Summary
Drought has a serious impact on agriculture worldwide. A plant's ability to adapt to rhizosphere drought stress requires reprogramming of root growth and development. Although physiological studies have documented the root adaption for tolerance to the drought stress, underlying molecular mechanisms is still incomplete, which is essential for crop engineering. Here, we identified OsNAC6‐mediated root structural adaptations, including increased root number and root diameter, which enhanced drought tolerance. Multiyear drought field tests demonstrated that the grain yield of OsNAC6 root‐specific overexpressing transgenic rice lines was less affected by drought stress than were nontransgenic controls. Genome‐wide analyses of loss‐ and gain‐of‐function mutants revealed that OsNAC6 up‐regulates the expression of direct target genes involved in membrane modification, nicotianamine (NA) biosynthesis, glutathione relocation, 3′‐phophoadenosine 5′‐phosphosulphate accumulation and glycosylation, which represent multiple drought tolerance pathways. Moreover, overexpression of NICOTIANAMINE SYNTHASE genes, direct targets of OsNAC6, promoted the accumulation of the metal chelator NA and, consequently, drought tolerance. Collectively, OsNAC6 orchestrates novel molecular drought tolerance mechanisms and has potential for the biotechnological development of high‐yielding crops under water‐limiting conditions.
Keywords: biotechnology, drought, NAC transcription factor, nicotianamine, rice, root
Introduction
Drought is a major environmental factor contributing to loss of crop yield worldwide, and in rice (Oryza sativa), this is due to drought‐induced phenomena such as delayed flowering time, a reduction in the number of spikelets and poor grain filling rate (Ekanayake et al., 1989; O'Toole and Namuco, 1983). Moreover, the proportion of agriculturally important areas with an inadequate water supply has increased substantially as a consequence of global warming and an explosive increase in human population (Mittler, 2006). Thus, the identification of plant drought tolerance mechanisms for deployment in crops is an important objective. To avoid and cope with drought stress, plants have evolved molecular mechanisms that coordinate the expression of suites of genes that protect them from drought‐induced damage, minimize loss of water and modulate their growth and development in arid environments (Shinozaki and Yamaguchi‐Shinozaki, 2007). Most drought‐inducible genes are regulated by drought‐responsive transcription factors (TFs), such as members of the AP2/ERF, MYB, bZIP and NAC families, which directly, or indirectly, regulate drought stress tolerance mechanisms (Abe et al., 2003; Fujita et al., 2004; Kang et al., 2002; Oh et al., 2009; Tran et al., 2004).
The NAC (NAM, ATAF and CUC) superfamily constitutes one of the largest plant‐specific TF families: 117 in Arabidopsis thaliana, 151 in rice, 163 in poplar (Populus trichocarpa) and 152 in both soybean (Glycine max) and tobacco (Nicotiana tabacum; Puranik et al., 2012). NAC TFs are involved in a wide range of abiotic and biotic stress responses. For example, A. thaliana, AtNAC72 (RD29), AtNAC109 and AtNAC55 contribute to drought tolerance by promoting the detoxification of aldehydes in the glyoxalase pathway (Fujita et al., 2004; Tran et al., 2004), while AtNAC2 is involved in responses to salt stress through ethylene and auxin signalling pathways (He et al., 2005). In rice, overexpression of OsNAC9, OsNAC45, OsNAC52 and OsNAC63 enhances tolerance to multiple abiotic stresses via the up‐regulation of genes involved in osmolyte production, detoxification activities, redox homeostasis and the protection of macromolecules (Hu et al., 2006; Redillas et al., 2012).
One key adaptation to drought stress involves changes in root growth and development in response to water‐deficit conditions (Sharp et al., 2004). Roots detect insufficient water availability in soils and release uncharacterized signals to induce resistance and/or adapt their architecture for optimal growth (Sieburth and Lee, 2010). Previous studies showed that rice inbred lines (IR20 × MGL‐2) with long and thick roots exhibit enhanced drought tolerance (Ekanayake et al., 1985). Moreover, overexpression of TaNAC2 and HRD (HARDY) in A. thaliana promotes primary and lateral root growth and thus increasing root numbers (Karaba et al., 2007; Mao et al., 2012), while overexpression of OsNAC5, OsNAC9 and OsNAC10 in rice roots activates radial root growth (Jeong et al., 2010, 2013; Redillas et al., 2012), all of which result in enhanced drought tolerance. Recently, mechanisms involving the phytohormone auxin, regulated by DEEPER ROOTING 1, were shown to confer drought tolerance to rice by altering root growth angle (Uga et al., 2013). Thus, modification of root architecture is closely associated with drought tolerance; however, the underlying molecular mechanisms that confer root‐mediated drought tolerance are not fully understood.
OsNAC6 is previously identified as a key regulator for rice stress responses (Nakashima et al., 2007; Ohnishi et al., 2005). Overexpression rice plants of OsNAC6 show various stress tolerances to drought, high salinity and blast disease. The OsNAC6 acts as a transcriptional activator and up‐regulates stress‐inducible genes including lipoxygenase and peroxidase for stress tolerance (Nakashima et al., 2007), indicating that the OsNAC6 is sufficient to confer stress tolerance in rice plant. Interestingly, the OsNAC6 controls root growth at early vegetative stage through chromatin modification (Chung et al., 2009). It suggests a possible connection between the root structure modifications by OsNAC6 and OsNAC6‐mediated drought tolerance.
In this study, we investigated the molecular mechanisms of OsNAC6‐mediated drought tolerance. Transgenic rice lines overexpressing OsNAC6 under the control of either the root‐specific or the constitutive promoters showed improved drought tolerance, whereas nac6 mutant exhibited drought susceptibility. In addition, multiyear field drought tests confirmed that root‐specific overexpression of OsNAC6 significantly enhanced drought tolerance. We further characterized OsNAC6‐mediated root phenotypes related to drought tolerance. RNA‐seq and ChIP‐seq analyses led to the identification of the direct target genes of OsNAC6, which together constitute the OsNAC6‐mediated drought tolerance pathways.
Results
OsNAC6 overexpression in roots is sufficient to confer drought tolerance
OsNAC6 is a drought‐responsive TF that is also regulated by the abscisic acid as well as by low temperature and salinity stresses (Figure S1; Jeong et al., 2010; Nakashima et al., 2007). To investigate its biological roles, we designed two different constructs for OsNAC6 overexpression in rice (Nipponbare): root‐specific RCc3::OsNAC6 and constitutive GOS2::OsNAC6. To eliminate somaclonal variation, successive field selection of T1–4 plants was performed to identify elite lines that grew normally, without stunting. Six independent homozygous lines (#7, 24 and 38 for RCc3::OsNAC6 and #18, 53 and 62 for GOS2::OsNAC6) were selected for further analysis.
To assess drought resistance, 4‐week‐old OsNAC6 overexpressors (T5 generation) and nontransgenic (NT, Nipponbare) plants were subjected to progressive drought stress by withholding water for 5 days under greenhouse conditions. NT plants showed drought‐associated visual symptoms, such as leaf rolling and wilting earlier than the transgenic plants (Figure 1a). Moreover, after re‐watering, both types of OsNAC6 overexpressors recovered better from the drought stress than the NT plants, which continued to wilt and finally died (Figure 1a). The RCc3::OsNAC6 lines showed high levels of OsNAC6 expression only in roots, while the GOS2::OsNAC6 lines showed high levels of OsNAC6 expression in both leaves and roots (Figure 1b). To independently confirm the conferred drought tolerance, we carried out a leaf chlorophyll fluorescence assay, measuring F v/F m (F v: variable fluorescence and F m: maximum fluorescence), an indicator of photochemical efficiency of photosystem II (PSII), which can be reduced by drought stress. NT leaves exhibited a rapid decrease in F v/F m values as early as 0.5 h after the onset of the drought treatment, while the transgenic leaves showed a delayed decrease in F v/F m values that were ~1.5‐fold higher than those of the NT (Figure 1c), indicating that PS II of the OsNAC6 overexpressors was less affected by drought stress. Notably, OsNAC6 overexpression in roots alone was sufficient to confer drought tolerance during the vegetative stage of growth.
Figure 1.
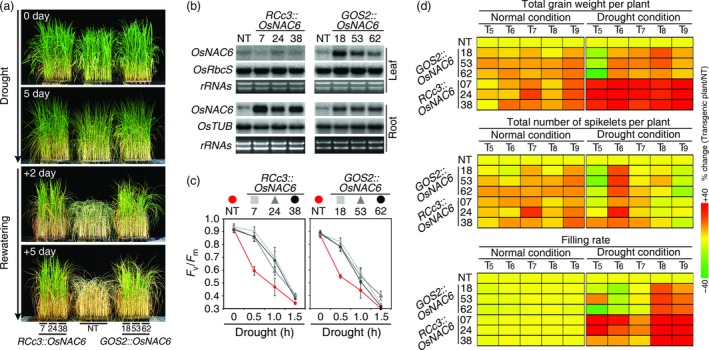
Drought tolerance of RCc3::OsNAC6 and GOS2::OsNAC6 transgenic plants. (a) Drought tolerance of three independent RCc3::OsNAC6 and GOS2::OsNAC6 lines (T5 generation) at a vegetative development stage. Four‐week‐old plants were exposed to drought for 5 days, followed by re‐watering. The number of days on the images indicates the duration of the drought and re‐watering. (b) RNA gel blot analysis using total RNA from leaves and roots of 4‐week‐old RCc3::OsNAC6, GOS2::OsNAC6 and NT plants, grown under normal growth condition. OsRbcS and OsTUB were used as internal controls. (c) Photochemical efficiency test, measuring the leaf chlorophyll fluorescence (F v/F m). Values for each time point represent the mean ± SE of three‐replicate experiments (n = 30 for each genotype). (d) Heatmap of agronomic traits of three independent homozygous lines (from T5 to T9 generation) grown in the field under normal and drought conditions. Mean values for each category (n = 30 for each condition of each line) are listed in Table S1. Change (%) of total grain weight, or total number of spikelets per line in each year, was calculated as (mean value of the agronomic trait per each line/mean value of agronomic trait of NT) × 100. Filling rate (%) was (total filled grain/[total filled grain + total unfilled grain]) × 100.
Multiyear field tests of the OsNAC6 overexpressors
As reproductive development is highly vulnerable to drought stress (Ekanayake et al., 1989; O'Toole and Namuco, 1983), and field tests represent a more informative approach to evaluate effective crop traits under agronomically relevant conditions (Nuccio et al., 2015), we performed drought studies of the OsNAC6 overexpressors in a rice paddy field and focused on the reproductive development over the course of 5 years (T5–9 generation). OsNAC6 overexpressors, along with NT plants, were transplanted in a paddy field in Gunwi, Korea, and grown to maturity. Yield parameters, such as total grain weight, the total number of spikelets and grain filling rate, were scored for 30 plants per transgenic event and for the NT control. Under normal growth conditions, total grain weight increased by 3%–25% in RCc3::OsNAC6 plants and by 3%–18% in GOS2::OsNAC6 compared with NT control plants (Figure 1d; Table S1). This increase in grain weight was mostly caused by an increase in the number of spikelets, rather than an increased filling rate (Figure 1d; Table S1), indicating that overexpression of OsNAC6 in roots affects reproductive development, especially grain yield, under normal growth conditions.
Under drought conditions (plants exposed to intermittent drought stress at the transition stage from vegetative to reproductive development), the total grain weight of the RCc3::OsNAC6 plants was 26%–74% greater than that of the NT controls, whereas GOS2::OsNAC6 plants showed similar values, or only a slight increase (−32% to 22%), compared with NT plants (Figure 1d; Table S1). Given the similar levels of drought tolerance shown by RCc3::OsNAC6 and GOS2::OsNAC6 plants at the vegetative stage, this difference in their total grain weight under drought conditions was unexpectedly large. We determined that this was mainly due to a higher grain filling rate in the RCc3::OsNAC6 plants than in either the NT or the GOS2::OsNAC6 plants under drought conditions (Figure 1d; Table S1). Taken together, these results indicate that root‐specific overexpression of OsNAC6 increases drought tolerance at the reproductive stage of growth under field drought conditions.
OsNAC6 expression in roots controls tiller development
To further investigate the higher spikelet number of the OsNAC6 overexpressors under normal growth conditions, we first grew RCc3::OsNAC6 and GOS2::OsNAC6 plants, together with NT control, until the panicle developmental stage (approximately 3‐month‐old plants) in rice paddy fields, and counted the number of tillers. The OsNAC6 overexpressors showed a slight increase (P > 0.05) in tiller number compared with NT plants (Figure 2a, c). However, as one tiller typically produces approximately 90 spikelets, the small difference in tiller number accounts for the substantially higher spikelet number in the OsNAC6 overexpressing lines. We also evaluated nac6, a null mutant (Hwayoung) that has a T‐DNA insertion in OsNAC6 (Chung et al., 2009), with NT (Hwayoung) plants, grown until the panicle stage in rice paddy fields. nac6 showed reduced grain productivity, mainly due to a reduced number of spikelets and poor grain filling rate (Table S2). In addition, nac6 produced significantly fewer (P < 0.05) tillers than NT plants (Figure 2b; Table S2), with averages over 2 years of 5 and 9, respectively (Figure 2c). This phenotype was rescued in complementation lines (nac6 COM), in which an OsNAC6 genomic region was inserted into the nac6 mutant (Figure S2a; Table S3). To further verify the role of OsNAC6 in tiller development, we grew OsNAC6 overexpressors and nac6 together with NT controls (Nipponbare and Hwayoung) under long‐day growth conditions in a greenhouse, under which rice plants produce more tillers (Figure 2i). NT (Nipponbare) plants had ~40 tillers at the panicle stage, whereas GOS2::OsNAC6 and RCc3::OsNAC6 transgenic lines had ~54 and ~49 tillers, respectively. In addition, NT (Hwayoung) plants produced ~43 tillers, whereas nac6 had ~25. The opposite tiller number phenotype in the OsNAC6 overexpressors compared with nac6 indicates that OsNAC6 regulates tiller development, and the root‐specific overexpression of OsNAC6 is sufficient to promote tiller development.
Figure 2.
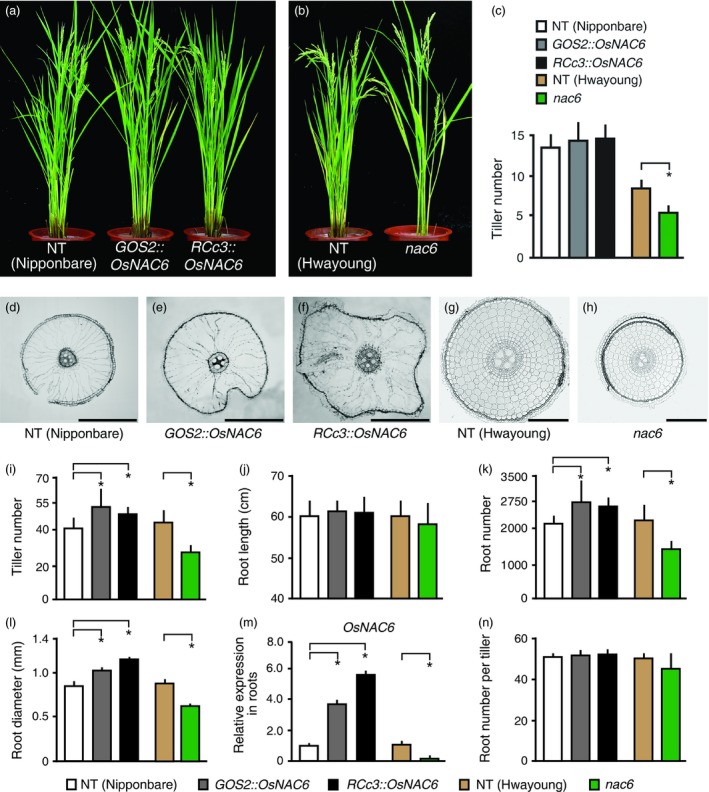
Phenotypes of RCc3::OsNAC6,GOS2::OsNAC6 and nac6 plants. (a–c) Phenotypes of 3‐month‐old OsNAC6 overexpressors and nac6 knockout mutants grown in a rice paddy field. Representative plants were transferred to pots for photographing. (a) OsNAC6 overexpressors. (b) nac6 mutants. (c) Tiller number of 3‐month‐old plants. Values shown are the mean + SD (n = 20 for each genotype). Asterisks indicate significant differences compared to NT control plants (P < 0.05, Student's t‐test). (d–m) Phenotypes of 3‐month‐old OsNAC6 overexpressors and nac6 knockout mutants grown under long‐day conditions in the greenhouse. (d–f) Cross sections of root maturation zones (~30 cm from the root apex). (g, h) Cross sections of roots in the elongation zone (~3 mm from the root apex). (i–m) Quantitative analysis of phenotypes. Values shown are the mean + SD (n = 5 plants for each genotype). (i) Tiller number. (j) Root length. (k) Crown root number. (l) Root diameter (~30 cm from the root apex; 50 roots from five plants for each genotype). (m) qRT‐PCR of OsNAC6 in 2‐week‐old roots. UBIQUITIN 1 expression was used as an internal control. Values shown as the mean + SD of two biological replicates, each of which had two technical replicates. (n) Crown root number per tiller. Asterisks indicate significant differences compared to NT control plants (P < 0.05, Student's t‐test). Scale bars, 500 μm in d–f and 100 μm in g and h.
Root development is regulated by OsNAC6
We next compared the roots of the OsNAC6 overexpressors and nac6 with those of NT controls grown at the panicle stage under long‐day conditions. Root length was similar among all the genotypes (Figure 2j; Figure S2b). However, root number and diameter were significantly different between OsNAC6 overexpressors and NT plants: NT plants had ~2,100 crown roots and an average root diameter of 0.8 mm, whereas the OsNAC6 overexpressors had ~2,600 crown roots and a 1.1 mm root diameter (Figure 2k, l). Conversely, nac6 had ~1,300 crown roots and an average root diameter of 0.8 mm (Figure 2k, l). We noted that large aerenchyma cells in roots of OsNAC6 overexpressors mainly contributed to their wider root diameter (Figure 2d–f) and that cell layers in the cortex regions of nac6 roots were substantially reduced in the elongation zones compared with NT plants (Figure 2g, h). These opposite root phenotypes were caused by expression level of the OsNAC6 in roots of nac6 mutants and OsNAC6 overexpressors (Figure 2m). As root number positively correlates with tiller number (Hockett, 1986), we evaluated the average root number per tiller, which was found to be similar among all the genotypes (Figure 2n). We therefore concluded that the root number variation was caused by OsNAC6 overexpression and the nac6 mutation leads to abnormal tiller development.
OsNAC6 is necessary for rice drought tolerance
To test whether the OsNAC6 is necessary for drought response, 4‐week‐old nac6, nac6 COM and WT (Hwayoung) plants were subjected to progressive drought stress and we monitored drought‐induced visual symptoms (Figure 3a). Leaf rolling and wilting were detected in nac6 earlier than nac6 COM and WT plants (Figure 3a). Moreover, after re‐watering, both nac6 COM and NT plants recovered better from the drought stress than the nac6 mutants, which continued to wilt and finally died (Figure 3a). To independently confirm the drought susceptibility of nac6 mutants, we carried out a leaf chlorophyll fluorescence assay (F v/F m). nac6 leaves exhibited a rapid decrease in F v/F m values at 1 h after the drought treatment, while the nac6 COM and NT leaves showed a delayed decrease in F v/F m values (Figure 3b), indicating that PS II of the nac6 mutants was more damaged by drought stress and nac6 COM plants showed normal drought response compared to WT plants. Collectively, OsNAC6 is necessary to confer rice drought tolerance.
Figure 3.
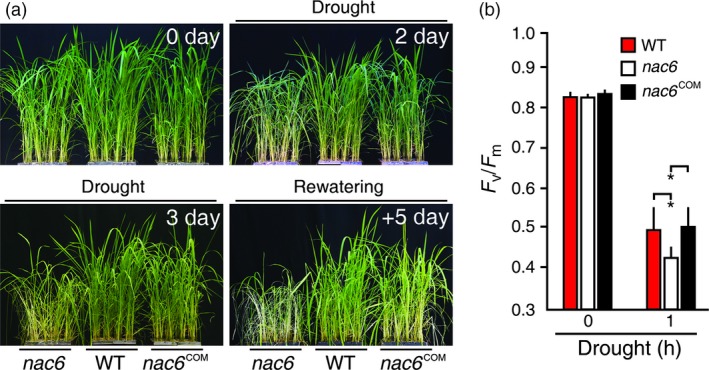
Drought susceptibility of nac6 mutant. (a) Drought response of nac6 and nac6 COM. Four‐week‐old plants were exposed to drought for 5 days, followed by re‐watering. The number of days on the images indicates the duration of the drought and re‐watering. (b) Photochemical efficiency (F v/F m) of 4‐week‐old nac6 and nac6 COM plants. Data are shown as the mean + SD of two replicates experiments (n = 20 for each genotype). Asterisks indicate significant differences compared to WT control plants (P < 0.05 by Student's t‐test).
OsNAC6 is predominantly expressed in the root endodermis, pericycle and phloem
To determine the expression patterns of OsNAC6 in roots, we generated transgenic rice plants (OsNAC6::GUS) harbouring an OsNAC6 promoter region driving expression of the β‐GLUCURONIDASE (GUS) gene. Four‐week‐old OsNAC6::GUS plants showed GUS activity in the root apical meristem that diminished in the root elongation zone (Figure 4a). In addition, in situ hybridization analysis in the elongation zone of crown roots revealed the expression of OsNAC6 in the endodermis and pericycle, as well as the vasculature, where it was predominantly expressed in the phloem (Figure 4b, c). The phloem‐dominant OsNAC6 expression was also observed in the base of shoots (Figure 4d–f). Expression in the endodermis and pericycle is consistent with OsNAC6 influencing root radial growth, while expression in the vasculature suggests an association with long‐distance regulation.
Figure 4.
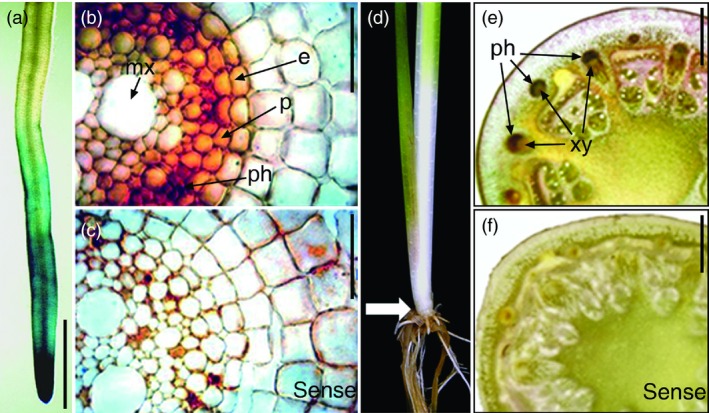
Expression patterns of OsNAC6. (a) Expression pattern in 4‐week‐old OsNAC6::GUS roots. (b, c) In situ hybridization analysis of OsNAC6 in the elongation zone of 3‐month‐old roots. (b) Antisense probe. Signals were detected in the endodermis, pericycle and phloem. (c) Sense probe. (d–f) In situ hybridization analysis of OsNAC6 at the base of 3‐month‐old shoots. (d) Position of a shoot cross section (arrow). (e) Antisense probe. Signals were detected in the phloem. (f) Sense probe. e, endodermis; mx, metaxylem; p, pericycle; ph, phloem; xy, xylem. Scale bars, 2 mm in a, e and f, and 100 μm in b and c.
Identification of OsNAC6‐regulated downstream genes
To identify molecular pathways by which OsNAC6 regulates root development and confers drought tolerance, we performed RNA‐seq analyses of five different roots from 2‐week‐old RCc3::OsNAC6, GOS2::OsNAC6, nac6 and two NT control plants (Nipponbare and Hwayoung). As OsNAC6 functions as a transcriptional activator (Nakashima et al., 2007), candidate target genes regulated by OsNAC6 were identified using the following cut‐off criteria: genes with ≥2‐fold higher expression in RCc3::OsNAC6 or GOS2::OsNAC6 (log2 ratio ≥1.0) than in NT and genes with ≥2‐fold lower expression in nac6 (log2 ratio ≤−1.0) than in NT. Accordingly, a total of 1,825 and 1,294 genes were up‐regulated in RCc3::OsNAC6 and GOS2::OsNAC6 lines, respectively, of which 479 genes were present in both sets (Figure 5a; Figure S3; Table S4). We expanded the analysis by comparing these genes with the 1,715 genes that were down‐regulated in nac6. Based on these RNA‐seq profiles, 51 genes were identified as potential key genes in OsNAC6‐mediated drought tolerance when up‐regulated in roots.
Figure 5.
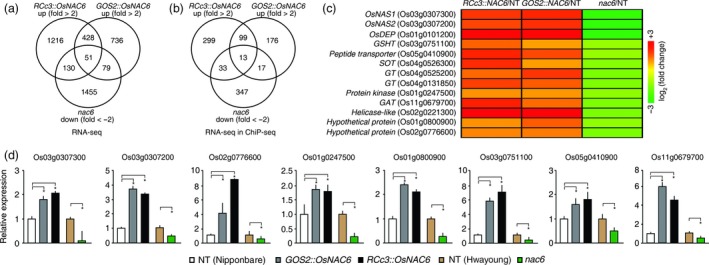
Transcriptomic analysis of RCc3::OsNAC6,GOS2::OsNAC6 and nac6 roots and a comparison between RNA‐seq and ChIP‐seq data. (a) Venn diagram of up‐regulated genes in roots of 2‐week‐old RCc3::OsNAC6 and GOS2::OsNAC6 relative to NT plants (cut‐off, ≥ 2.0‐fold) and down‐regulated genes in roots of nac6 relative to NT plants (cut‐off, ≤−2.0‐fold), using RNA‐seq. (b) Venn diagram showing the number of potential genes directly up‐regulated by OsNAC6 by comparing RNA‐seq with ChIP‐seq data. (c) Thirteen high‐confidence target genes up‐regulated by OsNAC6 and a heatmap of their expression levels based on RNA‐seq data. (d) qRT‐PCR verification of genes up‐regulated by OsNAC6. UBIQUITIN 1 expression was used as an internal control. Values shown as the mean + SD of two biological replicates, each of which had two technical replicates. Asterisks indicate significant differences compared to NT control plants (P < 0.05, Student's t‐test).
Among the 51 genes, putative direct targets were identified by considering the results of a chromatin immunoprecipitation (ChIP‐seq) analysis of roots from transgenic rice plants (RCc3::6xmyc‐OsNAC6; Figure S4). After removing background peaks, a total of 11,969 peaks were found to be associated with rice gene models. Among the 51 up‐regulated genes through the RNA‐seq analyses, 13 were found to be present in the ChIP‐seq profiles (Figure 5b; Table S4). The potential direct target genes included NICOTIANAMINE SYNTHASE 1 and NICOTIANAMINE SYNTHASE 2 (OsNAS1 and OsNAS2), DEHYDRASE‐ENLOASE PHOSPHATASE (OsDEP), GLUTATHIONE TRANSPORTER (GSHT), PEPTIDE TRANSPORTER, SULFOTRANSFERASE (SOT), GLUCOSYLTRANSFERASE (GT), GLYCEROL ACYLTRANSFERASE (GAT) and PROTEIN KINASE (Figure 5c). OsNAS1, OsNAS2 and OsDEP are key regulators of the biosynthesis of nicotianamine (NA), which acts as an Fe chelator and a potential antioxidant (Inoue et al., 2003; Itai et al., 2013; Lee et al., 2009, 2012). GSHT and SOT contribute to antioxidant activity through glutathione relocation and 3′‐phosphoadenosine 5′‐phosphate (PAP) biosynthesis, respectively (Klein and Papenbrock, 2004; Noctor et al., 2010). The accumulation of these antioxidants and PAP has been shown to alleviate drought‐induced oxidative damage (Cheng et al., 2015; Wilson et al., 2009). GAT is involved in membrane modification through lipid metabolism that affects abiotic stress tolerance (Li et al., 2007; Murata et al., 1992), while GT genes control the glycosylation of a structurally diverse range of substrates, including auxin, in association with water stress tolerance (Tognetti et al., 2010; Vogt and Jones, 2000). The expression of the target genes was verified by qRT‐PCR using independently isolated total RNAs from roots of the OsNAC6 overexpressors and nac6 (Figure 5d).
OsNAC6 up‐regulates NA biosynthesis in roots, thereby conferring drought stress tolerance
As OsNAS1, OsNAS2 and OsDEP were found to be direct targets of OsNAC6, we expanded our analysis of the RNA‐seq data to include genes in the whole NA biosynthesis pathway, including the methionine (Met) cycle (Figure 6a; Table S4). This revealed that the expression of OsNAS3, which encodes another enzyme in NA biosynthesis, was also up‐regulated in roots of the OsNAC6 overexpressors and down‐regulated in nac6 roots. Similarly, the expression of METHYLTHIORIBOSE KINASE (OsMTK1 and OsMTK2), OsDEP, METHYLTHIORIBUROSE‐1‐PHOSPHATE ISOMERASE (OsIDI2), ACIREDUCTONE DIOXYGENASE (OsIDI1) and AROMATIC AMINOTRANSFERASE (OsIDI4) genes, which encode key enzymes that generate S‐adenosyl‐Met, a NA precursor in Met cycle (Itai et al., 2013), were all up‐regulated in the OsNAC6 overexpressors and down‐regulated in nac6 (Figure 6a; Table S4). From this, we inferred that high NA accumulation in the roots of OsNAC6 overexpressors might promote root development, leading to drought tolerance.
Figure 6.
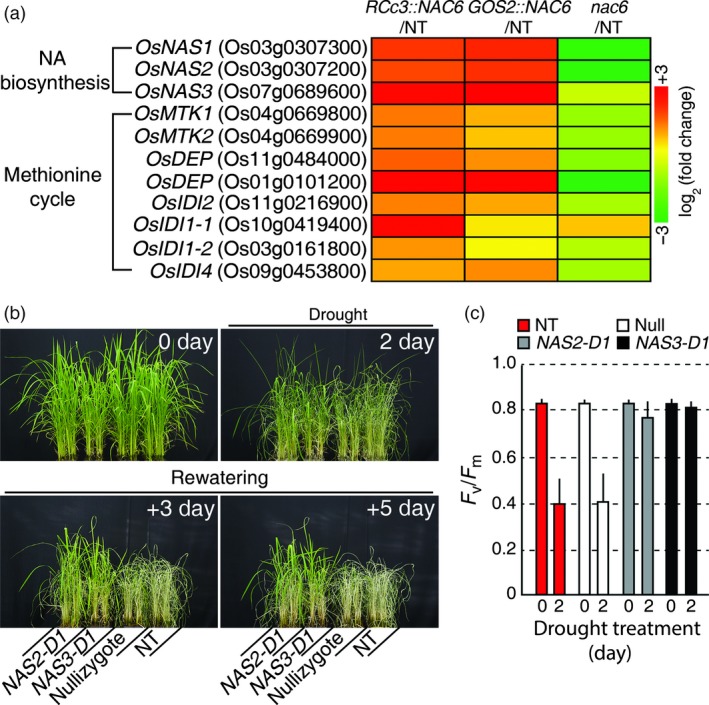
Drought tolerance of OsNAS2‐D1 and OsNAS3‐D1. (a) Expression levels of genes related to NA biosynthesis and the methionine cycle regulated by OsNAC6, based on RNA‐seq data. (b) Drought tolerance of OsNAS2‐D1 and OsNAS3‐D1 plants at a vegetative developmental stage. Four‐week‐old plants were exposed to drought for 5 days, followed by re‐watering. The numbers of days on the images indicate the duration of the drought (2 day means plants that were exposed to drought stress for 2 days) and re‐watering (+3 and +5 day mean plants that were re‐watered for three and 5 days, respectively). (c) Photochemical efficiency (F v/F m) of OsNAS2‐D1 and OsNAS3‐D1 rice plants, grown in a greenhouse at 28–30 °C for 4 weeks and drought treated for 2 days. Each value represents the mean + SE (n = 30 for each genotype).
To determine whether increased NA accumulation in rice plants affects their responses to drought, we exposed two activation‐tagged rice lines, OsNAS2‐D1 and OsNAS3‐D1, which have high levels of NA (Lee et al., 2009, 2012), together with nullizygotes (Null) from OsNAS2‐D1 and NT (Dongjin) plants to drought conditions during vegetative development. NT and Null plants showed drought‐associated visual symptoms, such as leaf rolling and wilting, earlier than OsNAS2‐D1 and OsNAS3‐D1 plants (Figure 6b). After re‐watering, OsNAS2‐D1 and OsNAS3‐D1 recovered, whereas the NT and Null plants continued to wilt and finally died (Figure 6b). To further confirm the drought tolerance, we measured F v/F m of leaves after 2 days of drought stress treatment. NT and Null leaves exhibited a sharp decrease in F v/F m values after 2 days of drought treatment, whereas there was no significant difference in the F v/F m values of leaves from OsNAS2‐D1 and OsNAS3‐D1 plants that were untreated or drought‐treated for 2 days (Figure 6c). These results suggested that OsNAC6‐mediated NA accumulation in roots is sufficient to confer drought tolerance.
Discussion
We demonstrated here that the rice transcription factor OsNAC6 is a regulator of drought tolerance pathways and represents a potentially valuable candidate for genetic engineering of drought‐tolerant high‐yielding crops. Root‐specific (RCc3::OsNAC6) and whole‐body (GOS2::OsNAC6) rice overexpression lines showed enhanced drought tolerance at the vegetative stage. Notably, only RCc3::OsNAC6 lines showed improved drought tolerance at the reproductive stage under field drought conditions. Similar observations were made when OsNAC5, OsNAC9 and OsNAC10 were overexpressed under the control of the RCc3 and GOS2 promoters (Jeong et al., 2010, 2013; Redillas et al., 2012). It is possible that whole‐body overexpression of OsNAC6 and its target genes may perturb reproductive development under drought conditions, resulting in a trade‐off in grain yield. This idea is supported by the observations that both UBIQUITIN promoter‐driven OsNAC6 plants and the stress‐inducible LIP9 promoter‐driven OsNAC6 plants exhibit low reproductive yields (Nakashima et al., 2007). Furthermore, these strong promoter‐driven OsNAC6 plants exhibit growth retardation at 14 days after germination, whereas the overexpression plants show no growth retardation at reproductive stage (Nakashima et al., 2007), indicating that the strong OsNAC6 expression causes vegetative growth retardation. Collectively, these data suggest that root‐specific overexpression of OsNAC6 represents a more effective approach than whole‐body overexpression.
Root structural adaptations to drought stresses were observed in RCc3::OsNAC6 and GOS2::OsNAC6 plants, which both showed higher numbers and a thicker diameter of roots, while the opposite phenotypes were observed in nac6 mutants. Root structural modification for enhanced drought tolerance is associated with root elongation, high number of root and increased radial root growth. Overexpression of TaNAC2 and HRD in A. thaliana or rice activates primary and lateral root elongation, which may promote the acquisition of water in deep soils (Karaba et al., 2007; Mao et al., 2012). Guidance of the roots to water sources in deep soils is regulated by DEEPER ROOTING 1, which modifies root growth angle (Uga et al., 2013). Overexpression of HRD also results in an increase in root number, which may enhance water uptake by increasing the total root surface area in contact with drying soils. Radial root growth, including the formation of larger aerenchyma, also enhances drought tolerance (Jeong et al., 2010, 2013; Redillas et al., 2012). In maize, the formation of root cortical aerenchyma promotes drought tolerance as it reduces the metabolic cost of soil exploration under water stress, permitting greater root growth and water acquisition from drying soil (Zhu et al., 2010). Moreover, the radial root growth maintains plant water potential under drought conditions (Karaba et al., 2007; Price et al., 1997). Taken together, modulation of root architecture by controlling root development can provide an effective strategy to combat to water‐deficit conditions, and our results suggest that such mechanisms occur in rice.
Our studies identified indirect and direct gene targets of OsNAC6. The RNA‐seq analyses revealed 51 up‐regulated genes by OsNAC6. After a comparison of these genes with ChIP‐seq data, we finally identified 13 up‐regulated genes as direct targets of OsNAC6, including OsNAS1, OsNAS2, OsDEP, GSHT, SOT, GT and GAT. These genes are involved in membrane modification, NA biosynthesis, glutathione relocation, PAP accumulation and glycosylation (Figure S5). In a previous study, OsNAC6 was found to directly regulate a peroxidase (AK104277) and a hypothetical protein (AK110725) gene, when analysed using DEX‐treated UBIQUITIN::OsNAC6‐GR plants (Nakashima et al., 2007). However, these genes were not identified through our root‐based RNA‐seq and ChIP‐seq analyses. This discrepancy may be due to differences in the tissues and promoters used in the two studies.
Drought inhibits photosynthesis due to stomatal closure, resulting in the increased production of reactive oxygen species (ROS) and the induction of drought‐mediated oxidative damage (Mittler, 2002). When ROS levels exceed the capacity of a plant to scavenge them, membrane damage can occur, due to the susceptibility of the unsaturated fatty acid components to the effects of ROS (Sharma et al., 2012). This was reported to be alleviated through the overexpression of GAT in Nicotiana tabacum by increasing the unsaturated fatty acid content of membranes, resulting in a stress‐tolerant phenotype (Murata et al., 1992). Similar phenotypes were observed when the A. thaliana glycerol‐3‐phosphate acyltransferase genes GPAT4 and GPAT8 were overexpressed, which altered the accumulation of cutin and suberin (Li et al., 2007). Consistent with these, OsNAC6 overexpression was observed to mediate the GAT up‐regulation and enhance drought tolerance (Figure S5).
To protect themselves from drought‐mediated oxidative stress, plants produce antioxidants or osmoprotectants (Mittler, 2002; Sharma et al., 2012). For example, the antioxidant glutathione scavenges ROS and plays a protective role in abiotic stresses (Cheng et al., 2015). OsNAC6 was found to directly up‐regulate GSHT expression in roots, suggesting that OsNAC6 overexpressors activate glutathione relocation via regulation of GSHT, which may have alleviated drought‐mediated oxidative stress (Figure S5). In addition, sulphur metabolites play important roles in drought tolerance, and among them, PAP produced by SOT is known to accumulate during drought in A. thaliana, thereby contributing to drought tolerance (Chan et al., 2012; Wilson et al., 2009). Expression of SOT was up‐regulated in the OsNAC6 overexpressors, suggesting that OsNAC6 overexpressors might accumulate PAP, giving rise to drought‐tolerant phenotypes (Figure S5).
Drought can also cause an increase in Fe concentration in plants that are producing ROS, resulting in drought‐mediated oxidative damage (Moran et al., 1994; Price and Hendry, 1991). Fe homeostasis is influenced by the Fe chelator nicotianamine (NA), which is produced by the enzymes OsNAS1 and OsNAS2 (Inoue et al., 2003; Lee et al., 2009, 2012). In roots of plants fed with Fe, OsNAS1 and OsNAS2 were reported to be specifically expressed in companion and pericycle cells (Inoue et al., 2003). This expression pattern was similar to that of OsNAC6, suggesting a role for OsNAC6 in a long‐distance Fe transport and homeostasis. Fe is an essential element for plant growth and development, especially through meristem‐specific callose deposition, which regulates cell‐to‐cell communication for root radial growth and normal root stem cell maintenance (Muller et al., 2015). However, excessive amount of Fe is highly toxic (Moran et al., 1994; Price and Hendry, 1991) and can perturb primary root elongation and lateral root formation via interaction with the auxin and ethylene signalling pathways (Giehl et al., 2012; Li et al., 2015; Ward et al., 2008). Despite being identified as one of the up‐regulated genes during drought conditions in A. thaliana, rice and wheat (Ergen et al., 2009; Shaik and Ramakrishna, 2013), the roles of NAS in drought tolerance mechanisms have not been reported to date. NA overproduction by OsNAC6‐mediated up‐regulation of NAS genes may promote the binding of excess Fe, thereby preventing the production of hydroxyl radicals, which consequently confers drought tolerance (Figure S5), indicating a role for NA in drought tolerance. In conclusion, overexpression of OsNAC6 not only improves drought tolerance but also increases grain yield, further indicating its potential importance for crop improvement.
Experimental procedures
Plasmid construction and rice transformation
Total RNA was extracted from 2‐week‐old japonica rice roots (Oryza sativa cv Nipponbare), grown in a greenhouse (16‐h light/8‐h dark cycle), and used to generate total cDNAs, from which the OsNAC6 (Os01g0884300) cDNA was amplified by PCR, using PrimeSTAR HS DNA Polymerase (Takara, Kusatsu, Japan) and the Reverse Transcription System (Promega, Madison, WI), according to the manufacturer's instructions. The primers were forward 5′‐CACCATGAGCGGCGGTCAGGACC‐3′ and reverse 5′‐CTAGAATGGCTTGCCCCAG‐3′. The PCR product was cloned into the entry vector, pENTR/SD (Invitrogen, Carlsbad, CA), and then ligated downstream of 2.2 kb of the GOS2 (Os07g0529800) promoter in the rice transformation vector, p700‐GOS2 (Jeong et al., 2010) for constitutive expression, or 1.3 kb of the RCc3 (Os02g0662000) promoter in the rice transformation vector, p700‐RCc3 (Jeong et al., 2010) for root‐specific expression, using the Gateway System (Invitrogen, Carlsbad, CA). The resulting vectors were named GOS2::OsNAC6 and RCc3::OsNAC6, respectively. To generate OsNAC6::GUS transgenic plants, a 2‐kb promoter region (upstream region of the ATG start codon) of OsNAC6 was amplified using PrimeSTAR HS DNA Polymerase (5′‐CTGCAGTGTGCAAACTTTCAATG TTGAC‐3′ and 5′‐GAATTCCTCTCTCCCCCTTCTCCGGT‐3′) and ligated upstream of the β‐glucuronidase (GUS) reporter gene in the rice transformation vector pCAMBIA1391Z using the EcoR1 and Pst1 restriction sites. Transgenic plants were obtained by Agrobacterium tumefaciens (LBA4404)‐mediated embryogenic callus (Nipponbare) transformation.
For complementation of the nac6 mutant, 5,081 bp of a OsNAC6 genomic fragment, including 2,240 bp of promoter (upstream region of the start ATG), 1,869 bp exons and introns (from ATG to stop codon) and 972 bp 3′ of then untranslated region, were amplified from genomic DNA of 2‐week‐old Nipponbare, using PrimeSTAR HS DNA Polymerase. The amplified genomic fragment was cloned into the rice transformation vector, pSB11 (Komori et al., 2007). Complementation lines were obtained by A. tumefaciens (LBA4404)‐mediated transformation of nac6 (Chung et al., 2009) embryogenic callus.
Stress treatments for RNA gel blot analysis
Rice (Nipponbare) seeds were germinated in soil and grown for 14 days in a greenhouse at 28–30 °C. For the drought treatment, seedlings were air‐dried under continuous light (~1000 μmol/m2/s), and for high‐salinity and ABA treatments, seedlings were transferred to a nutrient solution (Inoue et al., 2003) including 400 mm NaCl or 100 μm ABA, respectively. For low‐temperature treatments, seedlings were placed in a 4 °C cold chamber under continuous light (150 μmol/m2/s). Total RNA was extracted from these samples using TRIzol® reagent (Invitrogen, Carlsbad, CA), and 10 μg from each sample was fractionated on a 1.2% denatured agarose gel and blotted onto a Hybond N+ nylon membrane (Amersham Bioscience, Piscataway, NJ). A radiolabeled OsNAC6 cDNA fragment was used to probe the membrane, corresponding to a 376‐bp fragment of the 3′ untranslated region, which was generated by PCR amplification (5′‐CCTCCTCCAGGACATCCTCA‐3′ and 5′‐CGAATCAATCACCATGTACT‐3′). OsDip1 and OsRbcS probes were used as markers for the stress treatments (Jeong et al., 2013). To determine the OsNAC6 expression patterns and levels in the OsNAC6 overexpressors, total RNA was extracted from the roots and leaves of three homozygous T5 lines of RCc3::OsNAC6 and GOS2::OsNAC6 plants. Ten micrograms of each total RNA sample was used for RNA gel blot analysis, as above.
Drought stress treatment during vegetative development
Transgenic and NT control plants were germinated on Murashige and Skoog (MS) media (Duchefa, Haarlem, Netherlands) at 28 °C for 4 days, and eighteen seedlings of each transgenic lines and NT were transplanted into soil pots (4 × 4 × 6 cm; four plants per pot) and grown for 4 weeks in a greenhouse (16‐h light/8‐h dark cycle) at 28–30 °C. Each pot had the same size of holes in the bottom, and they were all placed in a single tray to synchronize watering. Drought stress was simultaneously applied to all the rice plants by first adding no water to the soil pots for 5 days and then re‐watering. Drought‐induced symptoms were monitored by imaging transgenic and NT plants at the indicated time points using a NEX‐5N camera (Sony, Tokyo, Japan).
Measurement of chlorophyll fluorescence
RCc3::OsNAC6, GOS2::OsNAC6 and NT plants were grown in greenhouse at 28–30 °C for 2 weeks. nac6, nac6 COM, OsNAS2‐D1, OsNAS3‐D1 and NT plants were grown in greenhouse at 28–30 °C for 4 weeks. Thirty leaves from ten seedlings were collected before each stress treatment. Samples were adapted in dark conditions for 10 min. To simulate drought, the leaf discs were air‐dried for the indicated time points at 28 °C. To measure F v/F m values, representing the activity of PSII, a PAM test was carried out with a pulse modulation fluorometer, Mini‐PAM (Walz, Effeltrich, Germany) as described previously (Redillas et al., 2012). The dark‐treated leaf was given a measuring light of 0.15 μmol photon m−2 s−1 for a minimal level of fluorescence and then a 0.8 s actinic light of 10 000 μmol photon m−2 s−1 for a maximal level of fluorescence.
Phenotypic and anatomical analysis of rice roots grown under long‐day conditions
RCc3::OsNAC6, GOS2::OsNAC6, nac6 and NT plants were transplanted to PVC tubes (1.2 m in length and 0.2 m in diameter) that were filled with natural paddy soil and placed into container filled with water. At the panicle development stage, roots were quantified and used for sectioning for anatomical analysis. The diameter of 50 individual roots from RCc3::OsNAC6, GOS2::OsNAC6, nac6 and NT plants (five plants for each genotype) was measured. Internal root anatomy was examined using a Technovit 7100 system, as previously described (Jang et al., 2011) with minor modifications. Technovit saturation was carried out for 3 days. Sections (3 μm) were generated using an ultramicrotome (MTX, RMC, USA), and images were captured with an Olympus DP70 camera mounted on Olympus BX 500 light microscope.
Agronomic trait analysis in rice paddy fields over a 5‐year period
The field experiments, including the use of fertilizers, drought treatments and analysis of agronomic traits, were as described previously (Oh et al., 2009). Briefly, to evaluate yield components of the transgenic plants under normal growth conditions, three independent homozygous lines from T5 (2009) to T9 (2013) for the RCc3::OsNAC6 and GOS2::OsNAC6 lines, together with NT, were planted in a rice paddy field at Gunwi (36˚06΄48.0˝N, 128˚38′38.0˝E), Kyungpook National University, Korea, and grown to maturity. A randomized design was employed for three replicates using three different 10 m2 plots. Yield parameters were scored for 30 plants per line, collected from three different plots. To evaluate the yield components of the transgenic plants under drought field conditions, we built rain‐off shelters to cover rice plants and made a semi‐field condition before drought treatment. Intermittent drought stress was applied twice during the panicle development by draining the water from the bottom of the container. When full leaf rolling was observed in the NT plants after the first drought treatment, they were irrigated overnight and subjected to a second round of drought stress until complete leaf rolling occurred again. After two drought stress treatments, the plants were irrigated until harvesting. Yield parameters were scored for 30 plants per line collected from three different plots corresponding to the drought field conditions. The results from three independent lines were compared with those of the NT controls, using one‐way ANOVA analysis.
Quantitative real‐time PCR analysis
For quantitative real‐time PCR (qRT‐PCR) experiments, a SuperScript™ III Platinum® One‐Step qRT‐PCR System (Invitrogen, Carlsbad, CA) was used to generate first‐strand cDNAs. qRT‐PCR was carried out using a Platinum® SYBR® Green qPCR SuperMix‐UDG (Invitrogen) and a Mx3000p Real‐Time PCR machine (Stratagene, La Jolla, CA). To validate the RNA‐seq data, total RNA was extracted from the roots of 2‐week‐old OsNAC6 overexpressors, nac6 and NT rice seedlings grown under normal growth conditions. Rice UBIQUITIN 1 (Os06g0681400) was used as an internal control, and two biological replicates, each with two technical replicates, were analysed. Gene‐specific primers used for qRT‐PCR are listed in Table S5.
In situ hybridization
Three‐month‐old crown roots were used for in situ hybridization using Technovit resin previously described (Jang et al., 2011), with minor modifications. Briefly, roots were incubated in FAA fixing solution (50% [v/v] ethanol, 5% [v/v] acetic acid and 3.7% [v/v] formaldehyde) for 3 h, then dehydrated and finally embedded with Technovit resin. The sections (3 μm) were made, and OsNAC6 probes were generated by in vitro transcription with a DIG RNA labeling Kit (Roche, Mannheim, Germany) targeting the 376‐bp 3′ untranslated region of the OsNAC6 mRNA, as described above.
RNA‐seq analysis
Total RNA was extracted from the roots of 2‐week‐old OsNAC6 overexpressors, nac6 and NT rice seedlings grown under normal growth conditions, using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. A modified TruSeq method was used to construct a strand‐specific RNA‐seq library with different index primers, and libraries were sequenced using an Illumina HiSeq 2000 system in the National Instrumentation Center of Environmental Management College of Agriculture and Life Science, Seoul National University. Genes were defined as being differentially expressed if their transcript abundance was ≥2‐fold higher in RCc3::OsNAC6 or GOS2::OsNAC6 compared to NT (Nipponbare) or ≥2‐fold lower in nac6 compared to NT (Hwayoung). These data can be found at http://www.ncbi.nlm.nih.gov/geo/ (Accession number: GSE81069).
ChIP‐seq analysis
To produce RCc3::6 × myc‐OsNAC6 plants, the coding sequence of OsNAC6 was amplified using PrimeSTAR DNA polymerase (Takara) with the OsNAC6‐B F‐primer (GGATCCATGAGCGGCGGTCAGGACC) and the OsNAC6‐N R‐primer (GCGGCCGCG CTAGAATGGCTTGCCCCAG). After digestion of the PCR products with BamHI and NotI, the coding sequence was ligated into the multiple cloning site of the pE3n vector (Dubin et al., 2008), which is flanked with a 6 × myc tag coding sequence. Finally, the 6 × myc‐OsNAC6 sequence from the pE3n‐OsNAC6 was subcloned into the p700‐RCc3 vector (Jeong et al., 2010) carrying a 1.3‐kb RCc3 promoter sequence, using the Gateway system. Chromatin immunoprecipitation (ChIP) was performed with roots of 2‐week‐old rice seedlings, as described previously (Chung et al., 2009). These data can be found at http://www.ncbi.nlm.nih.gov/geo/ (Accession number: GSE80986).
Conflict of interest
No conflict of interest declared.
Supporting information
Figure S1 Stress‐inducible and ABA‐dependent expression of OsNAC6. RNA gel‐blot analyses were performed with total RNA from 2‐week old roots and leaves, showing OsNAC6 transcript accumulation patterns in response to drought, high‐salinity, low‐temperature and ABA treatments. The blots were hybridized with an OsDIP1 (DEHYDRATION INDUCIBLE PROTEIN 1) probe as a positive control for various stresses. rRNAs were used to confirm equal loading of RNAs.
Figure S2 Phenotypes of nac6, nac6 COM , RCc3::OsNAC6, and GOS2::OsNAC6 plants. (a) NT, the nac6 knockout mutant, and nac6 COM plants grown in a rice paddy field for ~3 months. Representative plants were transferred to pots for photographing. (b) OsNAC6 overexpressors and nac6 knockout mutants, together with NT plants, were grown in PVC tubes under long‐day conditions in the greenhouse for ~3 months. After removing soils, images were captured using a NEX‐5N camera. Scale bar, 10 cm.
Figure S3 Transcriptomic analysis of RNA‐seq data. Clustering of genes up‐regulated by OsNAC6. Each cluster corresponds to each group described in Figure 5. The indicated scale is the log2 value of the normalized level of gene expression.
Figure S4 Analysis of myc‐OsNAC6 transcripts and myc‐OsNAC6 protein in roots of RCc3::6xmyc‐OsNAC6 transgenic plants. (a) Phenotypes of the NT control (Oryza sativa japonica cv. Ilmi) and RCc3::6xmyc‐OsNAC6 lines at the reproductive stage. (b) Expression level of myc‐OsNAC6 in RCc3::6xmyc‐OsNAC6 lines. UBIQUITIN 1 expression was used as an internal control. Values shown are the mean + SD of three biological replicates, each of which had two technical replicates. (c) Western blot (WB) and immunoprecipitation (IP) analyses of myc‐OsNAC6 in RCc3::6xmyc‐OsNAC6 lines using an anti‐myc Ab.
Figure S5 OsNAC6‐mediated drought tolerance pathways. The drought‐inducible OsNAC6 transcription factor controls target genes that are divided into 5 categories: membrane modification, nicotianamine biosynthesis, glutathione relocation, 3′‐phophoadenosine 5′‐phosphosulfate (PAP) accumulation, and glycosylation. The OsNAC6‐mediated multiple pathways modulate root adaptation to drought stress, and drought tolerance.
Table S1 Agronomic traits of OsNAC6 overexpressors.
Table S2 Agronomic traits of nac6 under normal conditions.
Table S3 Agronomic traits of nac6 complementation lines (nac6 COM) under normal conditions.
Table S5 List of gene specific primers for qRT‐PCR.
Table S4 Genes up‐regulated by OsNAC6 in Figure 5 and Figure S3.
Acknowledgements
We thank the Kyungpook National University for providing rice paddy fields and G. An (Kyung Hee University) for providing the OsNAS2‐D1 and OsNAS3‐D1 mutants. This research was supported by the Rural Development Administration under the Next‐Generation BioGreen 21 Program (PJ011829012016) and by the Basic Science Research Program through the National Research Foundation of Korea (NRF‐2014R1A2A1A11051690, NRF‐2014R1A6A3A04053795 and NRF‐2013R1A6A3A04060627).
References
- Abe, H. , Urao, T. , Ito, T. , Seki, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, K.X. , Wirtz, M. , Phua, S.Y. , Estavillo, G.M. and Pogson, B.J. (2012) Balancing metabolites in drought: the sulfur assimilation conundrum. Trends Plant Sci. 18, 18–29. [DOI] [PubMed] [Google Scholar]
- Cheng, M.C. , Ko, K. , Chang, W.L. , Kuo, W.C. , Chen, G.H. and Lin, T.P. (2015) Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J. 83, 926–939. [DOI] [PubMed] [Google Scholar]
- Chung, P.J. , Kim, Y.S. , Jeong, J.S. , Park, S.H. , Nahm, B.H. and Kim, J.K. (2009) The histone deacetylase OsHDAC1 epigenetically regulates the OsNAC6 gene that control seedling root growth in rice. Plant J. 59, 764–776. [DOI] [PubMed] [Google Scholar]
- Dubin, M.J. , Bowler, C. and Benvenuto, G. (2008) A modified gateway cloning strategy for overexpression tagged proteins in plants. Plant Methods 4, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekanayake, I.J. , O'Toole, J.C. , Garrity, D.P. and Masajo, T.M. (1985) Inheritance of root characters and their relations to drought resistance in rice. Crop Sci. 25, 927–933. [Google Scholar]
- Ekanayake, I.J. , De Datta, S.K. and Steponkus, P.L. (1989) Spikelet sterility and flowering response of rice to water stress at anthesis. Ann. Bot. 63, 257–264. [Google Scholar]
- Ergen, N.Z. , Thimmapuram, J. , Bohnert, H.J. and Budak, H. (2009) Transcriptome pathways unique to dehydration tolerant relatives of modern wheat. Funct. Integr. Genomics 9, 377–396. [DOI] [PubMed] [Google Scholar]
- Fujita, M. , Fujita, Y. , Maruyama, K. , Seki, M. , Hiratsu, K. , Ohme‐Takagi, M. , Tran, L.S. et al (2004) A dehydration‐induced NAC protein, RD26, is involved in a novel ABA‐dependent stress‐signaling pathway. Plant J. 39, 863–876. [DOI] [PubMed] [Google Scholar]
- Giehl, R.F.H. , Lima, J.E. and von Wiren, N. (2012) Localized iron supply triggers lateral root elongation in Arabidopsis by altering the AUX1‐mediated auxin distribution. Plant Cell 24, 33–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X.J. , Mu, R.L. , Cao, W.H. , Zhang, Z.G. , Zhang, J.S. and Chen, S.Y. (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916. [DOI] [PubMed] [Google Scholar]
- Hockett, E.A. (1986) Relationship of adventitious roots and agronomic characteristics in barley. Can. J. Plant Sci. 66, 257–280. [Google Scholar]
- Hu, H. , Dai, M. , Yao, J. , Xiao, B. , Li, X. , Zhang, Q. and Xiong, L. (2006) Overexpressing a NAM, ATAF and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl Acad. Sci. USA 103, 12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, H. , Higuchi, K. , Takahashi, M. , Nakanishi, H. , Mori, S. and Nishizawa, H.K. (2003) Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long‐distance transport of iron and differentially regulated by iron. Plant J. 36, 366–381. [DOI] [PubMed] [Google Scholar]
- Itai, R.N. , Ogo, Y. , Kobayashi, T. , Nakanishi, H. and Nishizawa, N.K. (2013) Rice genes involved in phytosiderophore biosynthesis are synchronously regulated during the early stages of iron deficiency in roots. Rice 6, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, G. , Yi, K. , Pires, N.D. , Menand, B. and Dolan, L. (2011) RSL genes are sufficient for rhizoid system development in early diverging land plants. Development 138, 2273–2281. [DOI] [PubMed] [Google Scholar]
- Jeong, J.S. , Kim, Y.S. , Baek, K.H. , Jung, H. , Ha, S.H. , Do, C.Y. , Kim, M. et al (2010) Root‐specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 153, 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, J.S. , Kim, Y.S. , Redillas, M.C. , Jang, G. , Jung, H. , Bang, S.W. , Choi, Y.D. et al (2013) OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114. [DOI] [PubMed] [Google Scholar]
- Kang, J.Y. , Choi, H.I. , Im, M.Y. and Kim, S.Y. (2002) Arabidopsis basic leucine zipper proteins that mediate stress‐responsive abscisic acid signaling. Plant Cell 14, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaba, A. , Dixit, S. , Greco, R. , Aharoni, A. , Trijatmiko, K.R. , Marsch‐Martinez, N. , Krishnan, K.N. et al (2007) Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proc. Natl Acad. Sci. USA 104, 15270–15275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, M. and Papenbrock, J. (2004) The multi‐protein family of Arabidopsis sulphotransferases and their relatives in other plant species. J. Exp. Bot. 55, 1809–1820. [DOI] [PubMed] [Google Scholar]
- Komori, T. , Imayama, T. , Kato, N. , Ishida, Y. , Ueki, J. and Komari, T. (2007) Current status of binary vectors and superbinary vectors. Plant Physiol. 145, 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Jeon, U.S. , Lee, S.J. , Kim, Y.K. , Persson, D.P. , Husted, S. , Schjorring, J.K. et al (2009) Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl Acad. Sci. USA 106, 22014–22019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S. , Kim, Y.S. , Jeon, U.S. , Kim, Y.K. , Schjorring, J.K. and An, G. (2012) Activation of rice nicotianamine synthase 2 (OsNAS2) enhances iron availability for biofortification. Mol. Cells 33, 269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Beisson, F. , Koo, A.J. , Molina, I. , Pollard, M. and Ohlrogge, J. (2007) Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin‐like monomers. Proc. Natl Acad. Sci. USA 104, 18339–18344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. , Xu, W. , Kronzucker, H.J. and Shi, W. (2015) Ethylene is critical to the maintenance of primary root growth and Fe homeostasis under Fe stress in Arabidopsis. J. Exp. Bot. 66, 2041–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, X. , Zhang, H. , Qian, X. , Li, A. , Zhao, G. and Jing, R. (2012) TaNAC2, a NAC‐type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler, R. (2002) Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Mittler, R. (2006) Abiotic stress, the field environment and stress combination. Trends Plant Sci. 11, 15–19. [DOI] [PubMed] [Google Scholar]
- Moran, J.F. , Becana, M. , Iturbe‐Ormaetxe, I. , Frechilla, S. , Klucas, R.V. and Aparicio‐Tejo, P. (1994) Drought induces oxidative stress in pea plants. Planta 194, 346–352. [Google Scholar]
- Muller, J. , Toev, T. , Heisters, M. , Teller, J. , Moore, K.L. , Hause, G. , Dinesh, D.C. et al (2015) Iron‐dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell 33, 216–230. [DOI] [PubMed] [Google Scholar]
- Murata, N. , Ishizaki‐Nishizawa, O. , Higashi, S. , Hayashi, H. , Tasaka, Y. and Nishida, I. (1992) Genetically engineered alteration in the chilling sensitivity of plants. Nature 356, 710–713. [Google Scholar]
- Nakashima, K. , Tran, L.S. , Van Nguyen, D. , Fujita, M. , Maruyama, K. , Todaka, D. , Ito, Y. et al (2007) Functional analysis of a NAC‐type transcription factor OsNAC6 involved in abiotic and biotic stress‐responsive gene expression in rice. Plant J. 51, 617–630. [DOI] [PubMed] [Google Scholar]
- Noctor, G. , Queval, G. , Mhamdi, A. , Chaouch, S. and Foyer, C.H. (2010) Glutathione. Arabidopsis book 9, e0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio, M.L. , Wu, J. , Mowers, R. , Zhou, H.P. , Meghji, M. , Primavesi, L.F. , Paul, M.J. et al (2015) Expression of trehalose‐6‐phosphate phosphatase in maize ears improves yield in well‐watered and drought conditions. Nat. Biotechnol. 33, 862–869. [DOI] [PubMed] [Google Scholar]
- Oh, S.J. , Kim, Y.S. , Kwon, C.W. , Park, H.K. , Jeong, J.S. and Kim, J.K. (2009) Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 150, 1368–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi, T. , Sugahara, S. , Yamada, T. , Kikuchi, K. , Yoshiba, Y. , Hirano, H.Y. and Tsutsumi, N. (2005) OsNAC6, a member of the NAC gene family, is induced by various stresses in rice. Genes Genet. Syst. 80, 135–139. [DOI] [PubMed] [Google Scholar]
- O'Toole, J.C. and Namuco, O.S. (1983) Role of panicle exsertion in water stress induced sterility. Crop Sci. 23, 1093–1097. [Google Scholar]
- Price, A.H. and Hendry, A.F. (1991) Iron‐catalysed oxygen radical formation and its possible contribution to drought damage in nine native grasses and three cereals. Plant Cell Environ. 14, 477–484. [Google Scholar]
- Price, A.H. , Tomos, A.D. and Virk, D.S. (1997) Genetic dissection of root growth in rice (Oryza sativa L.) I: a hydrophonic screen. Theor. Appl. Genet. 95, 132–142. [Google Scholar]
- Puranik, S. , Sahu, P.P. , Srivastava, P.S. and Prasad, M. (2012) NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. [DOI] [PubMed] [Google Scholar]
- Redillas, M.C. , Jeong, J.S. , Kim, Y.S. , Jung, H. , Bang, S.W. , Choi, Y.D. , Ha, S.H. et al (2012) The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. [DOI] [PubMed] [Google Scholar]
- Shaik, R. and Ramakrishna, W. (2013) Genes and co‐expression modules common to drought and bacterial stress responses in Arabidopsis and rice. PLoS ONE 8, e77261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, P. , Jha, A.B. , Dubey, R.S. and Pessarakli, M. (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot 2012, 1–26. [Google Scholar]
- Sharp, R.E. , Poroyko, V. , Hejlek, J.G. , Spollen, W.G. , Springer, G.K. , Bohnert, H.J. and Nguyen, H.T. (2004) Root growth maintenance during water deficits: physiology to functional genomics. J. Exp. Bot. 55, 2343–2351. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2007) Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. [DOI] [PubMed] [Google Scholar]
- Sieburth, L.E. and Lee, D.K. (2010) BYPASS1: how a tiny mutant tell a big story about root‐to‐shoot signaling. J. Integr. Plant Biol. 52, 77–85. [DOI] [PubMed] [Google Scholar]
- Tognetti, V.B. , Van Aken, O. , Morreel, K. , Vandenbroucke, K. , van de Cotte, B. , De Clercg, I. , Chiwocha, S. et al (2010) Perturbation of indole‐3‐butryric acid homeostasis by the UDP‐glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22, 2660–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, L.S. , Nakashima, K. , Sakuma, Y. , Simpson, S.D. , Fujita, Y. , Maruyama, K. , Fujita, M. et al (2004) Isolation and functional analysis of Arabidopsis stress‐inducible NAC transcription factors that bind to a drought‐responsive cis–element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga, Y. , Sugimoto, K. , Ogawa, S. , Rane, J. , Ishitani, M. , Hara, N. , Kitomi, Y. et al (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 45, 1097–1102. [DOI] [PubMed] [Google Scholar]
- Vogt, T. and Jones, P. (2000) Glycosyltransferases in plant natural product synthesis: characterization of a supergene family. Trends Plant Sci. 5, 380–386. [DOI] [PubMed] [Google Scholar]
- Ward, J.T. , Lahner, B. , Yakubova, E. , Salt, D.E. and Raghothama, K.G. (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 147, 1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, P.B. , Estavillo, G.M. , Field, K.J. , Pornsiriwong, W. , Carroll, A.J. , Howell, K.A. , Woo, N.S. et al (2009) The nucleotidase/phosphatase SAL1 is a negative regulator of drought tolerance in Arabidopsis. Plant J. 58, 299–317. [DOI] [PubMed] [Google Scholar]
- Zhu, J. , Brown, K.M. and Lynch, J.P. (2010) Root cortical aerenchyma improves the drought tolerance of maize (Zea mays L.). Plant Cell Environ. 33, 740–749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Stress‐inducible and ABA‐dependent expression of OsNAC6. RNA gel‐blot analyses were performed with total RNA from 2‐week old roots and leaves, showing OsNAC6 transcript accumulation patterns in response to drought, high‐salinity, low‐temperature and ABA treatments. The blots were hybridized with an OsDIP1 (DEHYDRATION INDUCIBLE PROTEIN 1) probe as a positive control for various stresses. rRNAs were used to confirm equal loading of RNAs.
Figure S2 Phenotypes of nac6, nac6 COM , RCc3::OsNAC6, and GOS2::OsNAC6 plants. (a) NT, the nac6 knockout mutant, and nac6 COM plants grown in a rice paddy field for ~3 months. Representative plants were transferred to pots for photographing. (b) OsNAC6 overexpressors and nac6 knockout mutants, together with NT plants, were grown in PVC tubes under long‐day conditions in the greenhouse for ~3 months. After removing soils, images were captured using a NEX‐5N camera. Scale bar, 10 cm.
Figure S3 Transcriptomic analysis of RNA‐seq data. Clustering of genes up‐regulated by OsNAC6. Each cluster corresponds to each group described in Figure 5. The indicated scale is the log2 value of the normalized level of gene expression.
Figure S4 Analysis of myc‐OsNAC6 transcripts and myc‐OsNAC6 protein in roots of RCc3::6xmyc‐OsNAC6 transgenic plants. (a) Phenotypes of the NT control (Oryza sativa japonica cv. Ilmi) and RCc3::6xmyc‐OsNAC6 lines at the reproductive stage. (b) Expression level of myc‐OsNAC6 in RCc3::6xmyc‐OsNAC6 lines. UBIQUITIN 1 expression was used as an internal control. Values shown are the mean + SD of three biological replicates, each of which had two technical replicates. (c) Western blot (WB) and immunoprecipitation (IP) analyses of myc‐OsNAC6 in RCc3::6xmyc‐OsNAC6 lines using an anti‐myc Ab.
Figure S5 OsNAC6‐mediated drought tolerance pathways. The drought‐inducible OsNAC6 transcription factor controls target genes that are divided into 5 categories: membrane modification, nicotianamine biosynthesis, glutathione relocation, 3′‐phophoadenosine 5′‐phosphosulfate (PAP) accumulation, and glycosylation. The OsNAC6‐mediated multiple pathways modulate root adaptation to drought stress, and drought tolerance.
Table S1 Agronomic traits of OsNAC6 overexpressors.
Table S2 Agronomic traits of nac6 under normal conditions.
Table S3 Agronomic traits of nac6 complementation lines (nac6 COM) under normal conditions.
Table S5 List of gene specific primers for qRT‐PCR.
Table S4 Genes up‐regulated by OsNAC6 in Figure 5 and Figure S3.


