubiquitination of ERAD substrates (Table 1). As in yeast, HRD1 functions in the degradation of both glycosylated (utilizing EDEM1, DERLIN-2, DERLIN-3, OS-9, XTP3-B, and SEL1L) and nonglycosylated substrates (utilizing BiP, HERP, and DERLIN-1) [30, 130–133]. While HRD1 activity for glycosylated substrates is generally thought to function in a SEL1L-dependent manner [123, 127], another HRD1 regulator, the ER membrane-resident protein FAM8A1, binds and regulates HRD1 function in an SEL1L-independent manner [134]. After substrate association with the HRD1 complex, AUP1, which contains a Cue domain homologous to that found in the yeast Cue1 protein, binds the complex and recruits UBE2G2 through its G2BR domain [127, 128, 134–137]. For some substrates, HRD1 also utilizes the Ubc6 homolog, UBE2J1 [138]. Which of these E2 ubiquitin-conjugating enzymes is the predominant enzyme for HRD1 is an open question. Mammalian HRD1 also binds to other factors that play a role in ERAD. For example, Hrd1 associates with UBXD2 and UBXD8, which function similarly to yeast Ubx2 in anchoring p97 to the ER membrane. Furthermore, UBXD2 recruits the mammalian Dsk2 homolog, called UBIQUILIN [127, 139–142], to the Hrd1 complex, whereas UBXD8 recruits a cytosolic chaperone, BAG6 [143, 144]. Recently, UBIQUILIN and BAG6 were shown to function as chaperones as well, suggesting that they play an important role as holdases for dislocated proteins prior to degradation [143,145]. Yet, another factor that binds the HRD1 complex and recruits p97 is VIMP (VCP-interacting membrane protein) [114]. VIMP localizes to the HRD1 complex via association with DERLIN-1 and subsequently binds p97. HRD1 and DERLIN-1 are also capable of binding p97, suggesting that they contribute to the maintenance of p97 residence at the ER membrane [115, 146].
The other ERAD-associated E3 complex in yeast centers around the nuclear membrane/ER membrane-localized E3, Doa10 [147]. While Hrd1 functions in collaboration with a variety of other proteins, Doa10 acts predominantly with three different components in a ubiquitination complex: Ubc6, Ubc7, and Cue1 [102, 147, 148]. Recent data suggest that Ubc6 initially monoubiquitinates the substrate, whereas Ubc7 creates the polyubiquitin chain by adding subsequent ubiquitin moieties [149]. In contrast with Hrd1, which ubiquitinates substrates containing misfolded lesions in the ER lumen and ER membrane, Doa10 substrates contain a cytosolic lesion (ERAD-C) [101]. As a result, cytosolic chaperones, such as an Hsp70, Ssa1, and the cytosolic Hsp40s, Ydj1 and Hlj1, are also important for the degradation of Doa10-dependent substrates [71,150–153]. Like Hrd1, Doa10 recruits Ubx2 to the complex, allowing for increased efficiency of substrate retrotranslocation by Cdc48 [73, 74]. In the mammalian system, TEB4/MARCH-VI [154, 155] is the Doa10 homolog [154, 156–158]. TEB4/MARCH-VI acts in a complex with the E2 ubiquitin-conjugating enzyme, UBE2G1 [154]. Similar to the situation observed in yeast, cytosolic chaperones, such as HSC70, DNAJB12, and HDJ2 [159–163], can help degrade some ERAD substrates and UBXD8 can aid in substrate retrotranslocation.
The mammalian E3, gp78 [164], is another major E3 ligase for ERAD substrates in mammalian cells. gp78 is an ER membrane protein that is similar to HRD1 and utilizes UBE2G2. This interaction is through an embedded G2BR domain [137]. While gp78 clearly functions as an E3 during ERAD, there is also evidence that it acts as an E4 in co-operation with other E3s [165]. Additional evidence suggests that gp78 participates in substrate retrotranslocation downstream of HRD1 [166]. The diversity in gp78 function may reflect substrate specificity and partner specificity, which represent an important area of future study. Regardless, similar to HRD1 and TEB4/MARCH-VI, gp78 also recruits UBXD2 or UBXD8 to help cement p97 at the ER membrane. Alternatively, gp78 can directly interact with p97 through its VIM (VCP-interacting motif) domain [115, 167].
One of the earliest ERAD-requiring E3s identified in mammalian cells was the cytosolic HSC70-interacting protein, CHIP. While several ERAD substrates were identified that require CHIP activity, the best-studied substrate is CFTR [161]. Interestingly, CHIP requires HSC70 as a cofactor for function, but CHIP can also act in collaboration with other E3s, such as PARKIN [161, 168, 169]. This may reflect the ability of CHIP to act cooperatively, or it may reflect an E4-type activity. For example, while RMA1 and CHIP both facilitate mutant CFTR degradation, the dependence on each E3 was linked to the subdomain in which the mutation resided [170]: RMA1 recognizes mutations early in the protein sequence and in the transmembrane domains of CFTR, whereas CHIP is believed to recognize cytoplasmic domains and the C-terminal second nucleotide-binding domain in CFTR.
Several additional E3 ubiquitin ligases have been identified that act on ERAD substrates in both yeast and mammalian cells (Table 1). Many of these enzymes are required for the degradation of only a few substrates, yet this might again reflect the fact that the universe of identified and characterized ERAD substrates is relatively small (see Future Work). In yeast, the cytosolic E3, Ubr1, is required along with Hrd1 and Doa10 during the degradation of Ste6* and CFTR, and employs the cytosolic E2, Ubc2 [171]. However, Ubr1 is better known for its role in degrading unstable N-end rule substrates [172]. In addition, Rsp5, a cytosolic/Golgi-resident E3, ubiquitinates overexpressed CPY* and other select substrates when cells are exposed to oxidative stress [173, 174]. Finally, in yeast, the nuclear membrane E3 complex, consisting of Asi1, Asi2, and Asi3, plays a role in the destruction of Erg11, a sterol synthesis component that resides in the ER [175, 176].
Roles for specialized E3s in ERAD have also been studied in mammalian cells. Some of these enzymes are important only under select conditions. For instance, SCFβ-TrCP and TMEM129 are utilized by viruses to ubiquitinate ER-localized factors associated with the immune system [177–179]. Other ligases recognize specific protein classes: the cytosolic E3s SCFFbx2 and SCFFbx6 target glycosylated ERAD substrates in the cytosol and modify them prior to proteasomal degradation [180, 181]. It is unknown if these E3s recognize retrotranslocated, glycosylated ER-resident proteins that have been missed by the cytoplasmic glycosidase, PNGase, which normally removes ER-catalyzed N-glycans prior to ERAD [182, 183]. Other E3s connected to the ERAD pathway include TRC8, RNF170, RNF185, TRIM13, SMURF1, NIXIN/ZNRF4, and NRDP1 (Table 1). One E3, RNF103, is an ER-resident ligase that has yet to be linked to substrate degradation. However, the factor regulates protein ubiquitination levels, interacts with p97 and DERLIN-1, and is autoubiquitinated [184]. Once again, this may reflect the dearth of characterized, potential ERAD substrates that can arise due to errors in secretory pathway folding.
The importance of ubiquitin chain extension (E4) and DUB enzymes during ERAD
For maximal binding to the 26S proteasome, a substrate needs a polyubiquitin chain of at least four ubiquitin molecules, but increasing the length beyond four molecules more modestly increases proteasome affinity [185, 186]. Some recent evidence indicates that the addition of multiple single ubiquitin moieties on a proteasomal substrate is sufficient for proteasome targeting [187–189]. As a result, varying the ubiquitin chain length could regulate degradation efficiency and modify the speed at which ubiquitinated substrates are degraded [190]. In general, however, the effect of chain length on the rate of substrate degradation has not been satisfactorily investigated. But, in order to control ubiquitin chain length, cells possess a multitude of factors that either increase (E4s) or decrease (DUBs) the length of the chain. While it is possible the extension of the ubiquitin chain is linked to increased degradation, trimming can rescue ubiquitinated substrates or — in some cases — facilitate degradation. For instance, if a substrate is deubiquitinated after p97/Cdc48 activity, it could aggregate in the cytoplasm or might subsequently be re-ubiquitinated by a cytoplasmic E3. Alternatively, if a substrate is deubiquitinated prior to p97/Cdc48 activity, it could remain in the ER membrane and potentially traffic to the Golgi or be re-ubiquitinated by ER-resident E3s. These different fates may be dictated by the timing of DUB activity. However, deubiquitination prior to p97/Cdc48 activity has also been suggested to allow substrate egress through the p97/Cdc48 hexamer [191]. Definitive proof of this model awaits the reconstitution and structural analysis of p97/Cdc48-dependent degradation. Regardless, of the ERAD-associated DUBs, several are associated with p97/Cdc48, whereas others reside in the ER membrane/cytoplasm (Table 1). This difference in localization/interaction partners could suggest alternative steps at which these DUBs act, but, to date, this has not been satisfactorily addressed.
The major DUB involved in ERAD in yeast is Otu1. Otu1 was not implicated in ERAD until it was shown to interact with Cdc48, which occurs through its ubiquitin regulatory X (UBX)-like domain [192]. Otu1 binding to the N-terminus of Cdc48 does not interfere with binding of the Cdc48 cofactors, Npl4 and Ufd1, which are vital to recognize ubiquitinated substrates [75, 193, 194]. These data suggest that different members of the Cdc48 hexameric ring bind Otu1 versus Npl4/Ufd1. While Otu1 exhibits deubiquitination activity both in vivo and in vitro, another ubiquitin chain modifier, Ufd3, does not appear to catalyze substrate deubiquitination [192]. However, in ufd3Δ cells, the level of free ubiquitin is significantly decreased [195]. The effect on free ubiquitin levels may be linked to the Ufd3-binding site on Cdc48, as Cdc48 cannot bind both Ufd3 and the E4, Ufd2, at the same time [192]. Interstingly, this site is different from the Otu1-binding site, as Cdc48 can associate with both the Otu1 and Ufd3 DUBs simultaneously. One view is that the binding of Otu1 and Ufd3 to the Cdc48 complex primes this enzyme to inhibit ubiquitination, and thus degradation, of ERAD substrates. How this critical step might be further regulated is unknown.
As might be anticipated, there are many more ERAD-associated DUBs in mammalian cells than in yeast. YOD1, the mammalian homolog of Otu1, also interacts with p97 through its UBX domain [191]. YOD1 deubiquitinates known ERAD substrates and associates with DERLIN-1 and UBXD8, two components of the HRD1 complex (see above). It was suggested that YOD1 co-operates with the p97 complex to efficiently retrotranslocate ubiquitinated substrates, which may be prevented from being threaded through the central p97 pore (also see above); alternatively, YOD1 may act upstream of retrotranslocation by controling ubiquitin chain length to optimize p97 recruitment [191,196].
An additional DUB in the mammalian ERAD pathway is ATAXIN-3. ATAXIN-3 aggregates upon extension of its polyglycine domain, which leads to Machado–Joseph Disease (also known as spinocerebellar ataxia type 3) [197]. ATAXIN-3 localizes to both the nucleus and the cytoplasm, where it interacts with substrate recognition factors for the 26S proteasome [198]. ATAXIN-3 was subsequently shown to bind p97 through an Arg/ Lys-rich region [199, 200]. Currently, the role that ATAXIN-3 plays in ERAD is not completely clear. Some evidence suggests that ATAXIN-3 removes the ubiquitin chain on ERAD substrates to increase protein half-life, which allows more time for substrate folding [201]. An alternative hypothesis is that ATAXIN-3 acts after retro-translocation to support the degradation of ERAD substrates [202]. One reason for this discrepancy may arise due to differences in methodology, which employed either overexpression studies in vivo or the use of purified complexes in in vitro reactions. Interestingly, ATAXIN-3 itself is modified by ubiquitination, which appears to increase DUB activity [203].
Other mammalian DUBs associated with the ERAD pathway include USP13, USP19, and USP25 (Table 1). USP13 interacts with p97, UFD1, NPL4, and UBXD8, and upon USP13 deletion, cells are more susceptible to ER stress. Moreover, deletion of the gene encoding USP13 leads to the accumulation of an ERAD substrate, TCRαGFP [204]. In another study, USP13 was shown to associate with gp78 and deubiquitinate a component of the ERAD complex, keeping the complex from becoming inactivated [205]. This result is in line with other studies implicating E3 and DUB activity in regulating the ERAD machinery in addition to or in contrast with the modification of ERAD substrates (also see below). In turn, USP19 has seven isoforms that arise from alternative splicing. Some of these isoforms contain a C-terminal transmembrane domain, which targets them to the ER membrane. The ER membrane-localized population rescues the degradation of two ERAD substrates, TCRα and ΔF508 CFTR [206]. Interestingly, expression of a catalytically inactive USP19 mutant partially rescued TCRα, suggesting a DUB-independent role during ERAD. However, Ye and colleagues have provided evidence that the role of USP19 during ERAD is dependent on its overexpression [207]. Of note, wild-type levels of USP19 are mostly cytosolic, lack association with ERAD factors, and fail to alter the degradation of ERAD substrates. Finally, USP25, which interacts with p97 as well as HRD1, decreases ubiquitination of the ERAD substrate CD3δ [208].
As mentioned above, a class of E3-like ligases, known as E4s, catalyzes the extension of ubiquitin chains. For ERAD, the best characterized member of this family is the yeast protein Ufd2. In the absence of Ufd2 activity, the length of ubiquitin chains assembled by the E1–E2–E3 complex onto a series of ERAD substrates is insufficient for protein turnover both in vitro and in vivo [69]. Owing to its interaction with Cdc48, Ufd2 facilitates the degradation of several substrates [72]. As noted in the previous section, Ufd2 competes with Ufd3 for binding to Cdc48, which modulates protein turnover [192]. Two mammalian Ufd2 homologs, UBE4A and UBE4B, exist, but little is known regarding their role in ERAD. Nevertheless, it has been shown that UBE4B can recognize K48 linkages and interact with p97 [209, 210].
Variations on a theme
Alternatives to Lys-48 ubiquitin linkage in ERAD
While the Lys-48 polyubiquitin linkage is predominantly used for proteasomal degradation, polyubiquitin chains appended to other lysine residues are recognized by the 26S proteasome. In yeast, ubiquitin chains containing Lys-6, Lys-11, Lys-23, Lys-29, and Lys-33 linkages all accumulate to varying levels when the proteasome is inhibited, whereas Lys-63 ubiquitin chain levels are unaffected [68]. Of the non-Lys-48 polyubiquitin chains, Lys-11-based chains are the most affected upon proteasomal inhibition; in addition, only Lys-11 chains increase when Rad23 and Dsk2 were deleted. Interestingly, the yeast E2, Ubc6, is autoubiquitinated with Lys-11 isopeptide moieties [68]; Doa10 and Ubc6 are responsible for a subpopulation of these cellular Lys-11 chains [68]. Lys-11-linked polyubiquitin chains are also induced upon ER stress [68]. When similar experiments were performed in mammalian cells, Lys-11, Lys-29, and Lys-48 linkages rose when the proteasome was inhibited [211]. Of these, the majority of the identified ubiquitin chain linkages that accumulated were Lys-11 and Lys-48. It is worth noting, however, that the accumulation of polyubiquitin moeities containing chains other than those attached via Lys-48 may represent mixed linkages. How these are built and assembled is an area of active research [212].
A mechanism for lysine-independent ubiquitination was first identified with viral E3s. For example, the viral E3 ligase, MIR1, promotes the ubiquitination and degradation of a modified MHCI that has an artificial glycine/alanine cytoplasmic domain containing only a single cysteine [213]. Since cysteine is the catalytic residue in E1s, E2s, and some E3s, this residue can clearly become ubiquitinated. Alternatively, another viral E3, mK3, can ubiquitinate lysine-less MHCI by conjugating ubiquitin to serine and threonine residues [214]. Yet another viral protein, the HIV protein, VPU, utilizes the SCFβ-TrCP E3 ligase to modify two substrates, tetherin and CD4, by utilizing lysine, serine, and threonine [215, 216]. Additional evidence obtained investigating the ubiquitination of tetherin by SCFβ-TrCP suggested that tyrosine residues in the cytoplasmic domain, or the free amino group in the N-terminus, might be targeted [217]. Since the exact nature of polyubiquitin chains in ERAD substrates is rarely investigated, it is possible that these phenomena are actually quite common. In fact, TCRα, which contains only five cytoplasmic residues (RLWSS), is modified by HRD1 on the serine side chains [218]. HRD1 also modifies serines, threonines, and lysines on the immunoglobulin Ns1 LC, which is a trapped ER luminal ERAD substrate [219]. Moreover, treatment of mini-HC (γ VH–CH1) and NHK α1-antitrypsin with sodium hydroxide decreased the amount of ubiquitin appendages, suggesting the presence of serine/threonine ubiquitin conjugates on these ERAD substrates. Finally, purified p97 protein complexes from cells treated with an ER stressor and a proteasome inhibitor harbored increased levels of sodium hydroxide-labile species [219]. These data suggest that serine/threonine ubiquitination rises during stress, and that this modification plays an important role to mitigate cellular stress in vivo.
ERAD substrates are not only modified with ubiquitin, but also there is evidence that two other protein modifications affect ERAD substrate stability: attachment of NEDD8 and SUMOylation. NEDD8 is a ubiquitin-like protein linked to cullins, which activate select E3 ligases [220]. A role for NEDD8 in ERAD has been suggested for the ERAD substrate ΔF508 CFTR [221]. During SUMOylation, SUMO (small ubiquitin-like modifier) is conjugated to lysines through the action of dedicated SUMO E1, E2, and E3 enzymes in a reaction analogous to the ubiquitin-conjugation cascade [222, 223]. One difference between the SUMO and ubiquitin pathways is that there are three SUMO isoforms in mammals, and only SUMO-2 and SUMO-3 form chains. While SUMOylation is required to regulate numerous cellular events, SUMO tags can also recruit an E3, RNF4, which leads to mixed SUMO/ubiquitin chains [224–226]. One prominent ERAD substrate modified by SUMOylation is CFTR [227]. While many E3s facilitate degradation of the disease-associated ΔF508 mutant form of CFTR (Table 1), SUMOylation is also evident. Specifically, ΔF508 is bound by the small heat-shock protein, HSP27, which recruits the E2 SUMO-conjugating enzyme, UBC9. The HSP27/UBC9 complex recognizes a non-native fold in ΔF508 CFTR, which triggers SUMOylation [228]. In turn, the poly-SUMO tag recruits RNF4, which polyubiquitinates CFTR [227]. There is still much more to be explored about this modification — and the co-ordinated interaction and assembly of ubiquitin and SUMO chains — and it is likely that a growing number of ERAD substrates will be discovered that are similarly SUMOylated.
Post-translational modifications on ERAD complexes
Although this review has focused on protein ubiquitination, Nα-acetylation has also been implicated in ERAD, at least in yeast. The importance of Nα-acetylation in ERAD came from a screen investigating the nature of the Doa10 complex [148]. Based on a genetic screen, nat3Δ mutants were found to stabilize a degron, Deg1, that derives from a Doa10-dependent substrate known as Mat2α. Nat3 resides in the NatB complex, which is partially responsible for Nα-acetylation in yeast [229]. Importantly, the degradation of this Nα-acetylated population of Deg1 is also Doa10-dependent [230]. However, the NatB requirement for Deg1 degradation was absent when wild-type Mat2α was examined [231]. Instead, Der1 requires Nα-acetylation and this activity is required for ERAD. Without Nα-acetylation, Der1 itself is ubiquitinated by Hrd1 and subsequently degraded.
As suggested in the preceding section, ERAD activity can also be regulated in response to cellular stress. For example, the ratio of components in the yeast Hrd1 complex is modified when cells are stressed chemically to induce the UPR or by constitutive UPR induction [96]. Inactivation of the Cdc48 complex similarly leads to altered ratios of Hrd1 complex components, which may reflect regulated complex assembly and perhaps increased ERAD efficiency [96]. In support of this hypothesis, autoubiquitination of Hrd1 in vitro is sufficient to recruit the Cdc48 complex, which can then engage and retrotranslocate a substrate into the cytosol [119]. Surprisingly, Hrd1 is also retrotranslocated from the ER in a reconstituted system, suggesting that a Hrd1 regulatory factor or a specific modification offsets this destructive event. In mammalian cells, a factor that may regulate the Hrd1 complex is the E2, UBE2J1. UBE2J1 activity decreases the levels of Hrd1 complex members SEL1L, EDEM, and OS-9 [135]. Without UBE2J1 activity, ERAD substrates were degraded more rapidly than in wild-type cells, which is believed to be due to greater amounts and activities of these ERAD-requiring components. Needless to say, numerous other systems probably exist to regulate the activity of the ERAD machinery, which must respond to changes in cellular stress as well as cellular differentiation and growth in vivo.
While the examples cited above suggest mechanisms to augment ERAD, negative ERAD regulators also exist. ERAD inhibition might allow more time for proteins to fold, perhaps after a stress response has been rectified. One negative regulator of ERAD in mammalian cells is the p97 cofactor SAKS1 [232, 233]. SAKS1 interacts with p97 through its UBX domain and also requires a functional ubiquitin-associating (UBA) domain to bind polyubiquitin chains [232]. When SAKS1 binds to both a polyubiquitinated substrate and p97, the degradation of an ERAD substrate and a misfolded cytosolic quality control (cytoQC) substrate was attenuated. Another negative regulator of ERAD in mammalian cells is the small p97/VCP-interacting protein (SVIP). SVIP localizes to the ER membrane, where it binds p97 and DERLIN-1 [234]. By associating with p97 and DERLIN-1, SVIP inhibits the ubiquitination and degradation of select gp78-dependent ERAD substrates. These results support the hypothesis that the p97/Cdc48 complex can be regulated by competition for cofactor binding.
Roles for unexpected factors in ERAD
As a result of the long-term search for factors involved in ERAD through genetic and proteomic approaches, it has become increasingly clear that cytosolic factors play important roles in the degradation of aberrant proteins in the ER. One such protein is the 26S proteasome-interacting E3/E4, Hul5 [82, 235]. Hul5 was first shown to be important for the degradation of cytoplasmic substrates [236]. However, by utilizing fusions of two known ERAD substrates, CPY* and Sec61-2, to a membrane-tethered product of the LEU2 gene (cytosolic 3-isopropylmalate dehydrogenase), Hul5 was found to facilitate the retrotranslocation of an ER membrane-resident protein [237]. Consistent with its activity as an E4, Hul5 was dispensable for the degradation of the CPY* or Sec61-2 portions of the fusion proteins, but instead elongated the ubiquitin chain on the transmembrane-anchored LEU2 moieties, which increased interaction with the Cdc48 complex. In the absence of Cdc48 complex interaction, the ER membrane-anchored LEU2 product was stabilized. Furthermore, as discussed earlier in the present study, two E3 ligases that reside in the Golgi and inner nuclear membrane, respectively, Rsp5 and the Asi complex, aid in the degradation of some substrates. Rsp5 degrades CPY* in a pathway known as Hrd1-independent proteolysis, whereas the Asi complex modifies the ER-resident enzyme, Erg11 [173–176]. It is unclear if Erg11 degradation represents a unique aspect of the ERAD pathway or if the Asi complex prevents Erg11 activity in the nuclear membrane. Overall, cross-talk between components of different cellular quality control pathways is important for the degradation of proteins, regardless of substrate localization.
Along with alternative E3s, molecular chaperones localized in the ER and the cytosol are implicated in ERAD. However, it is unclear whether these chaperones only serve as recognition factors for E3 binding to substrates or if they also serve to bridge the E3 to the substrate. In addition, evidence indicates that Hrd1 binds soluble ERAD substrates through its transmembrane domains during ubiquitination [119], and the integral membrane ERAD substrate, Hmg2, is recognized by Hrd1 transmembrane domains [238]. Some cytosolic chaperones help ubiquitinate transmembrane ERAD substrates. For example, the yeast Hsp70 (Ssa1) and Hsp40 chaperones (Ydj1 and Hlj1) affect the ubiquitination and degradation of several integral membrane substrates that display prominent cytoplasmic domains: Ste6* [71, 239], Pma1-D378S [152], and CFTR [150, 151]. Similar requirements for cytosolic chaperones during ubiquitination were seen in mammalian cells as well [159–161, 240, 241]. Owing to a lack of a requirement for these factors in the degradation of ER luminal substrates, it is believed that they do not function in the retrotranslocation complex, but instead catalyze ubiquitination and/or proteasome targeting.
Future work
Protein ubiquitination was first discovered as a mechanism that controls the steady-state levels of proteins, but only later was this pathway shown to regulate protein quality control [56, 242]. In turn, ER protein quality control was thought to require a luminal protease that could directly dispose of misfolded or improperly modified proteins in this compartment [243]. However, the degradation of the integral membrane CFTR protein [47, 49] and the reconstitution of soluble protein retrotranslocation [48, 244] indicated that the 26S proteasome was responsible for ER protein turnover. It logically followed that ERAD would require the cytoplasmic-localized ubiquitination machinery. Indeed, the first definitions of ERAD derived from the isolation of components required for substrate ubiquitination [38, 46, 88, 98, 99]. Since these early days, our knowledge of the intersection between the ERAD pathway and the ubiquitination machinery has rapidly expanded.
While the field has significantly matured, numerous questions have yet to be explored or remain unanswered. For example, lysines other than Lys-48 that are ubiquitinated have been encountered, and some appear to target substrates to the proteasome. Yet, it remains unresolved how these residues are processed and whether they represent a mechanism to control ERAD efficiency. The presence of mixed ubiquitin chains poses a particularly thorny problem, as their identification and production for in vitro analyses represent substantial technical hurdles. Are these alternate chains differentially recognized by components of the proteasome, ubiquitin-interacting factors, and DUBs? There exists a strong potential for specificity, as DUBS and ubiquitin elongation factors in the same family can recognize distinct ubiquitin linkages [245]. Moreover, the acquisition of different ubiquitin modifications, as well as SUMOylation, may represent responses to specific cellular conditions or stress responses. However, it remains unclear why the cell ‘chooses’ to build different linkages.
Another important direction for future research involves the expansion of the roles that post-translational modifications play in ERAD efficiency. In addition to the examples discussed above, ERAD facilitators can be tagged with other modifications. For example, Parkin is subject to S-nitrosylation [246], and BiP, the ER luminal Hsp70, is oxidized, ADP-ribosylated, and AMPylated [247–250]. Here too, these modifications may represent responses to cellular stress or alert the ER to conditions that will affect the efficacy of protein biogenesis. As an example, it is noteworthy that the UPR is activated before the proliferation of the ER that accompanies the transition of a plasma cell to a mature, antibody-producing B cell [251].
In theory, only bona fide ERAD substrates should be targeted for ubiquitination, whereas wild-type, folded proteins should escape this fate. Which characteristics of substrates lead to their recognition by the molecular chaperones that act upstream of the ubiquitination pathway or by the E3 ligases that directly recognize substrates? The structure (or misfolded state) of a protein should serve as a recognition device [252–254]. Another parameter that may control ERAD substrate recognition and subsequent ubiquitination is the appearance of surface-exposed hydrophobic side chains, which should be enclosed within the structure of properly folded proteins [119,255]. It is also possible that select post-translational modifications or amino acid motifs trigger ERAD when they are exposed [256,257].
Finally, the regulation of the ERAD machinery represents a fertile area for ongoing and future research. Although select examples have been described in this review, we know nothing about how ERAD is regulated in different tissues and how various disease states might alter ERAD efficiency. One report suggested that HER2+ breast cancer cells have a heightened requirement for ERAD, but this was not rigorously demonstrated [258]. Moreover, the pathway should respond to changes in cell, tissue, and organ development, and some hints on the requirement for HRD1 during these events have recently been uncovered [259–261]. But, given the fact that ~8000 proteins pass through the ER in a human cell, and there are numerous misfolded conformations that these proteins can attain, variations on the ERAD theme will most probably become the norm.
Acknowledgments
Funding
Work in the Brodsky Laboratory on ERAD is supported by grants GM75061 and DK79307 from the National Institutes of Health and grant BRODSK13XX0 from Cystic Fibrosis Foundation Therapeutics. G.M.P. acknowledges grants DK061296 from the National Institutes of Health and PRESTO15H0 from the Cystic Fibrosis Foundation.
Abbreviations
- CF
cystic fibrosis
- CFTR
cystic fibrosis transmembrane conductance regulator
- CHIP
C-terminus of Hsc70-interacting protein
- DUBs
deubiquitinating enzymes
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- ERAD-C
ER cytosolic lesion
- ERAD-L
ER luminal
- HC
heavy chain
- HCMV
human cytomegalovirus
- HECT
homologous to the E6AP carboxyl terminus
- HRD
Hmg CoA reductase degradation
- HSP
heat-shock protein
- LC
light chain
- MHCI
major histocompatability complex class I
- RING
really interesting new gene
- SCF
Skp1, Cullins, F-box proteins
- SUMO
small ubiquitin-like modifier
- SVIP
small p97/ VCP-interacting protein
- UBX
ubiquitin regulatory X
- UPR
unfolded protein response
- VIMP
VCP-interacting membrane protein
Footnotes
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Kanapin A, Batalov S, Davis MJ, Gough J, Grimmond S, Kawaji H, et al. Mouse proteome analysis. Genome Res. 2003;13:1335–1344. doi: 10.1101/gr.978703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Hudson DA, Gannon SA, Thorpe C. Oxidative protein folding: from thiol–disulfide exchange reactions to the redox poise of the endoplasmic reticulum. Free Radic Biol Med. 2015;80:171–182. doi: 10.1016/j.freeradbiomed.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodsky JL, Skach WR. Protein folding and quality control in the endoplasmic reticulum: recent lessons from yeast and mammalian cell systems. Curr Opin Cell Biol. 2011;23:464–475. doi: 10.1016/j.ceb.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silberstein S, Gilmore R. Biochemistry, molecular biology, and genetics of the oligosaccharyltransferase. FASEB J. 1996;10:849–858. [PubMed] [Google Scholar]
- 6.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 7.Orlean P, Menon AK. Thematic review series: lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: how we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Hentschel A, Zahedi RP, Ahrends R. Protein lipid modifications — more than just a greasy ballast. Proteomics. 2016;16:759–782. doi: 10.1002/pmic.201500353. [DOI] [PubMed] [Google Scholar]
- 9.Braakman I, Bulleid NJ. Protein folding and modification in the mammalian endoplasmic reticulum. Annu Rev Biochem. 2011;80:71–99. doi: 10.1146/annurev-biochem-062209-093836. [DOI] [PubMed] [Google Scholar]
- 10.Osborne AR, Rapoport TA, van den Berg B. Protein translocation by the Sec61/SecY channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- 11.Higy M, Junne T, Spiess M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry. 2004;43:12716–12722. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- 12.Jansens A, van Duijn E, Braakman I. Coordinated nonvectorial folding in a newly synthesized multidomain protein. Science. 2002;298:2401–2403. doi: 10.1126/science.1078376. [DOI] [PubMed] [Google Scholar]
- 13.Kim SJ, Yoon JS, Shishido H, Yang Z, Rooney LA, Barral JM, et al. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348:444–448. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- 14.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme: I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-W. [DOI] [PubMed] [Google Scholar]
- 15.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 16.Papp E, Száraz P, Korcsmáros T, Csermely P. Changes of endoplasmic reticulum chaperone complexes, redox state, and impaired protein disulfide reductase activity in misfolding α1-antitrypsin transgenic mice. FASEB J. 2006;20:1018–1020. doi: 10.1096/fj.05-5065fje. [DOI] [PubMed] [Google Scholar]
- 17.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 18.Ellgaard L, Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 19.Tagliavacca L, Wang Q, Kaufman RJ. ATP-dependent dissociation of non-disulfide-linked aggregates of coagulation factor VIII is a rate-limiting step for secretion. Biochemistry. 2000;39:1973–1981. doi: 10.1021/bi991896r. [DOI] [PubMed] [Google Scholar]
- 20.Molinari M, Galli C, Vanoni O, Arnold SM, Kaufman RJ. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Mol Cell. 2005;20:503–512. doi: 10.1016/j.molcel.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 21.Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- 22.Buck TM, Wright CM, Brodsky JL. The activities and function of molecular chaperones in the endoplasmic reticulum. Semin Cell Dev Biol. 2007;18:751–761. doi: 10.1016/j.semcdb.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, et al. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- 25.Blond-Elguindi S, Cwirla SE, Dower WJ, Lipshutz RJ, Sprang SR, Sambrook JF, et al. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 26.Rüdiger S, Buchberger A, Bukau B. Interaction of Hsp70 chaperones with substrates. Nat Struct Mol Biol. 1997;4:342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- 27.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol. 2004;14:20–28. doi: 10.1016/j.tcb.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 30.Vembar SS, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/S0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 32.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruggiano A, Foresti O, Carvalho P. ER-associated degradation: Protein quality control and beyond. J Cell Biol. 2014;204:869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casagrande R, Stern P, Diehn M, Shamu C, Osario M, Zúñiga M, et al. Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol Cell. 2000;5:729–735. doi: 10.1016/S1097-2765(00)80251-8. [DOI] [PubMed] [Google Scholar]
- 35.Adle DJ, Wei W, Smith N, Bies JJ, Lee J. Cadmium-mediated rescue from ER-associated degradation induces expression of its exporter. Proc Natl Acad Sci USA. 2009;106:10189–10194. doi: 10.1073/pnas.0812114106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tyler RE, Pearce MMP, Shaler TA, Olzmann JA, Greenblatt EJ, Kopito RR. Unassembled CD147 is an endogenous endoplasmic reticulum-associated degradation substrate. Mol Biol Cell. 2012;23:4668–4678. doi: 10.1091/mbc.E12-06-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen C-y, Malchus NS, Hehn B, Stelzer W, Avci D, Langosch D, et al. Signal peptide peptidase functions in ERAD to cleave the unfolded protein response regulator XBP1u. EMBO J. 2014;33:2492–2506. doi: 10.15252/embj.201488208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hampton R, Gardner R, Rine J. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol Biol Cell. 1996;7:2029–2044. doi: 10.1091/mbc.7.12.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/S0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- 40.Gauss R, Kanehara K, Carvalho P, Ng DTW, Aebi M. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol Cell. 2011;42:782–793. doi: 10.1016/j.molcel.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 41.Acconcia F, Sigismund S, Polo S. Ubiquitin in trafficking: the network at work. Exp Cell Res. 2009;315:1610–1618. doi: 10.1016/j.yexcr.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 42.Perrody E, Abrami L, Feldman M, Kunz B, Urbé S, van der Goot FG. Ubiquitin-dependent folding of the Wnt signaling coreceptor LRP6. eLife. 2016;5:e19083. doi: 10.7554/eLife.19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol Biol Cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 45.Rabinovich E, Kerem A, Fröhlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol Cell Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hiller MM, Finger A, Schweiger M, Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 47.Jensen TJ, Loo MA, Pind S, Williams DB, Goldberg AL, Riordan JR. Multiple proteolytic systems, including the proteasome, contribute to CFTR processing. Cell. 1995;83:129–135. doi: 10.1016/0092-8674(95)90241-4. [DOI] [PubMed] [Google Scholar]
- 48.Werner ED, Brodsky JL, McCracken AA. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc Natl Acad Sci USA. 1996;93:13797–13801. doi: 10.1073/pnas.93.24.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward CL, Omura S, Kopito RR. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 50.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith DM, Benaroudj N, Goldberg A. Proteasomes and their associated ATPases: a destructive combination. J Struct Biol. 2006;156:72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 52.Förster F, Unverdorben P, Sledz P, Baumeister W. Unveiling the long-held secrets of the 26S proteasome. Structure. 2013;21:1551–1562. doi: 10.1016/j.str.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 53.Brooks P, Fuertes G, Murray RZ, Suchira B, Knecht E, Rechsteiner MC, et al. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem J. 2000;346:155–161. doi: 10.1042/bj3460155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hershko A, Heller H, Elias S, Ciechanover A. Components of ubiquitin-protein ligase system. resolution, affinity purification, and role in protein breakdown. J Biol Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 55.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 56.Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 57.Lee WC, Lee M, Jung JW, Kim KP, Kim D. SCUD: Saccharomyces cerevisiae ubiquitination database. BMC Genomics. 2008;9:440. doi: 10.1186/1471-2164-9-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Wijk SJL, de Vries SJ, Kemmeren P, Huang A, Boelens R, Bonvin AMJJ, et al. A comprehensive framework of E2–RING E3 interactions of the human ubiquitin–proteasome system. Mol Syst Biol. 2009;5:295. doi: 10.1038/msb.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huibregtse JM, Scheffner M, Beaudenon S, Howley PM. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1–E2–E3 enzyme ubiquitin thioester cascade. Nature. 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 62.Cyr DM, Höhfeld J, Patterson C. Protein quality control: U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem Sci. 2002;27:368–375. doi: 10.1016/S0968-0004(02)02125-4. [DOI] [PubMed] [Google Scholar]
- 63.Borden KLB. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 64.Deshaies RJ, Joazeiro CAP. RING domain E3 ubiquitin ligases. Ann Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 65.Chau V, Tobias J, Bachmair A, Marriott D, Ecker D, Gonda D, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 66.Pickart CM. Ubiquitin in chains. Trends Biochem Sci. 2000;25:544–548. doi: 10.1016/S0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 67.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 68.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell. 1999;96:635–644. doi: 10.1016/S0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 70.Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, et al. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat Cell Biol. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- 71.Nakatsukasa K, Huyer G, Michaelis S, Brodsky JL. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 2008;132:101–112. doi: 10.1016/j.cell.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell. 2005;120:73–84. doi: 10.1016/j.cell.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 73.Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat Cell Biol. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- 74.Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nat Cell Biol. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- 75.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 78.Elsasser S, Chandler-Militello D, Müller B, Hanna J, Finley D. Rad23 and Rpn10 serve as alternative ubiquitin receptors for the proteasome. J Biol Chem. 2004;279:26817–26822. doi: 10.1074/jbc.M404020200. [DOI] [PubMed] [Google Scholar]
- 79.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 2001;20:5187–5196. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 81.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, Koonin EV, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 82.Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/S1097-2765(02)00638-X. [DOI] [PubMed] [Google Scholar]
- 83.Guterman A, Glickman MH. Complementary roles for Rpn11 and Ubp6 in deubiquitination and proteolysis by the proteasome. J Biol Chem. 2004;279:1729–1738. doi: 10.1074/jbc.M307050200. [DOI] [PubMed] [Google Scholar]
- 84.Rubin DM, Glickman MH, Larsen CN, Dhruvakumar S, Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 86.Ciehanover A, Hod Y, Hershko A. A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. Biochem Biophys Res Commun. 1978;81:1100–1105. doi: 10.1016/0006-291X(78)91249-4. [DOI] [PubMed] [Google Scholar]
- 87.Wilkinson K, Urban M, Haas A. Ubiquitin is the ATP-dependent proteolysis factor I of rabbit reticulocytes. J Biol Chem. 1980;255:7529–7532. [PubMed] [Google Scholar]
- 88.Sommer T, Jentsch S. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature. 1993;365:176–179. doi: 10.1038/365176a0. [DOI] [PubMed] [Google Scholar]
- 89.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MATα2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-Q. [DOI] [PubMed] [Google Scholar]
- 90.Vassal A, Boulet A, Decoster E, Faye G. QRI8, a novel ubiquitin-conjugating enzyme in Saccharomyces cerevisiae. Biochim Biophys Acta. 1992;1132:211–213. doi: 10.1016/0167-4781(92)90015-R. [DOI] [PubMed] [Google Scholar]
- 91.Skalnik DG, Narita H, Kent C, Simoni RD. The membrane domain of 3-hydroxy-3-methylglutaryl-coenzyme A reductase confers endoplasmic reticulum localization and sterol-regulated degradation onto beta-galactosidase. J Biol Chem. 1988;263:6836–6841. [PubMed] [Google Scholar]
- 92.Chun KT, Bar-Nun S, Simoni RD. The regulated degradation of 3-hydroxy-3-methylglutaryl-CoA reductase requires a short-lived protein and occurs in the endoplasmic reticulum. J Biol Chem. 1990;265:22004–22010. [PubMed] [Google Scholar]
- 93.Bays NW, Gardner RG, Seelig LP, Joazeiro CA, Hampton RY. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- 94.Gardner RG, Swarbrick GM, Bays NW, Cronin SR, Wilhovsky S, Seelig L, et al. Endoplasmic reticulum degradation requires lumen to cytosol signaling transmembrane control of Hrd1p by Hrd3p. J Cell Biol. 2000;151:69–82. doi: 10.1083/jcb.151.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hampton RY, Bhakta H. Ubiquitin-mediated regulation of 3-hydroxy-3-methylglutaryl-CoA reductase. Proc Natl Acad Sci USA. 1997;94:12944–12948. doi: 10.1073/pnas.94.24.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nakatsukasa K, Brodsky JL, Kamura T. A stalled retrotranslocation complex reveals physical linkage between substrate recognition and proteasomal degradation during ER-associated degradation. Mol Biol Cell. 2013;24:1765–1775. doi: 10.1091/mbc.E12-12-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gauss R, Sommer T, Jarosch E. The Hrd1p ligase complex forms a linchpin between ER-lumenal substrate selection and Cdc48p recruitment. EMBO J. 2006;25:1827–1835. doi: 10.1038/sj.emboj.7601088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wiertz EJHJ, Jones TR, Sun L, Bogyo M, Geuze HJ, Ploegh HL. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/S0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 99.Wiertz EJHJ, Tortorella D, Bogyo M, Yu J, Mothes W, Jones TR, et al. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 100.Bordallo J, Plemper RK, Finger A, Wolf DH. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell. 1998;9:209–222. doi: 10.1091/mbc.9.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vashist S, Ng DTW. Misfolded proteins are sorted by a sequential checkpoint mechanism of ER quality control. J Cell Biol. 2004;165:41–52. doi: 10.1083/jcb.200309132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Carvalho P, Goder V, Rapoport TA. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126:361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 103.Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- 104.Gauss R, Jarosch E, Sommer T, Hirsch C. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat Cell Biol. 2006;8:849–854. doi: 10.1038/ncb1445. [DOI] [PubMed] [Google Scholar]
- 105.Plemper RK, Bordallo J, Deak PM, Taxis C, Hitt R, Wolf DH. Genetic interactions of Hrd3p and Der3p/Hrd1p with Sec61p suggest a retro-translocation complex mediating protein transport for ER degradation. J Cell Sci. 1999;112:4123–4134. doi: 10.1242/jcs.112.22.4123. [DOI] [PubMed] [Google Scholar]
- 106.Vashistha N, Neal SE, Singh A, Carroll SM, Hampton RY. Direct and essential function for Hrd3 in ER-associated degradation. Proc Natl Acad Sci USA. 2016;113:5934–5939. doi: 10.1073/pnas.1603079113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Horn SC, Hanna J, Hirsch C, Volkwein C, Schütz A, Heinemann U, et al. Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol Cell. 2009;36:782–793. doi: 10.1016/j.molcel.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 108.Knop M, Finger A, Braun T, Hellmuth K, Wolf D. Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 1996;15:753–763. [PMC free article] [PubMed] [Google Scholar]
- 109.Stolz A, Schweizer RS, Schäfer A, Wolf DH. Dfm1 forms distinct complexes with Cdc48 and the ER ubiquitin ligases and is required for ERAD. Traffic. 2010;11:1363–1369. doi: 10.1111/j.1600-0854.2010.01093.x. [DOI] [PubMed] [Google Scholar]
- 110.Sato BK, Hampton RY. Yeast derlin Dfm1 interacts with Cdc48 and functions in ER homeostasis. Yeast. 2006;23:1053–1064. doi: 10.1002/yea.1407. [DOI] [PubMed] [Google Scholar]
- 111.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 112.Bagola K, von Delbrück M, Dittmar G, Scheffner M, Ziv I, Glickman MH, et al. Ubiquitin binding by a CUE domain regulates ubiquitin chain formation by ERAD E3 ligases. Mol Cell. 2013;50:528–539. doi: 10.1016/j.molcel.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 113.Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol. 2000;2:379–384. doi: 10.1038/35017001. [DOI] [PubMed] [Google Scholar]
- 114.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 115.Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lilley BN, Ploegh HL. Multiprotein complexes that link dislocation, ubiquitination, and extraction of misfolded proteins from the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14296–14301. doi: 10.1073/pnas.0505014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mehnert M, Sommer T, Jarosch E. Der1 promotes movement of misfolded proteins through the endoplasmic reticulum membrane. Nat Cell Biol. 2014;16:77–86. doi: 10.1038/ncb2882. [DOI] [PubMed] [Google Scholar]
- 118.Wahlman J, DeMartino GN, Skach WR, Bulleid NJ, Brodsky JL, Johnson AE. Real-time fluorescence detection of ERAD substrate retrotranslocation in a mammalian in vitro system. Cell. 2007;129:943–955. doi: 10.1016/j.cell.2007.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell. 2014;158:1375–1388. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Baldridge RD, Rapoport TA. Autoubiquitination of the HRD1 ligase triggers protein retrotranslocation in ERAD. Cell. 2016;166:394–407. doi: 10.1016/j.cell.2016.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kikkert M, Doolman R, Dai M, Avner R, Hassink G, van Voorden S, et al. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. J Biol Chem. 2004;279:3525–3534. doi: 10.1074/jbc.M307453200. [DOI] [PubMed] [Google Scholar]
- 122.Kaneko M, Ishiguro M, Niinuma Y, Uesugi M, Nomura Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Lett. 2002;532:147–152. doi: 10.1016/S0014-5793(02)03660-8. [DOI] [PubMed] [Google Scholar]
- 123.Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1–SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alcock F, Swanton E. Mammalian OS-9 is upregulated in response to endoplasmic reticulum stress and facilitates ubiquitination of misfolded glycoproteins. J Mol Biol. 2009;385:1032–1042. doi: 10.1016/j.jmb.2008.11.045. [DOI] [PubMed] [Google Scholar]
- 125.Rose MD, Misra LM, Vogel JP. KAR2, a karyogamy gene, is the yeast homolog of the mammalian BiP/GRP78 gene. Cell. 1989;57:1211–1221. doi: 10.1016/0092-8674(89)90058-5. [DOI] [PubMed] [Google Scholar]
- 126.Katsanis N, Fisher EMC. Identification, expression, and chromosomal localization of ubiquitin conjugating enzyme 7 (UBE2G2), a human homologue of the Saccharomyces cerevisiae Ubc7Gene. Genomics. 1998;51:128–131. doi: 10.1006/geno.1998.5263. [DOI] [PubMed] [Google Scholar]
- 127.Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci USA. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Klemm EJ, Spooner E, Ploegh HL. Dual role of ancient ubiquitous protein 1 (AUP1) in lipid droplet accumulation and endoplasmic reticulum (ER) protein quality control. J Biol Chem. 2011;286:37602–37614. doi: 10.1074/jbc.M111.284794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schulze A, Standera S, Buerger E, Kikkert M, van Voorden S, Wiertz E, et al. The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. J Mol Biol. 2005;354:1021–1027. doi: 10.1016/j.jmb.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 130.Okuda-Shimizu Y, Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol. 2006;172:383–393. doi: 10.1083/jcb.200507057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Olivari S, Cali T, Salo KEH, Paganetti P, Ruddock LW, Molinari M. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem Biophys Res Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- 133.Olivari S, Molinari M. Glycoprotein folding and the role of EDEM1, EDEM2 and EDEM3 in degradation of folding-defective glycoproteins. FEBS Lett. 2007;581:3658–3664. doi: 10.1016/j.febslet.2007.04.070. [DOI] [PubMed] [Google Scholar]
- 134.Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, et al. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hagiwara M, Ling J, Koenig PA, Ploegh HL. Posttranscriptional regulation of glycoprotein quality control in the endoplasmic reticulum is controlled by the E2 Ub-conjugating enzyme UBC6e. Mol Cell. 2016;63:753–767. doi: 10.1016/j.molcel.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Spandl J, Lohmann D, Kuerschner L, Moessinger C, Thiele C. Ancient ubiquitous protein 1 (AUP1) localizes to lipid droplets and binds the E2 ubiquitin conjugase G2 (Ube2g2) via its G2 binding region. J Biol Chem. 2011;286:5599–5606. doi: 10.1074/jbc.M110.190785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci USA. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Burr ML, Cano F, Svobodova S, Boyle LH, Boname JM, Lehner PJ. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc Natl Acad Sci USA. 2011;108:2034–2039. doi: 10.1073/pnas.1016229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liang J, Yin C, Doong H, Fang S, Peterhoff C, Nixon RA, et al. Characterization of erasin (UBXD2): a new ER protein that promotes ER-associated protein degradation. J Cell Sci. 2006;119:4011–4024. doi: 10.1242/jcs.03163. [DOI] [PubMed] [Google Scholar]
- 140.Lee JN, Zhang X, Feramisco JD, Gong Y, Ye J. Unsaturated fatty acids inhibit proteasomal degradation of insig-1 at a postubiquitination step. J Biol Chem. 2008;283:33772–33783. doi: 10.1074/jbc.M806108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lim PJ, Danner R, Liang J, Doong H, Harman C, Srinivasan D, et al. Ubiquilin and p97/VCP bind erasin, forming a complex involved in ERAD. J Cell Biol. 2009;187:201–217. doi: 10.1083/jcb.200903024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Seok Ko H, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566:110–114. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 143.Wang Q, Liu Y, Soetandyo N, Baek K, Hegde R, Ye Y. A ubiquitin ligase-associated chaperone holdase maintains polypeptides in soluble states for proteasome degradation. Mol Cell. 2011;42:758–770. doi: 10.1016/j.molcel.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu Y, Liu Y, Lee J-g, Ye Y. A ubiquitin-like domain recruits an oligomeric chaperone to a retrotranslocation complex in endoplasmic reticulum-associated degradation. J Biol Chem. 2013;288:18068–18076. doi: 10.1074/jbc.M112.449199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS. Ubiquilins chaperone and triage mitochondrial membrane proteins for degradation. Mol Cell. 2016;63:21–33. doi: 10.1016/j.molcel.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat Struct Mol Biol. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matα2 repressor degradation. Genes Dev. 2001;15:2660–2674. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ravid T, Kreft SG, Hochstrasser M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006;25:533–543. doi: 10.1038/sj.emboj.7600946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Weber A, Cohen I, Popp O, Dittmar G, Reiss Y, Sommer T, et al. Sequential poly-ubiquitylation by specialized conjugating enzymes expands the versatility of a quality control ubiquitin ligase. Mol Cell. 2016;63:827–839. doi: 10.1016/j.molcel.2016.07.020. [DOI] [PubMed] [Google Scholar]
- 150.Zhang Y, Nijbroek G, Sullivan ML, McCracken AA, Watkins SC, Michaelis S, et al. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol Biol Cell. 2001;12:1303–1314. doi: 10.1091/mbc.12.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Youker RT, Walsh P, Beilharz T, Lithgow T, Brodsky JL. Distinct roles for the Hsp40 and Hsp90 molecular chaperones during cystic fibrosis transmembrane conductance regulator degradation in yeast. Mol Biol Cell. 2004;15:4787–4797. doi: 10.1091/mbc.E04-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Han S, Liu Y, Chang A. Cytoplasmic Hsp70 promotes ubiquitination for endoplasmic reticulum-associated degradation of a misfolded mutant of the yeast plasma membrane ATPase, PMA1. J Biol Chem. 2007;282:26140–26149. doi: 10.1074/jbc.M701969200. [DOI] [PubMed] [Google Scholar]
- 153.Metzger MB, Maurer MJ, Dancy BM, Michaelis S. Degradation of a cytosolic protein requires endoplasmic reticulum-associated degradation machinery. J Biol Chem. 2008;283:32302–32316. doi: 10.1074/jbc.M806424200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hassink G, Kikkert M, van Voorden S, Shiow-Ju L, Spaapen R, van Laar T, et al. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem J. 2005;388:647–655. doi: 10.1042/BJ20041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kreft SG, Wang L, Hochstrasser M. Membrane topology of the yeast endoplasmic reticulum-localized ubiquitin ligase Doa10 and comparison with its human ortholog TEB4 (MARCH-VI) J Biol Chem. 2006;281:4646–4653. doi: 10.1074/jbc.M512215200. [DOI] [PubMed] [Google Scholar]
- 156.Foresti O, Ruggiano A, Hannibal-Bach HK, Ejsing CS, Carvalho P. Sterol homeostasis requires regulated degradation of squalene monooxygenase by the ubiquitin ligase Doa10/Teb4. eLife. 2013;2:e00953. doi: 10.7554/eLife.00953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zavacki AM, Arrojo e Drigo RA, Freitas BCG, Chung M, Harney JW, Egri P, et al. The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Mol Cell Biol. 2009;29:5339–5347. doi: 10.1128/MCB.01498-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zelcer N, Sharpe LJ, Loregger A, Kristiana I, Cook ECL, Phan L, et al. The E3 ubiquitin ligase MARCH6 degrades squalene monooxygenase and affects 3-hydroxy-3-methyl-glutaryl coenzyme A reductase and the cholesterol synthesis pathway. Mol Cell Biol. 2014;34:1262–1270. doi: 10.1128/MCB.01140-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Meacham GC, Lu Z, King S, Sorscher E, Tousson A, Cyr DM. The Hdj-2/Hsc70 chaperone pair facilitates early steps in CFTR biogenesis. EMBO J. 1999;18:1492–1505. doi: 10.1093/emboj/18.6.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fisher EA, Zhou M, Mitchell DM, Wu X, Omura S, Wang H, et al. The degradation of apolipoprotein B100 is mediated by the ubiquitin-proteasome pathway and involves heat shock protein 70. J Biol Chem. 1997;272:20427–20434. doi: 10.1074/jbc.272.33.20427. [DOI] [PubMed] [Google Scholar]
- 161.Younger JM, Ren HY, Chen L, Fan CY, Fields A, Patterson C, et al. A foldable CFTRΔF508 biogenic intermediate accumulates upon inhibition of the Hsc70–CHIP E3 ubiquitin ligase. J Cell Biol. 2004;167:1075–1085. doi: 10.1083/jcb.200410065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yamamoto Y-h, Kimura T, Momohara S, Takeuchi M, Tani T, Kimata Y, et al. A novel ER J-protein DNAJB12 accelerates ER-associated degradation of membrane proteins including CFTR. Cell Struct Funct. 2010;35:107–116. doi: 10.1247/csf.10023. [DOI] [PubMed] [Google Scholar]
- 163.Grove DE, Fan CY, Ren HY, Cyr DM. The endoplasmic reticulum–associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3–dependent degradation of nascent CFTRΔF508. Mol Biol Cell. 2011;22:301–314. doi: 10.1091/mbc.E10-09-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fang S, Ferrone M, Yang C, Jensen JP, Tiwari S, Weissman AM. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc Natl Acad Sci USA. 2001;98:14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Morito D, Hirao K, Oda Y, Hosokawa N, Tokunaga F, Cyr DM, et al. Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRΔF508. Mol Biol Cell. 2008;19:1328–1336. doi: 10.1091/mbc.E07-06-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhang T, Xu Y, Liu Y, Ye Y. Gp78 functions downstream of Hrd1 to promote degradation of misfolded proteins of the endoplasmic reticulum. Mol Biol Cell. 2015;26:4438–4450. doi: 10.1091/mbc.E15-06-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ballar P, Shen Y, Yang H, Fang S. The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. J Biol Chem. 2006;281:35359–35368. doi: 10.1074/jbc.M603355200. [DOI] [PubMed] [Google Scholar]
- 168.Imai Y, Soda M, Hatakeyama S, Akagi T, Hashikawa T, Nakayama KI, et al. CHIP is associated with parkin, a gene responsible for familial Parkinson’s disease, and enhances its ubiquitin ligase activity. Mol Cell. 2002;10:55–67. doi: 10.1016/S1097-2765(02)00583-X. [DOI] [PubMed] [Google Scholar]
- 169.Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- 170.Younger JM, Chen L, Ren HY, Rosser MFN, Turnbull EL, Fan CY, et al. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
- 171.Stolz A, Besser S, Hottmann H, Wolf DH. Previously unknown role for the ubiquitin ligase Ubr1 in endoplasmic reticulum-associated protein degradation. Proc Natl Acad Sci USA. 2013;110:15271–15276. doi: 10.1073/pnas.1304928110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bartel B, Wünning I, Varshavsky A. The recognition component of the N-end rule pathway. EMBO J. 1990;9:3179–3189. doi: 10.1002/j.1460-2075.1990.tb07516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Haynes CM, Caldwell S, Cooper AA. An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J Cell Biol. 2002;158:91–102. doi: 10.1083/jcb.200201053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Haynes CM, Titus EA, Cooper AA. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell. 2004;15:767–776. doi: 10.1016/j.molcel.2004.08.025. [DOI] [PubMed] [Google Scholar]
- 175.Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P. Quality control of inner nuclear membrane proteins by the Asi complex. Science. 2014;346:751–755. doi: 10.1126/science.1255638. [DOI] [PubMed] [Google Scholar]
- 176.Khmelinskii A, Blaszczak E, Pantazopoulou M, Fischer B, Omnus DJ, Le Dez G, et al. Protein quality control at the inner nuclear membrane. Nature. 2014;516:410–413. doi: 10.1038/nature14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Binette J, Dubé M, Mercier J, Halawani D, Latterich M, Cohen ÉA. Requirements for the selective degradation of CD4 receptor molecules by the human immunodeficiency virus type 1 Vpu protein in the endoplasmic reticulum. Retrovirology. 2007;4:75. doi: 10.1186/1742-4690-4-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mangeat B, Gers-Huber G, Lehmann M, Zufferey M, Luban J, Piguet V. HIV-1 Vpu neutralizes the antiviral factor tetherin/BST-2 by binding it and directing its beta-TrCP2-dependent degradation. PLoS Pathog. 2009;5:e1000574. doi: 10.1371/journal.ppat.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.van den Boomen DJH, Timms RT, Grice GL, Stagg HR, Skødt K, Dougan G, et al. TMEM129 is a derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proc Natl Acad Sci USA. 2014;111:11425–11430. doi: 10.1073/pnas.1409099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yoshida Y, Chiba T, Tokunaga F, Kawasaki H, Iwai K, Suzuki T, et al. E3 ubiquitin ligase that recognizes sugar chains. Nature. 2002;418:438–442. doi: 10.1038/nature00890. [DOI] [PubMed] [Google Scholar]
- 181.Yoshida Y, Tokunaga F, Chiba T, Iwai K, Tanaka K, Tai T. Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J Biol Chem. 2003;278:43877–43884. doi: 10.1074/jbc.M304157200. [DOI] [PubMed] [Google Scholar]
- 182.Blom D, Hirsch C, Stern P, Tortorella D, Ploegh HL. A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised. EMBO J. 2004;23:650–658. doi: 10.1038/sj.emboj.7600090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Suzuki T, Park H, Lennarz WJ. Cytoplasmic peptide: N-glycanase (PNGase) in eukaryotic cells: occurrence, primary structure, and potential functions. FASEB J. 2002;16:635–641. doi: 10.1096/fj.01-0889rev. [DOI] [PubMed] [Google Scholar]
- 184.Maruyama Y, Yamada M, Takahashi K, Yamada M. Ubiquitin ligase Kf-1 is involved in the endoplasmic reticulum-associated degradation pathway. Biochem Biophys Res Commun. 2008;374:737–741. doi: 10.1016/j.bbrc.2008.07.126. [DOI] [PubMed] [Google Scholar]
- 185.Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM. A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature. 2002;416:763–767. doi: 10.1038/416763a. [DOI] [PubMed] [Google Scholar]
- 186.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Boutet SC, Disatnik MH, Chan LS, Iori K, Rando TA. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 188.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kravtsova-Ivantsiv Y, Cohen S, Ciechanover A. Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-κB precursor. Mol Cell. 2009;33:496–504. doi: 10.1016/j.molcel.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 190.Lederkremer GZ, Glickman MH. A window of opportunity: timing protein degradation by trimming of sugars and ubiquitins. Trends Biochem Sci. 2005;30:297–303. doi: 10.1016/j.tibs.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 191.Ernst R, Mueller B, Ploegh HL, Schlieker C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol Cell. 2009;36:28–38. doi: 10.1016/j.molcel.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol Cell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 193.Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1–Npl4. EMBO J. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian Ufd1 and Npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Johnson ES, Ma PCM, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- 196.Liu Y, Ye Y. Roles of p97-associated deubiquitinases in protein quality control at the endoplasmic reticulum. Curr Protein Pept Sci. 2012;13:436–446. doi: 10.2174/138920312802430608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Koch P, Breuer P, Peitz M, Jungverdorben J, Kesavan J, Poppe D, et al. Excitation-induced ataxin-3 aggregation in neurons from patients with Machado-Joseph disease. Nature. 2011;480:543–546. doi: 10.1038/nature10671. [DOI] [PubMed] [Google Scholar]
- 198.Wang GH, Sawai N, Kotliarova S, Kanazawa I, Nukina N. Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum Mol Genet. 2000;9:1795–1803. doi: 10.1093/hmg/9.12.1795. [DOI] [PubMed] [Google Scholar]
- 199.Doss-Pepe EW, Stenroos ES, Johnson WG, Madura K. Ataxin-3 interactions with rad23 and valosin-containing protein and its associations with ubiquitin chains and the proteasome are consistent with a role in ubiquitin-mediated proteolysis. Mol Cell Biol. 2003;23:6469–6483. doi: 10.1128/MCB.23.18.6469-6483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Boeddrich A, Gaumer S, Haacke A, Tzvetkov N, Albrecht M, Evert BO, et al. An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J. 2006;25:1547–1558. doi: 10.1038/sj.emboj.7601043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zhong X, Pittman RN. Ataxin-3 binds VCP/p97 and regulates retrotranslocation of ERAD substrates. Hum Mol Genet. 2006;15:2409–2420. doi: 10.1093/hmg/ddl164. [DOI] [PubMed] [Google Scholar]
- 202.Wang Q, Li L, Ye Y. Regulation of retrotranslocation by p97-associated deubiquitinating enzyme ataxin-3. J Cell Biol. 2006;174:963–971. doi: 10.1083/jcb.200605100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Todi SV, Winborn BJ, Scaglione KM, Blount JR, Travis SM, Paulson HL. Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 2009;28:372–382. doi: 10.1038/emboj.2008.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu Y, Soetandyo N, Lee JG, Liu L, Xu Y, Clemons WM, Jr, et al. USP13 antagonizes gp78 to maintain functionality of a chaperone in ER-associated degradation. eLife. 2014;3:e01369. doi: 10.7554/eLife.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hassink GC, Zhao B, Sompallae R, Altun M, Gastaldello S, Zinin NV, et al. The ER-resident ubiquitin-specific protease 19 participates in the UPR and rescues ERAD substrates. EMBO Rep. 2009;10:755–761. doi: 10.1038/embor.2009.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lee JG, Kim W, Gygi S, Ye Y. Characterization of the deubiquitinating activity of USP19 and its role in endoplasmic reticulum-associated degradation. J Biol Chem. 2014;289:3510–3517. doi: 10.1074/jbc.M113.538934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Blount JR, Burr AA, Denuc A, Marfany G, Todi SV. Ubiquitin-specific protease 25 functions in endoplasmic reticulum-associated degradation. PLoS ONE. 2012;7:e36542. doi: 10.1371/journal.pone.0036542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 210.Kaneko C, Hatakeyama S, Matsumoto M, Yada M, Nakayama K, Nakayama KI. Characterization of the mouse gene for the U-box-type ubiquitin ligase UFD2a. Biochem Biophys Res Commun. 2003;300:297–304. doi: 10.1016/S0006-291X(02)02834-6. [DOI] [PubMed] [Google Scholar]
- 211.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci. 2012;125:539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 213.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 214.Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJHJ, Hansen TH. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol. 2007;177:613–624. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Magadán JG, Pérez-Victoria FJ, Sougrat R, Ye Y, Strebel K, Bonifacino JS. Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps. PLoS Pathog. 2010;6:e1000869. doi: 10.1371/journal.ppat.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Tokarev AA, Munguia J, Guatelli JC. Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu. J Virol. 2011;85:51–63. doi: 10.1128/JVI.01795-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Gustin JK, Douglas JL, Bai Y, Moses AV. Ubiquitination of BST-2 protein by HIV-1 Vpu protein does not require lysine, serine, or threonine residues within the BST-2 cytoplasmic domain. J Biol Chem. 2012;287:14837–14850. doi: 10.1074/jbc.M112.349928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ishikura S, Weissman AM, Bonifacino JS. Serine residues in the cytosolic tail of the T-cell antigen receptor α-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem. 2010;285:23916–23924. doi: 10.1074/jbc.M110.127936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shimizu Y, Okuda-Shimizu Y, Hendershot LM. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell. 2010;40:917–926. doi: 10.1016/j.molcel.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, et al. Nedd8 modification of cul-1 activates SCFpTrCP-dependent ubiquitination of iκbα. Mol Cell Biol. 2000;20:2326–2333. doi: 10.1128/MCB.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ramachandran S, Osterhaus SR, Parekh KR, Jacobi AM, Behlke MA, McCray PB., Jr SYVN1, NEDD8, and FBXO2 proteins regulate DeltaF508 cystic fibrosis transmembrane conductance regulator (CFTR) ubiquitin-mediated proteasomal degradation. J Biol Chem. 2016;291:25489–25504. doi: 10.1074/jbc.M116.754283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Eifler K, Vertegaal ACO. Mapping the SUMOylated landscape. FEBS J. 2015;282:3669–3680. doi: 10.1111/febs.13378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Flotho A, Melchior F. SUMOylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 224.Tatham MH, Geoffroy M-C, Shen L, Plechanovova A, Hattersley N, Jaffray EG, et al. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 225.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, et al. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 226.Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102–4112. doi: 10.1038/sj.emboj.7601839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ahner A, Gong X, Schmidt BZ, Peters KW, Rabeh WM, Thibodeau PH, et al. Small heat shock proteins target mutant cystic fibrosis transmembrane conductance regulator for degradation via a small ubiquitin-like modifier-dependent pathway. Mol Biol Cell. 2013;24:74–84. doi: 10.1091/mbc.E12-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gong X, Ahner A, Roldan A, Lukacs GL, Thibodeau PH, Frizzell RA. Non-native conformers of cystic fibrosis transmembrane conductance regulator NBD1 are recognized by Hsp27 and conjugated to SUMO-2 for degradation. J Biol Chem. 2016;291:2004–2017. doi: 10.1074/jbc.M115.685628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Polevoda B, Sherman F. N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol. 2003;325:595–622. doi: 10.1016/S0022-2836(02)01269-X. [DOI] [PubMed] [Google Scholar]
- 230.Hwang CS, Shemorry A, Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zattas D, Adle DJ, Rubenstein EM, Hochstrasser M. N-terminal acetylation of the yeast derlin Der1 is essential for Hrd1 ubiquitin-ligase activity toward luminal ER substrates. Mol Biol Cell. 2013;24:890–900. doi: 10.1091/mbc.E12-11-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.LaLonde DP, Bretscher A. The UBX protein SAKS1 negatively regulates endoplasmic reticulum-associated degradation and p97-dependent degradation. J Biol Chem. 2011;286:4892–4901. doi: 10.1074/jbc.M110.158030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.McNeill H, Knebel A, Arthur JSC, Cuenda A, Cohen P. A novel UBA and UBX domain protein that binds polyubiquitin and VCP and is a substrate for SAPKs. Biochem J. 2004;384:391–400. doi: 10.1042/BJ20041498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ballar P, Zhong Y, Nagahama M, Tagaya M, Shen Y, Fang S. Identification of SVIP as an endogenous inhibitor of endoplasmic reticulum-associated degradation. J Biol Chem. 2007;282:33908–33914. doi: 10.1074/jbc.M704446200. [DOI] [PubMed] [Google Scholar]
- 235.Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 236.Aviram S, Kornitzer D. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol Cell Biol. 2010;30:985–994. doi: 10.1128/MCB.00909-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kohlmann S, Schäfer A, Wolf DH. Ubiquitin ligase Hul5 is required for fragment-specific substrate degradation in endoplasmic reticulum-associated degradation. J Biol Chem. 2008;283:16374–16383. doi: 10.1074/jbc.M801702200. [DOI] [PubMed] [Google Scholar]
- 238.Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Huyer G, Piluek WF, Fansler Z, Kreft SG, Hochstrasser M, Brodsky JL, et al. Distinct machinery is required in saccharomyces cerevisiae for the endoplasmic reticulum-associated degradation of a multispanning membrane protein and a soluble luminal protein. J Biol Chem. 2004;279:38369–38378. doi: 10.1074/jbc.M402468200. [DOI] [PubMed] [Google Scholar]
- 240.Matsumura Y, David LL, Skach WR. Role of Hsc70 binding cycle in CFTR folding and endoplasmic reticulum–associated degradation. Mol Biol Cell. 2011;22:2797–2809. doi: 10.1091/mbc.E11-02-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Needham PG, Mikoluk K, Dhakarwal P, Khadem S, Snyder AC, Subramanya AR, et al. The thiazide-sensitive NaCl cotransporter is targeted for chaperone-dependent endoplasmic reticulum-associated degradation. J Biol Chem. 2011;286:43611–43621. doi: 10.1074/jbc.M111.288928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ciechanover A, Orian A, Schwartz AL. Ubiquitin-mediated proteolysis: biological regulation via destruction. BioEssays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 243.Needham PG, Brodsky JL. How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: the early history of ERAD. Biochim Biophys Acta. 2013;1833:2447–2457. doi: 10.1016/j.bbamcr.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.McCracken AA, Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chung KKK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, et al. S-nitrosylation of parkin regulates ubiquitination and compromises Parkinson’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 247.Wang J, Sevier CS. Formation and reversibility of BiP protein cysteine oxidation facilitate cell survival during and post oxidative stress. J Biol Chem. 2016;291:7541–7557. doi: 10.1074/jbc.M115.694810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Chambers JE, Petrova K, Tomba G, Vendruscolo M, Ron D. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J Cell Biol. 2012;198:371–385. doi: 10.1083/jcb.201202005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Hendershot LM, Ting J, Lee AS. Identity of the immunoglobulin heavy-chain-binding protein with the 78,000-dalton glucose-regulated protein and the role of posttranslational modifications in its binding function. Mol Cell Biol. 1988;8:4250–4256. doi: 10.1128/MCB.8.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Sanyal A, Chen AJ, Nakayasu ES, Lazar CS, Zbornik EA, Worby CA, et al. A novel link between Fic (filamentation induced by cAMP)-mediated adenylylation/AMPylation and the unfolded protein response. J Biol Chem. 2015;290:8482–8499. doi: 10.1074/jbc.M114.618348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.van Anken E, Romijn EP, Maggioni C, Mezghrani A, Sitia R, Braakman I, et al. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18:243–253. doi: 10.1016/S1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 252.Arteaga MF, Wang L, Ravid T, Hochstrasser M, Canessa CM. An amphipathic helix targets serum and glucocorticoid-induced kinase 1 to the endoplasmic reticulum-associated ubiquitin-conjugation machinery. Proc Natl Acad Sci USA. 2006;103:11178–11183. doi: 10.1073/pnas.0604816103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Furth N, Gertman O, Shiber A, Alfassy OS, Cohen I, Rosenberg MM, et al. Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol Biol Cell. 2011;22:4726–4739. doi: 10.1091/mbc.E11-05-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATα2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/S0092-8674(00)81421-X. [DOI] [PubMed] [Google Scholar]
- 255.Gilon T, Chomsky O, Kulka RG. Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J. 1998;17:2759–2766. doi: 10.1093/emboj/17.10.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Mbonye UR, Wada M, Rieke CJ, Tang HY, DeWitt DL, Smith WL. The 19-amino acid cassette of cyclooxygenase-2 mediates entry of the protein into the endoplasmic reticulum-associated degradation system. J Biol Chem. 2006;281:35770–35778. doi: 10.1074/jbc.M608281200. [DOI] [PubMed] [Google Scholar]
- 257.Shapira I, Charuvi D, Elkabetz Y, Hirschberg K, Bar-Nun S. Distinguishing between retention signals and degrons acting in ERAD. J Cell Sci. 2007;120:4377–4387. doi: 10.1242/jcs.011247. [DOI] [PubMed] [Google Scholar]
- 258.Singh N, Joshi R, Komurov K. HER2-mTOR signaling–driven breast cancer cells require ER-associated degradation to survive. Sci Signal. 2015;8:ra52. doi: 10.1126/scisignal.aaa6922. [DOI] [PubMed] [Google Scholar]
- 259.Francisco AB, Singh R, Li S, Vani AK, Yang L, Munroe RJ, et al. Deficiency of suppressor enhancer Lin12 1 like (SEL1L) in mice leads to systemic endoplasmic reticulum stress and embryonic lethality. J Biol Chem. 2010;285:13694–13703. doi: 10.1074/jbc.M109.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Sun S, Shi G, Han X, Francisco AB, Ji Y, Mendonça N, et al. Sel1l is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proc Natl Acad Sci USA. 2014;111:E582–E591. doi: 10.1073/pnas.1318114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Sha H, Sun S, Francisco AB, Ehrhardt N, Xue Z, Liu L, et al. The ER-associated degradation adaptor protein Sel1L regulates LPL secretion and lipid metabolism. Cell Metab. 2014;20:458–470. doi: 10.1016/j.cmet.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.McGrath JP, Jentsch S, Varshavsky A. UBA 1: an essential yeast gene encoding ubiquitin-activating enzyme. EMBO J. 1991;10:227–236. doi: 10.1002/j.1460-2075.1991.tb07940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Handley-Gearhart PM, Stephen AG, Trausch-Azar JS, Ciechanover A, Schwartz AL. Human ubiquitin-activating enzyme, E1.indication of potential nuclear and cytoplasmic subpopulations using epitope-tagged cDNA constructs. J Biol Chem. 1994;269:33171–33178. [PubMed] [Google Scholar]
- 264.Handley-Gearhart PM, Trausch-Azar J, Ciechanover A, Schwartz AL. Rescue of the complex temperature-sensitive phenotype of Chinese hamster ovary E36ts20 cells by expression of the human ubiquitin-activating enzyme cDNA. Biochem J. 1994;304:1015–1020. doi: 10.1042/bj3041015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Yamasaki S, Yagishita N, Sasaki T, Nakazawa M, Kato Y, Yamadera T, et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 2007;26:113–122. doi: 10.1038/sj.emboj.7601490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Biederer T, Volkwein C, Sommer T. Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin-proteasome pathway. EMBO J. 1996;15:2069–2076. [PMC free article] [PubMed] [Google Scholar]
- 267.Wang Q, Chang A. Eps1, a novel PDI-related protein involved in ER quality control in yeast. EMBO J. 1999;18:5972–5982. doi: 10.1093/emboj/18.21.5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Jungmann J, Reins HA, Schobert C, Jentsch S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature. 1993;361:369–371. doi: 10.1038/361369a0. [DOI] [PubMed] [Google Scholar]
- 269.Kaneko M, Koike H, Saito R, Kitamura Y, Okuma Y, Nomura Y. Loss of HRD1-mediated protein degradation causes amyloid precursor protein accumulation and amyloid-p generation. J Neurosci. 2010;30:3924–3932. doi: 10.1523/JNEUROSCI.2422-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lenk U, Yu H, Walter J, Gelman MS, Hartmann E, Kopito RR, et al. A role for mammalian Ubc6 homologues in ER-associated protein degradation. J Cell Sci. 2002;115:3007–3014. doi: 10.1242/jcs.115.14.3007. [DOI] [PubMed] [Google Scholar]
- 271.Flierman D, Coleman CS, Pickart CM, Rapoport TA, Chau V. E2-25K mediates US11-triggered retro-translocation of MHC class I heavy chains in a permeabilized cell system. Proc Natl Acad Sci USA. 2006;103:11589–11594. doi: 10.1073/pnas.0605215103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Yan L, Liu W, Zhang H, Liu C, Shang Y, Ye Y, et al. Ube2g2-gp78-mediated HERP polyubiquitylation is involved in ER stress recovery. J Cell Sci. 2014;127:1417–1427. doi: 10.1242/jcs.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Tiwari S, Weissman AM. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits involvement of ER-associated ubiquitin-conjugating enzymes (E2s) J Biol Chem. 2001;276:16193–16200. doi: 10.1074/jbc.M007640200. [DOI] [PubMed] [Google Scholar]
- 274.Webster JM, Tiwari S, Weissman AM, Wojcikiewicz RJ. Inositol 1,4,5-trisphosphate receptor ubiquitination is mediated by mammalian Ubc7, a component of the endoplasmic reticulum-associated degradation pathway, and is inhibited by chelation of intracellular Zn2+ J Biol Chem. 2003;278:38238–38246. doi: 10.1074/jbc.M305600200. [DOI] [PubMed] [Google Scholar]
- 275.Gardner RG, Shearer AG, Hampton RY. In vivo action of the HRD ubiquitin ligase complex: mechanisms of endoplasmic reticulum quality control and sterol regulation. Mol Cell Biol. 2001;21:4276–4291. doi: 10.1128/MCB.21.13.4276-4291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Yang H, Zhong X, Ballar P, Luo S, Shen Y, Rubinsztein DC, et al. Ubiquitin ligase Hrd1 enhances the degradation and suppresses the toxicity of polyglutamine-expanded huntingtin. Exp Cell Res. 2007;313:538–550. doi: 10.1016/j.yexcr.2006.10.031. [DOI] [PubMed] [Google Scholar]
- 277.Apodaca J, Kim I, Rao H. Cellular tolerance of prion protein PrP in yeast involves proteolysis and the unfolded protein response. Biochem Biophys Res Commun. 2006;347:319–326. doi: 10.1016/j.bbrc.2006.06.078. [DOI] [PubMed] [Google Scholar]
- 278.Plemper RK, Egner R, Kuchler K, Wolf DH. Endoplasmic reticulum degradation of a mutated ATP-binding cassette transporter Pdr5 proceeds in a concerted action of Sec61 and the proteasome. J Biol Chem. 1998;273:32848–32856. doi: 10.1074/jbc.273.49.32848. [DOI] [PubMed] [Google Scholar]
- 279.Avci D, Fuchs S, Schrul B, Fukumori A, Breker M, Frumkin I, et al. The yeast ER-intramembrane protease Ypf1 refines nutrient sensing by regulating transporter abundance. Mol Cell. 2014;56:630–640. doi: 10.1016/j.molcel.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 280.Habeck G, Ebner FA, Shimada-Kreft H, Kreft SG. The yeast ERAD-C ubiquitin ligase Doa10 recognizes an intramembrane degron. J Cell Biol. 2015;209:261–273. doi: 10.1083/jcb.201408088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Wang H, Li Q, Shen Y, Sun A, Zhu X, Fang S, et al. The ubiquitin ligase Hrd1 promotes degradation of the Z variant alpha 1-antitrypsin and increases its solubility. Mol Cell Biochem. 2011;346:137–145. doi: 10.1007/s11010-010-0600-9. [DOI] [PubMed] [Google Scholar]
- 282.Shmueli A, Tsai YC, Yang M, Braun MA, Weissman AM. Targeting of gp78 for ubiquitin-mediated proteasomal degradation by Hrd1: cross-talk between E3s in the endoplasmic reticulum. Biochem Biophys Res Commun. 2009;390:758–762. doi: 10.1016/j.bbrc.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Ballar P, Ors AU, Yang H, Fang S. Differential regulation of CFTRΔF508 degradation by ubiquitin ligases gp78 and Hrd1. Int J Biochem Cell Biol. 2010;42:167–173. doi: 10.1016/j.biocel.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 284.Kong S, Yang Y, Xu Y, Wang Y, Zhang Y, Melo-Cardenas J, et al. Endoplasmic reticulum-resident E3 ubiquitin ligase Hrd1 controls B-cell immunity through degradation of the death receptor CD95/Fas. Proc Natl Acad Sci USA. 2016;113:10394–10399. doi: 10.1073/pnas.1606742113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Lee KA, Hammerle LP, Andrews PS, Stokes MP, Mustelin T, Silva JC, et al. Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J Biol Chem. 2011;286:41530–41538. doi: 10.1074/jbc.M111.248856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Liang JS, Kim T, Fang S, Yamaguchi J, Weissman AM, Fisher EA, et al. Overexpression of the tumor autocrine motility factor receptor Gp78, a ubiquitin protein ligase, results in increased ubiquitinylation and decreased secretion of apolipoprotein B100 in HepG2 cells. J Biol Chem. 2003;278:23984–23988. doi: 10.1074/jbc.M302683200. [DOI] [PubMed] [Google Scholar]
- 287.Song B-L, Sever N, DeBose-Boyd RA. Gp78, a membrane-anchored ubiquitin ligase, associates with Insig-1 and couples sterol-regulated ubiquitination to degradation of HMG CoA reductase. Mol Cell. 2005;19:829–840. doi: 10.1016/j.molcel.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 288.Shen Y, Ballar P, Fang S. Ubiquitin ligase gp78 increases solubility and facilitates degradation of the Z variant of α-1-antitrypsin. Biochem Biophys Res Commun. 2006;349:1285–1293. doi: 10.1016/j.bbrc.2006.08.173. [DOI] [PubMed] [Google Scholar]
- 289.Tsai YC, Mendoza A, Mariano JM, Zhou M, Kostova Z, Chen B, et al. The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat Med. 2007;13:1504–1509. doi: 10.1038/nm1686. [DOI] [PubMed] [Google Scholar]
- 290.Kim SM, Acharya P, Engel JC, Correia MA. Liver cytochrome P450 3A ubiquitination in vivo by gp78/autocrine motility factor receptor and C terminus of Hsp70-interacting protein (CHIP) E3 ubiquitin ligases physiological and pharmacological relevance. J Biol Chem. 2010;285:35866–35877. doi: 10.1074/jbc.M110.167189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Höhfeld J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3:93–96. doi: 10.1038/35050618. [DOI] [PubMed] [Google Scholar]
- 292.Donnelly BF, Needham PG, Snyder AC, Roy A, Khadem S, Brodsky JL, et al. Hsp70 and Hsp90 multichaperone complexes sequentially regulate thiazide-sensitive cotransporter endoplasmic reticulum-associated degradation and biogenesis. J Biol Chem. 2013;288:13124–13135. doi: 10.1074/jbc.M113.455394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Stagg HR, Thomas M, van den Boomen D, Wiertz EJHJ, Drabkin HA, Gemmill RM, et al. The TRC8 E3 ligase ubiquitinates MHC class I molecules before dislocation from the ER. J Cell Biol. 2009;186:685–692. doi: 10.1083/jcb.200906110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lee JP, Brauweiler A, Rudolph M, Hooper JE, Drabkin HA, Gemmill RM. The TRC8 ubiquitin ligase is sterol regulated and interacts with lipid and protein biosynthetic pathways. Mol Cancer Res. 2010;8:93–106. doi: 10.1158/1541-7786.MCR-08-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Boname JM, Bloor S, Wandel MP, Nathan JA, Antrobus R, Dingwell KS, et al. Cleavage by signal peptide peptidase is required for the degradation of selected tail-anchored proteins. J Cell Biol. 2014;205:847–862. doi: 10.1083/jcb.201312009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Hsu JL, van den Boomen DJH, Tomasec P, Weekes MP, Antrobus R, Stanton RJ, et al. Plasma membrane profiling defines an expanded class of cell surface proteins selectively targeted for degradation by HCMV US2 in cooperation with UL141. PLoS Pathog. 2015;11:e1004811. doi: 10.1371/journal.ppat.1004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Lu JP, Wang Y, Sliter DA, Pearce MMP, Wojcikiewicz RJH. RNF170 protein, an endoplasmic reticulum membrane ubiquitin ligase, mediates inositol 1,4,5-trisphosphate receptor ubiquitination and degradation. J Biol Chem. 2011;286:24426–24433. doi: 10.1074/jbc.M111.251983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.El Khouri E, Le Pavec G, Toledano MB, Delaunay-Moisan A. RNF185 is a novel E3 ligase of endoplasmic reticulum-associated degradation (ERAD) that targets cystic fibrosis transmembrane conductance regulator (CFTR) J Biol Chem. 2013;288:31177–31191. doi: 10.1074/jbc.M113.470500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Ron I, Rapaport D, Horowitz M. Interaction between parkin and mutant glucocerebrosidase variants: a possible link between Parkinson disease and Gaucher disease. Hum Mol Genet. 2010;19:3771–3781. doi: 10.1093/hmg/ddq292. [DOI] [PubMed] [Google Scholar]
- 300.Lerner M, Corcoran M, Cepeda D, Nielsen ML, Zubarev R, Pontén F, et al. The RBCC gene RFP2 (Leu5) encodes a novel transmembrane E3 ubiquitin ligase involved in ERAD. Mol Biol Cell. 2007;18:1670–1682. doi: 10.1091/mbc.E06-03-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Altier C, Garcia-Caballero A, Simms B, You H, Chen L, Walcher J, et al. The Cavβ subunit prevents RFP2-mediated ubiquitination and proteasomal degradation of L-type channels. Nat Neurosci. 2011;14:173–180. doi: 10.1038/nn.2712. [DOI] [PubMed] [Google Scholar]
- 302.Guo X, Shen S, Song S, He S, Cui Y, Xing G, et al. The E3 ligase Smurf1 regulates Wolfram syndrome protein stability at the endoplasmic reticulum. J Biol Chem. 2011;286:18037–18047. doi: 10.1074/jbc.M111.225615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Neutzner A, Neutzner M, Benischke AS, Ryu SW, Frank S, Youle RJ, et al. A systematic search for endoplasmic reticulum (ER) membrane-associated RING finger proteins identifies Nixin/ZNRF4 as a regulator of calnexin stability and ER homeostasis. J Biol Chem. 2011;286:8633–8643. doi: 10.1074/jbc.M110.197459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Fry WHD, Simion C, Sweeney C, Carraway KL. Quantity control of the ErbB3 receptor tyrosine kinase at the endoplasmic reticulum. Mol Cell Biol. 2011;31:3009–3018. doi: 10.1128/MCB.05105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem. 2008;283:12797–12810. doi: 10.1074/jbc.M801122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, Shanks JR, et al. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–11049. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Goldfarb SB, Kashlan OB, Watkins JN, Suaud L, Yan W, Kleyman TR, et al. Differential effects of Hsc70 and Hsp70 on the intracellular trafficking and functional expression of epithelial sodium channels. Proc Natl Acad Sci USA. 2006;103:5817–5822. doi: 10.1073/pnas.0507903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Jo Y, Hartman IZ, DeBose-Boyd RA. Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet–associated endoplasmic reticulum membranes. Mol Biol Cell. 2013;24:169–183. doi: 10.1091/mbc.E12-07-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Tsuchiya Y, Morita T, Kim M, Iemura S-i, Natsume T, Yamamoto M, et al. Dual regulation of the transcriptional activity of Nrf1 by β-TrCP- and Hrd1-dependent degradation mechanisms. Mol Cell Biol. 2011;31:4500–4512. doi: 10.1128/MCB.05663-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Bernardi KM, Williams JM, Inoue T, Schultz A, Tsai B. A deubiquitinase negatively regulates retro-translocation of nonubiquitinated substrates. Mol Biol Cell. 2013;24:3545–3556. doi: 10.1091/mbc.E13-06-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]


