Abstract
The lack of physiological recordings from Caenorhabditis elegans embryos stands in stark contrast to the comprehensive anatomical and gene expression datasets already available. Using light-sheet fluorescence microscopy to address the challenges associated with functional imaging at this developmental stage, we recorded calcium dynamics in muscles and neurons and developed analysis strategies to relate activity and movement. In muscles, we found that the initiation of twitching was associated with a spreading calcium wave in a dorsal muscle bundle. Correlated activity in muscle bundles was linked with early twitching and eventual coordinated movement. To identify neuronal correlates of behavior, we monitored brainwide activity with subcellular resolution and identified a particularly active cell associated with muscle contractions. Finally, imaging neurons of a well-defined adult motor circuit, we found that reversals in the eggshell correlated with calcium transients in AVA interneurons.
Introduction
Spontaneous neural activity plays an important role in the formation and refinement of developing circuits in many parts of the nervous system. The retina has been a particularly powerful model for studying these phenomena because of its highly organized connections and well-defined cell-types with known physiology (1). With an invariant cell lineage and reproducible neuronal wiring diagram, the microscopic roundworm, Caenorhabditis elegans, offers the opportunity for a systems-level view of spontaneous activity during neurodevelopment. Fourteen hours after fertilization, the 222-cell nervous system of newly hatched larvae supports coordinated movement and even learning (2). Cell birth times are known (3) and process outgrowths are now being documented (4, 5, 6, 7), but functional recordings from muscles or neurons of C. elegans embryos have yet to be reported.
Although the nematode’s optically and genetically accessible nervous system is ideal for calcium imaging, the embryo’s small size, sensitivity to phototoxicity, and rapid movements throughout the 50 × 30 × 30 μm3 eggshell volume have traditionally complicated image acquisition. We addressed these issues with light-sheet fluorescence microscopy, where planar illumination and perpendicular detection enable rapid imaging with efficient optical sectioning and minimal photodamage (8). The inverted selective plane illumination microscope (iSPIM (5, 9, 10)) implementation is particularly well suited for C. elegans embryos because of its high spatial resolution (∼0.5 μm laterally, 1.5 μm axially) and compatibility with conventional sample mounting on coverslips. Using open-source control software (Materials and Methods; Fig. S1 in the Supporting Material) for iSPIM acquisition, we acquired dozens of images per s for up to 5 Hz volumetric imaging over several minutes across embryogenesis.
After recording unrestrained samples for functional imaging, considerable effort is still required to extract meaning from the raw four-dimensional datasets. The dynamic fluorescence signal must be segmented from the images, tracked in space and time, and mapped back to relevant anatomical and behavioral features. This has only recently been shown to be feasible for large neuronal populations in freely moving adult C. elegans (11, 12). The embryo poses unique challenges, as there is more movement in the axial dimension and increased postural diversity compared to a worm crawling on an agar surface. Here we address these analysis challenges to document spreading calcium waves in body wall muscles, record brainwide activity, and identify neural correlates of behavior during embryogenesis.
Materials and Methods
Strains
Animals were maintained on nematode growth medium seeded with Escherichia coli (OP50). The following strains were imaged: AQ2953 ljIs131[myo-3p::GCaMP3-SL2-tagRFP-T] (GCaMP in body wall muscles) (13), VG563 unc-13(e312); ljIs131 (from cross of AQ2953 ljIs131 and CB312 unc-13(e312)), ZIM294 mzmEx199[unc-31p::NLSGCaMP5K; unc-122p::GFP] (nuclear-localized GCaMP expressed pan-neuronally) (14), and TQ3032 xuEx1040[nmr-1p::G-CaMP3; nmr-1p::DsRed] (GCaMP in AVA interneurons and others) (15).
Light sheet microscopy
We used a fiber-coupled diSPIM (16) (Applied Scientific Instrumentation, Eugene, OR) to perform all imaging experiments. Laser excitation was coupled to a commercially available diSPIM scanhead. The two-dimensional MEMS mirror internal to the scanhead was used to create a light-sheet in the perpendicular scan direction to define an imaging volume (16). A lower objective (10×, 0.3 NA; Olympus, Melville, NY) was used to find and stage embryos and a pair of perpendicular water-dipping, long-working distance objectives (40×, 0.8 NA, Cat. No. MRD07420; Nikon, Melville, NY) were used to illuminate the sample and to detect the resulting fluorescence. Although this configuration can be used to collect orthogonal views, here we used a single imaging view for all experiments. A 488-nm long-pass filter in the emission arm of the microscope (Cat. No. LP02-488; Semrock, Rochester, NY) was used to eliminate excitation light (∼50–300 μW, measured after the objective) before detection on a scientific complementary metal-oxide-semiconductor camera (pco.edge 4.2 or ORCA-Flash 4.0; Hamamatsu, Hamamatsu City, Japan).
Micro-Manager
For data acquisition and instrument alignment we used the ASI diSPIM plugin within Micro-Manager (aka “μManager”; https://micro-manager.org/). Micro-Manager is a user-friendly, open-source software platform developed for easy integration of microscopy components and data acquisition (17). The diSPIM plugin features a graphical user interface for facile control of all diSPIM hardware, including alignment of the light-sheet with the imaging focal plane (via an automated calibration feature) and managing the acquisition settings (including laser intensity, number of volumes, number of planes/volume, intervolume period, imaging rate; see Fig. S1). This open-source plugin has been distributed with Micro-Manager since 2014 and ongoing development continues. Further documentation on the plugin is available at https://micro-manager.org/wiki/ASIdiSPIM_Plugin and http://dispim.org/software/micro-manager.
Imaging parameters
We acquired iSPIM volumes comprising 30–50 image planes. Exposure time per plane was 2.5–4.5 ms and interplane spacing was 1–1.2 μm. To maximize speed, embryos were oriented on the coverslip to minimize Z-steps (i.e., embryo long axis perpendicular to optical axis) and the camera was rotated to minimize readout time (i.e., embryo long axis perpendicular to camera chip readout direction), at 90° rotation as described in Wu et al. (5) and Kumar et al. (16). We imaged embryos at volumetric rates of up to 5 Hz ([2.5 ms exposure/plane + 1.5 ms camera readout] × 30 planes spanning 36 μm + 50 ms settle time = 170 ms/volume), but the associated phototoxicity (as determined by delayed hatch time) put an upper limit on the duration of these recordings. Depending on the strain and developmental stage, even 30 min of 2 Hz imaging could delay hatching at the low end of laser powers used here (i.e., 50 μW, measured after the objective).
Sample preparation
Embryos were obtained from gravid adults, placed on poly-L-lysine coverslips and imaged in M9 buffer as described in Kumar et al. (16). Before light-sheet imaging, the embryonic stage was determined with bright field illumination using the lower 10× objective.
Analysis: Body-wall muscles: two-fold embryos
Segmenting muscle quadrants. In the software ImageJ (National Institutes of Health, Bethesda, MD), the Simple Neurite Tracer (18) plugin was used to approximate the position of the body wall muscle quadrants in every third image stack (the same coordinates were used for three consecutive stacks). With Simple Neurite Tracer settings of σ = 2 and multiplier = 4, the first fold of each muscle bundle could be delineated with 1–3 paths (i.e., 2–4 clicks along the length of the bundle).
Intensity extraction. A custom MATLAB (The MathWorks, Natick, MA) script was used to define muscle bundle midlines and extract intensities. Owing to the vertical (y) orientation of the embryo, we identified the midline at every pixel of the y dimension. This was done by searching for a local maximum in an x,z plane centered on the Simple Neurite Tracer output coordinate and spanning seven slices in z and 41 pixels in x (i.e., a 7 × 6.7 μm plane). These values are plotted in Fig. 1 c. To calculate correlation coefficients (with corrcoef in MATLAB) between muscle bundles, GCaMP3 intensity was extracted from the bundle cross sections approximately halfway down the first fold.
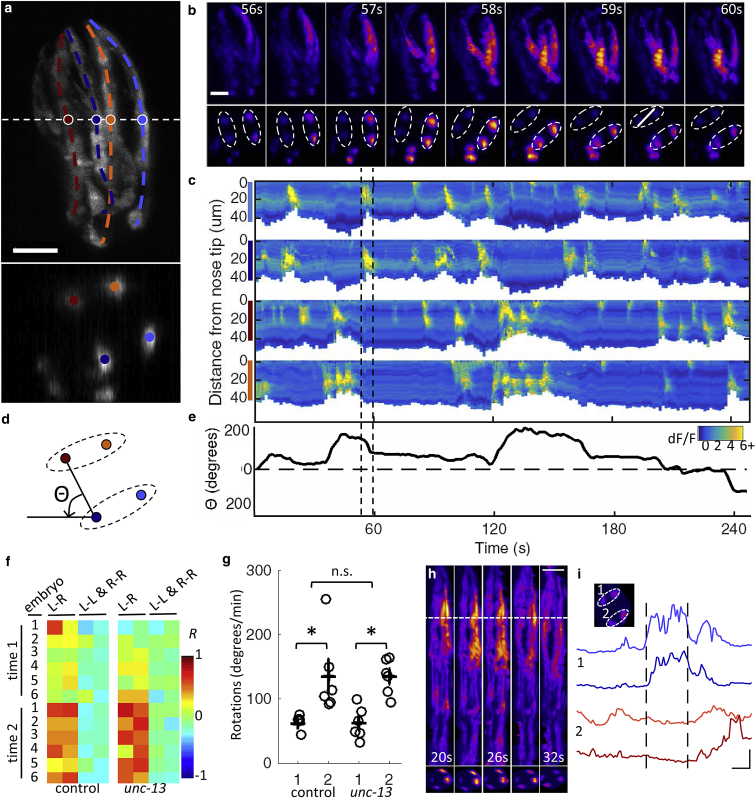
Figure 1.
High speed interrogation of muscular calcium dynamics in the posttwitching embryo. (a) Two-fold embryo expressing GCaMP3 from a myo-3 promoter, as seen in lateral maximum intensity projection (top) and cross section through the volume at dotted white line (bottom). Quadrants are colored identically in (a), (c), and (d). See also Movie S2. (b) Representative projections (top) and cross sections (bottom) at indicated time points, emphasizing spreading calcium waves (top, magenta) and rotation of animal (bottom). Dotted ellipses in bottom row encircle left and right muscle bundles. (c) Fluorescence intensity along each bundle over time. Dashed lines in (c) and (e) denote time window highlighted in (b). (d) Angular coordinate system from which rotation behavior is quantified in (e). (f) Correlation coefficients for fluorescence intensity in left-right and ipsilateral muscle bundle pairs. Each row corresponds to an embryo in one of two imaging sessions (time 1 or time 2). Imaging sessions were 3 min 20 s in duration and separated by 20 min during the 1.75- to two-fold transition. (g) Minimum to maximum angular change/min during time windows 1 and 2. (Open circles) Measurements derived from individual embryos; (black crosses) mean ± SE. (Asterisks and n.s.) Statistically distinguishable (paired t-test, p < 0.05) and indistinguishable (unpaired t-test, p > 0.05) groups, respectively. (h) Representative maximum intensity projections (left) and cross sections (right) at select time points in the untwisted reference frame. See also Movie S4. (i) Fluorescence traces for each bundle at location indicated by dotted white line in (h) (scale bars, 5 s (horizontal) and dF/F = 2 (vertical)). All other scale bars, 10 μm (horizontal). (Inset) Dotted ellipses encircle left and right muscle bundles in the cross section. Volumetric imaging was performed at 2 Hz for (b)–(e) and (h) and (i) and 1 Hz for (f) and (g). The ImageJ Fire Lookup Table was used in (b) and (h). To see this figure in color, go online.
Twitching quantification. In the software ImageJ, cross sections were taken at the embryo midpoint and the resulting image was despeckled (3 × 3 median filter) and smoothed (3 × 3 mean filter) before the use of “Find Maxima…” (with noise tolerance = 20) to locate the muscle bundles. The coordinates of local maxima were tracked across frames using u-track 2.0 (http://www.openmicroscopy.org/site/products/partner/u-track) (19), followed by manual confirmation and linking of partial traces.
Analysis: Three-fold embryos
Untwisting software (20) was used to build a two-dimensional lattice from the position of the GCaMP3-expressing contralateral ventral and dorsal muscle bundles. In the untwisted volumes, bundles were identified and linked across frames, as described for the two-fold embryo cross sections (Fig. 1 h).
Analysis: Pan-neuronal
As outlined below, local maxima were identified in each slice and then consolidated in the stack at putative nuclear centers based on position and intensity (Fig. S4). The marked nuclear centers were used to facilitate the manual tracking of cells across frames.
-
1)
Local maxima were identified in each slice manually using the “Find Maxima…” function of ImageJ (noise tolerance = 50).
-
2)
A custom MATLAB script linked maxima whose x,y positions in adjacent slices were separated by <1.4 μm (i.e., <8 pixels). Nuclear centers were then defined as the intensity peaks through the stack.
-
3)To facilitate cell tracking, segmented nuclear centers were marked as blue dots on the image. Nuclei were then manually linked across frames in ImageJ. After tracking, errors were identified (and subsequently corrected) by examining:
-
i)Extreme motion, as compared to other nuclei linked over that frame (i.e., if the movement was >4 SDs above the mean);
-
ii)Track convergence (i.e., if nuclear separation was <5.6 μm (i.e., <34 pixels); and
-
iii)Inconsistencies between independently derived tracks for the subset of cells with either multiple scorers or multiple attempts from one scorer (i.e., track divergence).
-
i)
A reviewer manually checked all flagged tracks. Intensity was extracted from a 7 × 7 pixel region centered on the nucleus.
Analysis: Command interneurons
Neurons were assigned canonical IDs based on their position relative to other nmr-1 expressing cells (21). AVA and AVE cell bodies were not distinguishable due to their close proximity. AVA/AVE, RIM, and PVC cell bodies were manually tracked in maximum intensity projections and cell centers were identified in stacks by examining intensity profiles through the volumes (Fig. S7). After manual confirmation of cell position, intensities were extracted from a 7 × 7 pixel region centered on the cell body. Forward and backward speed was calculated from AVA movement along the vector linking AVA with posteriorly positioned RIM (see Fig. 3 a).
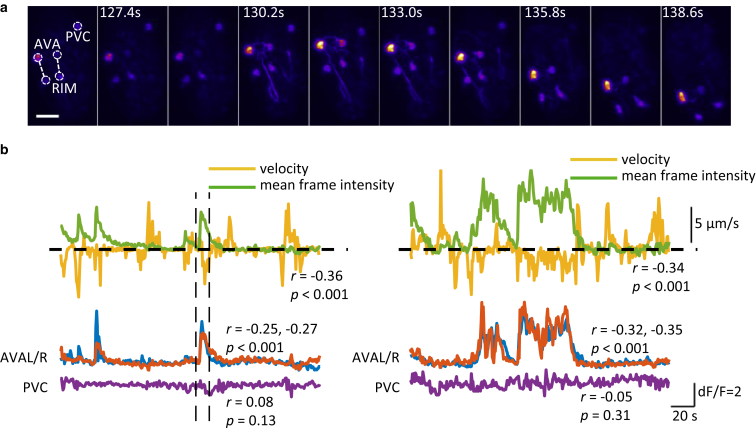
Figure 3.
Linking behavior to calcium activity in command interneurons. (a) (Left) Maximum intensity projections of a late three-fold embryo expressing GCaMP3 from an nmr-1 promoter. Velocity was calculated along the vector linking AVA and RIM. Scale bars, 10 μm (horizontal). The ImageJ Fire Lookup Table was used for display. See also Movie S7. (b) Velocity and mean fluorescence intensity of entire frame (above). Fluorescence intensity traces of tracked AVAL, AVAR, and PVC cell bodies from two embryos (left and right). (Vertical dashed lines) Period depicted in (a). Imaging was performed at 1.4 Hz (volumetric). Correlation coefficients for velocity and intensity are indicated in figure. To see this figure in color, go online.
Neural and muscular activity was reported as a fractional intensity change, dF/F = (F − Fo)/Fo, where F is the intensity value and Fo is the baseline for an individual muscle bundle, nucleus, or cell body, as defined by its lower 20th percentile intensity value.
Results
Embryonic muscle activity
The 14 h embryogenesis of C. elegans consists of two main phases, proliferation and organogenesis. Proliferation ends ∼5.5 h postfertilization at room temperature, at which point the embryo is a spheroid comprising ∼550 essentially undifferentiated cells. During organogenesis, cells terminally differentiate as the embryo elongates from lima-bean shape (∼6 h postfertilization) to two folds (∼8 h postfertilization) to three folds (∼9 h postfertilization). Neurite outgrowth coincides with this elongation. Spontaneous muscle contractions (twitching) begin ∼2 h after the transition from proliferation to organogenesis (∼8 h postfertilization), at the 1.75-fold stage, before the appearance of neuromuscular junctions (22). By this point, the body wall muscles have organized into four longitudinal bundles that extend the length of the animal in left and right, dorsal and ventral quadrants. Early spontaneous muscle twitching is essential for viability, as mutants lacking critical muscle genes fail to elongate from ovoid embryo to worm-shaped larva (23). To characterize the embryo’s first twitch, we imaged a strain expressing GCaMP3 (24) in muscle cells. Embryonic development was monitored with bright field illumination using the lower 10× objective. We commenced image acquisition several minutes before the embryo reached the 1.75-fold stage. Before twitching, we observed localized calcium transients that were not associated with muscle contraction (Movie S1). The first twitch was always a discrete event characterized by a large spreading calcium wave and contraction of one of the bundles (Movie S1). In all of the six embryos tested, the first bundle to contract was dorsally positioned—in two embryos it was dorsal left, and in four embryos it was dorsal right. Some degree of stereotypy in the onset of twitching is perhaps expected, given the invariance of embryogenesis up to this point.
We next investigated activity during early twitching by segmenting and tracking the muscle-localized GCaMP3 intensity. As large contiguous structures, the bundles were fairly easily defined in 1.75- and two-fold embryos with just a few mouse clicks in the Simple Neurite Tracer ImageJ plugin (18) (Materials and Methods). Based on the muscle quadrant coordinates so derived, a custom MATLAB script was used to identify midlines, extract intensity, and link bundles across frames. Examining GCaMP3 intensity shortly after the first twitch, in two-fold embryos, we counted multiple calcium waves per min in all quadrants (Fig. 1, a–c; Fig. S2; Movie S2). In addition to waves, we also observed bursts of fluorescence intensity that were localized within each bundle in both the first and second folds (Fig. S2). Calcium transients were associated with muscle contraction, as revealed by a shortening of the bundle (Fig. 1 c). We were able to link calcium transients to the motion of the animal using axial rotation as a quantitative behavioral metric (Fig. 1 d): the muscle bundles were automatically identified as local maxima from a cross section at the midpoint, and u-track 2.0 (19) with manual editing was used to link them across frames (Materials and Methods). Instances of accelerated angular velocity were associated with correlated events in left and right dorsal or ventral bundles (Fig. 1 e).
To further investigate the developmental time course of early contractions, we staged six 1.75-fold embryos and imaged 3 min 20 s of muscle activity within 10 min of the first twitch and then again 20 min later. GCaMP3 intensity was extracted from the bundle cross sections approximately halfway down the first fold. Postembryonically, left and right muscle bundle pairs are electrically coupled, but ipsilateral dorsal and ventral bundles are not (25). To test for bundle coupling, we calculated the correlation coefficient of fluorescence intensity between bundles. Strong positive correlations were observed between left-right pairs, particularly at the second time point (Fig. 1 f), suggesting the formation of interbundle junctions during the transition from the 1.75- to the two-fold stage. Consistent with a lack of electrical coupling, no significant positive correlations were observed in ipsilateral dorsal or ventral bundles (Fig. 1 f). Although negative correlations between these bundles are reminiscent of the alternating dorso-ventral muscle contractions underlying sinusoidal locomotion postembryonically (13, 26), the requisite neuromuscular junctions are not apparent in electron microscopy at this stage of embryogenesis (22). Consistent with the hypothesis that the earliest twitches are myogenic and not controlled by the nervous system, perturbing synaptic release with a mutant allele of unc-13 (27) did not impair the emergence of correlated activity in left-right muscle bundles (Fig. 1 f). Furthermore, despite severe locomotion deficits posthatching (28), movement of unc-13 mutant embryos was statistically indistinguishable from controls at the 1.75/two-fold stage (Fig. 1 g).
Twitching is initially driven by spontaneous muscle contractions, but hours after the first twitch, during the three-fold stage, neuromuscular junctions appear and movement becomes more coordinated, suggesting some degree of motor control (22). To visualize muscle dynamics in three-fold embryos, we imaged at rates up to 5 Hz (volumetric; Movie S3). Detailed examination of these dynamics is confounded by the convoluted embryo posture. To map calcium traces onto a common coordinate system, we adapted recently developed software for computationally straightening three-fold embryos (6). Although designed for a specific set of markers, body wall muscle fluorescence provided sufficient structure for the software to define a worm shape and untwist it. After untwisting, GCaMP intensity could be extracted from any position along the muscle bundle (Fig. 1, h and i; Movie S4). The ability to track muscle activity throughout embryogenesis provides a window into neurodevelopment, as spontaneous contractions shift to coordinated motor output presumably mediated by the nervous system.
Brainwide calcium imaging at subcellular resolution in a freely behaving embryo
We next attempted to characterize calcium flux in neurons directly. Recent technological advances in microscopy, combined with better fluorescent probes, have enabled near-brainwide calcium imaging in immobilized worms, flies, and flies, and fish (14, 29, 30). This was even recently demonstrated in freely behaving embryonic Drosophila (31) and adult C. elegans (11, 12). Despite the challenges associated with imaging a compact and entangled nervous system with significant movement in the axial dimension, the C. elegans embryo offers certain advantages for pan-neuronal imaging; it has fewer neurons than the adult (222 in the embryo vs. 302 in the adult hermaphrodite) and its behavior is naturally confined within the eggshell—nature’s own microfluidic device. To evaluate global brain dynamics with subcellular resolution in a freely behaving C. elegans embryo, we used a strain expressing nuclear-localized GCaMP5K (32) from a pan-neuronal promoter (14). Nuclear localization is essential for segmenting cells in the densely populated ganglia. Nuclear GCaMP will not report activity compartmentalized in neurites (33), but the large calcium permeable nucleus is at least a good proxy for the cell body (14), where most postembryonic calcium imaging studies have been performed.
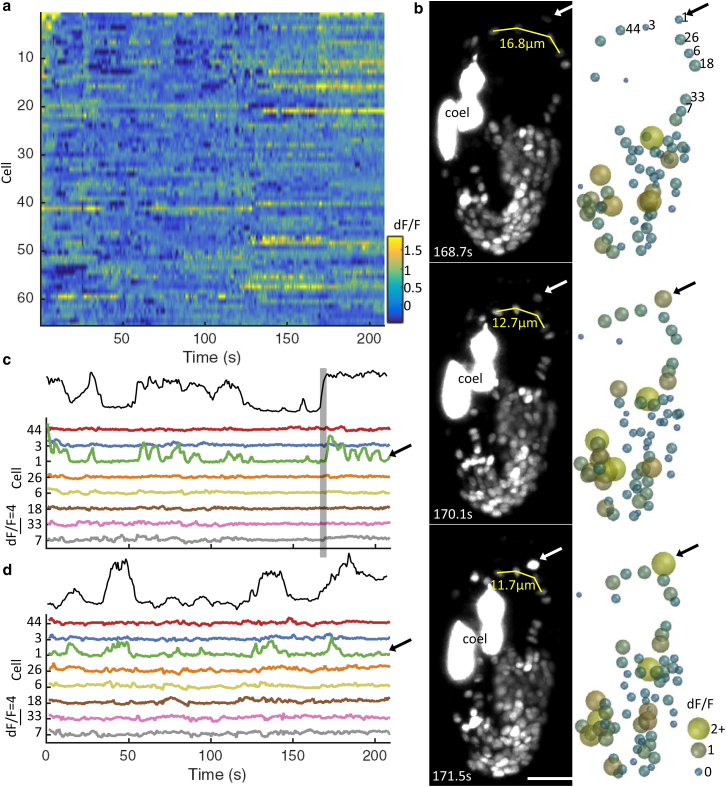
We recorded two 3.5-min sessions of activity in the early and mid three-fold stages, at a 1.4 Hz volumetric rate. To facilitate manual linking across frames, we first labeled nuclei centers based on the position and intensity of local maxima in each slice (Materials and Methods). We tracked 65 cells across all frames of imaging session 1 (Figs. 2 a, S3, and S4; Movie S5). From this dataset, one cell (cell 1 in Fig. 2) stood out as particularly active, with multiple large intensity spikes (Fig. 2 b). Its high level of activity relative to the nearest ventral nerve cord (VNC) motor neurons (positioned along the longitudinal axis of the embryo, i.e., cells 44, 3, 26, and 6) was readily apparent (Fig. 2 c) and suggests that imaging (Fig. S5) or motion artifacts cannot explain the observed intensity fluctuations. We also found that the intensity fluctuations observed in cell 1 were associated with a shortening of the vector connecting the VNC nuclei anterior and posterior to the active cell (Fig. 2, b and c). VNC motor neurons have been shown to have proprioceptive properties (34), but further work is needed to determine if this cell is eliciting and/or detecting the associated muscle contractions. The relative inactivity of other cells may arise from the incomplete representation of neural activity at the cell body at this developmental stage.
Figure 2.
Brainwide neuronal imaging in the three-fold embryo. (a) dF/F traces for neuronal nuclei tracked from an early three-fold embryo expressing nuclear-localized GCaMP5K from a unc-31 promoter. Cells are sorted by degree of correlation with the most active cell (i.e., neuron 1 in a–d). (b) Representative maximum intensity projections (left; coel, GFP-expressing coelomocytes) and corresponding schematic representations (right; color and size of nuclei correspond to dF/F). Scale bars, 10 μm (horizontal). See also Movie S5. (c) 1/length indicated in (b) and dF/F traces for specified neuronal nuclei. Note the correlation between length metric (solid) and activity of neuron 1. (Shaded band) denotes time window highlighted in (b). (d) Time point 2, 2 h after (c). Imaging was performed at 1.4 Hz (volumetric). To see this figure in color, go online.
Canonical cell identities were not known, but positional information could be used to identify the same cells in multiple imaging sessions across embryogenesis. Indeed, although increased movement with development made tracking more difficult (Fig. S6; Movie S6), we were able to follow many of the same cells 2 h later, including the particularly active cell described above. Again we observed that a shortening of the vector connecting its anterior and posterior nuclei correlated with calcium events that were larger and more frequent than in neighboring cells (Fig. 2 d). Although we could not assign a canonical ID to this particularly active cell, we did search other embryos for similarly positioned nuclei with calcium transients correlated with local muscle contractions. We identified one such candidate from five recordings of early three-fold embryos (Fig. S6). Monitoring large populations of neurons, at single-cell resolution, in freely behaving animals is essential for understanding nervous system control of motor output, yet—perhaps due to technical difficulties associated with image acquisition and analysis—such efforts remain scarce. Future work is aimed at strategies to unambiguously identify neurons within these large ensembles (an unsolved problem in general), to enable comparisons among animals and modeling of behavior at the cellular scale (35).
Reversals in the eggshell are associated with AVA activity
Using sparser labels, it is possible to track embryonic calcium activity in groups of cells whose identities are known. Cell ablation, calcium imaging, and optogenetic activation experiments have implicated bilaterally symmetrical AVA as the command interneurons driving reversals in larvae and adults (36, 37, 38). To determine if a similar circuit mediated reversals of the embryo, we tracked calcium dynamics in AVA using a strain driving GCaMP3 from the nmr-1 promoter, which expresses in six neuron classes, including AVA (21). In late-stage three-fold embryos, calcium events were apparent in cell bodies and processes in the nerve ring and ventral nerve cord (Movie S7). Measuring the mean intensity of the entire frame, we observed large fluctuations (dF/F > 0.6) in six of 17 embryos (Fig. S7). To identify the source of the signal and describe embryonic movement, we manually tracked AVA, RIM, and PVC cell bodies in three of these recordings. Movement along the vector linking AVA and RIM was used to calculate the velocity of the head along the longitudinal axis of the worm (Fig. 3 a). In two of the three embryos analyzed, there was a significant negative correlation between instantaneous velocity and mean intensity of that frame (Fig. 3 b). This significant negative correlation was also apparent using intensity derived from either of the AVA cell bodies (Fig. 3 b). However, there was no significant relationship between velocity and intensity derived from a PVC cell body (PVC is a class of interneurons in the tail contributing to stimulated forward movement in adults (36, 39)). Examining the traces, it is clear that the temporal coupling of backward movement and AVA activity in embryos is not as tight as in hatched animals crawling on agar (37, 40, 41). This could be the result of the immature state of the motor circuit or movement constraints imposed by the eggshell. Nonetheless, at some point before hatching, the C. elegans embryo is capable of coordinated behaviors associated with similar circuitry as in the adult.
Discussion
iSPIM allowed us to interrogate calcium flux in freely behaving C. elegans embryos with negligible phototoxicity, at subsecond temporal resolution, and submicron spatial resolution. With these capabilities we linked muscle and neural activity with movement. To study embryos with diverse GCaMP localization, we developed several custom semiautomated analysis pipelines that facilitated segmentation and tracking of dynamic changes in fluorescence intensity and subsequent correlation with behavioral metrics (rotation, movement) that were also derived from the underlying image data. We discovered that twitching initiates in dorsal muscle bundles and we documented the early correlated activity that drives axial rotation. We recorded brainwide calcium dynamics and showed that reversals in the eggshell are associated with activity of the reversal command interneurons, AVAL and AVAR.
Anticipated improvements in instrumentation to enable better optical sectioning (42), higher resolution, increased collection efficiency, and faster acquisition will further facilitate automation of cell segmentation and tracking over longer time periods. Documenting early muscle dynamics and twitches will lead to a better understanding of elongation, while examining the transition to coordinated movement will lead to insights into motor system development. In neurons, functional data will help to define the relationships among activity, process outgrowth, and synaptogenesis. While adult locomotion has been the focus of much research (43, 44, 45), movement of the embryo remains almost completely unexplored. Detailed behavioral characterization of the embryo in the post-twitching regime will help focus future calcium imaging experiments to periods when neural circuits are wiring up and coming online. We anticipate that functional data will add a valuable layer of information to cross reference with the rich anatomical and gene expression datasets already available in C. elegans (46).
Author Contributions
E.L.A. and A.K. conceived the project; E.L.A., A.K., and H.S. designed experiments; E.L.A. and A.K. acquired data; E.L.A., J.M., R.C., R.G., W.D., and H.S. analyzed data; A.K. built the system; J.S.D. and N.S. wrote the μ-Manager diSPIM plugin and tested it with assistance from A.K.; E.L.A. and H.S. wrote the article with input from all authors; and D.C.-R. and H.S. supervised the research.
Acknowledgments
We thank Applied Scientific Instrumentation for lending us a diSPIM frame, Mark Reinhardt and PCO for lending us scientific CMOS cameras, and Lynne Chang and Nikon for lending us objectives. We thank K. Joy, L. Joy, N. Kattapuram, and Y. Oyhama for help with manual tracking; V. Periwal (National Institute of Diabetes and Digestive and Kidney Diseases), J. Hawk (Yale University), and H. Vishwasrao (National Institutes of Health, NIH) for useful discussions regarding calcium imaging; and M. Kittelberger and K. Khodakhah for guidance and leadership in the Grass Lab. Some data were collected as part of the Marine Biological Laboratory Neurobiology course with the assistance of T. Graham and A. Thompson. We also thank the Research Center for Minority Institutions program and the Institute of Neurobiology at the University of Puerto Rico for providing a meeting and brainstorming platform.
Strains were provided by Manuel Zimmer and the Caenorhabditis Genetics Center (funded by NIH Office of Research Infrastructure Program No. P40-OD010440). E.L.A. and A.K. acknowledge support from the Grass Fellowship Program and D.C.-R. and H.S. acknowledge the Whitman Fellowship program at the Marine Biological Laboratory. This work was also supported by the intramural research program of the National Institute of Biomedical Imaging and Bioengineering and NIH grants No. U01-HD075602 and No. R24-OD016474 to D.C.-R. and A.K.
Editor: David Piston.
Footnotes
Seven figures and seven movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30250-3.
Supporting Material
Panels are synchronized to first twitch (at t = 100 s). Scale bar: 10 μm. Imaging was performed at 2 Hz (volumetric).
Lateral maximum intensity projection (top) and cross-sectional view (bottom, corresponding to white line in top panel). Scale bar: 10 μm. Imaging was performed at 2 Hz (volumetric).
Scale bar: 10 μm. Imaging was performed at 5 Hz (volumetric).
Maximum intensity projections before (left) and after (middle) computational untwisting. Yellow line denotes the position of the cross section shown at upper right. Scale bar: 10 μm. Imaging was performed at 2 Hz (volumetric).
Lateral maximum intensity projection (left) and corresponding schematic representations of dF/F (right; color and size scale corresponds to dF/F = 0, 1, and 2+). Scale bar: 10 μm. Imaging was performed at 1.4 Hz (volumetric).
Lateral maximum intensity projection (left) and corresponding schematic representations of dF/F (right; color and size scale corresponds to dF/F = 0, 1, and 2+). Scale bar: 10 μm. Imaging was performed at 1.4 Hz (volumetric).
The ImageJ Fire Lookup Table was used for display. Scale bar: 10 μm. Imaging was performed at 1.4 Hz (volumetric).
References
- 1.Blankenship A.G., Feller M.B. Mechanisms underlying spontaneous patterned activity in developing neural circuits. Nat. Rev. Neurosci. 2010;11:18–29. doi: 10.1038/nrn2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jin X., Pokala N., Bargmann C.I. Distinct circuits for the formation and retrieval of an imprinted olfactory memory. Cell. 2016;164:632–643. doi: 10.1016/j.cell.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sulston J.E., Schierenberg E., Thomson J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 4.Heiman M.G., Shaham S. DEX-1 and DYF-7 establish sensory dendrite length by anchoring dendritic tips during cell migration. Cell. 2009;137:344–355. doi: 10.1016/j.cell.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y., Ghitani A., Shroff H. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2011;108:17708–17713. doi: 10.1073/pnas.1108494108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen R.P., Bokinsky A., Shroff H. Untwisting the Caenorhabditis elegans embryo. eLife. 2015;4:e10070. doi: 10.7554/eLife.10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singhal A., Shaham S. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat. Commun. 2017;8:14100. doi: 10.1038/ncomms14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keller P.J., Ahrens M.B., Freeman J. Light-sheet imaging for systems neuroscience. Nat. Methods. 2015;12:27–29. doi: 10.1038/nmeth.3214. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y., Wawrzusin P., Shroff H. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat. Biotechnol. 2013;31:1032–1038. doi: 10.1038/nbt.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar A., Colon-Ramos D., Shroff H. Watching a roundworm develop with a sheet of light. Phys. Today. 2015;68:58–59. [Google Scholar]
- 11.Nguyen J.P., Shipley F.B., Leifer A.M. Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2016;113:E1074–E1081. doi: 10.1073/pnas.1507110112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkatachalam V., Ji N., Samuel A.D. Pan-neuronal imaging in roaming Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2016;113:E1082–E1088. doi: 10.1073/pnas.1507109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler V.J., Branicky R., Schafer W.R. A consistent muscle activation strategy underlies crawling and swimming in Caenorhabditis elegans. J. R. Soc. Interface. 2015;12:20140963. doi: 10.1098/rsif.2014.0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrödel T., Prevedel R., Vaziri A. Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light. Nat. Methods. 2013;10:1013–1020. doi: 10.1038/nmeth.2637. [DOI] [PubMed] [Google Scholar]
- 15.Piggott B.J., Liu J., Xu X.Z. The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans. Cell. 2011;147:922–933. doi: 10.1016/j.cell.2011.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A., Wu Y., Shroff H. Dual-view plane illumination microscopy for rapid and spatially isotropic imaging. Nat. Protoc. 2014;9:2555–2573. doi: 10.1038/nprot.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edelstein A.D., Tsuchida M.A., Stuurman N. Advanced methods of microscope control using μManager software. J. Biol. Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longair M.H., Baker D.A., Armstrong J.D. Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics. 2011;27:2453–2454. doi: 10.1093/bioinformatics/btr390. [DOI] [PubMed] [Google Scholar]
- 19.Jaqaman K., Loerke D., Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen R., Bokinsky A., Schroff H. An imaging and analysis toolset for the study of Caenorhabditis elegans neurodevelopment. Proc. SPIE. 2015;9334:93340C. [Google Scholar]
- 21.Brockie P.J., Mellem J.E., Maricq A.V. The C. elegans glutamate receptor subunit NMR-1 is required for slow NMDA-activated currents that regulate reversal frequency during locomotion. Neuron. 2001;31:617–630. doi: 10.1016/s0896-6273(01)00394-4. [DOI] [PubMed] [Google Scholar]
- 22.Durbin, R. M. 1987. Studies on the development and organization of the nervous system of Caenorhabditis elegans. Ph.D. thesis, MRC Laboratory of Molecular Biology, Cambridge, UK.
- 23.Williams B.D., Waterston R.H. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian L., Hires S.A., Looger L.L. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat. Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q., Chen B., Wang Z.W. Low conductance gap junctions mediate specific electrical coupling in body-wall muscle cells of Caenorhabditis elegans. J. Biol. Chem. 2006;281:7881–7889. doi: 10.1074/jbc.M512382200. [DOI] [PubMed] [Google Scholar]
- 26.Zhen M., Samuel A.D. C. elegans locomotion: small circuits, complex functions. Curr. Opin. Neurobiol. 2015;33:117–126. doi: 10.1016/j.conb.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 27.Richmond J.E., Davis W.S., Jorgensen E.M. UNC-13 is required for synaptic vesicle fusion in C. elegans. Nat. Neurosci. 1999;2:959–964. doi: 10.1038/14755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lemon W.C., Pulver S.R., Keller P.J. Whole-central nervous system functional imaging in larval Drosophila. Nat. Commun. 2015;6:7924. doi: 10.1038/ncomms8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahrens M.B., Orger M.B., Keller P.J. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat. Methods. 2013;10:413–420. doi: 10.1038/nmeth.2434. [DOI] [PubMed] [Google Scholar]
- 31.Chhetri R.K., Amat F., Keller P.J. Whole-animal functional and developmental imaging with isotropic spatial resolution. Nat. Methods. 2015;12:1171–1178. doi: 10.1038/nmeth.3632. [DOI] [PubMed] [Google Scholar]
- 32.Akerboom J., Chen T.W., Looger L.L. Optimization of a GCaMP calcium indicator for neural activity imaging. J. Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hendricks M., Ha H., Zhang Y. Compartmentalized calcium dynamics in a C. elegans interneuron encode head movement. Nature. 2012;487:99–103. doi: 10.1038/nature11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen Q., Po M.D., Samuel A.D. Proprioceptive coupling within motor neurons drives C. elegans forward locomotion. Neuron. 2012;76:750–761. doi: 10.1016/j.neuron.2012.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato S., Kaplan H.S., Zimmer M. Global brain dynamics embed the motor command sequence of Caenorhabditis elegans. Cell. 2015;163:656–669. doi: 10.1016/j.cell.2015.09.034. [DOI] [PubMed] [Google Scholar]
- 36.Chalfie M., Sulston J.E., Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chronis N., Zimmer M., Bargmann C.I. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat. Methods. 2007;4:727–731. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- 38.Schmitt C., Schultheis C., Gottschalk A. Specific expression of channelrhodopsin-2 in single neurons of Caenorhabditis elegans. PLoS One. 2012;7:e43164. doi: 10.1371/journal.pone.0043164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wicks S.R., Roehrig C.J., Rankin C.H. A dynamic network simulation of the nematode tap withdrawal circuit: predictions concerning synaptic function using behavioral criteria. J. Neurosci. 1996;16:4017–4031. doi: 10.1523/JNEUROSCI.16-12-04017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawano T., Po M.D., Zhen M. An imbalancing act: gap junctions reduce the backward motor circuit activity to bias C. elegans for forward locomotion. Neuron. 2011;72:572–586. doi: 10.1016/j.neuron.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Ben Arous J., Tanizawa Y., Schafer W.R. Automated imaging of neuronal activity in freely behaving Caenorhabditis elegans. J. Neurosci. Methods. 2010;187:229–234. doi: 10.1016/j.jneumeth.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 42.Kumar A., Christensen R., Shroff H. Using stage- and slit-scanning to improve contrast and optical sectioning in dual-view inverted light-sheet microscopy (diSPIM) Biol. Bull. 2016;231:26–39. doi: 10.1086/689589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stephens G.J., Johnson-Kerner B., Ryu W.S. Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput. Biol. 2008;4:e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yemini E., Jucikas T., Schafer W.R. A database of Caenorhabditis elegans behavioral phenotypes. Nat. Methods. 2013;10:877–879. doi: 10.1038/nmeth.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwarz R.F., Branicky R., Brown A.E. Changes in postural syntax characterize sensory modulation and natural variation of C. elegans locomotion. PLoS Comput. Biol. 2015;11:e1004322. doi: 10.1371/journal.pcbi.1004322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santella A., Catena R., Bao Z. WormGUIDES: an interactive single cell developmental atlas and tool for collaborative multidimensional data exploration. BMC Bioinformatics. 2015;16:189. doi: 10.1186/s12859-015-0627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Panels are synchronized to first twitch (at t = 100 s). Scale bar: 10 μm. Imaging was performed at 2 Hz (volumetric).
Lateral maximum intensity projection (top) and cross-sectional view (bottom, corresponding to white line in top panel). Scale bar: 10 μm. Imaging was performed at 2 Hz (volumetric).
Scale bar: 10 μm. Imaging was performed at 5 Hz (volumetric).
Maximum intensity projections before (left) and after (middle) computational untwisting. Yellow line denotes the position of the cross section shown at upper right. Scale bar: 10 μm. Imaging was performed at 2 Hz (volumetric).
Lateral maximum intensity projection (left) and corresponding schematic representations of dF/F (right; color and size scale corresponds to dF/F = 0, 1, and 2+). Scale bar: 10 μm. Imaging was performed at 1.4 Hz (volumetric).
Lateral maximum intensity projection (left) and corresponding schematic representations of dF/F (right; color and size scale corresponds to dF/F = 0, 1, and 2+). Scale bar: 10 μm. Imaging was performed at 1.4 Hz (volumetric).
The ImageJ Fire Lookup Table was used for display. Scale bar: 10 μm. Imaging was performed at 1.4 Hz (volumetric).