Abstract
Cytoskeletal structures characterized by actin filaments with uniform lengths, including the thin filaments of striated muscles and the spectrin-based membrane skeleton, use barbed and pointed-end capping proteins to control subunit addition/dissociation at filament ends. While several proteins cap the barbed end, tropomodulins (Tmods), a family of four closely related isoforms in vertebrates, are the only proteins known to specifically cap the pointed end. Tmods are ∼350 amino acids in length, and comprise alternating tropomyosin- and actin-binding sites (TMBS1, ABS1, TMBS2, and ABS2). Leiomodins (Lmods) are related in sequence to Tmods, but display important differences, including most notably the lack of TMBS2 and the presence of a C-terminal extension featuring a proline-rich domain and an actin-binding WASP-Homology 2 domain. The Lmod subfamily comprises three somewhat divergent isoforms expressed predominantly in muscle cells. Biochemically, Lmods differ from Tmods, acting as powerful nucleators of actin polymerization, not capping proteins. Structurally, Lmods and Tmods display crucial differences that correlate well with their different biochemical activities. Physiologically, loss of Lmods in striated muscle results in cardiomyopathy or nemaline myopathy, whereas complete loss of Tmods leads to failure of myofibril assembly and developmental defects. Yet, interpretation of some of the in vivo data has led to the idea that Tmods and Lmods are interchangeable or, at best, different variants of two subfamilies of pointed-end capping proteins. Here, we review and contrast the existing literature on Tmods and Lmods, and propose a model of Lmod function that attempts to reconcile the in vitro and in vivo data, whereby Lmods nucleate actin filaments that are subsequently capped by Tmods during sarcomere assembly, turnover, and repair.
Main Text
Introduction
Actin filaments assemble into diverse cytoskeletal structures via coordinated processes that include nucleation of new filaments, elongation through monomer addition at filament ends, capping of filament ends to terminate elongation, and depolymerization of aged filaments to recycle the monomer pool. In muscle cells, two related subfamilies of proteins, Tropomodulins (Tmods) and Leiomodins (Lmods), play essential roles in controlling thin (actin) filament assembly dynamics. Biochemically, Tmods reduce the rates of actin filament polymerization and depolymerization by specifically capping the pointed end of actin filaments in sarcomeres (1), whereas capping protein (or CapZ) exerts a similar role at the barbed end (2). In contrast, Lmods display strong nucleation activity in vitro, but do not appear to cap filaments (3). Both Tmods and Lmods bind tropomyosin (TM), which enhances Tmod’s pointed-end capping activity (1, 4, 5) and Lmod’s nucleation activity (3, 6, 7). In cells, Tmods regulate actin filament length and stability by capping pointed ends in cytoskeletal architectures as diverse as the spectrin-based membrane skeleton and the contractile sarcomeres of striated muscle myofibrils (reviewed in (8, 9, 10)). The function of Lmods have been studied primarily in cardiac and skeletal muscles, where they are important for the regulation of the length of thin filaments in sarcomeres and normal muscle contractility (11, 12, 13, 14, 15). Interest in this family of proteins has increased dramatically, owing to recent discoveries linking the expression of Lmod2 and Lmod3, respectively, to dilated cardiomyopathy (11, 12) and nemaline myopathy (13, 14, 16, 17, 18). Lmod1, on the other hand, has been linked to autoimmune disorders (19, 20), and a recent study finds that its deficiency impairs smooth muscle cytocontractility and causes megacystis microcolon intestinal hypoperistalsis syndrome (MMIHS) in humans and mice (21).
Our biochemical and structural understanding of the regulation of actin pointed-end assembly by Tmods agrees well with their function in vivo. In contrast, how Lmods’ potent nucleation activity is linked to their in vivo function is less clear. Here, we compare the biochemical, structural, and in vivo properties of Tmods and Lmods, and critically evaluate the experimental evidence for their functions in muscle cells. Our analysis leads us to propose a model in which Lmod nucleates actin filaments that are subsequently capped by Tmod during sarcomere assembly and muscle development, and during sarcomere turnover and repair in mature muscles.
Tmod and Lmod isoforms and tissue-specific expression
Tissue-specific Expression of Tmod and Lmod Isoforms
Vertebrates express four highly conserved Tmod and three less conserved Lmod isoforms in a tissue-specific manner (Fig. 1; Table 1; Fig. S1) (9). Tmod2 and Tmod4 are selectively expressed in neuronal tissue and skeletal muscle, respectively, whereas Tmod1 and Tmod3 are widely expressed, with Tmod1 being additionally enriched in mammalian erythrocytes, lens fibers, neurons, and smooth, cardiac, and skeletal muscles (22, 23, 24, 25, 26). Accordingly, Tmod1, Tmod2, Tmod3, and Tmod4 were originally known as erythrocyte, neuronal, ubiquitous, and skeletal Tmods (E-Tmod, N-Tmod, U-Tmod, and Sk-Tmod), respectively (9). Lmod2 and Lmod3 are specifically expressed in cardiac and skeletal muscles, whereas Lmod1 is highly enriched in smooth muscles, and is also present in some endocrine tissues (3, 11, 13, 16, 20, 25, 26, 27) (see also The Human Protein Atlas: http://www.proteinatlas.org). Lmod1 and Lmod2 have been referred to as the “smooth muscle” and “cardiac” Lmods (SM-Lmod and C-Lmod), respectively (25). Recently, Lmod1 expression was also demonstrated in the neurons of the central nervous system, including the CA3 region of the hippocampus, Purkinje cells in the cerebellum, and cortical neurons, where it acts as an autoimmune target in patients with nodding syndrome, an epileptic disorder triggered by infection with the parasite Onchocerca volvulus (19). Curiously, Lmod1 was originally known as 64 kDa autoantigen D1 (20, 28), based on reactivity with sera from patients with another autoimmune disease, Hashimoto’s thyroiditis (20).
Figure 1.
Tmod and Lmod isoforms. Sequence alignment of the four Tmod and three Lmod human isoforms is given. Amino acid conservation decreases from blue to white backgrounds. UniProt accession codes: hTmod1, P28289; hTmod2, Q9NZR1; hTmod3, Q9NYL9; hTmod4, Q9NZQ9; hLmod1, P29536; hLmod2, Q6P5Q4; hLmod3, Q0VAK6. Boxed regions include TM-binding sites 1 and 2 (TMBS1 and TMBS2, light green), actin-binding sites 1 and 2 (ABS1 and ABS2, magenta), proline-rich domain (PRD, cyan), and the WASP-Homology 2 domain (WH2, red). The region of Tmod that interacts with the DNase I-binding loop (D-loop) of actin, called here the D-loop-binding site (DBS), is highlighted inside an orange box. Note that Lmods lack TMBS2, DBS, and part of ABS1, whereas Tmods lack the PRD- and WH2-containing C-terminal extension. However, a recent study (41) suggests that ABS1 is N-terminally shifted in Lmods compared to Tmods (corresponding to human Lmod2 residues D43–E90, contoured magenta). Fig. S1 extends this alignment to other organisms. To see this figure in color, go online.
Table 1.
Tmod and Lmod Isoform Expression, Localizations, and Functions in Vertebrate Muscles
| Proteina | Tissue and Cell Expression | Developmental Expression in Mouse and Human | Cytoskeletal Localizations | Mouse Phenotype or Human Disease due to Loss of Tmod or Lmod | References |
|---|---|---|---|---|---|
| Tmod1b | cardiac muscle | E8.0 to adult | thin filament pointed ends in sarcomeres | mouse embryonic lethal E8.5–9.5; failure of embryonic cardiac myofibril assembly and contractility; aborted cardiac development; longer thin filaments after acute depletion or inhibition in cardiac myocytes | 8, 9, 29, 30, 75 |
| skeletal muscle | E9.5 to adult | thin filament pointed ends in sarcomeres; sarcoplasmic reticulum (SR) or T-tubule-associated compartment; costameres at sarcolemma | mouse skeletal muscle weakness; normal thin filament lengths due to compensation by Tmod3 and Tmod4; perturbed SR organization and Ca2+-handling due to Tmod3 relocation to thin filament pointed ends; longer thin filaments after acute depletion in adult skeletal muscle | 7, 8, 29, 63 | |
| smooth muscle | unknown | F-actin-associated puncta throughout cells | unknown | 26 | |
| Tmod3c | skeletal muscle | unknown | SR-associated actin cytoskeleton | mouse embryonic lethal E15.5–18.5 | 8, 9, 109, 110 |
| Tmod4 | skeletal muscle | unknown, to adult | thin filament pointed ends in sarcomeres | mouse skeletal muscle normal due to compensation by Tmod1 | 8, 9, 22, 63 |
| Lmod1d | cardiac muscle (embryonic) | E9.5–12.5 | unknown | unknown | 27 |
| Smooth muscle (vascular and visceral) | E13.5 to adult | diffuse staining in F-actin-rich regions between dense bodies | human and mouse postnatal or juvenile lethality due to MMIHS; aberrant actin filament assembly in bladder and intestinal smooth muscle and defective contractility | 21, 25, 26, 27 | |
| extraocular and sternothyroid striated muscle | unknown | A-band in sarcomeres | unknown | 25, 28 | |
| Lmod2 | cardiac muscle | mouse: E8.5 to adult human: fetal heart, adult |
thin filaments, near pointed ends and along the length of thin filaments in sarcomeres; A-band in sarcomeres | mouse juvenile lethality due to dilated cardiomyopathy, with shorter thin filaments and reduced contractile force in cardiomyocytes | 3, 7, 11, 12, 25, 27, 40 |
| skeletal muscle | E9.5 to adult | unknown | no apparent phenotype | 7, 11, 27, 63 | |
| Lmod3 | cardiac muscle | E12.5 to adult | unknown | mouse smaller heart; modestly impaired function | 13, 16, 27 |
| skeletal muscle | mouse: E10.5 to adult human: 14 weeks gestation to adult |
thin filaments, near pointed ends and along the length of thin filaments in sarcomeres; A-band in sarcomeres | Human and mouse neonatal lethality due to severe muscle weakness and respiratory insufficiency; nemaline myopathy, with shorter thin filaments and actin filament accumulation in nemaline bodies; fast fiber atrophy in mice | 13, 14, 16, 17, 27, 63 |
Tmod2 is not included as it is only expressed in neurons (9, 23). For simplicity, Tmod data on chicken and frog are not included.
Tmod1 is also expressed in nonmuscle cells, such as red blood cells and lens fiber cells (9).
Tmod3 is expressed ubiquitously, and is the sole Tmod in endothelial cells, polarized epithelial cells, megakaryocytes, and platelets (9, 110).
Lmod1 is also expressed in neurons of the central nervous system, including the CA3 region of the hippocampus, Purkinje cells in the cerebellum, and cortical neurons (19), as well as some endocrine glands (20). See also the Human Protein Atlas, http://www.proteinatlas.org/.
Developmental timing of Tmod and Lmod expression
The timing of Tmod and Lmod expression during muscle development differs (Table 1). Tmod1 transcripts and protein appear in the mouse myocardium early in development before looping of the heart tube, along with the initiation of myofibrillogenesis at embryonic day (E) 8.0 (chick HH stage 11). Lmod2 transcripts are expressed after looping of the heart tube, with the beginning of contractility around E8.5 (chick HH stage 14), whereas Lmod3 transcripts appear even later, at E12.5 (7, 11, 13, 29, 30). Lmod1 is expressed early in mouse heart development, at E9.5, and persists until E12.5, but becomes undetectable by E13.5, when it appears in developing vascular and visceral smooth muscle cells (27). In developing skeletal muscle, Tmod1 is first detected in mouse embryo somites at E9.5 (chick somites HH stage 17), with expression increasing robustly during development (7, 29). Lmod2 transcripts are also detected in mouse embryo somites at E9.5, with expression increasing by E10.5, whereas it decreases in chick at HH stage 19 (7, 11). Lmod3 transcripts are highly expressed in mouse embryo somites from at least E10.5, and persist at high levels throughout development (13). In Xenopus laevis, Lmod3 and Tmod4 transcripts are detected at the neurula stage in developing somites, before the initiation of myofibrillogenesis, and expression increases significantly during skeletal muscle development (15). The timing of Tmod3 expression during muscle development has not been reported. Thus, the developmental timing of expression of Tmods and Lmods varies in cardiac and skeletal muscles, a factor that should be taken into consideration when analyzing phenotypes due to loss of individual Tmod or Lmod isoforms.
Domain organization and biochemical activity of Tmods
Although Tmods and Lmods are related in sequence, they have radically different biochemical activities: filament pointed-end capping (1) and nucleation (3), respectively (Fig. 2). Sequence identity is 53–76% among the four human Tmod isoforms, falling to 32–39% among the three Lmod isoforms and 27–38% between members of the Tmod and Lmod subfamilies (Figs. 1 and S1) (9). The different activities of the two subfamilies correlate well with their different domain architectures. Thus, Tmods are ∼350 amino acids (aa) in length, and comprise alternating TM- and actin-binding sites (TMBS1, ABS1, TMBS2, and ABS2). TMBS1, ABS1, and TMBS2 are all contained within the N-terminal ∼160-aa region (4, 31, 32), thought to be mostly unstructured on its own (33). Studies using NMR and CD spectroscopy suggest that the intrinsically disordered TMBSs adopt a helical conformation upon binding to the N-terminal ∼35 aa of the TM coiled coil (32), substituting for end-to-end interactions of TM molecules along the length of the actin filament (34, 35). ABS2 is contained within the C-terminal region (residues ∼160 onward), which has a globular fold consisting mostly of a leucine-rich repeat (LRR) domain (36). The capping activity of Tmods is regulated by interaction with TM, with their pointed-end binding affinity increasing dramatically in the presence of TM (Kd of ∼0.1–0.2 μM for the pointed end versus ∼20 nM when the actin filaments are associated with TM) (1, 5, 22, 37) (Fig. 2, A–C). Unlike Tmod1 and Tmod4, Tmod2, and Tmod3 can also bind actin monomers (Kd ∼ 0.13 μM for Tmod3-ATP-actin) and nucleate actin polymerization in vitro (37, 38, 39), albeit very weakly compared to Lmods (3, 6, 16). Mutagenesis and truncation experiments indicate that Tmod3 binds monomers via ABS1, but requires both ABS1 and ABS2 for nucleation. However, in the presence of TM, actin monomer binding and nucleation are suppressed, and Tmod3 then acts as a high-affinity pointed-end capping protein, similar to Tmod1, suggesting that its cellular activities are dictated by the presence/absence of specific TM isoforms in cytoskeletal structures (37).
Figure 2.
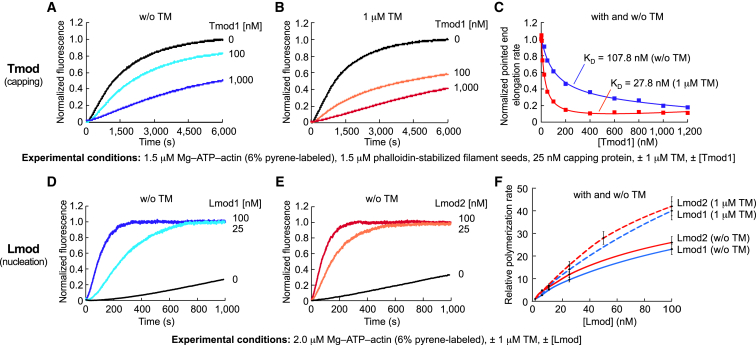
Biochemical activities of Tmod and Lmod. (A and B) Shown here are the time courses of pointed-end elongation of phalloidin-stabilized filament seeds as a function of Tmod1 concentration, and in the absence (A) or the presence (B) of TM. (C) Shown here are the normalized pointed-end elongation rates of filament seeds as a function of Tmod1 concentration, with or without TM. (D and E) Shown here are time courses of polymerization of 2 μM Mg-ATP-actin (6% pyrene-labeled) as a function of Lmod1 (D) or Lmod2 (E) concentration. (F) Shown here is the concentration dependence of polymerization rates of Lmod1 and Lmod2 in the absence (solid lines) or the presence (broken lines) of 1 μM TM, displayed as the mean of three experiments mean ± SE. Experimental conditions are listed separately for experiments in (A)–(F). Note that increasing Tmod concentrations lead to a decrease in polymerization, whereas increasing Lmod concentrations lead to a dramatic increase in polymerization compared to the actin control (black traces). Some of the data shown in this figure was reported in a different form in Rao et al. (5) and Boczkowska et al. (6). To see this figure in color, go online.
Domain organization and biochemical activity of Lmods
Domain organization of Lmods
Lmods display strong nucleation activity in vitro (3, 6, 16, 40) (Fig. 2, D–F), and are most notably distinguished from Tmods by the presence of a C-terminal extension, featuring a proline-rich domain (PRD) and a WASP-Homology 2 (WH2) domain (Fig. 1). Yet, closer inspection reveals several additional differences between Tmods and Lmods. Indeed, sequence conservation is relatively high only for TMBS1 and ABS2 (Fig. S1), and even ABS2 shows important differences that correlate with the different biochemical activities of the two subfamilies. This includes the absence in Lmods of the site responsible in Tmods for interaction with the DNase I-binding loop (D-loop) in actin (human Tmod1 residues 170–179, see below). A region visualized in a recent crystal structure (5) as encompassing Tmod’s ABS1 (human Tmod1 residues 58–99) is also poorly conserved in Lmods (Figs. 1 and S1), albeit a new study suggests that ABS1 is N-terminally shifted in Lmods compared to Tmods (41) (see below). Lmods bind TM via TMBS1 (3, 6, 40, 41, 42, 43), but lack the conserved TMBS2 of Tmods, with the corresponding region varying widely among Lmod isoforms, and displaying an abundance of negatively charged amino acids (Glu and Asp). Lmod1, in particular, presents a large insertion of >200-aa within this region, which curiously may contain a divergent TMBS, because a fragment of Lmod1 missing the first 30 aa still binds TM (26). Lmods also differ considerably within the C-terminal extension, which is markedly longer in Lmod2. Only the WH2 domain is well conserved within this extension, although all Lmods also present a less conserved PRD in between ABS2 and the WH2 domain.
Contribution of the N-terminal region to TM binding and nucleation
The role of the variable region N-terminal to ABS2 in Lmod’s activity is not well understood. In the absence of TM, this region contributes little to the overall nucleation activity of Lmod1 or Lmod2 (6). However, biochemical and cellular studies have shown that the interaction of Lmods with TM through TMBS1 enhances slightly Lmods’ nucleation activity (Fig. 2 F), and is also critical for proper localization in muscle sarcomeres (3, 6, 40, 41, 43). On the other hand, the region structurally visualized as ABS1 in Tmod (human Tmod1 residues P58-K99) is only partially conserved in Lmods (Fig. 1), and synthetic peptides encompassing the corresponding fragments of Lmod1 and Lmod2 did not bind actin (6). It was therefore concluded that Lmods lack ABS1. However, a recent study suggests that ABS1 in Lmods encompasses the sequence between TMBS1 and ABS1 of Tmods (41), corresponding to chicken Lmod2 residues D43–E90 (equivalent to human Lmod2 residues D43–G90) (Figs. 1 and S1). The newly proposed site extends 17-aa N-terminally to the site observed crystallographically in Tmod1 (Fig. 3, A and B), and comprises a region that is highly conserved among Tmods and Lmods (Fig. 1). Therefore, if these amino acids participate in actin binding in Lmods, they must do so also in Tmods. But, surprisingly, although eight of these amino acids were present in the crystal structure of Tmod1 (Fig. 3, A and B), they were disordered and did not appear to interact with actin. This, and the fact that the evidence for the newly proposed actin-binding site consists only of 1D-NMR (41), highlight the need for independent validation of this site.
Figure 3.
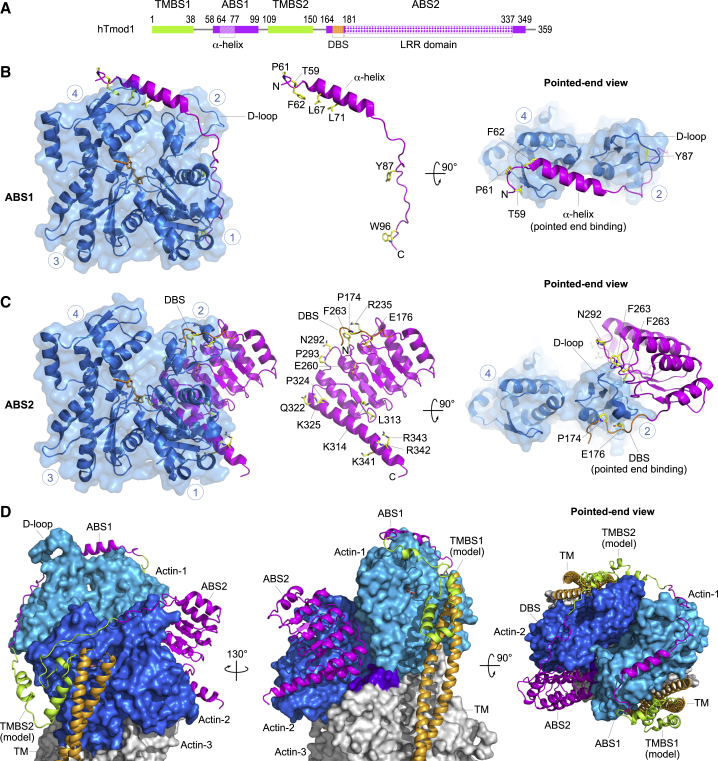
Tmod structures and pointed-end capping model. (A) Domain diagram of human Tmod1 (the other three Tmod isoforms have a very similar domain architecture). Domains are drawn to scale and colored and numbered according to Fig. 1. (B and C) Structures of Tmod ABS1 and ABS2 bound to actin (5), showing frontal and pointed end views. ABS1 and ABS2 are also shown separately in the middle, highlighting amino acids that have been mutated and shown to contribute toward Tmod’s pointed end-capping activity (5,31,38,44,55) (human Tmod1 numbering). Circled blue numbers indicate the four subdomains of actin. N and C denote the N- and C-terminal ends of the bound Tmod domains. (D) Model of Tmod at the pointed end is given (5). The structures of ABS1 and ABS2 in complex with actin were superimposed, respectively, onto the first and second subunits (marine and blue) of the filament structure (52). At this location, ABS2 also contacts the first and third (purple-colored surface) actin subunits of the filament. The structure of the two TM-binding sites (green) is unknown, and a tentative model was generated based on secondary structure prediction and energy minimization (5). TM is shown in the blocked state, which it assumes when bound to the filament with Ca2+-free troponin (53, 111). Of note, this model is consistent with TM’s ability to explore all three states on the filament (blocked, closed, and open) (53,54), without generating steric clashes with Tmod at the pointed end. To see this figure in color, go online.
ABS2 and not the C-terminal extension is the main source of Lmod's nucleation activity
Surprisingly, and contrary to Tmods, the isolated ABS2 of Lmods has significant nucleation activity on its own (6). On the other hand, the WH2 domain-containing C-terminal extension contributes to the nucleation activity (6), but not as importantly as originally thought (3, 7, 40). Indeed, Tmod1 fails to gain strong nucleation activity upon addition of the C-terminal extension of Lmod2, whereas both Lmod1 and Lmod2 retain strong nucleation activity after removal of this extension (6). Despite these considerations, Lmod1 and Lmod2 fragments extending from ABS2 to the C terminus display between two- and fourfold higher nucleation activity over a range of concentrations than their isolated ABS2 (6), emphasizing the nonnegligible contribution of the C-terminal extension to nucleation. As described below, this extension may fulfill additional roles; specifically, the binding site of the WH2 domain overlaps with actin-actin contacts in the filament, which may contribute to Lmod’s dissociation from the pointed end after nucleation. The substantial sequence differences among Lmod isoforms in the regions N- and C-terminal to ABS2 may also dictate muscle type-specific interactions that remain to be elucidated. It must be finally noted that the nucleation activity of the ABS2 of Lmod1 is higher than that of Lmod2, whereas Lmod2 has overall higher activity than Lmod1 (6), implying that the N- and C-terminal regions contribute more to the nucleation activity in Lmod2 than in Lmod1. The sources of these differences are not understood.
Does Lmod cap the pointed end?
Shorter Lmod2 fragments, 1–201 and 1–94 (comprising TMBS1 and the newly proposed ABS1), were found to have marginal TM-dependent pointed-end capping activity (41). Because these two fragments lack nucleation activity (6), their capping activity in bulk polymerization assays is not completely surprising, consistent with the observation of capping activity of analogous fragments of Tmod1 (31). The capping activity of Lmod2 fragment 1–201 was reduced by mutations in TMBS1 (L30E) and ABS1 (W73D), similar to the effect of analogous mutations in Tmod1 (33, 44). These results suggest that the isolated N-terminal region of Lmod2 can cap the pointed end of TM-decorated filaments in a way analogous to Tmod1’s N-terminal region (31). Although a larger Lmod2 fragment comprising ABS2 (residues 1–514) was also reported to cap pointed ends (7), other work indicates that this fragment has strong nucleating activity (6). The strong nucleation activity should mask any effect of Lmod fragments on pointed-end elongation in bulk polymerization assays (except for fragments lacking nucleation activity). Moreover, in competition experiments, even a fourfold excess of Tmod1 has no effect on the nucleation activity of Lmod2, with or without TM, suggesting that Lmods do not compete with Tmods for pointed-end binding and instead favor binding monomers for nucleation, whereas Tmods do not compete with Lmod for monomers and favor binding pointed ends (6) (Fig. 2). Therefore, the functional significance of the capping activity of Lmod2’s N-terminal fragments in vitro is unclear, as the nucleation activity of the full-length protein supersedes any capping activity under physiological conditions (3, 6, 16, 40).
Tmod structure-function relationships
Structural studies published during the last three years have significantly advanced our understanding of the functions of Tmods and Lmods. Until recently, the only structure available of any member of this family was that of the C-terminal globular domain of chicken Tmod1 determined at 1.45 Å resolution (36). On the other hand, NMR analysis had suggested that most of the N-terminal region was unstructured, except for a short helical segment within TMBS1 (residues 24–35) (33). Recently, two key structures were determined (5), corresponding to complexes of actin with human Tmod1 ABS1 (1.8 Å resolution) and ABS2 (2.3 Å resolution), both fused C-terminally to gelsolin segment 1 via a 9-aa Gly-Ser linker to facilitate crystallization (Fig. 3). Importantly, when not connected by a linker, both ABS1 and ABS2 bind to the complex of actin with gelsolin segment 1 with 1:1 stoichiometry and with similar affinities (Kd of 7.5 and 10.5 μM for ABS1 and ABS2, respectively) as they do to nonpolymerizable actin-latrunculin B (5, 6).
Complex of Tmod’s ABS1 with actin
The Tmod1 fragment used in crystallization comprised residues Q50–K101 around the ABS1 of human Tmod1. However, only residues P58–K99 were visualized in the electron density map (Fig. 3 B). In other words, residues E50–A57 and Q100–K101 were disordered, suggesting that they do not interact with actin. It was, therefore, concluded that ABS1 consisted of human Tmod1 residues P58–K99, comprising an α-helix (residues 64–77) and extended segments N- and C-terminal to this helix. Because of its extended conformation, ABS1 contacts three out of the four subdomains of actin (subdomains 4, 2, and 1, in that order), and bridges the pointed end of the actin monomer, inducing a slightly more closed conformation of the nucleotide-binding cleft than in most actin structures. In particular, ABS1 interacts with the D-loop in actin subdomain 2 (Fig. 3 B), a loop that mediates intersubunit contacts in the filament and whose conformation is intimately linked to polymerization (45, 46, 47, 48). Of note, prior work using zero-length chemical cross linking had also identified interactions between ABS1 and subdomains 1 and 2 of actin (38). Consistent with the findings of the crystal structure, residue mutations along the entire length of ABS1 (residues highlighted in Fig. 3 B), including residues absent in Lmods (Fig. 1), affect binding and capping of full-length Tmod (4, 5, 38, 44).
Complex of Tmod’s ABS2 with actin
The actin-bound structure of ABS2 (5) displays extra ordered residues at the N- and C-terminal ends compared to the original unbound structure of ABS2 (36) (Fig. 3 C). In particular, the segment Y170–N179 of ABS2 forms a loop that interacts with the D-loop, and is referred to here as the “D-loop-binding site” (DBS). Importantly, the DBS, which is missing in Lmods (Fig. 1 and see below), can only interact at the pointed end of the actin filament, because its binding interface is occluded by intersubunit contacts along the rest of the filament. As noted above, ABS1 also interacts with the D-loop in actin (Fig. 3 B). The fact that both ABS1 and ABS2 bind to surfaces exposed at the pointed end of the actin filament (Fig. 3, B and C) is a distinctive feature of Tmod among actin-binding proteins, which typically interact at the barbed end of the actin monomer and/or filament (49, 50), and correlates well with Tmod’s unique pointed-end capping activity. The rest of ABS2 consists of a four-and-a-half LRR repeat, whose hydrophobic core is shielded at both ends by helical caps, a common feature among LRR domains (51). The LRR portion of ABS2 binds on the back of the actin monomer (according to the classical view shown in Fig. 3 C, left), interacting with subdomains 2 and 1, in that order.
Model of Tmod at the pointed end of the actin filament
The structures of ABS1 and ABS2 can be unequivocally overlaid onto the pointed end of the high-resolution EM structure of the actin filament (52), resulting in a model in which a single Tmod molecule binds and caps the pointed end (5) (Fig. 3 D). This model is supported by numerous considerations. First, the pseudo-symmetric organization of Tmod, consisting of two TM- and actin-binding modules (TMBS1-ABS1 and TMBS2-ABS2), mirrors the pseudo-symmetric nature of the pointed end. Second, the structures show that Tmod can only bind at the pointed end, because both ABS1 and ABS2 cover surfaces on the actin monomer that are buried in the filament (contrary to Lmods, see below). Third, ABS1 and ABS2 must bind to two different subunits at the pointed end, because their binding surfaces on the actin monomer partially overlap. Fourth, ABS1 and ABS2 must bind to the first and second subunits at the pointed end of the filament, respectively, because swapping their positions also produces steric clashes. What is more, the resulting model places ABS2 in a cleft formed at the interface between the first three subunits of the filament (Fig. 3 D). As discussed below, the interacting surface of ABS2 at this site is highly conserved individually within the Tmod and Lmod subfamilies, but not across subfamilies, offering important clues about their specialized biochemical activities. Fifth, this model is consistent with the azimuthal sliding of TM on the surface of the actin filament (53, 54), whereby TM can explore three structural states (blocked, closed, and open) without generating steric clashes with Tmod at the pointed end, albeit likely influenced by Tmod-TM isoform specific interactions (see below). Sixth, this model is consistent with the results of numerous mutagenesis and capping studies (5, 31, 38, 44, 55, 56). One interesting feature of the model is that because of the relative structural independence and weak individual binding affinities of ABS1 and ABS2, they can be expected to bind and detach from the pointed end semiindependently, consistent with the observation of actin subunit exchange at the pointed end of the thin filaments in muscle sarcomeres (57, 58).
Lmod structure-function relationships
Complex of Lmod’s ABS2 with actin
Recent structural and biochemical studies provide insights into the role of the various Lmod domains in nucleation (Fig. 4). These include the 1.5 Å resolution crystal structure of ABS2 of Lmod1, two structures of a hybrid Tmod1-Lmod1 ABS2 construct crystallized alone (2.1 Å resolution) and in complex with actin (2.4 Å resolution) (6), and a 3.0 Å resolution structure of a complex of actin with a fragment of Lmod2 comprising from ABS2 to the C-terminal WH2 domain (59). The structures show that the highly conserved ABS2 of Lmods interacts with actin similarly to that of Tmods (5), with one important distinction; Lmods lack the DBS, which in Tmods is critical for pointed-end capping (Figs. 1, 3, and 4). Based on the structures and sequence analysis, 11 residues of Tmod1’s ABS2 were replaced by the corresponding residues in Lmod1, the strongest nucleator among the isolated ABS2s. These residues were selected based on two criteria: they are predicted to fall at the interface between actin subunits in the filament (Fig. 3 D), and they are conserved within the individual Lmod and Tmod subfamilies but not across subfamilies. Strikingly, whereas the wild-type ABS2 of Tmod displays TM-independent actin pointed-end capping activity (31), the resulting Tmod1 ABS2 mutant displays strong nucleation activity under conditions where neither full-length Tmod1 nor any of its fragments has such activity (6). This represents a rather unique example whereby the function of a conserved protein domain (capping in this case) can be converted to a different function (nucleation) by just a few structure-inspired point mutations, underscoring the notion that the Tmod and Lmod subfamilies have evolved to exert distinct cellular roles.
Figure 4.
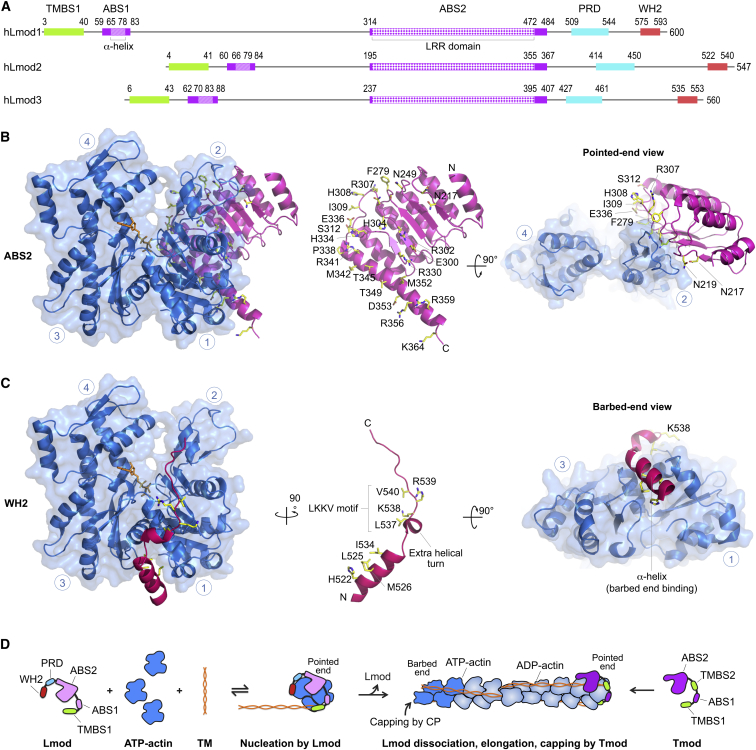
Lmod structures and nucleation model. (A) Domain diagram of the three Lmod isoforms expressed in humans. The diagrams are centered on ABS2, the most highly conserved region among isoforms. Domains are drawn to scale and colored and numbered according to Fig. 1. (B–C) Shown here are actin (blue) complexes with ABS2 (magenta) and the WH2 domain (red) from the structure of Lmod2 in complex with actin (PDB: 4RWT) (59). In addition to the frontal view, pointed end and barbed end views are shown for ABD2 and the WH2 domain, respectively. The two domains are also shown separately in the middle, highlighting amino acids that participate in interactions with actin (human Lmod2 numbering). Circled blue numbers indicate the four subdomains of actin. N and C denote the N- and C-terminal ends of the Lmod domains. Note that Lmod lacks DBS, which in Tmod interacts with the D-loop of actin (compare Figs. 2B, right, and 3C, right). (D) Shown here is the proposed nucleation mechanism of Lmod. Lmod contains two major actin-binding sites (ABS2 and the WH2 domain) and one tropomyosin-binding site, TMBS1. Although ABS1, present in all Tmod isoforms, is not well conserved in Lmods, it was recently proposed that a functional ABS1 may exist in Lmod (41), albeit N-terminally shifted compared to Tmods (see Fig. 1). Through its actin- and TM-binding sites, Lmod recruits at least three actin subunits and one TM molecule to form a polymerization nucleus. Lmod likely dissociates from the nucleus when it begins to elongate, which can be triggered by several factors: steric hindrance of the WH2 domain with actin subunits adding at the barbed end of the original nucleus and the lack of pointed-end capping elements present in Tmod, including most of ABS1, TMBS2, and DBS, which together contribute to the very high affinity of Tmod for pointed ends. Lmod dissociation would free both the pointed and barbed ends of the polymerization nucleus, allowing it to elongate while also freeing the way for Tmod to bind and cap the pointed end. To see this figure in color, go online.
The C-terminal extension of Lmod binds actin through the WH2 domain
The Lmod2-actin complex (59) also shows the interaction of the WH2 domain at the C terminus of Lmods with actin (Fig. 4 C). The interaction is similar to that of other WH2 domains (60, 61), with the WH2 domain of Lmod more closely resembling those of Cobl and Las17, in that it presents an extra helical turn in the loop between the N-terminal α-helix and the LKKV motif (62). The authors of the Lmod2-actin structure report an additional interaction with actin, involving the PRD and an adjacent α-helix in the linker between ABS2 and the WH2 domain (59). However, examination of the structure and deposited x-ray data reveals no reliable electron density for the region of Lmod2 between ABS2 and the WH2 domain, calling this claim into question.
Structural differences explain the functional differences between Tmods and Lmods
Taken together, the structures and biochemical data show that Lmods lack features that are necessary for pointed-end capping in Tmods, including a complete ABS1, TMBS2, and the DBS in ABS2. On the other hand, Lmods present features that are missing in Tmods, which favor nucleation while being incompatible with pointed-end capping, including the presence of a WH2 domain, which is a barbed end-binding element (Fig. 4 C), and specific amino acid substitutions in ABS2 that make it a strong nucleator. Because of these specializations, in face-to-face competition polymerization experiments that replicate cellular conditions (i.e., in the presence of actin monomers, barbed end-capped filaments, and TM), Lmod preferentially binds monomers and promotes polymerization through nucleation (6), whereas Tmod ignores monomers and binds pointed ends to decrease polymerization through capping (1, 5, 22, 37).
Lmod nucleation model
The structural and biochemical data suggest a model of Lmod nucleation (Fig. 4 D). Lmod binds up to three actin subunits to form a polymerization nucleus (3). The ABS2 of Lmod is expected to bind at the interface between the three actin subunits of the Lmod nucleus (6), analogous to that of Tmod (Fig. 3 D). The flexible N-terminal region of Lmod probably wraps around the first actin subunit at the pointed end of the nucleus, recruiting one TM coiled-coil. Although this interaction is also somewhat similar to that of Tmod at the pointed end (Fig. 3 D), the affinity is probably much lower, because Lmod lacks TMBS2 and has a truncated ABS1 (Fig. 1). The flexible C-terminal region of Lmod is anticipated to wrap around the third actin subunit of the nucleus, with the WH2 domain binding in the cleft between subdomains 1 and 3 at the barbed end of this subunit. This cleft participates in subunit-subunit contacts in the filament (52), such that the WH2 domain must dissociate for actin subunits to begin adding at the barbed end for elongation of the initial polymerization nucleus. Therefore, the incompatibility of WH2 domain binding with intersubunit contacts along the actin filament and the weaker affinity of the N-terminal region probably promote dissociation of Lmod from the elongating nucleus, freeing the way for Tmod binding. Of note, when Lmod is in excess compared to actin monomers, it can bind on the sides of the filament, as observed in cells (Fig. 5, A and B) (7, 16, 40, 63) and in cosedimentation assays (40). The structures suggest that binding of Lmod to the side of the filament can only happen through ABS2, because binding of both the N- and C-terminal flexible regions is sterically hindered along the length of the filament. In contrast, Tmods cannot bind along the sides of the filament (37, 64), because their ABS2 contains a pointed-end capping element, i.e., the DBS.
Figure 5.
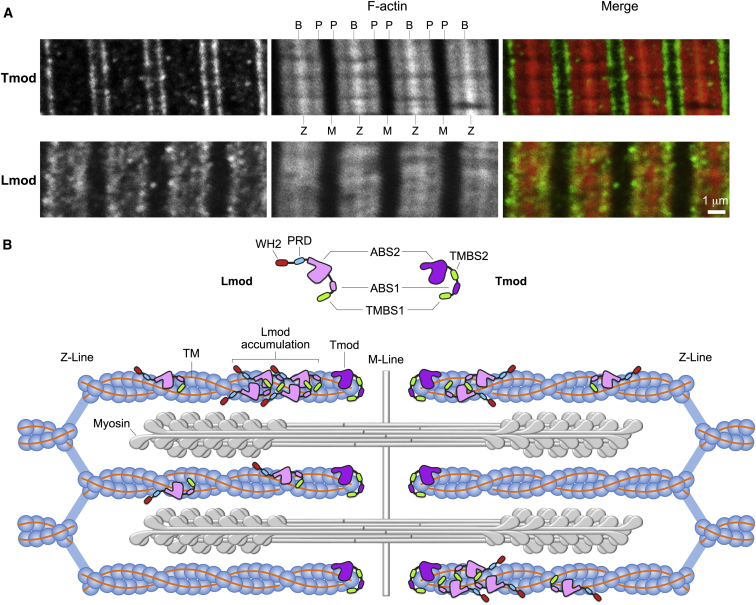
Tmod and Lmod localization in sarcomeres. (A) Shown here is immunofluorescence confocal microscopy of relaxed and stretched sarcomeres from mouse soleus muscle, showing Tmod stripes at the pointed ends of thin filaments and somewhat less crisp Lmod stripes that are closer to the Z-line. Lmod also localizes diffusely along the thin filaments. B and P indicate the barbed and pointed ends of the actin thin filaments, respectively, whereas Z and M indicate the sarcomeric Z- and M-lines, respectively. Longitudinal cryosections of mouse soleus muscle from 2 month-old mice were prepared and immunostained for Tmod (isoform 4, green) and rhodamine-phalloidin for thin filaments (red), as in Gokhin et al. (63,67), and distances measured as in Gokhin and Fowler (89). The Tmod and F-actin staining panels were modified from the wild-type images in figure 5B in reference (67). In unstretched sarcomeres, both Tmod and Lmod appear as single stripes at the M-line (data not shown). The bright Lmod stripes are 0.89 ± 0.07 μm away from the Z-line, ∼0.2-0.3 μm closer than the Tmod stripes, which are 1.24 ± 0.04 μm away from the Z-line, similar to thin filament lengths measured from phalloidin staining, 1.19 ± 0.08 μm (N = 48, 23, and 56 line scans for Lmod, Tmod, and thin filaments). Scale bars: 1 μm. (B) Shown here is a diagram of Tmod and Lmod localizations in striated muscle sarcomeres. Lmod is enriched along thin filaments and near pointed ends, but does not colocalize with Tmod, which caps the pointed end and binds to two TM coiled coils, one on each side of the actin filament. Lmod is also present along the thin filament, including the A-band region containing the myosin thick filaments. Although several Lmod molecules are indicated on a thin filament, the stoichiometry of Lmod to thin filament is unknown, and possibly not all thin filaments contain Lmod. To see this figure in color, go online.
Tmod localization and function in muscle sarcomeres
Tmods are associated with thin filament pointed ends in sarcomeres
There is clear agreement between the cellular and biochemical activities of Tmods, recognized as the only proteins that cap the pointed end (9). Thus, sarcomeric Tmods localize to thin filament pointed ends (Fig. 5, A and B; Table 1), either as a single isoform in mammalian cardiac muscle, Tmod1, or as two different isoforms in skeletal muscle, Tmod1 and Tmod4, with relative expression levels of ∼1:9 in mouse tibialis anterior muscle (fast), ∼1:4 in soleus muscle (slow), and ∼1:3 in diaphragm muscle (22, 23, 63, 65, 66, 67). In chicken, Tmod1 caps the pointed end of thin filaments in cardiac and slow skeletal muscles, while Tmod4 caps thin filament pointed ends in fast skeletal muscle after hatching (22). Furthermore, mutations or domain-masking antibodies that disrupt Tmod’s actin- or TM-binding activities in vitro also disrupt its localization and impair thin filament lengths and stability in striated muscle cells (55, 68, 69, 70, 71). In nonmuscle cells, which lack Tmod1 and Tmod4, the actomyosin stress fibers contain Tmod3, displaying a sarcomerelike striated pattern that alternates with α-actinin staining, consistent with pointed-end localization (72).
Tmods regulate thin filament lengths by capping pointed ends
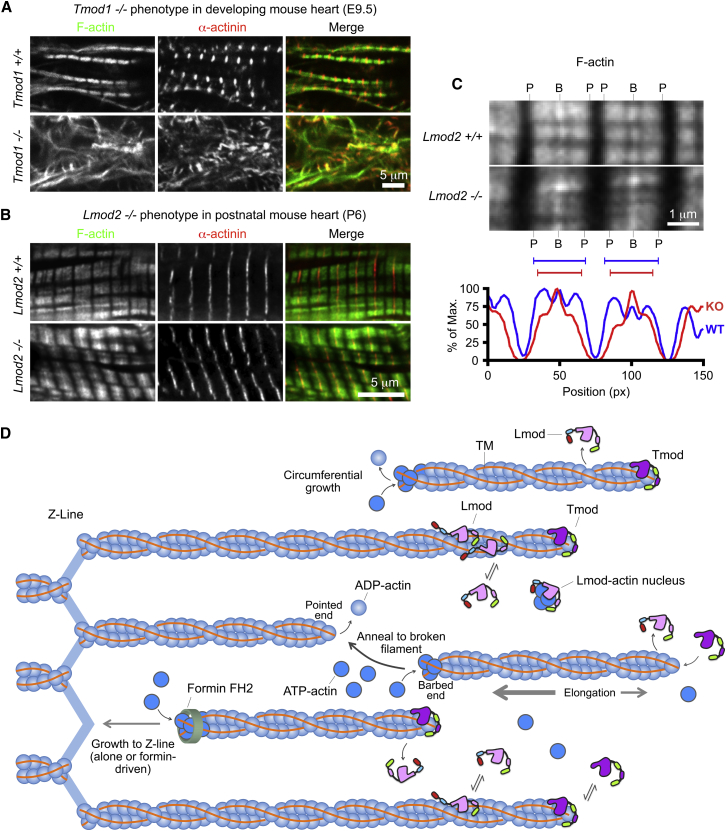
Tmod binding to the pointed end is dynamic in that it allows for controlled actin subunit addition and dissociation (1, 57, 58, 73, 74, 75). In this way, Tmod regulates thin filament lengths in inverse proportion to its expression level, as shown both in the sarcomeres of cardiac myocytes and Drosophila indirect flight muscle (7, 57, 58, 68, 73, 76, 77). Tmod capping is also required for thin filament assembly during striated muscle development. Thus, loss of Tmod1 in the mouse heart results in severe disorganization of the thin filaments, and accumulation of abnormal actin filament bundles, resulting in defective contractility, aborted cardiac development, and lethality at E9.5 (29, 30, 78) (Fig. 6 A; Table 1). Similarly, unc94/tmd-1 loss of function alleles in C. elegans and morpholino knockdown of Tmod4 in developing X. laevis skeletal muscle result in aberrant myofibril assembly and defective movement (15, 79, 80).
Figure 6.
Function of Tmod and Lmod in sarcomeres. (A) Deletion of Tmod1 in the developing mouse heart leads to failure of myofibril assembly, with aberrant F-actin bundles and α-actinin aggregates at embryonic (E) day 9.5. Shown is confocal microscopy of E9.5 embryonic mouse hearts stained for F-actin (green) and α-actinin (red) (unpublished data related to Fritz-Six et al. (29) and McKeown et al. (78)). (B) Deletion of Lmod2 in the developing mouse heart does not affect myofibril assembly, but leads to shorter thin filaments (figure modified from (11)). Wild-type or Lmod2-/- P6 mouse heart left ventricles stained for F-actin (green) and α-actinin (red), and imaged by deconvolution fluorescence microscopy. (C) Thin filaments are shorter in the absence of Lmod. When sarcomeres of the same length are compared, F-actin-stained I-bands are wider in wild-type than in Lmod2-/- sarcomeres. B and P indicate the barbed and pointed ends of the actin thin filaments, respectively. Line scans of F-actin intensity have a narrower profile in Lmod2-/- (red) than in wild-type (blue) sarcomeres, corresponding to ∼15% shorter thin filaments (11). (D) Given here is the model for Lmod nucleation of new filaments that are integrated into sarcomeres during normal filament turnover and repair in mature muscle, or during myofibrillogenesis in development. Lmod catalyzes the formation of an actin nucleus (dark blue, ATP-bound actin subunits). After Lmod dissociates from the actin nucleus (see Fig. 4D), actin subunits add rapidly to free barbed ends, either spontaneously or catalyzed by formins (green ring), leading to filament growth towards the Z-line. Actin hydrolyses ATP in the filament, and older filaments consist mainly of ADP-bound actin subunits (light blue). During sarcomere turnover and repair, the barbed ends could either anneal to the pointed ends of preexisting filaments damaged during contraction, or elongate to the Z-line where they are capped by CapZ and captured by α-actinin. Actin subunits also add slowly to the pointed ends of newly formed filaments, resulting in elongation toward the M-line, where the pointed end can be dynamically capped by Tmod. Cycles of Lmod nucleation, dissociation, and pause (possibly characterized by attachment of Lmod to thin filament sides) could permit a single Lmod molecule to assemble many new thin filaments. During myofibrillogenesis, new Lmod-nucleated filaments may also add to the periphery of sarcomeres, contributing to circumferential growth. In the case of Lmod overexpression, abundant nucleation generates an excess of free pointed ends, which are less frequently capped by Tmods. This can result in increased actin subunit addition at pointed ends and longer thin filaments. To see this figure in color, go online.
Tmod isoforms and thin filament lengths in skeletal muscle
Due to isoform compensation during muscle development, Tmod1 is dispensable for myofibril assembly and specification of thin filament lengths in mouse skeletal muscle, as Tmod4 (endogenously present) and additionally recruited Tmod3 substitute for Tmod1 at pointed ends during myofibril assembly (Table 1) (24, 81). However, muscle contractility is depressed in the absence of Tmod1, suggesting that whereas other isoforms can substitute for Tmod1 during development to assemble and maintain correct thin filament lengths, they cannot fully substitute for it functionally (see below) (81). Indeed, global deletion of Tmod4 does not affect muscle development, myofibril assembly, thin filament lengths, or contractility of adult muscle, as Tmod1 levels increase five- to sixfold during development, sufficient to cap all the pointed ends (63). However, acute siRNA knockdown of Tmod1 in adult Tmod4−/− tibialis anterior skeletal muscle leads to thin filaments that are longer by ∼15%, demonstrating a role for Tmods in controlling thin filament lengths in mature sarcomeres, in the absence of isoform compensation during development (63). Curiously, acute siRNA knockdown of Tmod1 alone in wild-type tibialis anterior muscle also leads to a ∼15% increase in thin filament lengths, with Tmod4 occupying the pointed ends of the longer filaments (63). In mouse models of Duchenne muscular dystrophy, calpain-mediated proteolysis of Tmod1 in tibialis anterior muscle, and both Tmod1 and Tmod4 in soleus muscle, results in similar ∼10–12% increases in thin filament lengths (67). Because Tmod1 and Tmod4 have similar capping activities in vitro (22), it is possible that acute loss of Tmod1 alone (∼10–25% of the total Tmod) impairs capping below a critical threshold required for filament length regulation. Alternatively, thin filament lengths in adult skeletal muscle sarcomeres may be additionally controlled by actin depolymerizing factors or Tmod-isoform-specific interactions near M-lines (73, 75, 82).
Tmod isoform regulation of skeletal muscle contraction
Skinned fiber mechanics and x-ray diffraction analysis show that reduced force generation after the substitution of Tmod1 by Tmod3 in Tmod1−/− skeletal muscles is due to fewer active myosin cross bridges and impaired TM strand movement upon thin filament activation (83). These observations imply that Tmods can influence the interaction of TM along the length of the thin filament from the pointed end, possibly acting in a TM- and Tmod-isoform-specific manner. Indeed, whereas Tmod3 and Tmod1 have similar capping activities in vitro (37), Tmod3 has weaker affinity for TM than Tmod1 (37, 71, 84), possibly explaining why Tmod3 cannot functionally substitute for Tmod1. Cryo-EM studies of Tmod-capped and TM-troponin-regulated thin filaments should further illuminate this mechanism.
Lmod localization and function in muscle sarcomeres
Compared to Tmods, it is more challenging to reconcile the proposed in vivo activities of Lmods with their biochemical activities. Biochemically, Lmods act as powerful nucleators of actin polymerization (Fig. 2), and this activity is enhanced by interaction with TM (3, 6, 7). However, the interpretation of the cellular and physiological data has led to the idea that Tmods and Lmods are interchangeable, with the main role of both subfamilies being pointed-end capping and the control of thin filament lengths (7, 11, 15, 16). Below, we present the existing evidence on Lmod localization and phenotypes due to loss of Lmod function, which altogether set apart the Lmod and Tmod subfamilies and emphasize the role of Lmods as filament nucleators, not capping proteins.
Lmods are associated with thin filaments near but not at pointed ends
Can the sarcomeric localization of Lmods shed light on their functions? The localization of Lmod2 and Lmod3 in striated muscles is consistent with a thin filament-associated protein, and superficially resembles that of Tmods; both proteins appear as a single stripe near M-lines in unstretched sarcomeres, and as a pair of stripes flanking the H-zone in stretched sarcomeres (Fig. 5, A and B; Table 1) (6, 7, 11, 15, 16, 40, 63). However, closer examination reveals that Lmod2 and Lmod3 do not colocalize precisely with Tmod; the Tmod stripes are closer to the M-line, whereas the stripes of Lmod2 and Lmod3 are closer to the Z-line (Fig. 5 A) (16, 40, 63). In some cases, Lmod2 and Lmod3 staining resembles A-band (myosin) staining (17, 40), as does the localization of Lmod1 in a subset of striated extraocular muscles (28). In addition to this predominant localization, Lmods also appear diffusely localized along the length of the thin filaments, except for the Z-line, in both cardiac and skeletal muscles (6, 7, 11, 16, 40) (Fig. 5, A and B). In skeletal muscle, the localization of the predominant Lmod3 stripe resembles that of the N-terminal end of nebulin, which also localizes closer to the Z-line than Tmod (Fig. 5 A) (10, 16, 63, 85). A functional interconnection between Lmod3 and nebulin is supported by the observation that they both bind kelch-like family member 40 (KLHL40), which protects nebulin from degradation and blocks Lmod3 ubiquitination and degradation (17). These observations indicate that Lmods may bind to the sides of thin filaments, consistent with structural features of Lmod that allow for side binding via ABS2 (Fig. 4 B) and cosedimentation of Lmod2 with F-actin in vitro (40), something not observed for Tmods (37, 64). The variety of Lmod staining patterns also suggests dynamic and regulated interactions with thin filament components. Thus, whereas Tmods are integral structural components of thin filaments, Lmods may be present at substoichiometric levels and may play a catalytic rather than a structural role. At present, the relative stoichiometry of Lmods to thin filaments is unknown.
Lmod1 Localization in Smooth Muscle
In smooth muscles, the lack of organized sarcomeres with precisely aligned thin filaments makes it challenging to determine the localization of Lmod1 with respect to pointed ends. Nevertheless, immunostaining of longitudinal cryosections of relaxed rat circular smooth muscle reveals distinct patterns of Lmod1 and Tmod1 (Table 1) (26). Lmod1 appears diffusely localized, whereas Tmod1 shows a punctate localization, likely corresponding to pointed-end clusters. On the other hand, in hypercontracted smooth muscles, Lmod1 and Tmod1 colocalize on the edges of contraction bands (26). Future studies, using superresolution electron microscopy, may help determine the precise localizations of Lmod1 and Tmod1 in smooth muscle cells.
Lmod2 regulates thin filament lengths by promoting actin assembly in sarcomeres of cardiac myocytes
Despite the different biochemical activities, domain organizations and sarcomeric localizations of Lmods and Tmods, functional studies have led to the idea that the two subfamilies compete with each other for pointed-end capping, and that Lmods might additionally actively promote pointed-end elongation (7, 11, 15), a function proposed earlier for SALS, an unrelated protein from Drosophila (58). Thus, overexpression of Lmod2 in cardiac myocytes reduces Tmod1 localization at pointed ends, resulting in slightly longer (∼6–8%) thin filaments (7, 41). Yet, overexpression of a Lmod2 construct lacking the WH2 domain (residues 1–514) does not lead to longer thin filaments (7), nor does overexpression of Lmod2 with a mutation in TMBS1 defective for TM binding (41). Conversely, genetic deletion of Lmod2 in the developing mouse heart results in ∼15% shorter thin filaments, without affecting sarcomere organization, Tmod1 levels or localization in cardiac myocytes (Fig. 6, B and C) (11). Lmod3 levels are unaffected by Lmod2 deletion in cardiac myocytes (11). Lmod2 appears to regulate thin filament lengths by enhancing actin assembly at pointed ends, based on adenovirus-mediated expression of GFP-Lmod2 in cultured Lmod2−/− neonatal cardiomyocytes, which increases the incorporation of rhodamine-actin at pointed ends and rescues normal thin filament length (11). Moreover, fluorescence-recovery-after-photobleaching experiments also show that GFP-Lmod2 increases mCherry-actin turnover near pointed ends but not barbed ends, again suggesting that Lmod2 promotes actin monomer incorporation in the vicinity of pointed ends.
Loss of Lmod2 Leads to Dilated Cardiomyopathy Due to Reduced Force Generation by Shorter Thin Filaments in Sarcomeres
Lmod2−/− mouse hearts with shorter thin filaments show reduced systolic performance after birth, progressing to dilated cardiomyopathy and juvenile lethality (Table 1) (11, 12). Lmod2−/− cultured neonatal cardiac myocytes also display reduced contractile force, suggesting that abnormal thin filament length regulation and force generation are the primary causes of cardiomyopathy (11). Longer thin filaments may be required to sustain cardiac force generation at longer sarcomere lengths during systolic function (11). The cardiac Lmod2 deletion phenotype is strikingly similar to a transgenic Tmod1 overexpression phenotype, characterized by shorter thin filaments, sarcomere disarray, and heart degeneration that progresses to dilated cardiomyopathy and results in death a few weeks after birth (77, 86). In other words, loss of Lmod2 appears to functionally phenocopy overexpression of Tmod1 in the mouse heart.
Unlike the loss of Tmod1 (Fig. 6 A), the loss of Lmod2 does not interfere with myofibril assembly during heart development, indicating that Lmod2 regulates thin filament lengths subsequent to sarcomere assembly (Fig. 6 B). Furthermore, while shorter thin filaments are detected as early as E12.5, sarcomere organization remains relatively normal until the development of cardiomyopathy symptoms at P15, indicating that sarcomere disarray is likely a degenerative phenotype, possibly due to increased mechanical load on the heart after birth (11). In addition to sarcomere disarray, another study also observed abnormal intercalated disk morphology and reduced expression of some intercalated disk genes at P25, which may reflect degenerative processes in the absence of Lmod2 (12). However, an additional role for Lmod in myofibril assembly in the developing heart cannot be excluded, because Lmod1 expression from E9.5–E12.5 (27) may compensate for loss of Lmod2 during this stage of development.
Loss of Lmod3 leads to skeletal muscle weakness and nemaline myopathy with shorter thin filaments
In humans, mutations in LMOD3 with absence of Lmod3 cause a rare congenital skeletal muscle nemaline myopathy in which sarcomeres have shorter thin filaments and reduced force generation at longer sarcomere lengths (Table 1) (16, 18). Sarcomere disarray and the replacement of Z-lines with nemaline bodies (abnormal, rod-shaped structures formed by Z-disc proteins) are prominent in severely affected patients, and in mouse, zebrafish and frog Lmod3-depletion models (13, 14, 15, 16). The loss of KLHL40, which leads to degradation of Lmod3 and nebulin, also results in nemaline myopathy in humans and mice (17). While sarcomere organization in KLHL40−/− mice is normal at P1, sarcomere disarray and nemaline bodies are evident by P8, and mice die before P22 (17). Thus, similar to the cardiac Lmod2−/− mouse phenotype (11), sarcomere disarray in the absence of skeletal muscle Lmod3 is likely due to muscle degeneration during muscle contraction rather than a developmental defect. However, it is puzzling that KLHL40 knockout mice did not show shorter thin filaments (17), unlike nebulin knockout mice (87, 88) or Lmod3-deficient patients (16). Because typical changes in filament lengths (∼100–200 nm) are smaller than the resolution of light microscopy, they can be missed, and thus future studies should benefit from the use of superresolution microscopy and automated quantification algorithms (89, 90).
Loss of Lmod1 leads to defective actin filament assembly and reduced contractility in visceral smooth muscle
Patients with mutations in the LMOD1 gene leading to the absence of Lmod1, and a Lmod1−/− mouse model, develop megacystis microcolon intestinal hypoperistalsis syndrome, caused by hypocontractility of bladder and intestinal smooth muscle that leads to functional obstruction of the urinary bladder and intestine (Table 1) (21). MMIHS appears to be a congenital disease due to defective smooth muscle contractility, because sporadic mutations in smooth muscle actin ACTG2 are among the most frequent causes of this syndrome (91), and at least one patient with a nonsense mutation in smooth muscle myosin MYH11 has also been reported (92). Although Lmod1 is abundant in both vascular and visceral smooth muscles (21, 25, 26, 27), Lmod1−/− mice exhibit pathological thinning and compaction of visceral (stomach, bladder, intestine), but surprisingly, not vascular (aorta, esophagus) smooth muscles (21). siRNA knockdown of Lmod1 in isolated intestinal smooth muscle cells results in decreased contractility, indicating that functional defects in vivo are intrinsic to smooth muscle cells. A striking feature of Lmod1−/− mouse visceral smooth muscle is an overall reduction in fluorescent-phalloidin staining for F-actin in cryosections, together with the formation of rodlike aggregates of actin filaments observed in transmission electron microscopy (21), resembling nemaline rods in Lmod3−/− mouse skeletal muscles (13, 16). This suggests that lack of Lmod1-mediated actin filament nucleation in contractile structures leads to inappropriate actin assembly in noncontractile Z-like structures. Thus, like Lmod2 in heart (11) and Lmod3 in skeletal muscle (13, 16), Lmod1 is a critical component of the contractile apparatus in smooth muscle.
Lmod and Tmod effects on thin filament and myofibril assembly may be coordinated via actin polymerization-dependent coregulation of gene expression
In X. laevis skeletal muscle, Lmod3 and Tmod4 unexpectedly appear to have overlapping functions, with overexpression of one protein compensating for loss of the other (15). Knockdowns of Lmod3 or Tmod4 lead to severe sarcomere disruption and muscle movement defects, which can be rescued by overexpression of either Lmod3 or Tmod4 (15). How can the disparate in vitro activities of Tmod4 (pointed-end capping) and Lmod3 (nucleation) substitute for one another in vivo? Whether off-target or gain-of-function effects can explain these results is a possibility that cannot be ruled out, because >10-fold overexpression levels were used in rescue experiments. Alternatively, Lmod3 and Tmod4 may functionally cross talk via actin polymerization-dependent regulation of sarcomeric gene expression by the myocardin-related transcription factor/serum response factor (MRTF/SRF) pathway (93, 94). MRTF is retained in the cytoplasm by binding actin monomers, and released upon actin polymerization, whereupon it enters the nucleus and interacts with SRF to activate the transcription of sarcomeric constituents (93). If increased expression of either Tmod4 or Lmod3 were to reduce the overall level of actin monomers, this would be expected to activate the MRTF/SRF pathway of sarcomeric gene expression and promote myofibril assembly. Lmod1 and Lmod3 are themselves MRTF/SRF-regulated genes (13, 27), and loss of Lmod3 in mouse skeletal muscle leads to decreased mRNA levels of sarcomeric proteins, consistent with downregulation of the MRTF/SRF transcriptional pathway (13). In support of a coregulatory circuit, loss of Lmod3 in nemaline myopathy patients results in reduced Tmod4 protein levels (16), whereas loss of Tmod4 in mouse skeletal muscle results in reduced Lmod3 mRNA and protein levels (63). In the future, it should be instructive to measure actin monomer/filament levels and MRTF nuclear/cytoplasmic localization as a function of Tmod4 or Lmod3 expression.
Model of Tmod versus Lmod function in thin filament assembly and length regulation
The knockout phenotypes of Tmods and Lmods in striated muscles indicate that Tmods restrict thin filament lengths by capping pointed ends, while Lmods promote longer thin filaments. How can Lmods’ in vitro nucleation activity be reconciled with the promotion of longer thin filaments in vivo? We note that catalyzed pointed-end elongation by proteins has never been observed in vitro, and that longer thin filaments in sarcomeres may be assembled via mechanisms not requiring catalyzed subunit addition at pointed ends. We propose that the effects of Lmod on thin filament length can be explained within the context of dynamic actin subunit exchange at pointed ends controlled by Tmod. In the case of Lmod overexpression (7, 41), new filaments would be nucleated whose barbed ends could anneal onto the transiently uncapped pointed ends of preexisting filaments, resulting in longer filaments, as proposed for the Drosophila formin DAAM (95). Alternatively, the pointed ends of the newly formed filaments could compete with preexisting pointed ends for limiting Tmod, resulting in reduced capping frequency, increased actin subunit addition at pointed ends, and net filament elongation, similar to the Tmod inhibition or depletion phenotypes (58, 63, 68). In the case of Lmod loss or depletion (11, 12, 16), decreased nucleation would lead to fewer pointed ends, resulting in increased Tmod capping frequency, reduced actin subunit addition at pointed ends, and shorter thin filaments, similar to Tmod overexpression phenotypes (7, 57, 58, 76, 77). Thus, the effects of Lmod depletion/overexpression on thin filament lengths could be an indirect consequence of Tmod’s regulation of pointed-end dynamics, with Lmod indirectly affecting Tmod capping by controlling the number of free pointed ends, rather than by directly competing with Tmod for binding to the pointed ends of preexisting filaments (Fig. 6 C).
What might be the role of Lmod nucleation for thin filament assembly and turnover in muscle? We propose that Lmod nucleates new filaments that are integrated into sarcomeres during myofibril maturation or repair, replacing immature filaments that turn over during development, or replacing filaments damaged during muscle contraction (Fig. 6 C). The elongation of these filaments may proceed from the barbed end, possibly driven by muscle-specific formins (96, 97) (see below), as observed in nonmuscle cells. Once filaments begin to elongate, Lmod likely dissociates from the pointed end, which is then capped by Tmod (Figs. 4 D and 6 C). Of note, thin filament lengths increase during embryonic heart development in mice (11); thus shorter filaments in the absence of Lmod2 may result from a failure to replace shorter immature with longer mature filaments (11). A function for Lmod2 in thin filament addition during circumferential growth of myofibrils is supported by the observation that Lmod2 knockdown in neonatal rat cardiomyocytes results in abnormally thin and misaligned myofibrils, whereas sarcomere size and H-zones appear normal (3). Alternatively, Lmod2 could nucleate new filaments in the vicinity of the M-line, whose free barbed ends anneal with the distal pointed ends of transiently uncapped preexisting filaments, or with uncapped distal ends created by breakage due to actomyosin contraction (40, 98) (Fig. 6 C). Consistent with this view, blebbistatin inhibition of myosin activity in cardiac myocytes leads to Lmod2 dissociation from sarcomeres, with no effect on Tmod1 (40). Thus, dilated cardiomyopathy in the absence of Lmod2, nemaline myopathy in the absence of Lmod3, and smooth muscle hypocontractility in MMIHS due to the absence of Lmod1, could be from a deficiency in nucleation of new filaments to replace thin filaments turning over during muscle development and/or contraction.
Summary and outstanding questions
Despite major advances during the last few years in our understanding of Lmod and Tmod function and structure, many questions remain, and conflicting evidence must be reconciled. Thus, it remains to be demonstrated whether Lmods contain a fully functional ABS1, and what is the full extent of this site in Tmods; does it also include the highly conserved sequence between TMBS1 and ABS1? In vitro polymerization experiments must be performed using near-physiological conditions, allowing Lmods and Tmods to compete with each other in the presence of TM, actin monomers, and CapZ-capped filaments (gelsolin, used in many experiments, is not an effective barbed end cap (5)). In addition to bulk polymerization assays, total internal reflection fluorescence microscopy can help visualize the interplay between Lmods and Tmods at the pointed end. Furthermore, whereas the biochemical activities of Lmod1 and Lmod2 have been extensively characterized, Lmod3 remains less well explored. The various Lmod isoforms display large differences in the WH2 domain-containing C-terminal tail and the region preceding ABS2, characterized by the presence of low-complexity sequences, which presumably give rise to intrinsically disordered regions. The role of these low-complexity regions and the functional significance of the differences between Lmod isoforms remain unknown. The sequence differences between Lmod isoforms likely translate into substantial variation in their biochemical activities, as well as different binding partners in cells. So far, actin, TM, and KLHL40 are the only known binding partners of Lmods, but additional, muscle-specific interactions are likely, which could be identified using modern proximity ligation assays (99). Such additional binding partners could control Lmod’s localization in sarcomeres, as well as the timing for nucleation during muscle development and repair. Physiologically, the roles of Lmod1 in nonmuscle cells, such as neurons, remain completely unknown.
Mechanisms that coordinate Lmod nucleation with Tmod capping may be required for precise thin filament length regulation, including during development and among different muscle types during contraction. Indeed, although the relative ratio of Tmods to pointed ends is rather constant (63, 65, 67), thin filament lengths and the relative ratio of Tmod1 to Tmod4 varies among striated muscle types (10, 63, 67, 85, 100, 101, 102). This suggests functional specialization of sarcomeric Tmods, whose biochemical and structural bases remain to be determined. It is also plausible that variations in Lmod stoichiometry or isoform composition contribute to controlling muscle-specific thin filament length and dynamics, a concept that remains to be explored. Tmod capping appears to be constitutive, as no regulatory mechanism has been identified. If Lmod nucleation is also constitutive, coordination between Tmod capping and Lmod nucleation may be achieved through gene expression, protein synthesis, and/or protein degradation, which will also require further investigation.
Lmods lack filament elongation activity in vitro (3), and may thus cooperate with one of several formins found in sarcomeres (recently reviewed in (103)) for filament assembly. While some formins can nucleate actin polymerization, they are more generally associated with inhibition of barbed end capping and acceleration of processive barbed end elongation from profilin-actin, and several formins have now been shown to team-up with other proteins for nucleation (62, 104). Among the sarcomeric formins, the most extensively studied are the FHOD- (96, 97, 105, 106, 107) and DAAM-related (95, 106, 108) formins, both of which have been implicated in myofibrillogenesis and the establishment of sarcomeric ultrastructure. However, FHOD-family formins lack nucleation activity in vitro (96), whereas DAAM is thought to mediate thin filament organization in sarcomeres but not polymerization (106). Conceivably, these and other sarcomeric formins could work synergistically with Lmods, whereby Lmods nucleate new filaments whose barbed ends are then elongated by formins (Fig. 6 D). This possibility should be explored in the future, particularly by turning on/off the activities of Lmods and formins using inhibitors and conditional mutations, followed by analysis of thin filament lengths and sarcomere ultrastructure, as well as through in vitro polymerization studies.
Going forward, it is important to establish a more complete understanding of how Lmods’ nucleation activity in vitro correlates with their functions in muscles and other cells, and how Lmods and Tmods functionally synergize with each other in cells. The identification of additional binding partners may help set the two subfamilies more clearly apart. The presence of four Tmod and three Lmod isoforms, with divergent properties and structures, is unlikely to be a fortuitous event; muscle cells must need these proteins for highly specialized functions. Future research in this area should tell us how.
Author Contributions
V.M.F. and R.D. participated equally in the preparation of the figures and manuscript.
Acknowledgments
We thank David Gokhin for comments and Roberta Nowak, Malgorzata Boczkowska, David Gokhin, Carol Gregorio, and Christopher Pappas for help with the compilation of the original data for Figs. 2, 5, and 6.
This work was funded by National Institutes of Health (NIH) grants No. R01 GM073791 to R.D. and No. R01 HL083464 to V.M.F.
Editor: Brian Salzberg.
Footnotes
One figure is available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30382-X.
Contributor Information
Velia M. Fowler, Email: velia@scripps.edu.
Roberto Dominguez, Email: droberto@mail.med.upenn.edu.
Supporting Material
References
- 1.Weber A., Pennise C.R., Fowler V.M. Tropomodulin caps the pointed ends of actin filaments. J. Cell Biol. 1994;127:1627–1635. doi: 10.1083/jcb.127.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edwards M., Zwolak A., Cooper J.A. Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 2014;15:677–689. doi: 10.1038/nrm3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chereau D., Boczkowska M., Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostyukova A.S., Choy A., Rapp B.A. Tropomodulin binds two tropomyosins: a novel model for actin filament capping. Biochemistry. 2006;45:12068–12075. doi: 10.1021/bi060899i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao J.N., Madasu Y., Dominguez R. Mechanism of actin filament pointed-end capping by tropomodulin. Science. 2014;345:463–467. doi: 10.1126/science.1256159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boczkowska M., Rebowski G., Dominguez R. How Leiomodin and Tropomodulin use a common fold for different actin assembly functions. Nat. Commun. 2015;6:8314. doi: 10.1038/ncomms9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukada T., Pappas C.T., Gregorio C.C. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J. Cell Sci. 2010;123:3136–3145. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gokhin D.S., Fowler V.M. Tropomodulin capping of actin filaments in striated muscle development and physiology. J. Biomed. Biotechnol. 2011;2011:103069. doi: 10.1155/2011/103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamashiro S., Gokhin D.S., Fowler V.M. Tropomodulins: pointed-end capping proteins that regulate actin filament architecture in diverse cell types. Cytoskeleton. 2012;69:337–370. doi: 10.1002/cm.21031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gokhin D.S., Fowler V.M. A two-segment model for thin filament architecture in skeletal muscle. Nat. Rev. Mol. Cell Biol. 2013;14:113–119. doi: 10.1038/nrm3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pappas C.T., Mayfield R.M., Gregorio C.C. Knockout of Lmod2 results in shorter thin filaments followed by dilated cardiomyopathy and juvenile lethality. Proc. Natl. Acad. Sci. USA. 2015;112:13573–13578. doi: 10.1073/pnas.1508273112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S., Mo K., Sun L.V. Lmod2 piggyBac mutant mice exhibit dilated cardiomyopathy. Cell Biosci. 2016;6:38. doi: 10.1186/s13578-016-0101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cenik B.K., Garg A., Liu N. Severe myopathy in mice lacking the MEF2/SRF-dependent gene leiomodin-3. J. Clin. Invest. 2015;125:1569–1578. doi: 10.1172/JCI80115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian L., Ding S., Xu T. Leiomodin-3-deficient mice display nemaline myopathy with fast-myofiber atrophy. Dis. Model. Mech. 2015;8:635–641. doi: 10.1242/dmm.019430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nworu C.U., Kraft R., Krieg P.A. Leiomodin 3 and tropomodulin 4 have overlapping functions during skeletal myofibrillogenesis. J. Cell Sci. 2015;128:239–250. doi: 10.1242/jcs.152702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuen M., Sandaradura S.A., Clarke N.F. Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy. J. Clin. Invest. 2014;124:4693–4708. doi: 10.1172/JCI75199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garg A., O’Rourke J., Olson E.N. KLHL40 deficiency destabilizes thin filament proteins and promotes nemaline myopathy. J. Clin. Invest. 2014;124:3529–3539. doi: 10.1172/JCI74994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sandaradura S., North K.N. LMOD3: the “missing link” in nemaline myopathy? Oncotarget. 2015;6:26548–26549. doi: 10.18632/oncotarget.5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson T.P., Tyagi R., Nath A. Nodding syndrome may be an autoimmune reaction to the parasitic worm Onchocerca volvulus. Sci. Transl. Med. 2017;9:eaaf6953. doi: 10.1126/scitranslmed.aaf6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Q., Ludgate M., Vassart G. Cloning and sequencing of a novel 64-kDa autoantigen recognized by patients with autoimmune thyroid disease. J. Clin. Endocrinol. Metab. 1991;72:1375–1381. doi: 10.1210/jcem-72-6-1375. [DOI] [PubMed] [Google Scholar]
- 21.Halim D., Wilson M.P., Miano J.M. Loss of LMOD1 impairs smooth muscle cytocontractility and causes megacystis microcolon intestinal hypoperistalsis syndrome in humans and mice. Proc. Natl. Acad. Sci. USA. 2017;114:E2739–E2747. doi: 10.1073/pnas.1620507114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almenar-Queralt A., Lee A., Fowler V.M. Identification of a novel tropomodulin isoform, skeletal tropomodulin, that caps actin filament pointed ends in fast skeletal muscle. J. Biol. Chem. 1999;274:28466–28475. doi: 10.1074/jbc.274.40.28466. [DOI] [PubMed] [Google Scholar]
- 23.Cox P.R., Zoghbi H.Y. Sequencing, expression analysis, and mapping of three unique human tropomodulin genes and their mouse orthologs. Genomics. 2000;63:97–107. doi: 10.1006/geno.1999.6061. [DOI] [PubMed] [Google Scholar]
- 24.Gokhin D.S., Fowler V.M. Cytoplasmic γ-actin and tropomodulin isoforms link to the sarcoplasmic reticulum in skeletal muscle fibers. J. Cell Biol. 2011;194:105–120. doi: 10.1083/jcb.201011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conley C.A., Fritz-Six K.L., Fowler V.M. Leiomodins: larger members of the tropomodulin (Tmod) gene family. Genomics. 2001;73:127–139. doi: 10.1006/geno.2000.6501. [DOI] [PubMed] [Google Scholar]
- 26.Conley C.A. Leiomodin and tropomodulin in smooth muscle. Am. J. Physiol. Cell Physiol. 2001;280:C1645–C1656. doi: 10.1152/ajpcell.2001.280.6.C1645. [DOI] [PubMed] [Google Scholar]
- 27.Nanda V., Miano J.M. Leiomodin 1, a new serum response factor-dependent target gene expressed preferentially in differentiated smooth muscle cells. J. Biol. Chem. 2012;287:2459–2467. doi: 10.1074/jbc.M111.302224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conley C.A., Fowler V.M. Localization of the human 64kD autoantigen D1 to myofibrils in a subset of extraocular muscle fibers. Curr. Eye Res. 1999;19:313–322. doi: 10.1076/ceyr.19.4.313.5304. [DOI] [PubMed] [Google Scholar]
- 29.Fritz-Six K.L., Cox P.R., Fowler V.M. Aberrant myofibril assembly in tropomodulin1 null mice leads to aborted heart development and embryonic lethality. J. Cell Biol. 2003;163:1033–1044. doi: 10.1083/jcb.200308164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chu X., Chen J., Sung L.A. E-Tmod capping of actin filaments at the slow-growing end is required to establish mouse embryonic circulation. Am. J. Physiol. Heart Circ. Physiol. 2003;284:H1827–H1838. doi: 10.1152/ajpheart.00947.2002. [DOI] [PubMed] [Google Scholar]
- 31.Fowler V.M., Greenfield N.J., Moyer J. Tropomodulin contains two actin filament pointed end-capping domains. J. Biol. Chem. 2003;278:40000–40009. doi: 10.1074/jbc.M306895200. [DOI] [PubMed] [Google Scholar]
- 32.Kostyukova A.S., Hitchcock-Degregori S.E., Greenfield N.J. Molecular basis of tropomyosin binding to tropomodulin, an actin-capping protein. J. Mol. Biol. 2007;372:608–618. doi: 10.1016/j.jmb.2007.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greenfield N.J., Kostyukova A.S., Hitchcock-DeGregori S.E. Structure and tropomyosin binding properties of the N-terminal capping domain of tropomodulin 1. Biophys. J. 2005;88:372–383. doi: 10.1529/biophysj.104.051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frye J., Klenchin V.A., Rayment I. Structure of the tropomyosin overlap complex from chicken smooth muscle: insight into the diversity of N-terminal recognition. Biochemistry. 2010;49:4908–4920. doi: 10.1021/bi100349a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao J.N., Rivera-Santiago R., Dominguez R. Structural analysis of smooth muscle tropomyosin α and β isoforms. J. Biol. Chem. 2012;287:3165–3174. doi: 10.1074/jbc.M111.307330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krieger I., Kostyukova A., Maéda Y. Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping. Biophys. J. 2002;83:2716–2725. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashiro S., Gokhin D.S., Fowler V.M. Differential actin-regulatory activities of Tropomodulin1 and Tropomodulin3 with diverse tropomyosin and actin isoforms. J. Biol. Chem. 2014;289:11616–11629. doi: 10.1074/jbc.M114.555128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashiro S., Speicher K.D., Fowler V.M. Mammalian tropomodulins nucleate actin polymerization via their actin monomer binding and filament pointed end-capping activities. J. Biol. Chem. 2010;285:33265–33280. doi: 10.1074/jbc.M110.144873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischer R.S., Yarmola E.G., Fowler V.M. Tropomodulin 3 binds to actin monomers. J. Biol. Chem. 2006;281:36454–36465. doi: 10.1074/jbc.M606315200. [DOI] [PubMed] [Google Scholar]
- 40.Skwarek-Maruszewska A., Boczkowska M., Lappalainen P. Different localizations and cellular behaviors of leiomodin and tropomodulin in mature cardiomyocyte sarcomeres. Mol. Biol. Cell. 2010;21:3352–3361. doi: 10.1091/mbc.E10-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ly T., Moroz N., Kostyukova A.S. The N-terminal tropomyosin- and actin-binding sites are important for leiomodin 2’s function. Mol. Biol. Cell. 2016;27:2565–2575. doi: 10.1091/mbc.E16-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kostyukova A.S. Leiomodin/tropomyosin interactions are isoform specific. Arch. Biochem. Biophys. 2007;465:227–230. doi: 10.1016/j.abb.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Colpan M., Tolkatchev D., Kostyukova A.S. Localization of the binding interface between leiomodin-2 and α-tropomyosin. Biochim. Biophys. Acta. 2016;1864:523–530. doi: 10.1016/j.bbapap.2016.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kostyukova A.S., Rapp B.A., Hitchcock-DeGregori S.E. Structural requirements of tropomodulin for tropomyosin binding and actin filament capping. Biochemistry. 2005;44:4905–4910. doi: 10.1021/bi047468p. [DOI] [PubMed] [Google Scholar]
- 45.Durer Z.A., Kudryashov D.S., Reisler E. Structural states and dynamics of the D-loop in actin. Biophys. J. 2012;103:930–939. doi: 10.1016/j.bpj.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hocky G.M., Baker J.L., Voth G.A. Cations stiffen actin filaments by adhering a key structural element to adjacent subunits. J. Phys. Chem. B. 2016;120:4558–4567. doi: 10.1021/acs.jpcb.6b02741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baek K., Liu X., Dominguez R. Modulation of actin structure and function by phosphorylation of Tyr-53 and profilin binding. Proc. Natl. Acad. Sci. USA. 2008;105:11748–11753. doi: 10.1073/pnas.0805852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hung R.J., Pak C.W., Terman J.R. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominguez R. Actin-binding proteins—a unifying hypothesis. Trends Biochem. Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 50.Dominguez R., Holmes K.C. Actin structure and function. Annu. Rev. Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bella J., Hindle K.L., Lovell S.C. The leucine-rich repeat structure. Cell. Mol. Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von der Ecken J., Müller M., Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehman W. Thin filament structure and the steric blocking model. Compr. Physiol. 2016;6:1043–1069. doi: 10.1002/cphy.c150030. [DOI] [PubMed] [Google Scholar]
- 54.McKillop D.F., Geeves M.A. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys. J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsukada T., Kotlyanskaya L., Kostyukova A.S. Identification of residues within tropomodulin-1 responsible for its localization at the pointed ends of the actin filaments in cardiac myocytes. J. Biol. Chem. 2011;286:2194–2204. doi: 10.1074/jbc.M110.186924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber A., Pennise C.R., Fowler V.M. Tropomodulin increases the critical concentration of barbed end-capped actin filaments by converting ADP.Pi-actin to ADP-actin at all pointed filament ends. J. Biol. Chem. 1999;274:34637–34645. doi: 10.1074/jbc.274.49.34637. [DOI] [PubMed] [Google Scholar]
- 57.Littlefield R., Almenar-Queralt A., Fowler V.M. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat. Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- 58.Bai J., Hartwig J.H., Perrimon N. SALS, a WH2-domain-containing protein, promotes sarcomeric actin filament elongation from pointed ends during Drosophila muscle growth. Dev. Cell. 2007;13:828–842. doi: 10.1016/j.devcel.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 59.Chen X., Ni F., Wang Q. Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells. Proc. Natl. Acad. Sci. USA. 2015;112:12687–12692. doi: 10.1073/pnas.1512464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chereau D., Kerff F., Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rebowski G., Namgoong S., Dominguez R. Structure of a longitudinal actin dimer assembled by tandem W domains: implications for actin filament nucleation. J. Mol. Biol. 2010;403:11–23. doi: 10.1016/j.jmb.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominguez R. The WH2 domain and actin nucleation: necessary but insufficient. Trends Biochem. Sci. 2016;41:478–490. doi: 10.1016/j.tibs.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gokhin D.S., Ochala J., Fowler V.M. Tropomodulin 1 directly controls thin filament length in both wild-type and tropomodulin 4-deficient skeletal muscle. Development. 2015;142:4351–4362. doi: 10.1242/dev.129171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fowler V.M. Tropomodulin: a cytoskeletal protein that binds to the end of erythrocyte tropomyosin and inhibits tropomyosin binding to actin. J. Cell Biol. 1990;111:471–481. doi: 10.1083/jcb.111.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fowler V.M., Sussmann M.A., Daniels M.P. Tropomodulin is associated with the free (pointed) ends of the thin filaments in rat skeletal muscle. J. Cell Biol. 1993;120:411–420. doi: 10.1083/jcb.120.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gregorio C.C., Fowler V.M. Mechanisms of thin filament assembly in embryonic chick cardiac myocytes: tropomodulin requires tropomyosin for assembly. J. Cell Biol. 1995;129:683–695. doi: 10.1083/jcb.129.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gokhin D.S., Tierney M.T., Fowler V.M. Calpain-mediated proteolysis of tropomodulin isoforms leads to thin filament elongation in dystrophic skeletal muscle. Mol. Biol. Cell. 2014;25:852–865. doi: 10.1091/mbc.E13-10-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gregorio C.C., Weber A., Fowler V.M. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 1995;377:83–86. doi: 10.1038/377083a0. [DOI] [PubMed] [Google Scholar]
- 69.Mudry R.E., Perry C.N., Gregorio C.C. The interaction of tropomodulin with tropomyosin stabilizes thin filaments in cardiac myocytes. J. Cell Biol. 2003;162:1057–1068. doi: 10.1083/jcb.200305031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moroz N.A., Novak S.M., Kostyukova A.S. Alteration of tropomyosin-binding properties of tropomodulin-1 affects its capping ability and localization in skeletal myocytes. J. Biol. Chem. 2013;288:4899–4907. doi: 10.1074/jbc.M112.434522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lewis R.A., Yamashiro S., Fowler V.M. Functional effects of mutations in the tropomyosin-binding sites of tropomodulin1 and tropomodulin3. Cytoskeleton. 2014;71:395–411. doi: 10.1002/cm.21179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu S., Dasbiswas K., Bershadsky A.D. Long-range self-organization of cytoskeletal myosin II filament stacks. Nat. Cell Biol. 2017;19:133–141. doi: 10.1038/ncb3466. [DOI] [PubMed] [Google Scholar]
- 73.Littlefield R.S., Fowler V.M. Thin filament length regulation in striated muscle sarcomeres: pointed-end dynamics go beyond a nebulin ruler. Semin. Cell Dev. Biol. 2008;19:511–519. doi: 10.1016/j.semcdb.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pappas C.T., Krieg P.A., Gregorio C.C. Nebulin regulates actin filament lengths by a stabilization mechanism. J. Cell Biol. 2010;189:859–870. doi: 10.1083/jcb.201001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ono S. Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton. 2010;67:677–692. doi: 10.1002/cm.20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mardahl-Dumesnil M., Fowler V.M. Thin filaments elongate from their pointed ends during myofibril assembly in Drosophila indirect flight muscle. J. Cell Biol. 2001;155:1043–1053. doi: 10.1083/jcb.200108026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sussman M.A., Baqué S., Kedes L. Altered expression of tropomodulin in cardiomyocytes disrupts the sarcomeric structure of myofibrils. Circ. Res. 1998;82:94–105. doi: 10.1161/01.res.82.1.94. [DOI] [PubMed] [Google Scholar]
- 78.McKeown C.R., Nowak R.B., Fowler V.M. Tropomodulin1 is required in the heart but not the yolk sac for mouse embryonic development. Circ. Res. 2008;103:1241–1248. doi: 10.1161/CIRCRESAHA.108.178749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stevenson T.O., Mercer K.B., Benian G.M. unc-94 encodes a tropomodulin in Caenorhabditis elegans. J. Mol. Biol. 2007;374:936–950. doi: 10.1016/j.jmb.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yamashiro S., Cox E.A., Ono S. Sarcomeric actin organization is synergistically promoted by tropomodulin, ADF/cofilin, AIP1 and profilin in C. elegans. J. Cell Sci. 2008;121:3867–3877. doi: 10.1242/jcs.040477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gokhin D.S., Lewis R.A., Fowler V.M. Tropomodulin isoforms regulate thin filament pointed-end capping and skeletal muscle physiology. J. Cell Biol. 2010;189:95–109. doi: 10.1083/jcb.201001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kremneva E., Makkonen M.H., Lappalainen P. Cofilin-2 controls actin filament length in muscle sarcomeres. Dev. Cell. 2014;31:215–226. doi: 10.1016/j.devcel.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ochala J., Gokhin D.S., Fowler V.M. Pointed-end capping by tropomodulin modulates actomyosin crossbridge formation in skeletal muscle fibers. FASEB J. 2014;28:408–415. doi: 10.1096/fj.13-239640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uversky V.N., Shah S.P., Kostyukova A.S. Systematic analysis of tropomodulin/tropomyosin interactions uncovers fine-tuned binding specificity of intrinsically disordered proteins. J. Mol. Recognit. 2011;24:647–655. doi: 10.1002/jmr.1093. [DOI] [PubMed] [Google Scholar]
- 85.Castillo A., Nowak R., Littlefield R.S. A nebulin ruler does not dictate thin filament lengths. Biophys. J. 2009;96:1856–1865. doi: 10.1016/j.bpj.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sussman M.A., Welch S., Molkentin J.D. Pathogenesis of dilated cardiomyopathy: molecular, structural, and population analyses in tropomodulin-overexpressing transgenic mice. Am. J. Pathol. 1999;155:2101–2113. doi: 10.1016/S0002-9440(10)65528-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bang M.L., Li X., Chen J. Nebulin-deficient mice exhibit shorter thin filament lengths and reduced contractile function in skeletal muscle. J. Cell Biol. 2006;173:905–916. doi: 10.1083/jcb.200603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Witt C.C., Burkart C., Labeit S. Nebulin regulates thin filament length, contractility, and Z-disk structure in vivo. EMBO J. 2006;25:3843–3855. doi: 10.1038/sj.emboj.7601242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gokhin D.S., Fowler V.M. Software-based measurement of thin filament lengths: an open-source GUI for distributed deconvolution analysis of fluorescence images. J. Microsc. 2017;265:11–20. doi: 10.1111/jmi.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Winter J.M., Joureau B., Ottenheijm C.A. Mutation-specific effects on thin filament length in thin filament myopathy. Ann. Neurol. 2016;79:959–969. doi: 10.1002/ana.24654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Halim D., Hofstra R.M., Alves M.M. ACTG2 variants impair actin polymerization in sporadic megacystis microcolon intestinal hypoperistalsis syndrome. Hum. Mol. Genet. 2016;25:571–583. doi: 10.1093/hmg/ddv497. [DOI] [PubMed] [Google Scholar]
- 92.Gauthier J., Ouled Amar Bencheikh B., Soucy J.F. A homozygous loss-of-function variant in MYH11 in a case with megacystis-microcolon-intestinal hypoperistalsis syndrome. Eur. J. Hum. Genet. 2015;23:1266–1268. doi: 10.1038/ejhg.2014.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Olson E.N., Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miralles F., Posern G., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 95.Molnár I., Migh E., Mihály J. DAAM is required for thin filament formation and Sarcomerogenesis during muscle development in Drosophila. PLoS Genet. 2014;10:e1004166. doi: 10.1371/journal.pgen.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Taniguchi K., Takeya R., Sumimoto H. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J. Biol. Chem. 2009;284:29873–29881. doi: 10.1074/jbc.M109.059303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iskratsch T., Lange S., Ehler E. Formin follows function: a muscle-specific isoform of FHOD3 is regulated by CK2 phosphorylation and promotes myofibril maintenance. J. Cell Biol. 2010;191:1159–1172. doi: 10.1083/jcb.201005060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Skwarek-Maruszewska A., Hotulainen P., Lappalainen P. Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J. Cell Sci. 2009;122:2119–2126. doi: 10.1242/jcs.046805. [DOI] [PubMed] [Google Scholar]
- 99.Weibrecht I., Leuchowius K.J., Söderberg O. Proximity ligation assays: a recent addition to the proteomics toolbox. Expert Rev. Proteomics. 2010;7:401–409. doi: 10.1586/epr.10.10. [DOI] [PubMed] [Google Scholar]
- 100.Gokhin D.S., Kim N.E., Fowler V.M. Thin-filament length correlates with fiber type in human skeletal muscle. Am. J. Physiol. Cell Physiol. 2012;302:C555–C565. doi: 10.1152/ajpcell.00299.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Granzier H.L., Akster H.A., Ter Keurs H.E. Effect of thin filament length on the force-sarcomere length relation of skeletal muscle. Am. J. Physiol. 1991;260:C1060–C1070. doi: 10.1152/ajpcell.1991.260.5.C1060. [DOI] [PubMed] [Google Scholar]
- 102.Ringkob T.P., Swartz D.R., Greaser M.L. Light microscopy and image analysis of thin filament lengths utilizing dual probes on beef, chicken, and rabbit myofibrils. J. Anim. Sci. 2004;82:1445–1453. doi: 10.2527/2004.8251445x. [DOI] [PubMed] [Google Scholar]
- 103.Sanger J.W., Wang J., Pruyne D. Assembly and maintenance of myofibrils in striated muscle. Handb. Exp. Pharmacol. 2017;235:39–75. doi: 10.1007/164_2016_53. [DOI] [PubMed] [Google Scholar]
- 104.Breitsprecher D., Goode B.L. Formins at a glance. J. Cell Sci. 2013;126:1–7. doi: 10.1242/jcs.107250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kanaya H., Takeya R., Sumimoto H. Fhos2, a novel formin-related actin-organizing protein, probably associates with the nestin intermediate filament. Genes Cells. 2005;10:665–678. doi: 10.1111/j.1365-2443.2005.00867.x. [DOI] [PubMed] [Google Scholar]
- 106.Rosado M., Barber C.F., Goode B.L. Critical roles for multiple formins during cardiac myofibril development and repair. Mol. Biol. Cell. 2014;25:811–827. doi: 10.1091/mbc.E13-08-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shwartz A., Dhanyasi N., Shilo B.Z. The Drosophila formin Fhos is a primary mediator of sarcomeric thin-filament array assembly. eLife. 2016;5:e16540. doi: 10.7554/eLife.16540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li D., Hallett M.A., Shou W. Dishevelled-associated activator of morphogenesis 1 (Daam1) is required for heart morphogenesis. Development. 2011;138:303–315. doi: 10.1242/dev.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sui Z., Nowak R.B., Fowler V.M. Tropomodulin3-null mice are embryonic lethal with anemia due to impaired erythroid terminal differentiation in the fetal liver. Blood. 2014;123:758–767. doi: 10.1182/blood-2013-03-492710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sui Z., Nowak R.B., Fowler V.M. Regulation of actin polymerization by tropomodulin-3 controls megakaryocyte actin organization and platelet biogenesis. Blood. 2015;126:520–530. doi: 10.1182/blood-2014-09-601484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yang S., Barbu-Tudoran L., Lehman W. Three-dimensional organization of troponin on cardiac muscle thin filaments in the relaxed state. Biophys. J. 2014;106:855–864. doi: 10.1016/j.bpj.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.