Abstract
The mitochondrial H+-ATP synthase is a primary hub of cellular homeostasis by providing the energy required to sustain cellular activity and regulating the production of signaling molecules that reprogram nuclear activity needed for adaption to changing cues. Herein, we summarize findings regarding the regulation of the activity of the H+-ATP synthase by its physiological inhibitor, the ATPase inhibitory factor 1 (IF1) and their functional role in cellular homeostasis. First, we outline the structure and the main molecular mechanisms that regulate the activity of the enzyme. Next, we describe the molecular biology of IF1 and summarize the regulation of IF1 expression and activity as an inhibitor of the H+-ATP synthase emphasizing the role of IF1 as a main driver of energy rewiring and cellular signaling in cancer. Findings in transgenic mice in vivo indicate that the overexpression of IF1 is sufficient to reprogram energy metabolism to an enhanced glycolysis and activate reactive oxygen species (ROS)-dependent signaling pathways that promote cell survival. These findings are placed in the context of mitohormesis, a program in which a mild mitochondrial stress triggers adaptive cytoprotective mechanisms that improve lifespan. In this regard, we emphasize the role played by the H+-ATP synthase in modulating signaling pathways that activate the mitohormetic response, namely ATP, ROS and target of rapamycin (TOR). Overall, we aim to highlight the relevant role of the H+-ATP synthase and of IF1 in cellular physiology and the need of additional studies to decipher their contributions to aging and age-related diseases.
Keywords: Mitochondria, ATPase inhibitory factor 1 (IF1), Metabolic reprogramming, Reactive oxygen species (ROS), Cancer, Lifespan
Introduction
Mitochondria have been traditionally regarded as the “power stations” of the cell since they produce most of cellular energy requirements in a process known as oxidative phosphorylation (OXPHOS). They are also primary hubs of intermediary metabolism because they generate precursors for the synthesis of macromolecules and synthesize heme and Fe-S clusters present in some proteins. However, this notion has been expanded in recent years after the demonstration of the role played by mitochondria in the execution of cell death [1–3] and in establishing pathways to communicate their status to the nucleus [4, 5] and to other compartments [6] to adapt cellular responses to different programs, such as proliferation, differentiation and stress responses [4, 7]. Mitochondrial communication is exerted by signaling molecules that emanate from the organelle and affect by different mechanisms cell fate and behavior. Some of the signaling molecules include Ca2+, reactive oxygen species (ROS), metabolites, such as citrate, ATP, phosphatidylethanolamine, and mitochondrial resident proteins and/or peptides encoded in mtDNA [7–15].
The dynamic fusion and fission events experienced by the mitochondrial reticulum are also involved in signaling [16–18]. The primary sensor of mitochondrial status is the mitochondrial membrane potential (ΔΨm) and preventing its collapse is essential for cellular viability [19]. The mitochondrial signalosome primarily triggers the reversible and/or irreversible modification of proteins and/or the allosteric regulation of enzymes that result in protein activation/inactivation of particular genetic and/or epigenetic programs for cellular adaptation. The list of programs affected by the mitochondrial signalosome is just being glimpse, and remarkably, some of them are responsible for promoting adaptive mechanisms that allow cells to withstand subsequent detrimental stresses, a concept coined as “mitohormesis” [20, 21].
A primary hub of mitochondrial energy generation and signaling is the OXPHOS system that integrates the complexes of the electron transport chain (ETC) and the H+-ATP synthase in defined supercomplexes and subdomains of the inner mitochondrial membrane (IMM) [22–24]. In OXPHOS, the electrons derived from biological oxidations are transferred to the complexes of the ETC to reduce molecular oxygen and to generate the proton gradient used for the synthesis of ATP by the H+-ATP synthase [25, 26]. Even under conditions of efficient electron transfer a small percentage of the electrons funneled into the ETC generate superoxide radical [9], which is rapidly converted into the signaling molecule hydrogen peroxide. Its production is largely increased in pathophysiological situations [7, 27, 28] or could contribute to mitohormesis [20, 21]. This review is mainly focused on the regulation of the activity of the H+-ATP synthase by its physiological inhibitor the ATPase inhibitory factor 1 (IF1) with emphasis in the different signaling pathways that contribute to mitohormesis as a result of its activation as inhibitor of the engine of OXPHOS.
The H+-ATP synthase: a core hub in mitochondrial structure and function
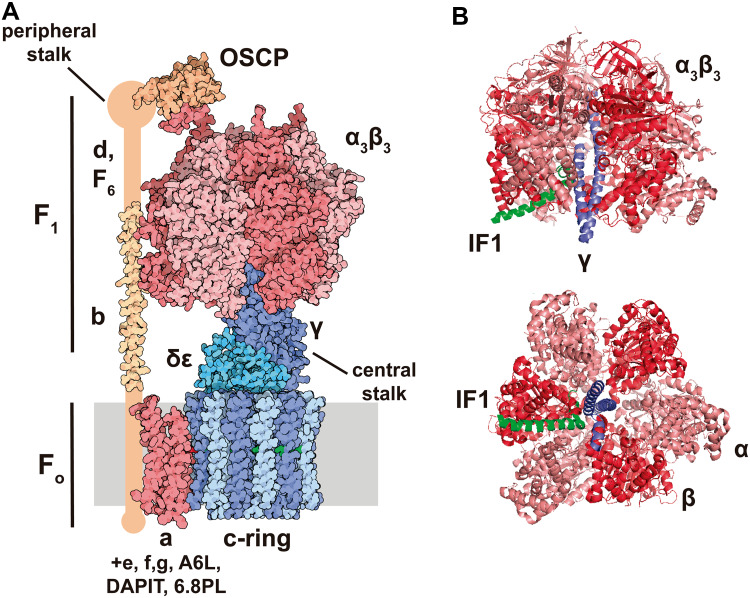
The H+-ATP synthase is a multisubunit complex built by two major functional domains: the membrane-embedded Fo-ATPase (a and 8c subunits) and the membrane-extrinsic catalytic F1-ATPase (3α, 3β, γ, δ and ε subunits) domains [25] (Fig. 1a). Both domains are linked by a central stalk that contains γ, δ and ε subunits of the F1-domain and a peripheral stalk that contains b, d, F6, and OSCP (oligomycin sensitivity-conferring protein) subunits [25] (Fig. 1a). Additional subunits have been described in the mitochondrial complex when the enzyme is purified in the presence of phospholipids: e, f, g, A6L, DAPIT (diabetes-associated protein in insulin-sensitive tissues) and the 6.8-kDa proteolipid (6.8PL). Their role in the activity of the enzyme remains to be fully characterized [25, 29]. ATP synthesis by the H+-ATP synthase is driven by the influx of protons into the matrix through the a subunit that promotes the rotation of the c-ring and of the attached central stalk, the later triggering the conformational changes in the F1-ATPase that drives ATP synthesis from ADP and Pi [25, 30]. The H+-ATP synthase is a reversible engine because ATP hydrolysis triggers rotation of the c-ring in the opposite direction generating a proton gradient across the inner membrane [25, 30].
Fig. 1.
Structure of the bovine H+-ATP synthase and binding site of IF1. a The soluble F1-ATPase domain is composed by the 3α3β subassembly (salmon/red) and γ (dark blue), δ and ε (light blue) subunits, while the membrane-embedded Fo domain is formed by subunit a (red) and a ring of 8c subunits (light/dark blue). Both domains are linked together by a central stalk (γ, δ, ε subunits of the F1 domain) and a peripheral stalk (b, d F6, A6L, OSCP subunits, in orange). The 3D structure of the peripheral stalk is not fully resolved. Except for a and A6L subunits, the remainder are encoded in the nucleus. Molecular reconstruction from PDB. b Lateral and basal view of the bovine F1 domain (α subunit is shown in salmon, β in red and γ in blue) complexed with a fragment of IF1 (green). IF1 binds to the αβ interface through residues 1–37 and also contacts the γ subunit. Images taken from [44] (PDB: 1OHH) and created with the PyMOL Molecular Graphics System
The assembly of the H+-ATP synthase in Saccharomyces cerevisiae comprises two separate and coordinated pathways that involve (1) the stepwise assembly of Atp6 (the homologue of a subunit) and Atp8 (the homologue of A6L subunit) with the peripheral stalk and (2) the assembly of the c-ring with the F1-ATPase domain [31, 32]. Later steps in the assembly process are mediated by the INA complex, a matrix-exposed inner membrane protein complex that facilitates the assembly of the peripheral stalk [33]. The assembly process in humans is similar to yeast [34]. Two of the “supernumerary” subunits, DAPIT [35] and 6.8PL [36], seem to be involved in the assembly process because its silencing in HeLa cells causes the loss of H+-ATP synthase complexes. Super-assemblies of the H+-ATP synthase [24] play an important role in determining the structure of the mitochondrial inner membrane because they contribute to the generation of cristae [37]. In fact, dimeric arrays of the H+-ATP synthase in the inner membrane of yeast [38] and mammalian mitochondria [39] generate the curvature of the IMM that promotes cristae formation [37, 40]. Recent findings suggest that while the e–g heterodimer promotes the local membrane curvature, the f subunit in the monomers establishes the contacts in the dimer interface [41].
The ATPase inhibitory factor 1 (IF1), which is a main regulator of the activity of the H+-ATP synthase (see below) has also been claimed to promote dimerization of the H+-ATP synthase [42–47]. However, other studies do not support this idea [48–50] or suggest that IF1 is just a bridge stabilizing two F1-ATPase domains with no crucial role in dimer formation [51]. Interestingly, recent findings support that the mitochondrial permeability transition pore (PTP) [3] contains dimers of the H+-ATP synthase [3, 52–54], consistent with the finding that subunit c of the Fo domain is required for the activity of the PTP [55, 56]. Interestingly, it has been suggested that the low probability of PTP opening at low pH might be due to the binding of IF1 [3] because the histidine reagent diethylpyrocarbonate is able to restore the ability to induce PTP opening and prevents IF1 binding to the inner membrane [3]. However, alternative explanations for IF1-mediated protection against cell death have already been provided in cellular [57] and in vivo mouse models [42, 58] by the mitohormetic role played by IF1.
The activity of the H+-ATP synthase is regulated by the cellular energy demand, the covalent modification of several of its subunits and by different metabolites and regulatory proteins that interact with the enzyme [59–62]. One of the key regulators of the H+-ATP synthase activity is Ca2+ [8]. Mitochondria are crucial organelles in intracellular Ca2+ buffering and signaling [63, 64]. S100A1 is a Ca2+-sensing protein that is expressed predominantly in cardiac muscle and interacts with the F1-ATPase in a Ca2+-dependent manner, increasing its activity [65]. Consistently, S100A1 knockout mice show reduced H+-ATP synthase activity in cardiomyocytes [65]. The Ca2+-inhibitor binding protein (CaBI) is another Ca2+-sensitive protein that dissociates from the H+-ATP synthase molecule in the presence of high intracellular Ca2+ levels and has been suggested as a candidate for the regulation of the H+-ATP synthase [61, 66].
The major Ca2+ store in the cell is the endoplasmic reticulum (ER). Contacts between mitochondria and the ER called mitochondria-associated membranes (MAMs), mediate the transfer of ions, metabolites, proteins and phospholipids, including Ca2+ [6]. One important complex involved in these contacts is the yeast ERMES (ER-mitochondria encounter structure) complex [67], which is composed by four core proteins resident both in the ER and in mitochondria. These proteins are functionally connected to phospholipid biosynthesis and Ca2+ signaling [67]. ERMES is naturally involved in establishing and maintaining contact sites between the two organelles [68]. Further studies have revealed that ERMES is involved in mitochondrial dynamics, inheritance, protein import, mtDNA inheritance and mitophagy [69]. No orthologues of the ERMES complex have been found in mammals.
DAPIT, a “supernumerary” subunit of the H+-ATP synthase, may also play a role in the regulation of the H+-ATP synthase since it is highly expressed in cells with high aerobic metabolism [70]. Knockdown of DAPIT results in a drastic reduction in the activity and content of H+-ATP synthase complexes in human cells [35]. In contrast to these findings, the overexpression of DAPIT in HEK293T cells impairs mitochondrial biogenesis and promotes the generation of ROS, both resulting in an enhanced aerobic glycolysis and the activation of the Wnt/β-catenin signaling pathway [71].
Besides S100A1, CaBI and DAPIT, the activity of the H+-ATP synthase is regulated by additional interacting proteins, including the antiapoptotic protein Bcl-xL (B-cell lymphoma-extra-large) [72], the hypoxia-induced protein G0/G1 switch gene 2 (G0s2) [73] and coupling factor B (FB) [29]. Moreover, different covalent modifications and metabolites produced in mitochondria regulate the activity of the H+-ATP synthase (see [62] for details).
The ATPase inhibitory factor 1 (IF1): a master regulator of the activity of the H+-ATP synthase
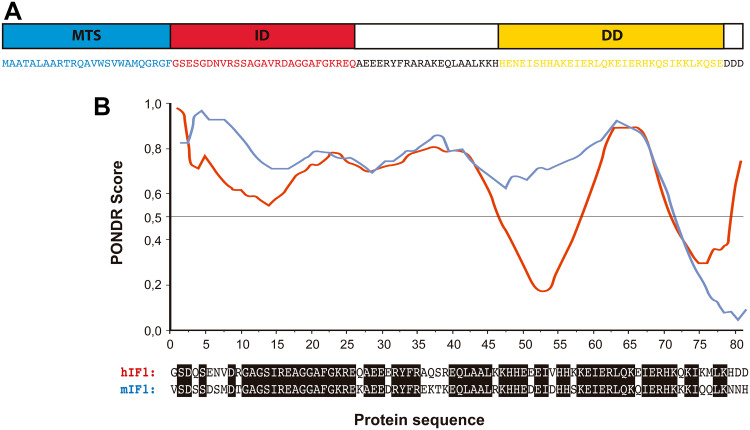
IF1 was first described in mitochondria from bovine heart as a small protein that inhibits the soluble F1 domain of the H+-ATP synthase [74] (Fig. 1b) with homologs in plants, yeasts, animals and humans (for review see [62]). The mammalian proteins present a high degree of sequence conservation and are able to inhibit the H+-ATP synthase of different species, including yeast [75]. In humans, IF1 is encoded in the nuclear ATPIF1 gene, located in chromosome 1, is translated as a precursor protein that upon mitochondrial import experiences the removal of the N-terminal 25-residue long pre-sequence [62]. The mature protein comprises a N-terminal intrinsically disordered domain [43] that interacts with the H+-ATP synthase and a C-terminal dimerization domain [76] (Fig. 2a). Bioinformatic predictions suggest that human and mouse homologues are also intrinsically disordered proteins (Fig. 2b). Intrinsically disordered proteins are important components of cellular signaling, since they can respond to intra- or extracellular cues by disorder-to-order conformational changes to exert a plethora of outcomes [77]. IF1 binding site is located in the catalytic interface between the α and β subunits of the F1 domain (Fig. 1b) as inferred using a truncated form of bovine IF1 (I1-60His) [78]. During the binding process, the N-terminus of I1-60His changes to an ordered α-helical structure [79] and once bound, IF1 blocks the rotation of the complex inhibiting the hydrolysis of ATP [80] (Fig. 1b).
Fig. 2.
IF1 is an intrinsically disordered protein. a Amino acid sequence of bovine IF1 and scheme of the main domains of the protein: the mitochondrial targeting sequence (MTS, blue), the N-terminal inhibitory domain (ID, red) and the α-helical coiled-coil dimerization domain (DD, yellow) are highlighted. b Prediction of the disordered regions of human (hIF1) and mouse IF1 (mIF1) using Predictor of Natural Disordered Regions (PONDR®) VLXT algorithm. The regions with PONDR score >0.5 are predicted as disordered
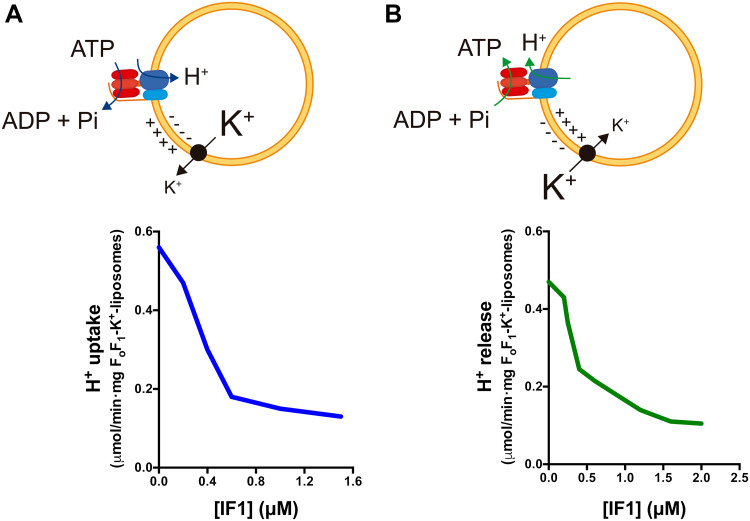
Recent findings point out that IF1 when bound to the enzyme inhibits both the ATP hydrolase and synthase activities [81] rather than acting solely as a unidirectional inhibitor of the hydrolase activity of the H+-ATP synthase [25, 46]. Indeed, inhibition of the H+-ATP synthase activity has been reported by the overexpression of IF1 or its constitutively active mutant H49K in cells in culture [57, 82, 83] and in neurons [58] and in hepatocytes [42] of transgenic mice in vivo. Consistently, it has been reported that IF1 inhibits the translocation of protons mediated by the H+-ATP synthase when operating in synthetic and hydrolytic modes [84] (Fig. 3). The IF1-mediated inhibition of the ATP synthetic activity can also be traced by the activation of signaling pathways sensing energy deprivation and the subsequent activation of glycolysis [42, 57, 58, 82, 83]. More direct confirmation that IF1 inhibits both activities of the enzyme was recently provided after showing that its binding to the synthase and inhibitory activities depend on its phosphorylation status [81].
Fig. 3.
IF1 inhibits H+ translocation through the H+-ATP synthase in both synthetic and hydrolytic modes. a H+ uptake is induced by valinomycin-mediated K+ release from FoF1-K+ liposomes with the H+-ATP synthase functioning in the hydrolytic mode. b H+ release is induced by valinomycin-mediated K+ uptake in FoF1-K+ liposomes with the H+-ATP synthase functioning in the synthetic mode. Rates of H+ uptake (a) and H+ release (b) are reduced when the liposomes are incubated with increasing concentrations of isolated IF1. Black circle valinomycin.
Adapted from [84]
Regulation of IF1 activity and expression
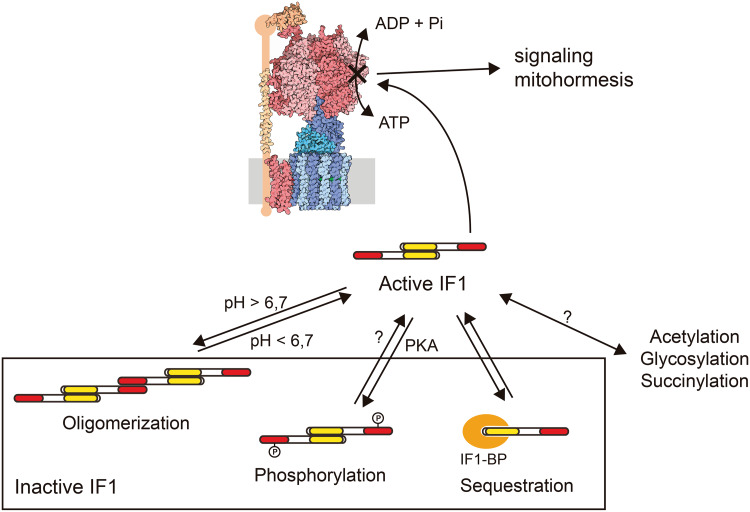
The regulation of IF1 activity as an inhibitor of the hydrolase activity of the H+-ATP synthase involves five histidine residues whose ionization status play an active role in the structure and oligomerization of IF1 [85]. When the mitochondrial matrix acidifies, i.e., when mitochondria become de-energized, such as in hypoxia and ischemia, IF1 tends to form inhibitory dimers [85]. At pH higher than 6.7, IF1 tends to form inactive tetramers by the interaction of two dimers through the N-terminus of the protein, hence masking the region that binds to the H+-ATP synthase [76, 85] (Fig. 4). Site-directed mutagenesis of H49 into a lysine generates the IF1-H49K mutant, which is active as inhibitor of the enzyme at pH higher than 6.7 [85] and has been used to effectively inhibit OXPHOS in a tissue-specific manner in transgenic mice [42, 58].
Fig. 4.
Post-translational regulation of IF1 activity. IF1 can be inhibited by pH-dependent oligomerization, phosphorylation mediated by a mitochondrial cAMP-dependent protein kinase A-like activity (PKA) or by sequestration by a mitochondrial membrane protein (IF1-BP). IF1 is also the substrate for acetylation, glycosylation and succinylation, although the physiological effects of these modifications remain to be fully characterized. The pool of active IF1 determines the population of inhibited H+-ATP synthase complexes, which might trigger retrograde signaling pathways
More recently, we have demonstrated that the inhibitory activity of IF1 depends on the phosphorylation status of a conserved serine 39 (S39) [81]. In the dephosphorylated state, IF1 binds to the H+-ATP synthase and inhibits both activities of the enzyme (Fig. 4). Phosphorylation of IF1 prevents its interaction with the H+-ATP synthase, abolishing its inhibitory activity (Fig. 4). IF1 is phosphorylated in vivo by the activity of a mitochondrial cAMP-dependent protein kinase [81]. Actually, PKA regulates diverse mitochondrial functions [62, 86], including an increase in the efficiency of OXPHOS [87–90].
A soluble adenylyl cyclase (sAC) has been described inside mitochondria [87, 91, 92], which is activated by Ca2+ [89] and bicarbonate [87]. In this regard, it is interesting to note that β-adrenergic stimulation of mouse heart in vivo also promotes an increase in cAMP levels inside mitochondria, the phosphorylation of IF1 and the subsequent activation of the H+-ATP synthase boosting ATP supply by OXPHOS [81]. We have recently summarized other physiologically relevant contexts for the regulation of IF1 activity by phosphorylation for tuning substrate availability and energy demand with OXPHOS activity in hypoxia, cell cycle progression and cancer [62, 81]. The mechanisms regulating the dephosphorylation of IF1 (if any) are unknown [62]. Nevertheless, we have suggested that the short half-life of the protein [83, 93] may rapidly reestablish the content of active dephospho-IF1 inside the organelle.
Besides the canonical binding site on the H+-ATP synthase, IF1 can also bind to OSCP [94] and to a small mitochondrial membrane protein that hampers its activity as an inhibitor of the H+-ATP synthase [95] (Fig. 4). Hence, IF1 oligomerization, phosphorylation and binding to a putative “receptor” may regulate the pool of active IF1 in mitochondria, allowing OXPHOS to adapt to different conditions of substrate availability and energy demand (Fig. 4). Moreover, additional covalent modifications of IF1 have been reported (Fig. 4) but their physiological relevance remains to be investigated [62].
Table 1 summarizes and compares the expression level of IF1 in different human tissues. As we have observed in mouse heart [81], not all the IF1 present in the tissue is active as an inhibitor under basal physiological conditions [81]. In fact, and based on the findings in heart, we suggest that a large fraction of IF1 is maintained functionally inactive (Fig. 4) in high energy-demanding tissues that show a high content of IF1 (brain, kidney and liver) to provide a reservoir of inhibitor that could prevent ATP wasting when OXPHOS is compromised [81]. On the other hand, the fraction of active IF1 inhibitor offers a pool of inactive H+-ATP synthase in the tissue that upon an enhanced metabolic demand could respond increasing the supply of ATP after the inactivation of IF1 (Fig. 4) [81]. We suggest that this pool of inactive H+-ATP synthase might contribute to retrograde signaling [62, 81] (Fig. 4).
Table 1.
IF1 expression in normal human tissues and in the corresponding carcinomas
| Organ/tissue | Normal | Carcinoma | Prognosis (High IF1) | References |
|---|---|---|---|---|
| Bladder | Negligible | High | High-risk | [107] |
| Brain (Neurons) | High | – | – | [58] |
| Brain (Glia) | Negligible | High | High-risk | [58, 113] |
| Breast | Negligible | High | Low-risk | [83] |
| Colon | Negligible | High | Low-risk | [57, 83, 109] |
| Endometrium | High | High | – | [83] |
| Kidney | High | High | – | [83] |
| Liver | High | High | High-risk | [83, 112] |
| Lung | Negligible | High | High-risk | [83, 108] |
| Ovary | Negligible | High | – | [83] |
| Stomach | High | High | High-risk/low-risk | [83, 109, 111] |
IF1 expression level is defined as negligible or high as assessed in western blots and/or immunohistochemistries. The relevance of IF1 as biomarker of survival and/or of disease recurrence was assessed in Kaplan–Meier plots using the cutoff to define potentially “high-risk” and “low-risk” groups as indicated in the indicated references. A high expression level of IF1 stratifies the cohort of patients as indicated. With the exception of data in reference [109] that has used mRNA levels, the rest of the studies used protein IF1 expression data
IF1 in cancer
Down-regulation of OXPHOS and the concurrent activation of aerobic glycolysis is a hallmark of cancer [96] and of undifferentiated cells [93, 97], including induced pluripotent stem cells (iPSCs) [98]. This metabolic phenotype is optimal because it supplies the anabolic intermediates needed for proliferation [99–101], and also protects from mitochondrial derived ROS [102] and cell death [103]. The analysis of large cohorts of different human carcinomas indicated that the partial reduction of OXPHOS in cancer cells is achieved by down-regulation of the expression of the catalytic subunit of the H+-ATP synthase (β-F1-ATPase) [104–106] (for additional references in other carcinomas see [99]). More recently, we have shown that metabolic rewiring to an enhanced glycolysis is also exerted by the overexpression of IF1 in carcinomas [57, 82, 83] (Table 1). Remarkably, the IF1 present in these carcinomas is found in its active dephosphorylated state [81].
Table 1 shows the changes in IF1 expression levels observed between normal tissues and the corresponding carcinomas. Tissues, such as bladder, breast, colon, lung and ovary show negligible expression level of IF1 under normal physiological conditions [57, 83, 107] (Table 1). However, upon transformation, the carcinomas arising in these tissues show very high levels of IF1 (Table 1). Interestingly, whereas in bladder [107] and non-small cell lung carcinomas [108] a high expression level of IF1 in the tumor predicts a worse patients’ prognosis, a low tumor expression of IF1 predicts a worse prognosis in colon and breast cancer patients [83]. Consistently, a recent study using mRNA expression levels also suggests that a high IF1 expression predicts the group of low-risk patients in colon cancer [109]. In agreement with a higher metastatic potential of breast cancer cells expressing a low level of IF1, it has been reported that lymph node metastasis of breast cancer patients had a lower expression level of IF1 when compared to the primary tumors [110]. The reasons for the differential behavior of IF1 in cancer progression in these carcinomas remain to be investigated.
High energy-demanding human tissues, such as brain, liver and kidney, and in addition stomach and endometrium, express the highest levels of IF1 under normal physiological conditions [82, 83] (Table 1). For most of these tissues (endometrium, kidney, liver and stomach) carcinogenesis does not promote a relevant increase in the tumor expression level of IF1 [83] (Table 1). However, in gastric carcinomas [111] and in hepatocarcinomas [112] the tumors that show a higher expression level of IF1 predict a worst patient prognosis. However, a recent study using mRNA expression levels suggests that a high IF1 expression predicts the group of low-risk patients [109]. Especial consideration deserves the brain which shows very high expression levels of IF1 in neurons and negligible in astrocytes [58] (Table 1). Interestingly, gliomas show a very high expression of IF1 that correlates with a poor patient prognosis [113] (Table 1).
The ATPIF1 proximal promoter contains a NFκB (nuclear factor kappa B) binding site [83]. It has been reported that binding of this transcription factor activates ATPIF1 transcription in a feedback loop that promotes hepatocarcinogenesis [112]. However, the accumulation of IF1 protein in other prevalent human carcinomas (colon, lung, breast and ovary) occurs with no relevant changes in ATPIF1 mRNA levels [83], suggesting that IF1 expression is mainly regulated at post-transcriptional levels in agreement with findings in other systems [114]. Indeed, IF1 has a very short half-life (~2–4 h) [83] and the control of its turnover is a relevant event in the differentiation of human mesenchymal stem cells [93]. The degradation of IF1 may involve several mitochondrial proteases [115], including serine-proteases [83, 93] and metalloproteases [116]. Mouse Ier3 (immediate early response 3) has been shown to physically interact with the C-terminal domain of IF1 and promote its degradation in a proteasome-independent way that requires ATP [116].
IF1 overexpression triggers mitochondrial hyperpolarization and the production of superoxide radical [57, 82]. Superoxide is rapidly converted into hydrogen peroxide, a ROS signaling molecule that activates nuclear programs of cell death protection. Mechanistically, ROS-mediated signaling induces the canonical NFκB pathway in IF1-overexpressing cancer cells [57, 83] and in neurons [58] promoting Bcl-xL-guided cell death protection. The nuclear signaling pathways activated in response to IF1 overexpression vary depending on the tissue. In fact, in liver it seems that the antioxidant response guided by Nrf2 [nuclear factor (erythroid-derived 2)-like 2] is crucial in the protection of hepatocytes from oxidative insults [42]. However, we should mention that the metabolic pre-conditioning afforded by IF1 overexpression in mouse liver is deleterious in the context of hepatocarcinogenesis [42], in agreement with recent findings in humans [112].
Since the H+-ATP synthase is a critical component of the PTP, IF1 by potentially regulating the oligomeric state of the H+-ATP synthase, might contribute to upgrade at the structural level the threshold to execute cell death [58, 117]. However, the overexpression of IF1-H49K mutant in the liver of transgenic mice, which promotes the formation of H+-ATP synthase dimers and protects the hepatocytes from oxidative insults, indicated that this protection is unrelated to differences in PTP opening and regulation [42]. Hence, we suggest that the cell death protection afforded by IF1 is primarily exerted by signaling mitohormesis [42, 57, 58].
Nucleo-mitochondria communication: mitohormesis
There is growing evidence supporting that a mild mitochondrial stress can protect the cell from subsequent insults, a concept termed mitohormesis [20, 21]. Mitohormesis is defined as an adaptive cellular response triggered by a mild mitochondrial malfunctioning that activates cytoprotective mechanisms to compensate the primary defect and results in long-lasting broad metabolic and molecular changes. These adaptive changes may finally lead to increased lifespan and healthspan. The mitohormetic response requires the existence of signaling pathways that sense mitochondrial function and inform the nucleus to trigger the adequate cellular programs to compensate stress [4]. The signaling pathways emanating from both mitochondria and the nucleus have been extensively reviewed elsewhere [4, 5].
A stress response pathway with well described cytoprotective effects is the mitochondrial unfolded protein response (UPRmt). Mitochondrial stress does not only involve a compromise in OXPHOS activity, arising from defects in mtDNA or poisoning of the ETC, but also by an accumulation of unfolded mitochondrial proteins [118]. Proteotoxic stresses that exceed the capacity of mitochondrial chaperones and proteases induce a transcriptional program in the nucleus in an attempt to restore proteostasis and adapt the cellular metabolism to the mitochondrial stress. This pathway has been thoroughly described in Caenorhabditis elegans [119–122] and is conserved in mammals [123–125]. In C. elegans, the sensor protein is ATFS-1 (activating transcription factor associated with Stress-1), which is normally imported into mitochondria and degraded in the matrix. Upon mitochondrial dysfunction, it is accumulated in the cytosol and tends to be imported into the nucleus, activating the transcription of genes encoding mitochondrial chaperones and proteases, proteins of the mitochondrial import machinery and genes involved in metabolism and ROS detoxification.
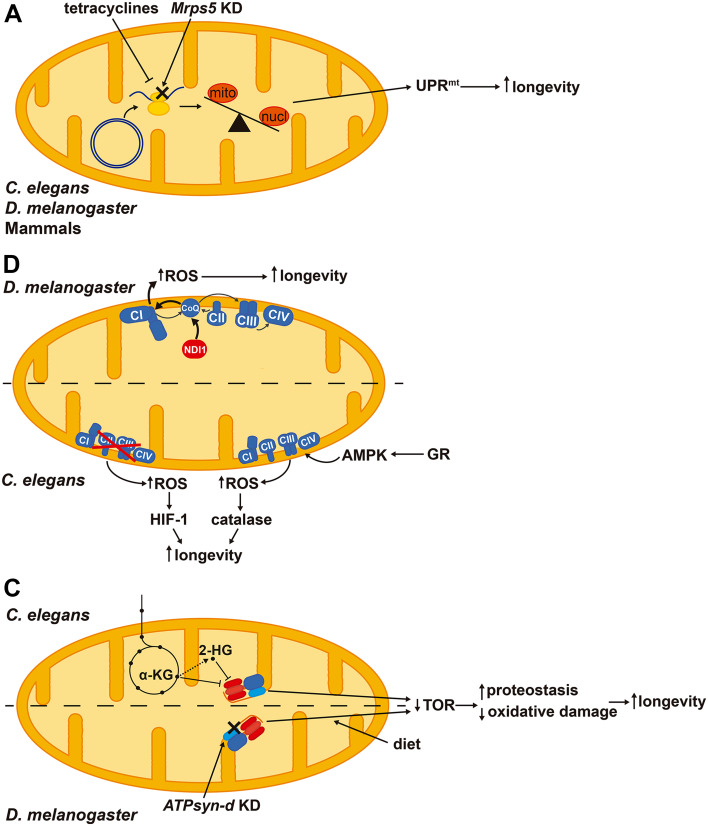
Activation of UPRmt may extend lifespan. Interestingly, knockdown of Mrps5 (mitochondrial ribosomal protein S5) and other ribosomal proteins in C. elegans impairs the synthesis of mtDNA-encoded ETC subunits and constitutively activates UPRmt extending lifespan [126] (Fig. 5a). In addition, doxycycline—an antibiotic that interferes with mitochondrial translation—also causes an imbalance between mitochondrial- and nuclear-encoded proteins activating UPRmt and extending lifespan in C. elegans and muscle fitness in Drosophila melanogaster [127] (Fig. 5a). Repletion of NAD+ levels in aged mice through the supplementation of its precursor nicotinamide riboside promotes longevity as well as increases the number and regenerative potential of muscle stem cells via UPRmt activation [128]. Importantly, inhibiting this pathway by silencing the prohibitin proteins blunts the pro-longevity effects of nicotinamide riboside [128]. Therefore, activation of UPRmt stress response improves mitochondrial homeostasis by enhancing proteotoxicity resistance and protects stem cells from senescence.
Fig. 5.
Signaling pathways that arise from mild mitochondrial stresses and contribute to lifespan extension in different model organisms. a Activation of the mitochondrial unfolded protein response (UPRmt) triggered by an imbalance between mitochondrial- and nuclear-encoded proteins extends longevity. b ROS derived from complex I (CI) by over-reduction of CoQ pool and reverse electron transport promote longevity in Drosophila melanogaster. Both reduction and enhancement of respiration in Caenorhabditis elegans trigger a ROS signal that promotes longevity via HIF-1 (hypoxia-inducible factor 1) or activating antioxidant defenses, respectively. c Inhibition of the H+-ATP synthase by α-ketoglutarate (α-KG) or 2-hydroxyglutarate (2-HG) increases longevity through repressing TOR (target of rapamycin) signaling and improving proteostasis. Diet composition interacts with the knockdown (KD) of D. melanogaster H+-ATP synthase subunit d (ATPsyn-d) and TOR signaling in extending lifespan. Mrps5: mitochondrial ribosomal protein S5, GR: glucose restriction, AMPK: AMP-dependent protein kinase
Increasing evidence suggests that the production of low levels of ROS may be protective for the cell, facing against the free-radical theory of aging and the long held supported protective role of antioxidants [21]. While high levels of ROS oxidize and destroy cellular structures [9], low amounts of ROS act as signaling molecules that can activate gene expression programs [4, 129]. Indeed, moderate ROS induce an antioxidant response mediated by Nrf2 [130], as well as mitochondrial biogenesis and expression of OXPHOS genes [131]. ROS have been reported to interact with two pro-longevity pathways in D. melanogaster, the UPRmt and antagonizing insulin signaling, which exert cell autonomous and systemic effects, respectively [132]. Interestingly, over-reduction of the coenzyme Q (CoQ) pool in D. melanogaster by the expression of NDI1—an alternative oxidoreductase that bypasses ETC complex I (CI)—enhances the production of superoxide radical and extends lifespan [133] (Fig. 5b). This effect is abolished when CI is genetically or pharmacologically inhibited suggesting that reverse electron transport may feed CI with electrons from CoQ to promote ROS production [133]. NDI1 expression, although increases superoxide levels, rescues the mitochondrial defects and the reduced lifespan in SOD2 and PINK1 knockdown flies, pointing out that the site of ROS production is important and might be beneficial for cellular homeostasis [133].
Likewise, reduction of mitochondrial activity has been related to extended lifespan in C. elegans. Silencing of subunits of the ETC complexes I, III and IV or atp-3 (homolog of OSCP) during development promotes longevity [134]. The effect of a mild inhibition of mitochondrial respiration on the extended lifespan appears to occur via increased ROS production, which activates HIF-1 (hypoxia-inducible factor 1)-mediated signaling [135] (Fig. 5b). Interestingly, treatment of worms with low levels of paraquat increases lifespan, while treatment with higher levels of the drug actually reduces lifespan, suggesting the existence of a hormetic response when the generated ROS are mild [135], in agreement with the idea that the amount of generated ROS influence lifespan [136].
The metabolic rate, and hence mitochondrial activity and ROS production have been linked to longevity in two controversial theories of aging [137, 138]. In yeast, an increased respiration rate is related to extended lifespan [139, 140] and calorie restriction—a well-known longevity-promoting intervention—increases mitochondrial respiration [141]. Similarly, calorie restriction extends lifespan in C. elegans [142] and in D. melanogaster [143] by inducing respiration. In case of the worm, promotion of mitochondrial respiration requires the activation of AMPK (AMP-dependent protein kinase) and also increases ROS production, triggering a cytoprotective antioxidant hormetic response [142] (Fig. 5b). Blunting the yeast TOR signaling pathway either by tor1 gene deletion or rapamycin treatment enhances the respiratory capacity and chemiosmotic coupling, thereby increasing mitochondrial membrane potential and ROS production [144]. Importantly, uncoupling of respiration in the tor1Δ mutants abolishes the pro-longevity effects of TOR inhibition, while triggering mitochondrial ROS production in wild-type strains with menadione is sufficient to extend lifespan [144]. In general, inhibition of mTOR signaling extends lifespan and healthspan in several organisms including mammals, and is believed as a main effector of dietary restriction [145]. Interventions that promote longevity, such as calorie restriction, mild mitochondrial dysfunction and impaired insulin/IGF1-signaling have been related to ROS-mediated signaling [21]. Regarding the link between mitochondrial activity and lifespan, different results have also been reported in mouse. For instance, in the conditional knockout model of the insulin receptor in adipose tissue an upregulated mitochondrial biogenesis and function correlates with an extended longevity apparently unrelated to an enhanced production of ROS [146]. In contrast, the knockout model of the ETC complex IV assembly factor Surf1 (surfeit locus protein 1)—which shows a reduction in cytochrome oxidase activity—is associated with a longer median lifespan and the induction of mitochondrial stress pathways comprising UPRmt, Nrf2 and mitochondrial biogenesis [147, 148].
Overall, and in agreement with previous suggestions [136], major thoughts that emerge from the implication of ROS in lifespan extension are that the amount being produced, the species generated, the site of production and the subsequent nuclear response determines ROS influence in lifespan extension.
Besides ROS, mitochondria also produce metabolites that can signal to the rest of the cell [4]. C. elegans respiratory mutants with extended lifespan generate high amounts of α-ketoacids [149], molecules structurally related to α-ketoglutarate, a metabolite that promotes longevity [59]. α-ketoglutarate binds to the α and β subunits of the H+-ATP synthase and inhibits the complex. By reducing ATP levels, α-ketoglutarate reduces TOR signaling, promoting autophagy and proteostasis that ultimately leads to lifespan extension (Fig. 5c) [59]. 2-hydroxyglutarate, an analog of α-ketoglutarate, also inhibits the H+-ATP synthase in C. elegans, reducing TOR signaling and extending lifespan [150] (Fig. 5c). Interestingly, treatment of wild-type C. elegans with the metabolites generated and extruded by respiratory mutant worms is sufficient to extend lifespan and this occurs at least by HIF-1-mediated signaling [149].
The mitochondrial H+-ATP synthase itself has been linked to lifespan because several of its subunits [atp-3, atp-4, atp-5 and asb-2, which, respectively, encode OSCP, F6, d and b subunit homologs and constitute the peripheral stalk of the complex (Fig. 1a)] were identified in RNAi screens for genes promoting longevity in C. elegans [134, 151]. Interestingly, a twofold upregulation of the D. melanogaster IF1 homolog (CG34423) has been reported in a fly model with extended lifespan [152]. Likewise, silencing of D. melanogaster ATPsyn-d (ATP synthase d subunit) reduces TOR signaling and activates genes involved in proteostasis and antioxidant defense, thus promoting a stress resistance phenotype that ultimately leads to lifespan extension [153] (Fig. 5c). Interestingly, this knockdown only promotes longevity when flies are fed with low carbohydrate-to-protein ratio, pointing out that longevity arises from an interaction between diet and the H+-ATP synthase [153]. This effect does not seem to be due to electron flux by different supercomplexes [154], because no changes in supercomplex assembly have been reported [153].
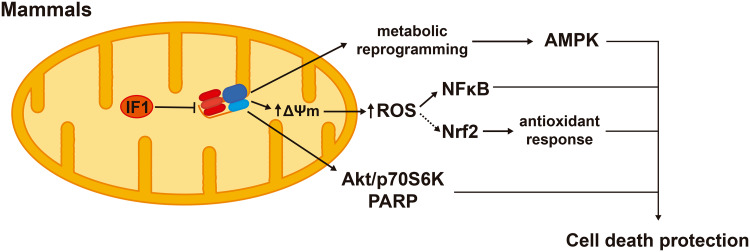
IF1 overexpression inhibits the H+-ATP synthase activity and reprograms energy metabolism to an enhanced glycolysis in cell lines [57] and in in vivo models [42, 58]. The IF1-mediated inhibition of the H+-ATP synthase represents a mild mitochondrial dysfunction that confers cytoprotection through the activation of cell survival, antioxidant and repair pathways. In colon cancer cells, IF1 triggers a mild ROS signal that promotes cell survival through NFκB signaling (Fig. 6) [57]. In the in vivo mouse model overexpressing hIF1-H49K in the liver, the transgene triggers the activation of the stress kinase AMPK [42]. In response to an oxidative insult, the transgenic mice deploy an increased antioxidant defense guided by Nrf2 when compared to controls, thus promoting hepatocyte survival [42] (Fig. 6). Likewise, the overexpression in vivo of hIF1-H49K in neurons triggers the activation of AMPK and affords neuroprotection against excitotoxic cell death through NFκB-mediated signaling and Akt (AKT serine/threonine kinase)/p70S6K (ribosomal protein S6 kinase, 70 kDa) and PARP (poly(ADP-ribose) polymerase) repair pathways [58] (Fig. 6). In these mouse models, the overexpression of IF1 is associated with a mild ROS signal as assessed by the carbonylation of cellular proteins and the concurrent activation of AMPK [42, 57, 58] that might reduce mTOR signaling. Hence, we speculate that mitochondrial retrograde signaling by limiting cellular ATP availability and enhancing the production of ROS through the IF1-mediated inhibition of the H+-ATP synthase contribute to preserve tissue homeostasis and healthy lifespan.
Fig. 6.
Signaling pathways modulated by IF1 that confer cell death protection. IF1-mediated inhibition of the H+-ATP synthase triggers metabolic reprogramming to an enhanced glycolysis and the activation of AMPK (AMP-dependent protein kinase). IF1 inhibition of the synthase also increases ROS production and the ROS-dependent activation of NFκB (nuclear factor kappa B). ROS might also mediate the enhanced Nrf2 [nuclear factor (erythroid-derived 2)-like 2]-guided response that prevents oxidative damage. In neurons, IF1 inhibition of the H+-ATP synthase activates Akt (AKT serine/threonine kinase), p70S6K (ribosomal protein S6 kinase, 70 kDa) and PARP (poly(ADP-ribose) polymerase) survival and repair pathways
Concluding remarks
The H+-ATP synthase is a crucial hub that integrates energy metabolism and intracellular signaling pathways that contribute to decide cellular fate. Regulation of the activity of the H+-ATP synthase is exerted by its physiological inhibitor IF1. IF1 phosphorylation/ dephosphorylation is regulated during the cell cycle, in hypoxia, by nutrient availability and upon an increase in energy demand of the tissue. Phosphorylation of IF1 by the activity of a mitochondrial cAMP-dependent protein kinase prevents the interaction of the inhibitor with the enzyme allowing ATP synthesis in energized mitochondria or ATP hydrolysis in case that mitochondria become de-energized. When IF1 is bound to the enzyme favors the generation of ROS from the ETC triggering a nuclear response that promotes cell survival by different mechanisms. The outcome of the IF1-mediated signaling depends on the genetic background of the cell. In normal post-mitotic cells, it allows withstanding subsequent detrimental stresses favoring normal tissue function. In malignant cells, IF1 propitiates the acquisition of cancer traits, such as increased proliferation, evasion from cell death and invasion capacity. Actually, IF1 is overexpressed in most prevalent human carcinomas being found essentially in the dephosphorylated state, and hence as an active inhibitor of the H+-ATP synthase. In mouse heart under basal conditions, IF1 is present both in the phosphorylated and dephosphorylated states. β-adrenergic stimulation triggers the phosphorylation of IF1 and the activation of ATP production within mitochondria, indicating that there is a fraction of H+-ATP synthase in the tissue that is inactive in the handling of ATP but might be active as a “lighthouse” that signals mitohormetic processes by regulating the generation of a mild ROS signal. The identification of the kinase and phosphatase that regulate the activity of IF1, as well as the quantification of dephosphorylated IF1 that is present under basal conditions in different tissues, will contribute to delineate the relevance of the H+-ATP synthase in tissue homeostasis. Inhibition of the H+-ATP synthase by metabolites and perhaps by other mechanisms contributes to extending lifespan in different organisms through reduced TOR signaling leading to increased proteostasis and reduced oxidative damage. ROS signaling appears to be a common pathway in lifespan extension. Deciphering the precise molecular pathways that are modulated by the H+-ATP synthase and contribute to tissue homeostasis and lifespan extension will be of great interest to develop new approaches for therapies in age-related diseases. The generation of mouse models with tissue-specific and regulated expression of IF1 and conditional IF1-knockout models will contribute to unveil the functional role of the H+-ATP synthase in cellular physiology and pathophysiology.
Acknowledgements
The authors gratefully acknowledge the work and ideas of many colleagues and collaborators, especially to Drs. María Sánchez-Aragó and Laura Formentini and to the excellent technical support provided by M. Chamorro and C. Núñez de Arenas over all these years. Work in the authors’ laboratory is supported by grants from the Ministerio de Economía y Competitividad (MINECO) (SAF2013-41945-R), Centro de Investigación Biomédica en Red de Enfermedades Raras (CIBERER) (CB06/07/0017), Fundación Ramón Areces (FRA) and Comunidad de Madrid (S2011/BMD-2402), Spain. P.B.E.M. and C.N.T. hold a predoctoral fellowship from Fundación La Caixa (Obra Social La Caixa) and FPI-MINECO and Fondo Social Europeo, respectively. The CBMSO receives an institutional grant from FRA. We apologize to authors whose work has not been cited owing to space limitations.
Footnotes
P. B. Esparza-Moltó, C. Nuevo-Tapioles equally contributed to this review.
References
- 1.Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell-free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/S0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- 2.Susin SA, Zamzami N, Kroemer G. Mitochondria as regulators of apoptosis: doubt no more. Biochim Biophys Acta. 1998;1366:151–165. doi: 10.1016/S0005-2728(98)00110-8. [DOI] [PubMed] [Google Scholar]
- 3.Bernardi P, Rasola A, Forte M, Lippe G. The mitochondrial permeability transition pore: channel formation by F-ATP synthase, integration in signal transduction, and role in pathophysiology. Physiol Rev. 2015;95:1111–1155. doi: 10.1152/physrev.00001.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandel NS. Evolution of mitochondria as signaling organelles. Cell Metab. 2015;22:204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Crisosto C, Bravo-Sagua R, Rodriguez-Pena M, Mera C, Castro PF, Quest AF, Rothermel BA, Cifuentes M, Lavandero S. ER-to-mitochondria miscommunication and metabolic diseases. Biochim Biophys Acta. 2015;1852:2096–2105. doi: 10.1016/j.bbadis.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glancy B, Balaban RS. Role of mitochondrial Ca2 + in the regulation of cellular energetics. BioChemistry. 2012;51:2959–2973. doi: 10.1021/bi2018909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Reyes I, Cuezva JM. The H(+)-ATP synthase: a gate to ROS-mediated cell death or cell survival. Biochim Biophys Acta. 2014;1837:1099–1112. doi: 10.1016/j.bbabio.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 10.Kaludercic N, Giorgio V. The dual function of reactive oxygen/nitrogen species in bioenergetics and cell death: the role of ATP synthase. Oxid Med Cell Longev. 2016;2016:3869610. doi: 10.1155/2016/3869610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg SE, Sena LA, Chandel NS. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr Opin Cell Biol. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, Kaech SM, Smiley JR, Means RE, Iwasaki A, Shadel GS. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Autreaux B, Toledano MB. ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 15.Guo B, Zhai D, Cabezas E, Welsh K, Nouraini S, Satterthwait AC, Reed JC. Humanin peptide suppresses apoptosis by interfering with Bax activation. Nature. 2003;423:456–461. doi: 10.1038/nature01627. [DOI] [PubMed] [Google Scholar]
- 16.Pernas L, Scorrano L. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu Rev Physiol. 2016;78:505–531. doi: 10.1146/annurev-physiol-021115-105011. [DOI] [PubMed] [Google Scholar]
- 17.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbe K, Murley A, Nunnari J. Determinants and functions of mitochondrial behavior. Annu Rev Cell Dev Biol. 2014;30:357–391. doi: 10.1146/annurev-cellbio-101011-155756. [DOI] [PubMed] [Google Scholar]
- 19.Zamzami N, Marchetti P, Castedo M, Zanin C, Vayssiere JL, Petit PX, Kroemer G. Reduction in mitochondrial potential constitutes an early irreversible step of programmed lymphocyte death in vivo. J Exp Med. 1995;181:1661–1672. doi: 10.1084/jem.181.5.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yun J, Finkel T. Mitohormesis. Cell Metab. 2014;19:757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ristow M. Unraveling the truth about antioxidants: mitohormesis explains ROS-induced health benefits. Nat Med. 2014;20:709–711. doi: 10.1038/nm.3624. [DOI] [PubMed] [Google Scholar]
- 22.Enriquez JA. Supramolecular organization of respiratory complexes. Annu Rev Physiol. 2016;78:533–561. doi: 10.1146/annurev-physiol-021115-105031. [DOI] [PubMed] [Google Scholar]
- 23.Cogliati S, Enriquez JA, Scorrano L. Mitochondrial cristae: where beauty meets functionality. Trends Biochem Sci. 2016;41:261–273. doi: 10.1016/j.tibs.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 24.Wittig I, Schagger H. Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim Biophys Acta. 2009;1787:672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 25.Walker JE. The ATP synthase: the understood, the uncertain and the unknown. Biochem Soc Trans. 2013;41:1–16. doi: 10.1042/BST20110773. [DOI] [PubMed] [Google Scholar]
- 26.Boyer PD. The ATP synthase. A splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 27.Boveris A, Chance B. The mitochondrial generation of hydrogen peroxide. General properties and effect of hyperbaric oxygen. Biochem J. 1973;134:707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brand MD, Affourtit C, Esteves TC, Green K, Lambert AJ, Miwa S, Pakay JL, Parker N. Mitochondrial superoxide: production, biological effects, and activation of uncoupling proteins. Free Radic Biol Med. 2004;37:755–767. doi: 10.1016/j.freeradbiomed.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 29.Lee J, Ding S, Walpole TB, Holding AN, Montgomery MG, Fearnley IM, Walker JE. Organization of subunits in the membrane domain of the bovine F-ATPase revealed by covalent cross-linking. J Biol Chem. 2015;290:13308–13320. doi: 10.1074/jbc.M115.645283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saita E, Suzuki T, Kinosita K, Jr, Yoshida M. Simple mechanism whereby the F1-ATPase motor rotates with near-perfect chemomechanical energy conversion. Proc Natl Acad Sci USA. 2015;112:9626–9631. doi: 10.1073/pnas.1422885112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng X, Neupert W, Tzagoloff A. The metalloprotease encoded by ATP23 has a dual function in processing and assembly of subunit 6 of mitochondrial ATPase. Mol Biol Cell. 2007;18:617–626. doi: 10.1091/mbc.E06-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rak M, Gokova S, Tzagoloff A. Modular assembly of yeast mitochondrial ATP synthase. EMBO J. 2011;30:920–930. doi: 10.1038/emboj.2010.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lytovchenko O, Naumenko N, Oeljeklaus S, Schmidt B, von der Malsburg K, Deckers M, Warscheid B, van der Laan M, Rehling P. The INA complex facilitates assembly of the peripheral stalk of the mitochondrial F1Fo-ATP synthase. EMBO J. 2014;33:1624–1638. doi: 10.15252/embj.201488076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujikawa M, Sugawara K, Tanabe T, Yoshida M. Assembly of human mitochondrial ATP synthase through two separate intermediates, F1-c-ring and b-e-g complex. FEBS Lett. 2015;589:2707–2712. doi: 10.1016/j.febslet.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Ohsakaya S, Fujikawa M, Hisabori T, Yoshida M. Knockdown of DAPIT (diabetes-associated protein in insulin-sensitive tissue) results in loss of ATP synthase in mitochondria. J Biol Chem. 2011;286:20292–20296. doi: 10.1074/jbc.M110.198523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fujikawa M, Ohsakaya S, Sugawara K, Yoshida M. Population of ATP synthase molecules in mitochondria is limited by available 6.8-kDa proteolipid protein (MLQ) Genes Cells. 2014;19:153–160. doi: 10.1111/gtc.12121. [DOI] [PubMed] [Google Scholar]
- 37.Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kuhlbrandt W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA. 2011;108:14121–14126. doi: 10.1073/pnas.1103621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buzhynskyy N, Sens P, Prima V, Sturgis JN, Scheuring S. Rows of ATP synthase dimers in native mitochondrial inner membranes. Biophys J. 2007;93:2870–2876. doi: 10.1529/biophysj.107.109728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strauss M, Hofhaus G, Schroder RR, Kuhlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paumard P, Arselin G, Vaillier J, Chaignepain S, Bathany K, Schmitter JM, Brethes D, Velours J. Two ATP synthases can be linked through subunits i in the inner mitochondrial membrane of Saccharomyces cerevisiae . BioChemistry. 2002;41:10390–10396. doi: 10.1021/bi025923g. [DOI] [PubMed] [Google Scholar]
- 41.Hahn A, Parey K, Bublitz M, Mills DJ, Zickermann V, Vonck J, Kuhlbrandt W, Meier T. Structure of a complete ATP synthase dimer reveals the molecular basis of inner mitochondrial membrane morphology. Mol Cell. 2016;63:445–456. doi: 10.1016/j.molcel.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santacatterina F, Sanchez-Cenizo L, Formentini L, Mobasher MA, Casas E, Rueda CB, Martinez-Reyes I, Nunez de Arenas C, Garcia-Bermudez J, Zapata JM, Sanchez-Arago M, Satrustegui J, Valverde AM, Cuezva JM. Down-regulation of oxidative phosphorylation in the liver by expression of the ATPase inhibitory factor 1 induces a tumor-promoter metabolic state. Oncotarget. 2016;7:490–508. doi: 10.18632/oncotarget.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon-Smith DJ, Carbajo RJ, Yang JC, Videler H, Runswick MJ, Walker JE, Neuhaus D. Solution structure of a C-terminal coiled-coil domain from bovine IF(1): the inhibitor protein of F(1) ATPase. J Mol Biol. 2001;308:325–339. doi: 10.1006/jmbi.2001.4570. [DOI] [PubMed] [Google Scholar]
- 44.Cabezon E, Montgomery MG, Leslie AG, Walker JE. The structure of bovine F1-ATPase in complex with its regulatory protein IF1. Nat Struct Biol. 2003;10:744–750. doi: 10.1038/nsb966. [DOI] [PubMed] [Google Scholar]
- 45.Garcia JJ, Morales-Rios E, Cortes-Hernandez P, Rodriguez-Zavala JS. The inhibitor protein (IF1) promotes dimerization of the mitochondrial F1F0-ATP synthase. BioChemistry. 2006;45:12695–12703. doi: 10.1021/bi060339j. [DOI] [PubMed] [Google Scholar]
- 46.Campanella M, Casswell E, Chong S, Farah Z, Wieckowski MR, Abramov AY, Tinker A, Duchen MR. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. doi: 10.1016/j.cmet.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Cabezon E, Arechaga I, Jonathan P, Butler G, Walker JE. Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1. J Biol Chem. 2000;275:28353–28355. doi: 10.1074/jbc.C000427200. [DOI] [PubMed] [Google Scholar]
- 48.Dienhart M, Pfeiffer K, Schagger H, Stuart RA. Formation of the yeast F1F0-ATP synthase dimeric complex does not require the ATPase inhibitor protein, Inh1. J Biol Chem. 2002;277:39289–39295. doi: 10.1074/jbc.M205720200. [DOI] [PubMed] [Google Scholar]
- 49.Tomasetig L, Di Pancrazio F, Harris DA, Mavelli I, Lippe G. Dimerization of F0F1ATP synthase from bovine heart is independent from the binding of the inhibitor protein IF1. Biochim Biophys Acta. 2002;1556:133–141. doi: 10.1016/S0005-2728(02)00344-4. [DOI] [PubMed] [Google Scholar]
- 50.Fujikawa M, Imamura H, Nakamura J, Yoshida M. Assessing the actual contribution of IF1, an inhibitor of mitochondrial FoF1, to ATP homeostasis, cell growth, mitochondrial morphology and cell viability. J Biol Chem. 2012;287:18781–18787. doi: 10.1074/jbc.M112.345793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minauro-Sanmiguel F, Wilkens S, Garcia JJ. Structure of dimeric mitochondrial ATP synthase: novel F0 bridging features and the structural basis of mitochondrial cristae biogenesis. Proc Natl Acad Sci USA. 2005;102:12356–12358. doi: 10.1073/pnas.0503893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giorgio V, von Stockum S, Antoniel M, Fabbro A, Fogolari F, Forte M, Glick GD, Petronilli V, Zoratti M, Szabo I, Lippe G, Bernardi P. Dimers of mitochondrial ATP synthase form the permeability transition pore. Proc Natl Acad Sci USA. 2013;110:5887–5892. doi: 10.1073/pnas.1217823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carraro M, Giorgio V, Sileikyte J, Sartori G, Forte M, Lippe G, Zoratti M, Szabo I, Bernardi P. Channel formation by yeast F-ATP synthase and the role of dimerization in the mitochondrial permeability transition. J Biol Chem. 2014;289:15980–15985. doi: 10.1074/jbc.C114.559633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Stockum S, Giorgio V, Trevisan E, Lippe G, Glick GD, Forte MA, Da-Re C, Checchetto V, Mazzotta G, Costa R, Szabo I, Bernardi P. F-ATPase of Drosophila melanogaster forms 53-picosiemen (53-pS) channels responsible for mitochondrial Ca2+-induced Ca2+ release. J Biol Chem. 2015;290:4537–4544. doi: 10.1074/jbc.C114.629766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonora M, Bononi A, De Marchi E, Giorgi C, Lebiedzinska M, Marchi S, Patergnani S, Rimessi A, Suski JM, Wojtala A, Wieckowski MR, Kroemer G, Galluzzi L, Pinton P. Role of the c subunit of the FO ATP synthase in mitochondrial permeability transition. Cell Cycle. 2013;12:674–683. doi: 10.4161/cc.23599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Alavian KN, Beutner G, Lazrove E, Sacchetti S, Park HA, Licznerski P, Li H, Nabili P, Hockensmith K, Graham M, Porter GA, Jr, Jonas EA. An uncoupling channel within the c-subunit ring of the F1FO ATP synthase is the mitochondrial permeability transition pore. Proc Natl Acad Sci USA. 2014;111:10580–10585. doi: 10.1073/pnas.1401591111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Formentini L, Sánchez-Aragó M, Sánchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 (IF1) triggers a ROS-mediated retrograde pro-survival and proliferative response. Mol Cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 58.Formentini L, Pereira MP, Sanchez-Cenizo L, Santacatterina F, Lucas JJ, Navarro C, Martinez-Serrano A, Cuezva JM. In vivo inhibition of the mitochondrial H+-ATP synthase in neurons promotes metabolic preconditioning. EMBO J. 2014;33:762–778. doi: 10.1002/embj.201386392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chin RM, Fu X, Pai MY, Vergnes L, Hwang H, Deng G, Diep S, Lomenick B, Meli VS, Monsalve GC, Hu E, Whelan SA, Wang JX, Jung G, Solis GM, Fazlollahi F, Kaweeteerawat C, Quach A, Nili M, Krall AS, Godwin HA, Chang HR, Faull KF, Guo F, Jiang M, Trauger SA, Saghatelian A, Braas D, Christofk HR, Clarke CF, Teitell MA, Petrascheck M, Reue K, Jung ME, Frand AR, Huang J. The metabolite alpha-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510:397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das AM, Harris DA. Control of mitochondrial ATP synthase in heart cells: inactive to active transitions caused by beating or positive inotropic agents. Cardiovasc Res. 1990;24:411–417. doi: 10.1093/cvr/24.5.411. [DOI] [PubMed] [Google Scholar]
- 61.Das AM. Regulation of the mitochondrial ATP-synthase in health and disease. Mol Genet Metab. 2003;79:71–82. doi: 10.1016/S1096-7192(03)00069-6. [DOI] [PubMed] [Google Scholar]
- 62.Garcia-Bermudez J, Cuezva JM. The ATPase Inhibitory Factor 1 (IF1): a master regulator of energy metabolism and of cell survival. Biochim Biophys Acta. 2016;1857:1167–1182. doi: 10.1016/j.bbabio.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 63.Rizzuto R, De Stefani D, Raffaello A, Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol. 2012;13:566–578. doi: 10.1038/nrm3412. [DOI] [PubMed] [Google Scholar]
- 64.Llorente-Folch I, Rueda CB, Pardo B, Szabadkai G, Duchen MR, Satrustegui J. The regulation of neuronal mitochondrial metabolism by calcium. J Physiol. 2015;593:3447–3462. doi: 10.1113/JP270254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boerries M, Most P, Gledhill JR, Walker JE, Katus HA, Koch WJ, Aebi U, Schoenenberger CA. Ca2+ -dependent interaction of S100A1 with F1-ATPase leads to an increased ATP content in cardiomyocytes. Mol Cell Biol. 2007;27:4365–4373. doi: 10.1128/MCB.02045-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yamada EW, Huzel NJ. The calcium-binding ATPase inhibitor protein from bovine heart mitochondria. Purification and properties. J Biol Chem. 1988;263:11498–11503. [PubMed] [Google Scholar]
- 67.Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Michel AH, Kornmann B. The ERMES complex and ER-mitochondria connections. Biochem Soc Trans. 2012;40:445–450. doi: 10.1042/BST20110758. [DOI] [PubMed] [Google Scholar]
- 69.Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kontro H, Hulmi JJ, Rahkila P, Kainulainen H. Cellular and tissue expression of DAPIT, a phylogenetically conserved peptide. Eur J Histochem. 2012;56:e18. doi: 10.4081/ejh.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kontro H, Cannino G, Rustin P, Dufour E, Kainulainen H. DAPIT Over-Expression Modulates Glucose Metabolism and Cell Behaviour in HEK293T Cells. PLoS One. 2015;10:e0131990. doi: 10.1371/journal.pone.0131990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Alavian KN, Li H, Collis L, Bonanni L, Zeng L, Sacchetti S, Lazrove E, Nabili P, Flaherty B, Graham M, Chen Y, Messerli SM, Mariggio MA, Rahner C, McNay E, Shore GC, Smith PJ, Hardwick JM, Jonas EA. Bcl-x(L) regulates metabolic efficiency of neurons through interaction with the mitochondrial F(1)F(O) ATP synthase. Nat Cell Biol. 2011;13:1224–1233. doi: 10.1038/ncb2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kioka H, Kato H, Fujikawa M, Tsukamoto O, Suzuki T, Imamura H, Nakano A, Higo S, Yamazaki S, Matsuzaki T, Takafuji K, Asanuma H, Asakura M, Minamino T, Shintani Y, Yoshida M, Noji H, Kitakaze M, Komuro I, Asano Y, Takashima S. Evaluation of intramitochondrial ATP levels identifies G0/G1 switch gene 2 as a positive regulator of oxidative phosphorylation. Proc Natl Acad Sci USA. 2014;111:273–278. doi: 10.1073/pnas.1318547111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pullman ME, Monroy GC. A naturally occurring inhibitor of mitochondrial adenosine triphosphatase. J Biol Chem. 1963;238:3762–3769. [PubMed] [Google Scholar]
- 75.Cabezon E, Butler PJ, Runswick MJ, Carbajo RJ, Walker JE. Homologous and heterologous inhibitory effects of ATPase inhibitor proteins on F-ATPases. J Biol Chem. 2002;277:41334–41341. doi: 10.1074/jbc.M207169200. [DOI] [PubMed] [Google Scholar]
- 76.Cabezon E, Runswick MJ, Leslie AG, Walker JE. The structure of bovine IF(1), the regulatory subunit of mitochondrial F-ATPase. Embo J. 2001;20:6990–6996. doi: 10.1093/emboj/20.24.6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gledhill JR, Montgomery MG, Leslie AG, Walker JE. How the regulatory protein, IF(1), inhibits F(1)-ATPase from bovine mitochondria. Proc Natl Acad Sci USA. 2007;104:15671–15676. doi: 10.1073/pnas.0707326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bason JV, Montgomery MG, Leslie AG, Walker JE. Pathway of binding of the intrinsically disordered mitochondrial inhibitor protein to F1-ATPase. Proc Natl Acad Sci USA. 2014;111:11305–11310. doi: 10.1073/pnas.1411560111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suzuki T, Tanaka K, Wakabayashi C, Saita E, Yoshida M. Chemomechanical coupling of human mitochondrial F1-ATPase motor. Nat Chem Biol. 2014;10:930–936. doi: 10.1038/nchembio.1635. [DOI] [PubMed] [Google Scholar]
- 81.Garcia-Bermudez J, Sanchez-Arago M, Soldevilla B, Del Arco A, Nuevo-Tapioles C, Cuezva JM. PKA phosphorylates the ATPase inhibitory factor 1 and inactivates its capacity to bind and inhibit the mitochondrial H-ATP synthase. Cell Rep. 2015;12:2143–2155. doi: 10.1016/j.celrep.2015.08.052. [DOI] [PubMed] [Google Scholar]
- 82.Sanchez-Cenizo L, Formentini L, Aldea M, Ortega AD, Garcia-Huerta P, Sanchez-Arago M, Cuezva JM. Up-regulation of the ATPase inhibitory factor 1 (IF1) of the mitochondrial H+-ATP synthase in human tumors mediates the metabolic shift of cancer cells to a Warburg phenotype. J Biol Chem. 2010;285:25308–25313. doi: 10.1074/jbc.M110.146480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sanchez-Arago M, Formentini L, Martinez-Reyes I, Garcia-Bermudez J, Santacatterina F, Sanchez-Cenizo L, Willers IM, Aldea M, Najera L, Juarranz A, Lopez EC, Clofent J, Navarro C, Espinosa E, Cuezva JM. Expression, regulation and clinical relevance of the ATPase inhibitory factor 1 in human cancers. Oncogenesis. 2013;2:e46. doi: 10.1038/oncsis.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zanotti F, Gnoni A, Mangiullo R, Papa S. Effect of the ATPase inhibitor protein IF1 on H + translocation in the mitochondrial ATP synthase complex. Biochem Biophys Res Commun. 2009;384:43–48. doi: 10.1016/j.bbrc.2009.04.046. [DOI] [PubMed] [Google Scholar]
- 85.Cabezon E, Butler PJ, Runswick MJ, Walker JE. Modulation of the oligomerization state of the bovine F1-ATPase inhibitor protein, IF1, by pH. J Biol Chem. 2000;275:25460–25464. doi: 10.1074/jbc.M003859200. [DOI] [PubMed] [Google Scholar]
- 86.Zhang F, Zhang L, Qi Y, Xu H. Mitochondrial cAMP signaling. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Acin-Perez R, Salazar E, Kamenetsky M, Buck J, Levin LR, Manfredi G. Cyclic AMP produced inside mitochondria regulates oxidative phosphorylation. Cell Metab. 2009;9:265–276. doi: 10.1016/j.cmet.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Acin-Perez R, Gatti DL, Bai Y, Manfredi G. Protein phosphorylation and prevention of cytochrome oxidase inhibition by ATP: coupled mechanisms of energy metabolism regulation. Cell Metab. 2011;13:712–719. doi: 10.1016/j.cmet.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Di Benedetto G, Scalzotto E, Mongillo M, Pozzan T. Mitochondrial Ca(2)(+) uptake induces cyclic AMP generation in the matrix and modulates organelle ATP levels. Cell Metab. 2013;17:965–975. doi: 10.1016/j.cmet.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 90.Sugawara K, Fujikawa M, Yoshida M. Screening of protein kinase inhibitors and knockdown experiments identified four kinases that affect mitochondrial ATP synthesis activity. FEBS Lett. 2013;587:3843–3847. doi: 10.1016/j.febslet.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 91.Di Benedetto G, Pendin D, Greotti E, Pizzo P, Pozzan T. Ca2 + and cAMP cross-talk in mitochondria. J Physiol. 2014;592:305–312. doi: 10.1113/jphysiol.2013.259135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lefkimmiatis K, Zaccolo M. cAMP signaling in subcellular compartments. Pharmacol Ther. 2014;143:295–304. doi: 10.1016/j.pharmthera.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sanchez-Arago M, Garcia-Bermudez J, Martinez-Reyes I, Santacatterina F, Cuezva JM. Degradation of IF1 controls energy metabolism during osteogenic differentiation of stem cells. EMBO Rep. 2013;14:638–644. doi: 10.1038/embor.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zanotti F, Raho G, Gaballo A, Papa S. Inhibitory and anchoring domains in the ATPase inhibitor protein IF1 of bovine heart mitochondrial ATP synthase. J Bioenerg Biomembr. 2004;36:447–457. doi: 10.1023/B:JOBB.0000047327.68173.9b. [DOI] [PubMed] [Google Scholar]
- 95.Lopez-Mediavilla C, Vigny H, Godinot C. Docking the mitochondrial inhibitor protein IF1 to a membrane receptor different from the F1-ATPase beta subunit. Eur J Biochem. 1993;215:487–496. doi: 10.1111/j.1432-1033.1993.tb18058.x. [DOI] [PubMed] [Google Scholar]
- 96.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124:269–270. [PubMed] [Google Scholar]
- 97.Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, Gil J, Beach D. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–299. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- 98.Xu X, Duan S, Yi F, Ocampo A, Liu GH, Izpisua Belmonte JC. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 99.Cuezva JM, Ortega AD, Willers I, Sanchez-Cenizo L, Aldea M, Sanchez-Arago M. The tumor suppressor function of mitochondria: translation into the clinics. Biochim Biophys Acta. 2009;1792:1145–1158. doi: 10.1016/j.bbadis.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 100.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 101.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 102.Brand KA, Hermfisse U. Aerobic glycolysis by proliferating cells: a protective strategy against reactive oxygen species. FASEB J. 1997;11:388–395. doi: 10.1096/fasebj.11.5.9141507. [DOI] [PubMed] [Google Scholar]
- 103.Santamaria G, Martinez-Diez M, Fabregat I, Cuezva JM. Efficient execution of cell death in non-glycolytic cells requires the generation of ROS controlled by the activity of mitochondrial H+-ATP synthase. Carcinogenesis. 2006;27:925–935. doi: 10.1093/carcin/bgi315. [DOI] [PubMed] [Google Scholar]
- 104.Cuezva JM, Krajewska M, de Heredia ML, Krajewski S, Santamaria G, Kim H, Zapata JM, Marusawa H, Chamorro M, Reed JC. The bioenergetic signature of cancer: a marker of tumor progression. Cancer Res. 2002;62:6674–6681. [PubMed] [Google Scholar]
- 105.Isidoro A, Casado E, Redondo A, Acebo P, Espinosa E, Alonso AM, Cejas P, Hardisson D, Fresno Vara JA, Belda-Iniesta C, Gonzalez-Baron M, Cuezva JM. Breast carcinomas fulfill the Warburg hypothesis and provide metabolic markers of cancer prognosis. Carcinogenesis. 2005;26:2095–2104. doi: 10.1093/carcin/bgi188. [DOI] [PubMed] [Google Scholar]
- 106.Lopez-Rios F, Sanchez-Arago M, Garcia-Garcia E, Ortega AD, Berrendero JR, Pozo-Rodriguez F, Lopez-Encuentra A, Ballestin C, Cuezva JM. Loss of the mitochondrial bioenergetic capacity underlies the glucose avidity of carcinomas. Cancer Res. 2007;67:9013–9017. doi: 10.1158/0008-5472.CAN-07-1678. [DOI] [PubMed] [Google Scholar]
- 107.Wei S, Fukuhara H, Kawada C, Kurabayashi A, Furihata M, Ogura S, Inoue K, Shuin T. silencing of ATPase inhibitory factor 1 inhibits cell growth via cell cycle arrest in bladder cancer. Pathobiology. 2015;82:224–232. doi: 10.1159/000439027. [DOI] [PubMed] [Google Scholar]
- 108.Gao YX, Chen L, Hu XG, Wu HB, Cui YH, Zhang X, Wang Y, Liu XD, Bian XW. ATPase inhibitory factor 1 expression is an independent prognostic factor in non-small cell lung cancer. Am J Cancer Res. 2016;6:1141–1148. [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang C, Min L, Liu J, Tian W, Han Y, Qu L, Shou C. Integrated analysis identified an intestinal-like and a diffuse-like gene sets that predict gastric cancer outcome. Tumour Biol. 2016 doi: 10.1007/s13277-016-5454-7. [DOI] [PubMed] [Google Scholar]
- 110.Kurbasic E, Sjostrom M, Krogh M, Folkesson E, Grabau D, Hansson K, Ryden L, Waldemarson S, James P, Nimeus E. Changes in glycoprotein expression between primary breast tumour and synchronous lymph node metastases or asynchronous distant metastases. Clin Proteomics. 2015;12:13. doi: 10.1186/s12014-015-9084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yin T, Lu L, Xiong Z, Wei S, Cui D. ATPase inhibitory factor 1 is a prognostic marker and contributes to proliferation and invasion of human gastric cancer cells. Biomed Pharmacother. 2015;70:90–96. doi: 10.1016/j.biopha.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 112.Song R, Song H, Liang Y, Yin D, Zhang H, Zheng T, Wang J, Lu Z, Song X, Pei T, Qin Y, Li Y, Xie C, Sun B, Shi H, Li S, Meng X, Yang G, Pan S, Zhu J, Qi S, Jiang H, Zhang Z, Liu L. Reciprocal activation between ATPase inhibitory factor 1 and NF-kappaB drives hepatocellular carcinoma angiogenesis and metastasis. Hepatology. 2014;60:1659–1673. doi: 10.1002/hep.27312. [DOI] [PubMed] [Google Scholar]
- 113.Wu J, Shan Q, Li P, Wu Y, Xie J, Wang X. ATPase inhibitory factor 1 is a potential prognostic marker for the migration and invasion of glioma. Oncol Lett. 2015;10:2075–2080. doi: 10.3892/ol.2015.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mourier A, Ruzzenente B, Brandt T, Kuhlbrandt W, Larsson NG. Loss of LRPPRC causes ATP synthase deficiency. Hum Mol Genet. 2014;23:2580–2592. doi: 10.1093/hmg/ddt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Quiros PM, Langer T, Lopez-Otin C. New roles for mitochondrial proteases in health, ageing and disease. Nat Rev Mol Cell Biol. 2015;16:345–359. doi: 10.1038/nrm3984. [DOI] [PubMed] [Google Scholar]
- 116.Shen L, Zhi L, Hu W, Wu MX. IEX-1 targets mitochondrial F1Fo-ATPase inhibitor for degradation. Cell Death Differ. 2009;16:603–612. doi: 10.1038/cdd.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Faccenda D, Tan CH, Seraphim A, Duchen MR, Campanella M. IF1 limits the apoptotic-signalling cascade by preventing mitochondrial remodelling. Cell Death Differ. 2013;20:686–697. doi: 10.1038/cdd.2012.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jovaisaite V, Mouchiroud L, Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol. 2014;217:137–143. doi: 10.1242/jeb.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Benedetti C, Haynes CM, Yang Y, Harding HP, Ron D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. Genetics. 2006;174:229–239. doi: 10.1534/genetics.106.061580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Haynes CM, Petrova K, Benedetti C, Yang Y, Ron D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans . Dev Cell. 2007;13:467–480. doi: 10.1016/j.devcel.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 122.Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Mol Cell. 2015;58:123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ. A mitochondrial specific stress response in mammalian cells. EMBO J. 2002;21:4411–4419. doi: 10.1093/emboj/cdf445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aldridge JE, Horibe T, Hoogenraad NJ. Discovery of genes activated by the mitochondrial unfolded protein response (mtUPR) and cognate promoter elements. PLoS One. 2007;2:e874. doi: 10.1371/journal.pone.0000874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dogan SA, Pujol C, Maiti P, Kukat A, Wang S, Hermans S, Senft K, Wibom R, Rugarli EI, Trifunovic A. Tissue-specific loss of DARS2 activates stress responses independently of respiratory chain deficiency in the heart. Cell Metab. 2014;19:458–469. doi: 10.1016/j.cmet.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 126.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moullan N, Mouchiroud L, Wang X, Ryu D, Williams EG, Mottis A, Jovaisaite V, Frochaux MV, Quiros PM, Deplancke B, Houtkooper RH, Auwerx J. Tetracyclines disturb mitochondrial function across eukaryotic models: a call for caution in biomedical research. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P, D’Amico D, Ropelle ER, Lutolf MP, Aebersold R, Schoonjans K, Menzies KJ, Auwerx J. NAD+ repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016 doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- 129.Shadel GS, Horvath TL. Mitochondrial ROS signaling in organismal homeostasis. Cell. 2015;163:560–569. doi: 10.1016/j.cell.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chae S, Ahn BY, Byun K, Cho YM, Yu MH, Lee B, Hwang D, Park KS. A systems approach for decoding mitochondrial retrograde signaling pathways. Sci Signal. 2013;6:rs4. doi: 10.1126/scisignal.2003266. [DOI] [PubMed] [Google Scholar]
- 132.Owusu-Ansah E, Song W, Perrimon N. Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell. 2013;155:699–712. doi: 10.1016/j.cell.2013.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Scialo F, Sriram A, Fernandez-Ayala D, Gubina N, Lohmus M, Nelson G, Logan A, Cooper HM, Navas P, Enriquez JA, Murphy MP, Sanz A. Mitochondrial ROS produced via reverse electron transport extend animal lifespan. Cell Metab. 2016;23:725–734. doi: 10.1016/j.cmet.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298:2398–2401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
- 135.Lee SJ, Hwang AB, Kenyon C. Inhibition of respiration extends C. elegans life span via reactive oxygen species that increase HIF-1 activity. Curr Biol. 2010;20:2131–2136. doi: 10.1016/j.cub.2010.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mookerjee SA, Divakaruni AS, Jastroch M, Brand MD. Mitochondrial uncoupling and lifespan. Mech Ageing Dev. 2010;131:463–472. doi: 10.1016/j.mad.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol. 2000;35:811–820. doi: 10.1016/S0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- 138.Harman D. Aging: a theory based on free radical and radiation chemistry. J Gerontol. 1956;11:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 139.Smith DL, Jr, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological lifespan of Saccharomyces cerevisiae independently of the Sirtuins. Aging Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 140.Ocampo A, Liu J, Schroeder EA, Shadel GS, Barrientos A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 142.Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–293. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 143.Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila . Cell. 2009;139:149–160. doi: 10.1016/j.cell.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pan Y, Schroeder EA, Ocampo A, Barrientos A, Shadel GS. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Johnson SC, Rabinovitch PS, Kaeberlein M. mTOR is a key modulator of ageing and age-related disease. Nature. 2013;493:338–345. doi: 10.1038/nature11861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Katic M, Kennedy AR, Leykin I, Norris A, McGettrick A, Gesta S, Russell SJ, Bluher M, Maratos-Flier E, Kahn CR. Mitochondrial gene expression and increased oxidative metabolism: role in increased lifespan of fat-specific insulin receptor knock-out mice. Aging Cell. 2007;6:827–839. doi: 10.1111/j.1474-9726.2007.00346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dell’agnello C, Leo S, Agostino A, Szabadkai G, Tiveron C, Zulian A, Prelle A, Roubertoux P, Rizzuto R, Zeviani M. Increased longevity and refractoriness to Ca(2+)-dependent neurodegeneration in Surf1 knockout mice. Hum Mol Genet. 2007;16:431–444. doi: 10.1093/hmg/ddl477. [DOI] [PubMed] [Google Scholar]
- 148.Pulliam DA, Deepa SS, Liu Y, Hill S, Lin AL, Bhattacharya A, Shi Y, Sloane L, Viscomi C, Zeviani M, Van Remmen H. Complex IV-deficient Surf1(-/-) mice initiate mitochondrial stress responses. Biochem J. 2014;462:359–371. doi: 10.1042/BJ20140291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mishur RJ, Khan M, Munkacsy E, Sharma L, Bokov A, Beam H, Radetskaya O, Borror M, Lane R, Bai Y, Rea SL. Mitochondrial metabolites extend lifespan. Aging Cell. 2016;15:336–348. doi: 10.1111/acel.12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fu X, Chin RM, Vergnes L, Hwang H, Deng G, Xing Y, Pai MY, Li S, Ta L, Fazlollahi F, Chen C, Prins RM, Teitell MA, Nathanson DA, Lai A, Faull KF, Jiang M, Clarke SG, Cloughesy TF, Graeber TG, Braas D, Christofk HR, Jung ME, Reue K, Huang J. 2-Hydroxyglutarate Inhibits ATP Synthase and mTOR Signaling. Cell Metab. 2015;22:508–515. doi: 10.1016/j.cmet.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim HJ, Morrow G, Westwood JT, Michaud S, Tanguay RM. Gene expression profiling implicates OXPHOS complexes in lifespan extension of flies over-expressing a small mitochondrial chaperone, Hsp22. Exp Gerontol. 2010;45:611–620. doi: 10.1016/j.exger.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 153.Sun X, Wheeler CT, Yolitz J, Laslo M, Alberico T, Sun Y, Song Q, Zou S. A mitochondrial ATP synthase subunit interacts with TOR signaling to modulate protein homeostasis and lifespan in Drosophila . Cell Rep. 2014;8:1781–1792. doi: 10.1016/j.celrep.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, Navas P, Cruz R, Carracedo A, Lopez-Otin C, Perez-Martos A, Fernandez-Silva P, Fernandez-Vizarra E, Enriquez JA. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]