Abstract
Exosomes are nanosized membrane particles that are secreted by cells that transmit information from cell to cell. The information within exosomes prominently includes their protein and RNA payloads. Exosomal micro-RNAs in particular can potently and fundamentally alter the transcriptome of recipient cells. Here we summarize what is known about exosome biogenesis, content, and transmission, with a focus on cardiovascular physiology and pathophysiology. We also highlight some of the questions currently under active investigation regarding these extracellular membrane vesicles and their potential in diagnostic and therapeutic applications.
Keywords: extracellular vesicles, microRNA, regenerative medicine, cardiovascular disease, stem cells, intracellular communication
INTRODUCTION
The cardiovascular system is a dynamic network of various cell types that collectively support the circulation of blood throughout the body. The heartbeat is initiated by electrical signals that chemically couple to contraction, propelling blood into vessels that finely control perfusion and fluid balance. The constant adaptation of these features to environmental challenges requires a tight and multifaceted system of cell-cell communication and regulation of the transcriptome. Much emphasis has traditionally been placed on cytokines, peptides, and nitric oxide as signaling mediators of cardiac contractility and vasomotor tone. We have, until recently, failed to recognize the role of extracellular vesicles (EVs), and particularly exosomes, as agents of cell-cell signaling both locally and remotely. EVs have been known to exist in biological fluids for more than seven decades. In 1940, Chargaff and others noted “lipoproteins of a very high particle weight… [that] readily form sediments in a strong centrifugal field… but remain in solution when subjected to weak centrifugal field” (1). The term used to describe these particles was thromboplastic substances. Despite the limited technology of the time, Chargaff & West (2) accurately predicted an important role of these particles in health and disease. Two decades later, Wolf and associates (3) isolated these thromboplastic particles from the subcellular fraction and resolved them using electron microscopy. Distinct small vesicles ranging from 20 to 50 nm were observed and were then termed platelet dust. The term exosomes was first coined by Johnstone and others to describe vesicles released from cultured maturing sheep reticulocytes (4, 5). These particles had enzymatic (e.g., acetylcholinesterase and transferrin) activity that was lost during reticulocyte-to-erythrocyte maturation. The prevailing view then was that these vesicles were a means to remove factors from the plasma membrane that were no longer needed by the mature erythrocyte (5).
With the advent of high-throughput proteomics and genomics and evolving understanding of paracrine mechanisms, particularly in cancer and development, the field of exosome biology experienced a paradigm shift; particles once thought to be involved in waste management are now widely accepted as highly conserved elements in a pathway of short- and long-range communication. All examined eukaryotic cell types—including hematopoietic cells, epithelial cells, neural cells, stem cells, adipocytes, and cancer cells—secrete exosomes in culture. Canonical exosome production has been demonstrated across all taxa of Eukaryota, from single-cell amoeboid protists (6) to fungi (7), plants (8), and animals (9–11). In vivo, exosomes can be isolated in great abundance from all body fluids, including blood, ascites, cerebrospinal fluid, saliva, milk, and even urine (12–19). Figure 1 represents bodily fluids that contain exosomes along with a schematic of the vesicle, its membrane contents, and molecular payload. These vesicles transmit a variety of signaling molecules in their payload, notably proteins, mRNA, and noncoding RNA—including, most notably, microRNAs (miRs) (20). Furthermore, the range of these particles and their ability to transfer their molecular payloads have been described in various models of health and disease. Indeed, there is now a wealth of evidence that exosomes can mediate autocrine, paracrine, and endocrine functions (21–23). The first area of clinical translation has been in diagnostics: Exosomes isolated from blood or urine can be used as markers of disease and prognosis in cancer (24) and perhaps also in CNS disease (25) and heart disease (26). Cell therapy for the objective of tissue regeneration has helped spur a third perspective regarding the role of exosomes in cardiovascular disease: Exosomes may mediate some or all cell-triggered therapeutic effects. This review summarizes what is currently known about exosomes in the cardiovascular space from the three aforementioned perspectives: markers of prognosis, functional role in pathology, and potential value in therapeutic applications. We also discuss current gaps in understanding and avenues of expanding prospective areas of investigation that would help propel this field to gain further insight and translational utility.
Figure 1.
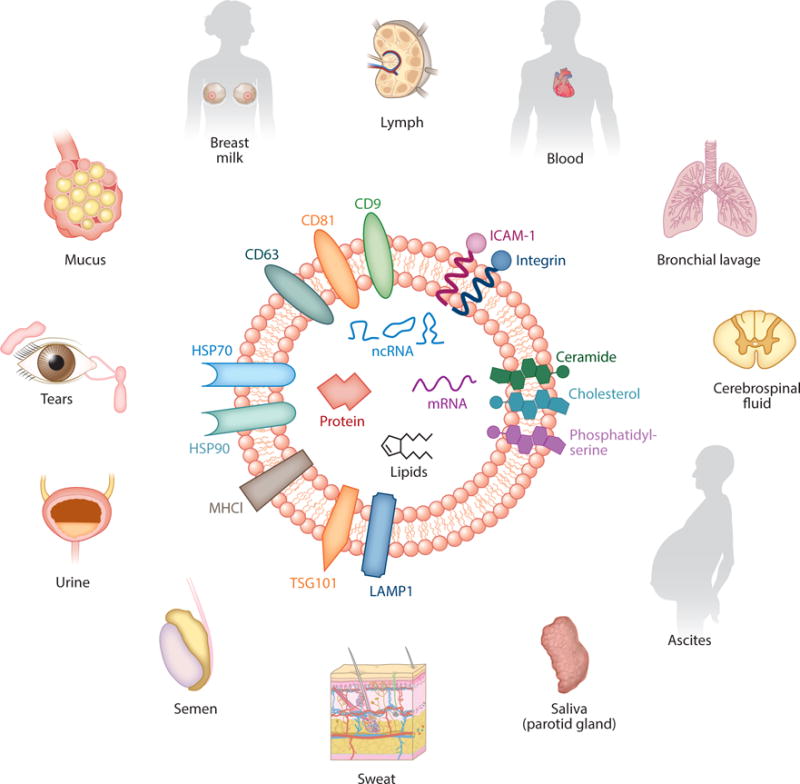
Exosomes are nanosized particles with a diameter range of 30–100 nm and are secreted by all cell types. Enriched in cholesterol, ceramide, and phosphatidylserine, these lipid bilayer particles have a lipid content different from that of the parent cell. Markers ubiquitous in most exosomes include tetraspanins (CD9, CD63, and CD81); heat shock proteins; adhesion molecules; and markers of the ESCRT (endosomal sorting complexes required for transport) pathway, including LAMP1 and TSG101.
EXOSOMES: CONSERVED MEDIATORS OF CELLULAR COMMUNICATION
Exosome Definition and Nomenclature
As is common with newly discovered entities, EVs of different classes have been termed inconsistently and nonsystematically. The initial definition of exosomes was 50–100-nm lipid bilayer particles released from cells. This definition has since been amended, with the size range expanding to incorporate particles as small as 20 nm in diameter (16) and those as large as 150 nm in diameter (27), although most studies use a size range of 30–100 nm. Particle size has become one of the mainstays of differentiating exosomes from a myriad of other particles, notably microvesicles, that tend to be larger (up to ~1,000 nm in diameter). In addition, exosomes were historically named after the cell of origin. For instance, cancer-derived exosomes were termed oncosomes, and dendritic cell—derived exosomes were termed dexosomes. This approach has led to the unfortunate and confusing introduction of at least 20 different terms describing lipid particles released by different cell types, with little regard for size or content.
As exosome biogenesis became better described, a second qualifier for exosome identity—their biological pathway of origin—emerged. Exosomes arise from intraluminal endosomal vesicles (as viewed under electron microscopy) that fuse with the endosome, a product of invagination of the plasma membrane. As the endosome matures and eventually merges with the plasma membrane, the contents released into the extracellular space become the exosomes (28–30). This ontogeny presents an advantage to EV characterization, as exosomes can be further identified by markers of their biogenesis. Specifically, exosomes contain remnants of the ESCRT (endosomal sorting complexes required for transport) pathway. However, the markers associated with ESCRT are not entirely exclusive. For instance, some of the markers conserved in exosomes, such as tetraspanins (notably CD63, CD81, and CD9), are also present on the plasma membrane, in the cytosol, and in vesicles derived from membrane shedding (31). The promiscuity that is seen with tetraspanins is likely due to the conserved and multifunctional capacities of these proteins, including cell activation and proliferation, cell adhesion and motility, and cell differentiation. Other exosome markers such as lysosomal-associated membrane proteins (LAMPs), including LAMP1 and LAMP2, are also abundantly present in the lysosomal compartment. Such abundance complicates the process of distinguishing them from lysosomal vesicles and autophagosomes (32).
Finally, the primary method for exosome isolation remains the same since its discovery more than seven decades ago: centrifugation. Among the various alternative strategies that have emerged since then are filtration; chromatography; bead isolation; and variations on existing centrifugation methods, including preparations using precipitation material that sediments lipid bilayer particles. However, the primary method remains purification by size. Therefore, given the overlap in the nomenclature regarding different classes of particles and the competing conventions, developing a unified nomenclature system is important. More discriminatory methods need to be developed to isolate bona fide exosomes that are free of membrane blebs and other EVs, so as to better understand the mechanisms and payload of exosome trafficking and downstream effects. Strides are being made in this regard. The Office of Strategic Coordination of the National Institutes of Health has launched a Common Fund program for extracellular RNA communication in an effort to define exosomes, to understand exosome biogenesis and transmission, and to investigate various facets of their clinical application (33). Part of that effort also involves unifying nomenclature and methods for exosome isolation and characterization.
Exosome Biogenesis and Release
Exosomes are a unique class of EVs by virtue of their biogenesis. Unlike apoptotic bodies (more broadly termed ectosomes), which form from the blebs of dead or dying cells, or microvesicles, which form from outward budding of the plasma membrane, exosomes arise from multivesicular endosomes (MVEs). MVEs, also known as multivesicular bodies (MVBs), form from invaginations of the plasma membrane that fuse with payload sorted in the endoplasmic reticulum and processed in the Golgi complex. This loaded “bag of marbles” then fuses to the plasma membrane for release as exosomes. However, not all vesicles in the MVEs are sorted for release. Some MVEs fuse with lysosomes as part of a degradation pathway (30, 34). MVEs are produced through lateral segregation of the molecular payload at the delimiting membrane of the endosome, followed by inward budding and pinching of vesicles into the endosomal lumen. Figure 2 provides a broad overview of the origins of EVs, including exosomes, apoptosomes, and microvesicles. Exosomes are a class of vesicles secreted via the classical secretory pathway that involves fusion of payload with MVEs. Why certain MVEs are destined for the lysosomal compartment versus fusion with plasma membrane remains poorly understood, although there is evidence that there are two populations of MVEs: cholesterol-rich secretory MVEs (sMVEs) and another cholesterol-poor population prone to fuse with lysosomes (lMVEs) (35) (Figure 2). Furthermore, lMVEs contain lysobisphosphatidic acid, which is absent in sMVEs (39). Microvesicles arise from membrane shedding from injured or transformed cells. Finally, apoptotic bodies, or apoptosomes, are vesicles shed from dying cells or cell bodies that remain after apoptosis. Consequently, the membrane lipid content of these EVs is more similar to that of the plasma membrane of the cell of origin. Exosomes, however, also include lipid from the Golgi complex, making them more distinct from the plasma membrane. The pathway of biogenesis is similar to that of ubiquitination and degradation of cellular products, with a few distinct features, including cholesterol content of the endosome (35).
Figure 2.
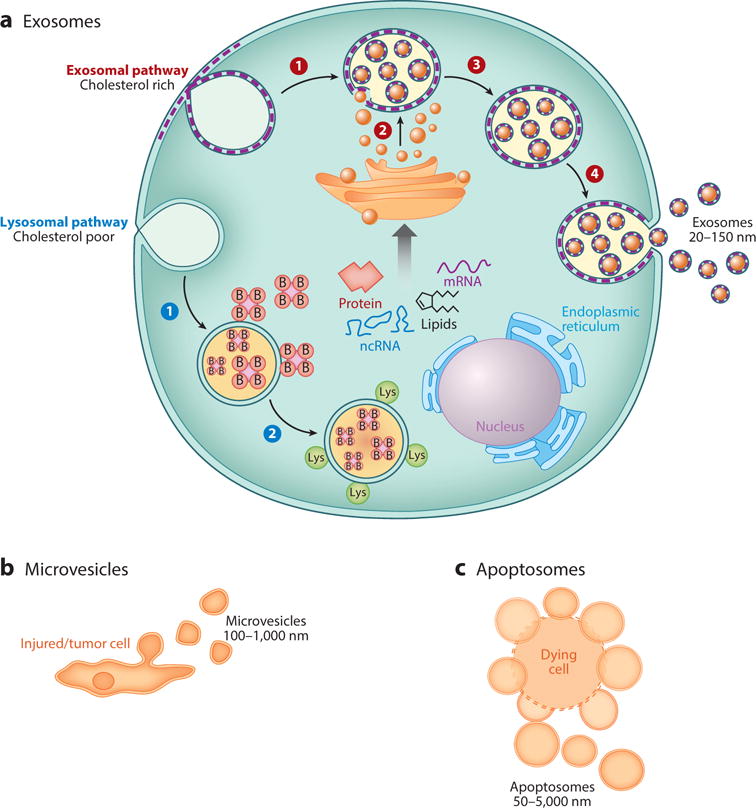
(a) Formation of two different multivesicular bodies (MVBs) through invagination of the plasma membrane. Exosome-associated MVBs are more enriched in cholesterol than are MVBs involved in the degradation pathway for ubiquitinated proteins. B represents the addition of ubiquitin to protein substrates; Lys denotes fusion of lysosomes. Steps 1–4 (red): Invagination of the plasma membrane to form a secretory endosome (1), followed by budding of payload into the endosomal membrane to form multivesicular endosomes (2). Maturation of the late endosome through acidification (3) triggers fusion with the plasma membrane and release of exosomes (4). Steps1 and 2(blue): Invagination of the plasma membrane to form lysosomal membrane (1), followed by fusion of ubiquitinated products for lysosomal degradation (2). Both pathways involve the ESCRT (endosomal sorting complexes required for transport) pathway for budding of molecular payload into the lumen ofa MVB as it matures to the late endosome or fuses with lysosomes for degradation. (b,c) Other extracellular vesicles and their mode of secretion include (b) microvesicles that shed from injured cells and (c) tumor cells and apoptosomes that bleb from dead and dying cells.
The cellular machinery involved in this process has been resolved, to some degree, using mutant yeast lines. Figure 3 represents members of the endosomal complex, including ESCRT 0, I, II, and III, which are involved in endosome biogenesis and exosome release. Complexes 0, I, and II recognize and sequester ubiquitinated protein products (for lysosomal degradation) at the endosomal delimiting membrane, whereas complex III is responsible for membrane budding and scission of intraluminal vesicles (36–38) (Figure 3).
Figure 3.
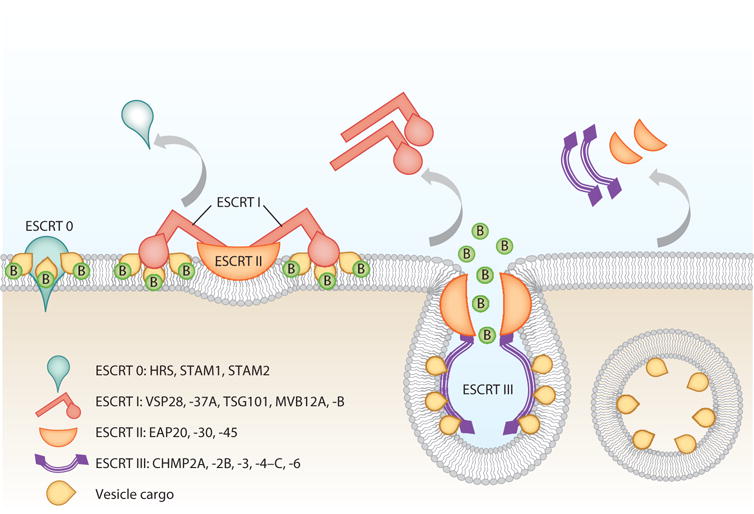
The ESCRT (endosomal sorting complexes required for transport) pathway is the mechanism by which molecular payload populates the multivesicular body (MVB). Although exosomes do not fully utilize the ESCRT machinery, here the four ESCRT complexes are shown. ESCRT 0 complex proteins (B represents a ubiquitin group bound to a protein substrate) recognize ubiquitinated protein products and concentrate them in microdomains by using clathrin molecules and recruit ESCRT I, which in turn recruits ESCRT II and triggers membrane involution. ESCRT III complexes form circular filaments and cause further invagination of the membrane and ultimately both membrane abscission and budding of the proteins into the lumen of the endosome and the elimination of ubiquitin outside the endosome.
Exosome Composition
Exosomes from different sources contain different structural proteins and lipids. The payloads are even more diverse. According to the current version of the exosome content database, ExoCarta (version 4; http://www.exocarta.org), from the 146 studies of different cell types and organisms, 4,563 proteins, 1,639 mRNAs, 764 miRNAs, and 194 species of lipids have been identified in exosomes (40). Nevertheless, this section focuses on the structurally conserved components of the vesicle and on the classes of signals incorporated into the payload.
Lipid composition
Exosomes are bounded by a lipid bilayer membrane consisting of cholesterol, diglycerides, sphingolipids, phospholipids, glycerophospholipids, and polyglycerophospholipids (41). The lipid composition of exosomes differs from that of the plasma membrane, which explains the difference in the physical properties, including the exceptional rigidity of exosome membranes compared with that of the plasma membrane. Some of these lipids also serve extrastructural functions, including trafficking during biogenesis (particularly trafficking of ceramide, which is critical for budding into the endosome) (42), recognition, and internalization (41). Aside from structural and trafficking components, exosomes also contain bioactive lipids, including prostaglandins, leukotrienes, and active enzymes that can generate these lipids (43).
Membrane proteins
The most commonly occurring proteins belong to classes of membrane transport and fusion proteins, including tetraspanins (e.g., CD63, CD9, and CD81), heat shock proteins (e.g., Hspa8, Hsp90), GTPases (e.g., EEF1A1, EEF2), and endosomal proteins and markers (e.g., Alix). Other proteins commonly found on exosomes include cytoskeletal, metabolic, signaling, and carrier proteins and albumin. Relative expression levels depend largely on the cell type of origin and may change under different environmental conditions. Other markers of note are major histocompatibility complexes (MHCs). MHC class I is ubiquitously expressed on all exosomes, whereas MHC II expression is confined to exosomes derived from antigen-presenting cells, including dendritic cells, macrophages, B cells, microglia, and intestinal epithelial cells.
Molecular cargo
Given the incredible diversity of cells, generalizations regarding exosome cargo should not be viewed dogmatically. The payload of each exosome population varies greatly according to the cell and tissue type of the secreting population. Nevertheless, certain features are common (Figure 1). Given the small size of exosomes, organelles such as ribosomes and mitochondria are generally absent; likewise, DNA is rarely found in exosomes in any abundance, unlike the case for cancer-derived EVs, which are typically larger than exosomes and contain double-stranded DNA (1). Proteins are present and vary greatly with the cell of origin. Exosomes can contain enzymes, transcription factors, and structural proteins (44–46). Perhaps the greatest source of signaling diversity is in the plentiful RNA content, which includes not only transcripts but also abundant noncoding RNA species such as miRs and transfer RNAs (20, 47). Exosome RNA content varies dramatically according to the cell type of origin, but it does not merely parrot the RNA profile of the secreting cell (20, 48–50). Poorly understood sorting processes are at play in determining what RNA species are packaged into exosomes (51). Furthermore, the unique signature of miRs derived from tumor exosomes has generated increasing interest in the use of exosomes as diagnostic and prognostic biomarkers (52). Perhaps the most important lingering question in exosome biology is the nature of the molecular determinants underlying selective packaging of signaling mediators and their release. Understanding how certain signals, including miRs, mRNA, and protein, are selectively packaged is critical to understanding the grander pathway of signal transduction and how cells alter the payload under different conditions such as stress and disease. An important first step is the recognition for heterogeneous nuclear ribonucleoprotein A2B1 in miR sorting into exosomes (115).
Exosome uptake
Exosomes enter recipient cells via a variety of mechanisms, which adds to their diversity. Mechanisms include receptor-mediated binding and activation of downstream signaling; protease-mediated cleavage of exosomes by recipient cells to liberate soluble ligands; lipid membrane fusion; internalization by receptor-mediated endocytosis; and uptake by immune cells through phagocytosis, pinocytosis, or micropinocytosis (53). The relative importance of the various uptake pathways differs dramatically according to the biological context. For example, systemically delivered exogenous tumor—derived exosomes distribute rapidly to the spleen and liver, followed by a more prolonged clearance phase through the hepatic and renal route (54). Increasing exosome dose broadens this distribution to the lungs, gastrointestinal tract, and kidneys (55). Intratumoral injection of exosomes, however, has shown significant and prolonged association with tumoral tissue (54).
EXOSOMES IN CARDIOVASCULAR PATHOPHYSIOLOGY
The concept of exosomes as propagators of disease is not a new one. Exosomes and other EVs play active roles in cancer cell signaling, including tumor growth, recruitment of vasculature, chemoresistance, and priming distant tissue for metastasis (56). Such involvement is perhaps not surprising because exosomes are highly conserved across the Eukaryota taxa (57) and thus play central roles in disease. Researchers have shown for multiple systems that exosome secretion and payload are sensitive to various stress stimuli, including infection (58).
Hypertrophy and Cardiac Remodeling
Various forms of stress in the heart—including hypertension, ischemia, valvular dysfunction, arrhythmias, and even transient stresses such as physical exertion and pregnancy—lead to an activated hypertrophic cellular response (59, 60). These responses are due in part to vesicle-mediated cellular cross talk among myocytes and other cells in the myocardium, including fibroblasts, endothelial cells, circulating blood elements, and inflammatory cells. Stressed adult cardiomyocytes upregulate their exosome secretion and alter exosome content to be enriched in proinflammatory and apoptotic factors, including HSP60 and TNF α (61). These factors result in further inflammation and death to nearby cardiomyocytes, driving even more inflammation and expanding scars.
Cardiac hypertrophy is characterized by myocyte enlargement, cardiac fibroblast proliferation, and secretion of extracellular matrix proteins and proinflammatory cytokines. Bang and others (62) demonstrated that fibroblasts secrete exosomes enriched in miR-21*; these exosomes contribute to cardiomyocyte hypertrophy. Cardiomyocytes in culture became hypertrophic upon exposure to conditioned media from fibroblasts or when cocultured with fibroblasts (63). As fibroblasts constitute the majority of the myocardium, the effect of pathogenic exosomes in mediating this phenotype becomes readily appreciable. Later studies further elucidated this mechanism and implicated exosomal transfer of miR-21* as a central driver of this hypertrophic effect. Taken up through an endocytic pathway, miR-21* in exosomes downregulates cardiomyocyte targets such as sorbin, SH3 domain—containing protein 2 (SORBS2), and PDZ and Lim domain 5 (PDLIM5) (64). Downregulation of these targets effectively triggered hypertrophy. The pericardial fluid of mice subjected to transverse aortic constriction exhibited increased levels of miR-21* (65). Much remains to be understood, however, regarding the mechanism whereby activated fibroblasts release deleterious exosomes. Whether ischemia is enough to change the exosome payload of these cells (as they transition from fibroblasts to myofibroblasts) or whether they are in turn influenced by EVs from plasma or endothelial cells remains to be seen. However, recent data from a kidney injury model suggest that epithelial cell—derived exosomes rich in TGF-β mRNA prime fibroblasts to secrete exosomes that are profibrotic (66). In the case of insult to the heart, the majority of EVs come from autocrine sources, plasma, and endothelial cells, which thus represent promising starting points of investigation.
Arrhythmia
Although the role of exosomes in arrhythmias is not established, a plausible case can be made for the involvement of exosomes. Exosomes secreted by ischemic cardiomyocytes, both from patients with coronary artery disease and in ischemic rat hearts, are enriched in miR-1 and miR-133 (67), which affect action potentials and cardiac conduction through targeting of Ca2+/calmodulin-dependent protein kinase II (68, 69). Other potentially proarrhythmogenic miRs that are upregulated in exosomes during ischemia include miR-328, which is enriched in platelet-derived exosomes (70) and targets L-type calcium channels (71, 72).
Cardiomyopathy
Exosomes also play a proinflammatory role in the heart during the onset of different cardiomyopathies. Sepsis-induced cardiomyopathy occurs when systemic bacterial infection ultimately leads to the depression of cardiac function, followed by multiple organ failure. Platelet-derived exosomes contribute to inflammation in a lipopolysaccharide (LPS)-induced mouse model of sepsis. In healthy volunteers, LPS triggers platelets to release exosomes enriched in NADPH oxidase, nitric oxide synthase, and disulfide isomerase and results in the downregulation of the anti-inflammatory miR-223 (which targets ICAM-1) (73). Exosomes derived from activated platelets induce apoptosis in rabbit aortic endothelial cells (73). Thus, platelets activated by exposure to bacterial toxins secrete both more exosomes and increasingly pathogenic exosomes that drive the vascular dysfunction seen in sepsis. In the cardiomyopathy associated with type 2 diabetes, hyperglycemia alters the exosomes released by stressed cardiomyocytes. Serum exosomes from type 2 diabetic rats are rich in miR-320 and are taken up by endothelial cells in vitro. MiR-320 inhibits endothelial cell proliferation and migration via targeting of IGF-1, Hsp20, and Ets2. Downregulation of these targets leads to inhibition of proliferation, of migration, and of tube formation (74). Addition of GW4869, an inhibitor of exosome biosynthesis, abrogated this effect (74). In conclusion, exosomes derived from injured cardiovascular tissue promote disease progression by a variety of propathological signals. Figure 4 illustrates the general model for disease-propagating exosomes (termed pathosomes) from injured, terminally differentiated tissue, including cardiomyocytes, fibroblasts, endothelial cells, and platelets. The available data are consistent with the emerging concept that pathosomes perpetuate responses to injury in cardiovascular tissue, ultimately leading to end-stage disease (Figure 4), but this concept has yet to be tested in detail.
Figure 4.
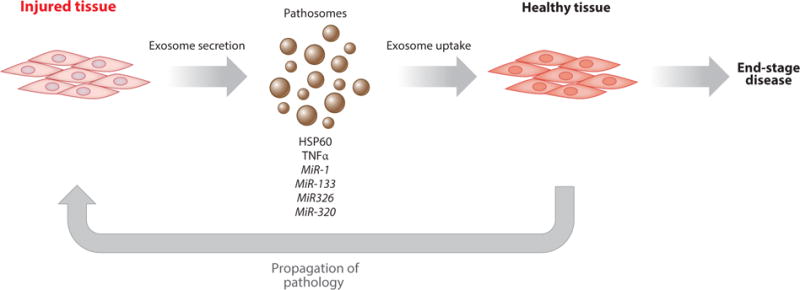
Tissue injured during insult releases exosomes containing signals and factors that promote further damage in distant tissue. Such signals and factors include prohypertrophic, proinflammatory, and proapoptotic/necrotic miRs, proteins, and mRNA.
EXOSOMES: NEXT-GENERATION THERAPEUTIC AGENTS?
Heart disease is the leading killer of men and women in the world today, with nearly 20 million deaths per year. Cardiovascular disease kills more Americans and imparts an economic burden on the US health care system greater than that for any other disease category, including all malignancies combined (75). A major driver of cardiovascular disease is myocardial infarction (MI), which affects >1 million Americans annually (76). Some MI survivors experience a steady decline in health, quality of life, and productivity by progression to chronic heart failure (HF) (77). Conventional therapy for HF can slow down the progression of this disease, but no current treatments can halt or reverse HF once it becomes symptomatic. Cell therapy has been touted for its potential to regenerate tissue previously thought to be permanently damaged, such as the post-MI heart (78–80). Canonical stem cell—mediated regeneration is motivated by the notion that injected multipotent cells engraft, proliferate, differentiate, and repopulate the injured region of the heart. However, multiple lines of evidence now indicate that most of the beneficial effects of transplanted cells are indirect. Stem cells secrete a variety of factors and molecules, including growth factors, antioxidants, proteasomes, miRs, and EVs. The recognition that exosomes may play an important role in mediating the benefits of cell therapy is an evolving paradigm, with evidence emerging only recently. If exosomes do turn out to be mechanistically important, they may be therapeutic candidates, supplanting the need for cell transplantation.
Mesenchymal Stem Cell Exosomes
Lai and colleagues showed that human embryonic stem cell—derived mesenchymal stem cells (MSCs) secrete exosomes 50–100 nm in size with therapeutic bioactivity, in that they reduced infarct size in a mouse ex vivo model of ischemia/reperfusion injury (81). Further findings by the same group demonstrated, in vivo in mice with MI, that systemic pretreatment withMSC exosomes reduced scars by 45%, increased ATP and NADH levels, and reduced oxidative stress in heart tissue (82). Exosomes derived from bone marrow MSCs blunt inflammation and attenuate vascular remodeling pathways in a mouse model of hypoxia-induced hypertension (HPH). Specifically, these exosomes suppressed phosphorylation of signal transducer and activator of transcription 3 (STAT3) and pulmonary levels of miR-204 (which are suppressed in HPH) (83). Therefore, MSC-derived exosomes exhibit anti-inflammatory properties and restore metabolic ability to injured tissue. However, more rigorous studies are needed to elucidate MSC exosomes’ capacity for regeneration and other pathways beyond cardioprotection.
Hematopoietic Stem Cell Exosomes
The therapeutic value of hematopoietic stem cells (HSCs) in the heart is hotly debated (84), but, to the extent that HSCs work, their effects appear to be paracrine. Sahoo and colleagues demonstrated that CD34+ cells (putative endothelial progenitors) secrete vesicles that express classical exosome markers such as CD63, phosphatidylserine, and TSG101 (85). These HSC-derived exosomes had angiogenic effects that were not reproduced by exosomes from CD34− cells. Motivated by the implication that miR-126 and miR-130a are involved in angiogenesis (86, 87), and by the decline in CD34+ cell potency with donor age (88), Mackie and others genetically modified CD34+ cells with sonic hedgehog (Shh), a known mediator of angiogenesis (89). The modified cells showed increased angiogenic properties, and Shh was enriched in these cells’ secreted exosomes (90). Taken together, the data are consistent with the notion that CD34+ cell—stimulated angiogenesis is mediated via secretion of exosomes and their distinctive payload.
Cardiac Progenitor Cell Exosomes
The identity of cardiac progenitor cells (CPCs) is controversial. Although CPCs were first argued to replenish the heart via canonical stem cell mechanisms, their effects now appear to be indirect. Exosomes from CPCs (91) are rich in matrix metalloproteinases (MMPs) and MMP inducers (e.g., EMMPRIN), which collectively dissolve the extracellular matrix and recruit more MMPs. Such exosomes also contain miR-451, an autophagy-regulating miR downregulated in hypertrophy and HF (92). CPC-derived exosomes also stimulated the migration of endothelial cells in vitro (93, 116) and inhibited cardiomyocyte death in a mouse ischemia/reperfusion model (94).
Cardiosphere-Derived Cell Exosomes
Cardiosphere-derived cells (CDCs) are a cardiac stromal cell population with regenerative effects (95) that are durable but mediated indirectly (96). At least 22 independent labs worldwide have reproduced the published methodology and verified CDCs’ identity and utility since their first description in 2007 (97–103). Conducted after systematic translational studies (91), the first human trial, CADUCEUS (104, 105), demonstrated the safety and efficacy of autologous CDC administration in patients with left ventricular dysfunction after MI. Contrast-enhanced magnetic resonance imaging revealed evidence for regrowth of viable heart tissue in a setting in which injury had been believed to be irreversible, consistent with therapeutic regeneration (106, 107). The mechanism of action is indirect (108); benefits persist long after injected CDCs have been cleared (109). Although CDCs secrete many growth factors and cytokines (110), exosomes mediate the salient benefits: CDCs treated with GW4869 lose their cardioprotective and regenerative properties, whereas CDC-secreted exosomes (but not those derived from inert fibroblasts) mimic the therapeutic effects of CDCs (111). The CDC exosomes are distinctive and enriched in miR-146a, which reproduced some (but not all) of the effects of CDC exosomes (103, 111). The observation that miR-146a failed to recapitulate some of the broader effects of exosome treatment, including the functional improvement, points to a broader combinatorial effect of multiple exosome payload factors in mediating the full panel of benefits. In an amplification process, CDC-derived exosomes altered the secretory profile of fibroblasts, rendering them therapeutically active (112).
CONCLUDING REMARKS
Although we focus above on cardiovascular examples, it seems plausible, if not likely, that exosomes touch upon virtually all biological processes. By virtue of being versatile carriers of molecular signals, exosomes can transmit signals that are both protective and pathological. Although the field is exploding, many questions remain. Will the recognition of exosomes and their roles usher in a new paradigm of endocrinology? How do exosomes traverse complex biological interfaces such as the intestinal lining and the blood-brain barrier? Can exosomes from plants (113) or parasites (114) modulate gene expression in animals, as has been claimed? What is the role of exosomes in propagating any given disease (or, conversely, in limiting its ill effects)? How can the therapeutic properties of exosomes best be harnessed? A final, fundamental question is the mechanism whereby exosomes, which are cleared fairly rapidly (54), exert lasting effects on cell behavior. We and others are currently testing the conjecture that payload RNAs, notably miRs, long noncoding RNAs, Y-RNAs, and piRNA, produce epigenetic modifications that impart lasting impacts on the target cell. Proteins and lipids may also play important roles in any given biological process. Understanding the roles of the various vesicular contents, and how each of the constituents acts, will be key to understanding the mechanism and scope of the diverse effects of exosomes.
Acknowledgments
E.M.’s work on exosomes is supported by the NIH (R01 HL124074).
Footnotes
DISCLOSURE STATEMENT
A.I. is an employee of Capricor. E.M. owns founder’s equity in Capricor and serves as unpaid advisor to the company. The authors are not aware of any other affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Thakur BK, Zhang H, Becker A, Matei I, Huang Y, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–69. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chargaff E, West R. The biological significance of the thromboplastic protein of blood. J Biol Chem. 1946;166:189–97. [PubMed] [Google Scholar]
- 3.Wolf P. The nature and significance of platelet products in human plasma. Br J Haematol. 1967;13:269–88. doi: 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- 4.Johnstone RM. Exosomes biological significance: a concise review. Blood Cells Mol Dis. 2006;36:315–21. doi: 10.1016/j.bcmd.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes) J Biol Chem. 1987;262:9412–20. [PubMed] [Google Scholar]
- 6.Kriebel PW, Barr VA, Rericha EC, Zhang G, Parent CA. Collective cell migration requires vesicular trafficking for chemoattractant delivery at the trailing edge. J Cell Biol. 2008;183:949–61. doi: 10.1083/jcb.200808105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albuquerque PC, Nakayasu ES, Rodrigues ML, Frases S, Casadevall A, et al. Vesicular transport in Histoplasma capsulatum: an effective mechanism for trans–cell wall transfer of proteins and lipids in ascomycetes. Cell Microbiol. 2008;10:1695–710. doi: 10.1111/j.1462-5822.2008.01160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An Q, van Bel AJ, Huckelhoven R. Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal Behav. 2007;2:4–7. doi: 10.4161/psb.2.1.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korkut C, Ataman B, Ramachandran P, Ashley J, Barria R, et al. Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell. 2009;139:393–404. doi: 10.1016/j.cell.2009.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liégeois S, Benedetto A, Garnier JM, Schwab Y, Labouesse M. The V0-ATPase mediates apical secretion of exosomes containing Hedgehog-related proteins in Caenorhabditis elegants. J Cell Biol. 2006;173:949–61. doi: 10.1083/jcb.200511072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med. 2012;10:5. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reinhardt TA, Lippolis JD, Nonnecke BJ, Sacco RE. Bovine milk exosome proteome. J Proteom. 2012;75:1486–92. doi: 10.1016/j.jprot.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 14.Taylor DD, Gercel-Taylor C. Tumour-derived exosomes and their role in cancer-associated T-cell signalling defects. Br J Cancer. 2005;92:305–11. doi: 10.1038/sj.bjc.6602316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Runz S, Keller S, Rupp C, Stoeck A, Issa Y, et al. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EPCAM. Gynecol Oncol. 2007;107:563–71. doi: 10.1016/j.ygyno.2007.08.064. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Llama P, Khositseth S, Gonzales PA, Star RA, Pisitkun T, Knepper MA. Tamm-Horsfall protein and urinary exosome isolation. Kidney Int. 2010;77:736–42. doi: 10.1038/ki.2009.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Skog J, Hsu CH, Lessard RT, Balaj L, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010;10:505–11. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179:1969–78. doi: 10.4049/jimmunol.179.3.1969. [DOI] [PubMed] [Google Scholar]
- 19.Levanen B, Bhakta NR, Torregrosa Paredes P, Barbeau R, Hiltbrunner S, et al. Altered microRNA profiles in bronchoalveolar lavage fluid exosomes in asthmatic patients. J Allergy Clin Immunol. 2013;131:894–903. doi: 10.1016/j.jaci.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–59. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 21.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim Biophys Acta. 2014;1841:108–20. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 22.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–57. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 23.Xu L, Yang BF, Ai J. MicroRNA transport: a new way in cell communication. J Cell Physiol. 2013;228:1713–19. doi: 10.1002/jcp.24344. [DOI] [PubMed] [Google Scholar]
- 24.Wittmann J, Jack HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806:200–7. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Rao P, Benito E, Fischer A. MicroRNAs as biomarkers for CNS disease. Front Mol Neurosci. 2013;6:39. doi: 10.3389/fnmol.2013.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oikonomou E, Siasos G, Tousoulis D, Kokkou E, Genimata V, et al. Diagnostic and therapeutic potentials of microRNAs in heart failure. Curr Top Med Chem. 2013;13:1548–58. doi: 10.2174/15680266113139990104. [DOI] [PubMed] [Google Scholar]
- 27.Schageman J, Zeringer E, Li M, Barta T, Lea K, et al. The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int. 2013;2013:253957. doi: 10.1155/2013/253957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pant S, Hilton H, Burczynski ME. The multifaceted exosome: biogenesis, role in normal and aberrant cellular function, and frontiers for pharmacological and biomarker opportunities. Biochem Pharmacol. 2012;83:1484–94. doi: 10.1016/j.bcp.2011.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanson PI, Shim S, Merrill SA. Cell biology of the ESCRT machinery. Curr Opin Cell Biol. 2009;21:568–74. doi: 10.1016/j.ceb.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayers JR, Audhya A. Vesicle formation within endosomes: An ESCRT marks the spot. Commun Integr Biol. 2012;5:50–56. doi: 10.4161/cib.18208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alcazar O, Hawkridge AM, Collier TS, Cousins SW, Bhattacharya SK, et al. Proteomics characterization of cell membrane blebs in human retinal pigment epithelium cells. Mol Cell Proteom. 2009;8:2201–11. doi: 10.1074/mcp.M900203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eskelinen EL. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol Aspects Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 33.NIH Office of Strategic Coordination. Extracellular RNA communication. 2011 https://commonfund. nih.gov/Exrna/index.
- 34.Futter CE, Pearse A, Hewlett LJ, Hopkins CR. Multivesicular endosomes containing internalized EGF-EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol. 1996;132:1011–23. doi: 10.1083/jcb.132.6.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Möbius W, Ohno-Iwashita Y, van Donselaar EG, Oorschot VM, et al. Immunoelectron microscopic localization of cholesterol using biotinylated and non-cytolytic perfringolysin O. J Histochem Cytochem. 2002;50:43–55. doi: 10.1177/002215540205000105. [DOI] [PubMed] [Google Scholar]
- 36.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–52. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 37.Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–87. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–66. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White IJ, Bailey LM, Aghakhani MR, Moss SE, Futter CE. EGF stimulates annexin 1–dependent inward vesiculation in a multivesicular endosome subpopulation. EMBOJ. 2006;25:1–12. doi: 10.1038/sj.emboj.7600759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–44. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subra C, Grand D, Laulagnier K, Stella A, Lambeau G, et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J Lipid Res. 2010;51:2105–20. doi: 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–47. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 43.Zakharova L, Svetlova M, Fomina AF. T cell exosomes induce cholesterol accumulation in human monocytes via phosphatidylserine receptor. J Cell Physiol. 2007;212:174–81. doi: 10.1002/jcp.21013. [DOI] [PubMed] [Google Scholar]
- 44.Henderson MC, Azorsa DO. The genomic and proteomic content of cancer cell—derived exosomes. Front Oncol. 2012;2:38. doi: 10.3389/fonc.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, et al. Genome-wide highresolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–53. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 47.Lotvall J, Valadi H. Cell to cell signalling via exosomes through esRNA. Cell Adh Migr. 2007;1:156–58. doi: 10.4161/cam.1.3.5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hessvik NP, Phuyal S, Brech A, Sandvig K, Llorente A. Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim Biophys Acta. 2012;1819:1154–63. doi: 10.1016/j.bbagrm.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 50.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40:10937–49. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Madrid F, Mittelbrunn M. Analysis of microRNA and protein transfer by exosomes during an immune synapse. Meth Mol Biol. 2013;1024:41–51. doi: 10.1007/978-1-62703-453-1_4. [DOI] [PubMed] [Google Scholar]
- 52.Jaiswal R, Gong J, Sambasivam S, Combes V, Mathys JM, et al. Microparticle-associated nucleic acids mediate trait dominance in cancer. FASEB J. 2012;26:420–29. doi: 10.1096/fj.11-186817. [DOI] [PubMed] [Google Scholar]
- 53.Turturici G, Tinnirello R, Sconzo G, Geraci F. Extracellular membrane vesicles as a mechanism of cell-to-cell communication: advantages and disadvantages. Am J Physiol Cell Physiol. 2014;306:C621–33. doi: 10.1152/ajpcell.00228.2013. [DOI] [PubMed] [Google Scholar]
- 54.Smyth T, Kullberg M, Malik N, Smith-Jones P, Graner MW, Anchordoquy TJ. Biodistribution and delivery efficiency of unmodified tumor-derived exosomes. J Control Release. 2015;199:145–55. doi: 10.1016/j.jconrel.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiklander OP, Nordin JZ, O’Loughlin A, Gustafsson Y, Corso G, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:26316. doi: 10.3402/jev.v4.26316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roma-Rodrigues C, Fernandes AR, Baptista PV. Exosome in tumour microenvironment: overview of the crosstalk between normal and cancer cells. BioMed Res Int. 2014;2014:179486. doi: 10.1155/2014/179486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lopez-Verrilli MA, Court FA. Exosomes: mediators of communication in eukaryotes. Biol Res. 2013;46:5–11. doi: 10.4067/S0716-97602013000100001. [DOI] [PubMed] [Google Scholar]
- 58.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110:3234–44. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sahoo S, Losordo DW. Exosomes and cardiac repair after myocardial infarction. Circ Res. 2014;114:333–44. doi: 10.1161/CIRCRESAHA.114.300639. [DOI] [PubMed] [Google Scholar]
- 60.Molkentin JD, Dorn GW., 2nd Cytoplasmic signaling pathways that regulate cardiac hypertrophy. Annu Rev Physiol. 2001;63:391–426. doi: 10.1146/annurev.physiol.63.1.391. [DOI] [PubMed] [Google Scholar]
- 61.Tian J, Guo X, Liu XM, Liu L, Weng QF, et al. Extracellular HSP60 induces inflammation through activating and up-regulating TLRs in cardiomyocytes. Cardiovasc Res. 2013;98:391–401. doi: 10.1093/cvr/cvt047. [DOI] [PubMed] [Google Scholar]
- 62.Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, et al. Cardiacfibroblast–derived microRNA passenger strand–enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Investig. 2014;124:2136–46. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fredj S, Bescond J, Louault C, Potreau D. Interactions between cardiac cells enhance cardiomyocyte hypertrophy and increase fibroblast proliferation. J Cell Physiol. 2005;202:891–99. doi: 10.1002/jcp.20197. [DOI] [PubMed] [Google Scholar]
- 64.Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, et al. Cardiac fibroblast–derived microRNA passenger strand–enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Investig. 2014;124:2136–46. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Queiros AM, Eschen C, Fliegner D, Kararigas G, Dworatzek E, et al. Sex-and estrogen-dependent regulation of a miRNA network in the healthy and hypertrophied heart. Int J Cardiol. 2013;169:331–38. doi: 10.1016/j.ijcard.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 66.Borges FT, Melo SA, Ozdemir BC, Kato N, Revuelta I, et al. TGF-β1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J Am Soc Nephrol. 2013;24:385–92. doi: 10.1681/ASN.2012101031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu C, Lu Y, Pan Z, Chu W, Luo X, et al. The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis by targeting HSP60, HSP70 and caspase-9 in cardiomyocytes. J Cell Sci. 2007;120:3045–52. doi: 10.1242/jcs.010728. [DOI] [PubMed] [Google Scholar]
- 68.Belevych AE, Sansom SE, Terentyeva R, Ho HT, Nishijima Y, et al. Microrna-1 and -133 increase arrhythmogenesis in heart failure by dissociating phosphatase activity from RyR2 complex. PLOS ONE. 2011;6:e28324. doi: 10.1371/journal.pone.0028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elton TS, Martin MM, Sansom SE, Belevych AE, Gyorke S, Terentyev D. MiRNAs got rhythm. Life Sci. 2011;88:373–83. doi: 10.1016/j.lfs.2010.11.022. [DOI] [PubMed] [Google Scholar]
- 70.Dempsey E, Feidhlim D, Maguire PB. Platelet derived exosomes are enriched for specific microRNAs which regulate Wnt signalling in endothelial cells. Presented at Am Soc Hematol Annu Meet; San Francisco, CA. Dec 6–9.2014. [Google Scholar]
- 71.Neppl RL, Wang DZ. The myriad essential roles of microRNAs in cardiovascular homeostasis and disease. Genes Dis. 2014;1:18–39. doi: 10.1016/j.gendis.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu Y, Zhang Y, Wang N, Pan Z, Gao X, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122:2378–87. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 73.Gambim MH, do Carmo Ade O, Marti L, Verissimo-Filho S, Lopes LR, Janiszewski M. Platelet-derived exosomes induce endothelial cell apoptosis through peroxynitrite generation: experimental evidence for a novel mechanism of septic vascular dysfunction. Crit Care. 2007;11:R107. doi: 10.1186/cc6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang X, Huang W, Liu G, Cai W, Millard RW, et al. Cardiomyocytes mediate anti-angiogenesis in type 2 diabetic rats through the exosomal transfer of miR-320 into endothelial cells. J Mol Cell Cardiol. 2014;74:139–50. doi: 10.1016/j.yjmcc.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.CDC. FastStats: leading causes of death. 2011 http://www.cdc.gov/nchs/fastats/leading-causes-of-death.htm.
- 76.Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD. Third universal definition of myocardial infarction. Nat Rev Cardiol. 2012;9:620–33. doi: 10.1038/nrcardio.2012.122. [DOI] [PubMed] [Google Scholar]
- 77.Jhund PS, McMurray JJV. Heart failure after acute myocardial infarction: a lost battle in the war on heart failure? Circulation. 2008;118:2019–21. doi: 10.1161/CIRCULATIONAHA.108.813493. [DOI] [PubMed] [Google Scholar]
- 78.Gerbin KA, Murry CE. The winding road to regenerating the human heart. Cardiovasc Pathol. 2015;24:133–40. doi: 10.1016/j.carpath.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim J, Shapiro L, Flynn A. The clinical application of mesenchymal stem cells and cardiac stem cells as a therapy for cardiovascular disease. Pharmacol Ther. 2015;151:8–15. doi: 10.1016/j.pharmthera.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 80.Wegener M, Bader A, Giri S. How to mend a broken heart: adult and induced pluripotent stem cell therapy for heart repair and regeneration. Drug Discov Today. 2015;20:667–85. doi: 10.1016/j.drudis.2015.02.010. [DOI] [PubMed] [Google Scholar]
- 81.Lai RC, Arslan F, Lee MM, Sze NS, Choo A, et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–22. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 82.Arslan F, Lai RC, Smeets MB, Akeroyd L, Choo A, et al. Mesenchymal stem cell—derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013;10:301–12. doi: 10.1016/j.scr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 83.Lee C, Mitsialis SA, Aslam M, Vitali SH, Vergadi E, et al. Exosomes mediate the cytoprotective action of mesenchymal stromal cells on hypoxia-induced pulmonary hypertension. Circulation. 2012;126:2601–11. doi: 10.1161/CIRCULATIONAHA.112.114173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Balmer GM, Riley PR. Harnessing the potential of adult cardiac stem cells: lessons from haematopoiesis, the embryo and the niche. J Cardiovasc Transl Res. 2012;5:631–40. doi: 10.1007/s12265-012-9386-3. [DOI] [PubMed] [Google Scholar]
- 85.Sahoo S, Klychko E, Thorne T, Misener S, Schultz KM, et al. Exosomes from human CD34+ stem cells mediate their proangiogenic paracrine activity. Circ Res. 2011;109:724–28. doi: 10.1161/CIRCRESAHA.111.253286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jakob P, Doerries C, Briand S, Mocharla P, Krankel N, et al. Loss of angiomiR-126 and 130a in angiogenic early outgrowth cells from patients with chronic heart failure: role for impaired in vivo neovascularization and cardiac repair capacity. Circulation. 2012;126:2962–75. doi: 10.1161/CIRCULATIONAHA.112.093906. [DOI] [PubMed] [Google Scholar]
- 87.Mocharla P, Briand S, Giannotti G, Dorries C, Jakob P, et al. AngiomiR-126 expression and secretion from circulating CD34+ and CD14+ PBMCs: role for proangiogenic effects and alterations in type 2 diabetics. Blood. 2013;121:226–36. doi: 10.1182/blood-2012-01-407106. [DOI] [PubMed] [Google Scholar]
- 88.Li TS, Kubo M, Ueda K, Murakami M, Mikamo A, Hamano K. Impaired angiogenic potency of bone marrow cells from patients with advanced age, anemia, and renal failure. J Thorac Cardiovasc Surg. 2010;139:459–65. doi: 10.1016/j.jtcvs.2009.07.053. [DOI] [PubMed] [Google Scholar]
- 89.Renault MA, Roncalli J, Tongers J, Thorne T, Klyachko E, et al. Sonic hedgehog induces angio-genesis via Rho kinase—dependent signaling in endothelial cells. J Mol Cell Cardiol. 2010;49:490–98. doi: 10.1016/j.yjmcc.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mackie AR, Klyachko E, Thorne T, Schultz KM, Millay M, et al. Sonic hedgehog—modified human CD34+ cells preserve cardiac function after acute myocardial infarction. Circ Res. 2012;111:312–21. doi: 10.1161/CIRCRESAHA.112.266015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Marban E. Breakthroughs in cell therapy for heart disease: focus on cardiosphere-derived cells. Mayo Clin Proc. 2014;89:850–58. doi: 10.1016/j.mayocp.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song L, Su M, Wang S, Zou Y, Wang X, et al. Mir-451 is decreased in hypertrophic cardiomyopathy and regulates autophagy by targeting TSC1. J Cell Mol Med. 2014;18:2266–74. doi: 10.1111/jcmm.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vrijsen KR, Sluijter JP, Schuchardt MW, van Balkom BW, Noort WA, et al. Cardiomyocyte progenitor cell—derived exosomes stimulate migration of endothelial cells. J Cell Mol Med. 2010;14:1064–70. doi: 10.1111/j.1582-4934.2010.01081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen L, Wang Y, Pan Y, Zhang L, Shen C, et al. Cardiac progenitor—derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury. Biochem Biophys Res Commun. 2013;431:566–71. doi: 10.1016/j.bbrc.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith RR, Barile L, Cho HC, Leppo MK, Hare JM, et al. Regenerative potential of cardiosphere-derived cells expanded from percutaneous endomyocardial biopsy specimens. Circulation. 2007;115:896–908. doi: 10.1161/CIRCULATIONAHA.106.655209. [DOI] [PubMed] [Google Scholar]
- 96.Chimenti I, Smith RR, Li T-S, Gerstenblith G, Messina E, et al. Relative roles of direct regeneration versus paracrine effects of human cardiosphere—derived cells transplanted into infarcted mice. Circ Res. 2010;106:971–80. doi: 10.1161/CIRCRESAHA.109.210682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aghila Rani KG, Kartha CC. Effects of epidermal growth factor on proliferation and migration of cardiosphere-derived cells expanded from adult human heart. Growth Factors. 2010;28:157–65. doi: 10.3109/08977190903512628. [DOI] [PubMed] [Google Scholar]
- 98.Gaetani R, Ledda M, Barile L, Chimenti I, De Carlo F, et al. Differentiation of human adult cardiac stem cells exposed to extremely low-frequency electromagnetic fields. Cardiovasc Res. 2009;82:411–20. doi: 10.1093/cvr/cvp067. [DOI] [PubMed] [Google Scholar]
- 99.Mishra R, Vijayan K, Colletti EJ, Harrington DA, Matthiesen TS, et al. Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation. 2011;123:364–73. doi: 10.1161/CIRCULATIONAHA.110.971622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takehara N, Tsutsumi Y, Tateishi K, Ogata T, Tanaka H, et al. Controlled delivery of basic fibroblast growth factor promotes human cardiosphere—derived cell engraftment to enhance cardiac repair for chronic myocardial infarction. J Am Coll Cardiol. 2008;52:1858–65. doi: 10.1016/j.jacc.2008.06.052. [DOI] [PubMed] [Google Scholar]
- 101.Tang YL, Zhu W, Cheng M, Chen L, Zhang J, et al. Hypoxic preconditioning enhances the benefit of cardiac progenitor cell therapy for treatment of myocardial infarction by inducing CXCR4 expression. Circ Res. 2009;104:1209–16. doi: 10.1161/CIRCRESAHA.109.197723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zakharova L, Mastroeni D, Mutlu N, Molina M, Goldman S, et al. Transplantation of cardiac progenitor cell sheet onto infarcted heart promotes cardiogenesis and improves function. Cardiovasc Res. 2010;87:40–49. doi: 10.1093/cvr/cvq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Barile L, Lionetti V, Cervio E, Matteucci M, Gherghiceanu M, et al. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc Res. 2014;103:530–41. doi: 10.1093/cvr/cvu167. [DOI] [PubMed] [Google Scholar]
- 104.Makkar RR, Smith RR, Cheng K, Malliaras K, Thomson LE, et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Malliaras K, Makkar RR, Smith RR, Cheng K, Wu E, et al. Intracoronary cardiosphere-derived cells after myocardial infarction: evidence of therapeutic regeneration in the final 1-year results of the CADUCEUS trial (cardiosphere-derived autologous stem cells to reverse ventricular dysfunction) J Am Coll Cardiol. 2014;63:110–22. doi: 10.1016/j.jacc.2013.08.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jeevanantham V, Butler M, Saad A, Abdel-Latif A, Zuba-Surma EK, Dawn B. Adult bone marrow cell therapy improves survival and induces long-term improvement in cardiac parameters: a systematic review and meta-analysis. Circulation. 2012;126:551–68. doi: 10.1161/CIRCULATIONAHA.111.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Preda MB, Valen G. Evaluation of gene and cell-based therapies for cardiac regeneration. Curr Stem Cell Res Ther. 2013;8:304–12. doi: 10.2174/1574888x11308040006. [DOI] [PubMed] [Google Scholar]
- 108.Chimenti I, Smith RR, Li TS, Gerstenblith G, Messina E, et al. Relative roles of direct regeneration versus paracrine effects of human cardiosphere—derived cells transplanted into infarcted mice. Circ Res. 2010;106:971–80. doi: 10.1161/CIRCRESAHA.109.210682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Malliaras K, Li TS, Luthringer D, Terrovitis J, Cheng K, et al. Safety and efficacy of allogeneic cell therapy in infarcted rats transplanted with mismatched cardiosphere-derived cells. Circulation. 2012;125:100–12. doi: 10.1161/CIRCULATIONAHA.111.042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stastna M, Chimenti I, Marban E, Van Eyk JE. Identification and functionality of proteomes secreted by rat cardiac stem cells and neonatal cardiomyocytes. Proteomics. 2010;10:245–53. doi: 10.1002/pmic.200900515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ibrahim AG, Cheng K, Marban E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Rep. 2014;2:606–19. doi: 10.1016/j.stemcr.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tseliou E, Fouad J, Reich H, Slipczuk L, de Couto G, et al. Fibroblasts rendered antifibrotic, antiapoptotic, and angiogenic by priming with cardiosphere-derived extracellular membrane vesicles. J Am Coll Cardiol. 2015;66:599–611. doi: 10.1016/j.jacc.2015.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mu J, Zhuang X, Wang Q, Jiang H, Deng ZB, et al. Interspecies communication between plant and mouse gut host cells through edible plant derived exosome-like nanoparticles. Mol Nutr Food Res. 2014;58:1561–73. doi: 10.1002/mnfr.201300729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 2014;5:5488. doi: 10.1038/ncomms6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gray WD, French KM, Ghosh-Choudhary S, Maxwell JT, Brown ME, et al. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ Res. 2015;116:255–63. doi: 10.1161/CIRCRESAHA.116.304360. [DOI] [PMC free article] [PubMed] [Google Scholar]


