Abstract
Extracellular matrix stiffness sensing by living cells is known to play a major role in a variety of cell mechanobiological processes, such as migration and differentiation. Various membrane and cytoplasmic proteins are involved in transmitting and transducing environmental signals to biochemical cascades. Protein kinases play a key role in regulating the activity of focal adhesion proteins. Recently, an interaction between mitogen-activated protein kinase (MAPK1) and vinculin was experimentally shown to mediate this process. Here, we adopt a molecular modeling approach to further investigate this interaction and its possible regulatory effects. Using a combination of data-driven flexible docking and molecular dynamics simulations guided by previous experimental studies, we predict the structure of the MAPK1-vinculin complex. Furthermore, by comparing the association of MAPK1 with open versus closed vinculin, we demonstrate that MAPK1 exhibits preferential binding toward the open conformation of vinculin, suggesting that the MAPK1-vinculin interaction is conformationally selective. Finally, we demonstrate that changes in the size of the D3-D4 cleft provide a structural basis for the conformational selectivity of the interaction.
Introduction
Focal adhesions are dynamic molecular assemblies that can sense the mechanical and chemical properties of the extracellular matrix (ECM) (1). The focal adhesion machinery serves to connect the cytoskeleton to the ECM and transduce extracellular mechanical stresses into biochemical responses within the cell through the process of mechanotransduction. One focal adhesion protein that plays important roles in mechanotransduction is vinculin, a protein that is critically involved in the regulation of integrin clustering, focal adhesion strength, and traction force generation in mature focal adhesions (2, 3).
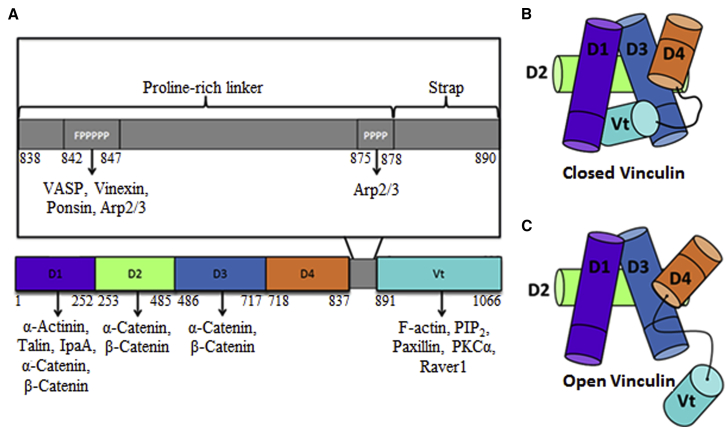
Vinculin is a 1066-amino acid (116 kDa) globular polypeptide that is directly involved in force transmission through focal adhesions and actin cytoskeleton stability (4). Structurally, vinculin consists of four domains, D1, D2, D3, and D4, and a vinculin tail domain Vt (Fig. 1 A). Domains D1–D3 are seven-helix bundles, whereas D4 is a four-helix bundle. The Vt domain is a five-helix bundle, and is connected to the head domain via a flexible proline-rich linker (PRL) and a strap (5). As a scaffolding protein, vinculin functions through its binding to other proteins, which include talin (6, 7), α-actinin (8, 9, 10), IpaA (11), α-catenin (12) and β-catenin (12), VASP (13), ponsin (14), vinexin (15), Arp2/3 (16), F-actin (17), paxillin (18, 19, 20), PIP2 (21, 22), PKCα (23, 24, 25), and Raver1 (26).
Figure 1.
Vinculin structural features, motifs, and binding partners. (A) Given here is a schematic of vinculin outlining the domains, motifs, and major binding partners of vinculin. Above the overall schematic of vinculin, a zoom-in on the proline-rich linker and strap is shown. Two motifs on the Proline-rich linker, the FPPPPP motif and the PPPP motif, are necessary for binding to various vinculin binding partners. The structures of closed vinculin (B) and open vinculin (C) are also depicted above. Interactions among domains D1, D3, and D4 with Vt block the binding sites of vinculin for many of its binding partners, rendering vinculin in an autoinhibited state. Intracellular or extracellular forces promote the release of the Vt domain from the vinculin head domain, inducing vinculin activation and binding to its binding partners. To see this figure in color, go online.
Critical to vinculin’s role in mechanosensing pathways is its interconversion between two conformations: an autoinhibited conformation (Fig. 1 B) and an activated conformation (Fig. 1 C). In its autoinhibited conformation, a high affinity interaction between Vt and head domains D1, D3, and D4 occludes the binding sites of vinculin for many of its binding partners, thereby attenuating vinculin activity (27, 28). Talin, as well as the presence of actomyosin-generated forces, is needed for the conversion of vinculin from the autoinhibited to the activated conformation (29, 30). Talin contains several buried vinculin binding sites (VBSs) that are inaccessible to vinculin. Actomyosin-mediated force application to focal adhesions stretches talin, inducing the exposure of the cryptic VBSs in its rod domain. This in turn leads to vinculin binding to talin, recruiting vinculin to focal adhesions, triggering vinculin activation (29, 31, 32). In the absence of any substrate or actomyosin-generated forces, talin is not stretched, and vinculin is retained in its autoinhibited conformation, which effectively inhibits vinculin activity (29, 33). Through this mechanism, intracellular or extracellular force application promotes the breakage of this autoinhibitory interaction, opening vinculin and activating substrate binding (30).
Mitogen-activated protein kinase 1 (MAPK1) is a serine/threonine kinase that plays critical roles in several cellular processes, including cell proliferation, survival, adhesion, migration, and stem cell differentiation. MAPK1 regulates these activities through its phosphorylation of hundreds of nuclear and cytosolic substrates in the cell, making it a master regulator of cell fate. MAPK1 engages its binding partners through two independent docking sites (34). The first one is the D-site recruitment site (DRS) of MAPK1, which interacts with the D-docking domain of its binding partners. The DRS of MAPK1 is situated opposite of the catalytic cleft of the kinase, and engages a wide variety of MAPK1 substrates and other molecules. The structure of the DRS is not regulated by MAPK1 activation status, and therefore proteins that engage the DRS of MAPK1 typically do not preferentially bind to activated or inhibited MAPK1. Binding partners may also engage through another regulated interface on MAPK1, called the “F-site recruitment site”. This site is located near the catalytic cleft of the kinase. Furthermore, the F-site recruitment site of MAPK1 is regulated by MAPK1 phosphorylation, becoming exposed upon dual phosphorylation (34). MAPK1 is considered to be a master regulator of stem cell differentiation, and responds to several mechanical and biochemical inputs to ultimately influence stem cell fate (35).
Stem cells are well known to differentiate based on the stiffness of the ECM (36). However, the molecular mechanisms through which this mechanotransduction occurs in stem cells remain largely uncharacterized. Recently, Holle et al. (37) identified a potential signaling axis through which this mechanotransduction could occur. They identified a direct interaction between the focal adhesion protein vinculin and the stem cell differentiation regulator MAPK1, and determined that this interaction was critical for the ECM stiffness-induced differentiation of human mesenchymal stem cells (hMSC) to myogenic lineages. They further showed that combined mutation of residues 762–768 of vinculin disrupted its binding to MAPK1. Ultimately, they hypothesized the following model for the molecular basis of stiffness-induced hMSC differentiation: Actomyosin forces generated within stem cells are transmitted through focal adhesions to the ECM, inducing ECM deformation. Stiffer ECM substrates will deform less than softer substrates. The strain induced by actomyosin forces is divided among the focal adhesion components and the ECM. Because stiffer ECM substrates deform less than softer substrates, for the same applied force focal adhesion-associated talin will experience more deformation (33), leading to the increased accessibility of normally cryptic VBSs within the talin rod domain, and thereby inducing greater vinculin activation on stiffer substrates (38). Assuming that MAPK1 exhibits conformational selectivity for vinculin by selectively interacting with open (activated) and not closed vinculin, higher ECM stiffness levels would induce higher levels of MAPK1-vinculin association. Vinculin interaction with MAPK1 then induces MAPK1 phosphorylation and activation through an uncharacterized mechanism. Differing ECM stiffnesses would therefore induce differing levels of MAPK1 activity, and ultimately regulate stem cell differentiation.
In this study, we were interested in understanding the structural basis for the conformational selectivity in binding between vinculin and MAPK1. Using a combination of flexible docking approaches and all-atomic molecular dynamics simulations guided by previous experimental data, we predicted a stable form of the MAPK1-vinculin complex. Furthermore, we analyzed the conformational regulation of this binding interaction, and discussed some potential functional consequences of such binding.
Materials and Methods
Structures
Full-length inactive rat MAPK1 (PDB: 4GT3) (39) was used for our docking and subsequent molecular dynamics experiments. ATP was removed from the structure to simplify modeling. Because the most complete structure of closed vinculin available (human vinculin; PDB: 1TR2) (40) lacked residues P843–P877 of the flexible PRL region, the homology modeling software SWISSMODEL (41) was used to complete the structure of closed vinculin. Additionally, to obtain the structure of full-length open vinculin, steered molecular dynamics on full-length closed vinculin was performed to separate the vinculin tail from the vinculin head domain. The methods used to obtain the structure of full-length open and closed vinculin are described further in Golji and Mofrad (28).
Docking
Flexible protein-protein docking of the full-length structures of inactive MAPK1 and open vinculin was performed using the easy interface of the HADDOCK webserver (42). As inputs for the docking, we specified residues D316, D319, L113, L119, H123, Y126, L155, T157, and T158, which constitute the DRS of MAPK1, as the MAPK1 active site. In addition, based on previous experimental work (37), vinculin residues 762–768 were specified as the vinculin active site. The top solutions from each of the top four generated clusters in HADDOCK were selected for further analysis using molecular dynamics.
Molecular dynamics simulations
To assess the relative strength and stability of each of the MAPK1-vinculin complexes generated by HADDOCK, we performed molecular dynamics simulations on the top solution from each of the top four generated clusters in HADDOCK. These simulations ran for 20 ns. From these four simulations, one structure was selected as the MAPK1-vinculin complex based on its agreement with prior experimental data (37). We then performed two additional 20 ns simulations of this structure using identical parameters, for a total of three independent trials. Additionally, three separate 10 ns simulations of MAPK1 with closed vinculin were performed. These simulations were prepared by aligning the structure of closed vinculin (derived from PDB: 1TR2) onto the generated MAPK1-open vinculin complex, and separating the two molecules in VMD until all steric clashes were removed. All structures were solvated and then ionized at a combined KCl concentration of 0.15 M. Structures were subsequently minimized for 100,000 steps and equilibrated for 0.5 ns using the NAMD molecular dynamics package and CHARMM27 force field (43, 44, 45, 46, 47). All simulations ran at an initial temperature of 310 K using the Nosé-Hoover thermostat (48, 49), and pressure was maintained at 1 atm using the Langevin piston (50). All equilibration and production run simulations were performed using a timestep of 2 fs. For all simulations, electrostatics of the system were determined using the particle mesh Ewald method (51). The van der Waals (VDW) interactions were modeled using a switching function to smoothly reduce the VDW force to zero at the cutoff distance of 1.2 nm. All visualizations and postprocessing of simulations were done with VMD (52).
Results
Stem cells are well known to sense and differentiate to a specific lineage in response to the stiffness of their ECM (36). Various scaffolding and signaling proteins mediate stem cell mechanotransduction, but the molecular mechanisms by which these proteins orchestrate differentiation pathways are not well-understood. Recently, a direct interaction between MAPK1 and vinculin was shown to at least partially mediate the ECM stiffness-induced differentiation of hMSCs to myogenic lineages (37). Furthermore, a predicted MAPK1 binding site on vinculin was mapped to vinculin residues 762–768, which reside on the D4 domain close to Vt.
The MAPK1-vinculin interaction should be conformationally selective in nature to transduce mechanical signals into stem cell differentiation cues (37). In particular, recent experimental observations (37) have inspired us to hypothesize that MAPK1 should preferentially engage with the open conformation of vinculin. To address this hypothesis, two aims were pursued in this study. First, we generated the predicted MAPK1-vinculin complex guided by the results of previous experimental studies. Next, using the structure of MAPK1-vinculin complex, we investigated the conformationally selective nature of the interaction. This task was accomplished by comparing the interaction of MAPK1 with the open and closed forms of vinculin.
Generation of the MAPK1-vinculin complex
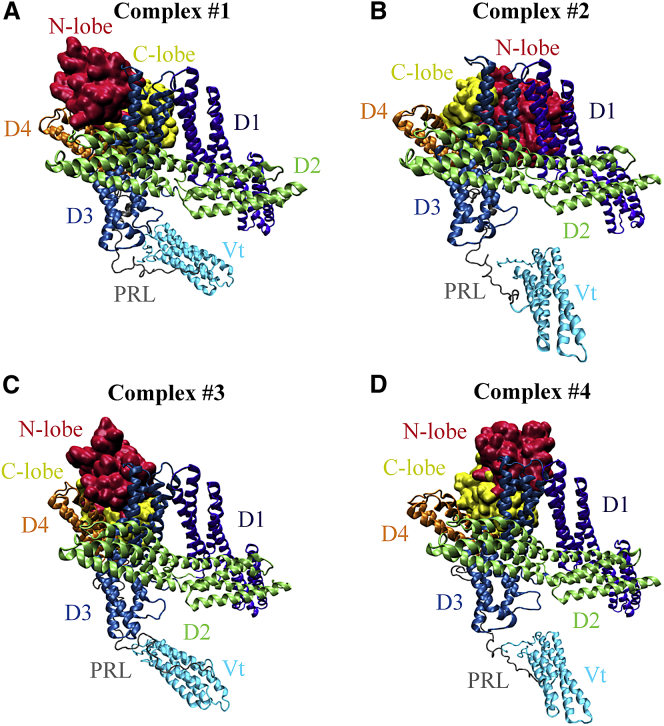
To generate the predicted MAPK1-vinculin complex, we performed flexible protein-protein docking using the HADDOCK webserver (42). Previous experimental work conducted by Holle et al. (37) was utilized to refine the positioning of MAPK1 and vinculin, and to improve the accuracy of the docking result. Previously, they demonstrated that combined mutation of residues 762–768 of the vinculin D4 domain impaired MAPK1 binding. These vinculin residues overlap with residues 758RRANRILLVA767, which constitutes a predicted MAPK1 DRS binding sequence (DRS binding motifs match the sequence (R/K)2-3-X2-6-ΦA-X-ΦB, where X is any amino acid, and Φ is any hydrophobic residue, typically isoleucine or leucine (34)). Therefore, for the initial docking of MAPK1 and vinculin, we specified the DRS of MAPK1 and residues 762–768 of vinculin as inputs for the docking. Representative docking solutions taken from the four top-ranked clusters are shown in Fig. 2, A–D, corresponding to complexes 1–4, respectively. Complexes are numbered from 1 to 4 according to their MAPK1-vinculin binding strength, as determined by the HADDOCK ranking algorithm.
Figure 2.
Results of MAPK1 and vinculin molecular docking using HADDOCK. Potential MAPK1-open vinculin complexes were generated using data-driven molecular docking with HADDOCK. Representative structures were taken from each of the top four ranked HADDOCK clusters, and are shown here. Complexes 1–4 are shown in (A)–(D), respectively. Unphosphorylated MAPK1 is shown in surface representation, and open vinculin is shown in ribbon representation. The N-lobe (red) and C-lobe (yellow) of MAPK1, as well as the D1 (violet), D2 (lime), D3 (blue), D4 (orange), PRL (gray), and Vt (cyan) domains of vinculin are separately colored. To see this figure in color, go online.
To confirm that complex 1 is the most favorable vinculin-MAPK1 complex, we applied another ranking approach. We ran 20 ns molecular dynamics simulations for each of the complexes shown. To obtain a plausible complex, we determined which complex most closely matched the previously reported experimentally predicted MAPK1 binding site on vinculin, residues 762–768 of D4. This was performed by selecting the complex that had the largest number of residues of the MAPK binding site engaged. Shown in Table 1 is a listing of each of the MAPK1 and vinculin residues that are at the MAPK1-vinculin interface of each complex.
Table 1.
List of Binding Interface Residues on MAPK1 and Vinculin for Each Simulated Complex
| Complex | Binding Interface Residues on MAPK1 | Binding Interface Residues on Vinculina | Number of Vinculin Residues Common to the Reported Binding Site |
|---|---|---|---|
| 1 | 10–14, 27–30, 36, 39, 43–48, 73–79, 81–86, 103–108, 112–117, 126, 133, 156–160, 162, 313–316, 318–319, 352, 354–356 | 600–609, 611–612, 615–619, 629, 700, 703, 707, 736, 746–747, 750–752, 754–755, 758, 761, 762, 764–765, 768, 771–772, 780, 784, 788, 791–792, 794–796, 799–800, 802, 803, 805 | 4/7 |
| 2 | 41, 44–48, 77, 85, 107, 122–123, 126, 156–160, 312–318, 324 | 603, 607, 654, 656–657, 660, 703, 707–708, 711–712, 715, 768, 769, 771–775, 780, 784, 788, 792, 795–796, 798–799, 802 | 1/7 |
| 3 | 9–14, 19–21, 24–29, 39, 41, 44–48, 77–79, 81–85, 104–105, 107, 122–123, 126, 156–160, 162, 313–316 | 604–609, 611–612, 615–620, 629, 747, 750–751, 754, 761, 764–765, 768, 781, 784, 788, 791–792, 794–796, 799–800, 802–805, 813 | 3/7 |
| 4 | 11–14, 25–28, 41, 43, 46, 48, 107, 122, 157, 279, 313 | 608, 611, 614–620, 689, 692–693, 696, 780, 784, 788, 792 | 0/7 |
Vinculin residues within the experimentally reported binding site (vinculin residues 762–768) are presented in bold.
As seen in Table 1, vinculin residues 761, 762, 764, 765, and 768 were engaged with MAPK1 in complex 1, vinculin residues 768 and 769 were engaged in complex 2, vinculin residues 764, 765, and 768 were engaged with MAPK1 in complex 3, and none of vinculin residues 762–768 were engaged with MAPK1 in complex 4. Therefore, we proposed complex 1 as the most accurate docking solution, and restricted further analyses to this complex.
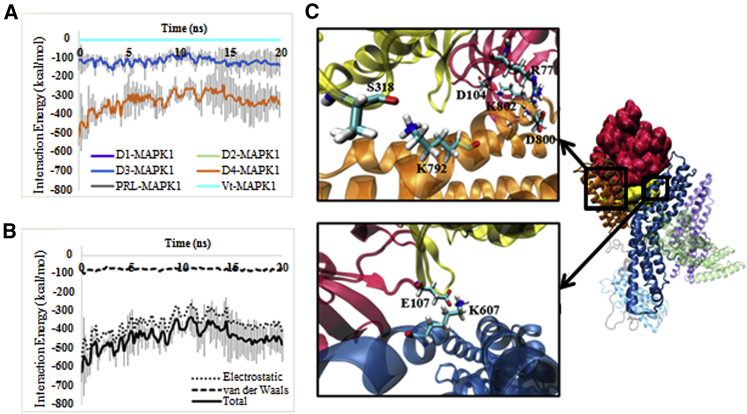
The properties of the proposed MAPK1-vinculin complex are described in further detail in Fig. 3. As seen in Fig. 3 A, the vinculin-MAPK1 interface is relatively strong, with a total interaction energy of ∼450 kcal/mol. This interface is primarily electrostatic in nature. Furthermore, MAPK1 interacts exclusively with domains D3 and D4 of vinculin (Fig. 3 B).
Figure 3.
Characterization of the MAPK1-open vinculin complex. (A) Shown here is the interaction energy between MAPK1 and individual vinculin domains. D1-MAPK1, D2-MAPK1, and PRL-MAPK1 energy plots overlap with the Vt-MAPK1 energy plot at 0 kcal/mol, and thus are not visible on the plot. MAPK1 interaction is exclusively restricted to D3 and D4 of vinculin. (B) Shown here is the interaction energy between MAPK1 and open vinculin, separated into electrostatic, VDW, and total (electrostatic + VDW) energies. For clarity purposes, error bars are only shown for the “Total” energy plot. MAPK1 interacts with vinculin largely through electrostatic contacts. (C) Shown here are the molecular details of the MAPK1-vinculin complex. Interacting residues are labeled and shown in licorice representation. The MAPK1 N-lobe (red) and C-lobe (yellow), as well as the D1 (violet), D2 (lime), D3 (blue), D4 (orange), PRL (gray), and Vt (cyan) domains of vinculin are separately colored. To see this figure in color, go online.
It should be noted that the reported binding energies throughout this article are a measure of binding strength. These are the nonbonded interaction energies calculated by the CHARMM27 force-field function consisting of VDWs and electrostatic terms. Particularly, in this study, the relative binding strengths between molecules are compared after the entropic barriers caused by water molecules and ions were passed. The structure of the proposed MAPK1-vinculin complex is shown in Fig. 3 C. In this structure, MAPK1 engages vinculin head domains D3 and D4 through four primary electrostatic interactions: an interaction between K792 of vinculin D4 and S318 of MAPK1, between D800 of vinculin D4 and R77 of MAPK1, between K802 of vinculin D4 and D104 of MAPK1, and between K607 of vinculin D3 and E107 of MAPK1. In this structure, the N-lobe of MAPK1 is situated within the D3–D4 cleft, and engages both D3 and D4 of vinculin, whereas the C-lobe is largely pointing away from the vinculin head domain.
MAPK1 selectively engages the open conformation of vinculin
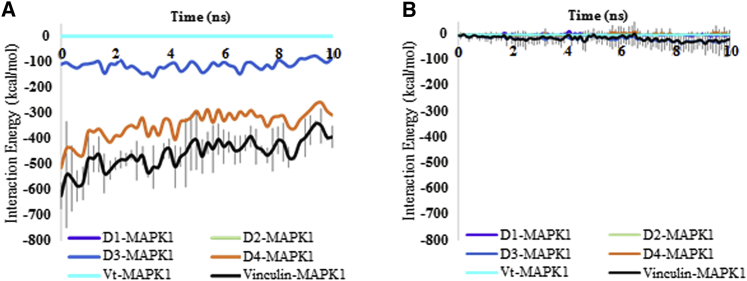
To test our hypothesis that MAPK1 preferentially engages the open conformation of vinculin, we performed three 10 ns simulations of MAPK1 with closed vinculin, using identical parameters for each of the three trials. To position closed vinculin and MAPK1 in our simulations, we superimposed the structure of closed vinculin onto open vinculin from our preexisting simulation of open vinculin in complex with MAPK1. We then separated the closed vinculin and MAPK1 molecules by 2.5 nm, the minimum distance that relieved structural overlap between closed vinculin and MAPK1, and ran the simulation. Shown in Fig. 4, A and B, is a comparison of the interaction energies between open vinculin with MAPK1 and closed vinculin with MAPK1, respectively.
Figure 4.
The MAPK1-vinculin signaling complex is mechanosensitive. The results of the (A) MAPK1-open vinculin and (B) MAPK1-closed vinculin simulations are compared. Shown are energy plots of the D1-MAPK1, D2-MAPK1, D3-MAPK1, D4-MAPK1, PRL-MAPK1, Vt-MAPK1, and full-length vinculin-MAPK1 interaction energies. For the sake of clarity, error bars are only shown for the full-length vinculin-MAPK1 energy plots. Several of these energy plots overlap at 0 kcal/mol, and thus are not visible on the figure. These results demonstrate a clear difference in the strength of interaction between MAPK1 and closed versus open vinculin. To see this figure in color, go online.
Interestingly, we observed dramatic differences in the interaction energy between MAPK1 with the closed and open conformations of vinculin. In our simulations, open vinculin engaged MAPK1 with an energy of ∼−450 kcal/mol, whereas closed vinculin was found to engage MAPK1 with an energy of ∼−25 kcal/mol. Furthermore, we observed no engagement between MAPK1 and D4 of closed vinculin (Fig. 4, A and B), and interactions between MAPK1 and closed vinculin were almost entirely restricted to D3. In contrast, MAPK1 in our open vinculin simulation was seen to engage both D3 (∼−120 kcal/mol) and D4 (∼−325 kcal/mol). Together, these results support our hypothesis that the interaction between MAPK1 and vinculin is conformationally sensitive.
Changes in the D3–D4 cleft size underlie MAPK1-vinculin conformational selectivity
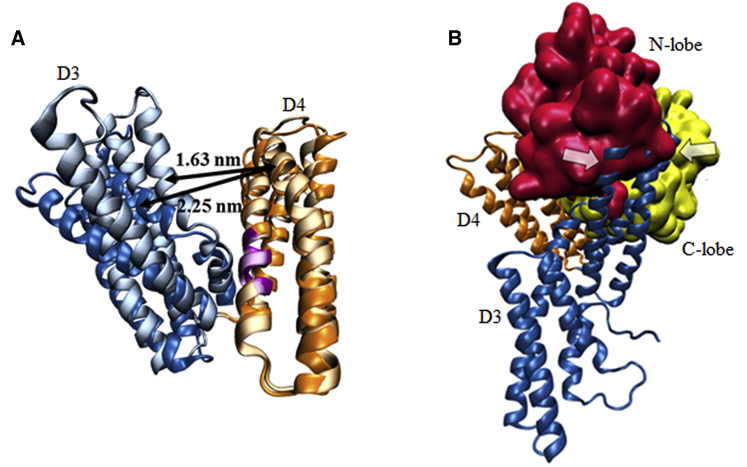
To gain further insight into the nature of the MAPK1-vinculin interaction, we analyzed the structural basis for the conformational selectivity of complex formation. According to our docking analysis, MAPK1 binds between domains D3 and D4 of open vinculin. Therefore, we hypothesized that D3–D4 interdomain rearrangements may occur upon vinculin inactivation, and may restrict MAPK1 from binding strongly to inactive vinculin. To test this, we measured the size of the D3–D4 cleft in closed vinculin and MAPK1-bound open vinculin (Fig. 5 A). This was done by measuring the distance between the arbitrarily chosen residues 607 of D3 and 754 of D4. As shown in Fig. 5 A, the D3–D4 cleft size differs significantly between open and closed vinculin, with a cleft size of 1.63 nm in closed vinculin and 2.25 nm in open vinculin. To determine whether this difference was sufficient to prevent MAPK1 binding to closed vinculin, we superimposed the structure of the MAPK1-open vinculin complex onto closed vinculin. As depicted in Fig. 5 B, when the structures are superimposed, MAPK1 clashes with closed vinculin (clashing region is indicated with arrows in Fig. 5 B), indicating that the MAPK1 binding interface present on open vinculin is sterically inaccessible on closed vinculin. Our results support the mechanoselectivity of the MAPK1-vinculin interaction, and suggest that opening of the D3–D4 cleft upon vinculin activation permits MAPK1 engagement of vinculin. Taken together, these results provide an important insight into the molecular basis for the conformational selectivity of the MAPK1-vinculin interaction.
Figure 5.
Changes in vinculin D3–D4 cleft size underlie mechanosensitivity. (A) Closed vinculin was superimposed onto the structure of open vinculin. Closed vinculin domains D3 and D4 are shown in light blue and light orange, respectively, and open vinculin domains D3 and D4 are shown in dark blue and dark orange, respectively. Closed vinculin residues 762–768 are highlighted in light purple, and open vinculin residues 762–768 are highlighted in dark purple. D3–D4 cleft size is shown, and was calculated by measuring the distance between residue 607 of D3 with 754 of D4. D3–D4 cleft size is considerably smaller in closed vinculin. (B) Closed vinculin was superimposed onto open vinculin from the structure of the open vinculin-MAPK1 complex. MAPK1 is shown in surface representation, and vinculin is shown in ribbon representation. The N-lobe (red) and C-lobe (yellow) of MAPK1, as well as the D3 (blue) and D4 (orange) domains of vinculin are shown. Indicated by arrows are the areas of overlap between MAPK1 from the MAPK1-open vinculin structure and closed vinculin. This indicates that the MAPK1 binding interface present on open vinculin is inaccessible to closed vinculin. To see this figure in color, go online.
Discussion
ECM stiffness cues play a major role in directing stem cell differentiation. Recently, a direct interaction between MAPK1 and vinculin was shown to mediate this process in hMSCs. In this study, we investigated the conformationally selective nature of the MAPK1-vinculin signaling axis. Previously, Holle et al. (37) demonstrated that an interaction between MAPK1 and vinculin regulates stem cell differentiation. They demonstrated that vinculin knockdown impaired stiffness-induced expression of the muscle transcription factor MyoD in hMSCs. Residues 762–768 of vinculin were shown to be important for interaction with MAPK1, as the combined mutation of these residues largely reduced stiffness-induced MyoD. Furthermore, vinculin knockdown impaired stiffness-induced MAPK1 phosphorylation, indicating that vinculin may mediate MAPK1 phosphorylation and activation, thereby regulating MyoD expression.
To gain a closer insight into the molecular details of MAPK1 interaction with vinculin, especially vinculin residues 762–768, we first sought to predict the structure of the MAPK1-vinculin complex. Although four out of seven of these residues were shown to be interfaced with MAPK1 in our proposed complex, we did not observe significant interaction of the remaining residues with MAPK1. One possible justification for this apparent discrepancy in our results may be related to the primarily hydrophobic nature of these residues. Because hydrophobic contacts between proteins are primarily dictated by entropic considerations, these contacts may take significantly more time to manifest than salt bridges. Although we were unable to extend our simulations further due to computational constraints, we predict that if our simulations were extended to longer timescales, we would observe engagement of these hydrophobic residues with MAPK1.
Our results imply that MAPK engages vinculin through a noncanonical binding interface. The canonical protein binding site on MAPK, the DRS, engages MAPK-interacting proteins through two spatially distinct regions, the common docking (CD) domain, and the hydrophobic cleft. The CD domain contains two negatively charged aspartate residues, D316 and D319, and the hydrophobic cleft is formed by residues L113, L119, H123, Y126, L155, T157, and T158. The associated canonical MAPK binding site on MAPK-interacting proteins, called the D-site, engages the DRS of MAPK, and is of the form (R/K)2-3-X2-6-ΦA-X-ΦB, where X is any amino acid, and Φ is any hydrophobic residue, usually isoleucine or leucine (34). Upon binding MAPK, residues (R/K)2-3 of the D-site interact with the negatively charged CD domain of MAPK, whereas residues ΦA and ΦB associate with the hydrophobic cleft. Although the D-site of proteins must match these sequence constraints, it should also satisfy certain structural constraints. Structurally, the 2–3 basic residues of the D-site typically reside on an α-helix, whereas the two necessary hydrophobic residues reside on a disordered structure. Because the two hydrophobic residues are always separated by only one residue and must be facing the same direction, they cannot reside on a helical motif, as α-helices have periodicities of 3.6, and not two. Although the D4 domain of vinculin contains the sequence 758RRANRILLVA767, which is the only sequence on vinculin that matches the consensus sequence (R/K)2-3-X2-6-ΦA-X-ΦB, it is structurally incompatible, as the hydrophobic residues reside on an α-helical motif. Engagement of MAPK through this site would therefore require unfolding of the stable four-helical D4 domain, which may not be physiologically relevant. This evidence provides indirect support for our claim that vinculin engages MAPK through a noncanonical binding site.
We proposed that D3–D4 interdomain changes may occur upon vinculin opening. This was also suggested by Bakolitsa et al. (5), who suggested that due to the weak nature of the D3–D4 interaction, upon vinculin opening, D3 and D4 may move with respect to each other. Such interdomain movement may not be possible in the closed conformation of vinculin, as simultaneous interactions between the vinculin tail domain and head domains D3 and D4 in closed vinculin may lock D3 and D4 in a conformation that cannot engage MAPK1.
The results of this study were generated through computational approaches. Future experimental work, such as mutagenesis studies to test our predicted vinculin-MAPK1 binding interface and affinity measurements between MAPK1 and full-length or Vt-lacking vinculin to test the conformational selectivity of MAPK1-vinculin binding, would be highly valuable and could help to validate and refine the predictions made in this study.
A direct interaction between vinculin and MAPK1 may not be the only mechanism through which focal adhesions regulate MAPK1 signaling and stem cell differentiation. Interestingly, expression of the vinculin tail domain in vinculin null cells partially reconstituted stiffness-induced MAPK1 signaling and MyoD expression (37). This effect may possibly be attributed to interactions between the vinculin tail and paxillin, a focal adhesion protein that has been previously demonstrated to regulate MAPK1 activity. It is currently unclear whether the interaction of the vinculin head region with MAPK1 and the activity of the vinculin tail domain represent two separate pathways through which stem cell mechanotransduction to occur, or if they are both involved in the same signaling axis. Furthermore, focal adhesion-associated paxillin has been shown to bind and induce activation of MAPK in a hepatocyte growth factor (HGF)-stimulated manner (53, 54). Paxillin is constitutively associated with MEK. In response to HGF binding to the c-MET receptor, paxillin-bound MEK is phosphorylated and activated, and MAPK is recruited to paxillin and brought in proximity with the paxillin-bound MEK, inducing MAPK phosphorylation and activation. Although this pathway has been shown to mediate HGF-stimulated MAPK activation at focal adhesions, it is unclear whether this pathway is capable of mechanosensing, and if it is capable of regulating stem cell differentiation.
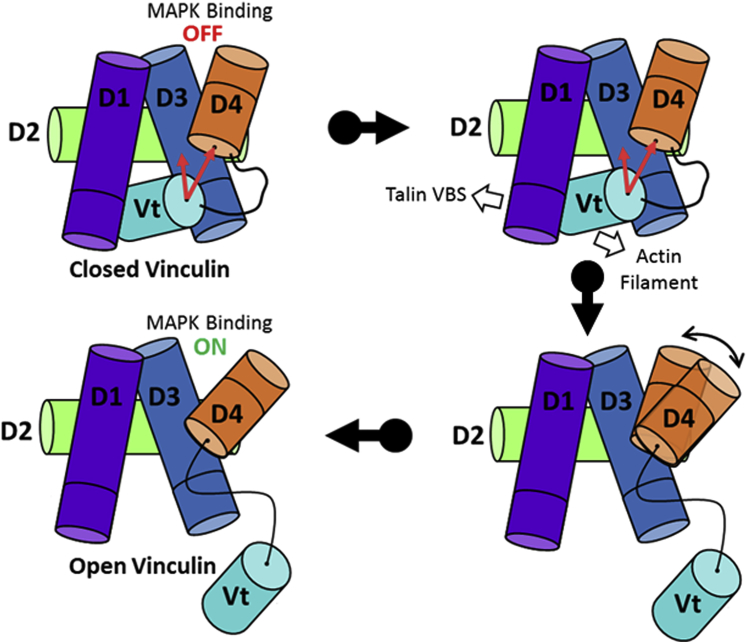
The results of this study ultimately support the following model for the structural basis of conformationally selective vinculin-MAPK1 binding (Fig. 6). When vinculin is in its closed conformation, D3-Vt and D4-Vt interactions constrain the D3–D4 cleft in a closed orientation. Upon force-dependent talin VBS exposure and vinculin engagement and activation, the vinculin tail is released from the head domain, and the D3-Vt/D4-Vt interactions present in closed vinculin are broken, allowing relative movement of the D3 and D4 domains due to the weak nature of the D3–D4 interaction. This consequently unlocks the D3–D4 cleft, allowing MAPK1 binding and activation.
Figure 6.
A model for the mechanoselectivity of vinculin-MAPK1 binding. Vinculin domains D1, D2, D3, D4, and Vt are shown in violet, lime, blue, orange, and cyan, respectively. The D3–D4 cleft of closed vinculin is stabilized in a MAPK1-inaccessible state through simultaneous interactions between D3 and D4 with Vt. Upon force-dependent talin engagement with vinculin, the Vt domain is released from the head domain, relieving its interaction with D3 and D4. The release of Vt from the head domain is proposed to increase the flexibility of the D3–D4 interface, releasing the steric constraints and permitting MAPK1 binding. To see this figure in color, go online.
Conclusion
Herein, we predicted the structure of the MAPK1-vinculin binding interface using a combination of flexible docking and molecular dynamics simulations. Our results confirmed that the MAPK1-vinculin interaction is mechanically regulated, and implicated a change in the vinculin D3–D4 cleft size upon vinculin activation as the basis for the conformational selectivity of MAPK1 binding toward open vinculin. Our results provide further support for a role for the vinculin-MAPK1 complex as a molecular strain gauge capable of cellular mechanosensing. The results of this study were generated solely through computational approaches and should be treated as predictions providing insight for further experimental studies.
Author Contributions
Conceived and designed the experiments: K.G., H.S., and M.R.K.M. performed the experiments: K.G. and H.S. Analyzed the data: K.G., H.S., and M.R.K.M. Contributed reagents/materials/analysis tools: M.R.K.M. Wrote the paper: K.G., H.S., and M.R.K.M.
Acknowledgments
This work was supported by National Science Foundation (NSF) grant No. CMMI 1538707 to M.R.K.M. In addition, this research used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation (NSF) grant No. ACI-1053575.
Editor: Catherine Galbraith.
References
- 1.Tan J.L., Tien J., Chen C.S. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc. Natl. Acad. Sci. USA. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carisey A., Tsang R., Ballestrem C. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 2013;23:271–281. doi: 10.1016/j.cub.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown C., Morham S.G., Naghavi M.H. Focal adhesion proteins talin-1 and vinculin negatively affect paxillin phosphorylation and limit retroviral infection. J. Mol. Biol. 2011;410:761–777. doi: 10.1016/j.jmb.2011.03.076. [DOI] [PubMed] [Google Scholar]
- 4.Ziegler W.H., Liddington R.C., Critchley D.R. The structure and regulation of vinculin. Trends Cell Biol. 2006;16:453–460. doi: 10.1016/j.tcb.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Bakolitsa C., Cohen D.M., Liddington R.C. Structural basis for vinculin activation at sites of cell adhesion. Nature. 2004;430:583–586. doi: 10.1038/nature02610. [DOI] [PubMed] [Google Scholar]
- 6.Burridge K., Mangeat P. An interaction between vinculin and talin. Nature. 1984;308:744–746. doi: 10.1038/308744a0. [DOI] [PubMed] [Google Scholar]
- 7.Cohen D.M., Chen H., Craig S.W. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 2005;280:17109–17117. doi: 10.1074/jbc.M414704200. [DOI] [PubMed] [Google Scholar]
- 8.Wachsstock D.H., Wilkins J.A., Lin S. Specific interaction of vinculin with α-actinin. Biochem. Biophys. Res. Commun. 1987;146:554–560. doi: 10.1016/0006-291x(87)90564-x. [DOI] [PubMed] [Google Scholar]
- 9.Shams H., Golji J., Mofrad M.R.K. Molecular trajectory of alpha-actinin activation. Biophys. J. 2012;103:2050–2059. doi: 10.1016/j.bpj.2012.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shams H., Golji J., Mofrad M.R.K. Dynamic regulation of α-actinin’s calponin homology domains on F-actin. Biophys. J. 2016;110:1444–1455. doi: 10.1016/j.bpj.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bourdet-Sicard R., Rüdiger M., Nhieu G.T. Binding of the Shigella protein IpaA to vinculin induces F-actin depolymerization. EMBO J. 1999;18:5853–5862. doi: 10.1093/emboj/18.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada S., Pokutta S., Nelson W.J. Deconstructing the cadherin-catenin-actin complex. Cell. 2005;123:889–901. doi: 10.1016/j.cell.2005.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brindle N.P., Holt M.R., Critchley D.R. The focal-adhesion vasodilator-stimulated phosphoprotein (VASP) binds to the proline-rich domain in vinculin. Biochem. J. 1996;318:753–757. doi: 10.1042/bj3180753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mandai K., Nakanishi H., Takai Y. Ponsin/SH3P12: an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. J. Cell Biol. 1999;144:1001–1017. doi: 10.1083/jcb.144.5.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kioka N., Sakata S., Aota S. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J. Cell Biol. 1999;144:59–69. doi: 10.1083/jcb.144.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeMali K.A., Barlow C.A., Burridge K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J. Cell Biol. 2002;159:881–891. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golji J., Mofrad M.R.K. The interaction of vinculin with actin. PLoS Comput. Biol. 2013;9:e1002995. doi: 10.1371/journal.pcbi.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner C.E., Glenney J.R., Jr., Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wood C.K., Turner C.E., Critchley D.R. Characterisation of the paxillin-binding site and the C-terminal focal adhesion targeting sequence in vinculin. J. Cell Sci. 1994;107:709–717. [PubMed] [Google Scholar]
- 20.Turner C.E., Miller J.T. Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak-binding region. J. Cell Sci. 1994;107:1583–1591. doi: 10.1242/jcs.107.6.1583. [DOI] [PubMed] [Google Scholar]
- 21.Fukami K., Endo T., Takenawa T. α-Actinin and vinculin are PIP2-binding proteins involved in signaling by tyrosine kinase. J. Biol. Chem. 1994;269:1518–1522. [PubMed] [Google Scholar]
- 22.Johnson R.P., Craig S.W. The carboxy-terminal tail domain of vinculin contains a cryptic binding site for acidic phospholipids. Biochem. Biophys. Res. Commun. 1995;210:159–164. doi: 10.1006/bbrc.1995.1641. [DOI] [PubMed] [Google Scholar]
- 23.Perez-Moreno M., Avila A., González-Mariscal L. Vinculin but not α-actinin is a target of PKC phosphorylation during junctional assembly induced by calcium. J. Cell Sci. 1998;111:3563–3571. doi: 10.1242/jcs.111.23.3563. [DOI] [PubMed] [Google Scholar]
- 24.Weekes J., Barry S.T., Critchley D.R. Acidic phospholipids inhibit the intramolecular association between the N- and C-terminal regions of vinculin, exposing actin-binding and protein kinase C phosphorylation sites. Biochem. J. 1996;314:827–832. doi: 10.1042/bj3140827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziegler W.H., Tigges U., Jockusch B.M. A lipid-regulated docking site on vinculin for protein kinase C. J. Biol. Chem. 2002;277:7396–7404. doi: 10.1074/jbc.M110008200. [DOI] [PubMed] [Google Scholar]
- 26.Hüttelmaier S., Illenberger S., Jockusch B.M. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J. Cell Biol. 2001;155:775–786. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Dokholyan N.V. Insights into allosteric control of vinculin function from its large scale conformational dynamics. J. Biol. Chem. 2006;281:29148–29154. doi: 10.1074/jbc.M605512200. [DOI] [PubMed] [Google Scholar]
- 28.Golji J., Mofrad M.R.K. A molecular dynamics investigation of vinculin activation. Biophys. J. 2010;99:1073–1081. doi: 10.1016/j.bpj.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Case L.B., Baird M.A., Waterman C.M. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat. Cell Biol. 2015;17:880–892. doi: 10.1038/ncb3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Golji J., Lam J., Mofrad M.R.K. Vinculin activation is necessary for complete talin binding. Biophys. J. 2011;100:332–340. doi: 10.1016/j.bpj.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S.E., Chunsrivirot S., Mofrad M.R. Molecular dynamics study of talin-vinculin binding. Biophys. J. 2008;95:2027–2036. doi: 10.1529/biophysj.107.124487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee S.E., Kamm R.D., Mofrad M.R.K. Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J. Biomech. 2007;40:2096–2106. doi: 10.1016/j.jbiomech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Golji J., Mofrad M.R.K. The talin dimer structure orientation is mechanically regulated. Biophys. J. 2014;107:1802–1809. doi: 10.1016/j.bpj.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roskoski R., Jr. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol. Res. 2012;66:105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 35.Jaiswal R.K., Jaiswal N., Pittenger M.F. Adult human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by mitogen-activated protein kinase. J. Biol. Chem. 2000;275:9645–9652. doi: 10.1074/jbc.275.13.9645. [DOI] [PubMed] [Google Scholar]
- 36.Engler A.J., Sen S., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 37.Holle A.W., Tang X., Engler A.J. In situ mechanotransduction via vinculin regulates stem cell differentiation. Stem Cells. 2013;31:2467–2477. doi: 10.1002/stem.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Engler A.J., Carag-Krieger C., Discher D.E. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J. Cell Sci. 2008;121:3794–3802. doi: 10.1242/jcs.029678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J., Shapiro P., Pozharski E. Structure of extracellular signal-regulated kinase 2 in complex with ATP and ADP. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2012;68:1434–1439. doi: 10.1107/S1744309112042972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borgon R.A., Vonrhein C., Izard T. Crystal structure of human vinculin. Structure. 2004;12:1189–1197. doi: 10.1016/j.str.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Schwede T., Kopp J., Peitsch M.C. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Zundert G.C.P., Rodrigues J.P.G.L.M., Bonvin A.M.J.J. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 43.Nelson M.T., Humphrey W., Kalé K.V. NAMD: a parallel, object-oriented molecular dynamics program. Int. J. High Perform. Comput. Appl. 1996;10:251–268. [Google Scholar]
- 44.MacKerell A.D., Bashford D., Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 45.MacKerell A.D., Jr., Feig M., Brooks C.L., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 46.Foloppe N., MacKerell A.D., Jr. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Comput. Chem. 2000;21:86–104. [Google Scholar]
- 47.MacKerell A.D., Banavali N.K. All-atom empirical force field for nucleic acids: II. Application to molecular dynamics simulations of DNA and RNA in solution. J. Comput. Chem. 2000;21:105–120. [Google Scholar]
- 48.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984;81:511. [Google Scholar]
- 49.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A Gen. Phys. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 50.Feller S.E., Zhang Y., Brooks B.R. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613. [Google Scholar]
- 51.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089. [Google Scholar]
- 52.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 53.Ishibe S., Joly D., Cantley L.G. Paxillin serves as an ERK-regulated scaffold for coordinating FAK and Rac activation in epithelial morphogenesis. Mol. Cell. 2004;16:257–267. doi: 10.1016/j.molcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 54.Kolch W. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 2005;6:827–837. doi: 10.1038/nrm1743. [DOI] [PubMed] [Google Scholar]