Abstract
Deficiency in cerebral amyloid β-protein (Aβ) clearance is implicated in the pathogenesis of the common late-onset forms of Alzheimer’s disease (AD). Accumulation of misfolded Aβ in the brain is believed to be a net result of imbalance between its production and removal. This in turn may trigger neuroinflammation, progressive synaptic loss, and ultimately cognitive decline. Clearance of cerebral Aβ is a complex process mediated by various systems and cell types, including vascular transport across the blood–brain barrier, glymphatic drainage, and engulfment and degradation by resident microglia and infiltrating innate immune cells. Recent studies have highlighted a new, unexpected role for peripheral monocytes and macrophages in restricting cerebral Aβ fibrils, and possibly soluble oligomers. In AD transgenic (ADtg) mice, monocyte ablation or inhibition of their migration into the brain exacerbated Aβ pathology, while blood enrichment with monocytes and their increased recruitment to plaque lesion sites greatly diminished Aβ burden. Profound neuroprotective effects in ADtg mice were further achieved through increased cerebral recruitment of myelomonocytes overexpressing Aβ-degrading enzymes. This review summarizes the literature on cellular and molecular mechanisms of cerebral Aβ clearance with an emphasis on the role of peripheral monocytes and macrophages in Aβ removal.
Keywords: Neurodegenerative diseases, Amyloid-β protein, Aβ-degrading enzymes, Innate immune cells, Myelomonocytes, Phagocytosis
Introduction
Alzheimer’s disease (AD) is a severe neurodegenerative disorder and the most common form of senile dementia, affecting over 5 million in the United States and 45 million worldwide [1, 2]. AD manifests as a progressive decline in cognitive function and behavior, invariably leading to death [3]. The epidemic of AD is especially damaging to the growing elderly population and the economy that supports them. This immense psychosocial and public health burden calls for a clearer understanding of disease pathophysiology to facilitate the development and implementation of more effective treatment strategies.
Over the past century, our understanding of the molecular mechanisms underlying the development of AD has greatly expanded. Though still pathological hallmarks, extracellular plaques and intracellular neurofibrillary tangles (NFTs) within the brain [4], comprised, respectively, of amyloid-β protein (Aβ) and hyperphosphorylated tau (pTau), no longer describe all pathogenic forms of these proteins. Beyond intracellular threads and tangles, misfolded tau may form extracellular assemblies that propagate through and disrupt synaptically dense regions [5, 6]. Meanwhile, extracellular and intracellular oligomers of Aβ were also found to be highly synaptotoxic and exist in a highly dynamic equilibrium between the small, soluble forms and the larger, insoluble intermediates and fibrils [7, 8]. Recent exploration of this disease outside the brain, in another central nervous system tissue, has further revealed Aβ pathology in the retina of AD patients, including those at early stages [9–13]. Converging data from genetic, physiologic, biochemical, and clinical studies demonstrate a strong association between Aβ accumulation and neuroinflammation, synaptic loss, impaired neuronal function, and ultimately, debilitating cognitive decline [3, 14]. Progressive accumulation and aggregation of Aβ peptides in the brain are thought to be a net result of imbalance between their production and clearance [15]. Moreover, the dramatic increase in cerebral Aβ far precedes the clinical impairment, beginning as early as 20 years prior to symptom manifestation [16]. Therefore, a common view is that any strategy that reduces Aβ levels in the brain, either by inhibiting its production/aggregation or by increasing its clearance, will be advantageous in preventing the development of AD.
Aβ denotes a group of endogenous peptides, typically of 36–43 amino acids. It derives from a larger transmembrane protein, the amyloid precursor protein (APP), in a complex proteolytic process, described extensively elsewhere [17]. The disease-associated (amyloidogenic) aggregation-prone Aβ1-40 (Aβ40) and Aβ1-42 (Aβ42) alloforms are generated through a sequential cleavage of APP by a β-secretase (BACE1) and a γ-secretase transmembrane complex. Mutations within the gene encoding APP and its Aβ coding sequence were found to cause early-onset, autosomal-dominant inherited forms of familial AD (FAD) [18]. Similarly, patients with Down syndrome (trisomy 21) who carry three copies of the APP gene develop AD-like Aβ and tau neuropathology, leading to cognitive decline [19]. In addition, inheritance of mutations within the genes encoding for presenelin-1 and -2 (PS1 and PS2), two components of the γ-secretase complex, invariably lead to FAD [20–22] (Table 1). These rare mutations and haplotypes result in either overproduction or increased aggregation of Aβ, and importantly, in favored generation of the more pathogenic Aβ42 alloforms [4, 23]. These findings strongly tie Aβ to the etiology of AD. Further support for this notion came recently from the identification of a protective APP mutation in non-demented Icelanders [24]. The A673T mutation in APP (alternatively called A2T mutation in Aβ) was shown to reduce amyloidogenic Aβ production and aggregation, providing protection against age-associated cognitive decline [24, 25].
Table 1.
Genes associated with Alzheimer’s disease
| Gene | Type | FREQa | Risk | Locus | Variants | ↑ Aβ prod. | ↓ Aβ clear. | Effects on Aβb | References |
|---|---|---|---|---|---|---|---|---|---|
| APP | FADc | Rare |

|
21q21.3 | Mutations Trisomy 21 | ✓ | – | ↑ Aβ42/40 ratio; ↑ Aβ42 aggregation | [4, 18, 21, 26–28] |
| PSEN1 | FADc | Rare |

|
14q24.3 | Mutations | ✓ | – | ↑ Aβ42/40 ratio | [20, 21, 23, 26, 28–31] |
| PSEN2 | FADc | Rare |

|
1q42.13 | Mutations | ✓ | – | ↑ Aβ42/40 ratio | [21–23, 26, 28, 31] |
| ABCA7 | LOAD | 16% |

|
19p13.3 |
rs3764650 rs3752246 rs4147929 |
✓ | ✓ | Understudied; ↑ Aβ secretion; ↓ MΦ/MG Aβ phagocytosis | [21, 32–39] |
| ADAM10 | LOAD | Rare |

|
15q21.3 |
Q170H R181G |
✓ | – | ↑ Aβ production; ↓ α-secretase activity | [40–43] |
| ACE | LOAD | 33–48% |

|
17q23.3 |
Indel; rs4219 rs1800764 rs4343 |
– | ✓ | Controversial; ↓ Aβ degradation; ↑ Aβ levels | [43–50] |
| APOE4 d | LOAD | 3%e |

|
19q13.2 |
ε4 Allelef ε2 Alleleg |
– | ✓ | ↓ Chaperone-mediated Aβ processing, clearance | [51–56] |
| BIN1 | LOAD | 45% |

|
2q14 |
rs744373 rs7561528 |
✓ | ✓ | ↑ Aβ production; May ↓ MΦ Aβ phagocytosis | [33, 34, 57–61] |
| CD2AP | LOAD | 3% |

|
6p12 |
rs9296559 rs9349407 |
– | ✓ | ↑Aβ plaque burden; ↓ Endosome/lysosome clearance | [33, 37, 57, 62, 63] |
| CD33 | LOAD | 30% |

|
19q13.3 |
rs3865444g rs3826656 |
– | ✓ | ↓ Mo/MG Aβ phagocytosis | [33, 34, 64–67] |
| CLU | LOAD | 38% |

|
8p21-p12 | rs9331896 | – | ✓ | ↓ Chaperone-mediated Aβ clearance | [32, 68–75] |
| CR1 | LOAD | 20% |

|
1q32 |
rs3818361 rs6656401 rs6701713 |
– | ✓ | ↓ Immune-mediated Aβ clearance; ↑ Aβ42 levels | [33, 34, 76–79] |
| EPHA1 | LOAD | 34% |

|
7q34 |
rs11771145g rs11767557g |
– | ✓ | Understudied; ↓ Immune-mediated Aβ clearance | [32–34, 80–82] |
| PICALM | LOAD | 36% |

|
11q14 |
rs3851179g rs541458g |
✓ | ✓ | ↓ Trafficking of Aβ across BBB; ↑ Aβ production | [83–89] |
| SIRT1 | LOAD | – |

|
10q21.3 | – | ✓ | ✓ | ↑ MG-dependent Aβ toxicity; ↓ α-secretase activity | [32, 90–95] |
| SORL1 | LOAD | 4% |

|
11q23.2-q24.2 |
rs12285364 rs2070045 rs2282649 |
✓ | ✓ | ↑ Aβ production; ↓ APP trafficking to endosomes | [94, 96–101] |
| TREM2 | LOAD | 6% |

|
6p21.1 | rs75932628 | – | ✓ | Controversial; ↓ Mo phagocytosis and immune response | [102–108] |
ABCA7 ATP-binding cassette, sub-family A (ABC1), member 7, ACE angiotensin-converting enzyme, APOE apolipoprotein E, APP amyloid precursor protein, BIN1 bridging integrator 1, CD2AP CD2-associated protein, CD33 sialic acid-binding immunoglobulin-like lectin 3, Clear. clearance, CLU clusterin (apolipoprotein J), CR1 complement component (3b/4b) receptor 1, EPHA1 EPH receptor A1, Exp. gene expression levels in AD, FAD early onset familial AD: inherited in an autosomal dominant fashion, Load late onset AD, Mo/MΦ monocytes/macrophages, MG microglia, PICALM phosphatidylinositol binding clathrin assembly protein, Prod. production, PSEN1 presenilin 1, PSEN2 presenilin 2, SIRT1 sirtuin 1, SNPs single nucleotide polymorphisms, SORL1 sortilin-related receptor 1, TREM2 triggering receptor expressed on myeloid cells 2
aApproximate frequency
bPostulated effects on Aβ and related immune response
cRare variants identified in LOAD
dStrongest genetic risk factor for LOAD
eCarriers of one or two APOε4 alleles
fDose-dependent effect of Apoε4 alleles
gReduced risk for AD
While FAD represents approximately 5% of all AD cases, the remaining majority of AD cases manifest later in life (typically over 65 years of age), and are termed sporadic or late-onset AD (LOAD). The etiology of LOAD is multifactorial: multiple genetic and environmental factors likely contribute to the development of disease. Strong support for the role of Aβ accumulation in both AD forms came from several clinical studies. While in FAD cases cerebral Aβ increase was explained by Aβ42 overproduction [109], deficient Aβ42 clearance was shown in the brains of LOAD patients [110]. Despite differences in etiology, FAD and LOAD are neuropathologically indistinguishable and present with similar clinical phenotypes [4].
Growing evidence indicates that Aβ exerts its neurotoxic effects in both an alloform- and conformation-dependent manner [7]. Small, soluble oligomeric forms of Aβ42 were shown to be especially neurotoxic [111–113] and more strongly predict cognitive decline than Aβ plaque load [114, 115]. Specifically, Aβ oligomers were shown to impact long-term potentiation, synaptic signaling and plasticity, dendritic morphology, and cognition in rodent models [113, 116–119]. Additionally, Aβ was shown to impair neuronal glucose transport [120] and accumulate within mitochondria [121], disrupting vital enzymatic activity and increasing free radical production [122]. Aβ fibrils can also induce inflammatory processes by binding to and activating microglia [123, 124] and peripheral monocytes [125–127]. This toxic microenvironment was further associated with impaired calcium regulation and energy metabolism throughout CNS tissues [128]. Beyond amyloid pathology in brain parenchyma, AD patients frequently exhibit cerebral amyloid angiopathy (CAA) along with reduced cerebral blood flow that can further compromise cognitive capacity [129]. This phenomenon was also found in retina microvasculature [13, 130]. In murine models of AD, it was recently found that vascular amyloid deposits hardened blood vessel walls and reduced blood flow [131].
Although the existence of Aβ plaques and NFTs establishes the definitive diagnosis of AD, many researches have challenged the predominant belief that Aβ is central to the development of disease. For example, studies have demonstrated that NFT pathology correlates more strongly than amyloid plaque load with brain atrophy and cognitive decline [132, 133]. In addition, clinical trials targeting cerebral Aβ plaque removal in symptomatic patients have largely failed to provide a clinical benefit and have consequently raised concerns regarding the role of Aβ in the etiology and treatment of AD [134]. Alternative theories of AD pathogenesis have also been postulated. For instance, different groups consider AD to be a combination of multiple disorders of diverse etiology [135], a by-product of normal aging [136, 137], or initiated by faulty immune activation [138]. Others have described AD as a metabolic disorder similar to diabetes, and even coined the term Type 3 Diabetes to highlight their shared molecular and cellular disturbances, such as insulin resistance, oxidative stress, and glycogen synthase kinase 3β activation [139]. These data are essential as the field continues to both expand and refine our understanding of AD pathogenesis and explore potential therapeutic avenues. However, this evidence does not preclude Aβ from playing a principal role in disease. Indeed, several studies have demonstrated that the presence of misfolded Aβ is sufficient to induce pTau and NFTs in vitro and in vivo [140–143]. Furthermore, overwhelming data from preclinical animal models have shown that targeting the production, aggregation, or immune-based removal of Aβ, and especially soluble Aβ42, preserved synapses and neuronal function as well as prevented cognitive decline [10, 144–146]. Importantly, a recent promising phase Ib human clinical trial, using a monoclonal antibody (aducanumab) to target the removal of both soluble oligomeric and fibrillar Aβ, has reinvigorated the field of Aβ-centered AD therapeutics. After 1 year of monthly aducanumab infusions, patients with prodromal or mild AD displayed a reduced cerebral Aβ plaque load and, by preliminary analyses, exhibited slowing of cognitive decline [147]. Taken together, it is no surprise that Aβ, in its various forms, remains the focus of AD research and a target for AD prevention and therapy.
In this review, we summarize various cellular and molecular, physiologic mechanisms of Aβ removal from the brain. Specifically, we cover Aβ transport across the blood–brain barrier (BBB), glymphatic clearance, cellular uptake, and enzymatic degradation. Large-scale genetic studies have further cemented the connection between Aβ accumulation, clearance by innate immune cells, and disease risk, and will be the topic of the following section. Finally, we place a particular emphasis on the growing evidence supporting a key role for microglia, and moreover, monocyte-derived macrophages in the physiological clearance of cerebral Aβ (see Fig. 1), and we examine their potential as targets for disease-modifying therapies.
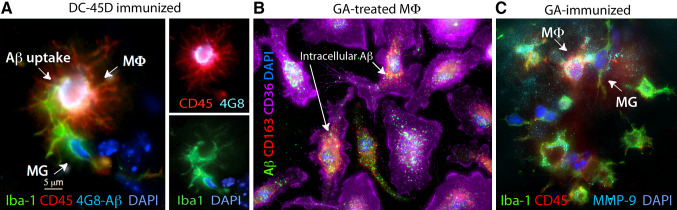
Fig. 1.
Cerebral Aβ clearance by peripheral monocyte-derived macrophages. a ADtg mice were immunized with dendritic cells (DCs) pulsed with an altered myelin-derived peptide (MOG45D). Brain-resident microglia (MG, Iba1+/CD45int-low), and moreover, blood-borne infiltrating Iba1+/CD45high macrophages (MΦ, red), are involved in the uptake of cerebral Aβ (4G8+; bright white areas), as shown in the hippocampal region from an immunized ADtg mouse. Image adopted from Koronyo-Hamaoui et al., J Neurochemistry [148]. b Phagocytosis of fibrillar Aβ42 (6E10) and co-localization within CD163+CD36high bone marrow-derived macrophages in cultures treated with glatiramer acetate (GA). c A GA-immunized ADtg mouse brain exhibiting increased expression of Aβ-degrading enzyme (MMP-9) by recruited blood-borne MΦ surrounding Aβ plaques. Microscopic images from Koronyo et al., Brain, [144]
Genes related to Aβ homeostasis and Alzheimer’s disease
Historically, the study of AD-related genes pertained to the rare, inheritable and early onset forms of the disease (termed FAD) [4, 22]. These early genetic studies identified FAD as a monogenic disorder resulting from mutations in APP, PS1, or PS2 leading to the amyloidogenic processing of APP and overproduction of synaptotoxic Aβ42 (Table 1) [4, 23]. In contrast, the far more common, late-onset AD (LOAD) is a multifactorial disease, with complex and heterogeneous interactions between genetic and environmental factors underlying its development [149, 150]. Importantly, insufficient cerebral Aβ clearance is thought to drive LOAD pathogenesis [110]. The strongest known susceptibility locus for LOAD encodes apolipoprotein E (ApoE) [51, 151]. Carriers of a single, and moreover, carriers of double APOE4 alleles have a significantly increased risk of developing AD [51, 151]. Apoε4 has been implicated in Aβ trafficking and neurovascular function, with possible additional effects on myeloid cell phenotype and ability to phagocytose Aβ (discussed further below) [52, 152–154]. Recent genome-wide association studies (GWAS), case-control and family-based studies, whole exome sequencing studies, and meta-analyses of large LOAD patient datasets have further identified over 20 novel risk factors with varying effect sizes and frequencies in the population (Table 1) [32–34, 76, 83, 96, 155]. Remarkably, a vast majority of these risk genes are associated with Aβ processing or trafficking as well as with a wide range of immunological responses, especially those related to myeloid cell-mediated Aβ clearance [156]. More specifically, LOAD risk genes have been demonstrated to impact inflammation (APOE, INDPP5D, CR1, TREM2, MS4A), complement activation (CLU, CR1), the HLA gene complex (HLA-DRB1, HLA-DRB5), and myeloid cell-mediated Aβ proteolysis (ACE, CD2AP) and phagocytosis (APOE, BIN1, INPP5D, CR1, ABCA7, TREM2) [21, 156]. In particular, polymorphisms in the genes CD33 (sialic acid-binding immunoglobulin-like lectin 3) and TREM2 (triggering receptor on myeloid cells 2) directly link impaired microglial and macrophage phagocytosis of Aβ to increased susceptibility to AD. The reported effect size of TREM2 variants on AD risk has varied in the literature [96, 155]: some investigations estimate an odds ratio of 3–4 (similar to the risk of carrying a single Apoε4 allele), while others show only a small to moderate effect [96, 151, 155, 157]. Nonetheless, TREM2 has remained in the spotlight for its effects on myelomonocytic cell phenotype and Aβ phagocytosis, which will be discussed in later sections. It has long been questioned whether AD-associated inflammation and myeloid cell dysfunction drive disease pathogenesis or instead represent a subsequent reaction to the associated neuropathology [123, 158]. Yet, these recent large-scale genetic studies, compiling data from thousands of AD subjects, illustrate unequivocally the principal role of immunological processes in development of AD, and for the first time, provide genetic evidence supporting the significance of the peripheral immune system.
Mechanisms of Aβ clearance
The key mechanisms of Aβ clearance were shown to involve either Aβ removal to the peripheral blood and lymphatic systems or degradation within the CNS tissues. Aβ reaches the peripheral circulation via chaperone-mediated transport across the blood brain barrier (BBB) [159], perivascular drainage [160], or through the glymphatic system [161, 162]. In the parenchyma, myelomonocytic cells were shown to phagocytose fibrillar Aβ, and perhaps their soluble oligomeric forms as well. These professional phagocytes, together with astrocytes and neurons, are jointly responsible for degradation and removal of amyloidogenic Aβ alloforms [123, 163]. Though each system likely contributes to Aβ clearance to varying extents, their summed effects are essential for Aβ homeostasis. This implies that perturbations of any singular process may underlie or predispose to pathologic Aβ accumulation, and consequently development of AD.
Extracellular enzymatic degradation of Aβ
Secreted peptidases are critical for the catabolism of Aβ peptides. These enzymes were reported to have an affinity for specific domains within the Aβ amino acid sequence and an ability to cleave and convert these peptides to shorter, more benign forms [164–167]. Table 2 describes major Aβ-degrading enzymes, their substrates, their cellular location and the cell types known to express and secrete them. The following paragraphs describe several Aβ-degrading enzymes that have been central in AD research.
Table 2.
Amyloid β-degrading enzymes in Alzheimer’s disease
| Enzyme | Type | Expression | Active site | Aβ substrate | References |
|---|---|---|---|---|---|
| NEP | Type II integral membrane zinc metalloprotease | Membrane-bound; neurons, Mo/MΦ, MG, astrocytes | Ext | sAβ40,42 | [166, 168–173] |
| IDE | Zinc metalloprotease | Cytosolic, cell surface, secreted; neurons, Mo/MΦ, MG, astrocytes | Ext and Int | sAβ40,42 | [165, 174–177] |
| MMP-2 | Matrixin; zinc metalloprotease | Membrane-bound, secreted; endothelial cells, Mo/MΦ, pyramidal neurons, astrocytes | Ext | sAβ | [178–182] |
| MMP-3 | Matrixin; Zinc metalloprotease | Secreted; endothelial cells, Mo/MΦ, MG, astrocytes | Ext | sAβ | [183, 184] |
| MMP-9 | Matrixin; zinc metalloprotease | Secreted; neurons, MG, astrocytes, Mo/MΦ | Ext | sAβ; fAβ; Mature plaques | [144, 167, 185–190] |
| ACE | Zinc metalloprotease | Membrane-bound, Secreted; Muscle and endothelial cells, lymphocytes and Mo/MΦ | Ext | sAβ40,42; fAβ40,42 | [145, 164, 191, 192] |
| ECE-1 | Zinc metalloprotease | Membrane-bound; endothelial cells, neurons, Mo/MΦ, MG, astrocytes | Ext | SynAβ40; Aβ in Ctx and Hip | [193–195] |
| Cathepsin B | Cysteine protease | Within lysosomes; various cell types | Int | Controversial; APP; Aβ40,42 | [196–201] |
| Cathepsin D | Aspartic protease | Within lysosomes; various cell types | Int | sAβ40,42 | [202–204] |
ACE angiotensin-converting enzyme, APP amyloid precursor protein, Ctx cortex, ECE-1 endothelin-converting enzyme 1, Ext. extracellular, fAβ fibrillar Aβ, Hip hippocampus, IDE insulin-degrading enzyme, Int. intracellular, Mo/MΦ monocytes/macrophages, MG microglia, MMP-2 matrix metalloproteinase 2, MMP-3 matrix metalloproteinase 3, MMP-9 matrix metalloproteinase 9, NEP neprilysin, oAβ oliogomeric Aβ, sAβ soluble Aβ, SynAβ synthetic Aβ
Angiotensin-converting enzyme (ACE)
ACE is a zinc-dependent peptidase with significant expression by endothelium throughout the body as well as by cortical neurons in the brain [205]. Most well known for transforming angiotensin-I to angiotensin-II and for its role in regulating hemodynamic stability and salt balance, ACE was also shown to degrade Aβ, and importantly, cleave Aβ42 into the less toxic Aβ40 alloform [164]. In post-mortem analyses, cortical and perivascular ACE expression was upregulated in the brains of AD patients and correlated with parenchymal plaque load and extent of perivascular amyloid deposition, respectively [206, 207]. Furthermore, lower levels of ACE protein and its activity were associated with lower CSF Aβ, indicating more prominent amyloid pathology in the parenchyma [44]. It was thus hypothesized that increased ACE activity in CNS tissues is a protective response to increasing amyloid pathology. While this claim is partially supported by both genetic studies in humans and physiologic studies in ADtg mice, there are inconsistencies within the literature. Both case-control studies and several large meta-analyses have identified an insertion within intron 16 of the gene ACE1 that reduces plasma ACE levels and increases risk for AD [45–47]. However, these findings were not always replicated [208]. Interestingly, AD patients homozygous for the insertion polymorphism had a greater risk of cognitive deterioration and clinical progression than other ACE genotypes [48], suggesting ACE activity may critically modulate the pathophysiology underlying neurodegeneration. Indeed, one long-term study of the ACE-inhibitor (ACE-I) captopril in ADtg mice supports the role of ACE in Aβ clearance, as both Aβ plaque load and Aβ42 levels were elevated after 11 months of treatment [209]. It is important to note, however, that studies of shorter duration did not report a measurable effect of other ACE-Is on Aβ pathology [205]. In ADtg mice, ACE overexpression by microglia and monocytes/macrophages lead to a dramatic reduction in cerebral Aβ levels and cognitive decline [145, 191], demonstrating great therapeutic potential discussed further below. Taken together, there is substantial, although inconsistent, evidence implicating ACE in the physiological clearance of Aβ that merits further investigation.
Insulin-degrading enzyme (IDE)
IDE is a zinc metalloprotease that is capable of degrading soluble Aβ40 and Aβ42 into non-toxic fragments [165, 174]. Although primarily localized in the cytosol, a small fraction of IDE is secreted by glial cells [175, 176, 185] or expressed on the cell surface of neurons [177], where it serves as a critical enzyme for extracellular Aβ degradation [165, 210]. Investigations in human and AD rodent models have yielded varying evidence regarding IDE mRNA expression, protein levels, and activity in the AD brain, most likely because the behavior of IDE is highly dependent on age [211–213], brain region [211–213], disease severity [212, 213], and APOE status [214]. In general, it seems IDE levels and activities are upregulated in response to Aβ exposure, with the exception of Apoε4/4 carriers, who exhibit reduced IDE expression.
Matrix metalloproteinase-9 (MMP-9)
MMP-9 is a secreted enzyme and member of the zinc metalloprotease (MMP) family. In general, MMPs are responsible for the degradation and maintenance of the extracellular matrix. MMP-9 has been shown to degrade compact plaques [186, 187] as well as soluble Aβ42 and Aβ40 [167]. In the CNS, MMP-9 is expressed by neurons [188], microglia [189], astrocytes [190], and infiltrating Iba+/CD45hi monocytes (Fig. 1C) [144, 148]. MMP-9 has also been shown to act as an α-secretase, favoring non-amyloidogenic processing of APP and the production of sAPPα [215]. In addition to its efficient degradation of Aβ, MMP-9 was shown to be involved in both TNFα-mediated pro-inflammatory and anti-inflammatory signaling in activated macrophages and microglia [216, 217]. Elevated levels of MMP-9 have been correlated with BBB breakdown, demyelination, and cell death in other CNS disorders like multiple sclerosis [218] and spinal cord injury [219]. These effects should be considered when modulating MMP-9 activity in vivo.
Neprilysin (NEP)
NEP is a type II integral membrane zinc metalloprotein with the bulk of its structure, including the active site, facing the extracellular space. NEP is expressed throughout the brain, predominantly on pre- and post-synaptic neuronal membranes [168, 169], and by microglia [170] and astrocytes [171]. NEP is considered the most potent Aβ-degrading enzyme [220, 221], preferentially cleaving oligomeric Aβ42 and Aβ40 [166, 172] but not fibrillar forms. NEP expression and activity has been shown to decline with age and disease in post-mortem human AD brain tissue [222], which may contribute to Aβ accumulation. Modeling this reduction by dampening NEP expression [223, 224] or activity [225] in ADtg mice resulted in elevated Aβ pathology and cognitive deficits. Conversely, the beneficial effects of NEP overexpression speak to the therapeutic potential of targeting neprilysin activity, discussed further below [146, 172].
Enzymatic degradation by innate immune cells
NEP, IDE, ACE and MMP-9 are Aβ-degrading enzymes expressed by innate immune cells and represent a crucial pathway by which these cells may eradicate pathogenic Aβ (Table 2). Expression of NEP, IDE, and MMP-9 was shown to decline in microglia of aged APP/PS1 mice, which may contribute to their functional impairment in later stages of AD [170]. This altered microglial phenotype was contingent on the presence of Aβ, as microglia from age-matched controls did not exhibit reduced enzyme expression. Microglial expression of NEP and IDE were also shown to be highly inducible in vitro and correlated with enhanced clearance of soluble Aβ42 [226].
For proteolytic processing of Aβ by monocyte-derived macrophages, the expression of MMP-9 appears to be especially important. APPSWE/PS1ΔE9 mice infused with CD115+ monocytes or immunized with the altered myelin-derived antigens, such as glatiramer acetate (GA) or myelin oligodendrocyte glycoprotein-derived peptide (MOG45D) displayed increased accumulation of MMP-9-secreting macrophages surrounding Aβ plaques (Fig. 1c), along with a marked reduction in Aβ neuropathology and cognitive impairment [144, 148]. GA stimulation of bone marrow-derived macrophages in vitro also dramatically induced MMP-9 expression [144]. Additionally, peripheral macrophages cultured on top of plaque-bearing brain sections of PDAPP mice cleared Aβ, in part, by upregulated expression of MMP-9 [185]. Interestingly, macrophages expressed MMP-9 in an ApoE-dependent manner. Apoε4 significantly dampened MMP-9 expression, suggesting an additional mechanism by which Apoε4 disrupts Aβ clearance [185].
Furthermore, ACE has a demonstrated ability to modulate the behavior of innate immune cells in ADtg murine models [191, 227]. In other disease models, targeted overexpression of ACE to myelomonocytic cells enhances their immune function, including their ability to clear cellular debris and promote tissue repair. Targeted ACE overexpression to myelomonocytes (ACE10/10 model) introduced to APPSWE/PS1ΔE9 transgenic mice resulted in increased infiltration of monocyte-derived macrophages that were tightly associated with Aβ plaques and displayed increased ability to phagocytose Aβ [145]. The net result was reduced soluble and insoluble Aβ levels, attenuated neuroinflammation, and improved cognitive performance. In contrast, inhibition of ACE catalytic domains in ACE10/10-ADtg mice exacerbated cerebral Aβ pathology [145]. Overall, the beneficial outcomes of ACE overexpression in myelomonocytes were most likely due to the summed effects of the enhanced immune response and proteolytic capacity endowed by ACE expression.
Intracellular degradation systems
Another important mechanism of Aβ catabolism is undertaken within cells that either absorb or engulf Aβ forms. Three such critical pathways—autophagy, endosomal/lysosomal degradation, and the ubiquitin–proteasome system (UPS)—prevent intracellular protein aggregation, and are thus instrumental in protecting against the neurotoxicity of cytosolic Aβ accumulation. In AD brains, however, these systems are considerably compromised [228–231]. Degradation targets for both the UPS and autophagy originate from the cytosol, although their identities differ between the two processes. Autophagy typically facilitates clearance of larger protein aggregates and damaged organelles, while the UPS degrades misfolded or damaged proteins. Furthermore, the UPS is more highly regulated than autophagy, requiring poly-ubiquitination of the target protein for its degradation. The lysosome, too, facilitates intracellular protein degradation, though the origin of these proteins may be either cytosolic or extracellular. Because the lysosome is a final common pathway for several systems, including autophagy, it is discussed separately below.
Lysosomal degradation
The lysosome is the final destination for both autophagic vacuoles and the endosomes formed by receptor-mediated endocytosis. The latter process occurs in neurons and glia through a distinct set of molecular chaperones, discussed in greater detail in the following sections. Each lysosome contains a cocktail of hydrolytic enzymes capable of degrading Aβ; however, the hydrolytic machinery is often overwhelmed in AD [228, 232–234]. As Aβ load exceeds the degradation capacity of the lysosome, aggregates may grow larger or leak into the cytosol [228, 232–234]. Aging [235] and the presence of Apoε4 [234, 236] particularly promote lysosomal instability. Intracellular Aβ negatively impacts multiple cellular and organelle functions, including proteasome inhibition, mitochondrial abnormalities, tau hyperphosphorylation, and presumably, the seeding of amyloid plaques following cell death [121, 237, 238].
Myeloid cells in particular may suffer AD-associated deficits in endosomal-lysosomal trafficking and Aβ processing. Microglia isolated from plaque-bearing sections of human AD tissue indicated that Aβ fibrils were located in the endoplasmic reticulum and deep invaginations of the cell membrane, instead of within endosomes or lysosomes [239]. Even non-diseased microglia cultured with fibrillar Aβ42 showed incomplete intracellular degradation, with non-degraded fibrils remaining in phagosomes for up to 20 days [239, 240]. This impairment was not seen in peripheral macrophage cultures under the same conditions. In fact, after 3 days of incubation with fibrillar Aβ, less than 30% of Aβ was retained in peritoneal macrophages, indicating successful degradation, while 80% remained associated with microglia [241]. One possible explanation for deficient microglial clearance is insufficient activity of lysosomal Aβ-degrading enzymes. In support of this notion, incubating microglia with mannose-6-phosphate tagged lysosomal enzymes rescued the clearance impairment. Mannose-6-phosphate typically targets hydrolytic enzymes to the lysosome from the Golgi apparatus, and this modification has been used to deliver extracellular enzymes to the lysosome in experimental conditions [242].
While healthy peripheral macrophages appear better equipped to degrade fibrillar Aβ than resident microglia, monocytes in AD patients exhibit lysosomal dysfunction [125, 229]. Specifically, more undigested Aβ molecules exist within monocytes isolated from AD patients compared to those from healthy age-matched controls, a deficit partially attributable to reduced expression and activity of cathepsin D and other major lysosomal enzymes [229, 243]. Upregulation of miR-128 was shown to target the transcripts of these enzymes and mediate their suppression. The discrepancy in lysosomal degradation capacity between microglia and infiltrating macrophages highlights their non-redundant roles in restricting Aβ pathology and as targets for future intervention.
Autophagy-mediated degradation
Three types of autophagy exist: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). While both macroautophagy and CMA dysfunction are implicated in AD [244, 245], the former is considered to be the predominant process, and will be referred to simply as “autophagy” throughout [245]. The mechanism of autophagy-mediated clearance involves isolation of cytoplasmic contents by a double-membrane vesicle called an autophagosome or autophagic vacuole (AV). Subsequent lysosomal fusion facilitates degradation of the AV and its contents [246], which may include Aβ and APP [247, 248]. In both AD patients and ADtg models, autophagy is markedly impaired, evidenced by the large accumulation of unprocessed, Aβ-rich AVs in dystrophic neurites [249, 250]. Indeed, deficits in the autophagy-lysosomal pathway occur early in the disease process, perhaps even preceding Aβ accumulation [230, 251]. Reduced expression of key autophagic proteins (beclin-1 and autophagy proteins 5 and 7) likely contribute to autophagic dysfunction, Aβ accumulation, and neuronal cell death [248, 252–254]. Furthermore, perturbations in signaling through the mammalian target of rapamycin (mTOR) pathway, the key regulator of autophagic activity, may also contribute to its impairment in AD. Under nutrient-rich conditions, heightened mTOR signaling suppresses autophagy by phosphorylating proteins necessary for AV formation and elongation [255]. Other pathologic conditions, such as cellular starvation, oxidative stress, organelle damage, and protein aggregation, inactivate mTOR and promote autophagy as a protective response [256, 257]. In the brains of AD patients, however, mTOR signaling was shown to be inappropriately active given the toxic environment [258]. Inhibition of the mTOR pathway has thus emerged as an attractive target for therapeutic intervention, with a demonstrated benefit on Aβ levels and cognition in murine models of AD [259].
Ubiquitin–proteasome system (UPS)
The UPS is a highly regulated degradation process for cytosolic short-lived and misfolded proteins. As such, it is an important protective mechanism against neurotoxic protein aggregates. Briefly, specific proteins are polyubiquinated by a series of ligases (E1, E2, and E3) for recognition and degradation by the 26S proteasome complex. Whether UPS dysfunction is a cause or consequence of AD-related degeneration remains unknown. In favor of the former, both ubiquitin conjugation and proteasome activity decline with age and in AD tissue [231, 260, 261]. Areas with reduced proteasome function overlap with those greatly impacted by AD: the hippocampus, nearby limbic structures, and the inferior parietal lobe [231]. Diminished activity of the 26S complex promotes Aβ deposition and perhaps its production as well through increased maturation and trafficking of APP [262, 263]. Taken together, this data could imply that declining proteasome function in aging and disease leaves the brain susceptible to Aβ aggregation. Nonetheless, multiple reports have demonstrated that Aβ accumulation, in fact, inhibits proteasome activity, possibly by directly binding to the 20S catalytic subunit [238, 263]. Aβ accumulation may then contribute to proteasome dysfunction rather than result from it, although these interactions need not be mutually exclusive.
Aβ clearance mediated by extracellular chaperones
Removal of Aβ into the peripheral circulation is thought to facilitate the majority of physiologic Aβ clearance [264]. Transport across the BBB requires a specialized transport system of molecular chaperones. Specifically, members of the LDL receptor (LDLR) family, such as the low-density lipoprotein-related protein 1 (LRP-1) and ATP-binding cassette (ABC) transporters, are primary receptors for Aβ efflux [264]. LRP-1-mediated transport requires the assistance of additional adaptor proteins, and this system in total will be the focus of this section. Transporters that mediate Aβ influx into the brain parenchyma, such as the receptor for advanced glycation endproducts (RAGE), will not be discussed.
Lipoprotein-related protein 1 (LRP-1)
Located on the abluminal surface of brain endothelial cells, LRP-1 binds either ApoE-Aβ complexes or Aβ alone [53, 265], subsequently stimulating endocytosis of either species. Notably, once Aβ is contained within endothelial cells, the luminal transport protein ABCB1 facilitates the removal of Aβ species into the vascular lumen [266]. Blocking LRP-1 expression in healthy, non-ADtg mice led to impaired Aβ clearance across the BBB, and consequently, greater Aβ deposition and cognitive deficits [267]. This study may recapitulate some of the consequences of declining LRP-1 expression reported in ADtg mice, AD patients, and aging adults [159, 265, 268]. Additionally, LRP-1 is expressed on neurons, astrocytes, and microglia, facilitating cellular Aβ uptake and lysosomal degradation within these cells [269–271].
Phosphatidylinositol binding clathrin assembly protein (PICALM)
PICALM is expressed on endothelial cells, and to a lesser extent, on neurons [272]. PICALM primarily functions as an adapter protein for the transcytosis of the Aβ-LRP-1 complex across the BBB. In addition to its role in Aβ clearance, recent reports show that single nucleotide polymorphisms (SNPs) in the upstream coding region for PICALM are major risk factors for AD [83, 273]. This may indicate that appropriate PICALM function is protective. In support of this, PICALM levels in cortical microvessels of subjects with advanced AD were half the levels measured in age-matched controls. Subjects with the lowest PICALM levels displayed the greatest Aβ burden and cognitive impairment [274].
Apolipoprotein E (ApoE)
Under physiologic conditions, ApoE is a carrier protein that maintains cholesterol and phospholipid homeostasis [275]. Major ApoE receptors include LDLR, LRP-1, the very low-density lipoprotein receptor (VLDLR), and ApoE receptor 2 (ApoER2) [276, 277]. However, the exact role of ApoE in AD pathogenesis remains elusive despite mounting evidence from genetic, physiologic, and clinical studies that unequivocally supports the carrier protein’s importance [51, 52, 151, 278–281]. In vitro studies have helped to elucidate the role of ApoE, demonstrating that it binds Aβ directly under certain conditions [282]. It is thought that the resulting ApoE-Aβ complexes bind to and are internalized by LRP-1 for delivery to the vasculature and removal from the brain [53, 68]. Supporting this amyloid-clearing role for ApoE, a recent study revealed that ApoE levels inversely correlated with cerebral Aβ load in non-demented healthy controls [283]. In contrast, however, ApoE was shown to compete with fibrillar or soluble Aβ for uptake and degradation by microglia and astrocytes, respectively [54, 284]. Taken together, the literature suggests distinct mechanisms by which ApoE enhances and hinders Aβ clearance. The effect likely depends on the specific Aβ conformation, the ApoE isoform and its lipidation state, as well as the relative ApoE receptor expression on the target cell [278].
Three isoforms of ApoE exist in humans: Apoε2, Apoε3, and Apoε4 [279]. Evidence suggests that the APOE2 allele may be protective against AD [151]; conversely, carrying one, or to a greater extent, two APOE4 alleles significantly increases the risk of developing AD and reduces the age of onset [51, 151, 282]. Furthermore, the APOE4/4 genotype is associated with accelerated and more pronounced cerebral amyloid pathology and CSF abnormalities [285]. Several pathogenic mechanisms may explain this increased risk associated with Apoε4. First, the rate of vascular Aβ clearance is diminished in those expressing Apoε4 compared to other isoforms [52, 53, 152], perhaps due to its reduced affinity for Aβ [286]. Additionally, Apoε4 can redirect the ApoE-Aβ complex to a different receptor, VLDLR, which has slower internalization kinetics than other LDLRs [53]. The net result is reduced internalization of Aβ by LRP-1, and ultimately reduced Aβ clearance [280, 281, 287]. Apoε4 may also promote damage to the BBB by upregulation of pro-inflammatory signaling through cyclophilin A [153]. In non-demented murine models, Apoε4 led to reduced cerebral blood flow and microvascular length, while also increasing BBB permeability [153]. A compromised BBB can reduce vascular Aβ clearance and predispose to further injury through leakage of toxic blood proteins [153, 288]. These destructive outcomes were not observed in mice expressing Apoε2 or Apoε3 [153, 289].
Apoε4 may hinder other important mechanisms of Aβ clearance, namely, intracellular catabolism by neurons and innate immune cells. Specifically, Apoε4 may interfere with these processes by inducing lysosomal leakage or by impeding myeloid cell-mediated clearance [185, 234, 236]. Though ApoE normally has an anti-inflammatory effect, this trait is markedly dampened by expression of Apoε4 on innate immune cells [154]. Crossing APOE4-targeted replacement mice with the 5XFAD ADtg model greatly increased microgliosis and astrogliosis surrounding Aβ plaques [290]. Similarly, in cell cultures of microglia and astrocytes isolated from these Apoε4-expressing mice, more pro-inflammatory cytokines were released from these cells in response to soluble oligomeric Aβ than from those expressing Apoε3 [154]. Furthermore, peripheral macrophages expressing Apoε4 exhibited a diminished capacity to phagocytose and clear Aβ when cultured on top of plaque-bearing brain sections of PDAPP mice [185]. It remains unclear whether Apoε4 influences AD predominantly through gain of toxic function, loss of protective function (i.e. vascular/immune cell dysfunction), or both. Further investigation is required to reveal the exact role of ApoE and its isoforms in AD, and the possible therapeutic potential of its manipulation.
Glymphatic clearance
The glymphatic system is a pathway of brain-wide waste clearance for small proteins and metabolites. In this pathway, CSF enters the periarterial space and, driven by arterial pulsations, enters the brain parenchyma to exchange with the interstitial fluid (ISF). Bulk flow of CSF/ISF, containing extracellular molecules such as Aβ, are then driven to perivenous spaces for recirculation in the CSF or clearance to peripheral lymphatics [161, 162]. Glymphatic activity is greatest during sleep, with Aβ clearance rates doubling those observed in periods of wakefulness [291]. The glymphatic system was named, in part, for acting as a surrogate to CNS lymphatic drainage, a system the brain traditionally lacked. However, a recent, seminal study has identified meningeal lymphatic vessels for the first time [292, 293]. This groundbreaking discovery calls for a re-evaluation of current notions of the neuroimmune connection, and raises exciting potential explanations of the pathophysiology of Aβ accumulation and defective clearance in some cases.
Water channels known as aquaporin 4 (AQP4) are the key elements in CSF-ISF exchange, and thus clearance through the glymphatic pathway. AQP4 is located on astrocytic end feet and encircles the vasculature. Mice lacking astrocytic AQP4 showed reduced CSF influx by ~70% and decreased interstitial Aβ clearance by ~55–65% [161, 162]. Advanced age also reduced glymphatic clearance rates in murine models, perhaps due to an age-dependent loss of AQP4 polarization [294]. Interstitial solutes may also be cleared directly into the CSF compartment through periarterial pathways flowing opposite to the glymphatic system. These two pathways may not be mutually exclusive; they might be two components of the same system, or their activities may vary in space and time throughout the CNS [264].
Myeloid cell-mediated phagocytosis
A growing body of evidence supports the emerging concept that activated inflammatory cells, mainly brain-resident microglia and infiltrating blood-borne monocyte-derived macrophages, are critical for the physiological clearance of Aβ [148, 295–298]. Microglia are tightly associated with Aβ deposits and senile plaques, and early studies have documented their involvement in cellular Aβ uptake [299, 300]. However, these investigations were lacking the capacity to distinguish activated microglia from blood-borne macrophages due to their similar immunophenotype and function. Recruited macrophages were thus inappropriately characterized as part of the microglial pool, and confusion ensued over their unique behavior [296, 301].
Today’s newer methodologies delineate subtle differences in marker expression, allowing for a more accurate categorization and attribution of function to these cell populations. For example, standard CD11b (MAC1), isolectin B4 (IB4), F4/80, or ionized calcium binding adapter molecule 1 (Iba1) markers in the brain are indistinguishably expressed by both infiltrating monocytes and resident microglia [302, 303]. Yet, the combination of one of these myelomonocytic markers with differential expression levels of CD45 [304, 305], P2RY12 [306], or Ly6C [303] can help differentiate these cell types (Fig. 1a, c). Other approaches may involve fluorescent labeling of peripheral innate immune cells (i.e. green or red fluorescent protein-labeled, GFP or RFP, respectively) or introducing genetic modifications, such as targeted NEP- or ACE overexpression in monocytic cells [145, 146, 191].
Other key developmental and functional differences between microglia and macrophages help distinguish these unique cell types. Microglia originate from hematopoietic stem cells of the yolk sac [307], while infiltrating monocyte-derived macrophages originate from bone marrow hematopoietic myelomonocytes [308]. In early post-natal life, microglia participate in synaptic pruning [309]. Later on, they are critical for maintaining CNS homeostasis, regulating immune surveillance, and responding to pathologic changes such as Aβ aggregation [310]. Less is known about CNS monocyte interactions under physiological conditions [311]. A comprehensive comparison of these two cell types is beyond the scope of this manuscript; however, detailed reviews on their unique embryology, development, and immune responses can be found elsewhere [303, 307, 308].
Heterogeneous populations of these immune cells exist in the brain, especially in the diseased state. Their demonstrated clearance capacity varies given the experimental paradigm and the phase of disease studied. Table 3 provides a summary of research on monocytes/macrophages in human AD subjects, while Table 4 briefly describes similar data in rodent models. The discussion that follows describes the phagocytic process mediated by microglia and monocyte-derived macrophages and the conditions in which they differ. When evaluating this data, it is important to keep in mind the difficulties involved in assessing peripheral monocytes and microglia as distinct cell types. Therefore, we cannot rule out the possibility that some investigations illustrating a role for microglia may also include effects of infiltrating monocytes.
Table 3.
Alzheimer’s disease-related impairments in human myeloid cells
| Study type | Study design | Altered protein/gene | Mo phenotype and Aβ clearance | References |
|---|---|---|---|---|
| HC Mo and MG | Pulse-chase analysis of cytokine impact on Aβ degradation |
↑IFN-γ, TNF-α ↑IL-4, IL-10, and TGF-β1 |
↓ Aβ degradation with pro-inflammatory cytokines; ↓ IDE ↑ Aβ degradation with anti-inflammatory and regulatory cytokines |
[312] |
| AD Mo | rt-PCR and flow cytometry analysis of CD33 expression | ↓ CD33 mRNA | ↓ CD33+ Mo in AD Patients; Positive correlation between number of CD33+ Mo and MMSE scores | [313] |
| Inflammatory profile; Mo analysis |
↑ HLA-DR and CD16 ↑ MCP-1 plasma levels ↓ CCR2 expression |
↓ Cerebral recruitment of Mo; ↑ Granularity by SSC | [314] | |
| Compared Mo from AD patients to HC |
↑ Inflammatory profile expressing CCR2, IL-6, IL-23, TLRs ↓ MGAT3 and TLR ↓ Cathepsin B, D, S ↓ Activity of β-Galactosidase, α-Manosidase β-Hexosaminidase |
↑ Apoptosis; ↓ Aβ phagocytosis by Mo; Impaired phenotype | [125, 127, 229, 243, 315] | |
| AD vs. MCI Mo | Histone acetylation; cytokine release; susceptibility to cell damage |
↑ Production of MIP2 and TNF-α ↓ Acetylation of H4K12 compared to MCI |
↑ Mo cell damage susceptibility in AD vs. MCI | [316] |
| AD peripheral blood | Microarray assessment of gene expression in blood; blood count |
Multiple early changes in gene expression >700 altered in blood from MCI, AD vs. HC |
↑ Mo number in AD vs. HC; ↑genes encoding cell adhesion molecules and other immune-related genes | [317] |
CCR2 C-C chemokine receptor type 2, CD33 Sialic acid-binding immunoglobulin-like lectin 3, H4K12 histone H4 at lysine 12, HC healthy control, HLA-DR human Leukocyte Antigen–antigen D Related (MHC class II surface receptor), IDE insulin degrading enzyme, IFN-γ interferon-γ, IL-4 interleukin-4, IL-6 interleukin-6, IL-10 interleukin-10, IL-23 interleukin-23, MCI mild cognitive impairment, MCP-1 monocyte chemoattractant protein-1, MG microglia, MGAT3 beta-1,4-mannosyl-glycoprotein 4-beta-N-acetylglucosaminyltransferase, MIP2 macrophage inflammatory protein 2, MMSE mini-mental state examination (Folstein test)—questionnaire used extensively in clinical and research settings to measure cognitive impairment, Mo/MΦ monocytes/macrophages, rt-PCR reverse transcription polymerase chain reaction, SSC side light-scatter characteristics (flow cytometry—measure of granularity and differentiation), TGF-β1 transforming growth factor-β1, TLRs toll-like receptors, TNF-α tumor necrosis factor-α
Table 4.
Studies in rodent models of Alzheimer’s disease implicating a role for peripheral myeloid cells in cerebral Aβ clearance
| Study Type | Study design | Mo infiltrationa | Aβ phagocytosis by Mo | Aβ levels | Neuroinflammation | Cognition | References |
|---|---|---|---|---|---|---|---|
| BM Transplantation | GFP-labeled BM cells in ADtg | ✓ | ✓ | ↓ | – | – | [144, 146, 295, 318, 319] |
| Blood Enrichment of BM-derived Mo | Treated ADtg mice with M-CSF or infusion of CD115+ GFP-labeled Mo | ✓ | ✓ | ↓ | ↓ | ↑ | [144, 146, 320, 321] |
| Immune Modulation | MOG45D-DC or GA immunization of ADtg | ✓ | ✓ | ↓ | ↓ | ↑ | [144, 148, 297] |
| Genetic Manipulation in Mo/MG | Infusion of GFP-labeled CD11b+ WT- or NEP-overexpressing Mo from healthy murine BM donors in ADtg | ✓ | – | ↓ | – | – | [146] |
| Targeted ACE overexpression of CD115+ Mo/MG in ADtg | ✓ | ✓ | ↓ | ↓ | ↑ | [145, 191, 227] | |
| Targeted blockade of TGF-β and Smad2/3 signaling in innate immune cells of ADtg | ✓ | ✓ | ↓ | ↓ | – | [322] | |
| Upregulation of TREM2 in ADtg | – | ✓ | ↓ | ↓ | ↑ | [323] | |
| TREM2 knockout in ADtg and stroke models | Χ (CD45hiLy6C+) | ✓ | ↓ | ↓ | – | [324, 325] | |
| SCARA1 upregulation | – | ✓ | ↓ | – | – | [326] | |
| Cultured WT macrophages on plaque-bearing sections of murine models | – |
✓ (APOE b) |
↓ | – | – | [185] | |
| CCL2 (MCP-1) and APP expression effects on Aβ clearance in primary BM-derived macrophages | – | ✓ | ↓ | – | – | [327] | |
| Ablation | Depletion of CD11c+ BM-derived myeloid cell or perivascular MΦ in ADtg | Χ | Χ | ↑ | – | – | [296, 297, 328, 329] |
| Inhibited Mo Infiltration | CCR2-deficient Mo in ADtg | Χ | Χ | ↑ | – | ↓ | [298, 330, 331] |
Aβ amyloid-beta protein, ACE angiotensin-converting enzyme, ADtg transgenic murine models of Alzheimer’s disease, APOE apolipoprotein E, APP amyloid-precursor protein, BM bone marrow, CCL2 C-C chemokine ligand 2, alternatively named monocyte chemotactic protein 1 (MCP-1), CCR2 C-C chemokine receptor type 2, GA glatiramer acetate, GFP green fluorescent protein, M-CSF macrophage colony-stimulating factor, MΦ macrophages, MG microglia, Mo monocytes, MOG45D-DC dendritic cells loaded with altered myelin oligodendrocyte glycoprotein-derived peptide (MOG45D; a weak agonist and a non-encephalitogenic variant of MOG(35–55) peptide), NEP neprilysin, SCARA1 class A1 scavenger receptor, TGF-β transforming growth factor-β, WT wild type
aIncreased Mo infiltration per Aβ plaques
b APOE-dependent effect
Microglia-mediated phagocytosis
Microglia aid in the normal development, function, and repair of the CNS. In response to injury or other pathological conditions, microglial processes and cell bodies migrate to lesion sites and initiate an immune response to contain and resolve particular insults [123, 124, 299]. Activated microglia are closely associated with senile plaques in both human and ADtg models. While microglia are capable of clearing Aβ in vitro [241, 300, 332–334], their in vivo clearance capacity has been questioned [335–337]. Successful Aβ internalization by microglia has been documented in some cases [338, 339], while others report incomplete processing [239, 240, 335–337]. In support of the latter, depletion of microglia in three different ADtg mouse models had no effect on fibrillar or soluble Aβ accumulation, indicating that microglia are not chiefly responsible for Aβ clearance in these models [335, 336]. Moreover, aging and toxic conditions in the AD brain render microglia chronically activated. This further reduces their phagocytic capacity and causes a prolonged neuroinflammatory response, including production of reactive oxygen species (ROS), cytokines [e.g. IL-1β, IL-6, TNFα, and transforming growth factor β (TGF-β)] and chemokines [e.g. macrophage inflammatory proteins (MIPs), monocyte chemotactic protein 1 (MCP-1), and C-C chemokine receptor types 3 and 5 (CCL3 and CCL5)] [340–342]. Elevated levels of these mediators have potent neurotoxic effects [343, 344] and correlate with increased Aβ pathology in certain brain regions of human AD patients and transgenic murine (APP/PS1) models [345]. Additionally, recent reports showed that microglia continue to participate in synaptic remodeling in aged mice [346], and can exacerbate synaptic dysfunction by modifying dendritic spine density and inappropriately engulfing endangered neurons [310, 347]. The aberrant microglial-mediated engulfment of dysfunctional synapses in ADtg models was mediated by components of the complement cascade (i.e. C1q, C3, CR3). Considering the recent genetic data linking certain SNPs in CR1 to the development of AD (Table 1), this work provides further support for the role of the immune system in AD.
Although this indicates a detrimental role for microglia in the AD brain, plaque-associated microglia have been shown to degrade scar tissue proteins with secreted proteases, clear cellular debris, and recruit the adaptive arm of the immune system to stimulate or regulate effective local immune responses [148, 348]. A recent investigation using in vivo two photon imaging also demonstrated that early on, microglia form a protective barrier around developing plaques, preventing accumulation of Aβ42 protofibrils and associated local neuritic damage [349]. Remarkably, a recent study demonstrated that stimulating hippocampal interneurons at frequencies consistent with gamma oscillations alters microglial phenotype and behavior in the 5XFAD model [350]. A 1-hour delivery of 40 Hz stimulation lowered global Aβ levels and modified microglial gene expression so that they more efficiently engulfed Aβ. Based on the available evidence, microglia cannot be labeled as either neuroprotective or neurotoxic. Instead, microglia co-exist in a range of functional states: ramified-resting under physiological conditions, classically and alternatively activated in response to injury, or dystrophic and neurotoxic in aging and chronic inflammation. These phenotypes are highly sensitive to the changes in CNS composition that accompany senescence and the neurodegeneration seen in AD [351]. Current research posits that in early stages of disease, healthy microglia comprise the first line of defense in restricting Aβ pathology, effectively clearing fibrillar and soluble Aβ through phagocytosis and proteolytic processing [123]. However, aged and diseased microglia in the AD brain have a markedly reduced capacity to do so [335, 337, 339, 349]. Taken together, it is not surprising that microglia have become candidates for potential disease-modifying therapies.
Monocyte/macrophage-mediated Aβ phagocytosis
Like microglia, monocyte-derived macrophages are professional phagocytes that support normal tissue function. However, microglial senescence in AD suggests that monocytes may have unique, complementary functions in the disease state, although this conclusion is highly controversial [303, 352–355]. Supporting evidence from genetic and physiological studies of human peripheral blood monocytes (PBMCs) highlights the importance of healthy, functional monocytes in mitigating disease (see summary of studies in Table 3). PBMCs isolated from AD patients exhibit poor differentiation, impaired phagocytosis, and increased pro-inflammatory cytokine production in response to soluble Aβ [125, 127, 229, 243, 315, 356] (Table 3). Further, rare variants of CD33 and TREM2, two genes negatively impacting the phagocytic and Aβ clearance capacity of monocytes, confer a greater risk of developing AD (Table 1) [33, 34, 64, 96, 155]. It remains to be elucidated whether the altered monocyte phenotype is a cause or consequence of disease.
Receptor-mediated Aβ phagocytosis: molecular machinery
Despite key differences highlighted previously, microglia and monocyte-derived macrophages do overlap in terms of phagocytic receptor expression and behavior [296, 301]. An extensive body of work describes under which conditions and by which mechanisms these cells are capable of engulfing distinct Aβ species. For example, microglia have been shown to phagocytose fibrillar Aβ40 and Aβ42 under in vitro [239, 299, 300, 334, 357], in vivo [358, 359], and ex vivo experimental conditions [357, 360]. However, the mechanism underlying soluble Aβ uptake is less clear. Some argue that microglia phagocytose soluble Aβ42 as they do fibrillar forms [332, 361], while others suggest uptake occurs through fluid-phase macropinocytosis [333]. These two processes may not be mutually exclusive; more precise methods of isolating distinct soluble oligomeric forms may reveal assembly dependent interactions with microglia. Similarly, studies using PBMCs isolated from healthy patients have demonstrated the ability of monocytes to effectively bind [356] and engulf soluble and fibrillar Aβ42 [125, 185, 315]. The following sections describe the major phagocytic receptors engaged in myeloid cell-mediated physiologic clearance of Aβ. Whenever possible, the discussion delineates between uptake of soluble oligomeric Aβ42, fibrillar Aβ42, and other conformations or alloforms. This distinction is particularly relevant given the varying toxicities of different Aβ species.
Toll-like receptors (TLRs)
TLRs are a family of pattern recognition receptors with distinct functions in the innate immune response. TLR2 and TLR4, in particular, were shown to be indirectly involved in Aβ phagocytosis through the formation of a receptor complex with CD14 and the subsequent activation of microglia and monocytes. Inhibiting or deleting any component of the CD14-TLR receptor complex in human monocytes or murine microglia diminished the production of pro-inflammatory cytokines and phagocytosis of fibrillar Aβ42 [362, 363].
Macrophage scavenger receptor 1 (SCARA1)
SCARA1 (alternatively named MSR-1, CD204, type-A1 scavenger receptor, and SR-A) is one of the principal receptors involved in Aβ uptake by immune cells. It is expressed on human and rodent macrophages [364], microglia [299, 332], and human monocytes [365]. SCARA1 can bind both soluble and fibrillar Aβ42 in vitro [326, 332, 365] and facilitate its subsequent uptake. Lack of functional SCARA1 in murine microglia and monocytes reduced Aβ42 uptake by a range of 50%-65% in several experimental preparations [326, 366]. Glatiramer acetate (GA), an altered myelin-derived antigen with demonstrated immunomodulatory benefits in ADtg mice [9, 144, 148, 348, 367], was shown to upregulate surface expression of SCARA1 on monocyte-derived macrophages and to increase Aβ uptake by this cell population [144]. Immunization with the FDA approved drug, GA, is an intriguing therapeutic strategy and will be discussed further below.
The importance of SCARA1 function in Aβ clearance has also been established in vivo. SCARA1-deficient APPSWE/PS1ΔE9 transgenic mice exhibited increased mortality and a significant elevation in surface area fraction stained for Aβ compared to control ADtg mice [326]. Increased microglial expression of SCARA1 around Aβ plaques has been demonstrated in multiple ADtg models [368, 369] as well as in human AD brains [370]. SCARA1 expression on CNS phagocytes appears to have a neuroprotective role in restricting toxic forms of Aβ and mitigating disease progression.
CD36
CD36 is a type B scavenger receptor expressed on the cell surface of monocytes, macrophages, astrocytes, and neurons [371]. CD36 has been shown to mediate phagocytosis of fibrillar Aβ42 through interactions with two distinct receptor complexes acting as a functional unit [334, 368, 371]. CD36-deficiency prevents microglial accumulation in response to stereotaxic intracerebral injections of fibrillar Aβ [372], and antagonists of CD36 effectively block phagocytosis of fibrillar Aβ42 in microglia cell lines [334]. Like SCARA1, expression of CD36 is substantially increased in monocyte-derived macrophages in response to GA stimulation, which may contribute to their superior Aβ clearance ability compared to untreated macrophages [144] (Fig. 1b; Table 4). CD36 was also shown to bind soluble Aβ42 directly [361, 373], although it may play a redundant role in soluble Aβ42 clearance [326]. Specific knockdown or inhibition of CD36 demonstrated a sustained ability of microglia to phagocytose soluble Aβ42 with continued expression of other scavenger receptors [332, 361].
CD36 adequately demonstrates the dichotomous role of microglia in AD pathogenesis. While CD36 confers neuroprotection through induction of Aβ removal, it also activates the NLRP3 inflammasome in microglia and stimulates pro-inflammatory cytokine release (i.e. interleukin IL-1β and ROS). Thus, microglia may contribute to the toxic environment that induces their own impairment [334, 361, 373, 374]. Moreover, a recent study has demonstrated that the soluble Aβ42-induced inflammatory milieu directly inhibits microglial phagocytosis of Aβ42 fibrils and downregulates CD36 expression in vitro [374]. In sum, it seems the ability of CD36 to initiate Aβ uptake is differentially regulated by multiple toxic species that accumulate in AD brains.
TREM2
The triggering receptor expressed on myeloid cells 2 protein is a single-pass type 1 transmembrane protein that is part of the immunoglobulin superfamily. Ligands of this receptor include anionic carbohydrates, phospholipids, and apolipoproteins such as ApoE [375–377]. TREM2, along with the protein DAP12, forms a signaling complex that is responsible for the activation of immune responses in myeloid cells including microglia, macrophages, and monocytes [378]. In AD, however, the predominant TREM2-expressing cell type has been contested [324, 376].
GWASs have recently implicated the R47H variant of TREM2 as an AD risk factor in multiple populations [96, 155]. In a post-mortem analysis of AD and control brains with and without the R47H variant, the mutation was associated with greater levels of pro-inflammatory markers and increased amyloid load in all brain areas examined [102]. Other TREM2 risk alleles have also been identified, including R62H and D87N [96, 155]. Remarkably, these mutations and others occur exclusively in the ligand-binding domain of the protein and diminish affinity of the mutant TREM2 to its ligands [377]. It was further demonstrated that myeloid cells can clear Aβ directly through TREM2-mediated uptake of lipoprotein-Aβ complexes, modeling the ApoE-Aβ interactions observed in vivo [377]. Moreover, monocytes isolated from AD patients with the R62H variant were unable to clear lipoprotein-Aβ complexes as efficiently as healthy controls. These findings imply that microglia and monocytes require a functional TREM2 protein to appropriately phagocytose Aβ.
Studies utilizing ADtg mouse models, however, point to a much more complex role for TREM2 than previously thought [323, 324, 376]. In one study, TREM2 knockout in APP/PS1 mice greatly ameliorated disease progression [324], while two other investigations successfully demonstrated TREM2-expressing immune cells containing AD pathology [323, 376]. The evidence appears contradictory; however, TREM2-modulated neuroinflammation and Aβ clearance may be highly context-dependent, influenced by the immune cell type and the inflammatory milieu in which it is expressed. In light of this, a recent study has shown that TREM2-deficient microglia and monocyte-derived macrophages phagocytose less fibrillar Aβ42 compared to wildtype cells, an impairment partially rescued by therapeutic anti-Aβ antibodies. Antibody-coated Aβ greatly enhanced phagocytosis by both TREM2 knockouts and wildtype cells, although clearance by mutant cells lagged behind controls under all conditions [325]. This finding has important implications for the efficacy of Aβ-targeted immunotherapies in patients with TREM2 mutations, yet further research is needed to fully elucidate these relationships.
CD33
CD33 is a member of the sialic acid-binding immunoglobulin-like lectins (SIGLECS) family, expressed on myeloid cells [65, 379]. In general, it is thought to dampen the immune response perhaps by inhibitory signaling through immunoreceptor tyrosine-based inhibition motifs (ITIM) [380]. In the brains of AD patients, CD33-positive microglia are enriched relative to age-matched controls and correlate with greater Aβ42 levels and plaque burden [65]. The diminished capacity of CD33-expressing microglia to phagocytose Aβ42 is thought to explain this relationship. In support of this, possession of the newly discovered rs3865444C risk allele [33, 34] results in a sevenfold increase in CD33 expression on monocytes with a significant reduction in ability to phagocytose Aβ42. Monocytes isolated from young individuals with the rs3865444C risk allele also displayed an Aβ42 phagocytic deficit [64]. Enriched monocytic CD33 expression actually mediated the relationship between this risk allele and higher amyloid plaque burden in AD brains [64]. Conversely, the protective rs3865444A allele dampens CD33 expression and increases the proportion of CD33 molecules that lack a SIGLEC-specific region responsible for phagocytosis inhibition [379]. These findings provide proof of impaired monocyte-mediated interactions with Aβ and enhanced disease risk. AD-related immune deficits are thus not solely driven by senescence or the disease process itself. Rather, monocyte phagocytic impairment may far precede Aβ deposition, as seen in these cases, and arguably predisposes to greater amyloid accumulation and lifetime risk.
Role of monocytes in AD: evidence and controversy
Despite the surging data favoring a critical role of monocytes in AD pathophysiology, it is important to acknowledge the contradictory evidence in the field surrounding monocyte-mediated Aβ clearance in chronic neurodegenerative diseases. Major questions remain. (1) Under what conditions do monocytes infiltrate the CNS? (2) Do monocytes and macrophages behave differently from microglia once in the CNS parenchyma, especially in their ability to resist misfolded Aβ forms? (3) Is the neuroprotection exhibited by monocytes a predominantly peripheral blood or a local effect? And (4) Is the effect cell-mediated, molecular or plasma-mediated, or both? The following sections address these controversies given the available literature and identify methodological discrepancies that may have generated some confusion.
Cerebral infiltration of monocytes in murine models of Alzheimer’s disease
Monocyte infiltration in AD was first documented by seminal studies transplanting GFP-labeled bone marrow cells into irradiated ADtg mice [295–297, 318]. Monocytes were shown to preferentially home to Aβ plaques and participate in their clearance [295–297, 318]. The applicability of these studies to normal physiology was later questioned due to the use of whole body irradiation (including brain) and bone marrow transplantation; the former in particular may artificially enhance monocyte infiltration into the brain parenchyma [355, 381]. Specifically, irradiation is known to induce transient BBB leakage, permitting greater passage of cells and blood contents. In addition, whole marrow transplantation increases the number of progenitor cells in the circulation.
To further elucidate the effects of irradiation, the GFP-transplantation paradigm was repeated, this time shielding the heads of recipient mice to conserve BBB integrity. This procedure reduced monocyte infiltration into the CNS, and called into question the conditions necessary for monocyte recruitment [330]. However, several investigations have successfully demonstrated spontaneous monocyte infiltration in the absence of irradiation, genetic manipulation, or chemotherapy (Table 4) [144, 146]. These experiments enriched the peripheral circulation with either CD11b+ or CD115+ monocytes from the bone marrow of young adult wildtype mice, rather than whole blood marrow, eliminating the additional confounder of increased progenitor cell numbers seen in earlier studies. Importantly, blood enrichment with GFP monocytes in age-matched wildtype (non-ADtg) animals did not cause recruitment of monocytes to the CNS [144, 146], implicating that a diseased-brain is a precondition for their cerebral recruitment. Taken together, brain irradiation is neither necessary nor sufficient for monocyte recruitment. Rather, several other conditions are consistently required, at least in ADtg models—namely, the presence of amyloid pathology, especially soluble oligomeric or fibrillar Aβ42 forms [382, 383], and binding of the monocytic surface receptor CCR2 to its ligand, MCP-1 [126, 298, 384, 385].
The mechanism by which cerebral amyloid accumulation induces monocyte infiltration is multifactorial. Vascular Aβ deposition can directly damage the vessel wall [386] and allow greater passage of monocytes into the parenchyma. Indeed, the presence of a leaky BBB was confirmed in 40–60% of AD patients [387, 388]. Furthermore, the Aβ-induced immune response alters the expression and production of inflammatory cytokines, chemokines, and their receptors [123, 311, 340–342]. The expression of MCP-1, a critical signaling factor for monocyte recruitment, is upregulated near Aβ plaques, on microglia, and on microvessels in the brains of AD patients and ADtg mice [127, 384, 385]. It is therefore postulated that the AD brain, and specifically chronically activated and overwhelmed microglia, solicit additional assistance from peripheral monocytes through MCP-1 signaling [126, 144, 148, 191, 297, 311, 389]. Other signaling cascades remain poorly understood.
Depletion or enrichment of myeloid cells: impact on cerebral Aβ burden
Modulation of monocyte recruitment to the CNS clearly demonstrates the significant contribution of monocyte-derived macrophages to Aβ clearance. Blocking CCR2 signaling [298, 330, 331] or selectively ablating these cells in the blood [296, 297, 328] greatly accelerates Aβ accumulation in ADtg models. Conversely, inducing monocyte recruitment by lipopolysaccharide (LPS) stimulation, immunization, or monocyte engraftment significantly reduces parenchymal and vascular amyloid pathology in transgenic mice [144–146, 148]. These investigations coupled with compelling in vitro data [144, 241] led to the conclusion that monocyte-derived macrophages, compared to their resident counterparts, possess a superior ability to clear fibrillar Aβ in AD (Fig. 1), resolving inflammation in spite of the toxic environment [240, 241, 296, 297, 300, 390].
Other studies utilizing microglial ablation techniques challenge this assumption. Crossing ADtg mice with the CD11b-HSVTK model, in which the thymidine kinase of the herpes simplex virus is expressed under the CD11b promoter, allows for elimination of local, proliferating myeloid cells upon intracerebroventricular administration of ganciclovir. Peripheral GFP-labeled macrophages can then repopulate the CNS, introduced by either transplantation [354] or parabiosis with an actin-enhanced GFP partner [353]. In both cases, repopulation did not augment plaque burden, insoluble Aβ, or soluble forms. Importantly, macrophages were diffusely spread across the parenchyma, in stark contrast to the plaque-associated microglia of control mice and the demonstrated plaque-homing abilities of monocytes in other models [353, 354]. Given the inability of re-populating monocytes to clear Aβ, these studies concluded that monocytes do not play a significant role in restricting amyloid pathology. However, it is possible that microglial depletion critically alters the delicate milieu required to induce monocyte phagocytic and anti-inflammatory properties. Indeed, the interplay between microglia, astrocytes, monocytes, and molecular mediators such as scar tissue proteins [i.e. chondroitin sulfate proteoglycans (CSPGs)], has been shown to attract these cells to the lesion sites and induce phenotypic shifts needed for protection in various disease states [144, 145, 148, 191, 297, 348, 367, 391]. Specifically, senescent, plaque-associated microglia are known to release MCP-1 required for monocyte recruitment [126, 298, 384, 385, 389, 392]. In addition, the impact of ganciclovir-induced neurotoxicity is poorly understood. From these repopulation studies, it is apparent that elimination of microglia impacts monocyte phenotype and function, and as such, these findings may not be representative of monocyte behavior in the natural progression of disease.
It is undeniable though that certain conditions do in fact enhance the migratory and Aβ clearing capacity of infiltrating monocytes over their resident counterparts. In particular, ADtg mice immunized with the myelin-derived peptides MOG45D or GA exhibited reduced Aβ levels and neuroinflammation, attributable to the increased recruitment of anti-inflammatory monocytes that directly engulfed Aβ [144, 148]. Other immunomodulatory approaches involving targeted overexpression of Aβ-degrading enzymes to [145, 146, 191] or genetic manipulation of [322] peripheral monocytes have demonstrated similar monocyte-mediated abrogation of Aβ deposition [Table 4]. These interventions may form the basis of promising, disease-modifying therapies that will be discussed further below.
Peripheral effects of monocytes on Aβ clearance
Recognition of the heterogeneity of different monocyte subtypes has emerged from recent studies that identified new functional biomarkers for myelomonocytic cells. An immunohistochemical and activity-based distinction has been proposed between murine monocyte subsets: an inflammatory (Ly6ChiCX3CR1intCCR2+) type pertaining to CNS recruitment and parenchymal Aβ clearance, and a patrolling (Ly6CloCx3CR1highCCR2−) type that remains associated with the vasculature [303, 308]. The discussion thus far has exclusively focused on the local effects of the inflammatory subset and their ability to reduce cerebral Aβ load in the parenchyma through cellular uptake and enzymatic degradation. However, mounting evidence suggests an additional role for patrolling monocytes and perivascular macrophages in the regulation of cerebral amyloid angiopathy (CAA), a disease process in which amyloid plaques accumulate within the walls of small cerebral blood vessels [129, 160]. CAA is seen in over 80% of AD patients and is frequently associated with microhemorrhages and cognitive deficits. Real-time in vivo imaging of APP/PS1 mice has elegantly demonstrated that patrolling monocytes are in fact attracted to and crawl along Aβ-positive veins, where they engulf Aβ and subsequently recirculate into the bloodstream [329]. To further confirm their role in perivascular Aβ clearance, depletion of patrolling monocytes substantially increased Aβ levels in the vasculature [328] as well as in the cortex and hippocampus [329]. A proposed equilibrium of Aβ clearance exists between the different CNS-associated compartments, including the brain parenchyma, perivascular spaces, CSF, and peripheral blood [328, 393, 394]. Thus, the recirculation of Aβ-containing monocytes to the periphery may effectively pull other Aβ species out of the parenchyma—a process termed the peripheral sink effect.
In addition to monocytes and macrophages in the perivascular space, recent data suggest that the activity of these cells in the peripheral blood may be pivotal for the regulation of neuroinflammation associated with AD and for inducing neuronal regeneration [144, 148, 163, 348, 367, 395–397]. Murine parabiosis studies, in which the vasculatures of two mice are joined, have effectively illustrated the impact of peripheral immune cells and, moreover, blood-soluble immune mediators on brain health. Joining the vasculatures of wildtype and ADtg mice, either before or after the onset of Aβ deposition, reduced Aβ plaque burden in the cortex and hippocampus of the ADtg parabiont, while also attenuating neuroinflammation, hyperphosphorylated tau, and neuronal apoptosis [397]. This was achieved in the absence of monocyte infiltration or CNS manipulation of known Aβ clearance pathways. It is therefore inferred that effective Aβ removal in murine models can be achieved by several mechanisms: either by blood enrichment of wildtype peripheral monocytes to boost infiltration and clearance of Aβ from brain parenchyma or by replacement and repair of the blood-soluble milieu to induce beneficial phenotypic changes in brain parenchymal cells that promote Aβ clearance. In support of the latter, earlier studies of parabiosis between young and old wildtype mice demonstrated increased synaptic plasticity, neurogenesis, and cognitive capacity in the older parabionts when sharing blood with young mice [395, 396]. This effect was attributed to the specific milieu in the blood of the younger mice rather than infiltration of peripheral immune cells [395]. Indeed, monocytes release small, soluble mediators, such as cytokines and chemokines, which can traverse the BBB and enter the brain parenchyma. Monocytes were also shown to promote anti-inflammatory behavior of surrounding microglia and astrocytes in several other disease models [144, 145, 148, 191, 297, 348, 367, 391]. Further investigation is greatly needed to understand the signaling that takes place in these models.
Therapeutic effects of peripheral monocytes and macrophages
Given the believed function of monocytes in AD etiology and the ease of access to the peripheral blood, modulation of monocyte phenotype and behavior represents a promising therapeutic target. Though not yet translated into clinical practice, recent investigations in murine models highlight the potential benefit of enhancing monocyte recruitment to the AD brain.
Stimulation with two distinct exogenous compounds has been successful in promoting Aβ clearance. Dietary curcumin, a major component of the spice turmeric, directly interacts with oligomeric and fibrillar Aβ42 [398] and may enhance Aβ phagocytosis by human PBMCs [315, 399]. Additionally, injections of the macrophage colony-stimulating factor (M-CSF) into APPSWE/PS1 mice prior to signs of cognitive impairment had a number of positive effects. These included increased circulating levels of CD45+/CD11b+/CD115+ monocytes and phagocytic activity of Aβ by Iba-1+ immune cells in brain parenchyma [320], leading to decreased size and density of Aβ plaques, and prevention of learning and memory deficits [321].
As mentioned above, peripheral immunization with DCs loaded with MOG45D (MOG45D-DC) or with GA also had profound effects on the function of innate immune cells, which consequently reduced various pathological features of AD. In ADtg mice, MOG45D-DC immunization increased CNS recruitment of anti-inflammatory macrophages, demonstrated by reduced TNF-α and increased IL-10 and TGF-β expression, that efficiently phagocytosed Aβ [148]. As a result, these mice showed restricted vascular and parenchymal Aβ deposits and reduced soluble Aβ42 levels, as well as increased expression of the Aβ-degrading enzyme MMP-9. GA immunization of ADtg mice yielded the same beneficial immunomodulatory and plaque-clearing effects [144, 297, 348], while also promoting neurogenesis and preservation of synapses and cognitive function [144]. In agreement with these findings, several other studies have shown that suppression of regulatory T-cells, either via peripheral blockade of the programmed cell death protein-1 (PD-1) [400] or of TGF-β signaling in monocyte-derived macrophages [322], enhanced monocyte recruitment to the brain in ADtg mice, and resulted in Aβ removal and improved cognitive performance [322, 400].
Other methods of immune modulation include adoptive transfer of healthy monocytes and bone marrow transplantation. Infusion of wildtype CD115+ monocytes to the peripheral blood of ADtg mice stimulated their spontaneous migration to amyloid lesions in the absence of irradiation, genetic manipulation, or chemotherapy. Treated mice exhibited reduced cerebral Aβ protein levels and astrogliosis, preserved pre-synaptic integrity, and ameliorated cognitive deficits [144]. Likewise, bone marrow transplants from wildtype donors increased monocyte recruitment to the CNS at sites of amyloid accumulation, while also reducing plaque burden [295, 297, 318].
Because peripheral monocytes were shown to cross the BBB and home to sites of Aβ accumulation, they can function as a delivery system of therapeutic agents. Targeted overexpression of either NEP or ACE has proven beneficial in abrogating AD progression in murine models. Injecting 9-month-old ADtg mice with NEP-expressing monocytes completely prevented further Aβ deposition when compared to untreated ADtg mice or those infused with monocytes containing inactive NEP [146]. Similarly, targeted overexpression of ACE to monocytic cells in the bigenic APP/PS1 mouse model of AD markedly reduced both soluble and insoluble levels of Aβ42, limited plaques and astrogliosis, and preserved cognitive function [145, 191].
Conclusion
Aβ clearance is a complex, multifactorial process, requiring the collaboration of various systems and cell types. Aβ can be removed to the peripheral circulatory or lymphatic systems by transport across the BBB or by absorption from the CSF and ISF. While innate immune cells are known to phagocytose and degrade fibrillar Aβ, these cells were only recently shown to engulf and clear soluble Aβ species as well. It is still unclear whether Aβ accumulation is a cause or consequence of disease. However, mounting evidence has shown that increased cerebral Aβ burden is the earliest pathognomonic event in AD. Moreover, soluble, oligomeric Aβ was shown to directly incite nerve and synaptic damage, leading to impaired neuronal function. In the late-onset, common cases of AD, Aβ buildup is attributed to defective clearance, rather than to its overproduction. The observed deficiency could result from impairments in any one of the removal processes or, more likely, a combination of minor clearance deficits and compounding risk factors that varies from patient to patient. Modulation of clearance mechanisms may be an important early strategy for curtailing Aβ accumulation and disease progression.
As our knowledge of AD continues to expand, so does a body of evidence that supports a key role for innate immune cells, especially monocyte-derived macrophages, in Aβ removal, local immune regulation, and repair. Bone marrow-derived monocytes can cross the BBB and clear Aβ through cellular uptake and enzymatic degradation, perhaps even more efficiently than resident microglia. The clearance process is, again, complex. Phagocytosis requires the coordination of many surface receptors (e.g. TLRs, integrins, scavenger receptors) for recognition and uptake, followed by intracellular trafficking, ultimately to lysosomes, for degradation. Monocytes, macrophages, and microglia also mediate extracellular Aβ degradation through surface expression or release of various proteases, such as ACE, IDE, NEP, and MMP-9. These functions were reported to be markedly impaired in peripheral monocytes isolated from AD patients. It is possible that the observed deficiency is a consequence of immune senescence and AD-related degeneration, or perhaps their dysfunction is a direct contributor to disease development. In support of the latter, possession of a rare variant of the AD-associated CD33 gene impacts the phagocytic capacity of monocytes isolated from young adult patients, indicating that this particular functional deficit is present throughout life. Other GWAS data have linked multiple immune-related risk factors to AD. Known relationships between the major risk gene TREM2 and monocyte/microglia phagocytic function offer a compelling demonstration of the immune system’s impact in AD.
Aggregates of misfolded Aβ are known to trigger a prolonged neuroinflammatory response that is tightly associated with synaptic dysfunction and cognitive decline. Enhancing cerebral recruitment of monocytes through either peripheral infusion or immunization with altered myelin-derived antigens was shown to temper these degenerative changes in murine models. Specifically, monocytes were able to efficiently clear Aβ and resolve the resulting astrogliosis and neuroinflammation, thereby preserving synaptic integrity and cognitive function. Immunomodulation approachs that enhance cerebral recruitment of neuroprotective monocytes hold great promise as disease-modifying therapeutic interventions and represent a valuable target for further application and translation.
Acknowledgements
This study was supported by the BrightFocus Foundation (formerly AHAF; M. K-H), The Coins for Alzheimer’s Research Trust (C.A.R.T; M. K-H) Fund, and The Cheryl and Haim Saban and The Marciano Family Foundations (M. K-H). We thank Ms. Hannah Schubloom and Ms. Mia Oviatt for editing assistance.
Abbreviations
- AD
Alzheimer’s disease
- ADtg
AD transgenic models
- Aβ
Amyloid-β protein
- AICD
Amyloid intracellular domain
- APP
Amyloid precursor protein
- ACE
Angiotensin-converting enzyme
- ApoE
Apolipoprotein E
- ApoER2
ApoE receptor 2 A
- AQP4
Aquaporin 4
- ABC
ATP-binding cassette
- AV
Autophagic vacuole
- BACE1
β-secretase 1
- BBB
Blood–brain barrier
- CCR2
c-c chemokine receptor type 2
- CNS
Central nervous system
- CAA
Cerebral amyloid angiopathy
- CSF
Cerebrospinal fluid
- CMA
Chaperone-mediated autophagy
- ECE-1
Endothelin-converting enzyme 1
- FAD
Familial Alzheimer’s disease
- GWAS
Genome-wide association study
- GA
Glatiramer acetate
- GFP
Green fluorescent protein
- HLA
Human leukocyte antigen
- pTau
Hyperphosphorylated tau
- ITIM
Immunoreceptor tyrosine-based inhibition motif
- IDE
Insulin-degrading enzyme
- IL-1β
Interleukin-1β
- IL-10
Interleukin-10
- ISF
Interstitial fluid
- Iba1
Ionized calcium binding adapter molecule 1
- LOAD
Late-onset AD
- LPS
Lipopolysaccharide
- LRP-1
Low-density lipoprotein-related protein 1
- LDLR
Low-density lipoprotein receptor
- M-CSF
Macrophage colony-stimulating factor
- SCARA1
Macrophage scavenger receptor 1
- mTOR
Mammalian target of rapamycin
- MMP-9
Matrix metalloproteinase-9
- MCP-1
Monocyte chemotactic protein 1
- MOG45D
Myelin oligodendrocyte glycoprotein-derived peptide
- NEP
Neprilysin
- NFTs
Neurofibrillary tangles
- PBMC
Peripheral blood monocyte
- PICALM
Phosphatidylinositol binding clathrin assembly protein
- PS1;PS2
Presenilin-1 and -2
- ptau
Hyperphosphorylated tau protein
- ROS
Reactive oxygen species
- RAGE
Receptor for advanced glycation end products
- RFP
Red fluorescent protein
- CD33
Sialic acid-binding immunoglobulin-like lectin 3
- SNP
Single nucleotide polymorphism
- TLR
Toll-like receptor
- TGF-β
Transforming growth factor β
- TREM2
Triggering receptor on myeloid cells 2
- TNFα
Tumor necrosis factor
- UPS
Ubiquitin–proteasome system
- VLDLR
Very low-density lipoprotein receptor
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
This paper is dedicated to the memory of Dr. Salomon Moni Hamaoui and Lillian Jones Black, both of whom died from Alzheimer’s disease.
L. Zuroff and D. Daley contributed equally to this manuscript.
References
- 1.Hebert LE, Weuve J, Scherr PA, Evans DA. Alzheimer disease in the United States (2010–2050) estimated using the 2010 census. Neurology. 2013;80(19):1778–1783. doi: 10.1212/WNL.0b013e31828726f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prince M, Wimo A, Guerchet M, Ali G, Wu Y, Prina M (2015) World Alzheimer Report 2015: the global impact of dementia. Alzheimer’s Disease International, London
- 3.Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298(5594):789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev. 2001;81(2):741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 5.Holmes BB, Diamond MI. Prion-like properties of Tau protein: the importance of extracellular Tau as a therapeutic target. J Biol Chem. 2014;289(29):19855–19861. doi: 10.1074/jbc.R114.549295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501(7465):45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8(6):595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meli G, Lecci A, Manca A, Krako N, Albertini V, Benussi L, Ghidoni R, Cattaneo A. Conformational targeting of intracellular Abeta oligomers demonstrates their pathological oligomerization inside the endoplasmic reticulum. Nat Commun. 2014;5:3867. doi: 10.1038/ncomms4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koronyo-Hamaoui M, Koronyo Y, Ljubimov AV, Miller CA, Ko MK, Black KL, Schwartz M, Farkas DL. Identification of amyloid plaques in retinas from Alzheimer’s patients and noninvasive in vivo optical imaging of retinal plaques in a mouse model. NeuroImage. 2011;54(Suppl 1):S204–217. doi: 10.1016/j.neuroimage.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koronyo Y, Salumbides BC, Black KL, Koronyo-Hamaoui M. Alzheimer’s disease in the retina: imaging retinal abeta plaques for early diagnosis and therapy assessment. Neurodegener Dis. 2012;10(1–4):285–293. doi: 10.1159/000335154. [DOI] [PubMed] [Google Scholar]
- 11.La Morgia C, Ross-Cisneros FN, Koronyo Y, Hannibal J, Gallassi R, Cantalupo G, Sambati L, Pan BX, Tozer KR, Barboni P, Provini F, Avanzini P, Carbonelli M, Pelosi A, Chui H, Liguori R, Baruzzi A, Koronyo-Hamaoui M, Sadun AA, Carelli V. Melanopsin retinal ganglion cell loss in Alzheimer disease. Ann Neurol. 2016;79(1):90–109. doi: 10.1002/ana.24548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai Y, Lu B, Ljubimov AV, Girman S, Ross-Cisneros FN, Sadun AA, Svendsen CN, Cohen RM, Wang S. Ocular changes in TgF344-AD rat model of Alzheimer’s disease. Investig Ophthalmol Vis Sci. 2014;55(1):523–534. doi: 10.1167/iovs.13-12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart NJ, Koronyo Y, Black KL, Koronyo-Hamaoui M. Ocular indicators of Alzheimer’s: exploring disease in the retina. Acta Neuropathol (Berl) 2016;132(6):767–787. doi: 10.1007/s00401-016-1613-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362(4):329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 15.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 16.Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, Iwatsubo T, Jack CR, Kaye J, Montine TJ, Park DC, Reiman EM, Rowe CC, Siemers E, Stern Y, Yaffe K, Carrillo MC, Thies B, Morrison-Bogorad M, Wagster MV, Phelps CH (2011) Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement J Alzheimer’s Assoc 7(3):280–292. doi:10.1016/j.jalz.2011.03.003 [DOI] [PMC free article] [PubMed]
- 17.Haass C, Kaether C, Thinakaran G, Sisodia S (2012) Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med 2(5):a006270. doi:10.1101/cshperspect.a006270 [DOI] [PMC free article] [PubMed]
- 18.Haass C, Hung AY, Selkoe DJ, Teplow DB. Mutations associated with a locus for familial Alzheimer’s disease result in alternative processing of amyloid beta-protein precursor. J Biol Chem. 1994;269(26):17741–17748. [PubMed] [Google Scholar]
- 19.Zigman WB, Devenny DA, Krinsky-McHale SJ, Jenkins EC, Urv TK, Wegiel J, Schupf N, Silverman W. Alzheimer’s disease in adults with down syndrome. Int Rev Res Ment Retard. 2008;36:103–145. doi: 10.1016/S0074-7750(08)00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borchelt DR, Thinakaran G, Eckman CB, Lee MK, Davenport F, Ratovitsky T, Prada CM, Kim G, Seekins S, Yager D, Slunt HH, Wang R, Seeger M, Levey AI, Gandy SE, Copeland NG, Jenkins NA, Price DL, Younkin SG, Sisodia SS. Familial Alzheimer’s disease-linked presenilin 1 variants elevate Abeta1-42/1–40 ratio in vitro and in vivo. Neuron. 1996;17(5):1005–1013. doi: 10.1016/S0896-6273(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 21.Karch CM, Goate AM. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol Psychiatry. 2015;77(1):43–51. doi: 10.1016/j.biopsych.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicolas G, Wallon D, Charbonnier C, Quenez O, Rousseau S, Richard AC, Rovelet-Lecrux A, Coutant S, Le Guennec K, Bacq D, Garnier JG, Olaso R, Boland A, Meyer V, Deleuze JF, Munter HM, Bourque G, Auld D, Montpetit A, Lathrop M, Guyant-Marechal L, Martinaud O, Pariente J, Rollin-Sillaire A, Pasquier F, Le Ber I, Sarazin M, Croisile B, Boutoleau-Bretonniere C, Thomas-Anterion C, Paquet C, Sauvee M, Moreaud O, Gabelle A, Sellal F, Ceccaldi M, Chamard L, Blanc F, Frebourg T, Campion D, Hannequin D. Screening of dementia genes by whole-exome sequencing in early-onset Alzheimer disease: input and lessons. Eur J Hum Genet. 2016;24(5):710–716. doi: 10.1038/ejhg.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheuner D, Eckman C, Jensen M, Song X, Citron M, Suzuki N, Bird TD, Hardy J, Hutton M, Kukull W, Larson E, Levy-Lahad E, Viitanen M, Peskind E, Poorkaj P, Schellenberg G, Tanzi R, Wasco W, Lannfelt L, Selkoe D, Younkin S. Secreted amyloid beta-protein similar to that in the senile plaques of Alzheimer’s disease is increased in vivo by the presenilin 1 and 2 and APP mutations linked to familial Alzheimer’s disease. Nat Med. 1996;2(8):864–870. doi: 10.1038/nm0896-864. [DOI] [PubMed] [Google Scholar]
- 24.Jonsson T, Atwal JK, Steinberg S, Snaedal J, Jonsson PV, Bjornsson S, Stefansson H, Sulem P, Gudbjartsson D, Maloney J, Hoyte K, Gustafson A, Liu Y, Lu Y, Bhangale T, Graham RR, Huttenlocher J, Bjornsdottir G, Andreassen OA, Jonsson EG, Palotie A, Behrens TW, Magnusson OT, Kong A, Thorsteinsdottir U, Watts RJ, Stefansson K. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488(7409):96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 25.Benilova I, Gallardo R, Ungureanu AA, Castillo Cano V, Snellinx A, Ramakers M, Bartic C, Rousseau F, Schymkowitz J, De Strooper B. The Alzheimer disease protective mutation A2T modulates kinetic and thermodynamic properties of amyloid-beta (Abeta) aggregation. J Biol Chem. 2014;289(45):30977–30989. doi: 10.1074/jbc.M114.599027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cruchaga C, Haller G, Chakraverty S, Mayo K, Vallania FL, Mitra RD, Faber K, Williamson J, Bird T, Diaz-Arrastia R, Foroud TM, Boeve BF, Graff-Radford NR, St Jean P, Lawson M, Ehm MG, Mayeux R, Goate AM. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PloS one. 2012;7(2):e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sleegers K, Brouwers N, Gijselinck I, Theuns J, Goossens D, Wauters J, Del-Favero J, Cruts M, van Duijn CM, Van Broeckhoven C. APP duplication is sufficient to cause early onset Alzheimer’s dementia with cerebral amyloid angiopathy. Brain J Neurol. 2006 doi: 10.1093/brain/awl203. [DOI] [PubMed] [Google Scholar]
- 28.Guerreiro RJ, Gustafson DR, Hardy J. The genetic architecture of Alzheimer’s disease: beyond APP, PSENs and APOE. Neurobiol Aging. 2012;33(3):437–456. doi: 10.1016/j.neurobiolaging.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutton M, Busfield F, Wragg M, Crook R, Perez-Tur J, Clark RF, Prihar G, Talbot C, Phillips H, Wright K, Baker M, Lendon C, Duff K, Martinez A, Houlden H, Nichols A, Karran E, Roberts G, Roques P, Rossor M, Venter JC, Adams MD, Cline RT, Phillips CA, Goate A, et al. Complete analysis of the presenilin 1 gene in early onset Alzheimer’s disease. Neuroreport. 1996;7(3):801–805. doi: 10.1097/00001756-199602290-00029. [DOI] [PubMed] [Google Scholar]
- 30.Barton AJ, Crook BW, Karran EH, Brown F, Dewar D, Mann DM, Pearson RC, Graham DI, Hardy J, Hutton M, Duff K, Goate AM, Clark RF, Roberts GW (1996) Alteration in brain presenilin 1 mRNA expression in early onset familial Alzheimer’s disease. Neurodegeneration 5(3):213–218 [DOI] [PubMed]
- 31.Chouraki V, Seshadri S. Genetics of Alzheimer’s disease. Adv Genet. 2014;87:245–294. doi: 10.1016/B978-0-12-800149-3.00005-6. [DOI] [PubMed] [Google Scholar]
- 32.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr., Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P (2013) Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature genetics 45(12):1452–1458. doi:10.1038/ng.2802 [DOI] [PMC free article] [PubMed]
- 33.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43(5):436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan SL, Kim WS, Kwok JB, Hill AF, Cappai R, Rye KA, Garner B. ATP-binding cassette transporter A7 regulates processing of amyloid precursor protein in vitro. J Neurochem. 2008;106(2):793–804. doi: 10.1111/j.1471-4159.2008.05433.x. [DOI] [PubMed] [Google Scholar]
- 36.Kim WS, Li H, Ruberu K, Chan S, Elliott DA, Low JK, Cheng D, Karl T, Garner B. Deletion of Abca7 increases cerebral amyloid-beta accumulation in the J20 mouse model of Alzheimer’s disease. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33(10):4387–4394. doi: 10.1523/JNEUROSCI.4165-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shulman JM, Chen K, Keenan BT, Chibnik LB, Fleisher A, Thiyyagura P, Roontiva A, McCabe C, Patsopoulos NA, Corneveaux JJ, Yu L, Huentelman MJ, Evans DA, Schneider JA, Reiman EM, De Jager PL, Bennett DA. Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA Neurol. 2013;70(9):1150–1157. doi: 10.1001/jamaneurol.2013.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh K, Abe-Dohmae S, Yokoyama S, St George-Hyslop P, Fraser PE. ATP-binding cassette transporter A7 (ABCA7) loss of function alters Alzheimer amyloid processing. J Biol Chem. 2015;290(40):24152–24165. doi: 10.1074/jbc.M115.655076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu Y, Hsiao JH, Paxinos G, Halliday GM, Kim WS. ABCA7 Mediates Phagocytic Clearance of Amyloid-beta in the. Brain J Alzheimer’s Dis JAD. 2016;54(2):569–584. doi: 10.3233/JAD-160456. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF. ADAM10 is the physiologically relevant, constitutive alpha-secretase of the amyloid precursor protein in primary neurons. Embo j. 2010;29(17):3020–3032. doi: 10.1038/emboj.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marcinkiewicz M, Seidah NG. Coordinated expression of beta-amyloid precursor protein and the putative beta-secretase BACE and alpha-secretase ADAM10 in mouse and human brain. J Neurochem. 2000;75(5):2133–2143. doi: 10.1046/j.1471-4159.2000.0752133.x. [DOI] [PubMed] [Google Scholar]
- 42.Suh J, Choi SH, Romano DM, Gannon MA, Lesinski AN, Kim DY, Tanzi RE. ADAM10 missense mutations potentiate beta-amyloid accumulation by impairing prodomain chaperone function. Neuron. 2013;80(2):385–401. doi: 10.1016/j.neuron.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim M, Suh J, Romano D, Truong MH, Mullin K, Hooli B, Norton D, Tesco G, Elliott K, Wagner SL, Moir RD, Becker KD, Tanzi RE. Potential late-onset Alzheimer’s disease-associated mutations in the ADAM10 gene attenuate {alpha}-secretase activity. Hum Mol Genet. 2009;18(20):3987–3996. doi: 10.1093/hmg/ddp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jochemsen HM, Teunissen CE, Ashby EL, van der Flier WM, Jones RE, Geerlings MI, Scheltens P, Kehoe PG, Muller M. The association of angiotensin-converting enzyme with biomarkers for Alzheimer’s disease. Alzheimer’s Res Ther. 2014;6(3):27. doi: 10.1186/alzrt257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kehoe PG, Russ C, McIlory S, Williams H, Holmans P, Holmes C, Liolitsa D, Vahidassr D, Powell J, McGleenon B, Liddell M, Plomin R, Dynan K, Williams N, Neal J, Cairns NJ, Wilcock G, Passmore P, Lovestone S, Williams J, Owen MJ. Variation in DCP1, encoding ACE, is associated with susceptibility to Alzheimer disease. Nat Genet. 1999;21(1):71–72. doi: 10.1038/5009. [DOI] [PubMed] [Google Scholar]
- 46.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 47.Lehmann DJ, Cortina-Borja M, Warden DR, Smith AD, Sleegers K, Prince JA, van Duijn CM, Kehoe PG. Large meta-analysis establishes the ACE insertion-deletion polymorphism as a marker of Alzheimer’s disease. Am J Epidemiol. 2005;162(4):305–317. doi: 10.1093/aje/kwi202. [DOI] [PubMed] [Google Scholar]
- 48.Chou PS, Wu MN, Chou MC, Chien I, Yang YH. Angiotensin-converting enzyme insertion/deletion polymorphism and the longitudinal progression of Alzheimer’s disease. Geriatr Gerontol Int. 2016 doi: 10.1111/ggi.12929. [DOI] [PubMed] [Google Scholar]
- 49.Kauwe JS, Bailey MH, Ridge PG, Perry R, Wadsworth ME, Hoyt KL, Staley LA, Karch CM, Harari O, Cruchaga C, Ainscough BJ, Bales K, Pickering EH, Bertelsen S, Fagan AM, Holtzman DM, Morris JC, Goate AM. Genome-wide association study of CSF levels of 59 alzheimer’s disease candidate proteins: significant associations with proteins involved in amyloid processing and inflammation. PLoS Genet. 2014;10(10):e1004758. doi: 10.1371/journal.pgen.1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edwards TL, Pericak-Vance M, Gilbert JR, Haines JL, Martin ER, Ritchie MD. An association analysis of Alzheimer disease candidate genes detects an ancestral risk haplotype clade in ACE and putative multilocus association between ACE, A2M, and LRRTM3. Am J Med Genet Part B Neuropsychiatr Genet. 2009;150b(5):721–735. doi: 10.1002/ajmg.b.30899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 52.Castellano JM, Kim J, Stewart FR, Jiang H, DeMattos RB, Patterson BW, Fagan AM, Morris JC, Mawuenyega KG, Cruchaga C, Goate AM, Bales KR, Paul SM, Bateman RJ, Holtzman DM. Human apoE isoforms differentially regulate brain amyloid-beta peptide clearance. Sci Transl Med. 2011;3(89):89ra57. doi: 10.1126/scitranslmed.3002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deane R, Sagare A, Hamm K, Parisi M, Lane S, Finn MB, Holtzman DM, Zlokovic BV. apoE isoform-specific disruption of amyloid beta peptide clearance from mouse brain. J Clin Invest. 2008;118(12):4002–4013. doi: 10.1172/JCI36663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verghese PB, Castellano JM, Garai K, Wang Y, Jiang H, Shah A, Bu G, Frieden C, Holtzman DM. ApoE influences amyloid-beta (Abeta) clearance despite minimal apoE/Abeta association in physiological conditions. Proc Natl Acad Sci USA. 2013;110(19):E1807–E1816. doi: 10.1073/pnas.1220484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bales KR, Verina T, Cummins DJ, Du Y, Dodel RC, Saura J, Fishman CE, DeLong CA, Piccardo P, Petegnief V, Ghetti B, Paul SM. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 1999;96(26):15233–15238. doi: 10.1073/pnas.96.26.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Conejero-Goldberg C, Gomar JJ, Bobes-Bascaran T, Hyde TM, Kleinman JE, Herman MM, Chen S, Davies P, Goldberg TE. APOE2 enhances neuroprotection against Alzheimer’s disease through multiple molecular mechanisms. Mol Psychiatry. 2014;19(11):1243–1250. doi: 10.1038/mp.2013.194. [DOI] [PubMed] [Google Scholar]
- 57.Beecham GW, Hamilton K, Naj AC, Martin ER, Huentelman M, Myers AJ, Corneveaux JJ, Hardy J, Vonsattel JP, Younkin SG, Bennett DA, De Jager PL, Larson EB, Crane PK, Kamboh MI, Kofler JK, Mash DC, Duque L, Gilbert JR, Gwirtsman H, Buxbaum JD, Kramer P, Dickson DW, Farrer LA, Frosch MP, Ghetti B, Haines JL, Hyman BT, Kukull WA, Mayeux RP, Pericak-Vance MA, Schneider JA, Trojanowski JQ, Reiman EM, Schellenberg GD, Montine TJ. Genome-wide association meta-analysis of neuropathologic features of Alzheimer’s disease and related dementias. PLoS Genet. 2014;10(9):e1004606. doi: 10.1371/journal.pgen.1004606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rezazadeh M, Khorrami A, Yeghaneh T, Talebi M, Kiani SJ, Heshmati Y, Gharesouran J. Genetic factors affecting late-onset alzheimer’s disease susceptibility. Neuromol Med. 2016;18(1):37–49. doi: 10.1007/s12017-015-8376-4. [DOI] [PubMed] [Google Scholar]
- 59.Miyagawa T, Ebinuma I, Morohashi Y, Hori Y, Young Chang M, Hattori H, Maehara T, Yokoshima S, Fukuyama T, Tsuji S, Iwatsubo T, Prendergast GC, Tomita T. BIN1 regulates BACE1 intracellular trafficking and amyloid-beta production. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw146. [DOI] [PubMed] [Google Scholar]
- 60.Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV, Geller F, Sottejeau Y, Harold D, Dourlen P, Grenier-Boley B, Kamatani Y, Delepine B, Demiautte F, Zelenika D, Zommer N, Hamdane M, Bellenguez C, Dartigues JF, Hauw JJ, Letronne F, Ayral AM, Sleegers K, Schellens A, Broeck LV, Engelborghs S, De Deyn PP, Vandenberghe R, O’Donovan M, Owen M, Epelbaum J, Mercken M, Karran E, Bantscheff M, Drewes G, Joberty G, Campion D, Octave JN, Berr C, Lathrop M, Callaerts P, Mann D, Williams J, Buee L, Dewachter I, Van Broeckhoven C, Amouyel P, Moechars D, Dermaut B, Lambert JC. Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry. 2013;18(11):1225–1234. doi: 10.1038/mp.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tan MS, Yu JT, Tan L. Bridging integrator 1 (BIN1): form, function, and Alzheimer’s disease. Trends Mol Med. 2013;19(10):594–603. doi: 10.1016/j.molmed.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 62.Cormont M, Meton I, Mari M, Monzo P, Keslair F, Gaskin C, McGraw TE, Le Marchand-Brustel Y. CD2AP/CMS regulates endosome morphology and traffic to the degradative pathway through its interaction with Rab4 and c-Cbl. Traffic. 2003;4(2):97–112. doi: 10.1034/j.1600-0854.2003.40205.x. [DOI] [PubMed] [Google Scholar]
- 63.Liao F, Jiang H, Srivatsan S, Xiao Q, Lefton KB, Yamada K, Mahan TE, Lee JM, Shaw AS, Holtzman DM. Effects of CD2-associated protein deficiency on amyloid-beta in neuroblastoma cells and in an APP transgenic mouse model. Mol Neurodegener. 2015;10:12. doi: 10.1186/s13024-015-0006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bradshaw EM, Chibnik LB, Keenan BT, Ottoboni L, Raj T, Tang A, Rosenkrantz LL, Imboywa S, Lee M, Von Korff A, Morris MC, Evans DA, Johnson K, Sperling RA, Schneider JA, Bennett DA, De Jager PL. CD33 Alzheimer’s disease locus: altered monocyte function and amyloid biology. Nat Neurosci. 2013;16(7):848–850. doi: 10.1038/nn.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Griciuc A, Serrano-Pozo A, Parrado AR, Lesinski AN, Asselin CN, Mullin K, Hooli B, Choi SH, Hyman BT, Tanzi RE. Alzheimer’s disease risk gene CD33 inhibits microglial uptake of amyloid beta. Neuron. 2013;78(4):631–643. doi: 10.1016/j.neuron.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mao YF, Guo ZY, Pu JL, Chen YX, Zhang BR. Association of CD33 and MS4A cluster variants with Alzheimer’s disease in East Asian populations. Neurosci Lett. 2015;609:235–239. doi: 10.1016/j.neulet.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 67.Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I, Jiang H, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, Hu-Lince D, Ryder M, Murphy A, Wagner SL, Blacker D, Becker KD, Tanzi RE. Genome-wide association analysis reveals putative Alzheimer’s disease susceptibility loci in addition to APOE. Am J Hum Genet. 2008;83(5):623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bell RD, Sagare AP, Friedman AE, Bedi GS, Holtzman DM, Deane R, Zlokovic BV. Transport pathways for clearance of human Alzheimer’s amyloid beta-peptide and apolipoproteins E and J in the mouse central nervous system. J Cereb Blood Flow Metab. 2007;27(5):909–918. doi: 10.1038/sj.jcbfm.9600419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oda T, Wals P, Osterburg HH, Johnson SA, Pasinetti GM, Morgan TE, Rozovsky I, Stine WB, Snyder SW, Holzman TF, et al. Clusterin (apoJ) alters the aggregation of amyloid beta-peptide (A beta 1–42) and forms slowly sedimenting A beta complexes that cause oxidative stress. Exp Neurol. 1995;136(1):22–31. doi: 10.1006/exnr.1995.1080. [DOI] [PubMed] [Google Scholar]
- 70.Matsubara E, Frangione B, Ghiso J. Characterization of apolipoprotein J-Alzheimer’s A beta interaction. J Biol Chem. 1995;270(13):7563–7567. doi: 10.1074/jbc.270.13.7563. [DOI] [PubMed] [Google Scholar]
- 71.Matsubara E, Soto C, Governale S, Frangione B, Ghiso J. Apolipoprotein J and Alzheimer’s amyloid beta solubility. Biochem J. 1996;316(Pt 2):671–679. doi: 10.1042/bj3160671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yerbury JJ, Poon S, Meehan S, Thompson B, Kumita JR, Dobson CM, Wilson MR. The extracellular chaperone clusterin influences amyloid formation and toxicity by interacting with prefibrillar structures. FASEB J Off Publ Feder Am Soc Exp Biol. 2007;21(10):2312–2322. doi: 10.1096/fj.06-7986com. [DOI] [PubMed] [Google Scholar]
- 73.Nuutinen T, Huuskonen J, Suuronen T, Ojala J, Miettinen R, Salminen A. Amyloid-beta 1–42 induced endocytosis and clusterin/apoJ protein accumulation in cultured human astrocytes. Neurochem Int. 2007;50(3):540–547. doi: 10.1016/j.neuint.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 74.Nuutinen T, Suuronen T, Kauppinen A, Salminen A. Clusterin: a forgotten player in Alzheimer’s disease. Brain Res Rev. 2009;61(2):89–104. doi: 10.1016/j.brainresrev.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 75.DeMattos RB, Cirrito JR, Parsadanian M, May PC, O’Dell MA, Taylor JW, Harmony JA, Aronow BJ, Bales KR, Paul SM, Holtzman DM. ApoE and clusterin cooperatively suppress Abeta levels and deposition: evidence that ApoE regulates extracellular Abeta metabolism in vivo. Neuron. 2004;41(2):193–202. doi: 10.1016/S0896-6273(03)00850-X. [DOI] [PubMed] [Google Scholar]
- 76.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 77.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PloS one. 2012;7(11):e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crehan H, Hardy J, Pocock J. Blockage of CR1 prevents activation of rodent microglia. Neurobiol Dis. 2013;54:139–149. doi: 10.1016/j.nbd.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 79.Giri M, Zhang M, Lu Y. Genes associated with Alzheimer’s disease: an overview and current status. Clinical interventions in aging. 2016;11:665–681. doi: 10.2147/CIA.S105769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schellenberg GD, Montine TJ. The genetics and neuropathology of Alzheimer’s disease. Acta Neuropathol (Berl) 2012;124(3):305–323. doi: 10.1007/s00401-012-0996-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakamoto A, Sugamoto Y, Tokunaga Y, Yoshimuta T, Hayashi K, Konno T, Kawashiri MA, Takeda Y, Yamagishi M. Expression profiling of the ephrin (EFN) and Eph receptor (EPH) family of genes in atherosclerosis-related human cells. J Int Med Res. 2011;39(2):522–527. doi: 10.1177/147323001103900220. [DOI] [PubMed] [Google Scholar]
- 82.Lai KO, Ip NY. Synapse development and plasticity: roles of ephrin/Eph receptor signaling. Curr Opin Neurobiol. 2009;19(3):275–283. doi: 10.1016/j.conb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 83.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miller SE, Sahlender DA, Graham SC, Honing S, Robinson MS, Peden AA, Owen DJ. The molecular basis for the endocytosis of small R-SNAREs by the clathrin adaptor CALM. Cell. 2011;147(5):1118–1131. doi: 10.1016/j.cell.2011.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller SE, Mathiasen S, Bright NA, Pierre F, Kelly BT, Kladt N, Schauss A, Merrifield CJ, Stamou D, Honing S, Owen DJ. CALM regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature. Dev Cell. 2015;33(2):163–175. doi: 10.1016/j.devcel.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao Q, Gil SC, Yan P, Wang Y, Han S, Gonzales E, Perez R, Cirrito JR, Lee JM. Role of phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) in intracellular amyloid precursor protein (APP) processing and amyloid plaque pathogenesis. J Biol Chem. 2012;287(25):21279–21289. doi: 10.1074/jbc.M111.338376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kanatsu K, Morohashi Y, Suzuki M, Kuroda H, Watanabe T, Tomita T, Iwatsubo T. Decreased CALM expression reduces Abeta42 to total Abeta ratio through clathrin-mediated endocytosis of gamma-secretase. Nature communications. 2014;5:3386. doi: 10.1038/ncomms4386. [DOI] [PubMed] [Google Scholar]
- 88.Tian Y, Chang JC, Fan EY, Flajolet M, Greengard P. Adaptor complex AP2/PICALM, through interaction with LC3, targets Alzheimer’s APP-CTF for terminal degradation via autophagy. Proc Natl Acad Sci USA. 2013;110(42):17071–17076. doi: 10.1073/pnas.1315110110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parikh I, Fardo DW, Estus S. Genetics of PICALM expression and Alzheimer’s disease. PloS One. 2014;9(3):e91242. doi: 10.1371/journal.pone.0091242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen J, Zhou Y, Mueller-Steiner S, Chen LF, Kwon H, Yi S, Mucke L, Gan L. SIRT1 protects against microglia-dependent amyloid-beta toxicity through inhibiting NF-kappaB signaling. J Biol Chem. 2005;280(48):40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- 91.Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P. Sadoshima J, Deng H, Pedrini S, Gandy S, Sauve AA, Pasinetti GM. Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Biol Chem. 2006;281(31):21745–21754. doi: 10.1074/jbc.M602909200. [DOI] [PubMed] [Google Scholar]
- 92.Jesko H, Wencel P, Strosznajder RP, Strosznajder JB. Sirtuins and their roles in brain aging and neurodegenerative disorders. Neurochem Res. 2016 doi: 10.1007/s11064-016-2110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Corpas R, Revilla S, Ursulet S, Castro-Freire M, Kaliman P, Petegnief V, Gimenez-Llort L, Sarkis C, Pallas M, Sanfeliu C. SIRT1 overexpression in mouse hippocampus induces cognitive enhancement through proteostatic and neurotrophic mechanisms. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-0087-9. [DOI] [PubMed] [Google Scholar]
- 94.Wang Z, Lei H, Zheng M, Li Y, Cui Y, Hao F. Meta-analysis of the association between Alzheimer disease and variants in GAB2, PICALM, and SORL1. Mol Neurobiol. 2016;53(9):6501–6510. doi: 10.1007/s12035-015-9546-y. [DOI] [PubMed] [Google Scholar]
- 95.Xue X, Zhang M, Lin Y, Xu E, Jia J. Association between the SORL1 rs2070045 polymorphism and late-onset Alzheimer’s disease: interaction with the ApoE genotype in the Chinese Han population. Neurosci Lett. 2014;559:94–98. doi: 10.1016/j.neulet.2013.11.042. [DOI] [PubMed] [Google Scholar]
- 96.Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368(2):107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Herskowitz JH, Offe K, Deshpande A, Kahn RA, Levey AI, Lah JJ. GGA1-mediated endocytic traffic of LR11/SorLA alters APP intracellular distribution and amyloid-beta production. Mol Biol Cell. 2012;23(14):2645–2657. doi: 10.1091/mbc.E12-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fjorback AW, Seaman M, Gustafsen C, Mehmedbasic A, Gokool S, Wu C, Militz D, Schmidt V, Madsen P, Nyengaard JR, Willnow TE, Christensen EI, Mobley WB, Nykjaer A, Andersen OM. Retromer binds the FANSHY sorting motif in SorLA to regulate amyloid precursor protein sorting and processing. J Neurosci Off J Soc Neurosci. 2012;32(4):1467–1480. doi: 10.1523/JNEUROSCI.2272-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schmidt V, Baum K, Lao A, Rateitschak K, Schmitz Y, Teichmann A, Wiesner B, Petersen CM, Nykjaer A, Wolf J, Wolkenhauer O, Willnow TE. Quantitative modelling of amyloidogenic processing and its influence by SORLA in Alzheimer’s disease. Embo j. 2012;31(1):187–200. doi: 10.1038/emboj.2011.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, Willnow TE. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem. 2007;282(45):32956–32964. doi: 10.1074/jbc.M705073200. [DOI] [PubMed] [Google Scholar]
- 101.Sager KL, Wuu J, Leurgans SE, Rees HD, Gearing M, Mufson EJ, Levey AI, Lah JJ. Neuronal LR11/sorLA expression is reduced in mild cognitive impairment. Ann Neurol. 2007;62(6):640–647. doi: 10.1002/ana.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Roussos P, Katsel P, Fam P, Tan W, Purohit DP, Haroutunian V. The triggering receptor expressed on myeloid cells 2 (TREM2) is associated with enhanced inflammation, neuropathological lesions and increased risk for Alzheimer’s dementia. Alzheimer’s Dement J Alzheimer’s Assoc. 2015;11(10):1163–1170. doi: 10.1016/j.jalz.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takahashi K, Prinz M, Stagi M, Chechneva O, Neumann H. TREM2-transduced myeloid precursors mediate nervous tissue debris clearance and facilitate recovery in an animal model of multiple sclerosis. PLoS Med. 2007;4(4):e124. doi: 10.1371/journal.pmed.0040124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takahashi K, Rochford CD, Neumann H. Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med. 2005;201(4):647–657. doi: 10.1084/jem.20041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hsieh CL, Koike M, Spusta SC, Niemi EC, Yenari M, Nakamura MC, Seaman WE. A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. J Neurochem. 2009;109(4):1144–1156. doi: 10.1111/j.1471-4159.2009.06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL (2006) Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. Journal of immunology (Baltimore, Md : 1950) 177 (4):2051–2055 [DOI] [PubMed]
- 107.Turnbull IR, Gilfillan S, Cella M, Aoshi T, Miller M, Piccio L, Hernandez M, Colonna M (2006) Cutting edge: TREM-2 attenuates macrophage activation. Journal of immunology (Baltimore, Md : 1950) 177 (6):3520–3524 [DOI] [PubMed]
- 108.Pottier C, Wallon D, Rousseau S, Rovelet-Lecrux A, Richard AC, Rollin-Sillaire A, Frebourg T, Campion D, Hannequin D. TREM2 R47H variant as a risk factor for early-onset Alzheimer’s disease. Journal of Alzheimer’s disease : JAD. 2013;35(1):45–49. doi: 10.3233/JAD-122311. [DOI] [PubMed] [Google Scholar]
- 109.Potter R, Patterson BW, Elbert DL, Ovod V, Kasten T, Sigurdson W, Mawuenyega K, Blazey T, Goate A, Chott R, Yarasheski KE, Holtzman DM, Morris JC, Benzinger TL, Bateman RJ. Increased in vivo amyloid-beta42 production, exchange, and loss in presenilin mutation carriers. Sci Transl Med. 2013;5(189):189ra177. doi: 10.1126/scitranslmed.3005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC, Yarasheski KE, Bateman RJ. Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science. 2010;330(6012):1774. doi: 10.1126/science.1197623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 112.Haass C, Selkoe DJ (2007) Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid beta-peptide. Nature reviews Molecular cell biology 8 (2):101–112. doi:10.1038/nrm2101 [DOI] [PubMed]
- 113.Lesné S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440(7082):352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 114.McLean CA, Cherny RA, Fraser FW, Fuller SJ, Smith MJ, Beyreuther K, Bush AI, Masters CL. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer’s disease. Ann Neurol. 1999;46(6):860–866. doi: 10.1002/1531-8249(199912)46:6<860::AID-ANA8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 115.Mc Donald JM, Savva GM, Brayne C, Welzel AT, Forster G, Shankar GM, Selkoe DJ, Ince PG, Walsh DM. The presence of sodium dodecyl sulphate-stable Abeta dimers is strongly associated with Alzheimer-type dementia. Brain : a journal of neurology. 2010 doi: 10.1093/brain/awq065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid beta protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416(6880):535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 117.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat Neurosci. 2005;8(1):79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 118.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, Regan CM, Walsh DM, Sabatini BL, Selkoe DJ. Amyloid-beta protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14(8):837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Viola KL, Klein WL. Amyloid beta oligomers in Alzheimer’s disease pathogenesis, treatment, and diagnosis. Acta Neuropathol (Berl) 2015;129(2):183–206. doi: 10.1007/s00401-015-1386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mark RJ, Pang Z, Geddes JW, Uchida K, Mattson MP. Amyloid beta-peptide impairs glucose transport in hippocampal and cortical neurons: involvement of membrane lipid peroxidation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1997;17(3):1046–1054. doi: 10.1523/JNEUROSCI.17-03-01046.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Caspersen C, Wang N, Yao J, Sosunov A, Chen X, Lustbader JW, Xu HW, Stern D, McKhann G, Yan SD (2005) Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer’s disease. FASEB J 19 (14):2040–2041. doi:10.1096/fj.05-3735fje [DOI] [PubMed]
- 122.Manczak M, Anekonda TS, Henson E, Park BS, Quinn J, Reddy PH. Mitochondria are a direct site of A beta accumulation in Alzheimer’s disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum Mol Genet. 2006;15(9):1437–1449. doi: 10.1093/hmg/ddl066. [DOI] [PubMed] [Google Scholar]
- 123.Heneka MT, Carson MJ, El Khoury J, Landreth GE, Brosseron F, Feinstein DL, Jacobs AH, Wyss-Coray T, Vitorica J, Ransohoff RM, Herrup K, Frautschy SA, Finsen B, Brown GC, Verkhratsky A, Yamanaka K, Koistinaho J, Latz E, Halle A, Petzold GC, Town T, Morgan D, Shinohara ML, Perry VH, Holmes C, Bazan NG, Brooks DJ, Hunot S, Joseph B, Deigendesch N, Garaschuk O, Boddeke E, Dinarello CA, Breitner JC, Cole GM, Golenbock DT, Kummer MP. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015;14(4):388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mosher KI, Wyss-Coray T. Microglial dysfunction in brain aging and Alzheimer’s disease. Biochem Pharmacol. 2014;88(4):594–604. doi: 10.1016/j.bcp.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fiala M, Lin J, Ringman J, Kermani-Arab V, Tsao G, Patel A, Lossinsky AS, Graves MC, Gustavson A, Sayre J, Sofroni E, Suarez T, Chiappelli F, Bernard G. Ineffective phagocytosis of amyloid-beta by macrophages of Alzheimer’s disease patients. Journal of Alzheimer’s disease : JAD. 2005;7(3):221–232. doi: 10.3233/JAD-2005-7304. [DOI] [PubMed] [Google Scholar]
- 126.Fiala M, Zhang L, Gan X, Sherry B, Taub D, Graves MC, Hama S, Way D, Weinand M, Witte M, Lorton D, Kuo YM, Roher AE. Amyloid-beta induces chemokine secretion and monocyte migration across a human blood–brain barrier model. Mol Med. 1998;4(7):480–489. [PMC free article] [PubMed] [Google Scholar]
- 127.Saresella M, Marventano I, Calabrese E, Piancone F, Rainone V, Gatti A, Alberoni M, Nemni R, Clerici M. A complex proinflammatory role for peripheral monocytes in Alzheimer’s disease. JAD. 2014;38(2):403–413. doi: 10.3233/JAD-131160. [DOI] [PubMed] [Google Scholar]
- 128.Smith MA, Perry G, Richey PL, Sayre LM, Anderson VE, Beal MF, Kowall N. Oxidative damage in Alzheimer’s. Nature. 1996;382(6587):120–121. doi: 10.1038/382120b0. [DOI] [PubMed] [Google Scholar]
- 129.Bell RD, Zlokovic BV. Neurovascular mechanisms and blood-brain barrier disorder in Alzheimer’s disease. Acta Neuropathol (Berl) 2009;118(1):103–113. doi: 10.1007/s00401-009-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Frost S, Guymer R, Aung KZ, Macaulay SL, Sohrabi HR, Bourgeat P, Salvado O, Rowe CC, Ames D, Masters CL, Martins RN, Kanagasingam Y, Group AT (2016) Alzheimer`s disease and the early signs of age-related macular degeneration. Curr Alzheimer Res 13(11):1259–1266. doi:10.2174/1567205013666160603003800 [DOI] [PubMed]
- 131.Kimbrough IF, Robel S, Roberson ED, Sontheimer H. Vascular amyloidosis impairs the gliovascular unit in a mouse model of Alzheimer’s disease. Brain J Neurol. 2015 doi: 10.1093/brain/awv327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Bennett DA, Schneider JA, Wilson RS, Bienias JL, Arnold SE. Neurofibrillary tangles mediate the association of amyloid load with clinical Alzheimer disease and level of cognitive function. Arch Neurol. 2004;61(3):378–384. doi: 10.1001/archneur.61.3.378. [DOI] [PubMed] [Google Scholar]
- 133.Murray ME, Lowe VJ, Graff-Radford NR, Liesinger AM, Cannon A, Przybelski SA, Rawal B, Parisi JE, Petersen RC, Kantarci K, Ross OA, Duara R, Knopman DS, Jack CR, Dickson DW. Clinicopathologic and 11C-Pittsburgh compound B implications of Thal amyloid phase across the Alzheimer’s disease spectrum. Brain J Neurol. 2015 doi: 10.1093/brain/awv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karran E, Hardy J. A critique of the drug discovery and phase 3 clinical programs targeting the amyloid hypothesis for Alzheimer disease. Ann Neurol. 2014;76(2):185–205. doi: 10.1002/ana.24188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Morris GP, Clark IA, Vissel B (2014) Inconsistencies and controversies surrounding the amyloid hypothesis of Alzheimer’s disease. Acta Neuropathol Commun 2:135. doi:10.1186/s40478-014-0135-5 [DOI] [PMC free article] [PubMed]
- 136.Chen M, Maleski JJ, Sawmiller DR. Scientific truth or false hope? Understanding Alzheimer’s disease from an aging perspective. JAD. 2011;24(1):3–10. doi: 10.3233/JAD-2010-101638. [DOI] [PubMed] [Google Scholar]
- 137.Herrup K. Reimagining Alzheimer’s disease—an age-based hypothesis. J Neurosci. 2010;30(50):16755–16762. doi: 10.1523/JNEUROSCI.4521-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Morris GP, Clark IA, Zinn R, Vissel B. Microglia: a new frontier for synaptic plasticity, learning and memory, and neurodegenerative disease research. Neurobiol Learn Mem. 2013;105:40–53. doi: 10.1016/j.nlm.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 139.Kandimalla R, Thirumala V, Reddy PH. Is Alzheimer’s disease a Type 3 diabetes? A critical appraisal. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbadis.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Götz J, Chen F, van Dorpe J, Nitsch RM. Formation of neurofibrillary tangles in P301l tau transgenic mice induced by Abeta 42 fibrils. Science. 2001;293(5534):1491–1495. doi: 10.1126/science.1062097. [DOI] [PubMed] [Google Scholar]
- 141.Lewis J, Dickson DW, Lin WL, Chisholm L, Corral A, Jones G, Yen SH, Sahara N, Skipper L, Yager D, Eckman C, Hardy J, Hutton M, McGowan E. Enhanced neurofibrillary degeneration in transgenic mice expressing mutant tau and APP. Science. 2001;293(5534):1487–1491. doi: 10.1126/science.1058189. [DOI] [PubMed] [Google Scholar]
- 142.Jin M, Shepardson N, Yang T, Chen G, Walsh D, Selkoe DJ. Soluble amyloid beta-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc Natl Acad Sci USA. 2011;108(14):5819–5824. doi: 10.1073/pnas.1017033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY. A three-dimensional human neural cell culture model of Alzheimer’s disease. Nature. 2014;515(7526):274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Koronyo Y, Salumbides BC, Sheyn J, Pelissier L, Li S, Ljubimov V, Moyseyev M, Daley D, Fuchs DT, Pham M, Black KL, Rentsendorj A, Koronyo-Hamaoui M. Therapeutic effects of glatiramer acetate and grafted CD115(+) monocytes in a mouse model of Alzheimer’s disease. Brain J Neurol. 2015 doi: 10.1093/brain/awv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bernstein KE, Koronyo Y, Salumbides BC, Sheyn J, Pelissier L, Lopes DH, Shah KH, Bernstein EA, Fuchs DT, Yu JJ, Pham M, Black KL, Shen XZ, Fuchs S, Koronyo-Hamaoui M. Angiotensin-converting enzyme overexpression in myelomonocytes prevents Alzheimer’s-like cognitive decline. J Clin Invest. 2014;124(3):1000–1012. doi: 10.1172/JCI66541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lebson L, Nash K, Kamath S, Herber D, Carty N, Lee DC, Li Q, Szekeres K, Jinwal U, Koren J, Dickey CA, Gottschall PE, Morgan D, Gordon MN. Trafficking CD11b-positive blood cells deliver therapeutic genes to the brain of amyloid-depositing transgenic mice. J Neurosci. 2010;30(29):9651–9658. doi: 10.1523/JNEUROSCI.0329-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sevigny J, Chiao P, Bussiere T, Weinreb PH, Williams L, Maier M, Dunstan R, Salloway S, Chen T, Ling Y, O’Gorman J, Qian F, Arastu M, Li M, Chollate S, Brennan MS, Quintero-Monzon O, Scannevin RH, Arnold HM, Engber T, Rhodes K, Ferrero J, Hang Y, Mikulskis A, Grimm J, Hock C, Nitsch RM, Sandrock A. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature. 2016;537(7618):50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- 148.Koronyo-Hamaoui M, Ko MK, Koronyo Y, Azoulay D, Seksenyan A, Kunis G, Pham M, Bakhsheshian J, Rogeri P, Black KL, Farkas DL, Schwartz M. Attenuation of AD-like neuropathology by harnessing peripheral immune cells: local elevation of IL-10 and MMP-9. J Neurochem. 2009;111(6):1409–1424. doi: 10.1111/j.1471-4159.2009.06402.x. [DOI] [PubMed] [Google Scholar]
- 149.Mormino EC, Sperling RA, Holmes AJ, Buckner RL, De Jager PL, Smoller JW, Sabuncu MR. Polygenic risk of Alzheimer disease is associated with early- and late-life processes. Neurology. 2016 doi: 10.1212/WNL.0000000000002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Grant WB, Campbell A, Itzhaki RF, Savory J. The significance of environmental factors in the etiology of Alzheimer’s disease. JAD. 2002;4(3):179–189. doi: 10.3233/JAD-2002-4308. [DOI] [PubMed] [Google Scholar]
- 151.Farrer LA, Cupples L, Haines JL, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein e genotype and alzheimer disease: a meta-analysis. JAMA. 1997;278(16):1349–1356. doi: 10.1001/jama.1997.03550160069041. [DOI] [PubMed] [Google Scholar]
- 152.Li J, Kanekiyo T, Shinohara M, Zhang Y, LaDu MJ, Xu H, Bu G. Differential regulation of amyloid-beta endocytic trafficking and lysosomal degradation by apolipoprotein E isoforms. J Biol Chem. 2012;287(53):44593–44601. doi: 10.1074/jbc.M112.420224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Bell RD, Winkler EA, Singh I, Sagare AP, Deane R, Wu Z, Holtzman DM, Betsholtz C, Armulik A, Sallstrom J, Berk BC, Zlokovic BV. Apolipoprotein E controls cerebrovascular integrity via cyclophilin A. Nature. 2012;485(7399):512–516. doi: 10.1038/nature11087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tai LM, Ghura S, Koster KP, Liakaite V, Maienschein-Cline M, Kanabar P, Collins N, Ben-Aissa M, Lei AZ, Bahroos N, Green SJ, Hendrickson B, Van Eldik LJ, LaDu MJ. APOE-modulated Abeta-induced neuroinflammation in Alzheimer’s disease: current landscape, novel data, and future perspective. J Neurochem. 2015;133(4):465–488. doi: 10.1111/jnc.13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368(2):117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Malik M, Parikh I, Vasquez JB, Smith C, Tai L, Bu G, LaDu MJ, Fardo DW, Rebeck GW, Estus S. Genetics ignite focus on microglial inflammation in Alzheimer’s disease. Mol Neurodegener. 2015;10:52. doi: 10.1186/s13024-015-0048-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Hooli BV, Parrado AR, Mullin K, Yip WK, Liu T, Roehr JT, Qiao D, Jessen F, Peters O, Becker T, Ramirez A, Lange C, Bertram L, Tanzi RE. The rare TREM2 R47H variant exerts only a modest effect on Alzheimer disease risk. Neurology. 2014;83(15):1353–1358. doi: 10.1212/WNL.0000000000000855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12(9):1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- 159.Shibata M, Yamada S, Kumar SR, Calero M, Bading J, Frangione B, Holtzman DM, Miller CA, Strickland DK, Ghiso J, Zlokovic BV. Clearance of Alzheimer’s amyloid-ss(1–40) peptide from brain by LDL receptor-related protein-1 at the blood-brain barrier. J Clin Invest. 2000;106(12):1489–1499. doi: 10.1172/JCI10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weller RO, Subash M, Preston SD, Mazanti I, Carare RO. Perivascular drainage of amyloid-beta peptides from the brain and its failure in cerebral amyloid angiopathy and Alzheimer’s disease. Brain Pathol. 2008;18(2):253–266. doi: 10.1111/j.1750-3639.2008.00133.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Iliff JJ, Nedergaard M. Is there a cerebral lymphatic system? Stroke. 2013 doi: 10.1161/STROKEAHA.112.678698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Iliff JJ, Wang M, Liao Y, Plogg BA, Peng W, Gundersen GA, Benveniste H, Vates GE, Deane R, Goldman SA, Nagelhus EA, Nedergaard M. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci Transl Med. 2012;4(147):147ra111. doi: 10.1126/scitranslmed.3003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Malm T, Koistinaho M, Muona A, Magga J, Koistinaho J. The role and therapeutic potential of monocytic cells in Alzheimer’s disease. Glia. 2010;58(8):889–900. doi: 10.1002/glia.20973. [DOI] [PubMed] [Google Scholar]
- 164.Hemming ML, Selkoe DJ. Amyloid beta-protein is degraded by cellular angiotensin-converting enzyme (ACE) and elevated by an ACE inhibitor. J Biol Chem. 2005;280(45):37644–37650. doi: 10.1074/jbc.M508460200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mukherjee A, Song E, Kihiko-Ehmann M, Goodman JP, Jr, Pyrek JS, Estus S, Hersh LB. Insulysin hydrolyzes amyloid beta peptides to products that are neither neurotoxic nor deposit on amyloid plaques. J Neurosci. 2000;20(23):8745–8749. doi: 10.1523/JNEUROSCI.20-23-08745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kanemitsu H, Tomiyama T, Mori H. Human neprilysin is capable of degrading amyloid beta peptide not only in the monomeric form but also the pathological oligomeric form. Neurosci Lett. 2003;350(2):113–116. doi: 10.1016/S0304-3940(03)00898-X. [DOI] [PubMed] [Google Scholar]
- 167.Yin KJ, Cirrito JR, Yan P, Hu X, Xiao Q, Pan X, Bateman R, Song H, Hsu FF, Turk J, Xu J, Hsu CY, Mills JC, Holtzman DM, Lee JM. Matrix metalloproteinases expressed by astrocytes mediate extracellular amyloid-beta peptide catabolism. J Neurosci. 2006;26(43):10939–10948. doi: 10.1523/JNEUROSCI.2085-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Iwata N, Takaki Y, Fukami S, Tsubuki S, Saido TC. Region-specific reduction of A beta-degrading endopeptidase, neprilysin, in mouse hippocampus upon aging. J Neurosci Res. 2002;70(3):493–500. doi: 10.1002/jnr.10390. [DOI] [PubMed] [Google Scholar]
- 169.Fukami S, Watanabe K, Iwata N, Haraoka J, Lu B, Gerard NP, Gerard C, Fraser P, Westaway D, St George-Hyslop P, Saido TC. Abeta-degrading endopeptidase, neprilysin, in mouse brain: synaptic and axonal localization inversely correlating with Abeta pathology. Neurosci Res. 2002;43(1):39–56. doi: 10.1016/S0168-0102(02)00015-9. [DOI] [PubMed] [Google Scholar]
- 170.Hickman SE, Allison EK, El Khoury J. Microglial dysfunction and defective beta-amyloid clearance pathways in aging Alzheimer’s disease mice. J Neurosci. 2008;28(33):8354–8360. doi: 10.1523/JNEUROSCI.0616-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fisk L, Nalivaeva NN, Boyle JP, Peers CS, Turner AJ. Effects of hypoxia and oxidative stress on expression of neprilysin in human neuroblastoma cells and rat cortical neurones and astrocytes. Neurochem Res. 2007;32(10):1741–1748. doi: 10.1007/s11064-007-9349-2. [DOI] [PubMed] [Google Scholar]
- 172.El-Amouri SS, Zhu H, Yu J, Marr R, Verma IM, Kindy MS. Neprilysin: an enzyme candidate to slow the progression of Alzheimer’s disease. Am J Pathol. 2008;172(5):1342–1354. doi: 10.2353/ajpath.2008.070620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Parthasarathy R, Chow KM, Derafshi Z, Fautsch MP, Hetling JR, Rodgers DW, Hersh LB, Pepperberg DR. Reduction of amyloid-beta levels in mouse eye tissues by intra-vitreally delivered neprilysin. Exp Eye Res. 2015;138:134–144. doi: 10.1016/j.exer.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kurochkin IV, Goto S (1994) Alzheimer’s beta-amyloid peptide specifically interacts with and is degraded by insulin degrading enzyme. FEBS Lett 345 (1):33–37 [DOI] [PubMed]
- 175.Qiu WQ, Walsh DM, Ye Z, Vekrellis K, Zhang J, Podlisny MB, Rosner MR, Safavi A, Hersh LB, Selkoe DJ. Insulin-degrading enzyme regulates extracellular levels of amyloid beta-protein by degradation. J Biol Chem. 1998;273(49):32730–32738. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- 176.Bulloj A, Leal MC, Xu H, Castano EM, Morelli L. Insulin-degrading enzyme sorting in exosomes: a secretory pathway for a key brain amyloid-beta degrading protease. JAD. 2010;19(1):79–95. doi: 10.3233/JAD-2010-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Vekrellis K, Ye Z, Qiu WQ, Walsh D, Hartley D, Chesneau V, Rosner MR, Selkoe DJ. Neurons regulate extracellular levels of amyloid beta-protein via proteolysis by insulin-degrading enzyme. J Neurosci. 2000;20(5):1657–1665. doi: 10.1523/JNEUROSCI.20-05-01657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Deb S, Zhang JW, Gottschall PE. Activated isoforms of MMP-2 are induced in U87 human glioma cells in response to beta-amyloid peptide. J Neurosci Res. 1999;55(1):44–53. doi: 10.1002/(SICI)1097-4547(19990101)55:1<44::AID-JNR6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 179.Fujioka H, Dairyo Y, Yasunaga K, Emoto K (2012) Neural functions of matrix metalloproteinases: plasticity, neurogenesis, and disease. Biochem Res Int 2012:789083. doi:10.1155/2012/789083 [DOI] [PMC free article] [PubMed]
- 180.Terni B, Ferrer I. Abnormal expression and distribution of MMP2 at initial stages of Alzheimer’s disease-related pathology. JAD. 2015;46(2):461–469. doi: 10.3233/JAD-142460. [DOI] [PubMed] [Google Scholar]
- 181.Li W, Poteet E, Xie L, Liu R, Wen Y, Yang SH. Regulation of matrix metalloproteinase 2 by oligomeric amyloid beta protein. Brain Res. 2011;1387:141–148. doi: 10.1016/j.brainres.2011.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Roher AE, Kasunic TC, Woods AS, Cotter RJ, Ball MJ, Fridman R. Proteolysis of A beta peptide from Alzheimer disease brain by gelatinase A. Biochem Biophys Res Commun. 1994;205(3):1755–1761. doi: 10.1006/bbrc.1994.2872. [DOI] [PubMed] [Google Scholar]
- 183.White AR, Du T, Laughton KM, Volitakis I, Sharples RA, Xilinas ME, Hoke DE, Holsinger RM, Evin G, Cherny RA, Hill AF, Barnham KJ, Li QX, Bush AI, Masters CL. Degradation of the Alzheimer disease amyloid beta-peptide by metal-dependent up-regulation of metalloprotease activity. J Biol Chem. 2006;281(26):17670–17680. doi: 10.1074/jbc.M602487200. [DOI] [PubMed] [Google Scholar]
- 184.Ito S, Kimura K, Haneda M, Ishida Y, Sawada M, Isobe K. Induction of matrix metalloproteinases (MMP3, MMP12 and MMP13) expression in the microglia by amyloid-beta stimulation via the PI3K/Akt pathway. Exp Gerontol. 2007;42(6):532–537. doi: 10.1016/j.exger.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 185.Zhao L, Lin S, Bales KR, Gelfanova V, Koger D, Delong C, Hale J, Liu F, Hunter JM, Paul SM. Macrophage-mediated degradation of beta-amyloid via an apolipoprotein E isoform-dependent mechanism. J Neurosci. 2009;29(11):3603–3612. doi: 10.1523/JNEUROSCI.5302-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yan P, Hu X, Song H, Yin K, Bateman RJ, Cirrito JR, Xiao Q, Hsu FF, Turk JW, Xu J, Hsu CY, Holtzman DM, Lee JM. Matrix metalloproteinase-9 degrades amyloid-beta fibrils in vitro and compact plaques in situ. J Biol Chem. 2006;281(34):24566–24574. doi: 10.1074/jbc.M602440200. [DOI] [PubMed] [Google Scholar]
- 187.Humpel C. Organotypic vibrosections from whole brain adult Alzheimer mice (overexpressing amyloid-precursor-protein with the Swedish-Dutch-Iowa mutations) as a model to study clearance of beta-amyloid plaques. Front Aging Neurosci. 2015;7:47. doi: 10.3389/fnagi.2015.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Backstrom JR, Lim GP, Cullen MJ, Tokes ZA. Matrix metalloproteinase-9 (MMP-9) is synthesized in neurons of the human hippocampus and is capable of degrading the amyloid-beta peptide (1–40) J Neurosci. 1996;16(24):7910–7919. doi: 10.1523/JNEUROSCI.16-24-07910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Gottschall PE, Yu X, Bing B. Increased production of gelatinase B (matrix metalloproteinase-9) and interleukin-6 by activated rat microglia in culture. J Neurosci Res. 1995;42(3):335–342. doi: 10.1002/jnr.490420307. [DOI] [PubMed] [Google Scholar]
- 190.Muir EM, Adcock KH, Morgenstern DA, Clayton R, von Stillfried N, Rhodes K, Ellis C, Fawcett JW, Rogers JH. Matrix metalloproteases and their inhibitors are produced by overlapping populations of activated astrocytes. Brain Res Mol Brain Res. 2002;100(1–2):103–117. doi: 10.1016/S0169-328X(02)00132-8. [DOI] [PubMed] [Google Scholar]
- 191.Koronyo-Hamaoui M, Shah K, Koronyo Y, Bernstein E, Giani JF, Janjulia T, Black KL, Shi PD, Gonzalez-Villalobos RA, Fuchs S, Shen XZ, Bernstein KE. ACE overexpression in myelomonocytic cells: effect on a mouse model of Alzheimer’s disease. Curr Hypertens Rep. 2014;16(7):444. doi: 10.1007/s11906-014-0444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Zou K, Maeda T, Watanabe A, Liu J, Liu S, Oba R, Satoh Y, Komano H, Michikawa M. Abeta42-to-Abeta40- and angiotensin-converting activities in different domains of angiotensin-converting enzyme. J Biol Chem. 2009;284(46):31914–31920. doi: 10.1074/jbc.M109.011437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Russell FD, Skepper JN, Davenport AP. Human endothelial cell storage granules: a novel intracellular site for isoforms of the endothelin-converting enzyme. Circ Res. 1998;83(3):314–321. doi: 10.1161/01.RES.83.3.314. [DOI] [PubMed] [Google Scholar]
- 194.Carty NC, Nash K, Lee D, Mercer M, Gottschall PE, Meyers C, Muzyczka N, Gordon MN, Morgan D. Adeno-associated viral (AAV) serotype 5 vector mediated gene delivery of endothelin-converting enzyme reduces Abeta deposits in APP + PS1 transgenic mice. Mol Ther. 2008;16(9):1580–1586. doi: 10.1038/mt.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Eckman EA, Reed DK, Eckman CB. Degradation of the Alzheimer’s amyloid beta peptide by endothelin-converting enzyme. J Biol Chem. 2001;276(27):24540–24548. doi: 10.1074/jbc.M007579200. [DOI] [PubMed] [Google Scholar]
- 196.Hook G, Yu J, Toneff T, Kindy M, Hook V. Brain pyroglutamate amyloid-beta is produced by cathepsin B and is reduced by the cysteine protease inhibitor E64d, representing a potential Alzheimer’s disease therapeutic. Journal of Alzheimer’s disease : JAD. 2014;41(1):129–149. doi: 10.3233/JAD-131370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hook V, Hook G, Kindy M. Pharmacogenetic features of cathepsin B inhibitors that improve memory deficit and reduce beta-amyloid related to Alzheimer’s disease. Biol Chem. 2010;391(8):861–872. doi: 10.1515/bc.2010.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Hook VY, Kindy M, Reinheckel T, Peters C, Hook G. Genetic cathepsin B deficiency reduces beta-amyloid in transgenic mice expressing human wild-type amyloid precursor protein. Biochem Biophys Res Commun. 2009;386(2):284–288. doi: 10.1016/j.bbrc.2009.05.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Mueller-Steiner S, Zhou Y, Arai H, Roberson ED, Sun B, Chen J, Wang X, Yu G, Esposito L, Mucke L, Gan L. Antiamyloidogenic and neuroprotective functions of cathepsin B: implications for Alzheimer’s disease. Neuron. 2006;51(6):703–714. doi: 10.1016/j.neuron.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 200.Cataldo AM, Nixon RA. Enzymatically active lysosomal proteases are associated with amyloid deposits in Alzheimer brain. Proc Natl Acad Sci USA. 1990;87(10):3861–3865. doi: 10.1073/pnas.87.10.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang C, Sun B, Zhou Y, Grubb A, Gan L. Cathepsin B degrades amyloid-beta in mice expressing wild-type human amyloid precursor protein. J Biol Chem. 2012;287(47):39834–39841. doi: 10.1074/jbc.M112.371641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hamazaki H. Cathepsin D is involved in the clearance of Alzheimer’s beta-amyloid protein. FEBS Lett. 1996;396(2–3):139–142. doi: 10.1016/0014-5793(96)01087-3. [DOI] [PubMed] [Google Scholar]
- 203.Higaki J, Catalano R, Guzzetta AW, Quon D, Nave JF, Tarnus C, D’Orchymont H, Cordell B. Processing of beta-amyloid precursor protein by cathepsin D. J Biol Chem. 1996;271(50):31885–31893. doi: 10.1074/jbc.271.50.31885. [DOI] [PubMed] [Google Scholar]
- 204.Ladror US, Snyder SW, Wang GT, Holzman TF, Krafft GA. Cleavage at the amino and carboxyl termini of Alzheimer’s amyloid-beta by cathepsin D. J Biol Chem. 1994;269(28):18422–18428. [PubMed] [Google Scholar]
- 205.Kehoe PG, Miners S, Love S. Angiotensins in Alzheimer’s disease—friend or foe? Trends Neurosci. 2009;32(12):619–628. doi: 10.1016/j.tins.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 206.Miners JS, Ashby E, Van Helmond Z, Chalmers KA. Palmer LE, Love S, Kehoe PG. Angiotensin-converting enzyme (ACE) levels and activity in Alzheimer’s disease, and relationship of perivascular ACE-1 to cerebral amyloid angiopathy. Neuropathol Appl Neurobiol. 2008;34(2):181–193. doi: 10.1111/j.1365-2990.2007.00885.x. [DOI] [PubMed] [Google Scholar]
- 207.Miners S, Ashby E, Baig S, Harrison R, Tayler H, Speedy E, Prince JA, Love S, Kehoe PG. Angiotensin-converting enzyme levels and activity in Alzheimer’s disease: differences in brain and CSF ACE and association with ACE1 genotypes. Am J Transl Res. 2009;1(2):163–177. [PMC free article] [PubMed] [Google Scholar]
- 208.Wang XB, Cui NH, Yang J, Qiu XP, Gao JJ, Yang N, Zheng F. Angiotensin-converting enzyme insertion/deletion polymorphism is not a major determining factor in the development of sporadic Alzheimer disease: evidence from an updated meta-analysis. PloS One. 2014;9(10):e111406. doi: 10.1371/journal.pone.0111406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zou K, Yamaguchi H, Akatsu H, Sakamoto T, Ko M, Mizoguchi K, Gong JS, Yu W, Yamamoto T, Kosaka K, Yanagisawa K, Michikawa M. Angiotensin-converting enzyme converts amyloid beta-protein 1–42 (Abeta(1–42)) to Abeta(1–40), and its inhibition enhances brain Abeta deposition. J Neurosci. 2007;27(32):8628–8635. doi: 10.1523/JNEUROSCI.1549-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Miller BC, Eckman EA, Sambamurti K, Dobbs N, Chow KM, Eckman CB, Hersh LB, Thiele DL. Amyloid-beta peptide levels in brain are inversely correlated with insulysin activity levels in vivo. Proc Natl Acad Sci USA. 2003;100(10):6221–6226. doi: 10.1073/pnas.1031520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Caccamo A, Oddo S, Sugarman MC, Akbari Y, LaFerla FM. Age- and region-dependent alterations in Abeta-degrading enzymes: implications for Abeta-induced disorders. Neurobiol Aging. 2005;26(5):645–654. doi: 10.1016/j.neurobiolaging.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 212.Miners JS, Baig S, Tayler H, Kehoe PG, Love S. Neprilysin and insulin-degrading enzyme levels are increased in Alzheimer disease in relation to disease severity. J Neuropathol Exp Neurol. 2009;68(8):902–914. doi: 10.1097/NEN.0b013e3181afe475. [DOI] [PubMed] [Google Scholar]
- 213.Miners JS, van Helmond Z, Kehoe PG, Love S. Changes with age in the activities of beta-secretase and the Abeta-degrading enzymes neprilysin, insulin-degrading enzyme and angiotensin-converting enzyme. Brain Pathol. 2010;20(4):794–802. doi: 10.1111/j.1750-3639.2010.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cook DG, Leverenz JB, McMillan PJ, Kulstad JJ, Ericksen S, Roth RA, Schellenberg GD, Jin LW, Kovacina KS, Craft S. Reduced hippocampal insulin-degrading enzyme in late-onset Alzheimer’s disease is associated with the apolipoprotein E-epsilon4 allele. Am J Pathol. 2003;162(1):313–319. doi: 10.1016/S0002-9440(10)63822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fragkouli A, Tsilibary EC, Tzinia AK. Neuroprotective role of MMP-9 overexpression in the brain of Alzheimer’s 5xFAD mice. Neurobiol Dis. 2014;70:179–189. doi: 10.1016/j.nbd.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 216.Chong YH, Sung JH, Shin SA, Chung JH, Suh YH. Effects of the beta-amyloid and carboxyl-terminal fragment of Alzheimer’s amyloid precursor protein on the production of the tumor necrosis factor-alpha and matrix metalloproteinase-9 by human monocytic THP-1. J Biol Chem. 2001;276(26):23511–23517. doi: 10.1074/jbc.M009466200. [DOI] [PubMed] [Google Scholar]
- 217.Lee EJ, Moon PG, Baek MC, Kim HS. Comparison of the effects of matrix metalloproteinase inhibitors on TNF-alpha release from activated microglia and TNF-alpha converting enzyme activity. Biomol Ther (Seoul) 2014;22(5):414–419. doi: 10.4062/biomolther.2014.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Yong VW, Power C, Forsyth P, Edwards DR. Metalloproteinases in biology and pathology of the nervous system. Nat Rev Neurosci. 2001;2(7):502–511. doi: 10.1038/35081571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.de Castro RC, Jr, Burns CL, McAdoo DJ, Romanic AM. Metalloproteinase increases in the injured rat spinal cord. Neuroreport. 2000;11(16):3551–3554. doi: 10.1097/00001756-200011090-00029. [DOI] [PubMed] [Google Scholar]
- 220.Shirotani K, Tsubuki S, Iwata N, Takaki Y, Harigaya W, Maruyama K, Kiryu-Seo S, Kiyama H, Iwata H, Tomita T, Iwatsubo T, Saido TC. Neprilysin degrades both amyloid beta peptides 1–40 and 1–42 most rapidly and efficiently among thiorphan- and phosphoramidon-sensitive endopeptidases. J Biol Chem. 2001;276(24):21895–21901. doi: 10.1074/jbc.M008511200. [DOI] [PubMed] [Google Scholar]
- 221.Iwata N, Tsubuki S, Takaki Y, Watanabe K, Sekiguchi M, Hosoki E, Kawashima-Morishima M, Lee HJ, Hama E, Sekine-Aizawa Y, Saido TC. Identification of the major Abeta1-42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat Med. 2000;6(2):143–150. doi: 10.1038/72237. [DOI] [PubMed] [Google Scholar]
- 222.Russo R, Borghi R, Markesbery W, Tabaton M, Piccini A (2005) Neprylisin decreases uniformly in Alzheimer’s disease and in normal aging. FEBS Lett 579 (27):6027–6030. doi:10.1016/j.febslet.2005.09.054 [DOI] [PubMed]
- 223.Iwata N, Tsubuki S, Takaki Y, Shirotani K, Lu B, Gerard NP, Gerard C, Hama E, Lee HJ, Saido TC. Metabolic regulation of brain Abeta by neprilysin. Science. 2001;292(5521):1550–1552. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- 224.Huttenrauch M, Baches S, Gerth J, Bayer TA, Weggen S, Wirths O. Neprilysin deficiency alters the neuropathological and behavioral phenotype in the 5XFAD mouse model of Alzheimer’s disease. JAD. 2015;44(4):1291–1302. doi: 10.3233/JAD-142463. [DOI] [PubMed] [Google Scholar]
- 225.Mouri A, Zou LB, Iwata N, Saido TC, Wang D, Wang MW, Noda Y, Nabeshima T. Inhibition of neprilysin by thiorphan (i.c.v.) causes an accumulation of amyloid beta and impairment of learning and memory. Behav Brain Res. 2006;168(1):83–91. doi: 10.1016/j.bbr.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 226.Shimizu E, Kawahara K, Kajizono M, Sawada M, Nakayama H. IL-4-induced selective clearance of oligomeric beta-amyloid peptide(1–42) by rat primary type 2 microglia. J Immunol (Baltimore, Md: 1950) 2008;181(9):6503–6513. doi: 10.4049/jimmunol.181.9.6503. [DOI] [PubMed] [Google Scholar]
- 227.Bernstein KE, Gonzalez-Villalobos RA, Giani JF, Shah K, Bernstein E, Janjulia T, Koronyo Y, Shi PD, Koronyo-Hamaoui M, Fuchs S, Shen XZ. Angiotensin-converting enzyme overexpression in myelocytes enhances the immune response. Biol Chem. 2014;395(10):1173–1178. doi: 10.1515/hsz-2013-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, Grutzendler J, De Camilli P, Ferguson SM. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci USA. 2015;112(28):E3699–E3708. doi: 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Tian L, Zhang K, Tian ZY, Wang T, Shang DS, Li B, Liu DX, Fang WG, Wang ZY, Chen YH. Decreased expression of cathepsin D in monocytes is related to the defective degradation of amyloid-beta in Alzheimer’s disease. JAD. 2014;42(2):511–520. doi: 10.3233/JAD-132192. [DOI] [PubMed] [Google Scholar]
- 230.Nixon RA, Yang DS. Autophagy failure in Alzheimer’s disease–locating the primary defect. Neurobiol Dis. 2011;43(1):38–45. doi: 10.1016/j.nbd.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer’s disease. J Neurochem. 2000;75(1):436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 232.Yang AJ, Chandswangbhuvana D, Margol L, Glabe CG. Loss of endosomal/lysosomal membrane impermeability is an early event in amyloid Abeta1-42 pathogenesis. J Neurosci Res. 1998;52(6):691–698. doi: 10.1002/(SICI)1097-4547(19980615)52:6<691::AID-JNR8>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 233.Umeda T, Tomiyama T, Sakama N, Tanaka S, Lambert MP, Klein WL, Mori H. Intraneuronal amyloid beta oligomers cause cell death via endoplasmic reticulum stress, endosomal/lysosomal leakage, and mitochondrial dysfunction in vivo. J Neurosci Res. 2011;89(7):1031–1042. doi: 10.1002/jnr.22640. [DOI] [PubMed] [Google Scholar]
- 234.Ji ZS, Mullendorff K, Cheng IH, Miranda RD, Huang Y, Mahley RW. Reactivity of apolipoprotein E4 and amyloid beta peptide: lysosomal stability and neurodegeneration. J Biol Chem. 2006;281(5):2683–2692. doi: 10.1074/jbc.M506646200. [DOI] [PubMed] [Google Scholar]
- 235.Brunk U, Brun A (1972) The effect of aging on lysosomal permeability in nerve cells of the central nervous system. An enzyme histochemical study in rat. Histochemie 30(4):315–324 [DOI] [PubMed]
- 236.Ji ZS, Miranda RD, Newhouse YM, Weisgraber KH, Huang Y, Mahley RW. Apolipoprotein E4 potentiates amyloid beta peptide-induced lysosomal leakage and apoptosis in neuronal cells. J Biol Chem. 2002;277(24):21821–21828. doi: 10.1074/jbc.M112109200. [DOI] [PubMed] [Google Scholar]
- 237.Echeverria V, Ducatenzeiler A, Dowd E, Janne J, Grant SM, Szyf M, Wandosell F, Avila J, Grimm H, Dunnett SB, Hartmann T, Alhonen L, Cuello AC. Altered mitogen-activated protein kinase signaling, tau hyperphosphorylation and mild spatial learning dysfunction in transgenic rats expressing the beta-amyloid peptide intracellularly in hippocampal and cortical neurons. Neuroscience. 2004;129(3):583–592. doi: 10.1016/j.neuroscience.2004.07.036. [DOI] [PubMed] [Google Scholar]
- 238.Oh S, Hong HS, Hwang E, Sim HJ, Lee W, Shin SJ, Mook-Jung I. Amyloid peptide attenuates the proteasome activity in neuronal cells. Mech Ageing Dev. 2005;126(12):1292–1299. doi: 10.1016/j.mad.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 239.Frackowiak J, Wisniewski HM, Wegiel J, Merz GS, Iqbal K, Wang KC. Ultrastructure of the microglia that phagocytose amyloid and the microglia that produce beta-amyloid fibrils. Acta Neuropathol (Berl) 1992;84(3):225–233. doi: 10.1007/BF00227813. [DOI] [PubMed] [Google Scholar]
- 240.Paresce DM, Chung H, Maxfield FR. Slow degradation of aggregates of the Alzheimer’s disease amyloid beta-protein by microglial cells. J Biol Chem. 1997;272(46):29390–29397. doi: 10.1074/jbc.272.46.29390. [DOI] [PubMed] [Google Scholar]
- 241.Majumdar A, Chung H, Dolios G, Wang R, Asamoah N, Lobel P, Maxfield FR. Degradation of fibrillar forms of Alzheimer’s amyloid beta-peptide by macrophages. Neurobiol Aging. 2008;29(5):707–715. doi: 10.1016/j.neurobiolaging.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Dahms NM, Lobel P, Kornfeld S. Mannose 6-phosphate receptors and lysosomal enzyme targeting. J Biol Chem. 1989;264(21):12115–12118. [PubMed] [Google Scholar]
- 243.Tiribuzi R, Crispoltoni L, Porcellati S, Di Lullo M, Florenzano F, Pirro M, Bagaglia F, Kawarai T, Zampolini M, Orlacchio A, Orlacchio A. miR128 up-regulation correlates with impaired amyloid beta(1–42) degradation in monocytes from patients with sporadic Alzheimer’s disease. Neurobiol Aging. 2014;35(2):345–356. doi: 10.1016/j.neurobiolaging.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 244.Koga H, Cuervo AM. Chaperone-mediated autophagy dysfunction in the pathogenesis of neurodegeneration. Neurobiol Dis. 2011;43(1):29–37. doi: 10.1016/j.nbd.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Zare-Shahabadi A, Masliah E, Johnson GV, Rezaei N. Autophagy in Alzheimer’s disease. Rev Neurosci. 2015;26(4):385–395. doi: 10.1515/revneuro-2014-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Stromhaug PE, Berg TO, Fengsrud M, Seglen PO. Purification and characterization of autophagosomes from rat hepatocytes. Biochem J. 1998;335(2):217–224. doi: 10.1042/bj3350217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Son SM, Jung ES, Shin HJ, Byun J, Mook-Jung I. Abeta-induced formation of autophagosomes is mediated by RAGE-CaMKKbeta-AMPK signaling. Neurobiol Aging. 2012;33(5):1006.e1011-1023. doi: 10.1016/j.neurobiolaging.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 248.Tian Y, Chang JC, Greengard P, Flajolet M. The convergence of endosomal and autophagosomal pathways: implications for APP-CTF degradation. Autophagy. 2014;10(4):694–696. doi: 10.4161/auto.27802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Boland B, Kumar A, Lee S, Platt FM, Wegiel J, Yu WH, Nixon RA. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28(27):6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007 doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 251.Correia SC, Resende R, Moreira PI, Pereira CM (2015) Alzheimer’s disease-related misfolded proteins and dysfunctional organelles on autophagy menu. DNA Cell Biol 34 (4):261–273. doi:10.1089/dna.2014.2757 [DOI] [PubMed]
- 252.Lipinski MM, Zheng B, Lu T, Yan Z, Py BF, Ng A, Xavier RJ, Li C, Yankner BA, Scherzer CR, Yuan J. Genome-wide analysis reveals mechanisms modulating autophagy in normal brain aging and in Alzheimer’s disease. Proc Natl Acad Sci USA. 2010;107(32):14164–14169. doi: 10.1073/pnas.1009485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Nilsson P, Loganathan K, Sekiguchi M, Matsuba Y, Hui K, Tsubuki S, Tanaka M, Iwata N, Saito T, Saido TC. Abeta secretion and plaque formation depend on autophagy. Cell Rep. 2013;5(1):61–69. doi: 10.1016/j.celrep.2013.08.042. [DOI] [PubMed] [Google Scholar]
- 254.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, Small S, Spencer B, Rockenstein E, Levine B, Wyss-Coray T. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. J Clin Invest. 2008;118(6):2190–2199. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Jung CH, Ro SH, Cao J, Otto NM, Kim DH (2010) mTOR regulation of autophagy. FEBS Lett 584 (7):1287–1295. doi:10.1016/j.febslet.2010.01.017 [DOI] [PMC free article] [PubMed]
- 256.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432(7020):1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 257.Scherz-Shouval R, Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol. 2007;17(9):422–427. doi: 10.1016/j.tcb.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 258.Li X, Alafuzoff I, Soininen H, Winblad B, Pei JJ. Levels of mTOR and its downstream targets 4E-BP1, eEF2, and eEF2 kinase in relationships with tau in Alzheimer’s disease brain. FEBS J. 2005;272(16):4211–4220. doi: 10.1111/j.1742-4658.2005.04833.x. [DOI] [PubMed] [Google Scholar]
- 259.Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, Richardson A, Strong R, Galvan V. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PloS one. 2010;5(4):e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Keller JN, Gee J, Ding Q. The proteasome in brain aging. Ageing Res Rev. 2002;1(2):279–293. doi: 10.1016/S1568-1637(01)00006-X. [DOI] [PubMed] [Google Scholar]
- 261.Lopez Salon M, Morelli L, Castano EM, Soto EF, Pasquini JM. Defective ubiquitination of cerebral proteins in Alzheimer’s disease. J Neurosci Res. 2000;62(2):302–310. doi: 10.1002/1097-4547(20001015)62:2<302::AID-JNR15>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 262.Hiltunen M, Lu A, Thomas AV, Romano DM, Kim M, Jones PB, Xie Z, Kounnas MZ, Wagner SL, Berezovska O, Hyman BT, Tesco G, Bertram L, Tanzi RE. Ubiquilin 1 modulates amyloid precursor protein trafficking and Abeta secretion. J Biol Chem. 2006;281(43):32240–32253. doi: 10.1074/jbc.M603106200. [DOI] [PubMed] [Google Scholar]
- 263.Tseng BP, Green KN, Chan JL, Blurton-Jones M, LaFerla FM. Abeta inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol Aging. 2008;29(11):1607–1618. doi: 10.1016/j.neurobiolaging.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tarasoff-Conway JM, Carare RO, Osorio RS, Glodzik L, Butler T, Fieremans E, Axel L, Rusinek H, Nicholson C, Zlokovic BV, Frangione B, Blennow K, Menard J, Zetterberg H, Wisniewski T, de Leon MJ. Clearance systems in the brain-implications for Alzheimer disease. Nat Rev Neurol. 2015;11(8):457–470. doi: 10.1038/nrneurol.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Deane R, Wu Z, Sagare A, Davis J, Du Yan S, Hamm K, Xu F, Parisi M, LaRue B, Hu HW, Spijkers P, Guo H, Song X, Lenting PJ, Van Nostrand WE, Zlokovic BV. LRP/amyloid beta-peptide interaction mediates differential brain efflux of Abeta isoforms. Neuron. 2004;43(3):333–344. doi: 10.1016/j.neuron.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 266.Elali A, Rivest S. The role of ABCB1 and ABCA1 in beta-amyloid clearance at the neurovascular unit in Alzheimer’s disease. Front Physiol. 2013;4:45. doi: 10.3389/fphys.2013.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Jaeger LB, Dohgu S, Hwang MC, Farr SA, Murphy MP, Fleegal-DeMotta MA, Lynch JL, Robinson SM, Niehoff ML, Johnson SN, Kumar VB, Banks WA. Testing the neurovascular hypothesis of Alzheimer’s disease: LRP-1 antisense reduces blood-brain barrier clearance, increases brain levels of amyloid-beta protein, and impairs cognition. JAD. 2009;17(3):553–570. doi: 10.3233/JAD-2009-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Silverberg GD, Messier AA, Miller MC, Machan JT, Majmudar SS, Stopa EG, Donahue JE, Johanson CE. Amyloid efflux transporter expression at the blood-brain barrier declines in normal aging. J Neuropathol Exp Neurol. 2010;69(10):1034–1043. doi: 10.1097/NEN.0b013e3181f46e25. [DOI] [PubMed] [Google Scholar]
- 269.Koistinaho M, Lin S, Wu X, Esterman M, Koger D, Hanson J, Higgs R, Liu F, Malkani S, Bales KR, Paul SM. Apolipoprotein E promotes astrocyte colocalization and degradation of deposited amyloid-beta peptides. Nat Med. 2004;10(7):719–726. doi: 10.1038/nm1058. [DOI] [PubMed] [Google Scholar]
- 270.Laporte V, Lombard Y, Levy-Benezra R, Tranchant C, Poindron P, Warter JM. Uptake of Abeta 1-40- and Abeta 1-42-coated yeast by microglial cells: a role for. LRP J Leukocyte Biol. 2004;76(2):451–461. doi: 10.1189/jlb.1203620. [DOI] [PubMed] [Google Scholar]
- 271.Kanekiyo T, Cirrito JR, Liu CC, Shinohara M, Li J, Schuler DR, Shinohara M, Holtzman DM, Bu G. Neuronal clearance of amyloid-beta by endocytic receptor LRP1. J Neurosci. 2013;33(49):19276–19283. doi: 10.1523/JNEUROSCI.3487-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Baig S, Joseph SA, Tayler H, Abraham R, Owen MJ, Williams J, Kehoe PG, Love S. Distribution and expression of picalm in Alzheimer disease. J Neuropathol Exp Neurol. 2010;69(10):1071–1077. doi: 10.1097/NEN.0b013e3181f52e01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Dickson DW, Graff-Radford NR, Petersen RC, Morgan K, Younkin SG. Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch Neurol. 2010;67(8):961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Zhao Z, Sagare AP, Ma Q, Halliday MR, Kong P, Kisler K, Winkler EA, Ramanathan A, Kanekiyo T, Bu G, Owens NC, Rege SV, Si G, Ahuja A, Zhu D, Miller CA, Schneider JA, Maeda M, Maeda T, Sugawara T, Ichida JK, Zlokovic BV. Central role for PICALM in amyloid-beta blood-brain barrier transcytosis and clearance. Nat Neurosci. 2015;18(7):978–987. doi: 10.1038/nn.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240(4852):622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 276.Beisiegel U, Weber W, Ihrke G, Herz J, Stanley KK. The LDL-receptor-related protein, LRP, is an apolipoprotein E-binding protein. Nature. 1989;341(6238):162–164. doi: 10.1038/341162a0. [DOI] [PubMed] [Google Scholar]
- 277.Kim DH, Iijima H, Goto K, Sakai J, Ishii H, Kim HJ, Suzuki H, Kondo H, Saeki S, Yamamoto T. Human apolipoprotein E receptor 2. A novel lipoprotein receptor of the low density lipoprotein receptor family predominantly expressed in brain. J Biol Chem. 1996;271(14):8373–8380. doi: 10.1074/jbc.271.14.8373. [DOI] [PubMed] [Google Scholar]
- 278.Kanekiyo T, Xu H, Bu G. ApoE and Abeta in Alzheimer’s disease: accidental encounters or partners? Neuron. 2014;81(4):740–754. doi: 10.1016/j.neuron.2014.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Raber J, Huang Y, Ashford JW. ApoE genotype accounts for the vast majority of AD risk and AD pathology. Neurobiol Aging. 2004;25(5):641–650. doi: 10.1016/j.neurobiolaging.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 280.Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, Mackey B, Olney J, McKeel D, Wozniak D, Paul SM. Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci USA. 2000;97(6):2892–2897. doi: 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, Roses AD. Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proc Natl Acad Sci USA. 1993;90(20):9649–9653. doi: 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci USA. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Shinohara M, Petersen RC, Dickson DW, Bu G. Brain regional correlation of amyloid-beta with synapses and apolipoprotein E in non-demented individuals: potential mechanisms underlying regional vulnerability to amyloid-beta accumulation. Acta Neuropathol (Berl) 2013;125(4):535–547. doi: 10.1007/s00401-013-1086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Mulder SD, Nielsen HM, Blankenstein MA, Eikelenboom P, Veerhuis R. Apolipoproteins E and J interfere with amyloid-beta uptake by primary human astrocytes and microglia in vitro. Glia. 2014;62(4):493–503. doi: 10.1002/glia.22619. [DOI] [PubMed] [Google Scholar]
- 285.Morris JC, Roe CM, Xiong C, Fagan AM, Goate AM, Holtzman DM, Mintun MA. APOE predicts amyloid-beta but not tau Alzheimer pathology in cognitively normal aging. Ann Neurol. 2010;67(1):122–131. doi: 10.1002/ana.21843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.LaDu MJ, Falduto MT, Manelli AM, Reardon CA, Getz GS, Frail DE. Isoform-specific binding of apolipoprotein E to beta-amyloid. J Biol Chem. 1994;269(38):23403–23406. [PubMed] [Google Scholar]
- 287.Wildsmith KR, Holley M, Savage JC, Skerrett R, Landreth GE (2013) Evidence for impaired amyloid beta clearance in Alzheimer’s disease. Alzheimer’s Res Ther 5(4):33. doi:10.1186/alzrt187 [DOI] [PMC free article] [PubMed]
- 288.Donahue JE, Johanson CE. Apolipoprotein E, amyloid-beta, and blood-brain barrier permeability in Alzheimer disease. J Neuropathol Exp Neurol. 2008;67(4):261–270. doi: 10.1097/NEN.0b013e31816a0dc8. [DOI] [PubMed] [Google Scholar]
- 289.Nishitsuji K, Hosono T, Nakamura T, Bu G, Michikawa M. Apolipoprotein E regulates the integrity of tight junctions in an isoform-dependent manner in an in vitro blood-brain barrier model. J Biol Chem. 2011;286(20):17536–17542. doi: 10.1074/jbc.M111.225532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Rodriguez GA, Tai LM, LaDu MJ, Rebeck GW. Human APOE4 increases microglia reactivity at Abeta plaques in a mouse model of Abeta deposition. J Neuroinflamm. 2014;11:111. doi: 10.1186/1742-2094-11-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Xie L, Kang H, Xu Q, Chen MJ, Liao Y, Thiyagarajan M, O’Donnell J, Christensen DJ, Nicholson C, Iliff JJ, Takano T, Deane R, Nedergaard M. Sleep drives metabolite clearance from the adult brain. Science. 2013;342(6156):373–377. doi: 10.1126/science.1241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Louveau A, Smirnov I, Keyes TJ, Eccles JD, Rouhani SJ, Peske JD, Derecki NC, Castle D, Mandell JW, Lee KS, Harris TH, Kipnis J. Structural and functional features of central nervous system lymphatic vessels. Nature. 2015;523(7560):337–341. doi: 10.1038/nature14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Aspelund A, Antila S, Proulx ST, Karlsen TV, Karaman S, Detmar M, Wiig H, Alitalo K. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J Exp Med. 2015;212(7):991–999. doi: 10.1084/jem.20142290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Kress BT, Iliff JJ, Xia M, Wang M, Wei HS, Zeppenfeld D, Xie L, Kang H, Xu Q, Liew JA, Plog BA, Ding F, Deane R, Nedergaard M. Impairment of paravascular clearance pathways in the aging brain. Ann Neurol. 2014;76(6):845–861. doi: 10.1002/ana.24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Malm TM, Koistinaho M, Parepalo M, Vatanen T, Ooka A, Karlsson S, Koistinaho J. Bone-marrow-derived cells contribute to the recruitment of microglial cells in response to beta-amyloid deposition in APP/PS1 double transgenic Alzheimer mice. Neurobiol Dis. 2005;18(1):134–142. doi: 10.1016/j.nbd.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 296.Simard AR, Soulet D, Gowing G, Julien JP, Rivest S. Bone marrow-derived microglia play a critical role in restricting senile plaque formation in Alzheimer’s disease. Neuron. 2006;49(4):489–502. doi: 10.1016/j.neuron.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 297.Butovsky O, Kunis G, Koronyo-Hamaoui M, Schwartz M. Selective ablation of bone marrow-derived dendritic cells increases amyloid plaques in a mouse Alzheimer’s disease model. Eur J Neurosci. 2007;26(2):413–416. doi: 10.1111/j.1460-9568.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- 298.El Khoury J, Toft M, Hickman SE, Means TK, Terada K, Geula C, Luster AD. Ccr2 deficiency impairs microglial accumulation and accelerates progression of Alzheimer-like disease. Nat Med. 2007;13(4):432–438. doi: 10.1038/nm1555. [DOI] [PubMed] [Google Scholar]
- 299.Paresce DM, Ghosh RN, Maxfield FR. Microglial cells internalize aggregates of the Alzheimer’s disease amyloid beta-protein via a scavenger receptor. Neuron. 1996;17(3):553–565. doi: 10.1016/S0896-6273(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 300.Chung H, Brazil MI, Soe TT, Maxfield FR. Uptake, degradation, and release of fibrillar and soluble forms of Alzheimer’s amyloid beta-peptide by microglial cells. J Biol Chem. 1999;274(45):32301–32308. doi: 10.1074/jbc.274.45.32301. [DOI] [PubMed] [Google Scholar]
- 301.Priller J, Flugel A, Wehner T, Boentert M, Haas CA, Prinz M, Fernandez-Klett F, Prass K, Bechmann I, de Boer BA, Frotscher M, Kreutzberg GW, Persons DA, Dirnagl U. Targeting gene-modified hematopoietic cells to the central nervous system: use of green fluorescent protein uncovers microglial engraftment. Nat Med. 2001;7(12):1356–1361. doi: 10.1038/nm1201-1356. [DOI] [PubMed] [Google Scholar]
- 302.Sedgwick JD, Schwender S, Imrich H, Dorries R, Butcher GW, ter Meulen V. Isolation and direct characterization of resident microglial cells from the normal and inflamed central nervous system. Proc Natl Acad Sci USA. 1991;88(16):7438–7442. doi: 10.1073/pnas.88.16.7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci. 2011;14(10):1227–1235. doi: 10.1038/nn.2923. [DOI] [PubMed] [Google Scholar]
- 304.Ford AL, Goodsall AL, Hickey WF, Sedgwick JD. Normal adult ramified microglia separated from other central nervous system macrophages by flow cytometric sorting. Phenotypic differences defined and direct ex vivo antigen presentation to myelin basic protein-reactive CD4 + T cells compared. J Immunol (Baltimore, Md: 1950) 1995;154(9):4309–4321. [PubMed] [Google Scholar]
- 305.Juedes AE, Ruddle NH. Resident and infiltrating central nervous system APCs regulate the emergence and resolution of experimental autoimmune encephalomyelitis. J Immunol (Baltimore, Md: 1950) 2001;166(8):5168–5175. doi: 10.4049/jimmunol.166.8.5168. [DOI] [PubMed] [Google Scholar]
- 306.Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, Koeglsperger T, Dake B, Wu PM, Doykan CE, Fanek Z, Liu L, Chen Z, Rothstein JD, Ransohoff RM, Gygi SP, Antel JP, Weiner HL. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci. 2014;17(1):131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci. 2014;15(5):300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 308.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327(5966):656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333(6048):1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 310.Chung WS, Welsh CA, Barres BA, Stevens B. Do glia drive synaptic and cognitive impairment in disease? Nat Neurosci. 2015;18(11):1539–1545. doi: 10.1038/nn.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Naert G, Rivest S. A deficiency in CCR2 + monocytes: the hidden side of Alzheimer’s disease. J Mol Cell Biol. 2013;5(5):284–293. doi: 10.1093/jmcb/mjt028. [DOI] [PubMed] [Google Scholar]
- 312.Yamamoto M, Kiyota T, Walsh SM, Liu J, Kipnis J, Ikezu T. Cytokine-mediated inhibition of fibrillar amyloid-beta peptide degradation by human mononuclear phagocytes. J Immunol (Baltimore, Md: 1950) 2008;181(6):3877–3886. doi: 10.4049/jimmunol.181.6.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Hu N, Tan MS, Sun L, Jiang T, Wang YL, Tan L, Zhang W, Yu JT, Tan L. Decreased expression of CD33 in peripheral mononuclear cells of Alzheimer’s disease patients. Neurosci Lett. 2014;563:51–54. doi: 10.1016/j.neulet.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 314.Zhang R, Miller RG, Madison C, Jin X, Honrada R, Harris W, Katz J, Forshew DA, McGrath MS. Systemic immune system alterations in early stages of Alzheimer’s disease. J Neuroimmunol. 2013;256(1–2):38–42. doi: 10.1016/j.jneuroim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Fiala M, Liu PT, Espinosa-Jeffrey A, Rosenthal MJ, Bernard G, Ringman JM, Sayre J, Zhang L, Zaghi J, Dejbakhsh S, Chiang B, Hui J, Mahanian M, Baghaee A, Hong P, Cashman J. Innate immunity and transcription of MGAT-III and Toll-like receptors in Alzheimer’s disease patients are improved by bisdemethoxycurcumin. Proc Natl Acad Sci USA. 2007;104(31):12849–12854. doi: 10.1073/pnas.0701267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Plagg B, Ehrlich D, Kniewallner KM, Marksteiner J, Humpel C (2015) Increased acetylation of histone H4 at lysine 12 (H4K12) in monocytes of transgenic alzheimer’s mice and in human patients. Curr Alzheimer Res 12(8):752–760 [DOI] [PMC free article] [PubMed]
- 317.Lunnon K, Ibrahim Z, Proitsi P, Lourdusamy A, Newhouse S, Sattlecker M, Furney S, Saleem M, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, Coppola G, Geschwind D, Simmons A, Lovestone S, Dobson R, Hodges A. Mitochondrial dysfunction and immune activation are detectable in early Alzheimer’s disease blood. JAD. 2012;30(3):685–710. doi: 10.3233/JAD-2012-111592. [DOI] [PubMed] [Google Scholar]
- 318.Stalder AK, Ermini F, Bondolfi L, Krenger W, Burbach GJ, Deller T, Coomaraswamy J, Staufenbiel M, Landmann R, Jucker M. Invasion of hematopoietic cells into the brain of amyloid precursor protein transgenic mice. J Neurosci. 2005;25(48):11125–11132. doi: 10.1523/JNEUROSCI.2545-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Keene CD, Chang RC, Lopez-Yglesias AH, Shalloway BR, Sokal I, Li X, Reed PJ, Keene LM, Montine KS, Breyer RM, Rockhill JK, Montine TJ. Suppressed accumulation of cerebral amyloid {beta} peptides in aged transgenic Alzheimer’s disease mice by transplantation with wild-type or prostaglandin E2 receptor subtype 2-null bone marrow. Am J Pathol. 2010;177(1):346–354. doi: 10.2353/ajpath.2010.090840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Naert G, Rivest S. Age-related changes in synaptic markers and monocyte subsets link the cognitive decline of APP(Swe)/PS1 mice. Front Cell Neurosci. 2012;6:51. doi: 10.3389/fncel.2012.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Boissonneault V, Filali M, Lessard M, Relton J, Wong G, Rivest S. Powerful beneficial effects of macrophage colony-stimulating factor on beta-amyloid deposition and cognitive impairment in Alzheimer’s disease. Brain J Neurol. 2009 doi: 10.1093/brain/awn331. [DOI] [PubMed] [Google Scholar]
- 322.Town T, Laouar Y, Pittenger C, Mori T, Szekely CA, Tan J, Duman RS, Flavell RA. Blocking TGF-beta-Smad2/3 innate immune signaling mitigates Alzheimer-like pathology. Nat Med. 2008;14(6):681–687. doi: 10.1038/nm1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Jiang T, Tan L, Zhu XC, Zhang QQ, Cao L, Tan MS, Gu LZ, Wang HF, Ding ZZ, Zhang YD, Yu JT. Upregulation of TREM2 ameliorates neuropathology and rescues spatial cognitive impairment in a transgenic mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2014;39(13):2949–2962. doi: 10.1038/npp.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Jay TR, Miller CM, Cheng PJ, Graham LC, Bemiller S, Broihier ML, Xu G, Margevicius D, Karlo JC, Sousa GL, Cotleur AC, Butovsky O, Bekris L, Staugaitis SM, Leverenz JB, Pimplikar SW, Landreth GE, Howell GR, Ransohoff RM, Lamb BT. TREM2 deficiency eliminates TREM2 + inflammatory macrophages and ameliorates pathology in Alzheimer’s disease mouse models. J Exp Med. 2015;212(3):287–295. doi: 10.1084/jem.20142322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Xiang X, Werner G, Bohrmann B, Liesz A, Mazaheri F, Capell A, Feederle R, Knuesel I, Kleinberger G, Haass C. TREM2 deficiency reduces the efficacy of immunotherapeutic amyloid clearance. EMBO Mol Med. 2016 doi: 10.15252/emmm.201606370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Frenkel D, Wilkinson K, Zhao L, Hickman SE, Means TK, Puckett L, Farfara D, Kingery ND, Weiner HL, El Khoury J. Scara1 deficiency impairs clearance of soluble amyloid-beta by mononuclear phagocytes and accelerates Alzheimer’s-like disease progression. Nature Commun. 2013;4:2030. doi: 10.1038/ncomms3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Yamamoto M, Kiyota T, Walsh SM, Ikezu T. Kinetic analysis of aggregated amyloid-beta peptide clearance in adult bone-marrow-derived macrophages from APP and CCL2 transgenic mice. J Neuroimmune Pharmacol. 2007;2(2):213–221. doi: 10.1007/s11481-006-9049-8. [DOI] [PubMed] [Google Scholar]
- 328.Hawkes CA, McLaurin J. Selective targeting of perivascular macrophages for clearance of beta-amyloid in cerebral amyloid angiopathy. Proc Natl Acad Sci USA. 2009;106(4):1261–1266. doi: 10.1073/pnas.0805453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Michaud JP, Bellavance MA, Prefontaine P, Rivest S. Real-time in vivo imaging reveals the ability of monocytes to clear vascular amyloid beta. Cell Rep. 2013;5(3):646–653. doi: 10.1016/j.celrep.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 330.Mildner A, Schlevogt B, Kierdorf K, Bottcher C, Erny D, Kummer MP, Quinn M, Bruck W, Bechmann I, Heneka MT, Priller J, Prinz M. Distinct and non-redundant roles of microglia and myeloid subsets in mouse models of Alzheimer’s disease. J Neurosci. 2011;31(31):11159–11171. doi: 10.1523/JNEUROSCI.6209-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Naert G, Rivest S. CC chemokine receptor 2 deficiency aggravates cognitive impairments and amyloid pathology in a transgenic mouse model of Alzheimer’s disease. J Neurosci. 2011;31(16):6208–6220. doi: 10.1523/JNEUROSCI.0299-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Yang CN, Shiao YJ, Shie FS, Guo BS, Chen PH, Cho CY, Chen YJ, Huang FL, Tsay HJ. Mechanism mediating oligomeric Abeta clearance by naive primary microglia. Neurobiol Dis. 2011;42(3):221–230. doi: 10.1016/j.nbd.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 333.Mandrekar S, Jiang Q, Lee CY, Koenigsknecht-Talboo J, Holtzman DM, Landreth GE. Microglia mediate the clearance of soluble Abeta through fluid phase macropinocytosis. J Neurosci. 2009;29(13):4252–4262. doi: 10.1523/JNEUROSCI.5572-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Koenigsknecht J, Landreth G. Microglial phagocytosis of fibrillar beta-amyloid through a beta1 integrin-dependent mechanism. J Neurosci. 2004;24(44):9838–9846. doi: 10.1523/JNEUROSCI.2557-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Grathwohl SA, Kalin RE, Bolmont T, Prokop S, Winkelmann G, Kaeser SA, Odenthal J, Radde R, Eldh T, Gandy S, Aguzzi A, Staufenbiel M, Mathews PM, Wolburg H, Heppner FL, Jucker M. Formation and maintenance of Alzheimer’s disease beta-amyloid plaques in the absence of microglia. Nat Neurosci. 2009;12(11):1361–1363. doi: 10.1038/nn.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Spangenberg EE, Green KN. Inflammation in Alzheimer’s disease: Lessons learned from microglia-depletion models. Brain Behav Immun. 2016 doi: 10.1016/j.bbi.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Krabbe G, Halle A, Matyash V, Rinnenthal JL, Eom GD, Bernhardt U, Miller KR, Prokop S, Kettenmann H, Heppner FL. Functional impairment of microglia coincides with Beta-amyloid deposition in mice with Alzheimer-like pathology. PloS One. 2013;8(4):e60921. doi: 10.1371/journal.pone.0060921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Bolmont T, Haiss F, Eicke D, Radde R, Mathis CA, Klunk WE, Kohsaka S, Jucker M, Calhoun ME. Dynamics of the microglial/amyloid interaction indicate a role in plaque maintenance. J Neurosci. 2008;28(16):4283–4292. doi: 10.1523/JNEUROSCI.4814-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Hellwig S, Masuch A, Nestel S, Katzmarski N, Meyer-Luehmann M, Biber K. Forebrain microglia from wild-type but not adult 5xFAD mice prevent amyloid-beta plaque formation in organotypic hippocampal slice cultures. Sci Rep. 2015;5:14624. doi: 10.1038/srep14624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Wyss-Coray T, Rogers J. Inflammation in Alzheimer disease-a brief review of the basic science and clinical literature. Cold Spring Harb Perspect Med. 2012;2(1):a006346. doi: 10.1101/cshperspect.a006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Akiyama H, Barger S, Barnum S, Bradt B, Bauer J, Cole GM, Cooper NR, Eikelenboom P, Emmerling M, Fiebich BL, Finch CE, Frautschy S, Griffin WS, Hampel H, Hull M, Landreth G, Lue L, Mrak R, Mackenzie IR, McGeer PL, O’Banion MK, Pachter J, Pasinetti G, Plata-Salaman C, Rogers J, Rydel R, Shen Y, Streit W, Strohmeyer R, Tooyoma I, Van Muiswinkel FL, Veerhuis R, Walker D, Webster S, Wegrzyniak B, Wenk G, Wyss-Coray T. Inflammation and Alzheimer’s disease. Neurobiol Aging. 2000;21(3):383–421. doi: 10.1016/S0197-4580(00)00124-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Xia MQ, Qin SX, Wu LJ, Mackay CR, Hyman BT. Immunohistochemical study of the beta-chemokine receptors CCR3 and CCR5 and their ligands in normal and Alzheimer’s disease brains. Am J Pathol. 1998;153(1):31–37. doi: 10.1016/S0002-9440(10)65542-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Takeuchi H, Jin S, Wang J, Zhang G, Kawanokuchi J, Kuno R, Sonobe Y, Mizuno T, Suzumura A. Tumor necrosis factor-alpha induces neurotoxicity via glutamate release from hemichannels of activated microglia in an autocrine manner. J Biol Chem. 2006;281(30):21362–21368. doi: 10.1074/jbc.M600504200. [DOI] [PubMed] [Google Scholar]
- 344.Wyss-Coray T, Mucke L. Inflammation in neurodegenerative disease–a double-edged sword. Neuron. 2002;35(3):419–432. doi: 10.1016/S0896-6273(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 345.Lopez-Gonzalez I, Schluter A, Aso E, Garcia-Esparcia P, Ansoleaga B, F LL, Carmona M, Moreno J, Fuso A, Portero-Otin M, Pamplona R, Pujol A, Ferrer I. Neuroinflammatory signals in Alzheimer disease and APP/PS1 transgenic mice: correlations with plaques, tangles, and oligomeric species. J Neuropathol Exp Neurol. 2015;74(4):319–344. doi: 10.1097/NEN.0000000000000176. [DOI] [PubMed] [Google Scholar]
- 346.Rice RA, Spangenberg EE, Yamate-Morgan H, Lee RJ, Arora RP, Hernandez MX, Tenner AJ, West BL, Green KN. Elimination of microglia improves functional outcomes following extensive neuronal loss in the hippocampus. J Neurosci. 2015;35(27):9977–9989. doi: 10.1523/JNEUROSCI.0336-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Spangenberg EE, Lee RJ, Najafi AR, Rice RA, Elmore MR, Blurton-Jones M, West BL, Green KN. Eliminating microglia in Alzheimer’s mice prevents neuronal loss without modulating amyloid-beta pathology. Brain J Neurol. 2016 doi: 10.1093/brain/aww016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Butovsky O, Koronyo-Hamaoui M, Kunis G, Ophir E, Landa G, Cohen H, Schwartz M. Glatiramer acetate fights against Alzheimer’s disease by inducing dendritic-like microglia expressing insulin-like growth factor 1. Proc Natl Acad Sci USA. 2006;103(31):11784–11789. doi: 10.1073/pnas.0604681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Condello C, Yuan P, Schain A, Grutzendler J. Microglia constitute a barrier that prevents neurotoxic protofibrillar Abeta42 hotspots around plaques. Nature Commun. 2015;6:6176. doi: 10.1038/ncomms7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Iaccarino HF, Singer AC, Martorell AJ, Rudenko A, Gao F, Gillingham TZ, Mathys H, Seo J, Kritskiy O, Abdurrob F, Adaikkan C, Canter RG, Rueda R, Brown EN, Boyden ES, Tsai LH. Gamma frequency entrainment attenuates amyloid load and modifies microglia. Nature. 2016;540(7632):230–235. doi: 10.1038/nature20587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Michell-Robinson MA, Touil H, Healy LM, Owen DR, Durafourt BA, Bar-Or A, Antel JP, Moore CS. Roles of microglia in brain development, tissue maintenance and repair. Brain J Neurol. 2015 doi: 10.1093/brain/awv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Mildner A, Schmidt H, Nitsche M, Merkler D, Hanisch UK, Mack M, Heikenwalder M, Bruck W, Priller J, Prinz M. Microglia in the adult brain arise from Ly-6ChiCCR2 + monocytes only under defined host conditions. Nat Neurosci. 2007;10(12):1544–1553. doi: 10.1038/nn2015. [DOI] [PubMed] [Google Scholar]
- 353.Prokop S, Miller KR, Drost N, Handrick S, Mathur V, Luo J, Wegner A, Wyss-Coray T, Heppner FL. Impact of peripheral myeloid cells on amyloid-beta pathology in Alzheimer’s disease-like mice. J Exp Med. 2015;212(11):1811–1818. doi: 10.1084/jem.20150479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Varvel NH, Grathwohl SA, Degenhardt K, Resch C, Bosch A, Jucker M, Neher JJ. Replacement of brain-resident myeloid cells does not alter cerebral amyloid-beta deposition in mouse models of Alzheimer’s disease. J Exp Med. 2015;212(11):1803–1809. doi: 10.1084/jem.20150478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ajami B, Bennett JL, Krieger C, Tetzlaff W, Rossi FM. Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci. 2007;10(12):1538–1543. doi: 10.1038/nn2014. [DOI] [PubMed] [Google Scholar]
- 356.Zaghi J, Goldenson B, Inayathullah M, Lossinsky AS, Masoumi A, Avagyan H, Mahanian M, Bernas M, Weinand M, Rosenthal MJ, Espinosa-Jeffrey A, de Vellis J, Teplow DB, Fiala M. Alzheimer disease macrophages shuttle amyloid-beta from neurons to vessels, contributing to amyloid angiopathy. Acta Neuropathol (Berl) 2009;117(2):111–124. doi: 10.1007/s00401-008-0481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Ard MD, Cole GM, Wei J, Mehrle AP, Fratkin JD. Scavenging of Alzheimer’s amyloid beta-protein by microglia in culture. J Neurosci Res. 1996;43(2):190–202. doi: 10.1002/(SICI)1097-4547(19960115)43:2<190::AID-JNR7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 358.Frautschy SA, Cole GM, Baird A. Phagocytosis and deposition of vascular beta-amyloid in rat brains injected with Alzheimer beta-amyloid. Am J Pathol. 1992;140(6):1389–1399. [PMC free article] [PubMed] [Google Scholar]
- 359.Weldon DT, Rogers SD, Ghilardi JR, Finke MP, Cleary JP, O’Hare E, Esler WP, Maggio JE, Mantyh PW. Fibrillar beta-amyloid induces microglial phagocytosis, expression of inducible nitric oxide synthase, and loss of a select population of neurons in the rat CNS in vivo. J Neurosci. 1998;18(6):2161–2173. doi: 10.1523/JNEUROSCI.18-06-02161.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Akiyama H, Schwab C, Kondo H, Mori H, Kametani F, Ikeda K, McGeer PL. Granules in glial cells of patients with Alzheimer’s disease are immunopositive for C-terminal sequences of beta-amyloid protein. Neurosci Lett. 1996;206(2–3):169–172. doi: 10.1016/S0304-3940(96)12474-5. [DOI] [PubMed] [Google Scholar]
- 361.Wilkinson K, Boyd JD, Glicksman M, Moore KJ, El Khoury J. A high content drug screen identifies ursolic acid as an inhibitor of amyloid beta protein interactions with its receptor CD36. J Biol Chem. 2011;286(40):34914–34922. doi: 10.1074/jbc.M111.232116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Udan ML, Ajit D, Crouse NR, Nichols MR. Toll-like receptors 2 and 4 mediate Abeta(1–42) activation of the innate immune response in a human monocytic cell line. J Neurochem. 2008;104(2):524–533. doi: 10.1111/j.1471-4159.2007.05001.x. [DOI] [PubMed] [Google Scholar]
- 363.Reed-Geaghan EG, Savage JC, Hise AG, Landreth GE. CD14 and toll-like receptors 2 and 4 are required for fibrillar A{beta}-stimulated microglial activation. J Neurosci. 2009;29(38):11982–11992. doi: 10.1523/JNEUROSCI.3158-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Hughes DA, Fraser IP, Gordon S. Murine macrophage scavenger receptor: in vivo expression and function as receptor for macrophage adhesion in lymphoid and non-lymphoid organs. Eur J Immunol. 1995;25(2):466–473. doi: 10.1002/eji.1830250224. [DOI] [PubMed] [Google Scholar]
- 365.El Khoury J, Hickman SE, Thomas CA, Cao L, Silverstein SC, Loike JD. Scavenger receptor-mediated adhesion of microglia to beta-amyloid fibrils. Nature. 1996;382(6593):716–719. doi: 10.1038/382716a0. [DOI] [PubMed] [Google Scholar]
- 366.Chung H, Brazil MI, Irizarry MC, Hyman BT, Maxfield FR. Uptake of fibrillar beta-amyloid by microglia isolated from MSR-A (type I and type II) knockout mice. Neuroreport. 2001;12(6):1151–1154. doi: 10.1097/00001756-200105080-00020. [DOI] [PubMed] [Google Scholar]
- 367.Bakalash S, Pham M, Koronyo Y, Salumbides BC, Kramerov A, Seidenberg H, Berel D, Black KL, Koronyo-Hamaoui M. Egr1 expression is induced following glatiramer acetate immunotherapy in rodent models of glaucoma and Alzheimer’s disease. Investig Ophthalmol Vis Sci. 2011;52(12):9033–9046. doi: 10.1167/iovs.11-7498. [DOI] [PubMed] [Google Scholar]
- 368.Wilkinson K, El Khoury J. Microglial scavenger receptors and their roles in the pathogenesis of Alzheimer’s disease. Int J Alzheimers Dis. 2012;2012:489456. doi: 10.1155/2012/489456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Bornemann KD, Wiederhold KH, Pauli C, Ermini F, Stalder M, Schnell L, Sommer B, Jucker M, Staufenbiel M. Abeta-induced inflammatory processes in microglia cells of APP23 transgenic mice. Am J Pathol. 2001;158(1):63–73. doi: 10.1016/S0002-9440(10)63945-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Christie RH, Freeman M, Hyman BT. Expression of the macrophage scavenger receptor, a multifunctional lipoprotein receptor, in microglia associated with senile plaques in Alzheimer’s disease. Am J Pathol. 1996;148(2):399–403. [PMC free article] [PubMed] [Google Scholar]
- 371.Yu Y, Ye RD. Microglial Abeta receptors in Alzheimer’s disease. Cell Mol Neurobiol. 2015;35(1):71–83. doi: 10.1007/s10571-014-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.El Khoury JB, Moore KJ, Means TK, Leung J, Terada K, Toft M, Freeman MW, Luster AD. CD36 mediates the innate host response to beta-amyloid. J Exp Med. 2003;197(12):1657–1666. doi: 10.1084/jem.20021546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Sheedy FJ, Grebe A, Rayner KJ, Kalantari P, Ramkhelawon B, Carpenter SB, Becker CE, Ediriweera HN, Mullick AE, Golenbock DT, Stuart LM, Latz E, Fitzgerald KA, Moore KJ. CD36 coordinates NLRP3 inflammasome activation by facilitating intracellular nucleation of soluble ligands into particulate ligands in sterile inflammation. Nat Immunol. 2013;14(8):812–820. doi: 10.1038/ni.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Pan XD, Zhu YG, Lin N, Zhang J, Ye QY, Huang HP, Chen XC. Microglial phagocytosis induced by fibrillar beta-amyloid is attenuated by oligomeric beta-amyloid: implications for Alzheimer’s disease. Mol Neurodegener. 2011;6:45. doi: 10.1186/1750-1326-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Daws MR, Sullam PM, Niemi EC, Chen TT, Tchao NK, Seaman WE. Pattern recognition by TREM-2: binding of anionic ligands. J Immunol (Baltimore, Md: 1950) 2003;171(2):594–599. doi: 10.4049/jimmunol.171.2.594. [DOI] [PubMed] [Google Scholar]
- 376.Wang Y, Cella M, Mallinson K, Ulrich JD, Young KL, Robinette ML, Gilfillan S, Krishnan GM, Sudhakar S, Zinselmeyer BH, Holtzman DM, Cirrito JR, Colonna M. TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell. 2015;160(6):1061–1071. doi: 10.1016/j.cell.2015.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron. 2016;91(2):328–340. doi: 10.1016/j.neuron.2016.06.015. [DOI] [PubMed] [Google Scholar]
- 378.Bouchon A, Hernandez-Munain C, Cella M, Colonna M. A DAP12-mediated pathway regulates expression of CC chemokine receptor 7 and maturation of human dendritic cells. J Exp Med. 2001;194(8):1111–1122. doi: 10.1084/jem.194.8.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, Estus S. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 2013;33(33):13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Pillai S, Netravali IA, Cariappa A, Mattoo H. Siglecs and immune regulation. Annu Rev Immunol. 2012;30:357–392. doi: 10.1146/annurev-immunol-020711-075018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Diserbo M, Agin A, Lamproglou I, Mauris J, Staali F, Multon E, Amourette C. Blood-brain barrier permeability after gamma whole-body irradiation: an in vivo microdialysis study. Can J Physiol Pharmacol. 2002;80(7):670–678. doi: 10.1139/y02-070. [DOI] [PubMed] [Google Scholar]
- 382.Wegiel J, Imaki H, Wang KC, Wegiel J, Rubenstein R. Cells of monocyte/microglial lineage are involved in both microvessel amyloidosis and fibrillar plaque formation in APPsw tg mice. Brain Res. 2004;1022(1–2):19–29. doi: 10.1016/j.brainres.2004.06.058. [DOI] [PubMed] [Google Scholar]
- 383.Wegiel J, Imaki H, Wang KC, Wegiel J, Wronska A, Osuchowski M, Rubenstein R. Origin and turnover of microglial cells in fibrillar plaques of APPsw transgenic mice. Acta Neuropathol (Berl) 2003;105(4):393–402. doi: 10.1007/s00401-002-0660-3. [DOI] [PubMed] [Google Scholar]
- 384.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29(6):313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Conductier G, Blondeau N, Guyon A, Nahon JL, Rovere C. The role of monocyte chemoattractant protein MCP1/CCL2 in neuroinflammatory diseases. J Neuroimmunol. 2010;224(1–2):93–100. doi: 10.1016/j.jneuroim.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 386.Perlmutter LS. Microvascular pathology and vascular basement membrane components in Alzheimer’s disease. Mol Neurobiol. 1994;9(1–3):33–40. doi: 10.1007/BF02816103. [DOI] [PubMed] [Google Scholar]
- 387.Algotsson A, Winblad B. The integrity of the blood-brain barrier in Alzheimer’s disease. Acta Neurol Scand. 2007;115(6):403–408. doi: 10.1111/j.1600-0404.2007.00823.x. [DOI] [PubMed] [Google Scholar]
- 388.Bowman GL, Kaye JA, Moore M, Waichunas D, Carlson NE, Quinn JF. Blood-brain barrier impairment in Alzheimer disease: stability and functional significance. Neurology. 2007;68(21):1809–1814. doi: 10.1212/01.wnl.0000262031.18018.1a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Meda L, Bernasconi S, Bonaiuto C, Sozzani S, Zhou D, Otvos L, Jr., Mantovani A, Rossi F, Cassatella MA. Beta-amyloid (25–35) peptide and IFN-gamma synergistically induce the production of the chemotactic cytokine MCP-1/JE in monocytes and microglial cells. J Immunol (Baltimore, Md: 1950) 1996;157(3):1213–1218. [PubMed] [Google Scholar]
- 390.London A, Cohen M, Schwartz M. Microglia and monocyte-derived macrophages: functionally distinct populations that act in concert in CNS plasticity and repair. Front Cell Neurosci. 2013;7:34. doi: 10.3389/fncel.2013.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Shechter R, Raposo C, London A, Sagi I, Schwartz M. The glial scar-monocyte interplay: a pivotal resolution phase in spinal cord repair. PloS One. 2011;6(12):e27969. doi: 10.1371/journal.pone.0027969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Huang D, Wujek J, Kidd G, He TT, Cardona A, Sasse ME, Stein EJ, Kish J, Tani M, Charo IF, Proudfoot AE, Rollins BJ, Handel T, Ransohoff RM. Chronic expression of monocyte chemoattractant protein-1 in the central nervous system causes delayed encephalopathy and impaired microglial function in mice. FASEB J. 2005;19(7):761–772. doi: 10.1096/fj.04-3104com. [DOI] [PubMed] [Google Scholar]
- 393.Deane R, Bell RD, Sagare A, Zlokovic BV. Clearance of amyloid-beta peptide across the blood-brain barrier: implication for therapies in Alzheimer’s disease. CNS Neurol Disord Drug Targ. 2009;8(1):16–30. doi: 10.2174/187152709787601867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Sehgal N, Gupta A, Valli RK, Joshi SD, Mills JT, Hamel E, Khanna P, Jain SC, Thakur SS, Ravindranath V. Withania somnifera reverses Alzheimer’s disease pathology by enhancing low-density lipoprotein receptor-related protein in liver. Proc Natl Acad Sci USA. 2012;109(9):3510–3515. doi: 10.1073/pnas.1112209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, Stan TM, Fainberg N, Ding Z, Eggel A, Lucin KM, Czirr E, Park JS, Couillard-Despres S, Aigner L, Li G, Peskind ER, Kaye JA, Quinn JF, Galasko DR, Xie XS, Rando TA, Wyss-Coray T. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477(7362):90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Villeda SA, Plambeck KE, Middeldorp J, Castellano JM, Mosher KI, Luo J, Smith LK, Bieri G, Lin K, Berdnik D, Wabl R, Udeochu J, Wheatley EG, Zou B, Simmons DA, Xie XS, Longo FM, Wyss-Coray T. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat Med. 2014;20(6):659–663. doi: 10.1038/nm.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Xiang Y, Bu XL, Liu YH, Zhu C, Shen LL, Jiao SS, Zhu XY, Giunta B, Tan J, Song WH, Zhou HD, Zhou XF, Wang YJ. Physiological amyloid-beta clearance in the periphery and its therapeutic potential for Alzheimer’s disease. Acta Neuropathol (Berl) 2015;130(4):487–499. doi: 10.1007/s00401-015-1477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Yang F, Lim GP, Begum AN, Ubeda OJ, Simmons MR, Ambegaokar SS, Chen PP, Kayed R, Glabe CG, Frautschy SA, Cole GM. Curcumin inhibits formation of amyloid beta oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J Biol Chem. 2005;280(7):5892–5901. doi: 10.1074/jbc.M404751200. [DOI] [PubMed] [Google Scholar]
- 399.Zhang L, Fiala M, Cashman J, Sayre J, Espinosa A, Mahanian M, Zaghi J, Badmaev V, Graves MC, Bernard G, Rosenthal M. Curcuminoids enhance amyloid-beta uptake by macrophages of Alzheimer’s disease patients. JAD. 2006;10(1):1–7. doi: 10.3233/JAD-2006-10101. [DOI] [PubMed] [Google Scholar]
- 400.Baruch K, Deczkowska A, Rosenzweig N, Tsitsou-Kampeli A, Sharif AM, Matcovitch-Natan O, Kertser A, David E, Amit I, Schwartz M. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer’s disease. Nat Med. 2016;22(2):135–137. doi: 10.1038/nm.4022. [DOI] [PubMed] [Google Scholar]