Abstract
Background and objectives
The calculated panel reactive of antibodies (cPRAs) necessary for kidney donor-pair exchange and highly sensitized programs are estimated using different panel reactive antibody (PRA) calculators based on big enough samples in Eurotransplant (EUTR), United Network for Organ Sharing (UNOS), and Canadian Transplant Registry (CTR) websites. However, those calculators can vary depending on the ethnic they are applied. Here, we develop a PRA calculator used in the Spanish Program of Transplant Access for Highly Sensitized patients (PATHI) and validate it with EUTR, UNOS, and CTR calculators.
Methods
The anti-human leukocyte antigen (HLA) antibody profile of 42 sensitized patients on waiting list was defined, and cPRA was calculated with different PRA calculators.
Results
Despite different allelic frequencies derived from population differences in donor panel from each calculator, no differences in cPRA between the four calculators were observed. The PATHI calculator includes anti-DQA1 antibody profiles in cPRA calculation; however, no improvement in total cPRA calculation of highly sensitized patients was demonstrated.
Interpretation and conclusion
The PATHI calculator provides cPRA results comparable with those from EUTR, UNOS, and CTR calculators and serves as a tool to develop valid calculators in geographical and ethnic areas different from Europe, USA, and Canada.
Keywords: anti-human leukocyte antigen antibodies, calculated panel reactive of antibody, kidney transplantation, highly sensitized patients, organ allocation, PATHI
Introduction
Kidney transplantation of highly sensitized patients remains a challenge. The presence of numerous preformed anti-human leukocyte antigen (HLA) antibodies makes it difficult to find a compatible donor. Therefore, these patients spend prolonged periods of time on the waiting list with associated increase in morbidity and mortality (1).
The way to measure the sensitization level has traditionally been the panel reactive antibodies (PRAs). The PRA is determined by testing the patient’s serum against a panel of HLA-typed donor cells and estimating the likelihood of finding a crossmatch compatible donor. However, PRA is labor intensive and depends on the composition of the panel limited by the number of donors available and the techniques used for anti-HLA antibodies detection.
The introduction of solid-phase assays using single HLA antigens manufactured by recombinant DNA technologies (Luminex) has increased the sensitivity and specificity to detect and identify HLA-specific antibodies compared to previous methods like complement-dependent cytotoxicity (CDC) (2, 3). The improvement of sensitivity not only allows a better definition of the anti-HLA antibody profile or forbidden HLA antigens but also identifies permissive antigens for transplantation.
The virtual crossmatch (vXM) could anticipate the results of complement-dependent cytotoxic crossmatch comparing HLA-typed donors with acceptable HLA antigens defined by the patient anti-HLA antibody profile (4).
The calculated panel reactive of antibody (cPRA) is based upon unacceptable HLA antigens (UHA), identified by the presence of HLA-specific antibodies in the sera of transplant candidates. It is calculated from the HLA antigen frequencies in a given population and represents the percentage of actual kidney donors that express at least one of the unacceptable antigens.
The different organ allocation systems such as Eurotransplant (EUTR), United Network for Organ Sharing (UNOS), and Canadian Transplant Registry (CTR) try to increase the chances of highly sensitized patients to be transplanted by giving them additional points or including them in special programs and perform vXM in transplant alerts. To avoid skewing in the allocation, due to different allelic frequencies among geographical areas (5), every transplant program ideally should use its own donor database to calculate the cPRA.
In 2014 the Spanish Ministry of Health, through the National Transplant Organization implemented the Spanish Program of Transplant Access for Highly Sensitized patients (PATHI), a specific program for highly sensitized renal patients. Here, we present the cPRA results obtained with the PRA calculator setup for the program and compare them with established PRA calculators from international sources to ascertain their results for a given patient population.
Materials and Methods
PATHI Calculator
The PATHI calculator has been designed with 250 genotypes (HLA loci A, B, C, DRB1, DRB3, DRB4, DRB5, and DQB1) from donors typed in Tissue Typing Laboratory at University Hospital of Albacete, Spain, whereas the DQA1 was deducted by linkage disequilibrium (6–8). Unlike other systems using the formula developed by Zachary and Braun (9) based on population frequency of antigens and haplotypes, PATHI uses the classical system for PRA calculation dividing the number of positive reactions by the total number of subjects tested.
Samples and Anti-HLA Testing
Serum samples from 42 sensitized patients on renal waiting list during 2015 from our institution (Hospital Universitario Marqués de Valdecilla) were studied for anti-HLA class I and class II antibody testing with the LABScreen® Mixed and Single Antigen assays (One Lambda, Canoga Park, CA, USA) according to the manufacturer’s protocol. We considered as positive specificities when raw mean fluorescence intensity (MFI) above 1,500 and/or baseline MFI above 1,000.
cPRA Comparison
Once the profile of UHA was defined for each patient, the cPRA was assessed using PATHI calculator for class I and class II antigens independently and for both classes combined. The results obtained were compared with those defined by cPRA from the EUTR and CTR webpage tool (10, 11) and the UNOS PRA calculator (12). The EUTR and UNOS calculators consider HLA-A, HLA-B, HLA-C, HLA-DR, and HLA-DQ frequencies whereas CTR and PATHI also include DQA1. The allele frequencies in PATHI calculator are shown in Table 1.
Table 1.
Allelic frequencies used for the panel reactive antibody PATHIa calculator.
| A locus | B locus | DR locus | |||
|---|---|---|---|---|---|
| A1 | 0.09226 | B7 | 0.07696 | DR1 | 0.08131 |
| A2 | 0.27889 | B8 | 0.05979 | DR103 | 0.02020 |
| A3 | 0.11456 | B13 | 0.01005 | DR2 | 0.12935 |
| A11 | 0.07696 | B18 | 0.08786 | DR3 | 0.16021 |
| A23 (9) | 0.02635 | B27 | 0.02635 | DR4 | 0.12822 |
| A24 (9) | 0.10557 | B35 | 0.11231 | DR5 | 0.12485 |
| A25 (10) | 0.02225 | B37 | 0.00000 | DR6 | 0.14692 |
| A26 (10) | 0.04501 | B38 (16) | 0.03460 | DR7 | 0.16813 |
| A29 (19) | 0.08131 | B39 (16) | 0.01005 | DR8 | 0.01410 |
| A30 (19) | 0.06192 | B41 | 0.01410 | DR9 | 0.00602 |
| A31 (19) | 0.01207 | B42 | 0.00200 | DR10 | 0.01005 |
| A32 (19) | 0.01816 | B44 (12) | 0.17054 | DR11 (5) | 0.11682 |
| A33 (19) | 0.02225 | B45 (12) | 0.02430 | DR12 (5) | 0.00803 |
| A34 (10) | 0.00000 | B46 | 0.00000 | DR13 (6) | 0.13282 |
| A36 | 0.00000 | B47 | 0.00401 | DR14 (6) | 0.01410 |
| A43 | 0.00000 | B48 | 0.00000 | DR15 (2) | 0.09889 |
| A66 (10) | 0.00401 | B49 (21) | 0.04501 | DR16 (2) | 0.03046 |
| A68 (28) | 0.02840 | B50 (21) | 0.02430 | DR17 (3) | 0.15620 |
| A69 (28) | 0.00602 | B51 (5) | 0.08131 | DR18 (3) | 0.00401 |
| A74 (19) | 0.00000 | B52 (5) | 0.01410 | ||
| A80 | 0.00200 | B53 | 0.01207 | ||
| B55 (22) | 0.00401 | ||||
| B56 (22) | 0.00200 | ||||
| B57 (17) | 0.02635 | ||||
| B58 (17) | 0.00803 | ||||
| B59 | 0.00000 | ||||
| B60 (40) | 0.01005 | ||||
| B61 (40) | 0.01005 | ||||
| B62 (15) | 0.04711 | ||||
| B63 (15) | 0.00401 | ||||
| B64 (14) | 0.01005 | ||||
| B65 (14) | 0.04292 | ||||
| B67 | 0.00000 | ||||
| B71 (70) | 0.00000 | ||||
| B72 (70) | 0.00200 | ||||
| B73 | 0.00000 | ||||
| B75 (15) | 0.00000 | ||||
| B76 (15) | 0.00000 | ||||
| B77 (15) | 0.00000 | ||||
| B78 | 0.00200 | ||||
| B81 | 0.00000 | ||||
| B82 | 0.00000 | ||||
aData obtained from 250 Spanish donors (details in Section “Material and Methods”).
Statistical Analysis
The Kruskal–Wallis and Spearman’s rho test were used to compare results from each calculator and to assess the correlation between the calculators. All the statistical analyses were performed using GraphPad Prism 5 software (San Diego, CA, USA).
Results
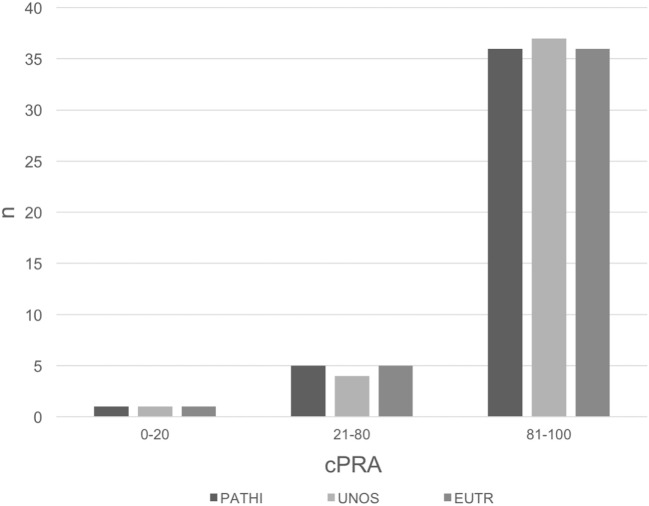
The distribution of cPRA calculated with PATHI in the 42 sensitized patients in our waiting list is summarized in Figure 1 where 33 of 42 presented >90% total cPRA. The total cPRA was calculated with different PRA calculators (10–12), but no differences were observed between calculators (Table 2). The correlation of cPRA calculations between PATHI and the other calculators was significant (Figure S1 in Supplementary Material). These calculators allow the estimation of independent class I and class II cPRA, but no differences were found in individual cPRA for either class I or class II antibodies (Table 2).
Figure 1.
Frequency distribution of calculated panel reactive of antibody (cPRA) with different panel reactive antibody calculators. Different ranges of cPRA were established: 0–20% (low sensitized), 21–80% (sensitized), and 81–100% (highly sensitized).
Table 2.
Comparison of class I, class II, and total calculated panel reactive of antibody (cPRA) with different calculators.
| cPRA, median (IQR) | PATHI | United Network for Organ Sharing | Eurotransplant | CTR | p Value |
|---|---|---|---|---|---|
| Class I | 91 (38.2–99) | 89 (37.2–99) | 89.8 (38.3–98.6) | 89.5 (38–99) | 0.99 |
| Class II | 86.5 (55.7–98) | 92.5 (60.5–98.2) | 92.4 (61.4–98.5) | 89 (60.5–98) | 0.90 |
| Total | 99 (94–100) | 99 (95–100) | 99 (94.6–99.9) | 99 (94.7–100) | 0.96 |
IQR, interquartile range.
Kruskal–Wallis test was applied.
Since PATHI calculator is used to evaluate sensitized patients in order to get access to the highly sensitized program, we compared the total cPRA in those patients with cPRA >90% (Figure S2 in Supplementary Material). A slightly better correlation of total cPRA including only these highly sensitized patients between PATHI and EUTR calculator was observed (r = 0.90 vs 0.87 vs 0.88, UNOS and CTR, respectively).
The main difference between PATHI and EUTR and UNOS calculators is that it includes HLA-DQA1 antigens. To assess the potential impact of HLA-DQA1 antigens in cPRA calculation, we compared the cPRA obtained with only DQA1 profile reaction and their DQB1 (Table 3) and DRB1 associations (Table S1 in Supplementary Material). The cPRA is much lower when DQA1 immunization is considered. For example, when a patient reacts against DQA1*02 and the calculator lacks DQA1 antigens, the assignment of DQB1 associations should be included as UHA (i.e., DQB1*02, DQB1*03, and DQB1*04). Consequently, the cPRA increases from 31% (PATHI-DQA1) to 83, 80.48, and 83% (in PATHI without DQA1, EUTR, and UNOS calculators, respectively, Table 3).
Table 3.
Comparison of calculated panel reactive of antibody (cPRA) assessing the effect of DQA1 anti-human leukocyte antigen profile and their associated DQB1 alleles.
| DQA1 unique reactive profile | cPRA PATHI (DQA1) (%) | Associated DQB1 | cPRA PATHI (%) | cPRA Eurotransplant (%) | cPRA United Network for Organ Sharing (%) |
|---|---|---|---|---|---|
| *01 | 58 | *05, *06 | 58 | 68.37 | 64 |
| *02 | 31 | *02, *03, *04 | 83 | 80.48 | 83 |
| *03 | 26 | *02, *03, *04 | 83 | 80.48 | 83 |
| *04 | 3 | *02, *04 | 55 | 39.75 | 45 |
| *05 | 50 | *02, *03:01 | 70 | 78.15 | 67 |
| *06 | 0 | *03:01 | 30 | 56.16 | 39 |
To further investigate the impact on different cPRA calculation including the DQA1-typed donors, we studied highly sensitized patients with anti-HLA antibody profile of DQA1 reaction. Three of 33 highly sensitized patients had different degrees of DQA1 reactivity (summarized in Table 4).
Table 4.
Cases of anti-human leukocyte antigen (HLA) antibody profile against DQA1 and the effect on DQ, class II, and global calculated panel reactive of antibody (cPRA).
| Case number | Specificities |
cPRA |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Anti-HLA class I antibodies |
Anti-HLA class II antibodies |
DQ cPRA |
Class II cPRA |
Total cPRA |
||||||||||
| A antigens | B antigens | DR antigens | DQB1 | DQA1 | PATHI | Eurotransplant (EUTR) | United Network for Organ Sharing (UNOS) | PATHI | EUTR | UNOS | PATHI | EUTR | UNOS | |
| Case 1 | *03:01, *04 | *02, *03 | 71 | 92 | 100 | |||||||||
| 1, 2, 23, 24, 25, 26, 29, 31, 32, 33, 34, 36, 43, 66, 68, 69, 74, 80 | 13, 35, 38, 44, 45, 46, 49, 50, 51, 52, 53, 56, 57, 58, 59, 62, 63, 73, 75, 76, 77, 82 | 1, 4, 7, 9, 10, 51, 53, 103 | *02, *03, *04 | 83 | 80.48 | 83 | 98 | 97.83 | 97 | 100 | 99.96 | 100 | ||
| Case 2 | *02, *03:01, *05 | *02 | 86 | 95 | 100 | |||||||||
| 1, 3, 11, 24, 25, 26, 29, 30, 31, 32, 33, 34, 36, 43, 66, 68, 69, 74, 80 | 7, 8, 13, 18, 27, 37, 38, 39, 41, 42, 44, 45, 46, 47, 48, 49, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 67, 71, 72, 73, 75, 76, 77, 78, 81, 82 | 1, 7, 9, 10, 12, 52, 103 | *02, *03, *04, *05 | 94 | 94.12 | 96 | 98 | 98.41 | 99 | 100 | 100 | 100 | ||
| Case 3 | *03, *04, *05 | *03 | 72 | 93 | 100 | |||||||||
| 1, 2, 3, 11, 24, 25, 26, 29, 30, 31, 33, 34, 36, 43, 66, 68, 69, 74, 80 | 7, 13, 18, 27, 35, 37, 41, 42, 49, 51, 52, 53, 54, 55, 56, 57, 58, 59, 67, 73, 78, 81, 82 | 1, 4, 7, 8, 9, 10, 13, 14, 53, 103 | *02, *03, *04, *05 | 94 | 94.12 | 96 | 98 | 98.41 | 99 | 100 | 100 | 100 | ||
The class II cPRA calculated with serologic DQ antigens was higher than with PATHI-DQA1 calculator: Case #1: 98, 97.83, and 97 vs 92%; Case #2: 98, 98.41, and 99 vs 95%, and Case #3: 98, 98.41, and 99 vs 93% (PATHI, EUTR, and UNOS vs PATHI-DQA1 calculators). Nevertheless, the reduction in the class II cPRA was abolished when total cPRA was estimated in highly sensitized patients: Case #1: 100, 99.96, and 100 vs 100%; Case #2: 100, 100, and 100 vs 100%, and Case #3: 100, 100, and 100 vs 100% (PATHI, EUTR, and UNOS vs PATHI-DQA1). Moreover, the use of both DQA1-DRB1 and DQA1-DQB1 association showed comparable results of cPRA between the calculators in highly sensitized patients (Table S2 in Supplementary Material).
Discussion
An accurate calculation of the cPRA is crucial for highly sensitized patients to be included in special allocation programs that increase their chances to be offered an organ. Different calculators have been developed with this purpose. The EUTR group established the Acceptable Mismatch program for patients with a cPRA >85% and at least 2 years on dialysis. The new Organ Procurement and Transplantation Network Kidney Allocation Scheme gives the highest priority for local, regional, and national sharing to candidates with cPRA values of 98, 99, and 100% respectively. In Spain, PATHI includes potential recipients with a cPRA >98% and at least 1 year on dialysis with additional points according to the time in dialysis, age, and distance between donation and transplant center.
The EUTR PRA calculator uses HLA typing data from 6,870 organ donors. The UNOS cPRA is determined using an established logarithm and based on ethnic frequencies and HLA frequencies derived from the HLA phenotypes of more than 14,000 deceased donors whereas CTR has more than 1,700 donors in 2011 (13).
PATHI calculator takes into account the HLA phenotypes of 250 donors. This seems to be a major limitation of our calculator. Nonetheless, its intended use is to serve as a tool to implement a cPRA calculator in those geographic areas with special HLA alleles in which the use of EUTR or UNOS calculator cannot be applied. We are aware that the frequency of some of the HLA antigens could be over or underestimated due to the small number of donors included, but the percentages of positive reactions seems not be skewed by sample size of donor panel, as recently discussed (14). However, with 250 typed donors, it is possible to obtain representative allelic frequencies of the population although the number would be insufficient in calculating haplotype frequencies that could not be used for the calculation of PRA according to the frequencies. Nevertheless, the classic PRA calculation method is based on crossmatch with panels of 30–50 cells; therefore, the method based on vXM significantly increases the sample size of the standard reference sample.
The PATHI calculator is very flexible and can be easily adapted to have a more accurate representation of a donor population by introducing the HLA typing of new donors.
Another potential advantage of PATHI calculator is the inclusion of HLA-DQA1, which is not considered in EUTR and UNOS calculators but included in the Canadian calculator from CTR (13). Unlike PATHI, DQA1 data of CTR are from typed donors, comparable cPRAs were found (Table 2; Figures S1 and S2 in Supplementary Material). These results could validate the use of linkage-based data to “real” HLA typing data in the formulation of new calculators. Although HLA-DQA1 typing is not usually performed in tissue typing laboratories, it would be nowadays advisable in those countries with active kidney donor-pair exchange and highly sensitized programs.
This fact is of the utmost importance, especially in the case of patients whose serum reacts clearly with HLA-DQA1 and not with HLA-DQB1 antigens. The cPRA can change considerably when considering only anti-HLA-DQA1 instead of anti-HLA-DQ antibodies (Table 4), although in highly sensitized patients similar cPRAs were obtained irrespectively of DQA1-DQB1 or DQA1-DRB1 association applied in the calculator (Table S2 in Supplementary Material). In cases of highly sensitized patients, the class I antigen specificities could overcome the improvement of PRA calculators that include DQA1-typed donors. Nevertheless, the importance of calculators based on DQA1-typed donors over total cPRA in highly sensitized patients reacting only with class II antigens should be considered, especially when PRA cutoff values are used as the main criterion to include highly sensitized patients in donor-pair exchange and highly sensitized programs. In fact, recently, a reduction in cPRA using a modified UNOS calculator including DQA1 antigens vs UNOS calculator was observed (15).
None of the calculators assessed takes into account the anti-HLA-DP antigens. Despite low grade of polymorphism of HLA-DP, the inclusion of DPA and DPB antigens in CTR has resulted in an increase in cPRA (14). It remains to be tested with our and other calculators.
The differences observed in PRA with the calculators may be due to the differences in the frequency of each antigen in the reference population rather than the method of calculation.
The PRA calculator used for allocation should be accurate enough to avoid positive CDC crossmatch with previous vXM negative but also have to define UHA precisely in order to avoid delayed time in organ allocation. The PATHI calculator for highly sensitized patients obtained the same results as UNOS and EUTR PRA calculators, offering a consistent tool to allocate highly sensitized patients in our program and demonstrating its potential use in other geographical settings with small numbers.
Ethics Statement
This study was carried out in accordance with the recommendations of Ethical Committee of Cantabria with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki.
Author Contributions
EA: contributed with drafting the work and the acquisition, analysis, and interpretation of the data. ML-H and JO: contributed with the interpretation of the data, revising and final approval of the version published. IR: contributed with the design of the work, analysis of the data, and drafting the work. DS: contributed with the design of the work, drafting the work, interpretation of the data, and final approval of the version published.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Maria Luz Valcárcel, Celestina Burgueño, Margarita Gonzalez, David Ramos, and Eva Cianca for their technical support.
Funding
The work was partially supported by grants from the Fondo de Investigaciones Sanitarias (PI1100990 and PI1400378 from ISCIII).
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2017.00540/full#supplementary-material.
Comparison of calculated panel reactive of antibody (cPRA) with different calculators. The cPRA measured with PATHI calculator was compared with United Network for Organ Sharing (UNOS) (A), Eurotransplant (EUTR) (B), and CTR (C) calculators.
Correlation of calculated panel reactive of antibody (cPRA) measured with different calculators in highly sensitized patients. The cPRA comparison was performed in patients with cPRA above 90%. The cPRA measured with PATHI calculator was compared with United Network for Organ Sharing (UNOS) (A), Eurotransplant (EUTR) (B), and CTR (C) calculators.
References
- 1.Iyer HS, Jackson AM, Zachary AA, Montgomery RA. Transplanting the highly sensitized patient: trials and tribulations. Curr Opin Nephrol Hypertens (2013) 22(6):681–8. 10.1097/MNH.0b013e328365b3b9 [DOI] [PubMed] [Google Scholar]
- 2.Picascia A, Sabia C, Grimaldi V, Montesano ML, Sommese L, Schiano C, et al. Lights and shadows of anti-HLA antibodies detected by solid-phase assay. Immunol Lett (2014) 162(1 Pt A):181–7. 10.1016/j.imlet.2014.08.014 [DOI] [PubMed] [Google Scholar]
- 3.Ferrari-Lacraz S, Tiercy JM, Villard J. Detection of anti-HLA antibodies by solid-phase assay in kidney transplantation: friend or foe? Tissue Antigens (2012) 79(5):315–25. 10.1111/j.1399-0039.2012.01853.x [DOI] [PubMed] [Google Scholar]
- 4.Ellis TM, Schiller JJ, Roza AM, Cronin DC, Shames BD, Johnson CP. Diagnostic accuracy of solid phase HLA antibody assays for prediction of crossmatch strength. Hum Immunol (2012) 73(7):706–10. 10.1016/j.humimm.2012.04.007 [DOI] [PubMed] [Google Scholar]
- 5.Fernandez Vina MA, Hollenbach JA, Lyke KE, Sztein MB, Maiers M, Klitz W, et al. Tracking human migrations by the analysis of the distribution of HLA alleles, lineages and haplotypes in closed and open populations. Philos Trans R Soc Lond B Biol Sci (2012) 367(1590):820–9. 10.1098/rstb.2011.0320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fernandez-Vina MA, Falco M, Gao X, Cerna M, Sun Y, Raimondi E, et al. DQA1*03 subtypes have different associations with DRB1 and DQB1 alleles. Hum Immunol (1994) 39(4):290–8. 10.1016/0198-8859(94)90272-0 [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Vina MA, Gao XJ, Moraes ME, Moraes JR, Salatiel I, Miller S, et al. Alleles at four HLA class II loci determined by oligonucleotide hybridization and their associations in five ethnic groups. Immunogenetics (1991) 34(5):299–312. 10.1007/BF00211994 [DOI] [PubMed] [Google Scholar]
- 8.Klitz W, Maiers M, Spellman S, Baxter-Lowe LA, Schmeckpeper B, Williams TM, et al. New HLA haplotype frequency reference standards: high-resolution and large sample typing of HLA DR-DQ haplotypes in a sample of European Americans. Tissue Antigens (2003) 62(4):296–307. 10.1034/j.1399-0039.2003.00103.x [DOI] [PubMed] [Google Scholar]
- 9.Zachary AA, Braun WE. Calculation of a predictive value for transplantation. Transplantation (1985) 39(3):316–8. 10.1097/00007890-198503000-00024 [DOI] [PubMed] [Google Scholar]
- 10.Eurotransplant. [cited 2017 Jan 10]. Available from: https://www.etrl.org/Virtual PRA/Default.aspx
- 11.Canadian Transplant Registry. [cited 2017 Jan 10]. Available from: http://www.pra-calculator.ca/
- 12.Organ Procurement and Transplantation Network. [cited 2017 Jan 10]. Available from: https://optn.transplant.hrsa.gov/resources/allocation-calculators/cpra-calculator/
- 13.Tinckam KJ, Liwski R, Pochinco D, Mousseau M, Grattan A, Nickerson P, et al. cPRA increases with DQA, DPA, and DPB unacceptable antigens in the Canadian cPRA calculator. Am J Transplant (2015) 15(12):3194–201. 10.1111/ajt.13355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baxter-Lowe LA, Cecka M, Kamoun M, Sinacore J, Melcher ML. Center-defined unacceptable HLA antigens facilitate transplants for sensitized patients in a multi-center kidney exchange program. Am J Transplant (2014) 14(7):1592–8. 10.1111/ajt.12734 [DOI] [PubMed] [Google Scholar]
- 15.Tambur AR, Leventhal JR, Walsh RC, Zitzner JR, Friedewald JJ. HLA-DQ barrier: effects on cPRA calculations. Transplantation (2013) 96(12):1065–72. 10.1097/TP.0b013e3182a452a5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of calculated panel reactive of antibody (cPRA) with different calculators. The cPRA measured with PATHI calculator was compared with United Network for Organ Sharing (UNOS) (A), Eurotransplant (EUTR) (B), and CTR (C) calculators.
Correlation of calculated panel reactive of antibody (cPRA) measured with different calculators in highly sensitized patients. The cPRA comparison was performed in patients with cPRA above 90%. The cPRA measured with PATHI calculator was compared with United Network for Organ Sharing (UNOS) (A), Eurotransplant (EUTR) (B), and CTR (C) calculators.