Abstract
Late embryogenesis abundant (LEA) proteins constitute a large protein family that is closely associated with resistance to abiotic stresses in multiple organisms and protect cells against drought and other stresses. Azotobacter vinelandii is a soil bacterium that forms desiccation-resistant cysts. This bacterium possesses two genes, here named lea1 and lea2, coding for avLEA1 and avLEA2 proteins, both containing 20-mer motifs characteristic of eukaryotic plant LEA proteins. In this study, we found that disruption of the lea1 gene caused a loss of the cysts’ viability after 3 months of desiccation, whereas at 6 months, wild-type or lea2 mutant strain cysts remained viable. Vegetative cells of the lea1 mutant were more sensitive to osmotic stress; cysts developed by this mutant were also more sensitive to high temperatures than cysts or vegetative cells of the wild type or of the lea2 mutant. Expression of lea1 was induced several fold during encystment. In addition, the protective effects of these proteins were assessed in Escherichia coli cells. We found that E. coli cells overexpressing avLEA1 were more tolerant to salt stress than control cells; finally, in vitro analysis showed that avLEA1 protein was able to prevent the freeze thaw-induced inactivation of lactate dehydrogenase. In conclusion, avLEA1 is essential for the survival of A. vinelandii in dry conditions and for protection against hyper-osmolarity, two major stress factors that bacteria must cope with for survival in the environment. This is the first report on the role of bacterial LEA proteins on the resistance of cysts to desiccation.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-017-0781-1) contains supplementary material, which is available to authorized users.
Keywords: Azotobacter, LEA proteins, Cysts desiccation, Osmotic stress, High temperatures
Introduction
Organisms are exposed to abiotic stress conditions, such as high or low temperatures, freezing, salinity and drought (Bray et al. 2000). Abiotic stress has a negative impact on living cells, affecting their sub-cellular structures, enzyme activity, displacement of membrane proteins, and loss of membrane integrity (Zhu et al. 1997). In the face of unfavourable and extreme conditions, organisms experience morphological, physiological, biochemical and molecular changes and develop responses that allow them to adapt to unfavourable situations (Yancey 2005). Late embryogenesis abundant (LEA) proteins are present in many organisms, including plants, bacteria and invertebrates, in which they play a role in resisting environmental conditions, such as freezing, drought or desiccation (Tunnacliffe and Wise 2007; Battaglia et al. 2008; Hand et al. 2011; Toxopeus et al. 2014). Several in vitro assays have shown that some LEA proteins are able to stabilize structures or molecules during freezing and drying (Tunnacliffe and Wise 2007; Battaglia et al. 2008), which is related to their ability to prevent enzyme aggregation under stress conditions (Nakayama et al. 2008; Boucher et al. 2010). Most LEA proteins are highly hydrophilic, with low cysteine, phenylalanine or tryptophan content (Dure et al. 1989; Wise 2003; Shih et al. 2008). LEA proteins are considered as fully disordered proteins with a high percentage of random coils in aqueous solution, but when water is removed, they form secondary structures, such as α helices, β sheets and hairpin loops (Tunnacliffe and Wise 2007; Garay-Arroyo et al. 2000). LEA proteins have been classified into six or more groups, according to similarities in their amino acid sequences and conserved motifs (Baker et al. 1988; Tunnacliffe and Wise 2007; Battaglia et al. 2008). Group 1 LEA proteins (G1LEA) possess a 20-amino acid motif (20-mer), which may be repeated up to eight times, as is the case of LEA proteins present in the crustacean Artemia franciscana (Campos et al. 2013; Toxopeus et al. 2014). LEA 20-mers are also similar to hydrophilic repeated motifs of general stress proteins of the bacterial GsiB superfamily (Stacy and Aalen 1998). Wu et al. (2011) found that the 20-mer motifs of G1LEA proteins are divisible into two sub-motifs; one sub-motif is conserved among archaea, bacteria and eukaryotes, whereas the other differs among prokaryotes-archaea and eukaryotes. Thus, the 20-mer seems to differ phylogenetically in eukaryotic and bacterial LEA proteins (Wu et al. 2011).
Azotobacter vinelandii is a gram-negative soil bacterium that produces the exopolysaccharide alginate, the intracellular poly-β-hydroxybutyrate (PHB) and a family of phenolic lipids known as alkylresorcinols (ARS). This bacterium has the ability to form metabolically dormant cells called cysts. One of the main features of the cyst is its ability to withstand desiccation, being able to survive storage in dry soil for more than 10 years (Vela 1974). In laboratory conditions, A. vinelandii cysts are produced in late stationary phase, or upon induction of vegetative cells with specific reagents, such as n-butanol or β-hydroxybutyrate (Lin and Sadoff 1968; Sutherland 1985).
The genome of A. vinelandii contains four genes encoding predicted LEA-like proteins (Campos et al. 2013; Setubal et al. 2009; Wu et al. 2011). These genes are annotated as Avin11010, Avin11020, Avin44390 and Avin02300. The protein encoded by Avin11010 contains four 20-mer repeats with identity to eukaryotic G1LEA proteins; therefore, it was thought to be an example of horizontal gene transfer between the domains of life (Wu et al. 2011). The proteins encoded by the other three genes contain only one 20-mer (Campos et al. 2013). The 20-mer present in the protein encoded by Avin11020 is shown here to contain the sub-motif present in eukaryotic LEA proteins, while the Avin44390 and Avin02300 encoded proteins possess the prokaryotic sub-motif and share a similarity to proteins of the GsiB family. Due to their resemblance to plant LEA proteins that accumulate in seeds during the dormancy period, where they are likely to be involved in desiccation tolerance, in the present study, we chose Avin11010 and Avin11020 to determine their role in desiccation tolerance of dormant cysts. We also determined the role of these genes under different abiotic stress conditions in cyst and vegetative cells of A. vinelandii. We demonstrated herein that the LEA protein encoded by Avin11010 is essential for the desiccation resistance (viability) of cysts over prolonged time, and in coping with abiotic stress conditions in vegetative cells.
Materials and methods
Biological material and growth conditions
Bacterial strains, plasmids, and oligonucleotides used in this study are listed in Table 1. For vegetative growth, A. vinelandii was cultured at 30 °C on Burk’s nitrogen-free salt medium (Kennedy et al. 1986), supplemented with 2% sucrose (BS), or on Burk’s nitrogen-free salt medium with 0.2% n-butanol as carbon source (BB), for encystment induction (Sadoff et al. 1971). Escherichia coli DH5α (Hanahan 1983) and BL21(DE3) were grown on Luria–Bertani (LB) medium at 37 °C (Miller 1972). Antibiotic concentrations used for A. vinelandii or E. coli were as follows: nalidixic acid (30 μg/mL), kanamycin (2 μg/mL), gentamycin (0.5 μg/mL), spectinomycin (100 μg/ mL), tetracycline (30 μg/mL) and ampicillin (100 μg/mL).
Table 1.
Bacterial strains, plasmids and oligonucleotides used in this work
| Strain plasmid or oligonucleotide | Description or sequence (5′–3′) | Referencea |
|---|---|---|
| Strains | ||
| A. vinelandii | ||
| AEIV | WT | [1] |
| lea1 | AEIV derivative harbouring pLEA1101 plasmid carrying a Av11010::Gm mutation | This work |
| lea2 | AEIV derivative harbouring pLEA1102 plasmid carrying a Av11020::Km mutation | This work |
| lea1-lea2 | AEIV derivative harbouring pLEADC carrying a Av11010-Av11020::Sp mutation | This work |
| lea1–2-plea1–2 | AEIV derivative with Av11010 and Av11020 genes co-integrated | This work |
| lea1-gusA | AEIV with transcriptional fusion from pUMATcgusAT carrying lea1::gusA | This work |
| lea2-gusA | AEIV with transcriptional fusion from pUMATcgusAT carrying lea2::gusA | This work |
| AEIV gacA | AEIV with a gacA::Gm mutation | [2] |
| E. coli | ||
| DH5α | supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | GIBCO-BRL |
| BL21(DE3) | F − ompT hsdS B (r − B m − B ) gal dcm | Invitrogen |
| BL(DE3)/pET22b+ | BL21 carrying the empty expression vector pET22b+ | This work |
| BL(DE3)/p avLEA1 | BL21 carrying the plasmid pavLEA1 for avLEA1 protein production | This work |
| Plasmids | ||
| pJET1.2/blunt | Cloning vector Ampr | Thermo Scientific |
| pET22b+ | Cloning and expression vector Ampr | Novagen |
| pBLS190 | Plasmid used to obtain Tcr cassette | [3] |
| pBSL97 | Plasmid used to obtain the Kmr cassette | [3] |
| pBSL98 | Plasmid used to obtain the Gmr cassette | [3] |
| pLEA11012 | pJET1.2/blunt containing a 1.2 kb PCR fragment from both, lea1 and lea2 genes | This work |
| pLEA1101 | pJET1.2/blunt containing a small fragment of 103 bp of lea1 | This work |
| pLEA1102 | pJET1.2/blunt containing a small fragment of 65 bp of lea2 | This work |
| pLEADC | pJET1.2/blunt containing lea1 and lea2 genes interrupted by Spr cassette | This work |
| pUMATc | pUMA derivative with a Tc cassette with the gusA gene for transcriptional fusions | [4] |
| pavLEA1-gusA | pJET1.2/blunt containing 178 bp corresponding to the lea1 promoter region | This work |
| pavLEA2-gusA | pJET1.2/blunt containing 195 bp corresponding to the lea2 promoter region | This work |
| pET22b+ | pET22b6XHis-Tag | Novagen |
| pavLEA1 | pET22b6XHis-Tag containing avLEA1 protein from A. vinelandii | This work |
| Oligonucleotide | ||
| Av1020F1 | CATGCGTAAAGCCGAAAGC | This work |
| Av1020R1 | GATGGGTTTTTTTGCGATAG | This work |
| Av10invF1 | CAAACAGGAAAACCGCGG | This work |
| Av10invR1 | GATCTCGCCGCCTTTCTG | This work |
| Av20invF1 | CGTGAACTCATCCAGGAAG | This work |
| Av20invR1 | CGCTTACACTGATCGAACC | This work |
| AvFTLEA1F1b | GAACCGCGGGGGGCGAAAAATAGGCGG | This work |
| AvFTLEA1R1b | CACCTGCAGGGAGCCTTTGTCCTTGCC | This work |
| AvFTLEA2F1 | CCAGAAAGGCGGCGAGAT | This work |
| AvFTLEA2R1 | TAGCCTTCCGGGCCGAGTT | This work |
| AvExpLEAF1c | GAACATATGAGCGTACGGGAGGCCGG | This work |
| AvExpLEAR1c | GAACTCGAGGCTCTTGCGGCTGCCCT | This work |
aReferences: [1] Svein Valla (Norwegian University of Science And Technology), [2] Manzo et al. 2011, [3] Alexeyev et al. 1995, [4] Muriel-Millán et al. 2015
bRestriction sites for SacII and PstI are cursive and underlined respectively
cRestriction sites for XhoI and NdeI are cursive underlined and bold respectively
Determination of cyst resistance to desiccation
For encystment induction and desiccation resistance, vegetative cells grown for 24 h on BS medium were centrifuged, washed and resuspended in Burk’s nitrogen-free salt medium. A sample of washed cells was plated on solid BB encysting medium. After 5 days of incubation, samples harvested from these plates containing 106 to 107 cysts (colony-forming units, CFU) were applied to Millipore membranes (0.2-μm pore size). The membranes were placed in sterile Eppendorf tubes and placed in sterile petri dishes. After 5 or more days, dried cysts in the filters were resuspended and plated on solid BS medium to determine viable counts.
In silico analysis of protein sequence
Sequence analysis of lea1 and lea2 was performed using the BLAST program on the NCBI web server (http://blast.ncbi.nlm.nih.gov/). Nucleotide translation and isoelectric point were performed using EXPASY tool (http://www.expasy.org/tools/protparam.html). Multiple sequence alignment was performed using CLUSTAL Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Hydropathy plots were constructed with the Kyte and Doolittle algorithm (http://ipsort.hgc.jp/) (Kyte and Doolittle 1982). Prediction of naturally disordered regions was done with PONDR (www.pondr.com).
Construction of lea mutants
The lea2 gene is located immediately upstream of the lea1 gene. We used primers Av1020F1/Av1020R1 and AEIV wild-type strain (AEIV) chromosomal DNA as a template to amplify by PCR (Phusion DNA Polymerase, Thermo Scientific, USA) a fragment of 1.12 kb containing lea1 and lea2 genes; this fragment was ligated into the pJET1.2/blunt vector (Thermo Scientific, USA) to produce pLEA11012 plasmid. Primers Av10invF1/Av10invR1 were used to delete by inverse PCR a 299-bp fragment of the lea1-coding region from plasmid pLEA11012. The linearized plasmid thus obtained was ligated to a Gmr cassette to produce plasmid pLEA1101 in which the lea1 gene was replaced by the Gmr cassette. Similarly, primers Av20invF1/Av20invR1 and plasmid pLEA11012 were used to remove by inverse PCR 89 bp of lea2 and were replaced by a Kmr cassette to produce pLEA1102. To remove both lea1 and lea2 genes from plasmid pLEA11012, a fragment of 636 bp was deleted by inverse PCR using primers Av10invF1/Av20invR1. In this case, the lea1-lea2 genes were replaced by a spectinomycin resistance cassette (Spr) yielding the plasmid pLEADC. A physical map of the lea1-lea2 region present in the four plasmids constructed is shown in Fig. 1S. A. vinelandii AEIV competent cells were transformed with plasmids pLEA1101, pLEA1102 and pLEADC, as described previously (Bali et al. 1992). Gentamycin-, kanamycin- and spectinomycin-resistant transformants were isolated and named lea1, lea2 and lea1-lea2 respectively; DNA from these mutant strains and Av1020F1/Av1020R1 primers was used to confirm by PCR the presence of the lea1–Gmr, lea2–Kmr and lea1–lea2–Spr mutations in the respective mutants. To construct the strain lea1–2–plea1–2, plasmid pLEA11012 that is unable to replicate in A. vinelandii was transformed into strain lea1–lea2 for integration into the chromosome. A transformant resistant to tetracycline was isolated and confirmed by PCR to carry plasmid pLEA11012 co-integrated into the chromosome as a result of a single-homologue recombination event. A schematic representation of the integration of plasmid pLEA11012 into the lea1–lea2 chromosome is shown in Fig. S1.
Osmotic stress tolerance assays
A. vinelandii strains were grown at 30 °C in 50 mL of BS medium in 250-mL Erlenmeyer flasks, supplemented with different concentrations of NaCl and sorbitol (Sigma-Aldrich, CA, USA) ranging from 0.1 to 1 M. After 48 h of growth, appropriate dilutions were plated onto BS plates to determine CFU.
E. coli BL(DE3)/pET22b+ and BL(DE3)/pavLEA1 strains were cultured at 37 °C in 20 mL of LB medium containing the appropriate antibiotics. Isopropyl-β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 1 mM was added when the cultures reached an OD600nm of 0.6 and were further incubated for 2 h. One hundred-microlitre samples from these cultures were spread on LB plates containing increasing concentrations of NaCl (0.1–0.5 mM). Cell viability was plotted as the percentage of CFU on the stressed plates, relative to the colony numbers of control appearing on the non-stressed plates (Soto et al. 1999) after overnight incubation at 37 °C.
Thermotolerance experiments
Test tubes containing 1 mL sample of A. vinelandii vegetative cells or cysts were incubated in water baths adjusted to temperatures ranging from 40 to 60 °C. After 15 min of incubation, the tubes were removed and cooled immediately; dilutions were plated onto BS plates. CFU were counted after 48 h of incubation at 30 °C.
Construction of transcriptional fusions
For the construction of transcriptional fusions of lea1 and lea2 genes with the gusA reporter gene, primers AvFTLEA1F1/AvFTLEA1R1 and AvFTLEA2F1/AvFTLEA2R1 were used to amplify PCR fragments of 178 and 195 bp, containing the promoter regions of lea1 and lea2 genes respectively. The amplified fragments were cloned into the SacII and PstI restriction sites of the pUMATgusAT vector (Muriel-Millán et al. 2015), to generate pavLEA1-gusA and pavLEA2-gusA plasmids respectively. These plasmids were transformed into AEIV strain; tetracycline-resistant derivatives were selected. The strains, containing the gene fusions integrated into the chromosome as a result of a double recombination event, were named lea1-gusA and lea2-gusA. The presence of the fusions into the chromosomes of A. vinelandii was confirmed by PCR (data not shown). β-Glucuronidase activities of at least three independent cultures were determined following the method described by Romero et al. (2013).
Expression and purification of avLEA1-6His protein
For construction of the expression vector pavLEA1, which contains avLEA1 fused with a C-terminal 6xHis-Tag (Merck, NJ, USA), primers AvExpLEAF1/AvExpLEAR1 (containing NdeI and XhoI restriction sites) and AEIV chromosomal DNA were used to amplify by PCR a DNA fragment of 396 bp corresponding to the lea1 gene lacking the ATG start codon and the stop codon; this fragment was cloned into the pET22b+ vector at NdeI and XhoI restriction sites to produce plasmid pavLEA1. E. coli strain BL21(DE3) was used to express the avLEA1 protein. Strain BL(DE3)/pavLEA1 was inoculated in LB medium supplemented with ampicillin (Sigma-Aldrich, CA, USA) and grown overnight at 37 °C. A sample of the overnight culture was used to inoculate 100 mL of fresh medium to 0.05 OD600nm. After 2 h at 37 °C, IPTG was added to a final concentration of 1 mM and further incubated for 2 h. Cell cultures were harvested by centrifugation. The recombinant protein was purified according to “The handbook for high-level expression and purification of 6xHis-Tagged proteins, 5th edition” (Qiagen, Hilden, Germany). Samples of the purified avLEA1 protein were analysed in SDS-PAGE using 12% acrylamide gels (Sambrook and Russell 2001) with Tris-glycine SDS running buffer. Detection of 6xHis-Tag purified protein was performed by immunoblotting, utilizing the SuperSignal West Pico Chemiluminescent Substrate Kit (Thermo-Scientific, Rockford, USA) (Fig. S2). Protein was quantified by the Bradford assay.
l-Lactate dehydrogenase activity assay
l-Lactate dehydrogenase (LDH) from rabbit muscle (Sigma-Aldrich, MO, USA) was dissolved (0.4 mg mL−1) in assay buffer, containing 100 mM dibasic sodium phosphate buffer pH 7.5, 2 mM pyruvate (Sigma-Aldrich, MA, USA) and 100 mM NADH (Roche, NJ, USA). LDH enzymatic activity was determined as follows. One hundred microlitres of the LDH solution was added to 500 μL demineralized water with or without purified avLEA1protein. The mixture was frozen for 15 min in dry ice and thawed for 15 min in a water bath at 25 °C. This freeze thaw cycle was repeated up to 10 times, after that. The LDH activity was then determined by NADH absorbance at OD340nm for 1 min due to the conversion of NADH into NAD at 25 °C using an Ultrospec 3300 Pro UV/Visible Spectrophotometer (Amersham-Biosciences, Freiburg, Germany). All of the values given were expressed as the percentage of the rate of the reaction measured for the untreated samples. Enzyme activity for each sample shown was determined in at least three independent tests. Bovine serum albumin (BSA), (Sigma-Aldrich, CA, USA) was used as a positive control because it is a protein with known cryoprotective activity.
Statistics
Each experiment was performed with three replicates. Statistical relevance was determined by Student’s t test or one-way ANOVA using Prism version 6 (GraphPad Software, USA).
Results
In silico analysis of A. vinelandii avLEA1 and avLEA2 proteins
The genome of A. vinelandii contains four genes encoding predicted LEA-like proteins (Campos et al. 2013). The protein encoded by Avin11010 contains four repeats of the 20-mer; the protein Avin11020 contains only one 20-mer (Campos et al. 2013). The 20-mer sequences of the protein encoded by Avin11010 are present in eukaryotic LEA proteins, but not in prokaryotes (Wu et al. 2011). To analyse the sequence of A. vinelandii LEA proteins, we performed a multiple alignment analysis of the 20-mers present in the four LEA proteins from A. vinelandii and LEA proteins from other prokaryotes, archaeae and eukaryotes (Fig. 1). As reported by Wu et al. 2011, the protein encoded by Avin11010 possesses eukaryotic sub-motifs (Fig. 1). Here, we found that the 20-mer present in the protein encoded by Avin11020 contains the sub-motif present in eukaryotic LEA proteins. The Avin44390-encoded LEA has an archaeae sub-motif, while Avin02300 possesses the prokaryotic sub-motif (Fig. 1). The 20-mer sequence of Avin02300 shares the highest identity to putative LEA proteins present in bacteria belonging to the genus Pseudomonas that are phylogenetically closely related to A. vinelandii (Setubal et al. 2009). The Avin11010 and Avin11020 genes were named lea1 and lea2 and their protein products avLEA1 and avLEA2 respectively. The full-length open reading frame (ORF) of lea1 is 402 bp, which encodes a protein of 133 amino acid residues with a calculated molecular mass of 14.3 kDa and a theoretical pI of 6.87. The lea2 gene encodes a 55-amino acid protein, with a calculated molecular mass of 5.9 kDa, and a theoretical pI of 9.9. The lea2 gene is located immediately upstream the lea1 gene separated by an intergenic region of 165 nucleotides.
Fig. 1.
Alignment of the 20-mer sequence of the A. vinelandii LEA proteins and LEA proteins from other prokaryotes, archaeae and eukaryotes, and sub-motifs present in all organisms (grey shading), Archaea sub-motif (underlined), Bacteria sub-motif (bold) and Eukarya (black shading). Accession numbers for LEA proteins used in this alignment are Artemia franciscana ADE45145; Arabidopsis thaliana NP_190749; A. vinelandii Avin11010, WP_012699753; Avin11020, WP_012699754; Avin44390, WP_012702924; Avin02300, WP_012698919; Anaeromyxobacter sp. WP_011985696; Methanocella paludicola BAI62068; Methylocystis parvus WP_026016492; Pseudomonas aeruginosa NP_250880; Pseudomonas fluorescens WP_003174480; Pseudomonas mendocina WP_003246794; Pseudomonas stutzeri WP_021209286; Pseudomonas syringae EGH33010
Kyte and Doolittle hydropathy plots revealed that both the avLEA1 and avLEA2 proteins are strongly hydrophilic, with a grand average of hydropathicity value of −1.6 and −1.3 respectively (GRAVY; Kyte and Doolittle 1982) (Fig. S1). In common with other LEA proteins, disordered prediction programs suggest that both avLEA1 and avLEA2 lack a fixed tertiary structure, being partially unfolded (Fig. S2).
avLEA1 is necessary to withstand desiccation on A. vinelandii cyst
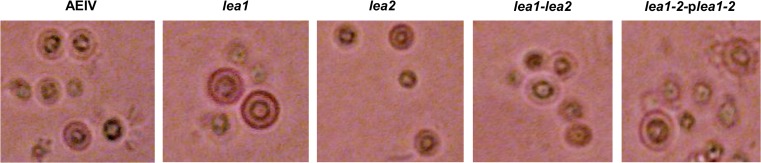
LEA proteins are associated with desiccation tolerance in prokaryotic and eukaryotic organisms. Cysts produced by A. vinelandii are resistant to desiccation. We therefore determined the role of avLEA1 and avLEA2 on the resistance to desiccation of A. vinelandii cysts. Strains carrying mutations in lea1 or lea2 genes, as well as a double lea1–lea2 mutant, were constructed as described in the “Materials and methods” section. These mutants and AEIV strain were induced for encystment by incubation of vegetative cells in solid BB medium plates over 5 days as described in the “Materials and methods” section. Samples of these cells (cysts) were harvested and used for light microscopy visualization after ARS staining. Cyst samples were also applied to Millipore membranes that were placed in sterile petri dishes for drying. After 5 days and for up to 6 months, the dried cysts on the filters were resuspended in BS medium, allowing the cysts to germinate; CFU were counted to determine the percentage of cysts resistant to desiccation. Both lea mutants and the double mutants were able to form cysts; no morphological differences between cysts formed by these strains and the wild-type strain were observed by optical microscopy (Fig. 2). Desiccation resistance assays were carried out in the cyst samples applied to Millipore membranes after 5 days and 1, 2, 3 and 6 months. Similarly to the cysts formed by the wild-type strain, cysts developed by all three lea mutants were resistant to desiccation after 5 days; however, cyst resistance to desiccation was diminished at 2 months and was lost at 3 months in the strain carrying the lea1 mutation. In contrast, the cysts formed by wild type and the lea2 mutant remained resistant to desiccation after 6 months (Table 2). We conclude that avLEA1 and avLEA2 are not necessary for the encystment process in A. vinelandii but avLEA1 is necessary for the cysts to withstand long periods of desiccation.
Fig. 2.
Light microscopy (bright field) of Fast Blue B-stained cysts of A. vinelandii AEIV strain and lea mutants. In all cases, cysts were developed after 5 days of incubation on BB medium plates
Table 2.
Cysts survival to desiccation of A. vinelandii lea1 and lea2 mutants
| Strain | Genotype | Desiccation resistance (%) | |||
|---|---|---|---|---|---|
| 5 days | 1 month | 3 months | 6 months | ||
| AEIV | WT | 9.7 ± 5.3 | 11.2 ± 4.1 | 13.1 ± 8.4 | 17.5 ± 3.2 |
| lea1 | Avin11010::Gm | 10.1 ± 3.9 | 4.2 ± 4.2 | 1.5 ± 2.3 | <0.0001 |
| lea2 | Avin11020::Km | 15.6 ± 3.5 | 10.3 ± 3.1 | 9.5 ± 6.1 | 18.3 ± 2.6 |
| lea1–lea2 | Avin11010-11020::Sp | 7.4 ± 4.6 | 3.5 ± 2.3 | 2.5 ± 1.3 | <0.0001 |
| lea1–2–plea1–2 | Avin11010-11020::Sp+pUMA | 11.5 ± 3.8 | 17.1 ± 6.5 | 21.4 ± 3.2 | 15.4 ± 4.1 |
| AEIV-gacA | AEIV with a gacA::Gm mutation | <0.0001 | <0.0001 | <0.0001 | <0.0001 |
Expression of avLEA1 and avLEA2 in vegetative and encysting cells
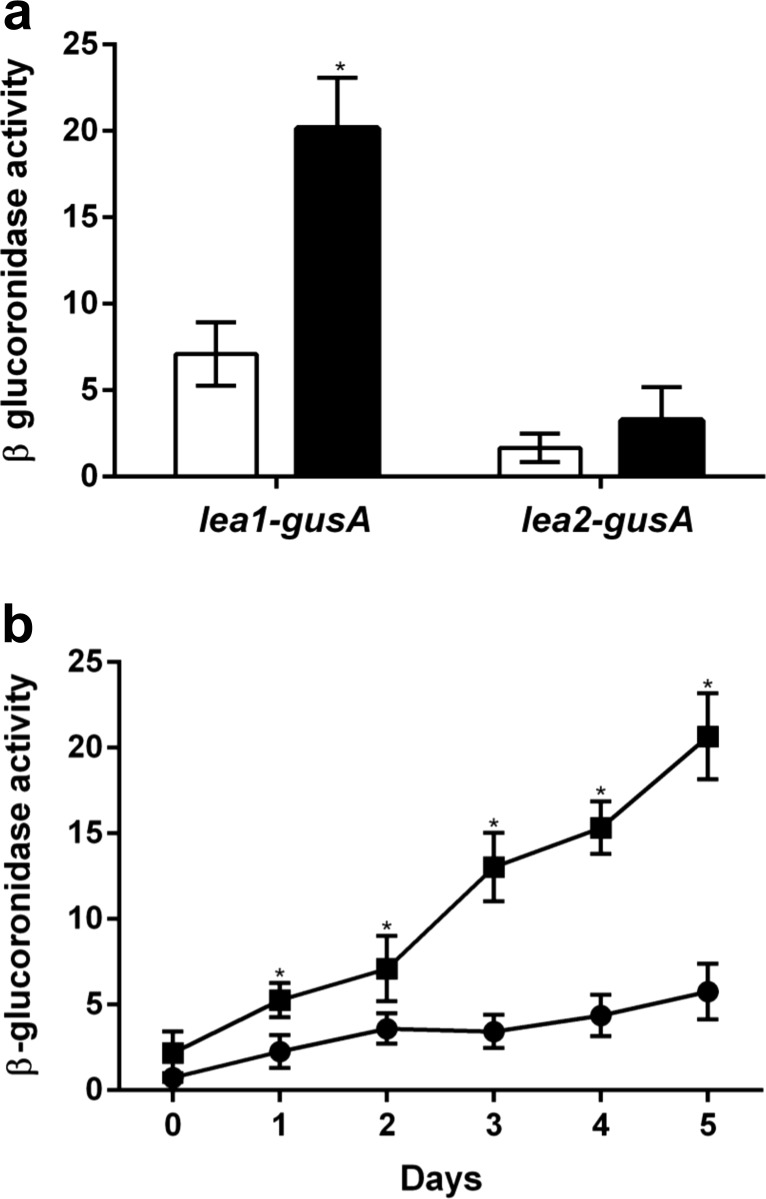
As shown above, avLEA1 but not avLEA2 contributes to the resistance of cyst to desiccation. Transcriptional lea1–gusA and lea2–gusA fusions were used to determine the expression of lea1 and lea2 genes in vegetative cells and in cells under encystment induction conditions. We observed that transcription of lea1 increased during encystment and was higher under this condition than in vegetative cells (Fig. 3a), while transcription of lea2 is similar in the two conditions tested and was threefold to fourfold lower than that of lea1 during the 5 days of the encystment process (Fig. 3b). These data indicate that the expression of the lea1 gene is induced under encystment conditions, a feature in agreement with its role in cyst resistance to desiccation.
Fig. 3.
a Expression of lea1 and lea2 under vegetative growth conditions (white bars) or under encystment induction conditions (black bars), determined as β-glucuronidase activity in AEIV derivatives containing lea1-gusA and lea2-gusA transcriptional fusions. b Expression of lea1-gusA (closed squares) and lea2-gusA (closed circles) in BB medium during the 5 days of the encystment process. One unit of β-glucuronidase activity corresponds to 1 nmol of substrate (X-Gluc) hydrolyzed per minute per milligram of protein. Error bars represent standard deviations of three independent experiments. Significant differences between lea1-gusA and lea2-gusA expression are indicated with asterisk evaluated with the Student’s t test at p < 0.05
avLEA1 confers tolerance to high temperatures in vegetative cells and in cysts
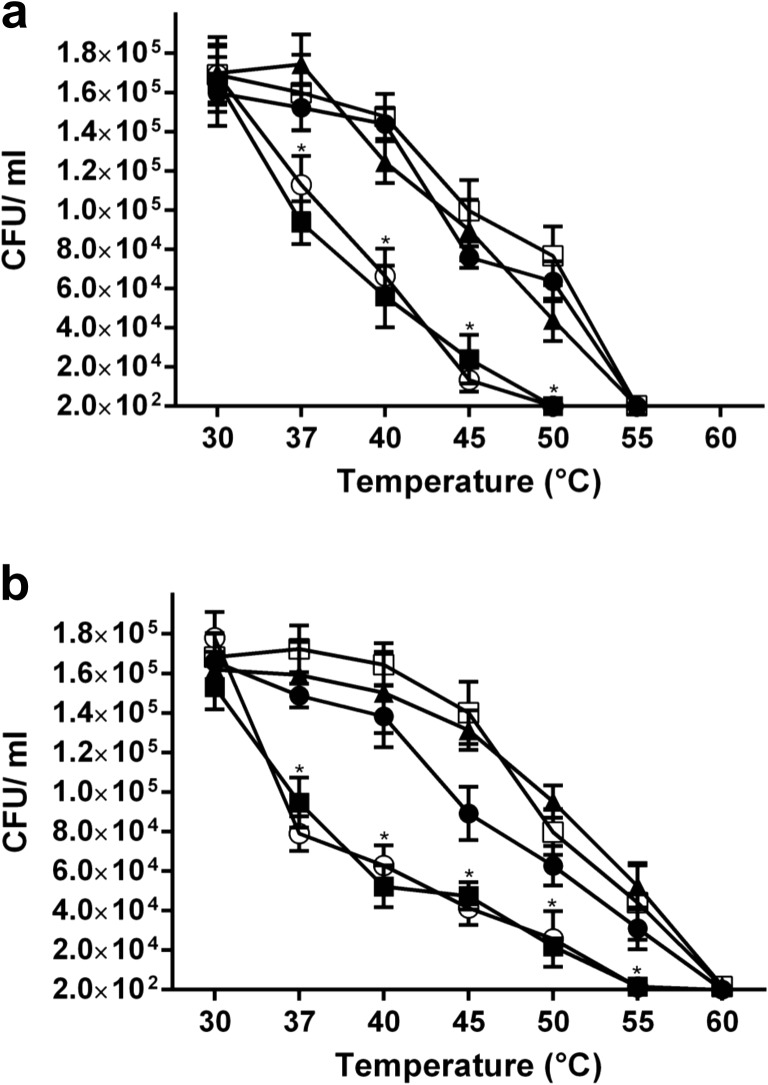
LEA proteins have been implicated in tolerance to high and low temperatures. To investigate the contribution of the avLEA1 and avLEA2 proteins in tolerance to high temperatures in A. vinelandii, we evaluated survival of vegetative cells and cysts of lea1 and lea2 mutants subjected to high temperatures. For vegetative cells, strains were grown for 24 h in BS medium at 30 °C and exposed during 15 min to temperatures ranging from 37 to 60 °C. As shown in Fig. 4a, survival diminished considerably as the cells were incubated at higher temperatures. The viability for the lea1 mutant was considerably affected at 45 and 50 °C. For the AEIV strain and the lea2 mutant, viability was completely lost at 55 °C. Similarly, when wild-type and lea mutant cysts were incubated for 15 min at temperatures ranging from 37 to 60 °C, a considerable loss of viability was observed, although cysts seem to be more resistant than vegetative cells. The strain carrying the lea1 mutation was not able to survive at 55 °C, whereas cysts produced by the wild type and lea2 mutant were able to germinate at 55 °C, but lost viability at 60 °C (Fig. 4b). These data indicate that, as was expected, encysted cells are more tolerant to high temperatures than vegetative cells and that the lea1 gene contributes to the tolerance of A. vinelandii to high temperatures.
Fig. 4.
Thermo tolerance of A. vinelandii cysts and vegetative cells developed by the lea1 and lea2 mutants. AEIV (open squares), lea1 mutant (closed squares), lea2 mutant (closed circles), lea1–lea2 double mutant (open circles) and lea1–2–plea1–2 complemented strain (closed triangles). Cells were grown at 30 °C in BS (a) or BB medium (b). One millilitre of these cultures was incubated during 15 min at temperature ranges from 30 to 60 °C. Cell viability of these samples was determined. Error bars indicate standard deviation of three independent experiments. Asterisk indicates significant difference at p < 0.05 with one-way ANOVA analysis
avLEA1 is required for osmotic stress tolerance in A. vinelandii
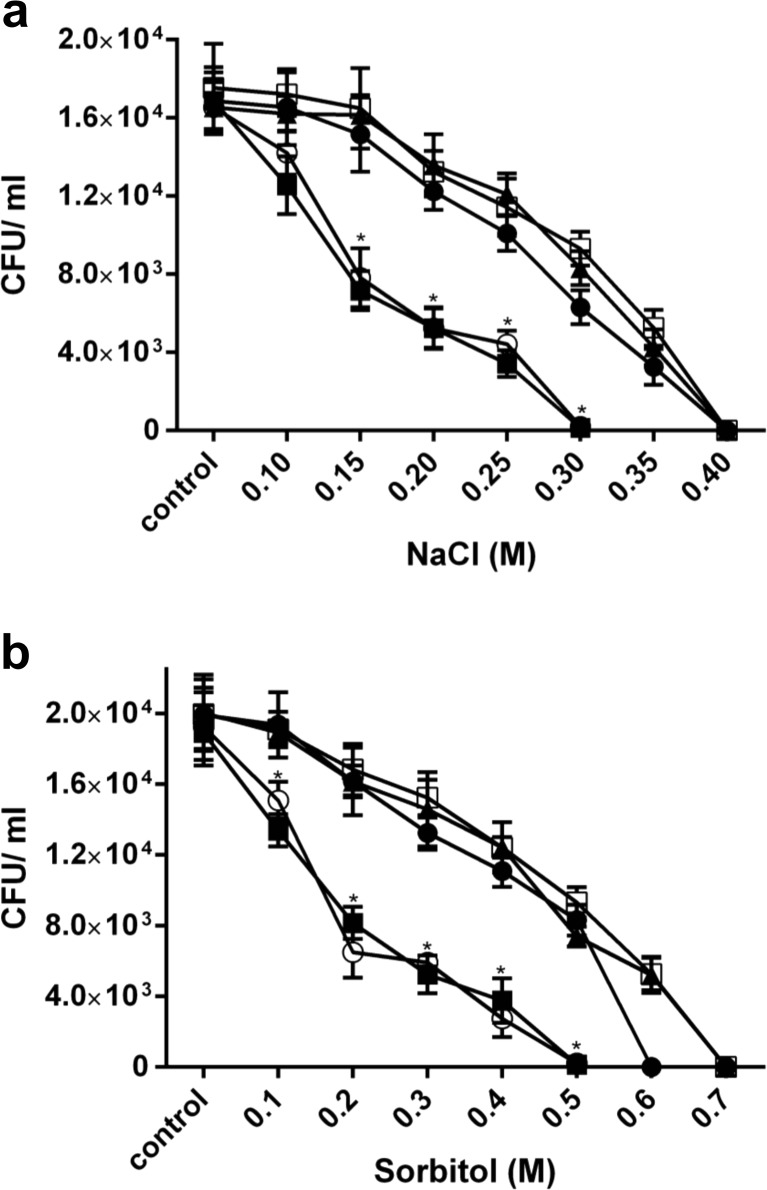
We determined the effect of the lea1 and lea2 mutations on the growth and survival of A. vinelandii under osmotic stress conditions. Mutant strain cultures were grown at different NaCl or sorbitol concentrations, and viable counts were performed after 48 h. The lea1 mutant was significantly more sensitive to osmotic stress when grown at 0.15 M NaCl than the AEIV strain and the lea2 mutant exposed to the same conditions (Fig. 5a). When the concentration of NaCl in the medium was increased to 0.3 M, lea1 cells were unable to survive, while the lea2 strain exhibited a tolerance similar to that of the AEIV strain. Similar results were obtained when an osmotically equivalent concentration of sorbitol (1 M) was present in the growth medium as the strains carrying the lea1 mutant were more sensitive to inhibiting concentrations of sorbitol than the wild type and the lea2 mutant (Fig. 5b). These results indicate that the lea1 gene is necessary for survival of A. vinelandii in high concentrations of osmotic agents such as NaCl and sorbitol.
Fig. 5.
Osmotic stress tolerance of A. vinelandii lea1 and lea2 mutants. Growth of AEIV (open squares), lea1 mutant (closed squares), lea2 mutant (closed circles), double mutant lea1–lea2 (open circles) and lea1–2plea1–2 complemented strain (closed triangles), in the presence of NaCl (a) or sorbitol (b). Cells were grown on liquid BS medium supplemented with increasing concentrations of NaCl or sorbitol; aliquots were plated onto BS agar plates. Error bars represent standard deviations of three independent experiments. Significant differences are indicated with asterisk evaluated with one-way ANOVA analysis at p < 0.05
avLEA1 protein has a protective effect on LDH activity under freeze thaw cycles in vitro
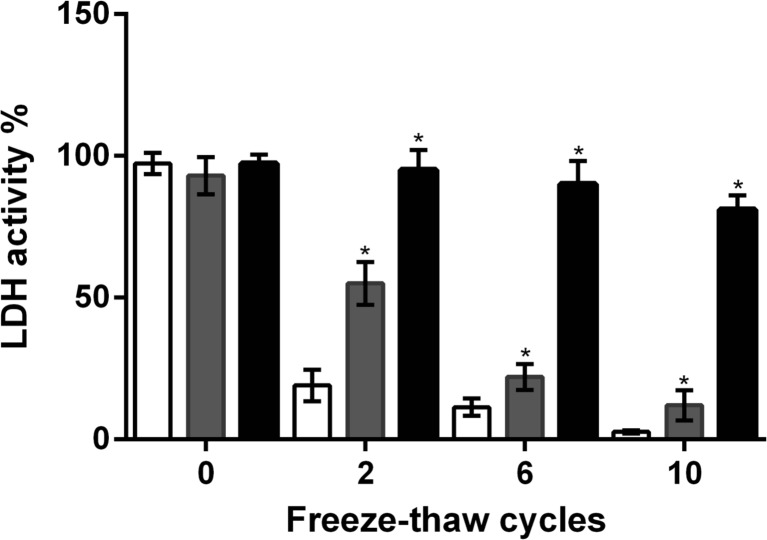
LDH enzymatic activity is known to be sensitive to water limitation upon freeze thaw cycles (Goyal et al. 2005). Some G1LEA proteins have been shown to protect the enzymatic activity of LDH when subjected to freeze thaw treatments (Reyes et al. 2008). An avLEA1 protein containing 6xHis-Tag was expressed in E. coli BL21(DE3) cells and purified as described in the “Materials and methods” section and Fig. 2S. Samples of the purified avLEA1 protein were used to evaluate its protective effect on the LDH enzymatic activity in vitro after freeze thaw cycles. Activity of LDH was monitored in the presence or absence of purified avLEA1. The avLEA1/LDH molar ratio used was 1:1. The LDH enzymatic activity was determined by the conversion of pyruvate, measured by a decrease in absorbance at 340 nm due to the oxidation of NADH. In the presence of avLEA1, the LDH activity remained at 52% after 2 cycles and 21% after 6 cycles, while in its absence, the remaining activity corresponded to 20 and 10% respectively (Fig. 6). In the presence of BSA, the LDH activity was totally protected. The ratio of BSA/avLEA1 was 1:1. This result indicates that like other LEA proteins, avLEA1 has a protective effect on LDH activity under repeated freeze thaw cycles.
Fig. 6.
Effect of avLEA1 on LDH activity after freeze thaw cycles. The LDH activity after 10 freeze thaw cycles was determined in the presence of purified avLEA1 (grey bars) or BSA protein (black bars); buffer (white bars) was used as a control. The data were expressed as percentage (%) of activity measured before the stress in the presence of the corresponding protein. The average of the enzyme activity and standard deviations were based on three independent experiments. Asterisk indicates significant difference evaluated with the Student’s t test at p < 0.05
Heterologous expression of avLEA1 improves salt tolerance but not heat resistance of E. coli
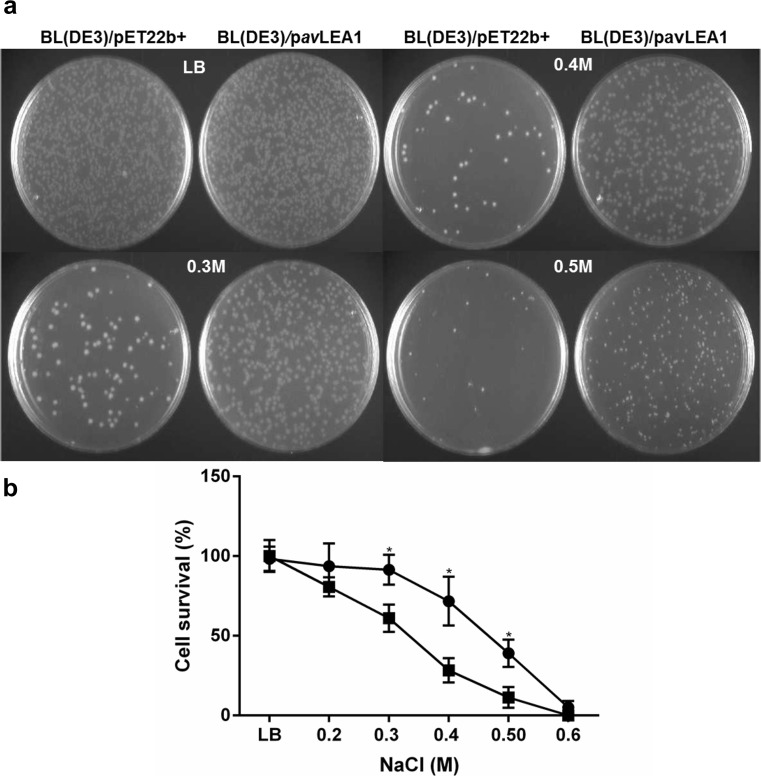
As shown above, avLEA1 protein is necessary to withstand osmotic stress caused by 0.3 M of NaCl in A. vinelandii. To determine the in vivo effect of avLEA1 overexpression in the tolerance of E. coli to ionic stress proper dilutions of E. coli cultures, BL(DE3)/pavLEA1 and control strain BL(DE3)/pET22b+ were spread on plates containing increasing concentrations of NaCl (from 0.2 to 0.6 M). As shown in Fig. 7, BL(DE3)/pavLEA1 and BL(DE3)/pET22b+ strains lost viability as the concentration of NaCl in the plates was increased; viability was completely lost for the two strains at 0.6 M of NaCl. However, viability of BL(DE3)/pavLEA1 was higher than that of the BL(DE3)/pET22b+ strain in NaCl concentrations 0.3 and 0.4 M. The survival of BL(DE3)/pET22b+ was 70 and 30% respectively, while in the BL(DE3)/pavLEA1 strain, 91 and 60% of the cells remained viable (Fig. 7). These results indicated that the overexpression of the avLEA1 protein enhances the tolerance of E. coli to high-salinity conditions.
Fig. 7.
Effect of avLEA1 expression on E. coli viability under osmotic stress (NaCl). a Growth of E. coli BL(DE3)/pavLEA1 and BL(DE3)/pET22b+ strains on LB plates with or without different NaCl concentrations ranging from 0.2 to 0.6 M. b Survival (%) of E. coli BL(DE3)/pavLEA1 (closed circles) and BL(DE3)/pET22b+ (closed squares). Survival was calculated as the percentage of CFU on the plates containing NaCl relative to CFU present in the control LB plates. The standard deviations were calculated from three independent experiments. Asterisk indicates significant difference evaluated with the Student’s t test at p < 0.05
The tolerance of E. coli strains BL(DE3)/pET22b+ and BL(DE3)/pavLEA1 subjected to heat stress was also determined by spreading dilutions of cultures on LB plates containing 1 mM IPTG and incubating them at 37, 45, 50 and 55 °C. The number of colonies grown at 37 °C was considered as the 100% viability control. No colonies were observed at 55 °C, and a similar loss of viability was observed for both strains at 45 and 50 °C, indicating that the avLEA1 protein did not confer tolerance to high temperatures (Data not shown).
Discussion
LEAs are highly hydrophilic, intrinsically unstructured proteins that, in vitro, reduce protein aggregation caused by desiccation and freezing. These proteins were first identified in plants (Dure et al. 1989; Dure 1993), but LEA homologues are also present in other eukaryotes, prokaryotes and archaea (Hand et al. 2011; Wu et al. 2011; Campos et al. 2013). LEA proteins are characterized by a 20-mer motif that possesses a sub-motif that differs among prokaryotes and eukaryotes. There are extensive correlative data linking the expression of LEA proteins with tolerance to water stress caused by desiccation, salinity or low temperatures in plants (Tunnacliffe and Wise 2007; Olvera-Carrillo et al. 2010). A relationship between LEA homologues and water deficit stress has also been reported in eukaryote organisms other than plants and in some prokaryotes (Garay-Arroyo et al. 2000; Solomon et al. 2000; Browne et al. 2002; Gal et al. 2004; Stacy and Aalen 1998; Battista et al. 2001; Denekamp et al. 2010; Hand et al. 2011; Wu et al. 2011; Hatanaka et al. 2013).
A cyst is a resting or dormancy stage of a microorganism, usually a bacterium, a protist or an invertebrate, that helps the organism to survive in unfavourable environmental conditions. Cysts allow these organisms to remain in a state of dormancy for long periods. In A. franciscana, a crustacean that produces cysts that survive desiccation and temperatures below −20 C (Crowe et al. 1981; Hengherr et al. 2011), LEA proteins belonging to group 1 have been shown to contribute to the desiccation and freeze tolerance of encysted embryos (Toxopeus et al. 2014). They also play an important role on the response to salt stress, as LEA-coding genes were found highly expressed under cyst conditions and in response to hyper saline stress in embryonic development (Wu et al. 2011).
A. vinelandii, the soil bacterium studied here, produces cysts that are resistant to desiccation, able to survive storage in dry soil for more than 10 years (Vela 1974). This bacterium also possesses four genes encoding proteins with identity to G1LEA proteins, two of them, lea1 and lea2, encoding for avLEA1 and avLEA2 proteins, which possess 20-mer sequences with the sub-motif present in LEA proteins from eukaryotic organisms. In this study, strains carrying deletion mutations of lea1 and lea2 were constructed to investigate their role in cyst tolerance to desiccation. We found that the avLEA1 protein contributes to the tolerance of the cysts to desiccation since the viability of cysts developed by the lea1 mutant was completely lost after 6 months of drying, while the cysts developed by the wild type or the lea2 mutant strain remained viable. As in A. vinelandii cysts, the contribution of LEA proteins to survival in drying conditions was also observed in Deinococcus radioduran, as the inactivation of two genes encoding for LEA proteins caused a loss of viability of 75% when cultures were exposed to drying for a period of 2 weeks (Battista et al. 2001). Also in Caenorhabditis elegans when the expression of LEA genes is silenced by RNA interference, there is a decrease in survival during induction of desiccation, osmotic and heat stress in this nematode (Gal et al. 2004).
The expression levels of lea1 during the encystment process were higher than in vegetative cells; moreover, lea2 was not highly induced on cysts or during the encystment process. Similarly to what we observed in A. vinelandii, a differential expression of LEA transcripts during development and cyst formation of A. franciscana was reported (Denekamp et al. 2010). Moreover, in C. elegans LEA genes, transcription is induced during dehydration at the stress-resistant juvenile stage (Gal et al. 2004).
This study also showed that avLEA1 but not avLEA2 conferred A. vinelandii resistance to osmotic and high-temperature condition stresses, suggesting that the difference in the number of 20-mers and/or the size in these two LEA proteins may cause a significant difference in their protective activity of subcellular structures and membranes. However, there are no reports that correlate the number of 20-mers present in a protein and their protective effects.
AEIV and OP A. vinelandii reference strains were isolated from cold soils from Wisconsin, USA, and from Trondheim, Norway, respectively. BLAST using the 20-mer of the avLEA1 or avLEA2 revealed the presence (in addition to Azotobacter croccocum and A. vinelandii) of genes encoding putative LEA proteins having the eukaryotic sub-motif in two other species of bacteria, Methylocystis parvus and Anaeromyxobacter sp. (Fig. 1). Interestingly, M. parvus belongs to Methylocystaceae taxa, associated with cold soils (Wunderlin et al. 2016), and both M. parvus and Anaeromyxobacter have been reported to be able to form cysts.
In general, the denaturation and dysfunction of enzymes or different proteins occur when cells are subjected to low- or high-temperature stress or dehydration (Goyal et al. 2005; Reyes et al. 2008). LEA proteins have been proposed to act as “molecular shields” (Goyal et al. 2005; Wise and Tunnacliffe 2004) avoiding the aggregation of proteins from irreversible interactions, protecting functions or enzyme activity. In the present study, the avLEA1 protein was shown to protect in vitro LDH activity against the inactivation induced by freeze thaw cycles; after 2 and 10 freeze thaw cycles, LDH activity remained twice that when LDH was freeze-thawed without the purified avLEA1. In Chlorella vulgaris, LEA proteins were found to protect a freeze-labile enzyme against freeze inactivation (Honjoh et al. 2000). Thus, we hypothesised that avLEA1 protein would play an important role in protecting the proteins and enzymes of A. vinelandii cells in vivo under high-temperature stress and in vitro when subjected to freeze thaw cycles.
Finally, the functional expression of LEA proteins in vivo in E. coli under abiotic stresses has been extensively investigated. One LEA4 gene from Brassica napus was overexpressed in E. coli and shown to confer salt and extreme temperature tolerance to this bacterium (Dalal et al. 2009). More recently, the heterologous expression of an atypical hydrophobic LEA protein from Oryza sativa L. in E. coli improved resistance against diverse abiotic stresses: high salinity, osmotic, freezing, heat and UV radiation (He et al. 2012). This study showed that overexpression of avLEA1 protein improved the tolerance of E. coli to NaCl osmotic stress; however, we did not observe protection of E. coli to high temperatures by avLEA1 protein. This unexpected result is at present difficult to explain, since other LEAs expressed in E. coli confer tolerance to high temperatures. One explanation is that the avLEA1 folding needed for the protective activity occurs at high temperatures in A. vinelandii but not in E. coli.
In conclusion, we demonstrated that avLEA1 protein is a key determinant on the tolerance of A. vinelandii towards abiotic stress such as desiccation, osmolarity and high temperatures since the survival of a lea1 mutant was severely impaired when the organism was exposed to stress conditions. This is the first report on the functional properties of an LEA protein in A. vinelandii.
Electronic supplementary material
Schematic representation of the plasmids constructed in this study, and the integration of plasmid pLEA11012 into the chromosome of strain lea1-lea2. Small arrows represent oligonucleotides used for the construction of the plasmids, and to confirm the presence of the mutations in the strains constructed (GIF 13 kb).
In silico analysis of avLEA1 and avLEA2. (a), Hydrophilic pattern of avLEA1 and avLEA2 using Kyte-Doolittle hydropathy plots; regions above a hydropathy score of zero are hydrophobic. (b), Prediction of protein disorder of avLEA1 and avLEA2 using web server PONDR and default parameters (GIF 120 kb).
SDS-PAGE and Immunoblotting analysis of avLEA1. Protein samples were analysed by SDS-PAGE (a), lane M Molecular mass marker; lane 1, soluble supernatant from the E. coli BL(DE3) /pavLEA1 without IPTG; or induced with 1 mM IPTG, lane 2; lane 3 purified recombinant avLEA1 protein. (b), Detection of avLEA1 protein containing 6xHis-Tag by Immunoblotting. (GIF 26 kb).
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-017-0781-1) contains supplementary material, which is available to authorized users.
References
- Alexeyev MF, Shokolenko IN, Croughan TP. Improved antibiotic-resistance gene cassettes and omega elements for Escherichia coli vector construction and in vitro deletion/insertion mutagenesis. Gene. 1995;160(1):63–67. doi: 10.1016/0378-1119(95)00108-I. [DOI] [PubMed] [Google Scholar]
- Baker J, Van DennSteele C, Dure L., 3rd Sequence and characterization of 6 LEA proteins and their genes from cotton. Plant Mol Biol. 1988;11(3):277–291. doi: 10.1007/BF00027385. [DOI] [PubMed] [Google Scholar]
- Bali A, Blanco G, Hill S, Kennedy C. Excretion of ammonium by a nifL mutant of Azotobacter vinelandii fixing nitrogen. Appl Environ Microbiol. 1992;58(5):1711–1718. doi: 10.1128/aem.58.5.1711-1718.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148(1):6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battista JR, Park MJ, McLemore AE. Inactivation of two homologues of proteins presumed to be involved in the desiccation tolerance of plants sensitizes Deinococcus radiodurans R1 to desiccation. Cryobiology. 2001;43(2):133–139. doi: 10.1006/cryo.2001.2357. [DOI] [PubMed] [Google Scholar]
- Boucher V, Buitink J, Lin X, Boudet J, Hoekstra FA, Hundertmark M, Renard D, Leprince O. MtPM25 is an atypical hydrophobic late embryogenesis-abundant protein that dissociates cold and desiccation-aggregated proteins. Plant Cell Environ. 2010;33(3):418–430. doi: 10.1111/j.1365-3040.2009.02093.x. [DOI] [PubMed] [Google Scholar]
- Bray EA, Bailey-Serres J, Weretilnyk E. Responses to abiotic stress. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and molecular biology of plants. Waldorf: American Society of Plant Biologists; 2000. pp. 1158–1203. [Google Scholar]
- Browne J, Tunnacliffe A, Burnell A. Anhydrobiosis: plant desiccation gene found in a nematode. Nature. 2002;416(6876):38. doi: 10.1038/416038a. [DOI] [PubMed] [Google Scholar]
- Campos F, Cuevas-Velazquez C, Fares MA, Reyes JL, Covarrubias AA. Group 1 LEA proteins, an ancestral plant protein group, are also present in other eukaryotes, and in the archaea and bacteria domains. Mol Gen Genomics. 2013;288(10):503–517. doi: 10.1007/s00438-013-0768-2. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, O’Dell SJ. Ice formation during freezing of Artemia cysts of variable water contents. Mol Physiol. 1981;1:145–152. [Google Scholar]
- Dalal M, Tayal D, Chinnusamy V, Bansai KC. Abiotic stress and ABA-inducible group 4 LAE from Brassica napus plays a key role in salt and drought tolerance. J Biotechnology. 2009;139:137–145. doi: 10.1016/j.jbiotec.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Denekamp NY, Reinhardt R, Kube M, Lubzens E. Late embryogenesis abundant (LEA) proteins in nondesiccated, encysted, and diapausing embryos of rotifers. Biol Reprod. 2010;82(4):714–724. doi: 10.1095/biolreprod.109.081091. [DOI] [PubMed] [Google Scholar]
- Dure L., 3rd A repeating 11-mer amino acid motif and plant desiccation. Plant J. 1993;3(3):363–369. doi: 10.1046/j.1365-313X.1993.t01-19-00999.x. [DOI] [PubMed] [Google Scholar]
- Dure L, 3rd, Crouch M, Harada J, Ho TH, Mundy J, Quatrano R, Thomas T, Sung ZR. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol. 1989;12(5):475–486. doi: 10.1007/BF00036962. [DOI] [PubMed] [Google Scholar]
- Gal TZ, Glazer I, Koltai H. An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett. 2004;577(1–2):21–26. doi: 10.1016/j.febslet.2004.09.049. [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem. 2000;275(8):5668–5674. doi: 10.1074/jbc.275.8.5668. [DOI] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Tunnacliffe A. LEA proteins prevent protein aggregation due to water stress. Biochem J. 2005;388(1):151–157. doi: 10.1042/BJ20041931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166(4):557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hand SC, Menze MA, Toner M, Boswell L, Moore D. LEA proteins during water stress: not just for plants anymore. Annu Rev Physiol. 2011;73(1):115–134. doi: 10.1146/annurev-physiol-012110-142203. [DOI] [PubMed] [Google Scholar]
- Hatanaka R, Hagiwara-Komoda Y, Furuki T, Kanamori Y, Fujita M, Cornette R, Sakurai M, Okuda T, Kikawada T. An abundant LEA protein in the anhydrobiotic midge, PvLEA4, acts as a molecular shield by limiting growth of aggregating protein particles. Insect Biochem Mol Biol. 2013;43(11):1055–1067. doi: 10.1016/j.ibmb.2013.08.004. [DOI] [PubMed] [Google Scholar]
- He S, Tan L, Hu Z, Chen G, Wang G, Hu T. Molecular characterization and functional analysis by heterologous expression in E. coli under diverse abiotic stresses for OslLEA5, the atypical hydrophobic LEA protein from Oryza sativa L. Mol Gen Genomics. 2012;287:39–54. doi: 10.1007/s00438-011-0660-x. [DOI] [PubMed] [Google Scholar]
- Hengherr S, Schill RO, Clegg JS. Mechanisms associated with cellular desiccation tolerance in the animal extremophile Artemia. Physiol Biochem Zool. 2011;84(3):249–257. doi: 10.1086/659314. [DOI] [PubMed] [Google Scholar]
- Honjoh KI, Matsumoto H, Shimizu H, Ooyama K, Tanaka K, Oda Y, Takata R, Joh T, Suga K, Miyamoto T, Lio M, Hatano S. Biosci Biotechnol Biochem. 2000;64(8):1656–1663. doi: 10.1271/bbb.64.1656. [DOI] [PubMed] [Google Scholar]
- Kennedy C, Gamal R, Hummprey R, Ramos J, Brigle K, Dean D. The nifH, nifM, and nifN genes of Azotobacter vinelandii: characterization by Tn5 mutagenesis and isolation from pLARF1gene bank. Mol Gen Genet. 1986;205(2):318–325. doi: 10.1007/BF00430445. [DOI] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lin LP, Sadoff HL. Encystment and polymer production by Azotobacter vinelandii in the presence of beta-hydroxybutyrate. J Bacteriol. 1968;95(6):2336–2343. doi: 10.1128/jb.95.6.2336-2343.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo J, Cocotl-Yanez M, Tzontecomani T, Martinez VM, Bustillos R, Velasquez C. Post-transcriptional regulation of the alginate biosynthetic gene algD by the Gac/Rsm system in Azotobacter vinelandii. J Mol Microbiol Biotechnol. 2011;21:147–159. doi: 10.1159/000334244. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in molecular genetics. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Muriel-Millán LF, Moreno S, Romero Y, Bedoya-Pérez LP, Castañeda M, Segura D, Espín G. The unphosphorylated EIIANtr protein represses the synthesis of alkylresorcinols in Azotobacter vinelandii. PLoS One. 2015;10(2):e0117184. doi: 10.1371/journal.pone.0117184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K, Okawa K, Kakizaki T, Inaba T. Evaluation of the protective activities of a late embryogenesis abundant (LEA) related protein, Cor15am, during various stresses in vitro. Biosci Biotechnol Biochem. 2008;72(6):1642–1645. doi: 10.1271/bbb.80214. [DOI] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Campos F, Reyes JL, Garciarrubio A, Covarrubias AA. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 2010;154(1):373–390. doi: 10.1104/pp.110.158964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Campos F, Wei H, Arora R, Yang Y, Karlson DT, Covarrubias AA. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 2008;31(12):1781–1790. doi: 10.1111/j.1365-3040.2008.01879.x. [DOI] [PubMed] [Google Scholar]
- Romero Y, Moreno S, Guzmán J, Espín G, Segura D. Sigma factor RpoS controls alkylresorcinol synthesis through ArpR, a LysR-type regulatory protein during encystment of Azotobacter vinelandii. J Bacteriol. 2013;195(8):1834–1844. doi: 10.1128/JB.01946-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoff HL, Berke E, Loperfido B. Physiological studies of encystment in Azotobacter vinelandii. J Bacteriol. 1971;105(1):185–189. doi: 10.1128/jb.105.1.185-189.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Setubal JC, dos Santos P, Goldman BS, Ertesvåg H, Espín G, Rubio LM, Valla S, Almeida NF, Balasubramanian D, Cromes L, Curatti L, Du Z, Godsy E, Goodner B, Hellner-Burris K, Hernandez JA, Houmiel K, Imperial J, Kennedy C, Larson TJ, Latreille P, Ligon LS, Lu J, Maerk M, Miller NM, Norton S, O’Carroll IP, Paulsen I, Raulfs EC, Roemer R, Rosser J, Segura D, Slater S, Stricklin SL, Studholme DJ, Sun J, Viana CJ, Wallin E, Wang B, Wheeler C, Zhu H, Dean DR, Dixon R, Wood D. Genome sequence of Azotobacter vinelandii, an obligate aerobe specialized to support diverse anaerobic metabolic processes. J Bacteriol. 2009;191(14):4534–4545. doi: 10.1128/JB.00504-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih MD, Hoekstra FA, Hsing YIC. Late embryogenesis abundant proteins. Adv Bot Res. 2008;48:2011–2055. [Google Scholar]
- Solomon A, Salomon R, Paperna I, Glazer I. Desiccation stress of entomopathogenic nematodes induces the accumulation n of a novel heat-stable protein. Parasitology. 2000;121(Pt 4):409–416. doi: 10.1017/S0031182099006563. [DOI] [PubMed] [Google Scholar]
- Soto A, Allona I, Collada C, Guevar MA, Casado R, Rodriguez-Cerezo E, Aragoncillo C, Gomez L. Heterologous expression of a plant small heat-shock protein enhances Escherichia coli viability under heat and cold stress. Plant Physiol. 1999;120(2):521–528. doi: 10.1104/pp.120.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacy RA, Aalen RB. Identification of sequence homology between the internal hydrophilic repeated motifs of group 1 late-embryogenesis-abundant proteins in plants and hydrophilic repeats of the general stress protein GsiB of Bacillus subtilis. Planta. 1998;206(3):476–478. doi: 10.1007/s004250050424. [DOI] [PubMed] [Google Scholar]
- Sutherland IW. Biosynthesis and composition of gram-negative bacterial extracellular and wall polysaccharides. Annu Rev Microbiol. 1985;39:243–270. doi: 10.1146/annurev.mi.39.100185.001331. [DOI] [PubMed] [Google Scholar]
- Toxopeus J, Warner AH, MacRae TH. Group 1 LEA proteins contribute to the desiccation and freeze tolerance of Artemia franciscana embryos during diapause. Cell Stress Chaperones. 2014;19(6):939–948. doi: 10.1007/s12192-014-0518-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A, Wise MJ. The continuing conundrum of the LEA proteins. Naturwissenschaften. 2007;94(10):791–812. doi: 10.1007/s00114-007-0254-y. [DOI] [PubMed] [Google Scholar]
- Vela GR. Survival of Azotobacter in dry soil. Appl Microbiol. 1974;28(1):77–79. doi: 10.1128/am.28.1.77-79.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ. LEA ping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics. 2003;4:52–70. doi: 10.1186/1471-2105-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise MJ, Tunnacliffe A. POPP the question: what do LEA proteins do? Trends Plant Sci. 2004;9(1):13–17. doi: 10.1016/j.tplants.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Wu G, Zhang H, Sun J, Liu F, Ge X, Chen WH, Yu J, Wang W. Diverse LEA (late embryogenesis abundant) and LEA-like genes and their responses to hypersaline stress in post-diapause embryonic development of Artemia franciscana. Comp Biochem Physiol B Biochem Mol Biol. 2011;160(1):32–39. doi: 10.1016/j.cbpb.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Wunderlin T, Ferrari B, Power M. Global and local scale variation in bacterial community structure of snow from the Swiss and Australian Alps. FEMS Microbiol Ecol. 2016;92(9):1–11. doi: 10.1093/femsec/fiw132. [DOI] [PubMed] [Google Scholar]
- Yancey PH. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol. 2005;208(Pt15):2819–2830. doi: 10.1242/jeb.01730. [DOI] [PubMed] [Google Scholar]
- Zhu J, Hasegawa P, Bressan R. Molecular aspects of osmotic stress in plants. Crit Rev Plant Sci. 1997;16:253–277. doi: 10.1080/07352689709701950. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the plasmids constructed in this study, and the integration of plasmid pLEA11012 into the chromosome of strain lea1-lea2. Small arrows represent oligonucleotides used for the construction of the plasmids, and to confirm the presence of the mutations in the strains constructed (GIF 13 kb).
In silico analysis of avLEA1 and avLEA2. (a), Hydrophilic pattern of avLEA1 and avLEA2 using Kyte-Doolittle hydropathy plots; regions above a hydropathy score of zero are hydrophobic. (b), Prediction of protein disorder of avLEA1 and avLEA2 using web server PONDR and default parameters (GIF 120 kb).
SDS-PAGE and Immunoblotting analysis of avLEA1. Protein samples were analysed by SDS-PAGE (a), lane M Molecular mass marker; lane 1, soluble supernatant from the E. coli BL(DE3) /pavLEA1 without IPTG; or induced with 1 mM IPTG, lane 2; lane 3 purified recombinant avLEA1 protein. (b), Detection of avLEA1 protein containing 6xHis-Tag by Immunoblotting. (GIF 26 kb).