Summary
Elucidating temporal windows of signaling activity required for synaptic and behavioral plasticity is crucial for understanding molecular mechanisms underlying these phenomena. Here, we developed photoactivatable autocamtide inhibitory peptide 2 (paAIP2), a genetically-encoded, light-inducible inhibitor of CaMKII activity. The photoactivation of paAIP2 in neurons for 1–2 min during the induction of LTP and structural LTP (sLTP) of dendritic spines inhibited these forms of plasticity in hippocampal slices of rodents. However, photoactivation ~1 min after the induction did not affect them, suggesting that the initial 1 min of CaMKII activation is sufficient for inducing LTP and sLTP. Furthermore, the photoactivation of paAIP2 expressed in amygdalar neurons of mice during an inhibitory avoidance task revealed that CaMKII activity during, but not after, training is required for the memory formation. Thus, we demonstrated that paAIP2 is useful to elucidate the temporal window of CaMKII activation required for synaptic plasticity and learning.
In brief
Murakoshi and Shin et al. developed a light-inducible inhibitor of CaMKII and demonstrated that a brief illumination of blue light to neurons expressing this inhibitor blocks synaptic plasticity and learning.
Introduction
Ca2+/Calmodulin Kinase II (CaMKII) is required for induction of Long-Term Potentiation (LTP) and the associated structural plasticity of dendritic spines (Lisman et al., 2012). A holoenzyme of CaMKII consists of 12 subunits and each subunit is activated by the association of Ca2+/Calmodulin (Chao et al., 2011; Lisman et al., 2012). Activation of two adjacent subunits leads to trans-autophosphorylation at Thr-286, which enables autonomous activity after Ca2+/calmodulin dissociation (Chao et al., 2011; Lisman et al., 2012; Zhabotinsky, 2000). Because of this calcium-independent activity, sustained CaMKII activity had been postulated as a mechanism of LTP maintenance. The duration of CaMKII activation during LTP has been measured using biochemical analysis, but the results are inconsistent, with some reports of more than ~1 hour (Barria et al., 1997; Fukunaga et al., 1993; Ouyang et al., 1997) and other reports of a much shorter time period (Lengyel et al., 2004). Recently, using 2-photon fluorescence lifetime imaging microscopy (2pFLIM) with a fluorescence resonance energy transfer (FRET)-based CaMKII sensor, we imaged CaMKII activity in dendritic spines during spine enlargement associated with LTP (refered to as structural LTP or sLTP) induced by 2-photon glutamate uncaging, and demonstrated that CaMKII activation persists for only ~1 min (Lee et al., 2009).
While the 2-photon FRET-FLIM imaging approach is useful to study the kinetics of CaMKII in single dendritic spines, its application to electrical stimulation-induced LTP has been complicated by the difficulty of locating stimulated spines among ~10,000 in each transfected neuron. As a complementary approach, the temporal requirement of CaMKII activation for LTP induction has been measured by using pharmacologic inhibitors. It has been shown that application of CaMKII inhibitors before LTP induction inhibits LTP expression, whereas application after LTP establishment does not inhibit LTP expression (Buard et al., 2010; Chen et al., 2001; Malinow et al., 1989; Otmakhov et al., 1997), consistent with the transient CaMKII activation found by 2-photon FRET-FLIM imaging (Lee et al., 2009). It also has been reported that the interaction between CaMKII and the N-methyl-D-aspartate receptor (NMDAR) is critical for both induction and expression of LTP using CN21, a peptide derived from a natural CaMKII inhibitor that inhibits CaMKII activation at low concentration and disrupts CaMKII-NMDAR interaction at high concentration (Sanhueza et al., 2011). However, the prolonged time required for bath applied pharmacologic inhibitors to reach intracellular compartments in brain slices precludes the precise measurement of the temporal requirement of CaMKII activation for LTP expression. In addition, it is difficult to distinguish between the requirement of basal kinase activity and stimulation-evoked activity with pharmacological analyses.
Here, in order to measure the precise temporal requirement of CaMKII activation for synaptic plasticity, we developed paAIP2, a photoactivatable CaMKII inhibitor. Its design is based on the light-oxygen-voltage domain 2 (LOV2)-Jα helix domain (404–539) of Phototropin1, which changes its conformation from a closed form to an open form upon blue light absorption (Wu et al., 2009). This light-controllable conformation change has been utilized for various types of photo-activatable proteins (Tischer and Weiner, 2014). Using a similar approach, we fused autocamtide inhibitory peptide 2 (AIP2), a potent CaMKII inhibitory peptide (Ishida et al., 1998), to LOV2-Jα, in order to inhibit CaMKII in a light-dependent manner. After validation of the paAIP2, we measured the temporal requirement of CaMKII activation for structural plasticity of dendritic spines and high-frequency stimulation (HFS)-induced LTP as well as an inhibitory avoidance (IA) task (Irvine et al., 2005; Yamagata et al., 2009).
Results
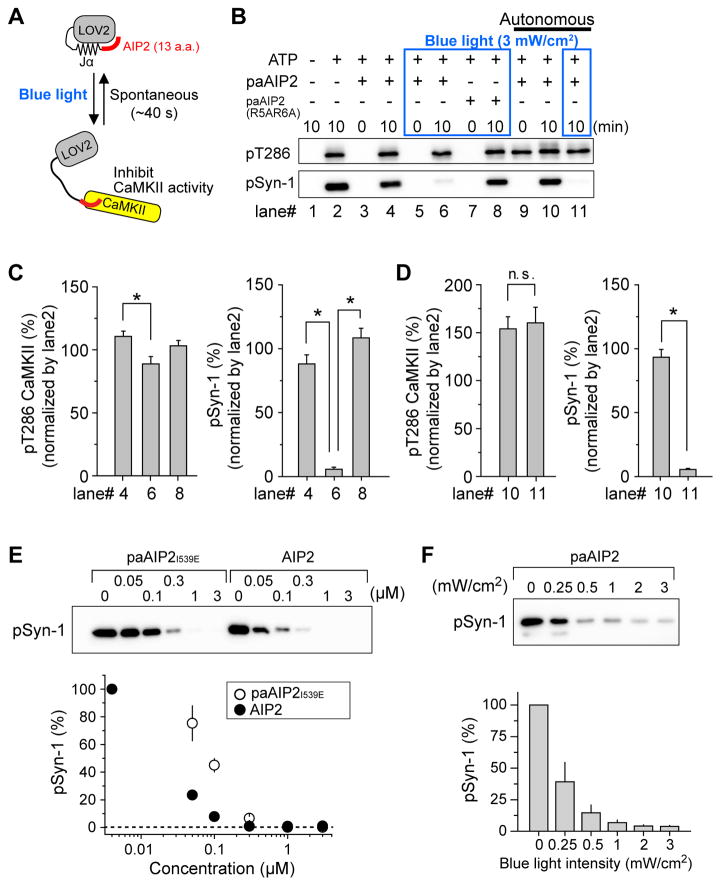
Photoactivatable AIP2 specifically inhibits CaMKII in a light-dependent manner
We designed paAIP2 so that LOV2-Jα in the closed conformation blocks the action of AIP2 due to steric hindrance, but upon light absorption, LOV2-Jα transitions to an open conformation and allows for AIP2-mediated inhibition of CaMKII. This “on-state” is reversed spontaneously as LOV2-Jα returns to its closed conformation (Wu et al., 2009), ending the inhibitory action of paAIP2 on CaMKII (Fig. 1A). To test the efficacy of paAIP2, we measured kinase activity of full-length CaMKIIα in the presence of paAIP2 in a cell-free assay. Autophosphorylation of T286 in CaMKIIα and phosphorylation of synapsin site 1 parent peptide (Syn1) (White et al., 1998), a CaMKIIα substrate peptide, were monitored by western blotting to assess CaMKII activity. Regardless of the presence of paAIP2, the phosphorylation of T286 and Syn1 similarly increases with the addition of ATP in the absence of blue light (Fig. 1B). In contrast, blue-light illumination at 3 mW/cm2 strongly inhibited phosphorylation of Syn1 in the presence of paAIP2, but not in its absence (Fig. 1B, 1C). When we used a low affinity paAIP2R5A/R6A, which has an approximately 104-fold lower affinity to CaMKII than that of wildtype AIP2 (Ishida et al., 1998), CaMKII activity was not inhibited even under blue light illumination. This result suggests that the inhibitory effect is due to the action of the AIP2 in paAIP2 (Fig. 1B, 1C). Similar to another peptide inhibitor CN-21 (Vest et al., 2007), paAIP2 did not inhibit autophosphorylation at Thr-286 but did inhibit phosphorylation of a CaMKII substrate (Fig. 1C). Ca2+-independent, autonomous activity of CaMKII was also inhibited by paAIP2 (Fig. 1D). PaAIP2 with a constitutively open LOV2I539E showed only a slightly reduced potency compared to AIP2 alone (Fig. 1E). The relationship between the degree of inhibition and the light intensity revealed that an intensity of ~1 mW/cm2 is sufficient to substantially inhibit CaMKII activity (Fig. 1F).
Fig. 1. A genetically encoded light-inducible CaMKII inhibitor, paAIP2.
A, Schematic of paAIP2. LOV2-Jα domain derived from phototropin1 was fused to autocamtide-2 related inhibitory peptide 2 (AIP2). The blue light absorption causes a reversible conformational change of LOV2 domain, leading to dissociation of the Jα helix-AIP2. The structurally released AIP2 binds to the kinase domain of CaMKII and inhibits its kinase activity.
B, PaAIP2 inhibits CaMKII activity in a blue-light dependent manner. Phosphorylation of full-length CaMKII at Thr286 and substrate peptide Syn-1 were detected by western blotting. ATP triggers CaMKII autophosphorylation and substrate phosphorylation (lane 1, 2). PaAIP2 does not inhibit phosphorylation in the absence of blue light (lane 3, 4) but does inhibit in the presence of blue light (lane 5, 6). A low affinity paAIP2 mutant (R5A/R6A in AIP2) fails to inhibit kinase activity in the presence of blue light (lane 7, 8). For lane 9–11, active CaMKII oligomer (autonomous state) was prepared as described in Methods section. Syn-1 peptide was mixed with active CaMKII oligomer and incubated for indicated time in the absence (lane 10) or presence (lane 11) of blue light. For blue light illumination (3 mW/cm2), the samples were continuously illuminated over the indicated time.
C, Quantification of (B, lane 4, 6, 8). For data analysis, the band intensity of lane 2 was normalized to 100%. Error bars indicate S.E.M. for six independent experiments. For the statistical test, one-way ANOVA with Bonferoni post-hoc test with *p < 0.05 was used.
D, Quantification of (B, lane 10, 11, p > 0.05, t-test, n.s., not significant). Error bars indicate S.E.M. for six independent experiments.
E, Inhibitory effect of AIP2 and paAIP2I539E (constitutively open form mutant) for active CaMKII was compared. Phosphorylation of purified Syn-1 peptide by active CaMKII1–325 in the presence of various concentration of AIP2 or paAIP2I539E was detected by western blotting. Error bars indicate S.E.M. for three independent experiments.
F, The light intensity dependency of CaMKII inhibition by paAIP2. Phosphorylation of purified Syn-1 peptide by active CaMKII1–325 in the presence of paAIP2 was detected by western blotting. Samples were continuously illuminated at indicated blue light intensity during the reaction (10 min). Error bars indicate S.E.M. for three independent experiments.
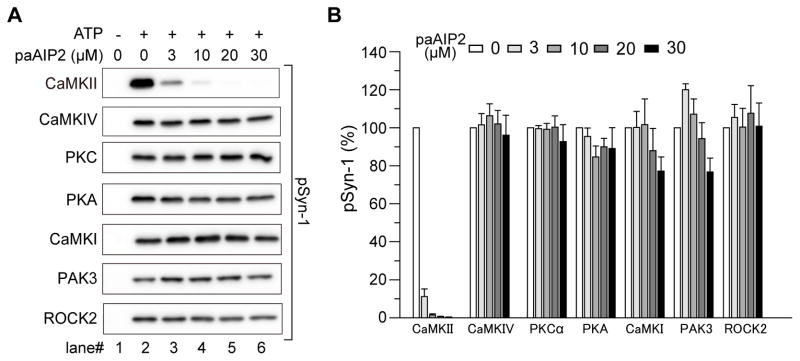
To test the specificity of paAIP2, we measured the activity of various purified kinase domains (i.e., constitutively active forms) derived from major kinases, CaMKIIα, CaMKIV, PKCα, PKA, CaMKI, PAK3, and ROCK2 for phosphorylating against Syn1. In the absence of paAIP2, all of these kinases phosphorylated Syn1 (Fig. 2). When paAIP2 with a constitutively open form of LOV2I593E was included in the solution (3–30 μM), the phosphorylation by CaMKIIα was inhibited with high efficacy and potency (> 85%), but not by other kinases, suggesting that paAIP2 is a specific inhibitor of CaMKII (Fig. 2). Notably unlike KN62 and KN93 (Enslen et al., 1994) but as with AIP2 (Ishida et al., 1995; Ishida et al., 1998), paAIP2 did not inhibit activity of CaMKI and CaMKIV. Consistent with the action of paAIP2 in a cell-free assay, immunoblot analysis demonstrated that paAIP2 expressed in dissociated neurons slightly inhibited autophosphorylation of CaMKII at T286 (Fig. S1A), but strongly inhibited phosphorylation of SynGAP, a target of CaMKII, in a light-dependent manner (Fig. S1B). In addition, the phosphorylation of Neuromodulin, a PKC substrate, and other PKC and PKA substrates were much less affected by paAIP2 (Fig. S1C–E). These results suggest that paAIP2 is a specific inhibitor of CaMKII.
Fig. 2. Specificity of paAIP2 inhibition.
A, Effect of paAIP2 under blue-light was tested on different kinases. Phosphorylation of purified Syn-1 peptide by various active kinases was detected by western blotting. PaAIP2 blocks CaMKII activity at various concentrations, while having no effect on other kinases.
B, Quantification of (A). For data analysis, kinase activity of lane 2 was normalized to 100% for each kinase. Error bars indicate S.E.M. for three independent experiments except PAK3 (n=4).
See also Figure S1.
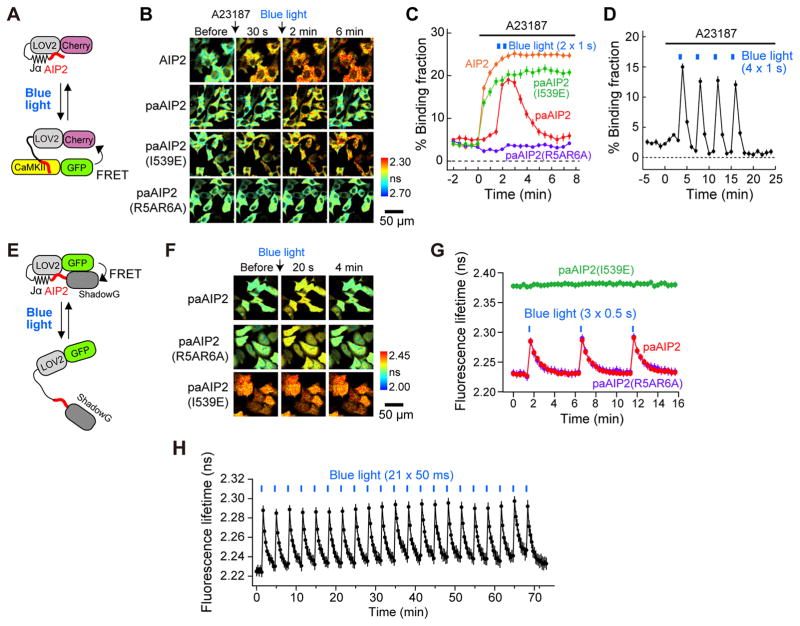
To test the kinetics of CaMKII inhibition in cells, we transfected mCherry-AIP2 or mCherry-paAIP2 and monomeric enhanced green fluorescence protein (mEGFP)-CaMKIIα into HeLa cells and measured their binding using 2pFLIM (Yasuda et al., 2006) (Fig. 3A–D). Elevation of intracellular Ca2+ levels by the application of ionophore (A23187) rapidly increased the binding between mEGFP-CaMKIIα and mCherry-AIP2 (Fig. 3B, C), consistent with findings that AIP2 is a pseudo-peptide competitive inhibitor (Ishida et al., 1995; Ishida et al., 1998). In contrast, mCherry-paAIP2 did not bind to mEGFP-CaMKIIα upon ionophore stimulation but rapidly bound to mEGFP-CaMKIIα upon light illumination, and dissociated over ~40 s (Fig. 3B, C). This could be repeated many times with blue-light pulses, demonstrating its reversibility (Fig. 3D). Constitutively active paAIP2I539E showed a similar time course to mCherry-AIP2, whereas low-affinity paAIP2R5A/R6A did not show any change in binding (Fig. 3C). These results show that paAIP2 binds to CaMKIIα in a lightdependent and reversible manner in HeLa cells.
Fig. 3. Characterization of paAIP2 in HeLa cells.
A, Schematic of the experiment to measure binding between CaMKII and paAIP2.
B, Fluorescence lifetime images of binding between CaMKII and paAIP2 mutants in HeLa cells. PaAIP2I539E (constitutively open form) and with a low affinity PaAIP2R5A/R6A are also shown. Cells expressing mEGFP-CaMKIIα and mCherry-paAIP2 were stimulated with 20 μM A23187 ionophore at time 0, and subsequently illuminated twice with blue light (473 nm, 100 mW/cm2) for 1 second with an interval of 30 s. Warmer colors indicate shorter lifetimes and higher levels of the binding between CaMKII and paAIP2. Scale bar, 50 μm.
C, The time course of the binding between mEGFP-CaMKIIα and mCherry-AIP2 or mCherry-paAIP2 (wildtype, I539E, R5A/R6A) in experiments described in B. The number of cells is 20 (AIP2), 20 (paAIP2), 22 (paAIP2 (I539E)) and 19 (paAIP2 (R5A/R6A)).
D, Binding between mEGFP-CaMKIIα and mCherry-paAIP2 (wildtype) in HeLa cells after ionophore with repetitive light stimulation.
E, Schematics of mEGFP-paAIP2-ShadowG used to characterize the conformational change of paAIP2. Opening of paAIP2 increases the distance between donor (mEGFP) and acceptor (ShadowG), leading to the decrease of FRET.
F, Fluorescence lifetime images of mEGFP-paAIP2-ShadowG in HeLa cells. PaAIP2 and its mutants (I539E and R5A/R6A) are also shown.
G, H, Characterization of the light-induced conformational change of paAIP2 and its mutants (I539E, R5A/R6A) in HeLa cells. Cells expressing mEGFP-paAIP2-ShadowG mutants were illuminated with blue light (473 nm, 100 mW/cm2) at the indicated times. The numbers of cells are 17 (paAIP2 in G), 18 (paAIP2I539E in G), 17 (paAIP2R5A/R6A in G), and 10 (paAIP2 in H).
See also Figure S2.
Next, since the association of CaMKII with the C-terminal tail of the GluN2B subunit of NMDA receptors plays an important role in autonomous CaMKII activity and LTP (Bayer et al., 2001; Zhou et al., 2007), we tested whether the interaction between CaMKII and the CaMKII interaction domain of GluN2B1277–1328 is affected by paAIP2 (Fig. S2). We monitored binding between mCherry-GluN2B1277–1328 and mEGFP-CaMKIIα in HeLa cells by 2pFLIM. Ionophore application rapidly increased the binding between mEGFP-CaMKIIα and mCherry-GluN2B1277–1328, suggesting that the CaMKII-GluN2B interaction requires CaMKII activation (Fig. S2B–C). However, in the presence of LOV2I539E-AIP2 or LOV2I539E-GluN2B, the binding between mCherry-GluN2B1277–1328 and mEGFP-CaMKIIα was significantly inhibited (Fig. S2C, D). In contrast, the low-affinity mutant LOV2I539E-AIP2R5A/R6A did not affect the CaMKII-GluN2B interaction (Fig. S2C, D). These results suggest that paAIP2 inhibits the CaMKII-GluN2B interaction in cells (Fig. S2C, D).
We further characterized the kinetics of the structural change of paAIP2 by measuring intra-molecular FRET of mEGFP-paAIP2-ShadowG in HeLa cells (Fig. 3E, F) (Murakoshi et al., 2015). As expected from the known conformation of LOV2-Jα, the constitutively open form of paAIP2I539E exhibited a longer fluorescence lifetime (lower FRET) (~2.38 ns) than that of wildtype (~2.23 ns) in dark conditions (Fig. 3F, G). Upon light illumination, paAIP2 displayed a rapid fluorescence lifetime increase (FRET decrease), which is recovered in ~1 minute (Fig. 3F, G). Constitutively active paAIP2I539E did not show further increase in the fluorescence lifetime (Fig. 3F, G). The fluorescence lifetime at the peak suggests that ~40% of paAIP2 opens under this condition. The low affinity paAIP2R5A/R6A exhibited similar open and close kinetics to paAIP2 (Fig. 3F, G). Furthermore, we confirmed that the photoactivation of paAIP2 can be repeated more than 20 times without any fatigue (Fig. 3H).
Temporal window of CaMKII activation necessary for structural plasticity of dendritic spines and LTP
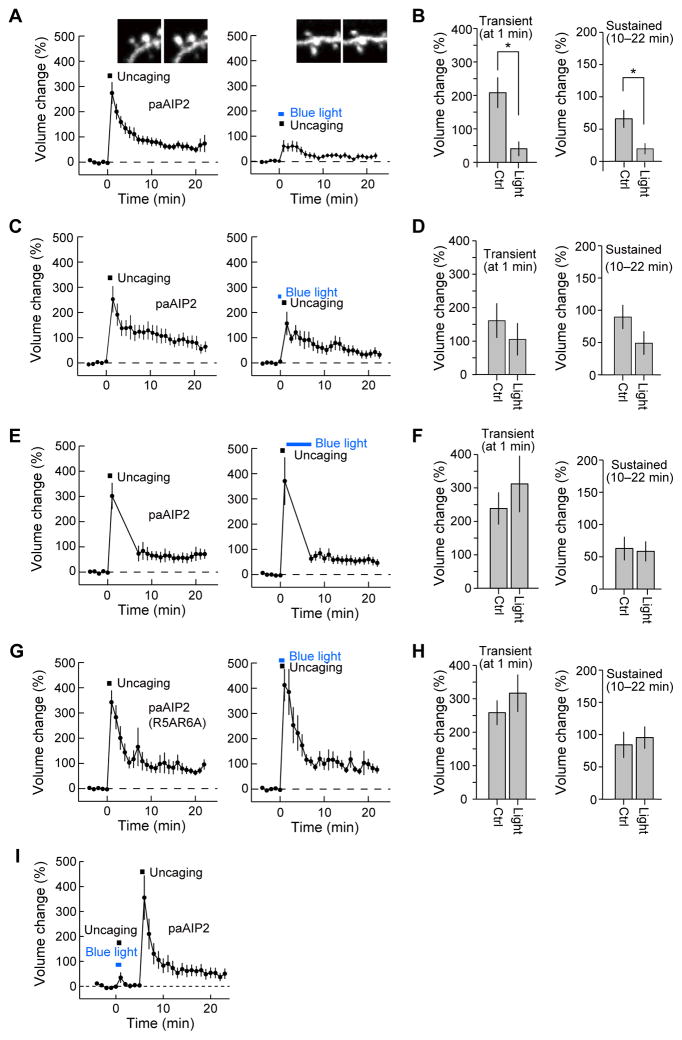
Next, we tested if blue light illumination inhibits CaMKII-dependent plasticity of dendritic spines in CA1 pyramidal neurons in cultured hippocampal slices (Fig. 4, 5). We biolistically transfected hippocampal slices with mEGFP-P2A-mCherry-P2A-paAIP2 plasmid where the DNA sequences coding mEGFP, mCherry, and paAIP2 were linked together with P2A elements so that these three proteins are expressed separately (Donnelly et al., 2001; McAllister, 2000). Since the excitation wavelength of paAIP2 overlaps with that of mEGFP, we used red fluorescence from mCherry for identifying neurons by epi-fluorescence microscopy. Fluorescence from mEGFP was used to image single dendritic spines under a 2-photon laser scanning microscope because of its superior photostability and brightness under 2-photon excitation than that of mCherry. To induce sLTP in a single dendritic spine, we applied a low frequency train of two-photon glutamate uncaging pulses (30 pulses at 0.5 Hz) to the spine in the absence of extracellular Mg2+ (Harvey and Svoboda, 2007; Lee et al., 2009; Matsuzaki et al., 2004). The spine volume increased rapidly by 300–400% following glutamate uncaging (transient phase) and relaxed to an elevated level of 70–100% for more than 20 min (sustained phase) (Fig. 4A), consistent with the previous reports (Harvey et al., 2008; Lee et al., 2009; Matsuzaki et al., 2004). This suggests that paAIP2 does not affect sLTP under 2-photon imaging (< 1.5 mW) and uncaging (~ 5 mW). Wide-field illumination by a blue laser (100 mW/cm2, 400 μmΦ) for 1 min during glutamate uncaging significantly inhibited both the transient and sustained spine enlargement (Fig. 4A, B) (Lee et al., 2009; Matsuzaki et al., 2004; Murakoshi et al., 2011), demonstrating that CaMKII-dependent plasticity can be inhibited by paAIP2 in a blue-light-dependent manner. In contrast, neither 1 minute long blue-light illumination immediately prior to uncaging nor 5 minute long blue-light illumination initiated 1 minute after uncaging was capable of inhibiting sustained phase spine volume changes (Fig. 4C–F), suggesting that CaMKII activity is only required in the early phase of structural plasticity induction. The low affinity mutant, paAIP2R5A/R6A, failed to inhibit spine enlargement (Fig. 4G, H), suggesting that blue-light-dependent inhibition of spine plasticity is due to the action of AIP2. Furthermore, we confirmed the reversibility of the inhibitory action of paAIP2 by two sets of glutamate uncaging stimulations delivered to the same spine. Glutamate uncaging with concomitant blue light illumination failed to produce sLTP, whereas the same glutamate uncaging protocol initiated 5 minutes later in the absence of blue-light illumination produced normal spine volume changes (Fig. 4I). Thus, the inhibitory effect of paAIP2 is reversible in cultured slices just as it is in HeLa cells. Finally, we characterized the relationship between the expression level of paAIP2 and changes in spine volume. The concentration of paAIP2 was measured from mEGFP fluorescence compared to purified mEGFP under the assumption that mEGFP and paAIP2 are equally expressed using the P2A sequence. We found that, through a broad range of expression levels (5–60 μM), paAIP2 inhibited spine structural plasticity to a similar degree (Fig. S3).
Fig. 4. PaAIP2 inhibits spine enlargement in a light-dependent manner.
A, C, E, G, I, Averaged time course of spine volume change induced with 2-photon glutamate uncaging (0.5 Hz, 30 pulses) in neurons expressing mEGFP-P2A-mCherry-P2A-paAIP2 (A, C, E, I) or its mutant (R5A/R6A) (G). Fluorescence intensity of mEGFP was used to measure the spine volume change in the stimulated spine. Experiments with (right panels) and without (left panels) blue light were performed in the same neurons. The numbers of samples (spines/neurons) are 14/10 (A, B), 12/12 (C, D), 12/9 (E, F), 12/8 (G, H) and 9/5 (I).
B, D, F, H. Transient volume change (volume change averaged over 1 min subtracted by that averaged over 10–22 min) and sustained volume change (volume change averaged over 10–22 min). For B, D, F, and H, bar graphs represent summarized data from A, C, E, and G, respectively. Stars denote statistical significance (p < 0.05, paired t-test).
See also Figure S3.
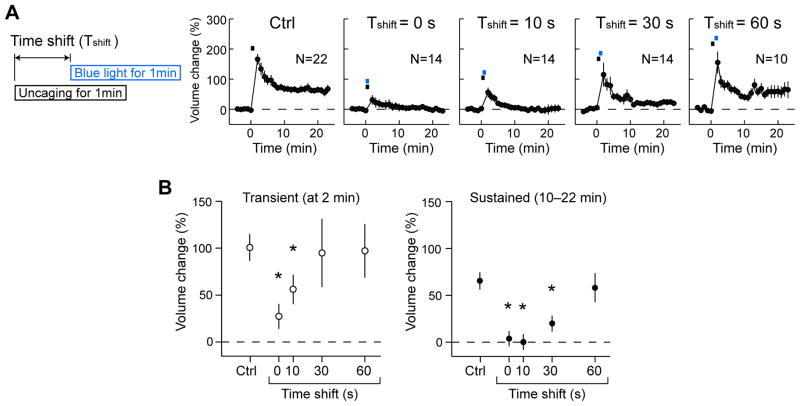
Fig. 5. Inhibition of spine enlargement under precise temporal control of CaMKII inhibition with paAIP2.
A, Averaged time course of spine volume change under different temporal inhibition of CaMKII. During single-spine stimulation with 2-photon glutamate uncaging (0.5 Hz, 30 pulses), neurons were illuminated with blue light for 1 min with different timing relative to the onset of glutamate uncaging (Time shift 0, 10, 30, 60 s). Fluorescence intensity of mEGFP was used to measure the spine volume change. The numbers of samples are indicated in the panels.
B, Transient volume change (volume change at 2 min subtracted by that averaged over 10–22 min) and sustained volume change (volume change averaged over 10–22 min) as a function of time shift. Stars denote statistical significance (p < 0.05, nonparametric one-way ANOVA followed by post-hoc tests using the Tukey’s least significant difference).
To further elucidate the precise temporal requirement of CaMKII activity for structural plasticity, we applied blue light for 1 min and glutamate uncaging for 1 min at different timing pairings (Fig. 5A). When blue light and glutamate uncaging were applied at the same time, both the transient and sustained phases of volume change were inhibited. However, by delaying the blue light application by 10–30 s, the transient phase, but not the sustained phase, was partially restored (Fig. 5A, B). When the blue light was shifted by 60 s, normal plasticity for both transient and sustained phases was observed (Fig. 5A, B). These results suggest that the transient and sustained phases of sLTP may have distinct temporal requirements for CaMKII activity.
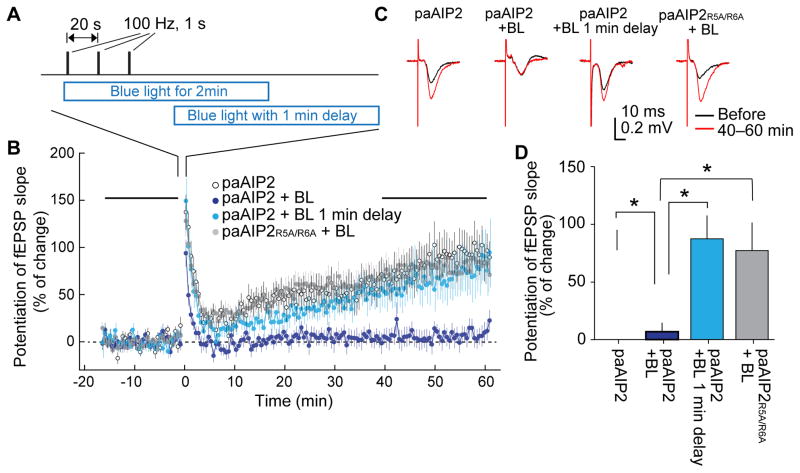
Next, to study the temporal requirement of CaMKII for a standard HFS-induced LTP, we injected adeno-associated virus (AAV) encoding mEGFP-P2A-paAIP2 under the control of the Camk2a promoter (AAV-mEGFP-P2A-paAIP2) into the hippocampus of mice and prepared acute hippocampal slices from the infected mice. The field excitatory postsynaptic potential (fEPSP) slope was recorded in these slices using an electrode located in CA1 while stimulating Schaffer-Collateral fibers with a bipolar electrode in the presence and absence of blue light (Fig. 6A). When HFS (100 Hz, 1 s, 3 times, 20 s intervals) was applied to the slice, we observed a robust potentiation of fEPSP slopes, indicating that LTP was induced (Fig. 6B, C, D). When blue light was illuminated for ~2 min during the LTP induction, LTP was abolished (Fig. 6B, C, D). However, when blue light was applied with a 1 min delay from the first stimulation, we observed normal LTP (Fig. 6B, C, D). When a low affinity paAIP2R5A/R6A mutant was used, normal LTP was observed even with blue light (Fig. 6B, C, D). Taken together, these experiments suggest that 1 min of CaMKII activation is sufficient to induce LTP.
Fig. 6. Inhibition of field LTP in acute hippocampal slices.
A. LTP induction protocol and the timing of blue light illumination (35 mW/cm2).
B. Averaged time course of field excitatory postsynaptic potential (fEPSP) slope measured with an electrode located at CA1 dendritic layer while stimulating Schaffer-Collateral fibers with a bipolar electrode. LTP was induced with the protocol in A at time zero. Blue light (BL) was applied as indicated in A.
C. Representative traces of fEPSP.
D. Quantification of LTP at 50–60 min after LTP induction. Stars denote statistical significance (p < 0.05, nonparametric one-way ANOVA followed by post-hoc tests using the Tukey’s least significant difference). Numbers of neurons/animals are: 21/19, 12/11, 18/12, 36/13 for paAIP2 without light, paAIP2 with blue light illumination, paAIP2 with blue light illumination with 1 min delay, and paAIP2R5A/R6A with blue light illumination, respectively.
The temporal window of CaMKII activation in amygdala required for inhibitory avoidance task
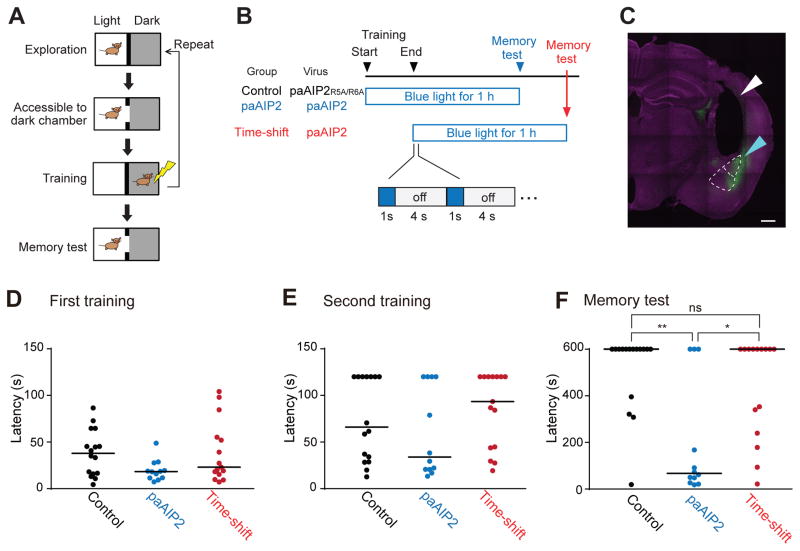
Finally, we tested the capability of paAIP2 to change an animal’s behavior in the inhibitory avoidance learning paradigm, in which mice are placed in the bright compartment of an apparatus with light and dark compartments, and the mice are given a brief foot shock (0.5 mA for 2 s when they enter the dark compartment (Fig. 7A). Learning to avoid the dark compartment in the inhibitory avoidance paradigm has been previously reported to be CaMKII-dependent (Irvine et al., 2005; Yamagata et al., 2009). Mice were subjected to double-trial training inhibitory avoidance learning in an inhibitory avoidance apparatus (Fig. 7A) and tested for the memory maintenance 1 hour after training. Animals were divided into three groups. For the first two groups (“control” and “paAIP2”; Fig. 7B), we bilaterally injected non-functional mutant AAV-mEGFP-P2A-paAIP2R5A/R6A (Control) or AAV-mEGFP-P2A-paAIP2 (paAIP2) into the amygdalae (lateral amygdala and basolateral amygdala) and implanted optical fibers (800 μm Φ) ~200 μm above the brain region (Fig. 7C, Fig. S4, Table S1). For both groups, blue light was applied through the optical fibers (cycles of 1 s light on at 30 mW and 4 s off) (Fig. 7B). The light application encompassed the entire training session and lasted for an hour. For the third group, AAV-mEGFP-P2A-paAIP2 was injected in the same region of the brain (amygdalae), but blue light illumination was initiated only after the training ceased and was maintained for the same duration (1h) (“time-shift”, Fig. 7B). All three groups of animals showed normal behavior and no significant difference in latency during the training (Fig. 7D and E). Following the light delivery, mice were subjected to a test of the acquired memory. We found that most of the mice in the control group avoided the dark chamber for more than 600 seconds, the maximum duration of the testing period (n = 16; median = 600 s) (Fig. 7F). In contrast, the paAIP2 group showed significant impairment: many animals crossed to the dark chamber before the 600 s cut-off point (n =12: p < 0.01; median = 67.2 s; Kruskal-Wallis test followed by Dunn’s test) (Fig. 7F). The animals in the time-shift group showed a latency comparable to that of the control (n = 15; median = 600 s; p > 0.99) (Fig. 7F). Taken together, we demonstrated that the activity of CaMKII in the amygdalae during the training session, but not after the training session, is necessary for memory formation.
Fig. 7. Inhibition of CaMKII by paAIP2 impairs memory formation in inhibitory avoidance learning.
A. A schematic of double-trial training inhibitory avoidance. Upon entering and exploring the light chamber within the inhibitory avoidance apparatus, a mouse is allowed to enter the dark chamber, after which a foot shock is administered. The training is repeated once more. Each training trial ends if the animal crosses or avoids the dark chamber for 120 s.
B. A schematic of light application during inhibitory avoidance. For the control and paAIP2 groups, blue light (0.2 Hz, 1s) is applied from the onset of training for an hour. For the time-shift group, light delivery begins after the training. Timelines are not drawn to scale.
C. The expression of mEGFP-P2A-paAIP2 in amygdala. The cyan and white triangles indicate the area of transgene expression and the fiber track above amygdala, respectively. The upper region demarcated by white dotted line indicates lateral amygdala (LA). The lower region indicates basolateral amygdala (BLA). Scale bar = 500 μm. Note that animals expressing detectable transgene expression either in LA or BLA in both hemispheres were included in the analyses D–F.
D, E. Cross latency for the first (A) and second (B) training trials. Three groups of mice show no statistically significant difference in cross latency in both training trials. Cutoff latency set at 120 s. Bars represent median. Dunn’s multiple comparisons test following Kruskal-Wallis test (n = 16, 12 and 15 for control, paAIP2, and time-shift group, respectively).
F. Cross latency for the memory test (1 h). Cutoff latency set at 600 s. Bars represent median. P-values are from Dunn’s multiple comparisons test following Kruskal-Wallis test.
Discussion
In this study, we developed paAIP2, a photo-inducible CaMKII inhibitor that enables CaMKII inhibition in a light-dependent manner. PaAIP2 presents several advantages over conventional pharmacological inhibition or genetic manipulation. First, it allows researchers to manipulate CaMKII activity with second to minute temporal resolution. Second, because it is genetically encoded, it enables the manipulation of CaMKII in a cell-type and brain-region specific manner. Third, it has high specificity for CaMKII: the specificity of paAIP2 is similar to that of other peptide inhibitors AIP2 (Ishida et al., 1995; Ishida et al., 1998) and CN-21 (Vest et al., 2007) and higher than that of commonly used small drugs such as KN62 and KN93, which inhibit CaMKI and CaMIV as well as CaMKII (Enslen et al., 1994). We applied this tool in several forms of synaptic plasticity and one form of learning and demonstrated that it is useful for dissecting the temporal window of CaMKII activation necessary for these CaMKII-dependent phenomena.
Our analyses of spine structural plasticity using paAIP2 suggest that ~10–30 s of CaMKII activation is sufficient for inducing transient spine enlargement, while ~60 s is required for sustained spine enlargement (Fig. 4, 5). These results confirmed our previous results showing that CaMKII activation lasts ~1 min during spine structural plasticity (Lee et al., 2009). Thus, these complementary studies revealed that both the duration of CaMKII activation during spine structural plasticity and the duration necessary for inducing the plasticity is ~1 min. The different temporal requirement between these two phases may be due to differences in the integration properties of downstream signaling. Previously we showed that Rho GTPases Rac, Rho, and Cdc42 are CaMKII-dependent, but they play differential roles in the different phases of structural plasticity (Hedrick et al., 2016; Murakoshi et al., 2011).
Our study also revealed the temporal requirement of CaMKII activation for HFS-induced LTP to be ~1 min, similar to that for spine structural plasticity (Fig. 6). While the process of glutamate uncaging-evoked spine structural plasticity and HFS-LTP has been thought to be similar (Matsuzaki et al., 2004), the kinetics of molecular activation during HFS-LTP has not been measured. These results suggest that similar signaling cascades are activated during glutamate uncaging-induced spine structural plasticity and HFS-LTP. Consistent with the relatively short window of CaMKII activation required for LTP and spine enlargement, acute inhibition of CaMKII using paAIP2 during the inhibitory avoidance task displayed that CaMKII activation in the amygdalae during the training session, but not after the training session, is necessary for this form of memory. Our studies on HFS-LTP and the inhibitory avoidance task demonstrate that our optogenetic kinase inhibitor allows for measurements of signaling activity in neuronal compartments that are not easily accessible with imaging methods. The light intensity required for inhibiting CaMKII with paAIP2 is orders of magnitude smaller than that required for channel rhodopsin 2 activation (1–100 mW/cm2 for paAIP2 vs ~1000 mW/cm2 for ChRh2 (Boyden et al., 2005)) and thus will not cause significant damage to the tissue.
One of the potential future directions is to combine paAIP2 with FRET-based signaling sensors. Previously we demonstrated that activity of small GTPase proteins Ras, Cdc42, RhoA, Rac can be monitored with FRET sensors in combination with 2pFLIM, and these molecules are activated downstream of CaMKII (Harvey et al., 2008; Murakoshi et al., 2011; Hedrick et al., 2016). By combining paAIP2 and these FRET sensors, we should be able to determine the temporal requirement of CaMKII activation for various downstream signaling molecules.
It is relatively straightforward to apply the strategy used in designing paAIP2, i.e., fusion of LOV2-Jα to a protein inhibitor peptide, to create photo-inducible inhibitors for other signaling proteins including the peptide inhibitors of protein kinase A (PKI) (Yi et al., 2014), and protein kinase C (Pseudo-substrate inhibitors). Such light-controllable tools will be useful for analyzing the temporal nature of intracellular signaling cascades.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ryohei Yasuda (ryohei.yasuda@mpfi.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All experimental protocols were approved by Max Planck Florida Institute IACUC, Duke University Medical Center or National Institutes of Natural Sciences and meet the guidelines of the National Institutes of Health guide for the Care and Use of Laboratory Animals.
Animals
For behavioral experiments, C57BL/6 male mice (males; Charles River) were individually housed under a reversed 12 h light/dark cycle with food and water ad libitum. Mice were between 3–5 months old and weighed approximately 27–35 g at the time of the behavior experiments. Mice used in electrophysiological experiments were between 2.5–3 months old and weighed approximately 24–28 g. Littermates were randomly assigned to experimental groups. Animals were not exposed to any previous procedures such as pharmacological treatments or altered diets. All animals exhibited a healthy and normal behavior.
Hippocampal Slices
Hippocampal slices were prepared from postnatal day 5–7 CD IGS rats (both males and females, from Charles River) as described (Stoppini et al., 1991) and cultured for 1–2 weeks prior to gene transfection. Briefly, we deeply anaesthetized the animal with isoflurane, after which the animal was quickly decapitated and the brain removed. The hippocampi were isolated and cut into 350 μm sections using a tissue chopper (McIlwain). Hippocampal slices were plated on tissue culture inserts (Millicell) fed by tissue medium (for 2.5 L: 20.95 g MEM, 1.19 g HEPES, 1.1 g NaHCO3, 5.8 g D-Glucose, 120 μL 25% ascorbic acid, 12.5 mL 200 mM L-Glutamine, 2.5 mL 1 mg/ml Insulin, 500 mL Horse Serum, 5 mL 1 M MgSO4, 2.5 mL 1 M CaCl2). Slices were incubated at 35°C in 3% CO2.
Primary neuronal culture
Hippocampi dissected from 4 newborn CD IGS rats (postnatal day 0; both males and females; Charles River) were triturated and plated into 35 mm dishes coated with 50 μg/ml PLL (Sigma) in culture medium composed of basal medium Eagle (BME) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen), 35 mM glucose (Sigma), 1 mM Glutamax (Sigma), 100 U/ml penicillin (Sigma), and 0.1 mg/ml streptomycin (Sigma). Proliferation of non-neuronal cells was inhibited by Cytosine arabinoside (2.5 μM) treatment on the second day after plating.
Cell Lines
Both HeLa cells and HEK293 cells (ATCC) were cultured in Dulbecco’s modified eagle medium supplemented with 10% fetal bovine serum at 37°C in 5% CO 2. Cells were used as an expression platform only, and were thus not rigorously tested for potential contamination from other cell lines.
METHOD DETAILS
Plasmids
PaAIP2 was constructed by fusing the codon optimized LOV2404–539 sequence to the amino terminus of autocamtide-2 related inhibitory peptide 2 (AIP2) sequence KKKLRRQEAFDAL (Ishida et al., 1998) with no amino acid linker. The mEGFP-P2A-mCherry-P2A-paAIP2 plasmid was constructed by inserting paAIP2 and mCherry (Shaner et al., 2004) together with P2A sequences ATNFSLLKQAGDVEENPGP into the pmEGFP-C1 vector which was constructed by introducing the A206K monomeric mutation in pEGFP-C1 (Clontech) (Zacharias et al., 2002). For construction of mEGFP-CaMKIIα plasmid, the full-length CaMKIIα sequence was inserted into pmEGFP-C1 with a linker amino acid SRLRSRA. For construction of the mCherry-paAIP2 plasmid, the paAIP2 sequence was inserted into pmCherry-C1 with a linker amino acid ASM. The mEGFP-paAIP2-mCherry was constructed by inserting paAIP2 and mCherry into the pmEGFP-C1 vector. The paAIP2R5A/R6A and paAIP2I539E (Harper et al., 2004) were prepared by introducing the mutations into AIP2 sequence using Site-Directed Mutagenesis kit (Stratagene). AAV-mEGFP-P2A-paAIP2 was prepared by inserting Camk2a promoter and mEGFP-P2A-paAIP2 into rAAV-9 vector. Virus was produced by University of North Carolina Vector Core.
Protein purification from bacteria
For construction of His-tagged super-folder GFP (sfGFP)-paAIP2 and their mutants (R5A/R6A, I539E), sfGFP (Pedelacq et al., 2006) and paAIP2 mutant were inserted into pRSET bacterial expression vector (Invitrogen) with a linker amino acid SM. For construction of His-tagged mCherry-Syn1, mCherry and Syn1 sequence encoding amino acid sequence LRRRLSDANF were cloned into the pRSET vector. Proteins were overexpressed in Escherichia coli (DH5α) and purified with a Ni+-nitrilotriacetate (NTA) column (HiTrap, GE Healthcare), and desalted with a desalting column (PD10, GE Healthcare) equilibrated with phosphate buffer saline (PBS). The concentration of the purified protein was measured by the absorbance of the fluorophore (sfGFP, A489 = 83,000 cm−1M−1; mCherry, A587 = 72,000 cm−1M−1) (Pedelacq et al., 2006; Shaner et al., 2004).
Protein purification from HEK293 cells
For construction of plasmids, CaMKIIα1–325, CaMKI1–319 (purchased from Kazusa), CaMKIV1–337 (a gift from Richard Maurer, Addgene plasmid # 45062) (Sun et al., 1994), PKA43–331 (synthesized), PKC338–618 (a gift from Alex Toker, Addgene plasmid #10805) (Hodges et al., 2002), PAK3247–544 (a gift from William Hahn & David Root, Addgene plasmid # 23439) (Johannessen et al., 2010), and ROCK287–390 (purchased from Kazusa) genes were inserted into the modified pEGFP-C1 vector with His-tagged mEGFP, respectively. For construction of full-length CaMKIIα, His-tag sequence was fused to the C-terminal of CaMKIIα gene, subsequently, CaMKIIα-His was inserted into the modified pEGFP-C1 vector by replacing EGFP. HEK293 cells were transfected with the plasmids using Lipofectamine2000 (Invitrogen). One day after transfection, cells were lysed and purified by NTA column, similarly to the protein purification from bacteria. The purified protein concentrations were measured by the absorbance of mEGFP (A488 = 58,000 cm−1M−1) (Murakoshi et al., 2015).
Kinase assay
Standard kinase assays were performed for the indicated time at room temperature with 20–40 nM purified full length CaMKIIα or CaMKIIα1–325, 2 μM mCherry-Syn1, 2 μM Calmodulin, 200 μM CaCl2, 200 μM ATP, 3 μM sfGFP-paAIP2 in a reaction buffer (50 mM Tris-HCl, pH 7.3, 10 mM MgCl2, 2 mM DTT). For PKA, PKC, PAK3, and ROCK2, the experiments were done in the absence of calmodulin and CaCl2. For the preparation of autonomous CaMKII, 15 nM full length CaMKIIα was activated for 1 min in the reaction buffer containing 2 μM Calmodulin, 200 μM CaCl2, 200 μM ATP. Subsequently, the reaction was stopped by adding 1 mM EDTA. At indicated lanes, samples were continuously illuminated with a light-emitting diode (LED) (M455L2-C1; Thorlabs) at 3 mW/cm2 (450 nm) during incubation. The reactions were stopped by adding SDS sample buffer, and then analyzed by western blotting. The following antibodies were used: anti-CaMKII (ab52476; Abcam); anti-phosphoT286 CaMKII (p1005-286; PhosphoSolutions); anti-phospho-PKA substrate (RRXS*/T*) (#9624; Cell Signaling Technology) for phosphorylated mCherry-Syn1 detection; HRP-anti-mouse and HRP-anti-rabbit (Zymax).
Cell preparations
CA1 pyramidal neurons within hippocampal slices were transfected with biolistic gene transfer (McAllister, 2000) using gold beads (8–12 mg) coated with plasmids containing 30 μg of total cDNA, and imaged 1–3 days after transfection.
HeLa cells were transfected with the plasmids using Lipofectamine2000 (Invitrogen), and imaged 1 day after transfection. For CaMKII activation, 20 μM of A23187 ionophore (Calbiochem) was used. For paAIP2 activation, laser illumination at a wavelength of 473 nm (100 mW/cm2) through the bottom dish was used.
2-photon fluorescence microscopy
Two-photon imaging was performed using a custom-built two-photon microscope. Monomeric EGFP was excited with a Ti:Sapphire laser (Spectraphysics, Maitai) tuned at a wavelength of 920 nm. The fluorescence was collected by an objective lens (60x, 0.9 NA, Olympus) and detected with photoelectron multiplier tubes (Hamamatsu, H7422-40p) placed after wavelength filters (Chroma, HQ520/60m-2p for green). The signal was acquired using a data acquisition board (National instruments, PCI-6110) and Scanimage software (Pologruto et al., 2003).
2-photon glutamate uncaging and blue light illumination
A second Ti:Sapphire laser tuned at a wavelength of 720 nm was used to uncage 4-Methoxy-7-nitroindolinyl-caged-L-glutamate (MNI-caged glutamate, Tocris) in extracellular solution with a train of 4–6 ms, 5 mW pulses (30 times at 0.5 Hz) near the spine of interest. At indicated time, samples were continuously illuminated with a blue laser (Shanghai Laser) at a wavelength of 473 nm (100 mW/cm2) from the bottom of the sample. Experiments were performed in Mg2+-free artificial cerebral spinal fluid (ACSF; 127 mM NaCl, 2.5 mM KCl, 4 mM CaCl2, 25 mM NaHCO3, 1.25 mM NaH2PO4 and 25 mM glucose) containing 1 μM tetrodotoxin (TTX) and 1 mM MNI-caged L-glutamate aerated with 95% O2 and 5% CO2 at 25–27°C, as described previously (Lee et al., 2009).
2-photon fluorescence lifetime imaging
Details of FRET imaging using a custom-built 2-photon fluorescence lifetime imaging microscope have been described previously (Murakoshi et al., 2008; Yasuda et al., 2006). For fluorescence lifetime imaging, a PMT with low transfer time spread (Hamamatsu, H7422-40p) was used. Fluorescence lifetime images were obtained using a time-correlated single photon counting board (SPC-150; Becker & Hickl) controlled with a custom software (Yasuda et al., 2006).
Electrophysiology
AAV encoding mEGFP-P2A-paAIP2 or its mutant mEGFP-P2A-paAIP2R5A/R6A was injected in the hippocampus of C57BL/6 male mice at ~2 months of age. After 2–3 weeks, acute slices were prepared from the infected mice and subjected to fEPSP recording in ACSF containing 2 mM CaCl2, 2 mM MgCl2 and 10 μM picrotoxin. A glass electrode (resistance ~3 MΩ) containing 1 M NaCl was located in the dendritic layer of CA1 (~100–200 μm away from the soma) while stimulating Schaffer Collateral fibers with current square pulses (0.1 ms) using a concentric bipolar stimulation electrode (FHC). The initial slope of the EPSP was monitored with custom software. The stimulation strength was set to ~50% saturation. LTP was induced by applying 3 sets of high frequency stimuli (100 Hz, 1 s) with 20 s intervals (Fig. 6A). Blue light (mercury arc lamp through 470/40 filter: 35 mW/cm2) was applied as described (Fig. 6A).
Neuronal cultures and treatments
At DIV14 primary neuronal cultures were infected with 1 μl/ml AAV9 particles containing mEGFP-P2A-paAIP2 (titer: 1.5 × 1013 genome copies/μl; produced at University of North Carolina Vector Core). On DIV21, cultures were pretreated for 2–4 hours with 1 μM TTX, and subsequently placed in HEPES-buffered ACSF (containing: 130 mM NaCl, 20 mM HEPES, 2 mM NaHCO3, 25 mM D-glucose, 2.5 mM KCl, 1.25 mM NaH2PO4, pH 7.3) supplemented with 0.8 mM MgCl2 and 1.8 mM CaCl2, followed by specific treatments: 50 μM NMDA for 1 minute or 25 μM NMDA + 2 μM PDBu for 5 minutes or 50 μM Forskolin + 100 μM IBMX for 5 minutes. To activate the paAIP2 for 2 minutes prior and during stimulation we used a DPSS laser (473 nm) at 100 mW intensity that allowed continuous illumination of the entire 35 mm dish area. Subsequently, cells were immediately extracted with T-PER extraction buffer (ThermoFisher/Pierce), supplemented with inhibitors for proteases and phosphatases (Roche). The lysates were centrifuged at 15000 g for 15 minutes at 4°C and the supernatants were used for further analysis. Protein concentrations were determined using the BCA method (ThermoFisher/Pierce).
SDS-PAGE and immunoblotting
Samples were prepared for standard SDS-PAGE and separated on 4–15% acrylamide gradient gels (Mini-PROTEAN TGX precast gels, Bio-Rad) then transferred to 0.45 μm pore size PVDF membranes (Millipore) in semidry transfer buffer (containing 25 mM Tris, 200 mM glycine and 20% methanol). Membranes were blocked with 5% nonfat milk (Great Value) in TBS-T (Tris Buffered Saline with 0.1% Tween-20) for 1hour at room temperature, then incubated for 24–48 hours at 4°C with primary antibodies diluted in 5% BSA in TBS-T. The following commercially available antibodies were used: rabbit anti-phospho-Thr286 CaMKIIα (Abcam); rabbit anti-phospho-Ser1123 SynGAP (Abcam); rabbit anti-GFP (Life Technologies); rabbit anti-phospho-Neurogranin (Ser36)/Neuromodulin (Ser41) (Upstate); rabbit anti-phospho-(Ser) PKC substrate (Cell Signaling Technology); rabbit anti-phospho-(Ser/Thr) PKA substrate (Cell Signaling Technology) and mouse anti-actin (Sigma). Membranes were washed 3 times for 15 minutes in TBS-T, followed by incubation for 2 hours at room temperature with HRP-conjugated goat anti-rabbit or rabbit anti-mouse secondary antibodies (Bio-Rad), diluted 1:5000 in 5% nonfat milk in TBS-T. Membranes were then incubated with Pierce ECL Plus western blotting substrate or Pierce ECL western blotting substrate (for β-actin) for detection of western blotted proteins. We used the Image Quant LAS4000 Imaging System (GE Healthcare) to visualize protein bands. Reported values for phosphorylated proteins were normalized to actin, used as loading control.
Stereotactic Viral Injection and Optical Fiber Implantation
AAV9 particles for CaMKIIp-mEGFP-P2A-paAIP2 (~1.6 × 1013 infections units/ml) and its functional mutant CaMKIIp-mEGFP-P2A-paAIP2R5AR6A (~1.1 × 1013 infections units/ml) were packaged, purified and concentrated by the University of North Carolina Vector Core. Virus (1 μl/site) was bilaterally injected into the basolateral amygdala (BLA) (From bregma, anteroposterior [AP] −1.65 mm, mediolateral [ML] ±3.75 mm, dorsoventral [DV] −3.8 mm) using a Picospritzer III (Parker) at a rate of 0.2 μl/min. Optical fiber implantation was performed as previously described (Sparta et al., 2011) with slight modifications: custom-made chronic implantable optical fibers (800 μm core Φ, 0.39 NA) housed inside a ferrule (2.5 mm OD, Precision Fiber Products) were placed ~ 0.2 mm above BLA (AP −1.65 mm, ML ±3.75 mm, DV −3.6 mm) and the implant was secured to the skull using dental cement (Lang Dental). Optical fiber implantation was performed 1–2 weeks after viral injection. Behavioral experiments were conducted 4–6 weeks from viral injection.
To activate paAIP2, blue light (0.2 Hz, 1 s, 30 mW output/site) from a laser (473 nm Blue DPSS Laser, Shanghai Laser & Optics Century) was delivered bilaterally through a custom-made bifurcating optical patch cable (200 μm core Φ, 0.22 NA) connected to the implanted optical fibers.
Double-Trial Training Inhibitory Avoidance Learning
For three days (30–60 min/day) before experiment, mice were habituated to the tethering procedure using a dummy optical patch cable. Behavior experiments were performed following the tethering procedure.
The inhibitory avoidance task was performed following a previous publication with modifications (Irvine et al., 2005). The training was performed in the following steps. A mouse was placed inside the light chamber (500 lux) within the inhibitory avoidance apparatus, which was custom-made to allow free movement of an animal tethered to a patch cable. The mouse was allowed to explore and adapt to the light chamber for 1 min, after which the door opened. Once it crossed to the dark chamber, the door was closed. “Crossing” was defined as the simultaneous detection of three body points (nose-point, center point and tail-base) of the mouse beyond a small entry area bordering light chamber within dark chamber. Cross latency was defined as the time from door opening to crossing. After a delay of 6 s, a mild foot shock (0.5 mA for 2 s) was administered to the mouse. 10 s after the foot shock, the first training trial was finished. After a brief delay (~ 2 min) for cleaning the apparatus, the training trial was repeated. If the mouse crossed to the dark chamber again within 120 s (cut-off point), the door was closed and the mouse received another foot shock. If the mouse stayed in the light chamber for 120 s, the training was finished. Mice were returned to the home cage once the training was completed. After the 1h laser delivery, short-term memory was assessed by measuring the cross latency to the dark chamber (600 s cut-off point). Twenty-four hours from the double-trial training, long-term memory was assessed (600 s cut-off point). The results of the long-term memory tests were not conclusive and thus not presented. Mice were excluded from the analysis if they did not cross for 120 s in the first training trial. Behavioral data was recorded via an infrared camera interfaced with Ethovision software (Noldus Information Technologies). After the experiments, infected areas in BLA and LA were analyzed from mEGFP fluorescence in histology images. Initial analysis included animals expressing the mEGFP in one hemisphere only (Table S1). These animals were excluded from the final analysis (Fig. 7). The main conclusion was not changed after the removal. Statistical tests were performed with one-way Kruskal-Wallis test followed by Dunn’s multiple comparisons test.
Histology, Immunohistochemistry and Microscopy
Mice were perfused with phosphate-buffered saline (PBS), followed by 4% paraformaldehyde in PBS. 70 μm brain sections were stained for DAPI and mounted. Brain images were obtained on a Zeiss LSM 780 confocal microscope using a 10x objective (z stack, tiled). Acquired images were used to assess the expression of transgene in amygdala and placement of the optical fiber track.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using the Graphpad Prism 7 software. Statistical parameters including the types of the statistical tests used, exact value of n, precision measures (mean ± SEM) and statistical significance are reported in the figures and figure legends. The significance threshold was placed at α = 0.05 (n.s., p > 0.05; 3, p < 0.05; 33, p < 0.01; 333, p < 0.001). n denotes the number of cells for HeLa cell experiments, number of slices for LTP experiments and the numbers of spines/neurons for spine imaging experiments.
Supplementary Material
Highlights.
PaAIP2 is a genetically-encoded, light-inducible inhibitor of CaMKII
PaAIP2 inhibits CaMKII with high specificity and with second to minute temporal resolution.
PaAIP2 measures the temporal window of CaMKII required for synaptic plasticity
PaAIP2 measures the kinetics of CaMKII activity during learning and memory
Acknowledgments
We thank C. Hall, J. Lisman, S.H. Soderling, S. Raghavachari, L. Colgan, Y. Murakoshi, W. B. Gan and N. Hedrick for discussion and comments on the manuscript. We also thank A. Wan, D. Kloetzer, J. Nabekura, T. Ohba, M. Onda, and A. Sato for their general assistance. This study was funded by Max Planck Society, National Institute of Health (R01MH111486, R01MH080047, DP1NS096787) (R.Y,) and the JST PRESTO Program, MEXT/Japan Society for the Promotion of Sciences (No. 26115718, No. 26650067, No. 16H01437 “Resonance Bio”, No. 16H01287, No. 16K15225, 15H05373), the Okazaki ORION project, Nakajima Foundation, the Uehara Memorial Foundation, the Research Foundation for Opto-Science and Technology, the Brain Science Foundation, the Mochida Memorial Foundation and Yamada Science Foundation, the Mitsubishi Foundation (H.M.).
Footnotes
Author Contributions
HM created paAIP2, HM and RY designed the experiments, HM, MES, ACE and EMS performed biochemical experiments, HM and ACE performed imaging experiments, PPB and MES performed electrophysiological experiments, MES performed behavioral experiments and HM, MES and RY wrote the paper. All the authors discussed the results and commented on the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barria A, Muller D, Derkach V, Griffith LC, Soderling TR. Regulatory phosphorylation of AMPA-type glutamate receptors by CaM-KII during long-term potentiation. Science. 1997;276:2042–2045. doi: 10.1126/science.276.5321.2042. [DOI] [PubMed] [Google Scholar]
- Bayer KU, De Koninck P, Leonard AS, Hell JW, Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411:801–805. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Buard I, Coultrap SJ, Freund RK, Lee YS, Dell’Acqua ML, Silva AJ, Bayer KU. CaMKII “autonomy” is required for initiating but not for maintaining neuronal long-term information storage. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2010;30:8214–8220. doi: 10.1523/JNEUROSCI.1469-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao LH, Stratton MM, Lee IH, Rosenberg OS, Levitz J, Mandell DJ, Kortemme T, Groves JT, Schulman H, Kuriyan J. A Mechanism for Tunable Autoinhibition in the Structure of a Human Ca2+/Calmodulin- Dependent Kinase II Holoenzyme. Cell. 2011;146:732–745. doi: 10.1016/j.cell.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HX, Otmakhov N, Strack S, Colbran RJ, Lisman JE. Is persistent activity of calcium/calmodulin-dependent kinase required for the maintenance of LTP? J Neurophysiol. 2001;85:1368–1376. doi: 10.1152/jn.2001.85.4.1368. [DOI] [PubMed] [Google Scholar]
- Donnelly ML, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD. Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J Gen Virol. 2001;82:1013–1025. doi: 10.1099/0022-1317-82-5-1013. [DOI] [PubMed] [Google Scholar]
- Enslen H, Sun P, Brickey D, Soderling SH, Klamo E, Soderling TR. Characterization of Ca2+/calmodulin-dependent protein kinase IV. Role in transcriptional regulation. J Biol Chem. 1994;269:15520–15527. [PubMed] [Google Scholar]
- Fukunaga K, Stoppini L, Miyamoto E, Muller D. Long-term potentiation is associated with an increased activity of Ca2+/calmodulin-dependent protein kinase II. J Biol Chem. 1993;268:7863–7867. [PubMed] [Google Scholar]
- Harper SM, Christie JM, Gardner KH. Disruption of the LOV-Jalpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- Harvey CD, Svoboda K. Locally dynamic synaptic learning rules in pyramidal neuron dendrites. Nature. 2007;450:1195–1200. doi: 10.1038/nature06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CD, Yasuda R, Zhong H, Svoboda K. The spread of Ras activity triggered by activation of a single dendritic spine. Science. 2008;321:136–140. doi: 10.1126/science.1159675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick NG, Harward SC, Hall CE, Murakoshi H, McNamara JO, Yasuda R. Rho GTPase complementation underlies BDNF-dependent homo- and heterosynaptic plasticity. Nature. 2016;538:104–108. doi: 10.1038/nature19784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges RR, Kazlauskas A, Toker A, Dartt DA. Effect of overexpression of protein kinase C alpha on rat lacrimal gland protein secretion. Adv Exp Med Biol. 2002;506:237–241. doi: 10.1007/978-1-4615-0717-8_32. [DOI] [PubMed] [Google Scholar]
- Irvine EE, Vernon J, Giese KP. AlphaCaMKII autophosphorylation contributes to rapid learning but is not necessary for memory. Nat Neurosci. 2005;8:411–412. doi: 10.1038/nn1431. [DOI] [PubMed] [Google Scholar]
- Ishida A, Kameshita I, Okuno S, Kitani T, Fujisawa H. A novel highly specific and potent inhibitor of calmodulin-dependent protein kinase II. Biochem Biophys Res Commun. 1995;212:806–812. doi: 10.1006/bbrc.1995.2040. [DOI] [PubMed] [Google Scholar]
- Ishida A, Shigeri Y, Tatsu Y, Uegaki K, Kameshita I, Okuno S, Kitani T, Yumoto N, Fujisawa H. Critical amino acid residues of AIP, a highly specific inhibitory peptide of calmodulin-dependent protein kinase II. FEBS Lett. 1998;427:115–118. doi: 10.1016/s0014-5793(98)00405-0. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengyel I, Voss K, Cammarota M, Bradshaw K, Brent V, Murphy KP, Giese KP, Rostas JA, Bliss TV. Autonomous activity of CaMKII is only transiently increased following the induction of long-term potentiation in the rat hippocampus. Eur J Neurosci. 2004;20:3063–3072. doi: 10.1111/j.1460-9568.2004.03748.x. [DOI] [PubMed] [Google Scholar]
- Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci. 2012;13:169–182. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinow R, Schulman H, Tsien RW. Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science. 1989;245:862–866. doi: 10.1126/science.2549638. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GC, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister AK. Biolistic transfection of neurons. Sci STKE. 2000;2000:pl1. doi: 10.1126/stke.2000.51.pl1. [DOI] [PubMed] [Google Scholar]
- Murakoshi H, Lee SJ, Yasuda R. Highly sensitive and quantitative FRET-FLIM imaging in single dendritic spines using improved non-radiative YFP. Brain Cell Biol. 2008;36:31–42. doi: 10.1007/s11068-008-9024-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Shibata AC, Nakahata Y, Nabekura J. A dark green fluorescent protein as an acceptor for measurement of Forster resonance energy transfer. Sci Rep. 2015;5:15334. doi: 10.1038/srep15334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakoshi H, Wang H, Yasuda R. Local, persistent activation of Rho GTPases during plasticity of single dendritic spines. Nature. 2011;472:100–104. doi: 10.1038/nature09823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otmakhov N, Griffith LC, Lisman JE. Postsynaptic inhibitors of calcium/calmodulin-dependent protein kinase type II block induction but not maintenance of pairing-induced long-term potentiation. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1997;17:5357–5365. doi: 10.1523/JNEUROSCI.17-14-05357.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Kantor D, Harris KM, Schuman EM, Kennedy MB. Visualization of the distribution of autophosphorylated calcium/calmodulin-dependent protein kinase II after tetanic stimulation in the CA1 area of the hippocampus. The Journal of neuroscience: the official journal of the Society for Neuroscience. 1997;17:5416–5427. doi: 10.1523/JNEUROSCI.17-14-05416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- Pologruto TA, Sabatini BL, Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanhueza M, Fernandez-Villalobos G, Stein IS, Kasumova G, Zhang P, Bayer KU, Otmakhov N, Hell JW, Lisman J. Role of the CaMKII/NMDA receptor complex in the maintenance of synaptic strength. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:9170–9178. doi: 10.1523/JNEUROSCI.1250-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Sparta DR, Stamatakis AM, Phillips JL, Hovelso N, van Zessen R, Stuber GD. Construction of implantable optical fibers for long-term optogenetic manipulation of neural circuits. Nat Protoc. 2011;7:12–23. doi: 10.1038/nprot.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppini L, Buchs PA, Muller D. A simple method for organotypic cultures of nervous tissue. J Neurosci Methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- Sun P, Enslen H, Myung PS, Maurer RA. Differential activation of CREB by Ca2+/calmodulin-dependent protein kinases type II and type IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- Tischer D, Weiner OD. Illuminating cell signalling with optogenetic tools. Nat Rev Mol Cell Biol. 2014;15:551–558. doi: 10.1038/nrm3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vest RS, Davies KD, O’Leary H, Port JD, Bayer KU. Dual mechanism of a natural CaMKII inhibitor. Mol Biol Cell. 2007;18:5024–5033. doi: 10.1091/mbc.E07-02-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RR, Kwon YG, Taing M, Lawrence DS, Edelman AM. Definition of optimal substrate recognition motifs of Ca2+-calmodulin-dependent protein kinases IV and II reveals shared and distinctive features. J Biol Chem. 1998;273:3166–3172. doi: 10.1074/jbc.273.6.3166. [DOI] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata Y, Kobayashi S, Umeda T, Inoue A, Sakagami H, Fukaya M, Watanabe M, Hatanaka N, Totsuka M, Yagi T, et al. Kinase-dead knock-in mouse reveals an essential role of kinase activity of Ca2+/calmodulin-dependent protein kinase IIalpha in dendritic spine enlargement, long-term potentiation, and learning. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2009;29:7607–7618. doi: 10.1523/JNEUROSCI.0707-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda R, Harvey CD, Zhong H, Sobczyk A, van Aelst L, Svoboda K. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- Yi JJ, Wang H, Vilela M, Danuser G, Hahn KM. Manipulation of endogenous kinase activity in living cells using photoswitchable inhibitory peptides. ACS Synth Biol. 2014;3:788–795. doi: 10.1021/sb5001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of Lipid-Modified Monomeric GFPs into Membrane Microdomains of Live Cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- Zhabotinsky AM. Bistability in the Ca2+/calmodulin-dependent protein kinase-phosphatase system. Biophys J. 2000;79:2211–2221. doi: 10.1016/S0006-3495(00)76469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Takahashi E, Li W, Halt A, Wiltgen B, Ehninger D, Li GD, Hell JW, Kennedy MB, Silva AJ. Interactions between the NR2B receptor and CaMKII modulate synaptic plasticity and spatial learning. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2007;27:13843–13853. doi: 10.1523/JNEUROSCI.4486-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.