Abstract
Peritoneal Dialysis (PD) is considered the best option for a cost-effective mid-term dialysis in patients with Chronic Renal Failure. However, functional failure of the peritoneal membrane (PM) force many patients to stop PD treatment and start haemodialysis. Currently, PM functionality is monitored by the peritoneal equilibration test, a tedious technique that often show changes when the membrane damage is advanced. As in other pathologies, the identification and characterization of extracellular vesicles (EVs) in the peritoneal dialysis efflux (PDE) may represent a non-invasive alternative to identify biomarkers of membrane failure. Using size-exclusion chromatography, we isolated EVs from PDE in a group of patients. Vesicles were characterized by the presence of tetraspanin markers, nanoparticle tracking analysis profile, cryo-electron microscopy and mass spectrometry. Here, we report the isolation and characterization of PDE-EVs. Based on mass spectrometry, we have found a set of well-conserved proteins among patients. Interestingly, the peptide profile also revealed remarkable changes between newly enrolled and longer-treated PD patients. These results are the first step to the identification of PDE-EVs based new markers of PM damage, which could support clinicians in their decision-making in a non-invasive manner.
Introduction
Peritoneal Dialysis (PD) is a renal replacement technique based on the semipermeable characteristics of the peritoneal membrane (PM). This membrane is composed by a monolayer of mesothelial cells and an interstitial matrix with a high number of capillaries that, in the presence of hyperosmotic PD fluids, permits the removal of small, medium and, to a lesser extent, large molecules, as well as water ultrafiltration. Prolonged exposure to PD fluids, the low pH of the solutions, as well as episodes of peritonitis or haemoperitoneum can cause detachment of mesothelial cells, fibrosis and neovascularization of the PM, resulting in functional degradation. Although the mechanisms of peritoneal fibrosis are still under investigation, one of the most accepted hypotheses is the epithelial to mesenchymal cell transition[1,2], involving factors such as vascular endothelial growth factor (VEGF) or tumour growth factor-β (TGF -β) (reviewed in[3]). Thus, despite PD is considered the best alternative for cost-effective sustainability of dialysis treatment[4,5], different changes ultimately lead to the failure of ultrafiltration of the PM, causing many patients to discontinue their treatment.
Monitoring the PM's functional state is therefore of outstanding importance for patients' management. Currently, PM is monitored based on the 4-hour lasting peritoneal equilibration test (PET). PET data inform about the permeability and transfer characteristics of the PM, estimating the water transport secondary to osmotic changes in the peritoneal cavity. These data allow clinicians to estimate the peritoneal transport, set the dose and type of PD required for each patient, and monitor the function of the PM. However, PET data render a delayed vision of the status of the PM, as the functional failure only occurs in advanced fibrotic lesions. Thus, monitoring early changes may help to identify and prevent functional worsening of the PM, thus helping the clinician to apply the appropriate therapeutic tools to extend their functionality. In this sense, efforts have been made on the proteomic analysis of peritoneal dialysis efflux (PDE)[6–8].
In recent years, the study of extracellular vesicles (EVs) has gained enormous interest in the diagnostic and therapeutic scenarios[9]. EVs are lipid-bilayered vesicles of 50 to 200 nm in diameter produced by most cells, mainly containing proteins, RNAs and metabolites[10]. EVs’ main function is related to cellular communication[10,11], but as their specific composition varies depending on the physiological and functional state of the producing cells, they have been extensively reported as potential biomarkers in a variety of diseases, including those of the renal system[12]. It is conceivable that the cells of the PM respond to the dialysis treatment by secreting EVs, and that these EVs change their composition reflecting the physiological state of the compartment of origin.
Here, our aim was to identify, isolate and characterize PDE-EVs of patients on PD. The results show that PDE is a non-invasive feasible material to isolate EVs using conventional, clinically applicable techniques. Analyses of PDE-EVs content permitted the identification of specific peptide profiles that changed according to time on dialysis. Thus, the study of EVs present in the PDE opens a new line of research to find non-invasive potential biomarkers for the early detection of PM damage in PD patients.
Materials and methods
Patients
The Ethical Committee of “Germans Trias i Pujol” Hospital approved the study, and all subjects gave their written consent according to the Declaration of Helsinki[13]. Inclusion criteria were patients over 18 years old diagnosed of a renal disease requiring PD as chronic renal replacement therapy. Patients starting PD due to heart failure, or those showing a peritonitis episode in the previous two months were excluded. Also, patients showing changes in the peritoneal membrane transport type compared to their initial PET or patients showing ultrafiltration failure were also excluded. Nine patients (56% female) from our PD unit were considered for the study. No patients presented any peritonitis episodes in the 2 months previous to the study. Renal diseases included: 2 renal polycystic disease, 2 tubulointersticial nephritis, 4 glomerulonephritis and 1 unknown aetiology. Seven patients were on Continuous Ambulatory Peritoneal Dialysis (CAPD) and 2 patients were on Automated Peritoneal Dialysis (APD). Seven patients were treated with icodextrin. Clinical and laboratory variables, Peritoneal Equilibration Test (PET), type of peritoneal dialysis solution, and total Kt/V as well as peritoneal Kt/V and renal Kt/V were evaluated.
Peritoneal equilibration test
The Peritoneal Dialysis Unit routinely perform PET monitoring to each patient one month after the begining of the treatment, and then repeat the test approximately every 6 months. In this study, samples used for EV analyses were obtained at the same time that a routine PET was perfomed. All patients included in the study were stable as for the PET functional result (ie, no changes were detected from their initial test). Samples were obtained between June and September 2014.
Peritoneal equilibration tests were performed with 3.87% glucose solution. The test bag was drained and reinfused at 60 min as reported[14]. A blood sample was withdrawn at 240 min and dialysate samples were taken from the pre-infusion bag, and at 0, 60, 120, and 240 min. Urea, creatinine, glucose, sodium, and potassium were analysed in all samples; urate, phosphate, total protein, albumin, were analysed in blood and dialysate samples at 240 min (Cobas 711 Roche diagnostics, Switzerland). A correction was applied for plasma water concentration for small solutes in the blood sample and, in the dialysate sample, creatinine concentration was corrected for the presence of glucose.
Calculations
Dialysate to plasma (D/P) ratios for urea and creatinine, and dialysate to baseline dialysate ratios (D/Do) for glucose were calculated. The mass transfer area coefficients (MTAC) for urea, creatinine, glucose, urate, phosphate, and potassium were calculated according to Waniewski et al. [15], using F = 0.5. Peritoneal clearances of total protein and albumin were also calculated. All parameters were corrected for 1.73 m2 surface area.
Ultrafiltration in PET at 240 min was calculated as the difference between the drained volumes and the initial volume, as follows:
where Uf, ultrafiltration; t, time (min); V, volume (mL).
Statistical analyses of patient data
Data are presented as median (rank). Quantitative data of the two groups were compared using U-Mann-Whitney test, while qualitative data of the groups were analysed using Fisher’s test. (SPSS, version 18.0, Chicago, IL, USA). Statistical significance was defined as p<0.05.
Isolation of EVs from PDE
Isolation of EVs from the concentrated PDE was based on a modification of a previous method described by our group[16]. Five hundred mL PDE were centrifuged at 3,000 g for 5 min immediately after collection. The supernatant was filtered through a 0.2 μm filter and concentrated using a Centricon plus-70 filter unit (100 kDa cut-off; Millipore, Bedford, MA). In brief, supernatants were loaded onto the Centricon filter and centrifuged at 2,800 g for 30 min. This step was repeated using one filter unit for each sample until the total volume was processed. The retained volume (ranging from 0.8 mL to 2 mL) of concentrated PDE was loaded onto a size-exclusion chromatography (SEC) column.
Size-exclusion chromatography
Up to 2 mL of concentrated PDE samples were loaded onto 12 mL of Sepharose-CL2B (Sigma-Aldrich, St. Louis, MO, USA) columns equilibrated in citrate buffer (phosphate-buffered saline, PBS/0.32% citrate) and eluted with PBS. Immediately after, up to 20 fractions of approximately 0.5 mL each were collected and keep at -80°C until further use.
Protein concentration
The protein concentration was measured by Bradford assay (10 μL of sample; Bio-Rad laboratories, USA) with a standard linear curve based on bovine serum albumin (BSA) (Sigma Aldrich).
Flow cytometry
Flow cytometry was used to identify fractions containing EVs according to their tetraspanin content and performed as reported before[16]. Antibodies anti-CD9 (1:10, Clone VJ1/20), anti-CD63 (1:10, Clone TEA 3/18), or polyclonal isotype (1:5000, Abcam (ab37355), Cambridge, UK) were added to samples an incubated at 4°C for 30 min. After two washes, beads were incubated with FITC-conjugated secondary antibody (SouthernBiotech, Birmingham, AL) and analysed in a FacsVerse flow cytometer (BD Biosciences, San Jose, CA). Approximately 10,000 beads/sample were acquired and analysed using the Flow Jo software (Tree Star, Ashland, OR). In all samples, the top three tetraspanin-containing chromatographic fractions (those containing EVs) were pooled and used in experiments thereafter.
Sodium dodecyl sulphate-polyacrylamide gel electrophoresis
Protein content profile of EV fractions was determined using sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Ten μL of each sample were diluted in the same volume of Laemmli buffer (2x; Bio-Rad) with β-mercaptoethanol (5%; Bio-Rad) and boiled at 95°C for 10 min. Then, 20 μL of the mix were loaded into a gel (Mini-Protean TGX gel; 10% polyacrylamide; Bio-Rad). Electrophoresis was performed for 1 hour at 150V. Gels were the stained with SilverQuest (Invitrogen) following the manufacturer's instructions.
Nanoparticle tracking analysis
To determine the concentration and size distribution of EVs, nanoparticle tracking analysis (NTA) was performed in a Nanosight LM10 (Malvern Instruments Ltd, Malvern, UK) with charge-coupled device (CCD) camera (model F-033) and a 638 nm laser. Data were analysed with the NTA V3.0 software. Samples were diluted 10- to 40-fold with 0.2 μm-filtered PBS to yield 40 to 120 particles/frame as recommended by the manufacturer. Up to 3 videos of 60 seconds each were recorded for each sample with the camera shutter at 30.02 ms, gain set at 650 and camera level at 16. Blur and Max Jump Distance were set automatically, detection threshold was set to 5.
Cryo-electron microscopy
Ten μL of pooled tetraspanin-peak fractions were used for cryoelectron microscopy (cryo-EM). Each sample was laid on Formvar-Carbon EM grids, frozen and immediately analysed with Jeol JEM 2011 transmission electron microscope equipped with a 626 Gatan cryoholder operating at an accelerating voltage of 200 kV. The samples, maintained at -182°C during imaging, were recorded on a Gatan Ultrascan cooled CCD camera under low electron dose conditions to minimize electron bean radiation. The ImageJ software (NIH) was used to measure EVs size.
Mass spectrometry analysis
Protein content of PDE-EV-enriched fractions was analysed by liquid chromatography followed by mass spectrometry (LC-MS/MS) on a LTQ Orbitrap Velos (Thermo Fisher, Carlsbad, CA). Samples were reduced with DTT, alkylated with ioidoacetamide and precipitated with trichloroacetic acid. The samples were then washed with acetone and reconstituted in urea before an overnight digestion with trypsin.
Proteomic data processing and analysis
Raw data files were analysed with Max Quant software[17] (version 1.5.3.30) against Uniprot human database (downloaded on December 11, 2015, 70,076 proteins). Parameters set for single protein identification include: (i) minimum peptide length of 7; (ii) maximum false discovery rate (FDR) for peptides and proteins of 1%; (iii) minimum peptides per protein of 1 and minimum unique peptides per protein of 0; (iv) the minimum score for modified peptides was set to 40; (v) main search error of 4 ppm. In addition, in all searches cysteine carbamidomethylation was established as a fixed modification and methionine oxidation and acetylation of the N-terminus were established as variable modifications, with a maximum number of modifications per peptide set to 5. Proteins identified as potential contaminants, those only identified by site or by a reverse sequence, as well as proteins with less than 2 unique peptides were not further considered.
Further analyses of proteins were made using the Intensity-Based Absolute Quantification (iBAQ) values obtained from MaxQuant, and analysed using Perseus software[18] (version 1.5.6.0), InteractiVenn [19] and the EVs specific databases EVpedia [20], Exocarta [21] and Vesiclepedia [22].
iBAQ values were logarithmized to perform the subsequent analysis such as correlation plots, hierarchical clustering analysis (HCA), Principal Component Analysis (PCA) and volcano plot. Gene Ontology (GO) terms for biological process and cellular components were annotated using Perseus. For PCA, data imputation to substitute non-quantified values with low valid intensities based on normal distribution (down-shift of 1.8 and distribution width of 0.3) was performed. Non-supervised HCA was also done after data imputation. Additional HCA was performed considering only the 63 "core" proteins shared by all samples, in both cases after data normalization with z-score and using Euclidean distance in columns and rows. A volcano plot was used to identify the most significant proteins by plotting fold-change difference of log2 iBAQ on x axis and -log2 (p-value) on y axis. The two-sided unpaired t-test was performed with FDR set at 0.05 and s0 at 0.1.
Results
Clinical and epidemiological characteristics of dialysis patients
The study included 9 patients divided in two groups depending on the time on PD: patients with less than 10 months on PD (Newly-Enrolled Patients or NEPs), and patients on PD for more than 18 months (Longer-Treated Patients or LTPs). Clinical data are summarized in Table 1 and detailed per patient in S1 Table. Only 1 patient of the LTP group had type 2 diabetes mellitus, while 3 NEPs and 4 LTPs had hypertension. Regarding the modality of PD, CAPD was used in all NEPs and in 3 patients LTPs. Two LTPs patients used APD. No statistical differences were observed in any of these parameters between both groups.
Table 1. Basal characteristics of the patients.
| n = 9 patients | NEPs <10 months (n = 4 patients) |
LTPs >18 months (n = 5 patients) |
P-valueb |
|---|---|---|---|
| Age (years) | 53.5 (42.0–62.0)a | 54.0 (27.0–75.0)a | 0.806 |
| Time on PD (months) | 7.0 (5.0–10.0)a | 24.0 (21.0–67.0)a | 0.0001 |
| DM (n) | 0 | 1 | 1.000 |
| HTA (n) | 3 | 4 | 1.000 |
| CAPD/APD (n) | 4/0 | 3/2 | 0.444 |
| Icodextrin (n) | 3 | 4 | 1.000 |
aMedian (rank)
b p-values for quantitative data were calculated using U-Mann-Whitney test while qualitative data of the groups were analysed using Fisher’s test.
DM, diabetes mellitus; HTA, hypertension; CAPD, Continuous Ambulatory Peritoneal Dialysis; APD, Automated Peritoneal Dialysis.
Based on PET results (summarized in Table 2 and detailed in S2 Table), 2 NEP patients were classified as "medium transport" and the other 2 NEP patients as "high transport". In the LTP group, 3 patients were classified as "low transport" and 2 patients as "medium transport". The median 4-hour ultrafiltration was 402 (676–82) mL, and the median of total Kt/V was 2.14 (2.37–1.78). Again, no statistically significant differences were found.
Table 2. PET characteristics of the patients.
| n = 9 patients | NEPs <10 months (n = 4 patients) |
LTPs >18 months (n = 5 patients) |
P-value b |
|---|---|---|---|
| D/P creatinine | 0.75 (0.64–0.89) a | 0.56 (0.47–0.75) a | 0.190 |
| D/P urea | 0.84 (0.75–0.87) a | 0.79 (0.76–0.89) a | 1.000 |
| D/D0 glucose | 0.22 (0.20–0.26) a | 0.35 (0.24–0.39) a | 0.063 |
| UF 240 min (mL) | 310.0 (82.0–482.0) a | 577.0 (116.0–676.0) a | 0.286 |
a Median (rank)
b p-values were calculated using U-Mann-Whitney test
Isolation of PDE-EVs from PD patients
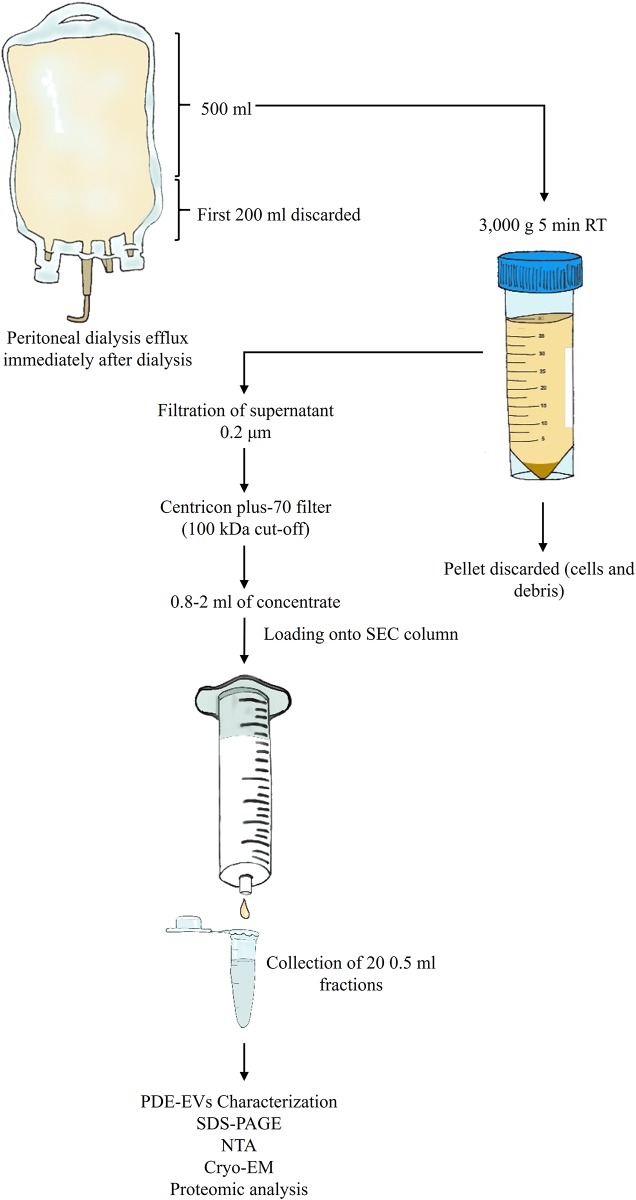
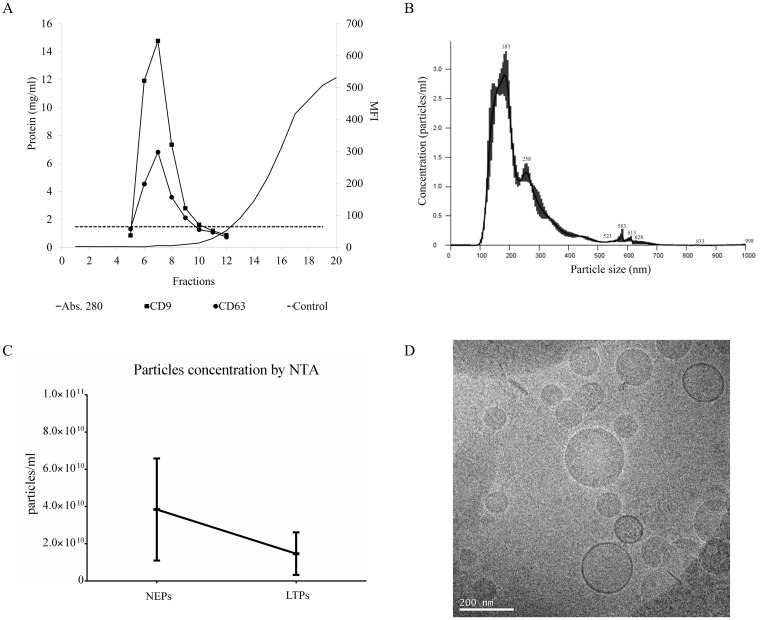
Peritoneal efflux-derived EVs were isolated from patients following a modification of the SEC method (Fig 1). As shown, PDE samples (500 mL) were cleaned from debris, ultra-filtered using a 100 kDa ultrafiltration unit, and loaded into SEC columns. Collected fractions containing higher amount of proteins eluted well-after fraction 10 (Fig 2A). When the same chromatographic fractions were analysed for their tetraspanin markers, CD9 and CD63 expression were found mostly in fractions 6 to 10 (Fig 2A) among the different samples, indicating the presence of PDE-EVs in those fractions.
Fig 1. Schematic representation of peritoneal dialysis efflux sample processing and EV isolation.
Fig 2. Characterization of PDE-EVs.
PDE concentrated samples were further separated using SEC. Up to 20 fractions were recovered and analysed in each sample. In plot A, fractions were analysed for their protein content by BCA (black line). Protein concentration from the different EV-enriched fractions was measured by absorbance at 280nm and calculated using a BSA standard curve. Also, the expression of the EV markers CD9 (black squares) and CD63 (white circles) was determined by flow cytometry. The dotted line represents the isotype control. The left axis represents the total protein content (mg/ml) and the right axis shows the median fluorescence intensity (MFI). For each sample, the three fractions with the highest CD9 and CD63 MFI were pooled for further analyses. A representative plot from 9 experiments is shown. Plot B shows a representative NTA of PDE-EVs (n = 9). Plot C depicts particle concentration determinations, also performed by NTA analyses, in n = 4 NEPs and n = 5 LTPs. Finally, pooled PDE-EVs were visualized by cryo-EM (Fig 2D).
Then, NTA analyses of these fractions showed that PDE-EVs had a modal distribution mainly ranging from 100 to 200 nm (Fig 2B). Regarding particles' concentration, a faint non-significant reduction in the number of detected particles was found in LTPs compared to NEPs (Fig 2C).
Further confirmation of the presence of PDE-EVs in tetraspanin fractions was obtained using Cryo-EM. Images revealed membrane-limited round shaped vesicles (Fig 2D). All together these data indicated that PDE-EVs could be obtained from NEPs and LTPs undergoing PD.
Proteomic analysis of PDE-EV fractions
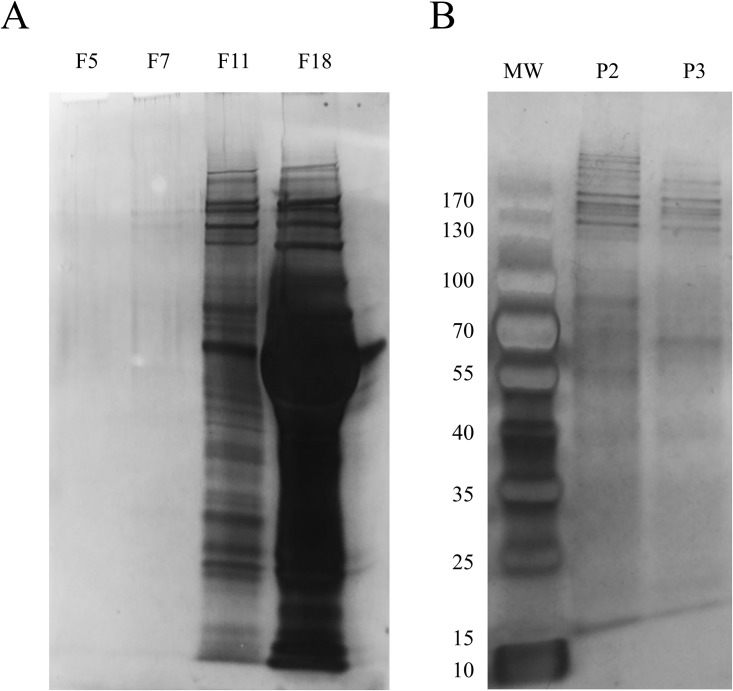
As EVs fractions contain low protein amounts, a preliminary protein content analysis was performed using SDS-PAGE experiments to further characterize the PDE-EVs profile. Different fractions from one patient were analysed and silver-stained gels revealed the presence of some bands in fraction 7 (F7, EV-peak fraction), whilst in fractions 11 and 18 (F11, F18, protein fractions) the number and intensity of the bands increased, clearly revealing the presence of bulk proteins (Fig 3A). When pooled EV fractions from each sample (as detected in Fig 2A) were analysed, silver stained gels showed clearer bands although still less abundant compared to protein-containing fractions (Fig 3B, P2 and P3).
Fig 3. Protein profiling SEC fractions by SDS-PAGE.
(A) Silver staining SDS-PAGE of several SEC fractions, including a pre-tetraspanin fraction (F5), a high tetraspanin-containing fraction (F7) and non-EV protein proximal (F11) and distal (F18) fractions. In plot B, pooled tetraspanin-rich fractions from two different experiments (P2 and P3) are shown. Molecular weight markers are also depicted.
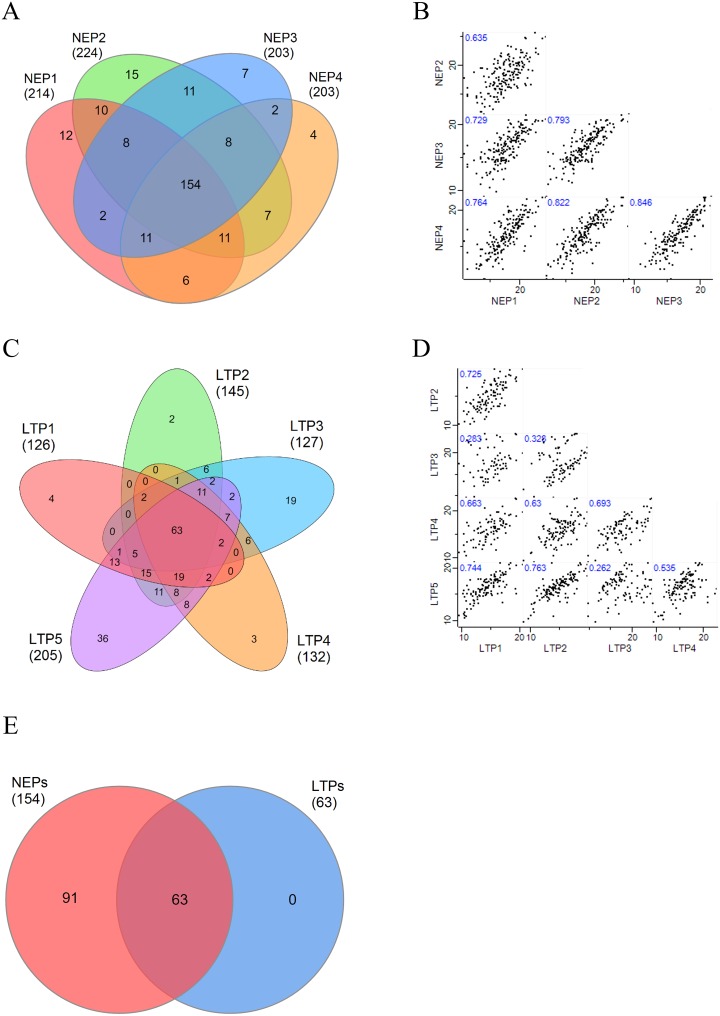
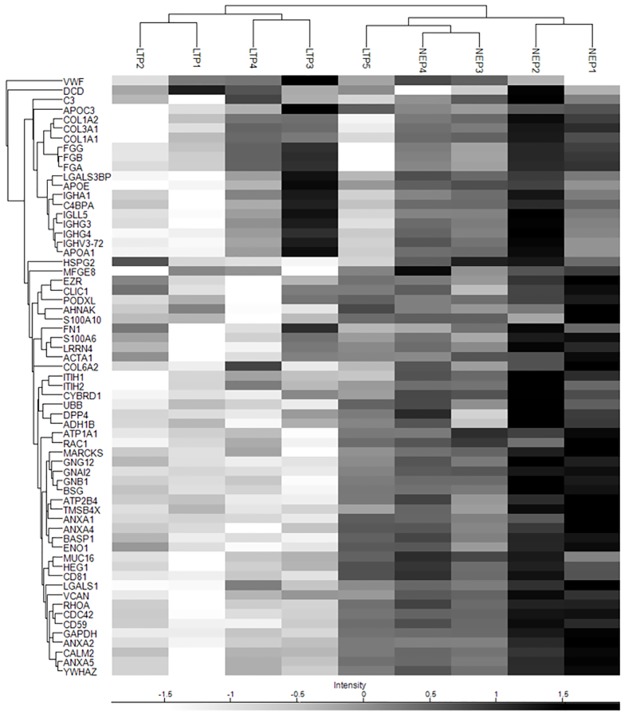
Peritoneal efflux-derived EVs obtained from each NEP (n = 4) and each LTP (n = 5) were further studied to determine their specific peptide profiles using LC-MS/MS. Only proteins identified by at least 2 unique peptides were considered. Overall, a total of 274 proteins were identified. Among NEPs samples, a mean of 211 proteins were identified (211±8), from which 73% (154 proteins) were found in all patients (Fig 4A), revealing a high intragroup similarity. This was further confirmed by multi-scatter plot showing a Pearson Correlation mean “r” value of 0.76±0.08 (mean±sd) (Fig 4B). Regarding LTPs, a mean of 147 proteins (147±23) were identified, from which only 43% (63 proteins) were shared among all LTPs (Fig 4C), with a Pearson Correlation mean “r” value of 0.56±0.20 (Fig 4D). Interestingly, all 63 proteins shared by LTPs were identified also in all NEPs (Fig 4E). These "core" proteins (listed in Table 3) included proteins unequivocally related to EVs, such as CD81, Galectin 3-binding protein (LGALS3BP), Ezrin (EZR) and several members of the Apolipoprotein (APO) and Annexin (ANXA) families, among others.
Fig 4. Protein analyses from PDE-EVs.
Venn diagrams showing overlapping proteins from n = 4 NEPs (A) and n = 5 LTPs (C) are shown. Correlation multi-scatter plots to analyse the correlation within NEPs (B) and LTPs (D) samples. Pearson Correlation “r” values are labelled on each plot. (E) Venn diagram of the proteins shared by all NEPs compared to the proteins shared by all LTPs.
Table 3. Proteins found in all PDE-EVs samples.
Sequence coverage, number of matched peptides, expression fold change between NEPs and LTPs and MS/MS counts are shown for each protein, according to MaxQuant processing of mass-spectrometry data. The proteins are listed in the same order as shown in the clustering analysis in Fig 5. All the proteins present a q-value lower than 10−3.
| Uniprot entry | Protein name | Gene | Sequence coverage (%) | Matched peptides | Fold Change (NEP/LTP) | Total MS/MS count | MS/MS count | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NEP1 | NEP2 | NEP3 | NEP4 | LTP1 | LTP2 | LTP3 | LTP4 | LTP5 | |||||||
| P04275 | von Willebrand factor;von Willebrand antigen 2 | VWF | 25.9 | 60 | -1.057 | 453 | 1 | 9 | 47 | 70 | 91 | 20 | 122 | 62 | 31 |
| P81605 | Dermcidin;Survival-promoting peptide;DCD-1 | DCD | 20.0 | 2 | -0.514 | 23 | 1 | 2 | 2 | 1 | 5 | 3 | 2 | 3 | 4 |
| P01024 | Complement C3;Complement C3 beta chain;C3-beta-c;Complement C3 alpha chain;C3a anaphylatoxin;Acylation stimulating protein;Complement C3b alpha chain;Complement C3c alpha chain fragment 1;Complement C3dg fragment;Complement C3g fragment;Complement C3d fragment;Complement C3f fragment;Complement C3c alpha chain fragment 2 | C3 | 53.9 | 73 | 1.536 | 1426 | 125 | 192 | 175 | 118 | 73 | 263 | 61 | 306 | 113 |
| P02656 | Apolipoprotein C-III | APOC3 | 39.3 | 3 | 1.579 | 69 | 9 | 5 | 4 | 6 | 2 | 1 | 27 | 3 | 12 |
| P08123 | Collagen alpha-2(I) chain | COL1A2 | 15.0 | 15 | 2.431 | 311 | 49 | 40 | 44 | 34 | 34 | 15 | 20 | 61 | 14 |
| P02461 | Collagen alpha-1(III) chain | COL3A1 | 12.6 | 13 | 2.734 | 274 | 57 | 39 | 37 | 32 | 16 | 14 | 24 | 44 | 11 |
| P02452 | Collagen alpha-1(I) chain | COL1A1 | 20.0 | 23 | 2.940 | 343 | 54 | 44 | 54 | 45 | 37 | 8 | 27 | 56 | 18 |
| P02679 | Fibrinogen gamma chain | FGG | 55.0 | 26 | 2.877 | 1213 | 219 | 148 | 96 | 137 | 101 | 66 | 190 | 244 | 12 |
| P02675 | Fibrinogen beta chain;Fibrinopeptide B;Fibrinogen beta chain | FGB | 75.6 | 38 | 2.804 | 2020 | 489 | 257 | 116 | 181 | 106 | 68 | 359 | 429 | 15 |
| P02671 | Fibrinogen alpha chain;Fibrinopeptide A;Fibrinogen alpha chain | FGA | 40.1 | 28 | 2.921 | 639 | 158 | 87 | 50 | 53 | 30 | 27 | 106 | 125 | 3 |
| Q08380 | Galectin-3-binding protein | LGALS3BP | 37.4 | 15 | 3.795 | 251 | 20 | 36 | 48 | 47 | 1 | 1 | 67 | 17 | 14 |
| P02649 | Apolipoprotein E | APOE | 61.2 | 18 | 3.297 | 256 | 22 | 25 | 53 | 40 | 3 | 1 | 72 | 12 | 28 |
| P01876 | Ig alpha-1 chain C region | IGHA1 | 53.5 | 13 | 2.999 | 541 | 48 | 115 | 78 | 52 | 3 | 27 | 132 | 62 | 24 |
| P04003 | C4b-binding protein alpha chain | C4BPA | 57.0 | 27 | 3.843 | 513 | 59 | 92 | 88 | 52 | 1 | 30 | 150 | 21 | 20 |
| B9A064;P0CG04 | Immunoglobulin lambda-like polypeptide 5;Ig lambda-1 chain C regions | IGLL5;IGLC1 | 40.4 | 7 | 2.775 | 262 | 25 | 57 | 27 | 30 | 9 | 14 | 63 | 25 | 12 |
| P01860 | Ig gamma-3 chain C region | IGHG3 | 34.0 | 12 | 2.937 | 198 | 16 | 51 | 23 | 28 | 3 | 5 | 45 | 23 | 4 |
| P01861 | Ig gamma-4 chain C region | IGHG4 | 47.4 | 10 | 3.250 | 45 | 2 | 22 | 2 | 2 | 1 | 1 | 8 | 4 | 3 |
| A0A0B4J1Y9 | IGHV3-72 | 51.5 | 4 | 2.773 | 56 | 5 | 13 | 7 | 9 | 3 | 2 | 8 | 5 | 4 | |
| P02647 | Apolipoprotein A-I;Proapolipoprotein A-I;Truncated apolipoprotein A-I | APOA1 | 58.1 | 16 | 3.159 | 257 | 16 | 53 | 42 | 33 | 2 | 1 | 78 | 25 | 7 |
| P98160 | Basement membrane-specific heparan sulfate proteoglycan core protein;Endorepellin;LG3 peptide | HSPG2 | 10.1 | 32 | 3.238 | 111 | 6 | 13 | 26 | 14 | 3 | 41 | 1 | 2 | 5 |
| Q08431 | Lactadherin;Lactadherin short form;Medin | MFGE8 | 47.5 | 15 | 2.354 | 138 | 16 | 5 | 10 | 37 | 31 | 3 | 2 | 12 | 22 |
| P15311 | Ezrin | EZR | 58.5 | 34 | 1.641 | 607 | 141 | 42 | 45 | 57 | 56 | 126 | 26 | 22 | 92 |
| O00299 | Chloride intracellular channel protein 1 | CLIC1 | 66.0 | 11 | 1.771 | 128 | 29 | 9 | 5 | 16 | 9 | 36 | 5 | 2 | 17 |
| O00592 | Podocalyxin | PODXL | 12.0 | 7 | 2.582 | 57 | 13 | 3 | 7 | 5 | 8 | 4 | 3 | 1 | 13 |
| Q09666 | Neuroblast differentiation-associated protein AHNAK | AHNAK | 13.8 | 25 | 1.953 | 88 | 37 | 3 | 4 | 7 | 12 | 6 | 1 | 1 | 17 |
| P60903 | Protein S100-A10 | S100A10 | 35.1 | 3 | 1.981 | 93 | 31 | 6 | 12 | 11 | 8 | 9 | 1 | 2 | 13 |
| P02751 | Fibronectin;Anastellin;Ugl-Y1;Ugl-Y2;Ugl-Y3 | FN1 | 48.8 | 79 | 1.726 | 1930 | 247 | 240 | 260 | 131 | 69 | 486 | 247 | 67 | 183 |
| P06703 | Protein S100;Protein S100-A6 | S100A6 | 28.2 | 3 | 1.828 | 63 | 9 | 5 | 8 | 8 | 2 | 13 | 6 | 5 | 7 |
| Q8WUT4 | Leucine-rich repeat neuronal protein 4 | LRRN4 | 28.8 | 16 | 2.023 | 368 | 70 | 35 | 34 | 44 | 19 | 66 | 20 | 12 | 68 |
| P68133;P68032;P63267;P62736 | Actin, alpha skeletal muscle;Actin, alpha cardiac muscle 1;Actin, gamma-enteric smooth muscle;Actin, aortic smooth muscle | ACTA1;ACTC1;ACTG2;ACTA2 | 34.0 | 11 | 2.057 | 129 | 30 | 12 | 20 | 9 | 4 | 25 | 9 | 8 | 12 |
| P12110 | Collagen alpha-2(VI) chain | COL6A2 | 17.7 | 14 | 2.363 | 49 | 6 | 5 | 3 | 7 | 3 | 4 | 1 | 15 | 5 |
| P19827 | Inter-alpha-trypsin inhibitor heavy chain H1 | ITIH1 | 30.6 | 18 | 2.358 | 461 | 85 | 54 | 62 | 68 | 45 | 40 | 11 | 55 | 41 |
| P19823 | Inter-alpha-trypsin inhibitor heavy chain H2 | ITIH2 | 28.3 | 22 | 1.711 | 379 | 40 | 44 | 51 | 52 | 33 | 32 | 17 | 46 | 64 |
| Q53TN4 | Cytochrome b reductase 1 | CYBRD1 | 8.7 | 2 | 2.267 | 16 | 2 | 2 | 3 | 3 | 1 | 1 | 2 | 1 | 1 |
| P62987;P62979;P0CG47;P0CG48 | Ubiquitin-60S ribosomal protein L40;Ubiquitin;60S ribosomal protein L40;Ubiquitin-40S ribosomal protein S27a;Ubiquitin;40S ribosomal protein S27a;Polyubiquitin-B;Ubiquitin;Polyubiquitin-C;Ubiquitin | UBB;RPS27A;UBC;UBA52;UBBP4 | 46.2 | 4 | 1.752 | 97 | 10 | 12 | 11 | 15 | 10 | 9 | 4 | 10 | 16 |
| P27487 | Dipeptidyl peptidase 4;Dipeptidyl peptidase 4 membrane form;Dipeptidyl peptidase 4 soluble form | DPP4 | 32.6 | 25 | 1.964 | 272 | 50 | 29 | 13 | 58 | 20 | 30 | 9 | 10 | 53 |
| P00325;P07327;P00326 | Alcohol dehydrogenase 1B;Alcohol dehydrogenase 1A;Alcohol dehydrogenase 1C | ADH1B;ADH1A;ADH1C | 38.7 | 12 | 2.519 | 92 | 14 | 26 | 3 | 15 | 9 | 6 | 4 | 1 | 14 |
| P05023 | Sodium/potassium-transporting ATPase subunit alpha-1 | ATP1A1 | 25.6 | 21 | 2.949 | 136 | 31 | 12 | 30 | 12 | 11 | 9 | 1 | 7 | 23 |
| P63000;P60763 | Ras-related C3 botulinum toxin substrate 1;Ras-related C3 botulinum toxin substrate 3 | RAC1;RAC3 | 25.5 | 5 | 2.461 | 39 | 9 | 2 | 7 | 6 | 3 | 1 | 1 | 2 | 8 |
| P29966 | Myristoylated alanine-rich C-kinase substrate | MARCKS | 48.2 | 8 | 2.686 | 76 | 12 | 11 | 8 | 9 | 6 | 10 | 2 | 7 | 11 |
| Q9UBI6 | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-12 | GNG12 | 66.7 | 4 | 2.523 | 87 | 16 | 9 | 8 | 13 | 9 | 13 | 3 | 4 | 12 |
| P04899 | Guanine nucleotide-binding protein G(i) subunit alpha-2 | GNAI2 | 58.3 | 15 | 2.312 | 281 | 56 | 23 | 48 | 39 | 18 | 27 | 4 | 15 | 51 |
| P62873 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | GNB1 | 56.8 | 14 | 2.303 | 139 | 32 | 14 | 21 | 15 | 8 | 14 | 1 | 8 | 26 |
| P35613 | Basigin | BSG | 44.3 | 6 | 2.373 | 164 | 39 | 18 | 18 | 15 | 12 | 27 | 1 | 8 | 26 |
| P23634 | Plasma membrane calcium-transporting ATPase 4 | ATP2B4 | 18.3 | 17 | 2.764 | 119 | 26 | 12 | 9 | 18 | 19 | 16 | 2 | 2 | 15 |
| P62328 | Thymosin beta-4;Hematopoietic system regulatory peptide | TMSB4X | 47.7 | 3 | 1.548 | 29 | 3 | 2 | 3 | 3 | 4 | 7 | 1 | 2 | 4 |
| P04083 | Annexin A1 | ANXA1 | 61.8 | 18 | 2.241 | 261 | 80 | 19 | 23 | 25 | 28 | 26 | 6 | 16 | 38 |
| P09525 | Annexin A4;Annexin | ANXA4 | 41.7 | 11 | 3.180 | 87 | 23 | 10 | 9 | 13 | 2 | 3 | 2 | 2 | 23 |
| P80723 | Brain acid soluble protein 1 | BASP1 | 75.3 | 10 | 2.798 | 64 | 10 | 7 | 8 | 11 | 2 | 8 | 1 | 4 | 13 |
| P06733 | Alpha-enolase;Enolase | ENO1 | 32.7 | 10 | 2.614 | 78 | 17 | 8 | 6 | 10 | 7 | 10 | 1 | 1 | 18 |
| Q8WXI7 | Mucin-16 | MUC16 | 12.7 | 49 | 2.319 | 773 | 66 | 77 | 130 | 164 | 25 | 67 | 30 | 57 | 157 |
| Q9ULI3 | Protein HEG homolog 1 | HEG1 | 10.3 | 11 | 2.959 | 61 | 9 | 7 | 5 | 15 | 1 | 1 | 0 | 3 | 20 |
| P60033 | Tetraspanin;CD81 antigen | CD81 | 35.8 | 3 | 2.154 | 179 | 26 | 18 | 23 | 24 | 12 | 22 | 6 | 19 | 29 |
| P09382 | Galectin-1 | LGALS1 | 57.0 | 6 | 2.722 | 82 | 27 | 9 | 7 | 11 | 2 | 2 | 0 | 9 | 15 |
| P13611 | Versican core protein | VCAN | 5.4 | 15 | 2.328 | 263 | 76 | 22 | 35 | 30 | 10 | 21 | 13 | 24 | 32 |
| P61586;P08134 | Transforming protein RhoA;Rho-related GTP-binding protein RhoC | RHOA;RHOC | 42.0 | 7 | 2.927 | 88 | 18 | 7 | 14 | 16 | 1 | 8 | 1 | 7 | 16 |
| P60953 | Cell division control protein 42 homolog | CDC42 | 25.1 | 4 | 2.664 | 80 | 18 | 11 | 11 | 9 | 5 | 10 | 1 | 4 | 11 |
| P13987 | CD59 glycoprotein | CD59 | 29.6 | 4 | 3.278 | 86 | 16 | 11 | 10 | 13 | 0 | 14 | 2 | 3 | 17 |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | 36.5 | 6 | 1.898 | 70 | 16 | 5 | 10 | 9 | 5 | 8 | 2 | 4 | 11 |
| P07355;A6NMY6 | Annexin A2;Annexin;Putative annexin A2-like protein | ANXA2;ANXA2P2 | 69.0 | 23 | 2.285 | 557 | 132 | 48 | 68 | 52 | 48 | 72 | 21 | 40 | 76 |
| P62158;P27482 | Calmodulin | CALM2;CALM1;CALM3 | 42.2 | 8 | 1.912 | 123 | 30 | 9 | 15 | 15 | 2 | 17 | 3 | 9 | 23 |
| P08758 | Annexin A5;Annexin | ANXA5 | 70.9 | 19 | 3.374 | 286 | 106 | 28 | 33 | 31 | 5 | 10 | 6 | 24 | 43 |
| P63104 | 14-3-3 protein zeta/delta | YWHAZ | 51.8 | 12 | 2.729 | 163 | 46 | 15 | 16 | 21 | 6 | 17 | 6 | 13 | 23 |
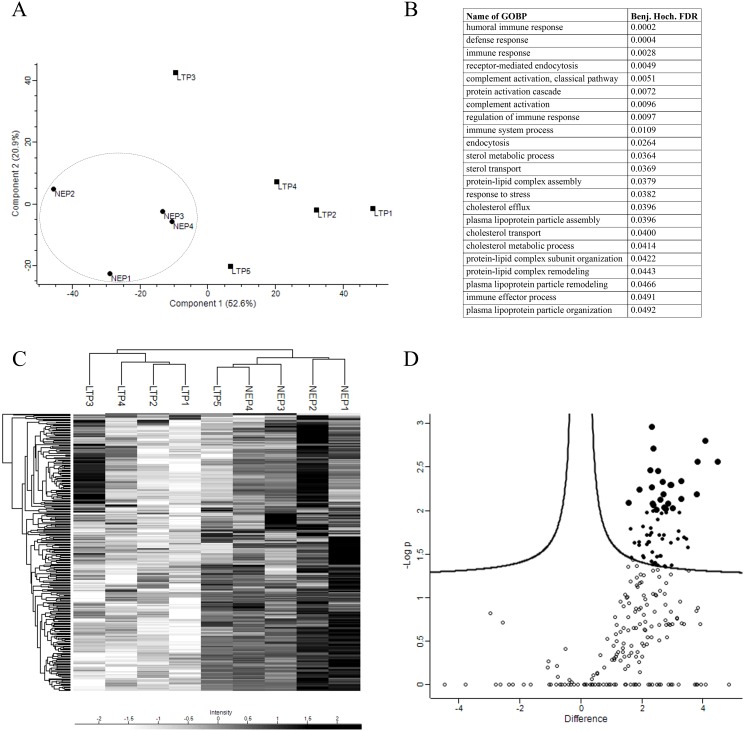
To further evidence the differences between both groups, a PCA was performed. Based on component 1, which accounts for the 52.6% of the variability between the samples, both groups were segregated based on their time on PD (Fig 6A). The Gene Ontology biological processes analysis of component 1 revealed that most enriched terms in NEP in comparison to LTP are those related to the immune system (Fig 6B). Additionally, HCA of the 274 proteins detected among all samples also clustered most patients based on their time on PD (Fig 6C). Finally, a volcano plot comparing the protein expression between the two groups evidenced the statistically significant proteins showing a significantly different level of expression (Fig 6D). These analyses revealed that up to 67 proteins were significantly overexpressed in NEP than LTP (p-value <0.05).
Fig 6. Proteins analyses from PDE-EVs.
(A) Two dimensional scatter plot of Principal Component Analysis (PCA) showing component 1 and 2, which account for 52.6% and 20.9%, respectively, the variability of all the 274 proteins. NEPs (circles) and LTPs (squares) are separated by component 1. A dashed line circle indicates grouped NEPs. (B) Table with Gene Ontology biological process enriched terms for component 1 with their corresponding Benjamini-Hochberg FDR values is shown (all the listed terms have a Benj. Hoch. FDR <0.05). (C) HCA associated with a heat map of the 274 proteins (rows) and the samples (columns). (D) A volcano plot was performed to determine significantly differentially expressed proteins between groups. Each circle represents a protein, being statistically significant proteins with this parameters shown as filled circles. Proteins with p-value <0.01 are represented as bigger filled circles.
In addition, a HCA performed exclusively on the 63 "core" proteins identified in all samples resulted in the segregation of 8 out of 9 patients based on time in PD, in a very similar way to the HCA performed with all the proteins (Fig 5).
Fig 5. Hierarchical clustering analysis of the 63 “core” proteins.
Samples and the 63 proteins shared by all samples were clustered with HCA associated with a heat map. Names of the codifying genes are shown.
Discussion
In this study we report, for the first time to our best knowledge, the presence, isolation and characterization of PDE-EVs from patients on PD.
Peritoneal dialysis is a convenient treatment for end-stage kidney disease patients waiting for a kidney transplant. Studies have reported better survival rates, quality of life and independence of PD patients compared to haemodialysis patients[23,24]. However, during the treatment, fibrotic changes reduce the ultrafiltration capacity of the PM, meaning that many patients have to discontinue treatment.
Current monitoring of the PM function (the PET), requires patients' attendance to the dialysis centre, is time-consuming and only shows alterations when the PM is in an advanced state of fibrosis. Time-delays on identifying PMs' dysfunction may carry dangerous complications, and even lead to the death of the patient. Therefore, finding early biomarkers of PM dysfunction that minimally disturb patients’ daily life may help to overcome these limitations, contribute keeping functional PMs for longer periods and improve patients’ management. Several studies have searched for biomarkers in PDE correlating with PM function, detection of fibrosis, and/or the failure of the technique (reviewed in[25]). Proteomic studies of mesothelial cell lines[7] and transcriptome analysis in rats[26] have reported differences between the protein and miRNA content, respectively, of cells exposed or non-exposed to peritoneal liquids.
It is of current acceptance that information contained in EVs may serve as biomarkers of pathological situations. Biomarkers for kidney pathology have been described in urine EVs[12,27], and serum/plasma EVs have been also related to multiple pathologies[28]. It may be therefore envisaged that PDE-EVs may also provide useful information about the state and function of the PM. Such information could help the clinician to accommodate the treatment to enhance the optimal functionalism of the PM.
PDE-EVs were equally identified in all patients from both NEPs and LTPs groups. SEC-isolated vesicles contained in the tetraspanin rich fractions had a size and morphology compatible with EV, as shown by NTA and cryo-EM analyses. As reported before in urine[29] and plasma samples[30], our results point to SEC as an efficient technique to isolate EVs also from PDE samples. Importantly, SEC permits the segregation of EVs from the bulk of proteins found in samples, thus allowing more accurate analyses of the EV-protein content and enabling the search for minimally expressed proteins. In line, SDS-PAGE results confirmed that EVs were cleanly separated from other major components of the PDE, and preliminary proteomic analysis of SEC-isolated EVs identified a number of well-defined EV-related proteins. Importantly, all these results are in accordance with the recommendations of the International Society for Extracellular Vesicles (ISEV) to identify EVs in a given sample [31], thus validating SEC to isolate EVs also from PDE.
Having identified EVs in all samples, and to further explore possible differences in this pilot study, patients were distributed in two arbitrary groups based on their median time on PD. Both groups did not show major differences in any of the parameters analysed, nor in the PET test. However, a slightly (not significant) reduced number of EVs and also a reduced number of proteins were identified in the PDE-EVs from the LTP group compared to NEPs., It was also interesting to note that a "core" of proteins were identified in both groups, although showing some differences in their level of expression. These "core" proteins included most proteins unequivocally related to EVs. Whether these differences may anticipate a possible worsening of the ultrafiltration capacity of the membrane not detected by PET analyses need further investigation and validation in a wider cohort of patients.
Since this study has consistently demonstrated that EVs can be isolated from concentrated PDE, it seems reasonable to think that these EVs could be used as a source of biomarkers. In addition, the non-invasive origin of the sample and the reduced inconvenience for patients point to the analysis of PDE-EVs as a next step in the definition of early biomarkers of ultrafiltration failure in peritoneal dialysis.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
Thanks to Marco A. Fernández (Flow Cytometry Unit, IGTP) and Pablo Castro Hartmann (Electron Microscopy Unit, UAB). Also, Carolina Gálvez-Montón for graphics and Hernando del Portillo (ICREA at ISGLOBAL-IGTP) for the NTA instrument. Antibodies to CD9 and CD63 were a gift from Dr. Yáñez-Mó (Unidad de Investigación, Hospital Sta Cristina, IIS-IP; Departamento Biología Molecular/CBM-SO, UAM) and Dr. Francisco Sánchez-Madrid (Servicio de Inmunología, Hospital Universitario de la Princesa, IIS-IP, UAM; Cell-cell Communication Laboratory, CNIC).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the PI16/00072 project, integrated in the National R + D + I and funded by the ISCIII and the European Regional Development Fund http://www.isciii.es), “Suport Grups de Recerca” programme of Generalitat de Catalunya (2014SGR804, Group REMAR, http://agaur.gencat.cat), Instituto de Salud Carlos III-Red de Investigación Renal (REDinREN) (RD16/0009 Feder Funds, http://www.isciii.es, http://redinren.org), and Fundació Cellex. MF is sponsored by the Beatriu de Pinós-B contract (2014BP B00118, http://agaur.gencat.cat) from Agència de Gestió d’Ajuts Universitaris i de Recerca (AGAUR) – Generalitat de Catalunya. FEB is sponsored by the “Researchers Stabilization Program” from the Spanish “Sistema Nacional de Salud” (SNS- ISCIII, http://www.isciii.es) and “Direcció d’Estratègia i Coordinació” Catalan Health Department (CES07/015, http://salutweb.gencat.cat). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Aroeira L, Loureiro J. Characterization of epithelial-to-mesenchymal transition of mesothelial cells in a mouse model of chronic peritoneal exposure to high glucose dialysate. Perit Dial. 2008;28:29–33. [PubMed] [Google Scholar]
- 2.Yáñez-Mo M, Lara-Pezzi E, Selgas R, Ramirez-Huesca M, Dominguez-Jimenez C, Jimenez-Heffernan JA, et al. Peritoneal dialysis and epithelial-to-mesenchymal transition of mesothelial cells. N Engl J Med. 2003;348(5):403–13. 10.1056/NEJMoa020809 [DOI] [PubMed] [Google Scholar]
- 3.Aguilera A, Yáñez-Mo M, Selgas R, Sánchez-Madrid F, López-Cabrera M. Epithelial to mesenchymal transition as a triggering factor of peritoneal membrane fibrosis and angiogenesis in peritoneal dialysis patients. Curr Opin Investig Drugs. 2005. March;6(3):262–8. [PubMed] [Google Scholar]
- 4.Treharne C, Liu FX, Arici M, Crowe L, Farooqui U. Peritoneal dialysis and in-centre haemodialysis: A cost-utility analysis from a UK payer perspective. Appl Health Econ Health Policy. 2014;12(4):409–20. 10.1007/s40258-014-0108-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atapour A, Eshaghian A, Taheri D, Dolatkhah S. Hemodialysis Versus Peritoneal Dialysis, Which is Cost-effective? Saudi J Kidney Dis Transpl. 2015;26(5):962–5. 10.4103/1319-2442.164578 [DOI] [PubMed] [Google Scholar]
- 6.Sritippayawan S, Chiangjong W, Semangoen T, Aiyasanon N, Jaetanawanitch P, Sinchaikul S, et al. Proteomic Analysis of Peritoneal Dialysate Fluid in Patients with Different Types of Peritoneal Membranes—Journal of Proteome Research (ACS Publications). J Proteome Res [Internet]. 2007;6(11):4356–62. Available from: 10.1021/pr0702969 [DOI] [PubMed] [Google Scholar]
- 7.Lechner M, Kratochwill K, Lichtenauer A, Rehulka P, Mayer B, Aufricht C, et al. A proteomic view on the role of glucose in peritoneal dialysis. J Proteome Res. 2010;9(5):2472–9. 10.1021/pr9011574 [DOI] [PubMed] [Google Scholar]
- 8.Wang HY, Tian YF, Chien CC, Kan WC, Liao PC, Wu HY, et al. Differential proteomic characterization between normal peritoneal fluid and diabetic peritoneal dialysate. Nephrol Dial Transplant. 2010;25(6):1955–63. 10.1093/ndt/gfp696 [DOI] [PubMed] [Google Scholar]
- 9.Fais S, O’Driscoll L, Borras FE, Buzas E, Camussi G, Cappello F, et al. Evidence-Based Clinical Use of Nanoscale Extracellular Vesicles in Nanomedicine. ACS Nano. 2016;10(4):3886–99. 10.1021/acsnano.5b08015 [DOI] [PubMed] [Google Scholar]
- 10.Yáñez-Mó M, Siljander PR-M, Andreu Z, Zavec AB, Borràs FE, Buzas EI, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell vesicles [Internet]. 2015;4:27066 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4433489&tool=pmcentrez&rendertype=abstract 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simons M, Raposo G. Exosomes—vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–81. 10.1016/j.ceb.2009.03.007 [DOI] [PubMed] [Google Scholar]
- 12.Gàmez-Valero A, Lozano-Ramos SI, Bancu I, Lauzurica-Valdemoros R, Borràs FE. Urinary Extracellular Vesicles as Source of Biomarkers in Kidney Diseases. Front Immunol [Internet]. 2015;6(January):6 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4311634&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynöe N, Sandlund M, Dahlqvist G, Jacobsson L. Informed consent: study of quality of information given to participants in a clinical trial. BMJ. 1991;303(6803):610–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mujais S, Nolph K, Gokal R, Blake P, Burkart J, Coles G, et al. Evaluation and Management of Ultrafiltration Residual Renal Therapy. Society. 2000;20:5–21. [PubMed] [Google Scholar]
- 15.Waniewski J, Werynski A, Heimbürger O, Lindholm B. Simple models for description of small-solute transport in peritoneal dialysis. Blood Purif. 1991;9:129–41. [DOI] [PubMed] [Google Scholar]
- 16.Lozano-Ramos I, Bancu I, Oliveira-Tercero A, Armengol MP, Menezes-Neto A, Del Portillo HA, et al. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J Extracell vesicles [Internet]. 2015;4:27369 Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4449362&tool=pmcentrez&rendertype=abstract 10.3402/jev.v4.27369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol [Internet]. 2008;26(12):1367–72. Available from: 10.1038/nbt.1511\nhttp://www.ncbi.nlm.nih.gov/pubmed/19029910 [DOI] [PubMed] [Google Scholar]
- 18.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods [Internet]. 2016;13(9):731–40. Available from: http://www.nature.com/doifinder/10.1038/nmeth.3901%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/27348712 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 19.Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R. InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics [Internet]. 2015;16(1):169 Available from: http://www.biomedcentral.com/1471-2105/16/169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH, et al. EVpedia: A community web portal for extracellular vesicles research. Bioinformatics. 2015;31(6):933–9. 10.1093/bioinformatics/btu741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9(21):4997–5000. 10.1002/pmic.200900351 [DOI] [PubMed] [Google Scholar]
- 22.Kalra H, Simpson RJ, Ji H, Aikawa E, Altevogt P, Askenase P, et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012;10(12):8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heaf JG, Wehberg S. Relative survival of peritoneal dialysis and haemodialysis patients: Effect of cohort and mode of dialysis initiation. PLoS One. 2014;9(3):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukowsky LR, Mehrotra R, Kheifets L, Arah O a, Nissenson AR, Kalantar-Zadeh K. Comparing mortality of peritoneal and hemodialysis patients in the first 2 years of dialysis therapy: a marginal structural model analysis. Clin J Am Soc Nephrol [Internet]. 2013;8(8):619–28. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23307879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopes Barreto D, Krediet RT. Current status and practical use of effluent biomarkers in peritoneal dialysis patients. Am J Kidney Dis [Internet]. Elsevier Inc.; 2013;62(4):823–33. Available from: 10.1053/j.ajkd.2013.01.031 10.1053/j.ajkd.2013.01.031 [DOI] [PubMed] [Google Scholar]
- 26.Lin F, Wu X, Zhang H, You X, Zhang Z, Shao R, et al. A microrna screen to identify regulators of peritoneal fibrosis in a rat model of peritoneal dialysis. BMC Nephrol [Internet]. 2015;16(1):48 Available from: http://www.scopus.com/inward/record.url?eid=2-s2.0-84927761541&partnerID=tZOtx3y1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Erdbrugger U, Le TH. Extracellular Vesicles in Renal Diseases: More than Novel Biomarkers? J Am Soc Nephrol [Internet]. 2015;1–15. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26251351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics—Clin Appl. 2015;9(3–4):358–67. 10.1002/prca.201400114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lozano-Ramos I, Bancu I, Olivera-Tercero A, Armengol M, de Menezes-Neto A, del Portillo HA, et al. Size exclusion chromatography-based enrichment of extracelluar vesicles from urine samples. J Extracell Vesicles. 2015;accepted f:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gámez-Valero A, Monguió-Tortajada M, Carreras-Planella L, Franquesa M, Beyer K, Borràs FE. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci Rep [Internet]. Nature Publishing Group; 2016;6(September):33641 Available from: http://www.nature.com/articles/srep33641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lener T, Gioma M, Aigner L, Börger V, Buzas E, Camussi G, et al. Applying extracellular vesicles based therapeutics in clinical trials—an ISEV position paper. J Extracell Vesicles [Internet]. 2015;4(January 2016). Available from: http://www.journalofextracellularvesicles.net/index.php/jev/article/view/30087/xml_44 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.