Abstract
Therapeutic response to metformin, a first‐line drug for type 2 diabetes (T2D), is highly variable, in part likely due to genetic factors. To date, metformin pharmacogenetic studies have mainly focused on the impact of variants in metformin transporter genes, with inconsistent results. To clarify the significance of these variants in glycemic response to metformin in T2D, we performed a large‐scale meta‐analysis across the cohorts of the Metformin Genetics Consortium (MetGen). Nine candidate polymorphisms in five transporter genes (organic cation transporter [OCT]1, OCT2, multidrug and toxin extrusion transporter [MATE]1, MATE2‐K, and OCTN1) were analyzed in up to 7,968 individuals. None of the variants showed a significant effect on metformin response in the primary analysis, or in the exploratory secondary analyses, when patients were stratified according to possible confounding genotypes or prescribed a daily dose of metformin. Our results suggest that candidate transporter gene variants have little contribution to variability in glycemic response to metformin in T2D.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Published studies on the impact of the polymorphisms in organic cation transporter genes on metformin response have been inconsistent and mostly hindered by small sample size.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ This study explored whether variations in metformin transporter genes affect glycemic response to metformin through a large‐scale meta‐analysis.
WHAT THIS STUDY ADDS TO OUR KNOWLEDGE
☑ Nine candidate variants in membrane transporter genes showed no significant effect on metformin response, assessed as HbA1c reduction, in patients with T2D.
HOW THIS MIGHT CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE
☑ Candidate variants in transporter genes, despite their role in metformin pharmacokinetics, showed no relevant contribution to variability in metformin efficacy and might not be useful as predictors for individualized treatment with metformin.
Metformin is the first‐line pharmacological therapy for type 2 diabetes (T2D) and the most widely prescribed antidiabetic drug. The glycemic response to metformin is, however, highly variable. In patients receiving metformin as an initial treatment for T2D, less than two‐thirds achieve acceptable glycemic control or a target HbA1c <7% (53 mmol/mol).1, 2 Genetic factors play an important role in the variable glycemic response to metformin, with up to 34% of variance in HbA1c reduction explained by common genetic variants, each conferring a small to moderate impact.3 As a result, a large sample size is required for pharmacogenetic studies aiming to discover these common metformin response variants.
Previously published studies of metformin pharmacogenetics have mostly focused on the candidate genes involved in drug pharmacokinetics, with the expectation that these might have a large clinical effect. For example, polymorphisms in the transporters, organic anion‐transporting polypeptide 1B1 and breast cancer resistance protein, have been associated with large effect sizes with the pharmacokinetics and pharmacodynamics of several drugs, including statins.4 Metformin is not metabolized and is excreted unchanged by the kidneys. Its primary mode of action seems to be an increase of hepatic insulin sensitivity through inhibition of gluconeogenesis,5 although there is increasing recognition of its role in the gut.6 As metformin is an organic cation at physiologic pH, cation‐selective carrier proteins mediate its transport across cell membranes in the intestine, liver, and kidneys. Organic cation transporter 1 (OCT1; encoded by SLC22A1) is expressed on the sinusoidal membrane of hepatocytes and is the main transporter of metformin into the liver.7 Organic cation transporter 2 (OCT2; encoded by SLC22A2) is expressed primarily at the basolateral membrane in the kidney tubular cells and facilitates the uptake of metformin from the blood into the kidneys.8 The multidrug and toxin extrusion transporter 1 (MATE1; encoded by SLC47A1) and MATE2‐K (encoded by SLC47A2), are H+/drug antiporters located in the apical membrane of the renal tubular cells, and facilitate metformin excretion from tubular cells into urine.9, 10, 11 A recent study showed that metformin is also a substrate of carnitine/cation transporter 1 (OCTN1; encoded by SLC22A4).12 OCTN1 is highly expressed at the apical membranes in renal proximal tubules and could also contribute to metformin elimination.13, 14 To date, several polymorphisms in these transporter genes have been associated with the pharmacokinetics and pharmacodynamics of metformin in healthy volunteers15, 16, 17, 18 and with metformin response in T2D.18, 19, 20, 21, 22, 23, 24, 25 In addition, a few studies have reported gene‐gene interactions between polymorphisms in transporter genes.18, 22, 26 However, the results of these studies have been inconsistent and the impact of the established pharmacokinetic variants on metformin clinical response in T2D is uncertain.27 Apart from the different measures of glycemic response used in these studies, the small sample size and reporting bias is another potential explanation for the observed inconsistency.
To clarify the role of genetic variants in these transporters on metformin clinical response, we performed a large‐scale meta‐analysis of the impact of known candidate variants on metformin efficacy in T2D, across the cohorts of recently established Metformin Genetics Consortium (MetGen). This resource now has in excess of 10,000 individuals in whom metformin response can be defined, and offers a unique opportunity for a highly powered pharmacogenetic meta‐analysis of glycemic response to metformin.
RESULTS
We studied the effect of nine candidate variants in transporter genes OCT1,15, 16, 19, 20, 23, 28 OCT2,17, 23, 25, 29, 30 MATE1,18, 21, 23, 25, 28, 31 MATE2‐K,18, 24 and OCTN114, 28 (Table 1) on metformin glycemic response in 7,968 MetGen participants of European ancestry. Of these, the definition of metformin response could be aligned for a meta‐analysis in 7,656 participants, of whom 5,836 were initiated on metformin monotherapy and 1,820 were initiated on metformin as add‐on treatment for sulfonylureas (dual therapy; Table 2). Forest plots for the meta‐analyses of the individual variants in the monotherapy group are presented in Figure 1. There was no significant heterogeneity between the studies for any polymorphism. None of the variants was significantly associated with glycemic response to metformin (Figure 1). Similarly, when patients on monotherapy and dual therapy were analyzed together, no single‐nucleotide polymorphism (SNP) showed significant association with HbA1c reduction (Supplementary Table S1 online). The results from the HOME and SDDS studies, two MetGen cohorts in which metformin was added to insulin therapy, did not show significant impact on metformin response assessed as HbA1c reduction (Supplementary Table S2 online).
Table 1.
Single‐nucleotide polymorphisms explored in the meta‐analysis
| Gene | dbSNP ID | Nucleotide change | Amino acid change | MAFa | Reference |
|---|---|---|---|---|---|
| OCT1 (SLC22A1) | rs12208357 | c.181C>T | R61C | 0.06 | Shu et al., 200715; Shu et al., 200816; Tzvetkov et al., 200928; Zhou et al., 200919; Christensen et al., 201123 |
| rs72552763 | c.1260GAT>del | M420del | 0.1915, 45 | Shu et al., 200715; Shu et al., 200816; Tzvetkov et al., 200928; Zhou et al., 200919; Christensen et al., 201123 | |
| rs622342 | Intron A>C | 0.38 | Becker et al., 200920; Christensen et al., 201123 | ||
| OCT2 (SLC22A2) | rs316019 | c.808G>T | A270S | 0.11 | Song et al., 200829; Wang et al., 200830; Chen et al., 200917; Christensen et al., 201123; Tkáč et al., 201325 |
| MATE1 (SLC47A1) | rs2289669 | Intron G>A | 0.42 | Becker et al., 200921; Tzvetkov et al., 200928; Jablonski et al., 201031; Christensen et al., 201123; Tkáč et al., 201325 | |
| rs2252281 | g.‐66T>C | 0.41 | Stocker et al., 201318 | ||
| MATE2‐K (SLC47A2) | rs12943590 | g.‐130G>A | 0.27 | Choi et al., 201124; Stocker et al., 201318 | |
| OCTN1 (SLC22A4) | rs272893 | c.917C>T | T306I | 0.41 | Yoon et al., 201314 |
| rs1050152 | c.1507C>T | L503F | 0.39 | Tzvetkov et al., 200928 |
Minor alleles are shown in bold. dbSNP, single nucleotide polymorphism database; ID, identification; MAF, minor allele frequency; MATE1, multidrug and toxin extrusion transporter; OCT1, organic cation transporter.
Minor allele frequencies from 1000 Genome Project Phase 3 EUR population (www.ncbi.nlm.nih.gov/projects/SNP).
Table 2.
Characteristics of cohort participants included in the meta‐analysis
| Characteristic | DCS | GoDARTS | Kosice | PMT1‐EU | PMT2‐EU | Riga | Rotterdam | Sarajevo |
|---|---|---|---|---|---|---|---|---|
| No. of participants | 1,380 | 3,170 | 148 | 125 | 2,358 | 64 | 323 | 87 |
| Age, years | 62.1 ± 10.6 | 61.9 ± 11.1 | 57.5 ± 10.4 | 57.2 ± 12.9 | 66.2 ± 9.8 | 59.7 ± 10.7 | 69.7 ± 10.6 | 58.2 ± 9.0 |
| Sex, male, % | 757 (55) | 1,813 (57) | 72 (49) | 61 (49) | 1,283 (54) | 19 (30) | 147 (46) | 37 (43) |
| BMI, kg/m2 | 30.9 ± 5.2 | 31.7 ± 5.9 | 31.5 ± 4.6 | 37.7 ± 9.1 | 32.3 ± 6.8 | 34.4 ± 5.2 | 29.5 ± 4.7 | 31.2 ± 4.3 |
| Pretreatment HbA1c, % | 7.3 ± 1.3 | 8.9 ± 1.4 | 7.6 ± 1.1 | 7.8 ± 1.3 | 7.8 ± 1.5 | 7.1 ± 1.1 | 7.8 ± 1.3 | 7.8 ± 1.4 |
| On‐treatment HbA1c | 6.6 ± 0.8 | 7.2 ± 1.1 | 7.0 ± 0.8 | 6.7 ± 0.8 | 6.5 ± 0.8 | 6.4 ± 0.5 | 6.7 ± 0.7 | 6.7 ± 0.7 |
| Creatinine clearance, ml/mina | 105 ± 38 | 93.2 ± 35.4 | 104 ± 36 | 80.2 ± 20.1 | 100.0 ± 38.1 | 122.4 ± 44.5 | 85.2 ± 32.6 | 109.5 ± 30.4 |
| Metformin daily dose, mg | 1,089 ± 597 | 1,321 ± 515 | 1,400 ± 540 | 913 ± 326 | 932 ± 490 | 1,704 ± 579 | 800 ± 480 | 1,200 ± 608 |
| Adherence | – | 82.4 ± 16.2 | – | >80b | 88.9 ± 20.4 | – | 79 ± 34 | – |
| Metformin monotherapy, % | 85 | 69 | 100 | 100 | 75 | 100 | 88 | 100 |
BMI, body mass index.
aCreatinine clearance was estimated using the Cockcroft‐Gault formula, except for the PMT1‐EU study in which the MDRD formula was used. bPatients needed to have ≥80% adherence to be included in the study.
Figure 1.
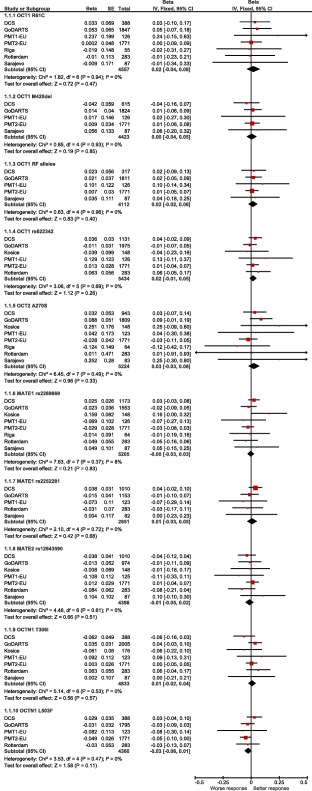
Effects of candidate variants in transporter genes on metformin glycemic response assessed as HbA1c reduction in patients on metformin monotherapy. Beta values obtained from individual studies are presented with 95% confidence interval (CI); arrowheads indicate the CI exceeding the limits of the graph. Overall betas are presented as black diamonds. The organic cation transporter 1 (OCT1) reduced‐function (RF) alleles denote combined genotype for R61C and M420del – number of RF alleles. MATE1, multidrug and toxin extrusion transporter 1. [Color figure can be viewed at wileyonlinelibrary.com]
As the transport of metformin could depend on its concentration, we next explored possible gene by dose interactions, using a dose as a proxy of metformin concentration. In the meta‐analysis of SNP × dose interaction effects, none of the interactions showed significant effect in the monotherapy group (Table 3) or the total group when patients on dual therapy were added to the analysis (Supplementary Table S3a online). Likewise, no significant associations were found in the separate meta‐analyses of the effects of variants on metformin response in individuals treated with low (≤1,000 mg) or high daily doses of metformin (>1,000 mg; Table 4 and Supplementary Table S3b online).
Table 3.
Meta‐analysis results of single‐nucleotide polymorphism × dose interaction effects in participants on metformin monotherapy
| SNP | Effect allele | No. of studies | No. of patients | Continuous dose | Dichotomous dose | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | SE | P value | I2 | P(Q) | Beta | SE | P value | I2 | P(Q) | ||||
| OCT1 R61C | T | 5 | 4,376 | 0.005 | 0.019 | 0.816 | 0.0 | 0.965 | ‐0.005 | 0.078 | 0.946 | 0.0 | 0.720 |
| OCT1 M420del | del | 4 | 4,297 | 0.006 | 0.014 | 0.669 | 0.0 | 0.502 | 0.043 | 0.089 | 0.631 | 61.8 | 0.049 |
| OCT1 RF allelesa | 4 | 3,986 | 0.001 | 0.012 | 0.923 | 0.0 | 0.659 | 0.041 | 0.049 | 0.394 | 44.0 | 0.148 | |
| OCT1 rs622342 | C | 5 | 5,273 | 0.028 | 0.010 | 0.006 | 39.8 | 0.156 | 0.092 | 0.037 | 0.014 | 0.0 | 0.413 |
| OCT2 A270S | T | 6 | 5,002 | 0.008 | 0.017 | 0.647 | 30.8 | 0.205 | ‐0.003 | 0.056 | 0.952 | 23.0 | 0.261 |
| MATE1 rs2289669 | A | 6 | 4,980 | 0.014 | 0.008 | 0.094 | 0.2 | 0.415 | 0.081 | 0.036 | 0.024 | 0.0 | 0.485 |
| MATE1 rs2252281 | C | 4 | 2,528 | 0.004 | 0.015 | 0.791 | 0.0 | 0.505 | 0.059 | 0.043 | 0.177 | 0.0 | 0.484 |
| MATE2 rs12943590 | A | 6 | 4,273 | ‐0.008 | 0.011 | 0.467 | 0.0 | 0.663 | 0.007 | 0.044 | 0.875 | 0.0 | 0.827 |
| OCTN1 T306I | T | 6 | 4,647 | ‐0.005 | 0.010 | 0.624 | 0.0 | 0.466 | ‐0.006 | 0.037 | 0.879 | 0.0 | 0.875 |
| OCTN1 L503F | T | 4 | 4,202 | 0.025 | 0.016 | 0.110 | 60.4 | 0.056 | 0.120 | 0.085 | 0.161 | 76.0 | 0.006 |
I2, heterogeneity index; MATE, multidrug and toxin extrusion transporter; OCT1, organic cation transporter 1; P(Q), P value for Cochrane's Q statistic; RF, reduced function; SNP, single‐nucleotide polymorphism.
Combined genotype for R61C and M420del – number of reduced‐function alleles. A positive beta is a greater glycemic response to metformin associated with the effect allele.
Table 4.
Meta‐analysis results for the effects of candidate variants in transporter genes on metformin glycemic response in participants on metformin monotherapy, stratified by metformin dose
| SNP | Effect allele | No. of studies | Dose ≤1,000 mg | Dose >1,000 mg | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of patients | Beta | SE | P value | I2 | P(Q) | No. of patients | Beta | SE | P value | I2 | P(Q) | |||
| OCT1 R61C | T | 5 | 3,015 | 0.019 | 0.038 | 0.619 | 0.0 | 0.855 | 1,361 | 0.000 | 0.000 | 1.000 | 0.0 | 0.719 |
| OCT1 M420del | del | 4 | 2,903 | ‐0.001 | 0.050 | 0.987 | 55.6 | 0.080 | 1,394 | 0.013 | 0.040 | 0.747 | 44.9 | 0.142 |
| OCT1 RF allelesa | 4 | 2,690 | 0.006 | 0.025 | 0.812 | 0.2 | 0.391 | 1,296 | 0.032 | 0.039 | 0.412 | 26.1 | 0.255 | |
| OCT1 rs622342 | C | 5 | 3,571 | ‐0.004 | 0.019 | 0.837 | 0.0 | 0.523 | 1,702 | 0.055 | 0.030 | 0.064 | 0.0 | 0.679 |
| OCT2 A270S | T | 6 | 3,388 | 0.013 | 0.029 | 0.659 | 0.0 | 0.749 | 1,614 | 0.062 | 0.082 | 0.453 | 50.9 | 0.070 |
| MATE1 rs2289669 | A | 6 | 3,428 | ‐0.014 | 0.018 | 0.452 | 38.1 | 0.152 | 1,552 | 0.060 | 0.030 | 0.046 | 20.0 | 0.282 |
| MATE1 rs2252281 | C | 4 | 1,629 | 0.004 | 0.026 | 0.877 | 0.0 | 0.750 | 899 | ‐0.006 | 0.031 | 0.843 | 32.2 | 0.219 |
| MATE2 rs12943590 | A | 6 | 3,009 | ‐0.012 | 0.023 | 0.592 | 0.0 | 0.799 | 1,264 | ‐0.070 | 0.088 | 0.431 | 93.1 | 0.000 |
| OCTN1 T306I | T | 6 | 3,116 | 0.017 | 0.020 | 0.401 | 0.0 | 0.728 | 1,531 | 0.038 | 0.033 | 0.251 | 0.0 | 0.971 |
| OCTN1 L503F | T | 4 | 2,889 | ‐0.044 | 0.020 | 0.028 | 35.8 | 0.197 | 1,313 | ‐0.029 | 0.036 | 0.434 | 48.4 | 0.121 |
I2, heterogeneity index; MATE, multidrug and toxin extrusion transporter; OCT1, organic cation transporter 1; P(Q), P value for Cochrane's Q statistic; RF, reduced function; SNP, single‐nucleotide polymorphism.
Combined genotype for R61C and M420del – number of reduced‐function alleles. A positive beta is a greater glycemic response to metformin associated with the effect allele.
We next explored the potential interactions between these transporters that might affect metformin response. The interactions were tested by examining the impact of one SNP in two separate subgroups of participants, homozygotes for the reference, and variant allele of a potential confounding SNP. Tables 5 and 6 show the results from meta‐analyses of participants on monotherapy for transporters in the liver and kidneys, respectively. Supplementary Tables S4a and S4b online show the meta‐analyses results for all participants, including both monotherapy and dual therapy. These exploratory subgroup analyses revealed no significant interactions between the SNPs that affect metformin glycemic response.
Table 5.
Meta‐analysis results for the effects of individual single‐nucleotide polymorphisms in the subgroups of patients homozygous for the wild‐type or variant allele of possible confounding single‐nucleotide polymorphisms – interactions between metformin liver transporters – monotherapy group
| Subgroup | SNP | Effect allele | No. of studies | No. of patients | Beta | SE | P value | I2 | P(Q) |
|---|---|---|---|---|---|---|---|---|---|
| 0 OCT1 RF allelesa | MATE1 rs2289669 | A | 5 | 1,736 | 0.004 | 0.029 | 0.879 | 0.0 | 0.468 |
| MATE1 rs2252281 | C | 4 | 891 | 0.028 | 0.045 | 0.542 | 0.0 | 0.976 | |
| 2 OCT1 RF alleles | MATE1 rs2289669 | A | 5 | 179 | ‐0.095 | 0.108 | 0.380 | 80.6 | 0.000 |
| MATE1 rs2252281 | C | 4 | 102 | 0.167 | 0.136 | 0.221 | 66.5 | 0.030 | |
| OCT1 rs622342 wt/wt | MATE1 rs2289669 | A | 6 | 1,899 | 0.007 | 0.026 | 0.789 | 0.0 | 0.429 |
| MATE1 rs2252281 | C | 4 | 997 | 0.046 | 0.036 | 0.204 | 0.0 | 0.799 | |
| OCT1 rs622342 v/v | MATE1 rs2289669 | A | 6 | 596 | 0.084 | 0.063 | 0.178 | 61.7 | 0.023 |
| MATE1 rs2252281 | C | 4 | 329 | 0.060 | 0.051 | 0.243 | 35.5 | 0.200 | |
| MATE1 rs2289669 wt/wt | OCT1 RF alleles | 5 | 1,179 | 0.025 | 0.036 | 0.494 | 31.6 | 0.211 | |
| OCT1 rs622342 | C | 6 | 1,608 | 0.035 | 0.049 | 0.477 | 53.2 | 0.058 | |
| MATE1 rs2289669 v/v | OCT1 RF alleles | 5 | 292 | ‐0.008 | 0.080 | 0.921 | 0.0 | 0.779 | |
| OCT1 rs622342 | C | 6 | 527 | 0.024 | 0.053 | 0.653 | 0.0 | 0.505 | |
| MATE1 rs2252281 wt/wt | OCT1 RF alleles | 4 | 606 | 0.064 | 0.052 | 0.213 | 0.0 | 0.837 | |
| OCT1 rs622342 | C | 4 | 956 | 0.040 | 0.035 | 0.250 | 0.0 | 0.870 | |
| MATE1 rs2252281 v/v | OCT1 RF alleles | 4 | 256 | ‐0.005 | 0.092 | 0.957 | 49.1 | 0.117 | |
| OCT1 rs622342 | C | 4 | 373 | 0.019 | 0.054 | 0.734 | 0.0 | 0.575 |
I2, heterogeneity index; MATE, multidrug and toxin extrusion transporter; OCT1, organic cation transporter 1; P(Q), P value for Cochrane's Q statistic; RF, reduced function; SNP, single‐nucleotide polymorphism; v/v, homozygous variant allele carriers; wt/wt, homozygous wild‐type allele carriers.
Combined genotype for R61C and M420del ‐ number of RF alleles. A positive beta is a greater glycemic response to metformin associated with the effect allele.
Table 6.
Meta‐analysis results for the effects of individual single‐nucleotide polymorphisms in the subgroups of patients homozygous for the wild‐type or variant allele of possible confounding single‐nucleotide polymorphisms ‐ interactions between metformin kidney transporters ‐ monotherapy group
| Subgroup | SNP | Effect allele | No. of studies | No. of patients | Beta | SE | P value | I2 | P(Q) |
|---|---|---|---|---|---|---|---|---|---|
| OCT2 A270S wt/wt | MATE1 rs2289669 | A | 7 | 3,869 | 0.008 | 0.018 | 0.673 | 41.9 | 0.112 |
| MATE1 rs2252281 | C | 5 | 1,835 | ‐0.008 | 0.027 | 0.756 | 0.0 | 0.964 | |
| MATE2 rs12943590 | A | 7 | 3,176 | ‐0.011 | 0.023 | 0.642 | 0.0 | 0.481 | |
| OCTN1 T306I | T | 7 | 3,642 | 0.030 | 0.019 | 0.118 | 0.0 | 0.660 | |
| OCTN1 L503F | T | 5 | 3,437 | ‐0.040 | 0.019 | 0.034 | 0.0 | 0.720 | |
| OCT2 A270S v/v | MATE1 rs2289669 | A | 3 | 59 | 0.045 | 0.210 | 0.831 | 0.0 | 0.950 |
| MATE1 rs2252281 | C | 2 | 35 | 0.575 | 0.175 | 0.001 | 31.8 | 0.226 | |
| MATE2 rs12943590 | A | 3 | 48 | 0.170 | 0.337 | 0.615 | 0.0 | 0.557 | |
| OCTN1 T306I | T | 3 | 51 | 0.164 | 0.212 | 0.440 | 34.6 | 0.217 | |
| OCTN1 L503F | T | 3 | 50 | ‐0.017 | 0.206 | 0.934 | 20.3 | 0.285 | |
| MATE1 rs2289669 wt/wt | OCT2 A270S | T | 7 | 1,598 | 0.021 | 0.042 | 0.613 | 0.0 | 0.568 |
| MATE2 rs12943590 | A | 7 | 1,288 | ‐0.066 | 0.071 | 0.347 | 58.9 | 0.024 | |
| OCTN1 T306I | T | 7 | 1,412 | 0.044 | 0.054 | 0.418 | 63.4 | 0.012 | |
| OCTN1 L503F | T | 5 | 1,328 | ‐0.069 | 0.029 | 0.015 | 0.0 | 0.884 | |
| MATE1 rs2289669 v/v | OCT2 A270S | T | 6 | 495 | ‐0.066 | 0.146 | 0.655 | 65.0 | 0.014 |
| MATE2 rs12943590 | A | 7 | 363 | 0.065 | 0.052 | 0.215 | 0.0 | 0.529 | |
| OCTN1 T306I | T | 7 | 410 | 0.138 | 0.053 | 0.009 | 6.4 | 0.379 | |
| OCTN1 L503F | T | 5 | 365 | ‐0.156 | 0.055 | 0.004 | 0.0 | 0.612 | |
| MATE1 rs2252281 wt/wt | OCT2 A270S | T | 5 | 862 | ‐0.014 | 0.056 | 0.802 | 0.0 | 0.942 |
| MATE2 rs12943590 | A | 5 | 720 | 0.006 | 0.046 | 0.902 | 9.2 | 0.354 | |
| OCTN1 T306I | T | 5 | 726 | ‐0.037 | 0.042 | 0.382 | 47.5 | 0.107 | |
| OCTN1 L503F | T | 4 | 662 | ‐0.017 | 0.046 | 0.711 | 19.9 | 0.290 | |
| MATE1 rs2252281 v/v | OCT2 A270S | T | 4 | 329 | 0.129 | 0.076 | 0.087 | 0.0 | 0.592 |
| MATE2 rs12943590 | A | 5 | 276 | ‐0.042 | 0.061 | 0.494 | 0.0 | 0.829 | |
| OCTN1 T306I | T | 5 | 283 | 0.013 | 0.067 | 0.853 | 22.1 | 0.274 | |
| OCTN1 L503F | T | 4 | 262 | ‐0.011 | 0.074 | 0.884 | 0.0 | 0.650 | |
| MATE2 rs12943590 wt/wt | OCT2 A270S | T | 7 | 1,964 | 0.001 | 0.036 | 0.978 | 22.4 | 0.259 |
| MATE1 rs2289669 | A | 7 | 1,980 | ‐0.010 | 0.023 | 0.673 | 0.0 | 0.454 | |
| MATE1 rs2252281 | C | 5 | 1,014 | 0.051 | 0.034 | 0.138 | 0.0 | 0.661 | |
| OCTN1 T306I | T | 7 | 1,828 | 0.043 | 0.026 | 0.095 | 39.7 | 0.127 | |
| OCTN1 L503F | T | 5 | 1,636 | ‐0.050 | 0.027 | 0.062 | 26.4 | 0.246 | |
| MATE2 rs12943590 v/v | OCT2 A270S | T | 7 | 298 | ‐0.017 | 0.088 | 0.849 | 0.0 | 0.830 |
| MATE1 rs2289669 | A | 7 | 300 | 0.183 | 0.115 | 0.112 | 51.3 | 0.055 | |
| MATE1 rs2252281 | C | 5 | 139 | 0.089 | 0.097 | 0.357 | 0.0 | 0.534 | |
| OCTN1 T306I | T | 7 | 279 | ‐0.071 | 0.056 | 0.203 | 0.0 | 0.744 | |
| OCTN1 L503F | T | 5 | 241 | 0.080 | 0.058 | 0.168 | 0.0 | 0.455 | |
| OCTN1 T306I wt/wt | OCT2 A270S | T | 7 | 1,806 | 0.081 | 0.041 | 0.048 | 0.0 | 0.557 |
| MATE1 rs2289669 | A | 7 | 1,728 | ‐0.020 | 0.028 | 0.469 | 0.0 | 0.554 | |
| MATE1 rs2252281 | C | 5 | 827 | ‐0.001 | 0.040 | 0.976 | 35.2 | 0.187 | |
| MATE2 rs12943590 | A | 7 | 1,426 | 0.018 | 0.033 | 0.581 | 0.0 | 0.630 | |
| OCTN1 T306I v/v | OCT2 A270S | T | 6 | 634 | ‐0.017 | 0.067 | 0.803 | 31.7 | 0.198 |
| MATE1 rs2289669 | A | 7 | 609 | ‐0.023 | 0.040 | 0.571 | 25.7 | 0.232 | |
| MATE1 rs2252281 | C | 5 | 267 | 0.049 | 0.122 | 0.690 | 63.7 | 0.027 | |
| MATE2 rs12943590 | A | 7 | 541 | ‐0.145 | 0.094 | 0.121 | 63.1 | 0.012 | |
| OCTN1 L503F wt/wt | OCT2 A270S | T | 5 | 1,367 | ‐0.040 | 0.047 | 0.394 | 41.8 | 0.143 |
| MATE1 rs2289669 | A | 5 | 1,274 | ‐0.019 | 0.031 | 0.548 | 0.0 | 0.724 | |
| MATE1 rs2252281 | C | 4 | 583 | ‐0.059 | 0.063 | 0.344 | 42.2 | 0.158 | |
| MATE2 rs12943590 | A | 5 | 1,114 | ‐0.049 | 0.057 | 0.394 | 53.5 | 0.072 | |
| OCTN1 L503F v/v | OCT2 A270S | T | 5 | 816 | 0.106 | 0.063 | 0.095 | 40.2 | 0.153 |
| MATE1 rs2289669 | A | 5 | 768 | ‐0.042 | 0.045 | 0.354 | 0.0 | 0.982 | |
| MATE1 rs2252281 | C | 4 | 337 | 0.017 | 0.065 | 0.793 | 5.6 | 0.365 | |
| MATE2 rs12943590 | A | 5 | 584 | 0.031 | 0.053 | 0.561 | 0.0 | 0.857 |
A positive beta is a greater glycemic response to metformin associated with the effect allele.
I2, heterogeneity index; MATE, multidrug and toxin extrusion transporter; OCT1, organic cation transporter 1; P(Q), P value for Cochrane's Q statistic; SNP, single‐nucleotide polymorphism; v/v, homozygous variant allele carriers; wt/wt, homozygous wild‐type allele carriers.
In the supplemental locus‐wise meta‐analysis of the association of all common SNPs in transporter gene regions with metformin glycemic response, none of the variants showed significant signal after correction for multiple testing (P > 1.6 × 10−4; Supplementary Figure S1).
DISCUSSION
Despite the established role of cation‐selective transporters in metformin pharmacokinetics, polymorphisms in these transporters showed no significant impact on glycemic response to metformin in this large study of patients with T2D. This meta‐analysis had 80% power to detect an allelic effect of HbA1c reduction >0.14% (1.5 mmol/mol) for any of the candidate SNPs at the nominal significance level of α = 0.005. Thus, none of the SNPs reported as being associated with metformin response in previous literature is likely to have an allelic impact on HbA1c reduction of >0.14%. Furthermore, it is unlikely that other SNPs, such as cis‐regulatory variants, in these genes could have a significant impact on metformin glycemic response, as shown by a locus‐wise meta‐analysis of all common SNPs within genes' proximity in 6,964 patients.
Our findings contrast to most of those previously reported in healthy subjects,15, 16, 17, 18 although it is in keeping with a recent study showing no impact of the OCT1 genotype on the glucose production in fasting healthy subjects.32 This may reflect the fact that our study was conducted on patients with T2D and, as such, we were able to assess HbA1c reduction rather than other surrogates of metformin response. Some18, 20, 21, 23, 24, 25 but not all19 previous studies have reported an association of a variant altering metformin transport and glycemic response to metformin in patients with T2D. These studies have varied in size but have generally been small, and have varied in their definition of glycemic response to metformin and analytical approaches. In this MetGen meta‐analysis, we included all studies that have previously explored associations between transporter variants and glycemic response to metformin in patients with T2D of European ancestry with both positive and negative findings,18, 19, 20, 21, 22, 23, 24, 25 and also other cohorts for which results have not been published. In this way, we have reduced the chance of reporting bias and maximized the power. In addition, we have reduced heterogeneity by aligning metformin response definitions, models, and covariates for all studies included in the meta‐analysis. Our results suggest that metformin transporters do not have a significant role in how patients with T2D respond to metformin therapy.
Transporters exhibit asymmetry in their kinetic properties; thus, for facilitated transporters that are bidirectional, the direction of the transport will depend on the substrate concentration.33, 34 Systemic plasma levels of metformin are dependent on dose; therefore, in our secondary analyses, we analyzed dose × SNP interactions and assessed the effect of the studied variants separately in individuals prescribed low (≤1,000 mg) or high (>1,000 mg) doses of the drug. We did not find significant impact of the analyzed interactions on metformin glycemic response, and, accordingly, significant association between any variant and response in the dose‐stratified analysis. However, we used the prescribed dose as a proxy of metformin concentration. There were differences in the characteristics of patients between the cohorts, and, for instance, older age and lower estimated glomerular filtration rate in the Rotterdam cohort could result in higher metformin serum levels, despite being prescribed lower doses. In addition, data on drug adherence were available only in four studies, although all studies were adjusted for creatinine clearance and other known clinical covariates, which could influence metformin response.
Studies of small cohorts have previously reported gene‐gene interactions affecting glycemic response to metformin.18, 22 Here, we explored whether such interactions could explain the lack of association between metformin response and transporter variants. The exploratory analyses of SNP × SNP interactions, assessed as the impact of studied polymorphisms on metformin response in the subgroups of patients homozygous for possibly confounding SNPs, did not show any significant effects. However, these subgroup analyses had substantially less power to detect moderate effects due to the smaller sample sizes, especially for the rarer SNPs. Larger studies would be needed to detect these effects and to explore possible but more complex multiple gene‐gene interactions.
This is the largest metformin pharmacogenetic study reported to date, despite a few limitations due to the need to align cohorts. Our finding that there is no significant role for metformin transporter variants in mediating glycemic response to metformin challenges our understanding of metformin action in patients with T2D chronically treated with metformin. For example, there is increasing recognition that metformin works presystemically in the gut, via a number of mechanisms, to improve glycemia.6, 35, 36, 37 Indeed, a recent delayed release metformin achieves low systemic metformin concentrations but is effective at lowering blood glucose in patients with T2D.38 It is also possible that the influence of the analyzed transporter gene variants is less prominent clinically than expected due to the redundancy of transporters in vivo.27 If one or more transporters have reduced function, other transporters may take over their roles and mediate metformin uptake or efflux from the organs. In addition, there might be more membrane transporters of metformin yet to be identified that could have a role in its absorption, distribution, and elimination. In the current study, we focused on the candidate variants in the membrane transporter genes that have been associated previously with metformin pharmacokinetics or response. However, in addition to liver and kidney transporters, transporters in the intestine may play a significant role in metformin levels. Recent studies have suggested that other transporters, which were not the subject of this study, play an important role in metformin absorption,34, 39, 40, 41 and there still may be unidentified cation transporters in the intestine involved in metformin absorption, which could also affect metformin response. It should also be noted that variants in transporters in various tissues may play opposing roles. For example, OCT1 could mediate basolateral flux of metformin from enterocytes to the portal circulation7, 42 and across the sinusoidal membrane of hepatocytes.7 Thus, a reduced function OCT1 genetic variant may result in increased concentrations in enterocytes and decreased concentrations in hepatocytes.
Although the current study demonstrates that genetic variants in transporters that play a role in metformin pharmacokinetics have no significant effect on metformin glycemic response in large cohorts of patients with diabetes, there are some limitations to our study. First, despite the large sample size used in the current study, we did not have the statistical power to detect an allelic effect size for HbA1c reduction smaller than 0.06–0.14% (0.7–1.5 mmol/mol, depending on SNP frequency), however, these small effect sizes may be unlikely to have any clinical importance. Second, the meta‐analysis included population‐based observational studies, which could be confounded by a number of factors, such as patient adherence or frequency of HbA1c measurements in the cohort. However, in four cohorts, including two largest cohorts, GoDARTS and PMT2‐EU, adherence could be calculated from the drug dispensing records, and was added as a covariate in the model. Likewise, although we could not completely harmonize the definition of on‐treatment HbA1c due to different frequency of HbA1c measurements available in the cohorts, four studies, including two of the largest studies, used minimal HbA1c achieved within 18 months as outcome. Third, concomitant medications, which affect metformin transport, are well established to cause changes in its pharmacokinetics,43 and may obscure effects of genetic variants on metformin response. Information on prescription and over‐the‐counter drugs that affect metformin pharmacokinetics, such as cimetidine, were not gathered in the current study. In addition, several previous studies have shown effects of polymorphisms in transporters on metformin response in multi‐ethnic cohorts, including many individuals from non‐European ancestries.18, 24 Genetic variants may have different effect sizes on drug response in individuals from different ethnic backgrounds. Further studies are needed to test the effects of these variants on metformin response in individuals from non‐European backgrounds. Finally, recent studies suggest that genetic variants in transcription factors that affect expression of several metformin transporters may have larger effect sizes on metformin response than genetic variants in the individual transporters themselves,44 underscoring the need to understand not only the mechanisms of metformin transport in various tissues, but the proteins that modulate their activity and expression.
In conclusion, in our large meta‐analysis, including almost 8,000 individuals across 10 international cohorts of the MetGen Consortium, variants in metformin transporter genes have shown no relevant contribution to variability in metformin response in patients with T2D, although we cannot rule out gene‐concentration or more complex multiple gene‐gene interactions that may be required to account for transporter redundancy. As has been recognized now for a number of years in disease and complex trait genetics, this study shows the importance of large sample sizes, usually only available to international consortia, for robust pharmacogenetic studies. Future even larger consortia efforts are required to corroborate these findings and to unravel genetic variations that could be used as better predictors for personalized metformin therapy.
METHODS
Studies
The MetGen Consortium consists of research groups from Europe and the United States with available data for studies of metformin pharmacogenetics from population observational studies and clinical trials. The 10 MetGen cohorts with available data for the study of metformin transporter gene variants are presented in Supplementary Table S5 online. The studies were approved by relevant institutional review boards, and all participants gave written informed consent.
Single‐nucleotide polymorphism selection and genotyping
Nine SNPs in five transporter genes, reported to be associated with metformin response or pharmacokinetics in previous studies, were included in the meta‐analysis (Table 1). The numbers of available SNPs, genotyping methods, and the minor allele frequencies in each cohort are provided in Supplementary Table S6 online. All SNPs were in line with Hardy‐Weinberg equilibrium (P > 0.01). The variants were first analyzed individually for association with metformin response. The reduced‐function OCT1 polymorphisms, R61C and M420del, were analyzed individually, and also together as a combined genotype, according to the number of haplotypes carrying reduced‐function alleles: 0, 1, or 2, in line with previous studies in patients of European ancestry.23, 45, 46
Assessment of metformin response
Metformin response was defined as a reduction in HbA1c during treatment with metformin: pretreatment minus on‐treatment HbA1c. As such, a positive β in the regression models indicates an association of the effect allele with greater glycemic response to metformin. Inclusion criteria for all participants included in the meta‐analysis were continuous treatment with metformin for at least 3 months, pretreatment HbA1c measured within 6 months prior to metformin therapy and <14%, measurement of on‐treatment HbA1c within 18 months of metformin commencement, and no treatment with other glucose‐lowering agents except stable sulfonylurea treatment before and during metformin therapy. Hence, two cohorts from randomized controlled trials, HOME and SDDS studies, in which metformin was added to insulin therapy, were not included in the meta‐analysis, and the results are shown separately.
On‐treatment HbA1c was defined as minimal HbA1c measured within 18 months of metformin commencement in the GoDARTS, PMT2‐EU, Riga, and Rotterdam studies, as HbA1c measured after 6 months (Košice and Sarajevo studies) or 12 months (DCS study) of metformin treatment, and as HbA1c measured within the first 3–9 months of metformin therapy in the PMT1‐EU study.
Statistical analysis
In each cohort, the effects of individual SNPs on metformin response were assessed in additive genetic model using linear regression with reduction of HbA1c as outcome (primary analysis). The pretreatment HbA1c, metformin daily dose, adherence, creatinine clearance, baseline gap (time between pretreatment HbA1c measurement and start of metformin therapy), and treatment group (metformin prescribed as monotherapy or dual therapy – metformin added to stable sulfonylurea treatment), were added as covariates, when available/applicable (Supplementary Table S7 online). In the HOME study, analyses were adjusted for pretreatment HbA1c, metformin daily dose and creatinine clearance, and in the SDDS study, pretreatment HbA1c and randomization group23 were added as covariates.
As the effects of transporter SNPs could depend on metformin level, or they could be confounded by the effect of other variants, secondary analyses were performed to explore possible gene‐dose and gene‐gene interactions. For gene‐dose interactions, we first examined interaction models that included SNP × dose interaction term with dose as a continuous variable and then dose coded as a dichotomized variable (low or high dose) in the following model: HbA1c reduction ∼ pre‐treatment HbA1c + adherence + creatinine clearance + baseline gap + treatment group + dose + SNP + SNP × dose.
Next, we assessed the association of SNPs with metformin response separately for patients taking low (≤1,000 mg) or high doses (>1,000 mg) of metformin. The cutoff value of 1,000 mg was chosen based on the median dose in the largest cohort. The analyses were performed using the same basic regression model: HbA1c reduction ∼ pre‐treatment HbA1c + adherence + creatinine clearance + baseline gap + treatment group + dose + SNP.
To examine potential interactions between the variants, additional exploratory analyses of the effects of individual SNPs were carried out in the subgroups of individuals who were homozygous for wild‐type allele and of the individuals who were homozygous for variant allele of possibly confounding SNPs, assuming that impact of the variants could be more pronounced in more extreme genotype groups. Possible 1 × 1 interactions between SNPs in metformin liver transporters (OCT1 and MATE1), and SNPs in metformin kidney transporters (OCT2, MATE1, MATE2‐K, and OCTN1) were explored.
All analyses were carried out for the total group of patients treated with metformin, including monotherapy and dual therapy group (metformin added to stable sulfonylurea therapy), and also separately for the metformin monotherapy group. The results from individual studies were combined in the meta‐analysis using PLINK software under fixed‐effects model (http://pngu.mgh.harvard.edu/purcell/plink/).47 For gene‐dose interaction models, estimates of SNP × dose interaction terms were pooled in the meta‐analysis.48 Heterogeneity across studies was assessed using the Cochran Q test and the I2 heterogeneity index. In case the substantial heterogeneity was detected (P value for Cochran Q test <0.10), the results of the random‐effects model were presented. Power analyses were performed using Quanto (http://biostats.usc.edu/Quanto.html). Forest plots were created using RevMan 5.3 software (Review Manager, The Nordic Cochrane Centre, The Cochrane Collaboration).
As the primary analysis included nine SNPs and one combined OCT1 genotype, a nominal statistical significance threshold was set to 5 × 10−3 (P = 0.05/10), and a study‐wise statistical significance threshold to 2 × 10−4 to correct for the overall number of tests performed (P = 0.05/236).
In addition to our main hypothesis, we performed a supplemental, locus‐wise meta‐analysis of all common SNPs within the proximity of the transporter genes to examine the possibility that original associations reported in previous studies might be driven by other SNPs in these gene regions. A total of 3,471 variants with minor allele frequency ≥1% were tested for association with metformin glycemic response in 6,964 participants from two MetGen cohorts with available genome‐wide association study imputed data, the GoDARTS and PMT2‐EU. The details of genotyping, quality control, imputation, association analysis, and meta‐analysis were described in our previous study.49 The experiment‐wise significance threshold for this supplemental analysis was derived from 10,000 permutations of the phenotype in the GoDARTS data,47 which gave a 95% cutoff for significance of P = 1.6 × 10−4.
AUTHOR CONTRIBUTIONS
T.D., K.Z., K.M.G., and E.R.P. wrote the manuscript. T.D., K.M.G., and E.R.P. designed the research. T.D., K.Z., S.W.Y., N.v.L., C.E.d.K., M.J., S.G., L.Z., M.M.H.C., M.O., R.T., M.K., M.M.H., A.A.v.d.H., L.K., V.P., A.K., K.B., J.K., S.S., I.T., B.H.S., C.N.A.P., L.M.t.H., K.M.G., and E.R.P. performed the research. T.D., K.Z., S.W.Y., N.v.L., C.E.d.K., M.J., S.G., L.Z., M.M.H.C., and M.O. analyzed the data.
CONFLICT OF INTEREST
The authors declared no conflict of interest.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
DCS: The DCS study is supported by the Diabetes Care System West‐Friesland, the Netherlands. This part of the study was funded by the Netherlands Organisation for Health Research and Development (Priority Medicines Elderly Programme 113102006) and by the Innovative Medicines Initiative Joint Undertaking under grant agreement number 115317 (DIRECT), resources of which are composed of financial contribution from the European Union's Seventh Framework Programme (FP7/2007‐2013) and EFPIA companies' in kind contribution (http://www.direct‐diabetes.org/). GoDARTS: We are grateful to all the participants who took part in this study, to the general practitioners, to the Scottish School of Primary Care for their help in recruiting the participants, and to the whole team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists, and nurses. The Wellcome Trust United Kingdom Type 2 Diabetes Case Control Collection (GoDARTS) cohort collection was funded by The Wellcome Trust and informatics support was provided by the Chief Scientist Office, Scotland. E.R.P. holds a Wellcome Trust New Investigator Award (102820/Z/13/Z).
Košice: The study was supported by research grants VEGA 1/0389/14 and VEGA 1/0027/16 from the Ministry of Education, Science, Research and Sport, Slovak Republic. PMT1 & PMT2: K.M.G., S.W.Y., and S.G. would like to acknowledge support from NIH grants (GM117163, T32 GM007175). The genotyping of the PMT1‐EU was supported by the National Institutes of Health (U19 GM061390) and the RIKEN Institute. The genotyping of the GERA cohort (called PMT2‐EU in this study) was supported by grant RC2 AG036607 from the National Institutes of Health; development of the RPGEH and GERA cohort was supported by grants from the Robert Wood Johnson Foundation, the Ellison Medical Foundation, the Wayne and Gladys Valley Foundation, and Kaiser Permanente. We would like to acknowledge BioVU (Vanderbilt DNA databank), The Marshfield Clinic Personalized Medicine Research Project (PMRP), and Kaiser Permanente South East Georgia for collecting the clinical information for PMT1‐EU. Sarajevo: The study was supported by grants received from the Council of Ministers/Ministry of Civil Affairs of Bosnia and Herzegovina and the Federal Ministry for Education and Science of Bosnia and Herzegovina awarded to S.S., and by a European Foundation for the Study of Diabetes Albert Renold Travel Fellowship to T.D. SDDS: K.B. acknowledges support from AJ Andersen and Wife's Foundation (J. No. 01737‐0005), the AP Moeller Foundation for the Advancement of Medical Science (J. No. 09034), and the Region of Southern Denmark (J. No. 09/12913).
References
- 1. Hermann, L.S. , Scherstén, B. , Bitzén, P.O. , Kjellström, T. , Lindgärde, F. & Melander, A. Therapeutic comparison of metformin and sulfonylurea, alone and in various combinations. A double‐blind controlled study. Diabetes Care 17, 1100–1109 (1994). [DOI] [PubMed] [Google Scholar]
- 2. Kahn, S.E. et al Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N. Engl. J. Med. 355, 2427–2443 (2006). [DOI] [PubMed] [Google Scholar]
- 3. Zhou, K. et al Heritability of variation in glycaemic response to metformin: a genome‐wide complex trait analysis. Lancet Diabetes Endocrinol. 2, 481–487 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giacomini, K.M. et al International Transporter Consortium commentary on clinically important transporter polymorphisms. Clin. Pharmacol. Ther. 94, 23–26 (2013). [DOI] [PubMed] [Google Scholar]
- 5. Foretz, M. , Guigas, B. , Bertrand, L. , Pollak, M. & Viollet, B. Metformin: from mechanisms of action to therapies. Cell. Metab. 20, 953–966 (2014). [DOI] [PubMed] [Google Scholar]
- 6. McCreight, L.J. , Bailey, C.J. & Pearson, E.R. Metformin and the gastrointestinal tract. Diabetologia 59, 426–435 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang, D.S. , Jonker, J.W. , Kato, Y. , Kusuhara, H. , Schinkel, A.H. & Sugiyama, Y. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J. Pharmacol. Exp. Ther. 302, 510–515 (2002). [DOI] [PubMed] [Google Scholar]
- 8. Kimura, N. , Okuda, M. & Inui, K. Metformin transport by renal basolateral organic cation transporter hOCT2. Pharm. Res. 22, 255–259 (2005). [DOI] [PubMed] [Google Scholar]
- 9. Otsuka, M. , Matsumoto, T. , Morimoto, R. , Arioka, S. , Omote, H. & Moriyama, Y. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc. Natl. Acad. Sci. USA 102, 17923–17928 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Masuda, S. et al Identification and functional characterization of a new human kidney‐specific H+/organic cation antiporter, kidney‐specific multidrug and toxin extrusion 2. J. Am. Soc. Nephrol. 17, 2127–2135 (2006). [DOI] [PubMed] [Google Scholar]
- 11. Kusuhara, H. et al Effects of a MATE protein inhibitor, pyrimethamine, on the renal elimination of metformin at oral microdose and at therapeutic dose in healthy subjects. Clin. Pharmacol. Ther. 89, 837–844 (2011). [DOI] [PubMed] [Google Scholar]
- 12. Nakamichi, N. et al Involvement of carnitine/organic cation transporter OCTN1/SLC22A4 in gastrointestinal absorption of metformin. J. Pharm. Sci. 102, 3407–3417 (2013). [DOI] [PubMed] [Google Scholar]
- 13. Tamai, I. et al Involvement of OCTN1 (SLC22A4) in pH‐dependent transport of organic cations. Mol. Pharm. 1, 57–66 (2004). [DOI] [PubMed] [Google Scholar]
- 14. Yoon, H. , Cho, H.Y. , Yoo, H.D. , Kim, S.M. & Lee, Y.B. Influences of organic cation transporter polymorphisms on the population pharmacokinetics of metformin in healthy subjects. AAPS J. 15, 571–580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shu, Y. et al Effect of genetic variation in the organic cation transporter 1 (OCT1) on metformin action. J. Clin. Invest. 117, 1422–1431 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shu, Y. et al Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin. Pharmacol. Ther. 83, 273–280 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen, Y. et al Effect of genetic variation in the organic cation transporter 2 on the renal elimination of metformin. Pharmacogenet. Genomics 19, 497–504 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stocker, S.L. et al The effect of novel promoter variants in MATE1 and MATE2 on the pharmacokinetics and pharmacodynamics of metformin. Clin. Pharmacol. Ther. 93, 186–194 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhou, K. et al Reduced‐function SLC22A1 polymorphisms encoding organic cation transporter 1 and glycemic response to metformin: a GoDARTS study. Diabetes 58, 1434–1439 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Becker, M.L. , Visser, L.E. , van Schaik, R.H. , Hofman, A. , Uitterlinden, A.G. & Stricker, B.H. Genetic variation in the organic cation transporter 1 is associated with metformin response in patients with diabetes mellitus. Pharmacogenomics J. 9, 242–247 (2009). [DOI] [PubMed] [Google Scholar]
- 21. Becker, M.L. , Visser, L.E. , van Schaik, R.H. , Hofman, A. , Uitterlinden, A.G. & Stricker, B.H. Genetic variation in the multidrug and toxin extrusion 1 transporter protein influences the glucose‐lowering effect of metformin in patients with diabetes: a preliminary study. Diabetes 58, 745–749 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Becker, M.L. , Visser, L.E. , van Schaik, R.H. , Hofman, A. , Uitterlinden, A.G. & Stricker, B.H. Interaction between polymorphisms in the OCT1 and MATE1 transporter and metformin response. Pharmacogenet. Genomics 20, 38–44 (2010). [DOI] [PubMed] [Google Scholar]
- 23. Christensen, M.M. et al The pharmacogenetics of metformin and its impact on plasma metformin steady‐state levels and glycosylated hemoglobin A1c. Pharmacogenet. Genomics 21, 837–850 (2011). [DOI] [PubMed] [Google Scholar]
- 24. Choi, J.H. et al A common 5′‐UTR variant in MATE2‐K is associated with poor response to metformin. Clin. Pharmacol. Ther. 90, 674–684 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tkáč, I. et al Pharmacogenomic association between a variant in SLC47A1 gene and therapeutic response to metformin in type 2 diabetes. Diabetes Obes. Metab. 15, 189–191 (2013). [DOI] [PubMed] [Google Scholar]
- 26. Christensen, M.M. et al A gene‐gene interaction between polymorphisms in the OCT2 and MATE1 genes influences the renal clearance of metformin. Pharmacogenet. Genomics 23, 526–534 (2013). [DOI] [PubMed] [Google Scholar]
- 27. Todd, J.N. & Florez, J.C. An update on the pharmacogenomics of metformin: progress, problems and potential. Pharmacogenomics 15, 529–539 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tzvetkov, M.V. et al The effects of genetic polymorphisms in the organic cation transporters OCT1, OCT2, and OCT3 on the renal clearance of metformin. Clin. Pharmacol. Ther. 86, 299–306 (2009). [DOI] [PubMed] [Google Scholar]
- 29. Song, I.S. et al Genetic variants of the organic cation transporter 2 influence the disposition of metformin. Clin. Pharmacol. Ther. 84, 559–562 (2008). [DOI] [PubMed] [Google Scholar]
- 30. Wang, Z.J. , Yin, O.Q. , Tomlinson, B. & Chow, M.S. OCT2 polymorphisms and in‐vivo renal functional consequence: studies with metformin and cimetidine. Pharmacogenet. Genomics 18, 637–645 (2008). [DOI] [PubMed] [Google Scholar]
- 31. Jablonski, K.A. et al Common variants in 40 genes assessed for diabetes incidence and response to metformin and lifestyle intervention in the diabetes prevention program. Diabetes 59, 2672–2681 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christensen, M.M. et al Endogenous glucose production increases in response to metformin treatment in the glycogen‐depleted state in humans: a randomised trial. Diabetologia 58, 2494–2502 (2015). [DOI] [PubMed] [Google Scholar]
- 33. Koepsell, H. , Lips, K. & Volk, C. Polyspecific organic cation transporters: structure, function, physiological roles, and biopharmaceutical implications. Pharm. Res. 24, 1227–1251 (2007). [DOI] [PubMed] [Google Scholar]
- 34. Chen, E.C. et al Targeted disruption of organic cation transporter 3 attenuates the pharmacologic response to metformin. Mol. Pharmacol. 88, 75–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Napolitano, A. et al Novel gut‐based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One 9, e100778 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Duca, F.A. et al Metformin activates a duodenal Ampk‐dependent pathway to lower hepatic glucose production in rats. Nat. Med. 21, 506–511 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Forslund, K. et al Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buse, J.B. et al The primary glucose‐lowering effect of metformin resides in the gut, not the circulation: results from short‐term pharmacokinetic and 12‐week dose‐ranging studies. Diabetes Care 39, 198–205 (2016). [DOI] [PubMed] [Google Scholar]
- 39. Han, T.K. , Proctor, W.R. , Costales, C.L. , Cai, H. , Everett, R.S. & Thakker, D.R. Four cation‐selective transporters contribute to apical uptake and accumulation of metformin in Caco‐2 cell monolayers. J. Pharmacol. Exp. Ther. 352, 519–528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou, M. , Xia, L. & Wang, J. Metformin transport by a newly cloned proton‐stimulated organic cation transporter (plasma membrane monoamine transporter) expressed in human intestine. Drug Metab. Dispos. 35, 1956–1962 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liang, X. et al Metformin is a substrate and inhibitor of the human thiamine transporter, THTR‐2 (SLC19A3). Mol. Pharm. 12, 4301–4310 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Müller, J. , Lips, K.S. , Metzner, L. , Neubert, R.H. , Koepsell, H. & Brandsch, M. Drug specificity and intestinal membrane localization of human organic cation transporters (OCT). Biochem. Pharmacol. 70, 1851–1860 (2005). [DOI] [PubMed] [Google Scholar]
- 43. Stage, T.B. , Brøsen, K. & Christensen, M.M. A comprehensive review of drug‐drug interactions with metformin. Clin. Pharmacokinet. 54, 811–824 (2015). [DOI] [PubMed] [Google Scholar]
- 44. Goswami, S. et al Genetic variants in transcription factors are associated with the pharmacokinetics and pharmacodynamics of metformin. Clin. Pharmacol. Ther. 96, 370–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tzvetkov, M.V. , Saadatmand, A.R. , Bokelmann, K. , Meineke, I. , Kaiser, R. & Brockmöller, J. Effects of OCT1 polymorphisms on the cellular uptake, plasma concentrations and efficacy of the 5‐HT(3) antagonists tropisetron and ondansetron. Pharmacogenomics J. 12, 22–29 (2012). [DOI] [PubMed] [Google Scholar]
- 46. Dujic, T. , Zhou, K. , Donnelly, L.A. , Tavendale, R. , Palmer, C.N. & Pearson, E.R. Association of organic cation transporter 1 with intolerance to metformin in type 2 diabetes: a GoDARTS study. Diabetes 64, 1786–1793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Purcell, S. et al PLINK: a tool set for whole‐genome association and population‐based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Manning, A.K. et al Meta‐analysis of gene‐environment interaction: joint estimation of SNP and SNP x environment regression coefficients. Genet. Epidemiol. 35, 11–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou, K. et al Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat. Genet. 48, 1055–1059 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


