Abstract
Chemical kinetic modeling has previously been used to predict that fast-translating codons can enhance cotranslational protein folding by helping to avoid misfolded intermediates. Consistent with this prediction, protein aggregation in yeast and worms was observed to increase when translation was globally slowed down, possibly due to increased cotranslational misfolding. Observation of similar behavior in molecular simulations would confirm predictions from the simpler chemical kinetic model and provide a molecular perspective on cotranslational folding, misfolding, and the impact of translation speed on these processes. All-atom simulations cannot reach the timescales relevant to protein synthesis, and most conventional structure-based coarse-grained models do not allow for nonnative structure formation. Here, we introduce a protocol to incorporate misfolding using the functional forms of publicly available force fields. With this model we create two artificial proteins that are capable of undergoing structural transitions between a native and a misfolded conformation and simulate their synthesis by the ribosome. Consistent with the chemical kinetic predictions, we find that rapid synthesis of misfolding-prone nascent-chain segments increases the fraction of folded proteins by kinetically partitioning more molecules through on-pathway intermediates, decreasing the likelihood of sampling misfolded conformations. Novel to this study, to our knowledge, we observe that differences in protein dynamics, arising from different translation-elongation schedules, can persist long after the nascent protein has been released from the ribosome, and that a sufficient level of energetic frustration is needed for fast-translating codons to be beneficial for folding. These results provide further evidence that fast-translating codons can be as biologically important as pause sites in coordinating cotranslational folding.
Introduction
Molecular simulations of structure-based, coarse-grained protein models (i.e., Gō models (1)) have advanced our understanding of the general principles of protein folding (2, 3, 4) and have provided molecular interpretations of experimental results (5, 6). In some cases, these models have made predictions in quantitative agreement with experimental data (7, 8). The numerous variants of such models are alike in that residues that are in contact in an experimentally determined structure are assigned attractive interactions in the force field. Because the parameterization of Gō models is based on a single structure, most versions of these models do not allow for nonnative tertiary structure formation. Protein misfolding, which involves the formation of nonnative conformations, is relevant to a wide range of biological phenomena, including the specific activity of enzymes and the aggregation of proteins. Structure-based coarse-grained models able to describe protein misfolding are needed to address these processes.
Several methods have been devised to model transitions among different ordered structures using structure-based models. In the approach of Best et al. (9), the Hamiltonians encoding different structures are combined through a parallel-programming method. In the approach by Okazaki et al. (10), a new energy term is introduced in the force field to shift the equilibrium between two different ordered conformations. These approaches have been used to study protein allosteric movements (10, 11, 12) and fold switching in metamorphic proteins (13, 14). Here, we present a method that avoids the need for simulating multiple Hamiltonians or adding additional energy terms by assigning different energies to the native and misfolded contacts while maintaining the structural fidelity of each structure. We then apply this method to the process of protein synthesis on the ribosome, with the aim of understanding how codon translation rates influence the likelihood of cotranslational folding and misfolding.
Cotranslational protein folding is likely to occur in an appreciable fraction of the proteome of organisms (15) and can be influenced by the variable speed at which the ribosome translates mRNA codons into amino acids that are added to the growing polypeptide (16, 17, 18, 19, 20, 21). Under quasiequilibrium conditions, cotranslational folding is determined by the physical forces responsible for domain stability as well as interactions with the ribosome and surrounding macromolecules (17, 22, 23, 24). Under nonequilibrium conditions, when nascent-chain structural rearrangements occur on similar or slower timescales compared to elongation, the nascent polypeptide explores a smaller number of conformations that are dependent on earlier stages of elongation and thereby on the rates at which codons are translated.
The relationship between elongation rates and cotranslational events is beginning to be quantitatively understood through theoretical, computational and experimental efforts (16, 20, 25, 26, 27, 28, 29, 30). Kinetic modeling predicted that increasing the polypeptide elongation speed can promote cotranslational folding by reducing the number of misfolding events during the time that the ribosome dwells in the misfolding-prone nascent-chain length regime, contrary to the common view that slow synthesis favors native structure formation (31). Consistent with this hypothesis, recent experiments (32, 33) and the qualitative trends observed in atomistic molecular dynamics simulations of a small protein (28) imply that slow-translating codons are not always beneficial for cotranslational folding and can promote protein aggregation.
Using our modeling approach, we created two artificial misfolding-prone proteins and simulated their synthesis by the ribosome using Langevin dynamics. Consistent with the chemical kinetic predictions, we find that rapid elongation through the proteins’ misfolded region can either facilitate the formation of on-pathway intermediates or delay the formation of misfolded structures until the domain has fully emerged from the tunnel, where folding to the native conformation outcompetes misfolding. Beyond the previous predictions of the kinetic model, we show how the distribution of folding pathways is altered by fast translation, that posttranslational protein dynamics can depend on the translation-elongation schedule, and that a sufficient level of energetic frustration is needed before fast-translation becomes beneficial for folding.
Materials and Methods
Constructing the multibasin Gō model
The protocol for constructing a multibasin Gō model, as well as the details about the coarse-grained ribosome and nascent chain are provided in the Supporting Material.
Starting structures and simulation protocols
To construct the two misfolding-prone protein models, Protein Data Bank (PDB) PDBs:1RYK and 2BTG (multibasin model 1) and PDBs:1ZZV and 1ND9 (multibasin model 2) were defined as the native and misfolded structures, respectively. Selection of these PDBs was discussed in the Results and Discussion. An Escherichia coli ribosome was converted into a coarse-grained model using the crystal structure with PDB:5AFI. A 60-amino acid (aa) residue unstructured linker (sequence = 5′-AVQLA LAALI SALEK EVVIL LALVK ALGAL LLLLA ALAAL AAIDA LELVA ATALL LLLVA-3′) was generated in an extended conformation using the CHARMM simulation software package (https://www.charmm.org/) and attached to the C terminus of the nascent chain. The linker sequence was obtained as the consensus sequence of C-terminal unstructured regions from the database of cytosolic E. coli proteins reported in Ciryam et al. (15). Atomistic structures were coarse-grained using one bead per amino acid and three or four beads per nucleotide. The ribosome-nascent-chain complex was assembled using in-house code developed for single-structure Gō models, as described previously (34).
Langevin dynamics simulations were performed with a friction coefficient of 0.05 ps−1 using a modified version of the CHARMM software that includes the Debye-Hückel and double-well functional forms for the electrostatic and angle potential terms, respectively. This friction coefficient minimizes the folding time in simulations of isolated proteins. The Langevin equations of motion were integrated every 0.015 ps and the temperature was set to 310 K, which is the optimal growth temperature of E. coli.
The two multibasin Gō-models, built using the combination of PDBs:1RYK and 2BTG or PDBs:1ZZV and 1ND9, were simulated with replica exchange (REX) simulations either off the ribosome or attached to arrested ribosomes at different nascent chain lengths. Selected nascent chain lengths were used in stalled ribosome simulations as reported in the Supporting Material (Figs. S1 and S9) to provide a description of the structural ensembles populated on the ribosome, while reducing the computational burden. For each REX run, 20 replicas were used ranging in temperature from 300 to 550 K, and 100,000 exchanges were made, with the first 20,000 eliminated from the analysis to allow for system equilibration. Four-thousand integration time-steps were carried out between attempted REXs.
For each translation-elongation schedule, 300 continuous synthesis simulations were performed at 310 K starting from a nascent chain length of 40 residues and ending at 129 or 140 residues. An unstructured linker was attached to the C terminus and synthesized from nascent chain length 70–129 (multibasin model 1) or 81–140 (multibasin model 2). The unstructured linker allows the protein domain to fully emerge outside the ribosome exit tunnel, thereby mimicking the synthesis of a domain within a multidomain protein. Synthesis simulations were carried out at different amino acid addition rates. For the uniformly fast and slow synthesis simulations, residues (Cα beads) were added to the nascent chain every 0.15 and 15 ns, respectively. All translation-elongation schedules used in this study are reported in the Supporting Material (Fig. S4).
Every 7 ns, on average, a residue was added to the nascent chain in our simulations, which is a much faster timescale compared to the average experimental value of ∼100 ms measured in E. coli (35). The use of such residue addition times was nevertheless necessary to compensate for the accelerated dynamics typical of protein coarse-grained models (36, 37). After previous simulation studies of cotranslational folding, the elongation timescales used in our simulations are obtained by rescaling the average experimental elongation timescale by a factor of 106 to 108 (8, 26, 29). The acceleration factor can also be obtained by comparing the rates of protein folding in experiments and coarse-grained simulations. Two studies addressed the relationship between experimental and simulated protein folding timescales using coarse-grained models and found that the simulated folding rates were 104 and 105 faster compared to experiments (38, 39). Albeit smaller than the rescaling factors we used, these estimates do not account for the larger spread of experimental folding rates compared to simulations and moreover, they can depend sensitively on the friction used in the Langevin equation. Because of the larger spread of experimental data, from the mentioned studies we could estimate larger acceleration factors, within the range of the ones we used in this work.
Analyses
Conformations and related free energies and probabilities were characterized in the simulations based on selected structural quantities including the root mean square deviation (RMSD) RMSDk, where k indicates the experimental native (n) or misfolded (m) structure, and the fraction of contacts Qk that are similar to those of structure k. (The latter quantity is commonly known as the fraction of native contacts, but for clarity with the terminology adopted for n and m, we do not use this expression.) A contact between residues I and J is present in a given system configuration if the distance between Cα(I)-Cα(J) is within 0.5 Å of the same distance in the kth crystal structure.
From arrested ribosome simulations, the probabilities p(Qn,Qm|L) were calculated using a bin width of 0.07 and combined using the formula , where the summation is performed on a set of nascent chain lengths selected as described before. Maxima on the surface S correspond to frequently visited conformations and were used to partition S into structurally distinct conformations as described in the Results and Discussion.
Pathways were defined as the time series of states populated as a function of nascent chain length. The states defined by S(Qn,Qm) were used to identify all pathways. The edges of the rectangles that partition S (Fig. 3 a) were decreased by 0.05, to reduce the noise caused by trajectory recrossings at their borders. This operation preserved the overall state definitions and generated small gaps where conformations were assigned to the most recently visited state. To reduce the extremely large number of possible pathways, a clustering was performed. Two pathways were assigned to the same representative pathway if the difference between them was <20%. The difference metric was defined as , where Nt is the number of time points in the trajectory, ℓ and ℓ′ are two pathways, and δ is the Kronecker function. The same procedure was used to calculate the number of transitions between states. The software tools VMD (http://www.ks.uiuc.edu/Research/vmd/) and gnuplot (http://www.gnuplot.info/) were used to generate the figures (40).
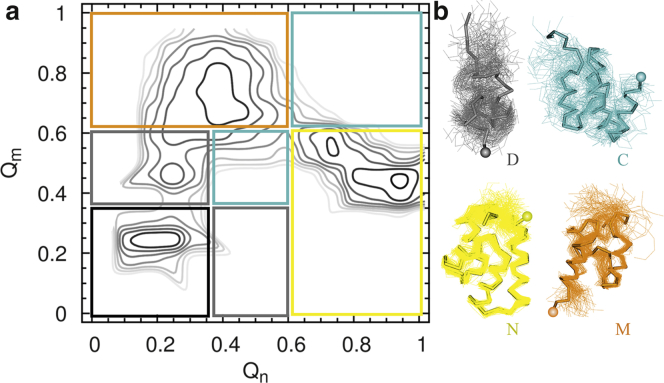
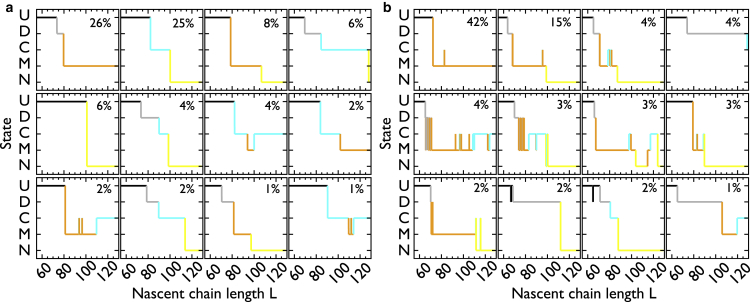
Figure 3.
Nascent-protein structural states populated during arrested-ribosome REX simulations. (a) Shown here is a contour map of the probability function S(Qn, Qm) of finding conformations with fractions of contacts Qn and Qm during arrested ribosome simulations at 310 K. S is computed by summing the probabilities of observing Qn and Qm at selected nascent chain lengths between 40 and 129 residues (see Materials and Methods). Darker contour lines indicate higher probability values. The partitioning of the (Qn, Qm) space is indicated by the colored boxes and is based on the position of the maxima and minima of S. (b) Shown here is an α-trace representation of the disordered (gray), combination (cyan), misfolded (orange), and native (yellow) conformations characteristic of the states demarcated in (a) (same color coding). For clarity, the unfolded state (black) is not shown in (b). Colored spheres indicate the C-terminal end of the domain. The ribosome and the disordered linker are not shown.
Statistical significance of the difference between state probabilities pI was assessed using the two-tailed Student’s t-test. Error bars on the state probabilities correspond to the 95% confidence intervals.
Results and Discussion
A coarse-grained model that allows for protein misfolding
To study the general features of protein misfolding, a multibasin coarse-grained protein model was created as detailed in the Supporting Material and was subsequently applied to the study the co- and post-translational folding of misfolding-prone proteins. In this approach, a Gō-based force field is initially generated based on a structure representing the native state (denoted n) (1, 41). This force field is subsequently augmented with information based on a conformation representing a misfolded state (denoted m), thereby capturing the competition between the two potential energy surfaces. Rather than combining the two energy surfaces into a single surface displaying two minima (9, 10), we sum the residue-residue interaction potentials in a single nonbonded term (Eq. S1) (11, 12, 42), as this procedure is easy to implement and does not assume any potential energy surface topology a priori. Here, we apply this approach to compact structures with different shapes and sequence lengths.
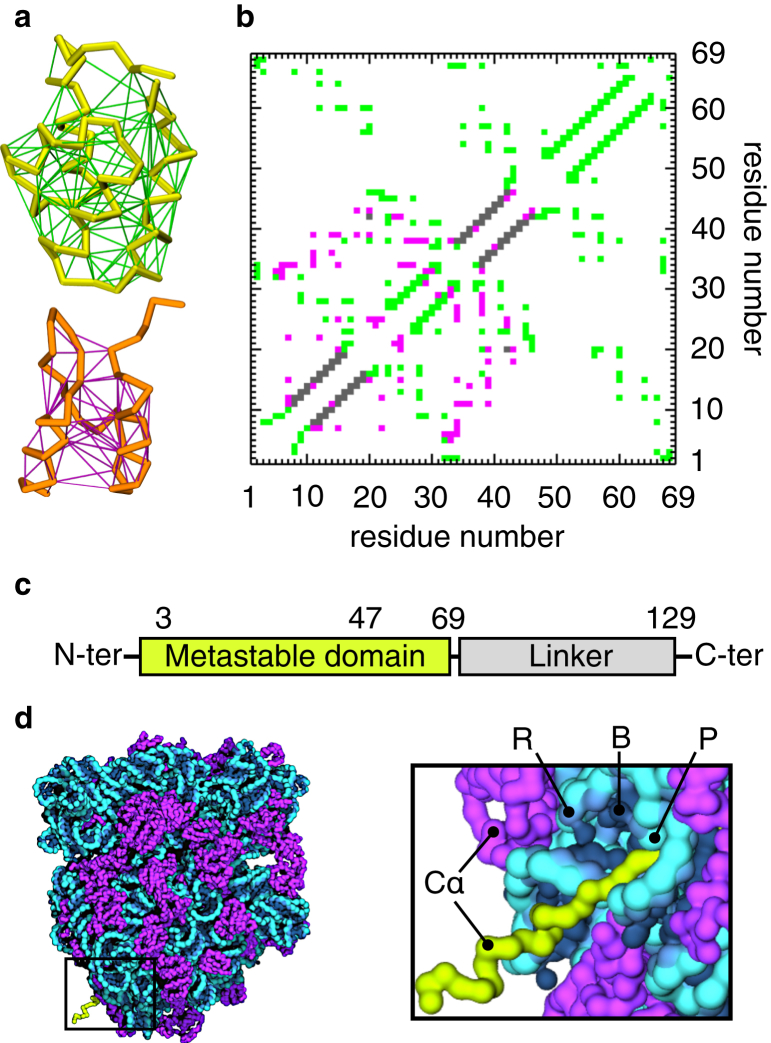
It is difficult to experimentally characterize the structure of soluble misfolded proteins because of their heterogeneous, transient nature and their tendency to aggregate. Therefore, we chose two well-resolved and ordered structures n and m to create multibasin model 1, composed of Nn = 69 and Nm = 45 residues, respectively, and built the artificial misfolding-prone protein depicted in Fig. 1. These structures were selected randomly from the PDB, requiring that they be E. coli proteins <100 residues in length, that they have different topologies, and that Nm < Nn. The last two conditions were imposed to allow the modeling of off-pathway misfolding events, which have been detected experimentally (43, 44). The interaction networks that stabilize structures n and m were represented as contact maps Z(n) and Z(m), respectively. Z(m) is embedded within Z(n) at a specific position, s, defined as the position at which the first element of Z(m) occurs within Z(n) after the optimization procedure described in the Materials and Methods. This procedure aims to merge the two contact maps such that the number of interresidue contacts that are common to both n and m is minimized. The resulting combined contact map has some contacts that are unique to each structure n or m and others that are shared (gray pixels in Fig. 1 b). The 32 shared contacts, found when Z(m) is embedded within Z(n) at its optimal position of s = 3 residues, are all short-ranged, intrahelix interactions with interresidue distances differing by <1 Å. These shared contacts are then uniquely assigned to either the n or m structure. The contacts found in the merged contact map have Lennard-Jones well depths equal to either εn or εm (Eq. S3). These parameters influence the relative stability of the n- and m-like conformations in the coarse-grained model.
Figure 1.
The multibasin coarse-grained model for protein misfolding on the ribosome. (a) Shown here is a Cα-trace representation of the native and misfolded structures generated from PDBs:1RYK (top) and 2BTG (bottom). The networks of stabilizing Cα-Cα interactions (contacts) are depicted in green and magenta. (b) Shown here is a contact map of the multibasin model. Each colored pixel represents a stabilizing interaction between two residues (see term Unel in Eq. S1). The misfolded-state contact map (magenta) is embedded in the native-state contact map (green) starting at position s = 3 through a procedure that minimizes the number of shared contacts between the two structures (gray). (c) Given here is an illustration of the full-length misfolding-prone construct. The native structure is composed of residues 1–69, whereas the misfolding-prone region is between residues 3 and 47. The unstructured linker starts at residue 70. (d) Shown here is a coarse-grained representation of the ribosome-nascent chain complex used in the simulations (left). Coarse-grained interaction sites are centered on the Cα atom of amino acids (yellow and magenta) and on the phosphorus atom (P, cyan), ribose center (R, light blue) and base-ring centers (B, dark blue) of nucleotides (right). Folding and misfolding events are energetically driven by the network of contacts in (b), as the metastable domain (yellow) is being synthesized by the ribosome.
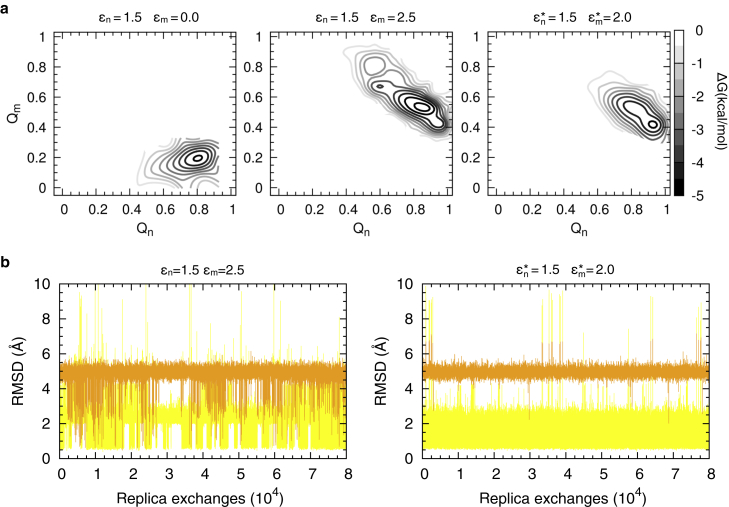
The εn and εm values are chosen following a three-step procedure (see Materials and Methods for details). In the first step, εm was set to 0.001 (i.e., misfolding cannot occur) and εm tuned to match the experimental folding free energy of n, resulting in an εn value of 1.5 kcal/mol at 310 K, the temperature of E. coli optimal growth (this temperature is used in all subsequent simulations). Fig. 2 a illustrates the protein’s free energy surface as a function of the fraction of contacts (Qn, Qm) for the simulation at (εn, εm) = (1.5, 0.001) kcal/mol, where n is stable and m is never populated. The only visible basin is centered around (Qn, Qm) = (0.8, 0.2), corresponding to a well-formed native state.
Figure 2.
Parameterization of the force field to obtain the target misfolding population. (a) Shown here are the free energy surfaces of the multibasin model in bulk solution at 310 K and different values of the parameters εn, εm (indicated above each panel), where n and m label, respectively, the experimental structures PDBs:1RYK and 2BTG. Qn and Qm are the fraction of contacts formed as in n and m during REX simulations. (b) Given here is the RMSD between Cα positions of the simulated protein at 310 K and PDBs:1RYK (yellow) or 2BTG (orange) as a function of the number of REXs. RMSDs were calculated by structurally aligning the trajectory snapshots to the residues belonging to α-helices and β-sheets of either n or m (secondary structures were identified using the web tool STRIDE; http://webclu.bio.wzw.tum.de/stride/).
In the second step, misfolding is introduced by increasing εm. The goal is to have a protein that predominantly populates its folded state at equilibrium, with a small misfolded population. Therefore, using target native and misfolded populations, respectively, of p0,n = 0.95 and p0,m = 0.05, and a number of contacts ηn = 79 and ηm = 48, we estimate that εm = 2.5 kcal/mol will achieve this level of misfolding based on Eq. S5 (see Materials and Methods). The increase of εm from 0.0 to 2.5 kcal/mol perturbs the system’s equilibrium and dynamic properties, producing a shift of the native-state basin from (Qn, Qm) = (0.8, 0.2) to (Qn, Qm) = (0.9, 0.5) (Fig. 2 a). Although the most probable Qm value increases from 0.2 to 0.5, this basin still corresponds to the native state as evidenced by an average root mean square deviation (RMSD), RMSDn = 2.0 Å and average RMSDm = 5.0 Å, where RMSDk is the RMSD from the experimental structure k. A larger Qm value results from those shared contacts that are present in both the native and misfolded states (gray pixels in Fig. 1 b). Transitions to the misfolded state are observed in simulations with εm = 2.5 kcal/mol (Fig. 2 b), resulting in native (all conformations having RMSDn ≤ 4.0 Å or Qn ≥ 0.6) and nonnative populations of 0.8 and 0.2, respectively. Both n- and m-like conformations are explored with high structural fidelity, reaching RMSD values ≤3 Å from their corresponding n and m structures (Fig. 2 b).
As the third step in the procedure, aimed at decreasing the probability of observing nonnative states (misfolded and additional compact states), we lowered εm, thereby correcting for the fact that Eq. S5 does not account for the formation of additional states. The final, tuned energy values (εn∗, εm∗) = (1.5, 2.0) kcal/mol produce a multibasin model with native and nonnative state populations of 0.99 and 0.01, respectively, which are close to the target populations (Fig. 2 a).
Folding kinetics consists of fast and slow phases
We characterized the folding kinetics of the protein whose tuned force field parameters are (εn∗, εm∗) by running 800 independent temperature-quench simulations (22). In each simulation the protein was first heated to 1000 K for 30 ns, then instantaneously cooled to 310 K and simulated for an additional 100 ns. All 800 trajectories were confirmed to be unfolded after the initial heating phase based on their low Qn and Qm values. We then computed the fraction of trajectories that were folded at each time point using the native state definition Qn ≥ 0.6 and Qm ≤ 0.6, and observed a fast and slow folding phase (Fig. S2). The fast phase has a characteristic folding time of 1.0 ns, similar to that found in Gō models of other proteins (22, 34), with ∼56% of the trajectories folding on this timescale. Such multiphase kinetics is common in energetically frustrated proteins or large proteins, and has been observed experimentally (45, 46) as well as in coarse-grained protein models with transferable force fields (47) and in Gō models that cannot populate misfolded states (34). For some proteins, the slow-folding timescale can be on the order of minutes (45). In the rest of this article, when we refer to the folding time of the model protein, we are referring to the 1.0 ns value of the fast phase.
The misfolding-prone protein explores a heterogeneous structural ensemble on the ribosome
To understand how translation kinetics impacts the cotranslational folding of this protein, we first characterized the states the nascent chain populates on arrested ribosomes, as these states provide a reference to which the continuous-synthesis simulation results can be compared. The misfolding-prone domain was attached to an unstructured C-terminal linker that, at its full length, allows the domain to emerge from the exit tunnel and fold (Fig. 1 c), thereby mimicking the behavior of an N-terminal domain that is part of a multidomain protein. Arrested ribosome-nascent chain (RNC) complexes were simulated using REX (48) at nascent chain lengths, L, between 40 and 129 residues, using the tuned (εn∗, εm∗) force-field values. We did not simulate nascent chain lengths shorter than 40 residues because the nascent protein populates only unfolded conformations at those lengths (Fig. S1).
As L increases from 40 to 129 residues the protein explores different conformations at equilibrium (Fig. S1). At the shortest nascent-chain lengths the protein is unfolded, but at intermediate and longer lengths the conformations are, respectively, most similar to the misfolded and native structures. We characterized the conformational ensembles explored during the arrested RNC simulations using the probability function S(Qn, Qm) of finding a conformation with fractions of contacts equal to Qn and Qm (Fig. 3 a). S(Qn, Qm) is an average over the probabilities at the different L values shown in Fig. S1. The maxima and minima of S(Qn, Qm) allow us to partition this space into distinct conformational subpopulations corresponding to the native (N), misfolded (M), combination (C), disordered (D), and unfolded (U) states (Fig. 3 b). States N and M occupy the regions of S that have high values of Qn or Qm, implying that they resemble, respectively, the experimental structures n or m. Indeed, we find that the RMSD between state N from the experimental structure n is lower (0.8–3 Å) than the RMSD between N from the experimental structure m (4.5–5 Å; Fig. S3). We find the opposite result for state M (data not shown). The U and D states are the least compact, and for this reason they occupy regions with the lowest (Qn, Qm) combinations. State U is largely devoid of secondary and tertiary contacts, whereas state D preferentially occupies the gray rectangle at higher Qm values in Fig. 3 a because D is populated at L < 65 (Fig. S1), when the majority of contacts permitted to form belong to the contact map Z(m). Because other contacts are prevented from forming due to the steric confinement of the ribosome exit tunnel, state D differs from state M. The two regions enclosed within the cyan rectangles in Fig. 3 a correspond to state C, which has structural features of both n and m, stabilized by a roughly equal number of contacts from each structure. The most likely conformation is the one having an N-terminal segment similar to the m structure whereas the C-terminal is folded similarly to the n structure. However, state C is heterogeneous, as different combinations of the n and m structures are allowed to form by the force field.
The state definitions illustrated in Fig. 3 account for the formation of isolated secondary structural elements within the ribosome exit tunnel. However, because these secondary structures form only transiently during the arrested ribosome simulations they only contribute marginally to the probability S(Qn, Qm). This partitioning of the conformational space provides a reduced set of states that we use to study the behavior of the nascent protein during continuous synthesis simulations.
Rapid synthesis through misfolding-prone segments increases cotranslational folding
To determine if fast elongation improves cotranslational folding, we performed synthesis simulations of the misfolding-prone nascent chain at various elongation schedules {τaa(L)}, where τaa(L) indicates the time of elongating the nascent chain from length L to L+1. The τaa(L) values used in our simulations are smaller than the experimentally measured times of amino acid addition in E. coli. As described in greater detail in the Materials and Methods, we used faster elongation timescales to take into account the accelerated dynamics inherent to coarse-grained simulations at low viscosity (20). Results from these simulations are presented in Fig. 4 as the probability of observing a state I at nascent chain length L, where I is one of the states U, D, C, M, or N (Fig. 3). As in the arrested ribosome simulations, we started the continuous synthesis simulations at a nascent chain length of 40 residues to reduce the simulation time. This choice does not affect our conclusions, because below 60 residues the nascent protein is unfolded (black data points in Fig. 4).
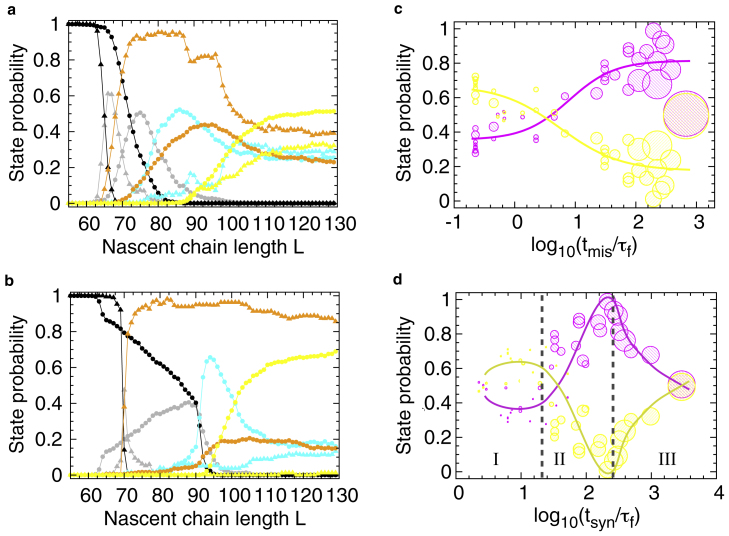
Figure 4.
The effect of different elongation schedules on cotranslational folding and misfolding. The probabilities of populating states U (black), D (gray), M (orange), C (cyan), and N (yellow) as a function of nascent chain length L, when one amino acid is added every τaa(L) ns (elongation schedules s1 to s40 are reported in Fig. S4 c). In all panels, lines are meant as a guide to the eye and the standard errors about the mean are smaller than the symbols. (a) State probabilities from the uniformly slow (s30) and uniformly fast (s2) elongation schedules are shown, respectively, as triangles and circles. Each data point is an average over 300 independent synthesis trajectories. (b) Same as (a), except the state probabilities resulting from the nonuniform, slow schedule s39 (triangles) and nonuniform, fast schedule s5 (circles) are shown. (c) Shown here are the native (yellow) and nonnative (M and C; magenta) state probabilities at the end of synthesis (i.e., at L = 129 residues) as a function of the reduced time Tmis = tmis/τf. The value tmis is the time necessary to synthesize residues 70–90 and τf = 1.0 ns is the mean folding time of the fast-kinetic phase of this protein off the ribosome. The data point sizes are proportional to the reduced total time Tsyn = tsyn/τf needed to synthesize the entire protein (from residue 40–129). Each data point results from one of the elongation schedules shown in Fig. S4 c. (d) Same as (c), but as a function of the reduced time Tsyn = tsyn/τf. The value tsyn is the time necessary to synthesize the entire protein from residues 40–129. Circle sizes are proportional to the time Tmis needed to synthesize the misfolding-prone segment. Vertical dashed lines indicate boundaries among regions I, II, and III.
We find that when the protein is synthesized uniformly slowly (τaa(L) = 15 ns at all L), the states are populated at shorter nascent chain lengths compared to when elongation is 100-fold faster (Fig. 4 a). State D starts to be populated at L = 63, state M at L = 65, and state C at L = 77 residues when synthesis is uniformly slow (schedule s30 in Fig. S4 c), whereas at a uniformly faster elongation (schedule s2 in Fig. S4 c), state D appears at L = 66, and either M or C at L = 71 residues. For both elongation schedules, N starts to be appreciably populated at = 89 residues, consistent with earlier simulation results (22). The positions at which the state probabilities achieve a maximum also shift to longer nascent chain lengths when elongation is faster (Fig. 4 a). At the longest nascent chain length of 129 residues, the uniformly fast elongation schedule promotes the formation of 59% more nativelike molecules compared to the uniformly slow schedule (pN = 0.51 vs. 0.32 in Fig. 4 a; p value < 10-4 Student’s t-test), whereas the probabilities of populating M and C decrease. Thus, uniformly faster elongation increases the probability of cotranslational folding whereas slower elongation promotes misfolding in this artificial protein.
Comparing the state probabilities during the uniformly slow- and fast-synthesis simulations, we noticed that the probability of misfolding pM (orange symbols in Fig. 4 a) is dramatically altered between the nascent chain lengths of 70–90 residues, where, according to the equilibrium (arrested) ribosome simulations, the nascent protein populates the misfolded state (see Fig. S1). This suggests that the nascent chain conformations explored at these lengths are particularly sensitive to changes in elongation rates and therefore that this region may be a major cause of downstream changes in the probability of cotranslational folding pN. To test this hypothesis, we simulated protein synthesis using a slow rate at residue positions 70–90, whereas the remaining positions were synthesized at the fast rate (schedule s6 in Fig. S4 c). Slowing down elongation in this region causes the fraction of nativelike molecules at the final nascent chain length to decrease by 63% compared to the uniformly fast elongation (pN = 0.51 decreases to 0.19 in Fig. S4 a; p value <10-4 Student’s t-test). A similar decrease (Fig. S4 b) is obtained even when the region of slower elongation is reduced to only a single residue located at position 80 (schedule s37 in Fig. S4 c). Thus, rapidly synthesizing through the misfolding-prone region promotes this protein’s cotranslational folding.
A general trend in cotranslational folding versus synthesis time
We next examined if this effect could be observed using a more diverse set of elongation schedules. We simulated the protein’s synthesis using 36 additional elongation schedules (Fig. S4 c), which can be classified into three groups: 1) uniform schedules, 2) schedules that differ in the misfolding-prone region or adjacent segments, and 3) schedules with a single slow-translated residue. We plotted the probability of cotranslational folding as a function of the reduced time Tmis = tmis/τf, where tmis is the time that the ribosome dwelled in the misfolding-prone region (i.e., residues 70–90) and τf = 1.0 ns is the folding time of the fast phase off the ribosome. Because the protein kinetics are accelerated as a consequence of both the coarse-grained representation and low-friction simulations, the absolute time tmis cannot be compared directly to experiments unless the acceleration factor is known (20). It is therefore useful to rescale the simulated timescale tmis by τf to eliminate this intrinsic acceleration.
We find that the probability of domain folding at the final nascent chain length increases as Tmis decreases, whereas the probability of populating nonnative states decreases in an anticorrelated fashion (continuous lines in Fig. 4 c). When synthesis is very fast, eventually becoming comparable to or faster than the folding time of this protein (Tmis < 4 in Fig. 4 c), the likelihood of folding is larger than the likelihood of misfolding. The size of the data points in Fig. 4 c is proportional to the time Tsyn necessary to synthesize the entire protein. The majority of points with large or small sizes are distributed, respectively, at large and small Tmis values. Thus, for this protein, cotranslational folding is promoted, on average, by fast synthesis across the misfolding-prone region, as well as across the entire polypeptide.
Even though the native and nonnative state probabilities follow a sigmoidal relationship as a function of Tmis (solid lines in Fig. 4 c), there are deviations that are explained in part by the different total synthesis times Tsyn. For example, the largest magenta and yellow data points in Fig. 4 c, which correspond to the outcomes of the slowest-elongation schedule (s40 in Fig. S4), deviate from the general trends. In this case, although slow synthesis across the misfolding-prone region initially increases the misfolded population up to 0.95, the slow elongation of the remaining protein segment affords the nascent chain enough time to transition from states M or C to N, which is the thermodynamically favored state at the end of synthesis. This suggests that the state probabilities are also sensitive to the kinetics of protein synthesis across the segments adjacent to the misfolding-prone region.
The interplay between elongation timescales across the different protein segments is more clearly illustrated when the state probabilities are plotted as a function of Tsyn (Fig. 4 d). The regime that promotes folding by speeding up elongation is visible at intermediate values of Tsyn (region II in Fig. 4 d), consistent with Fig. 4 c. This is also the region where the state probabilities show the largest variability (between 0 and 0.69). Two additional regions are distinguishable in which speeding up synthesis either penalizes folding or has a neutral effect. For relatively large Tsyn values (region III in Fig. 4 d), the probability of cotranslational folding decreases as the elongation speed is increased because, as noted earlier, the nascent protein is closer to thermodynamic equilibrium. For relatively small values of Tsyn (region I in Fig. 4 d), the state probabilities remain almost constant as a function of the total synthesis time, with a more populated native state compared to the misfolded state. In region I, the overall fast synthesis allows the nascent protein to populate unstructured conformations at later stages of elongation (nascent chains >92 residues) and consequently to evolve toward the native state more easily than toward the misfolded state.
The data in Fig. 4, c and d, indicate that Tsyn and Tmis can independently affect cotranslational folding, suggesting that elongation schedules with a similar total synthesis time but different distributions of elongation rates can produce very different cotranslational behaviors. To test this hypothesis we selected two schedules, the nonuniform, fast schedule s5 (Fig. S4 c) and the uniformly fast schedule, and compared their synthesis outcomes. Schedule s5 differs from the uniformly fast schedule in that it has a faster elongation rate in the misfolding-prone region and slower elongation rate for L > 90 residues, but a similar Tsyn value (within 3%). At the end of synthesis, the cotranslational folding probability of the nonuniform, fast schedule (pN = 0.69, yellow circles in Fig. 4 b) is 35% higher than the uniformly fast schedule (pN = 0.51, yellow circles in Fig. 4 a). An additional comparison between the uniformly slow schedule and the nonuniform, slow schedule s39 (Fig. S4 c), which differ by <4% in Tsyn, reveals that the folding probability decreases from 0.32 to 0.01 at the end of synthesis (yellow triangles in Fig. 4, a and b), essentially abolishing cotranslational folding. (In the schedule s39, τaa(L) is relatively large in the misfolding-prone region and relatively small in the region L > 90 to achieve a Tsyn similar to the uniformly slow case.) Thus, even when two schedules have similar total synthesis times, the populations of native and nonnative states can be very sensitive to how the fast- and slow-elongation rates are distributed during synthesis, especially in the misfolding-prone region.
These simulation results are consistent with in vivo experiments that measured either an increase of protein aggregation upon slowing down translation (49) or a decrease in protein misfolding upon speeding up translation (32). The degradation of γB-crystallin was observed to decrease upon synonymous codon substitutions that increased the mRNA translation speed (32). Based on our results, we speculate that some of the slow-to-fast synonymous codon substitutions might have been introduced in misfolding-prone regions of γB-crystallin, thereby promoting cotranslational folding. In another experiment, fast-to-slow synonymous codon mutations were introduced in the nucleotide binding domain 1 of the cystic fibrosis transmembrane conductance regulator (49). By affording this domain enough time to populate long-lived nonnative conformations, slower elongation may have negatively impacted folding of this topologically complex protein, in agreement with our results.
Fast elongation promotes folding by reducing the number of pathways involving misfolded states
The folding pathways of the nascent protein were characterized to determine the molecular mechanisms by which faster elongation promotes folding and penalizes the formation of misfolded conformations. Using the clustering procedure described in the Materials and Methods, we calculated a set of representative cotranslational folding pathways, accounting for 87 and 85% of the uniformly fast and slow synthesis trajectories (Fig. 5).
Figure 5.
Representative folding pathways during continuous synthesis simulations. (a) Show here is the structural state versus nascent chain length resulting from the uniformly fast elongation schedule. Each pathway differs by the series of states (U, D, C, M, and N) that the nascent chain populates as a function of L. The percentage of the 300 trajectories that is represented by each pathway is indicated in the upper-right corner of each panel. (b) Same as (a), but for the uniformly slow elongation schedule.
The most probable pathway in the uniformly fast (Fig. 5 a) and slow synthesis (Fig. 5 b) trajectories leads to state M at the end of synthesis. This pathway is characterized by a long-lived M state that does not transition to any other states, indicating that the N-terminal region of the nascent protein can populate kinetically trapped misfolded conformations as soon as the protein emerges from the ribosome exit tunnel. This pathway accounts for 26% of the fast-synthesis pathways and 42% of the slow-synthesis pathways (Fig. 5), suggesting that the fast schedule may be responsible for promoting folding by decreasing the number of molecules forming misfolded states during synthesis. To test this hypothesis, we calculated the fraction of trajectories that visit state M at least once during the simulation. We find that the number of pathways involving state M decreases from 91 to 57% when the elongation speed is increased. Taken together, these observations suggest that speeding up elongation helps folding by reducing the number of initial transitions U(D)→M and the likelihood to populate M at subsequent elongation stages.
Concomitant with the decrease in misfolding pathways, Fig. 5, a and b, show that speeding up elongation increases the number of pathways leading to N by the end of synthesis, in agreement with the results in Fig. 4 a. The most probable series of state transitions responsible for the formation of N is the transition U(D)→C→N, which occurs in 37% of the fast pathways and 5% of the slow ones. Two additional mechanisms are U(D)→N and the pathway to N mediated by previous formation of M. Whereas the former mechanism occurs in 6% of the fast pathways and 0% of the slow ones, the latter occurs in 9% of the fast pathways and 24% of the slow ones. Therefore, speeding up synthesis promotes folding pathways through state C and decreases those involving state M. Such phenomenon in the chemical and biological sciences is referred to as kinetic partitioning, where kinetics plays an important role in driving a system into different states (50). Thus, fast rates of amino-acid addition can influence the kinetic partitioning of nascent proteins to favor folded structures at the expense of misfolded conformations.
To further understand the role that the elongation rates play in selecting the molecular pathways to the folded state, we analyzed the trajectories generated with two schedules that achieve high folding probabilities of 0.62 (schedule s26) and 0.69 (schedule s5) by the end of synthesis (Fig. S4 c). Under schedule s26, in which the misfolding-prone region elongates two times faster than s2, pathway U→N is the most probable (Fig. S5), whereas the most probable pathway for schedule s5 is U→D→C→N (Fig. S6). From this analysis we conclude that the nascent protein can reach the folded state following multiple pathways, and that the elongation schedule determines which of these pathways is most likely during synthesis.
Fast synthesis helps protein folding above a threshold-level of energetic frustration
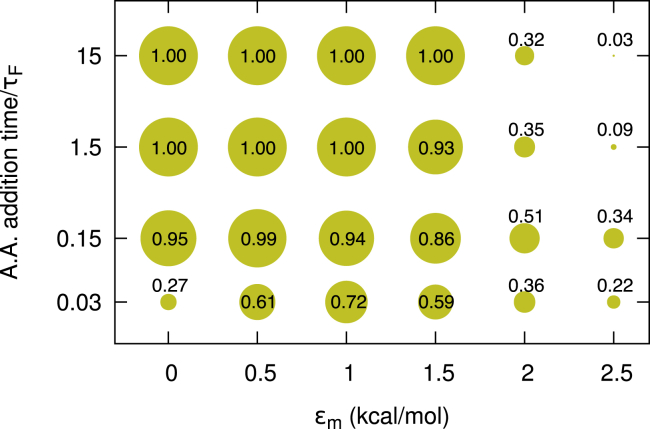
We next explored how changing the stability of the misfolded state and the rate of nascent protein elongation affect the amount of correctly folded protein produced at the end of synthesis. Specifically, we varied εm between 0.001 and 2.5 kcal/mol and held εn constant at 1.5 kcal/mol, thereby modulating the degree of frustration in the system. We find that at or below an εm of 1.5 kcal/mol, fast elongation decreases the cotranslational folding probability, whereas above this value fast elongation increases cotranslational folding (Fig. 6). Although this threshold value of εm will most likely be protein dependent, these results indicate that a particular level of energetic frustration is required for fast elongation to promote folding. We speculate that strengthening the misfolded state interactions results in slower interconversion rates between the states, thereby shifting the system into a kinetically controlled regime during protein synthesis.
Figure 6.
The probability of cotranslational folding probability as a function of elongation kinetics and interresidue interaction strength εm. One-hundred protein synthesis simulations were carried out for each value of εm and uniform amino acid addition time, while keeping εn fixed at the tuned value of 1.5 kcal/mol. The amino acid addition time is rescaled by τf = 10 ns, the mean folding time of this protein off the ribosome. The size of each point, and the number close to it, represent the fraction of folded molecules (state N in Fig. 3) at the end of synthesis (residue 129). Speeding up elongation increases the fraction of folded molecules when εm > 1.5 kcal/mol (e.g., going from 15 to 0.15 at εm = 2.0 or 2.5 kcal/mol).
Robustness with respect to changes in εn
To ensure our main conclusion is robust with respect to the choice of εn, we varied εn between 1.3 and 1.5 kcal/mol and εm between 1.3 and 2.2 kcal/mol, and synthesized the protein at the uniformly fast and 10-fold slower elongation schedules (Schedules s2 and s10 in Fig. S4 c). We computed the difference in the resulting folding probabilities at the last codon position. If fast translation promotes folding, then this difference will be positive. We find that at all εn values there exist multiple εm values for which fast translation promotes folding (Fig. S10), indicating this scenario can arise regardless of the choice of the model protein’s residue-residue interaction energies.
Posttranslational protein behavior can be influenced by cotranslational events
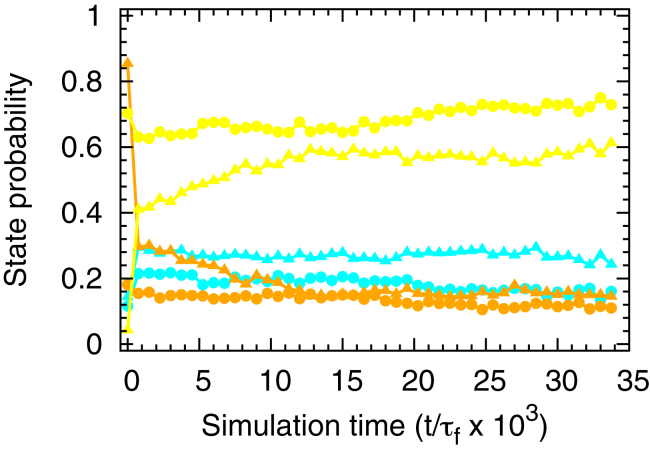
To examine if the elongation history affects the posttranslational behavior of our model protein, we continued the simulations after the nascent protein had completed synthesis and had been released from the ribosome. As before, we rescaled the time-dependent results from the posttranslational simulations by the fast-phase folding time of this protein (1.0 ns) to provide a reduced timescale that is unaffected by the accelerated dynamics characteristic of coarse-grained models. Posttranslational simulations were started from the ensemble of nascent-chain conformations reached at the end of the nonuniform, fast and nonuniform, slow synthesis simulations (schedules s5 and s39 in Fig. S4 c) because they resulted in the highest and lowest cotranslational folding probabilities (0.69 vs. 0.01; Fig. 4 b). During the posttranslational simulations we observe that, whereas the fraction of folded molecules increases over time, differences between the folding probabilities persist during the 35-μs posttranslational simulation (Fig. 7). We also simulated the protein’s posttranslational behavior starting from the conformational ensembles populated at the end of the uniformly fast and slow elongation schedules. For this case, there is very little difference in the protein’s postsynthesis behavior during the 9-μs simulation (Fig. S7), likely due to a smaller difference between the starting folding probabilities (0.51 vs. 0.32; Fig. 4 a).
Figure 7.
Posttranslational protein dynamics. Shown here is the time evolution of the probability of populating state M (orange), C (cyan), and N (yellow) after the nascent chain has been released from the ribosome. The simulation time t is rescaled by the mean folding time of the fast-kinetic phase of this protein off the ribosome (τf = 10 ns). The postsynthesis simulations are started from the conformations that the nascent protein attains at the end of the two synthesis simulations reported in Fig. 4b. Circles and triangles denote, respectively, simulations started after the nonuniform, fast elongation schedule (s5 in Fig. S4 c) and the nonuniform, slow elongation schedule (s39 in Fig. S4 c). The probabilities of populating states U and D are close to zero, and are therefore not plotted.
These results demonstrate that in some cases the influence of the translation-elongation schedule can persist long after a protein has been synthesized. This implies that the fraction of posttranslational proteins that are misfolded can be modulated through the appropriate choice of the elongation schedule. Consistent with our results, experiments have found that synonymous codon substitutions altered an antibody’s posttranslational binding affinity and aggregation propensity (51), suggesting that the antibody’s cotranslational folding was perturbed by altered translation kinetics.
The same effects are observed in another artificial misfolding-prone protein
To test whether the main conclusions of this study can apply to other proteins, we constructed an additional misfolding-prone system by combining the single-basin Gō models of PDBs:1ZZV (structure n) and 1ND9 (structure m). The contact map of the resulting coarse-grained model (Fig. S8) is characterized by the optimal value s = 1 (see definition in the Materials and Methods). The force-field parameters εn and εm (see Eqs. S2 and S3), which describe the strengths of the contacts in Fig. S8, were tuned to values of 1.6 and 1.48 kcal/mol, respectively. The resulting coarse-grained protein model can populate a folded state composed of 80 residues with a α/β topology, or a misfolded state composed of 49 residues with α-helical topology. In REX simulations without the ribosome, the probabilities of the native and misfolded states are, respectively, 0.98 and 0.02 at 310 K.
The protein predominantly populates the misfolded state at nascent chain lengths between 70 and 105 residues, according to the arrested-ribosome simulations (Fig. S9, a and b). Therefore, we simulated synthesis of this protein using two elongation schedules whose rates differ only in the misfolding-prone region (Fig. S9 c). When elongation is faster, we observe a fourfold increase in the probability of cotranslational folding compared to when synthesis is slower (0.36 versus 0.08, Fig. S9 d). These results indicate that fast-translating codons can help proteins with different native and misfolded topologies to fold cotranslationally.
The ratio of simulation timescales are relevant to one-third of the E. coli proteome
A question of fundamental importance concerning the validity of our results is whether the ratio of timescales of amino acid addition to domain folding is realistic, because this ratio influences how far from equilibrium cotranslational folding is in our simulations, and consequently the importance of kinetic control to the folding process. In our simulations, amino-acid addition occurred on timescales ranging from 0.03 to 15 ns, and the mean folding time of the fast-kinetic phase was 1.0 ns off the ribosome (Fig. S2). Thus, this ratio varies from 0.03 to 15 in our simulations. To estimate the fraction of the cytosolic E. coli proteome that falls within this range we utilized the estimated domain folding times from the structural and kinetic database in Ciryam et al. (15), which reports the folding times for 1236 domains from 758 cytosolic proteins. Assuming the E. coli’s average codon translation time is 0.10 s, allowing us to compute the ratio of amino-acid addition to folding time of each domain in the database. We find that 37% of the proteome falls within the simulated ratio of timescales, indicating that by this metric, our simulations are kinetically relevant to an appreciable fraction of E. coli proteins.
Conclusions
In this study, we have introduced, to our knowledge, a new protocol for incorporating misfolding into structure-based coarse-grained models of proteins. This method allows information from two experimental structures to be incorporated into the parameterization of the force field, to maintain the structural fidelity of the alternative tertiary structures and to tune the relative stability of these ordered states. Our approach can be used with publicly available force fields (e.g., CHARMM, AMBER), without the need for modification to their functional forms or implementation of multi-Hamiltonian simulations. Although we modeled artificial misfolding-prone proteins, our approach can be applied to real misfolding-prone proteins, provided high-resolution structures of both the native and misfolded states are available. A key challenge in this latter area is that soluble, misfolded structures have not been characterized at high enough resolution in the experimental literature to be used as inputs for our modeling strategy.
We applied this coarse-grained methodology to understand at the molecular level how fast-translation can promote protein folding, complementing the predictions obtained from a previous kinetic model (31). We simulated the synthesis of two proteins, each able to populate different native and misfolded states, using a variety of translation-elongation schedules. We also simulated the posttranslational behavior of one of them. Our results indicate that because these nascent proteins can be trapped in long-lived nonnative states, the outcome of synthesis (state probabilities and folding pathways) strongly depends on the distribution of amino acid-addition rates during elongation. Specifically, in the nonequilibrium regime, the simulated nascent proteins are more likely to reach the native state at the end of synthesis if elongation across the misfolding-prone segments is fast, consistent with the chemical kinetic predictions (31). This trend is also observed when the entire protein is synthesized quickly, which is qualitatively consistent with lower levels of aggregation observed experimentally upon speedup of translation in yeast and worms (47). Fast elongation rates are beneficial not only for folding in difficult-to-fold regions, but a suitable combination of fast and slow elongation rates can also maximize cotranslational folding. Conversely, we observe that even a single slow-translating codon may result in very high levels of nascent-protein misfolding (Fig. S4 b) whose effect can persist after the protein has been released from the ribosome (Fig. 7).
Deviations from these trends are mainly observed when synthesis of the nascent protein is slow enough to switch the protein’s behavior from a nonequilibrium regime (i.e., kinetic control) to a quasi-equilibrium regime (i.e., thermodynamic control). Because the kinetic regime that a nascent chain experiences is a complex function of the various timescales associated with translation and cotranslational folding (26), not all proteins that are misfolding-prone will benefit from fast elongation. However, we have estimated that up to 37% of cytosolic E. coli proteins have values of the ratio of folding time to elongation time that fall within the range of ratios used in our simulations. This suggests that the kinetic partitioning mechanism we have observed in our simulations may be relevant to an appreciable subset of proteins.
The results of our simulations confirm the kinetic partitioning mechanism identified previously in a chemical kinetic model. By utilizing a coarse-grained description of the nascent protein and the ribosome, we have explored this phenomenon using a more realistic model, which is able to capture energetic and structural details of cotranslational folding, and that results in a multiplicity of folding pathways that are absent in the kinetic model. The coarse-grained model has provided a number of additional insights including a molecular view of the coupling of cotranslational folding and elongation schedules, how individual pathway probabilities are affected by 40 different elongation schedules, how increasing energetic frustration shifts the protein from a regime in which accelerating translation elongation hinders folding to one in which it helps, and the impact of elongation on posttranslational protein behavior.
How broadly our conclusions apply to other proteins depends on additional factors, including the structural details of the native and misfolded states and the location of the misfolding-prone segment along the protein’s sequence. Therefore, we do not expect that all misfolding-prone proteins will benefit from fast translation elongation rates. For example, a protein with a misfolding segment located at its C terminus would not benefit from the kinetic partitioning mechanism we have identified because both the misfolding-prone region and the fully synthesized domain would simultaneously emerge from the exit tunnel. An interesting question for future research is to understand if the use of fast-translating codons is a wide-spread evolutionary strategy to prevent aggregation caused by cotranslational protein misfolding. Answering this question will require either the application of our approach to a large, diverse set of different proteins or high-throughput experimental methods.
Author Contributions
E.P.O. and F.T. designed research. F.T. performed the research. E.P.O. and F.T. contributed analytical tools. F.T. analyzed data. E.P.O. and F.T. wrote the paper.
Acknowledgments
F.T. thanks Giordano Fumagalli for the support provided with the statistical analysis.
Editor: Rohit Pappu.
Footnotes
Supporting Materials and Methods and ten figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30393-4.
Supporting Material
References
- 1.Ueda Y., Taketomi H., Go N. Studies on protein folding, unfolding, and fluctuations by computer simulation. II. A. Three-dimensional lattice model of lysozyme. Biopolymers. 1978;17:1531–1548. [Google Scholar]
- 2.Hills R.D., Jr., Brooks C.L., 3rd Insights from coarse-grained Gō models for protein folding and dynamics. Int. J. Mol. Sci. 2009;10:889–905. doi: 10.3390/ijms10030889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryngelson J.D., Onuchic J.N., Wolynes P.G. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 4.Thirumalai D., Klimov D.K. Deciphering the timescales and mechanisms of protein folding using minimal off-lattice models. Curr. Opin. Struct. Biol. 1999;9:197–207. doi: 10.1016/S0959-440X(99)80028-1. [DOI] [PubMed] [Google Scholar]
- 5.Roy M., Chavez L.L., Jennings P.A. The native energy landscape for interleukin-1β. Modulation of the population ensemble through native-state topology. J. Mol. Biol. 2005;348:335–347. doi: 10.1016/j.jmb.2005.02.059. [DOI] [PubMed] [Google Scholar]
- 6.Craig P.O., Lätzer J., Wolynes P.G. Prediction of native-state hydrogen exchange from perfectly funneled energy landscapes. J. Am. Chem. Soc. 2011;133:17463–17472. doi: 10.1021/ja207506z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng W., Borgia A., Best R.B. Probing the action of chemical denaturant on an intrinsically disordered protein by simulation and experiment. J. Am. Chem. Soc. 2016;138:11702–11713. doi: 10.1021/jacs.6b05443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O’Brien E.P., Ziv G., Thirumalai D. Effects of denaturants and osmolytes on proteins are accurately predicted by the molecular transfer model. Proc. Natl. Acad. Sci. USA. 2008;105:13403–13408. doi: 10.1073/pnas.0802113105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Best R.B., Chen Y.-G., Hummer G. Slow protein conformational dynamics from multiple experimental structures: the helix/sheet transition of arc repressor. Structure. 2005;13:1755–1763. doi: 10.1016/j.str.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Okazaki K., Koga N., Wolynes P.G. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: structure-based molecular dynamics simulations. Proc. Natl. Acad. Sci. USA. 2006;103:11844–11849. doi: 10.1073/pnas.0604375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schug A., Whitford P.C., Onuchic J.N. Mutations as trapdoors to two competing native conformations of the Rop-dimer. Proc. Natl. Acad. Sci. USA. 2007;104:17674–17679. doi: 10.1073/pnas.0706077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuckerman D.M. Simulation of an ensemble of conformational transitions in a united-residue model of calmodulin. J. Phys. Chem. B. 2004;108:5127–5137. [Google Scholar]
- 13.Camilloni C., Sutto L. Lymphotactin: how a protein can adopt two folds. J. Chem. Phys. 2009;131:245105. doi: 10.1063/1.3276284. [DOI] [PubMed] [Google Scholar]
- 14.Ramírez-Sarmiento C.A., Noel J.K., Artsimovitch I. Interdomain contacts control native state switching of RfaH on a dual-funneled landscape. PLoS Comput. Biol. 2015;11:e1004379. doi: 10.1371/journal.pcbi.1004379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciryam P., Morimoto R.I., O’Brien E.P. In vivo translation rates can substantially delay the cotranslational folding of the Escherichia coli cytosolic proteome. Proc. Natl. Acad. Sci. USA. 2013;110:E132–E140. doi: 10.1073/pnas.1213624110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Brien E.P., Ciryam P., Dobson C.M. Understanding the influence of codon translation rates on cotranslational protein folding. Acc. Chem. Res. 2014;47:1536–1544. doi: 10.1021/ar5000117. [DOI] [PubMed] [Google Scholar]
- 17.Nilsson O.B., Hedman R., von Heijne G. Cotranslational protein folding inside the ribosome exit tunnel. Cell Reports. 2015;12:1533–1540. doi: 10.1016/j.celrep.2015.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedyukina D.V., Cavagnero S. Protein folding at the exit tunnel. Annu. Rev. Biophys. 2011;40:337–359. doi: 10.1146/annurev-biophys-042910-155338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Brien E.P., Vendruscolo M., Dobson C.M. Prediction of variable translation rate effects on cotranslational protein folding. Nat. Commun. 2012;3:868. doi: 10.1038/ncomms1850. [DOI] [PubMed] [Google Scholar]
- 20.Trovato F., O’Brien E.P. Insights into cotranslational nascent protein behavior from computer simulations. Annu. Rev. Biophys. 2016;45:345–369. doi: 10.1146/annurev-biophys-070915-094153. [DOI] [PubMed] [Google Scholar]
- 21.Yu C.-H., Dang Y., Liu Y. Codon usage influences the local rate of translation elongation to regulate co-translational protein folding. Mol. Cell. 2015;59:744–754. doi: 10.1016/j.molcel.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Brien E.P., Christodoulou J., Dobson C.M. New scenarios of protein folding can occur on the ribosome. J. Am. Chem. Soc. 2011;133:513–526. doi: 10.1021/ja107863z. [DOI] [PubMed] [Google Scholar]
- 23.Trovato F., Tozzini V. Diffusion within the cytoplasm: a mesoscale model of interacting macromolecules. Biophys. J. 2014;107:2579–2591. doi: 10.1016/j.bpj.2014.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheung M.S. Where soft matter meets living matter--protein structure, stability, and folding in the cell. Curr. Opin. Struct. Biol. 2013;23:212–217. doi: 10.1016/j.sbi.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyeon C., Thirumalai D. Capturing the essence of folding and functions of biomolecules using coarse-grained models. Nat. Commun. 2011;2:487. doi: 10.1038/ncomms1481. [DOI] [PubMed] [Google Scholar]
- 26.Sharma A.K., Bukau B., O’Brien E.P. Physical origins of codon positions that strongly influence cotranslational folding: a framework for controlling nascent-protein folding. J. Am. Chem. Soc. 2016;138:1180–1195. doi: 10.1021/jacs.5b08145. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka T., Hori N., Takada S. How co-translational folding of multi-domain protein is affected by elongation schedule: molecular simulations. PLOS Comput. Biol. 2015;11:e1004356. doi: 10.1371/journal.pcbi.1004356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang E., Wang J., Xiao Y. Computational evidence that fast translation speed can increase the probability of cotranslational protein folding. Sci. Rep. 2015;5:15316. doi: 10.1038/srep15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elcock A.H. Molecular simulations of cotranslational protein folding: fragment stabilities, folding cooperativity, and trapping in the ribosome. PLOS Comput. Biol. 2006;2:e98. doi: 10.1371/journal.pcbi.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B., Miller T.F., 3rd Long-timescale dynamics and regulation of sec-facilitated protein translocation. Cell Reports. 2012;2:927–937. doi: 10.1016/j.celrep.2012.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Brien E.P., Vendruscolo M., Dobson C.M. Kinetic modelling indicates that fast-translating codons can coordinate cotranslational protein folding by avoiding misfolded intermediates. Nat. Commun. 2014;5:2988. doi: 10.1038/ncomms3988. [DOI] [PubMed] [Google Scholar]
- 32.Buhr F., Jha S., Komar A.A. Synonymous codons direct cotranslational folding toward different protein conformations. Mol. Cell. 2016;61:341–351. doi: 10.1016/j.molcel.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nedialkova D.D., Leidel S.A. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Brien E.P., Christodoulou J., Dobson C.M. Trigger factor slows co-translational folding through kinetic trapping while sterically protecting the nascent chain from aberrant cytosolic interactions. J. Am. Chem. Soc. 2012;134:10920–10932. doi: 10.1021/ja302305u. [DOI] [PubMed] [Google Scholar]
- 35.Chaney J.L., Clark P.L. Roles for synonymous codon usage in protein biogenesis. Annu. Rev. Biophys. 2015;44:143–166. doi: 10.1146/annurev-biophys-060414-034333. [DOI] [PubMed] [Google Scholar]
- 36.Tozzini V. Coarse-grained models for proteins. Curr. Opin. Struct. Biol. 2005;15:144–150. doi: 10.1016/j.sbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Trovato F., Nifosì R., Tozzini V. A minimalist model of protein diffusion and interactions: the green fluorescent protein within the cytoplasm. Macromolecules. 2013;46:8311–8322. [Google Scholar]
- 38.Kouza M., Li M.S., Thirumalai D. Effect of finite size on cooperativity and rates of protein folding. J. Phys. Chem. A. 2006;110:671–676. doi: 10.1021/jp053770b. [DOI] [PubMed] [Google Scholar]
- 39.Wallin S., Chan H.S. Conformational entropic barriers in topology-dependent protein folding: perspectives from a simple native-centric polymer model. J. Phys. Condens. Matter. 2009;21 329801–329801. [Google Scholar]
- 40.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 41.Karanicolas J., Brooks C.L., 3rd The origins of asymmetry in the folding transition states of protein L and protein G. Protein Sci. 2002;11:2351–2361. doi: 10.1110/ps.0205402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chu J.-W., Voth G.A. Coarse-grained free energy functions for studying protein conformational changes: a double-well network model. Biophys. J. 2007;93:3860–3871. doi: 10.1529/biophysj.107.112060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanazono Y., Takeda K., Miki K. Structural studies of the N-terminal fragments of the WW domain: insights into co-translational folding of a β-sheet protein. Sci. Rep. 2016;6:34654. doi: 10.1038/srep34654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chow C.C., Chow C., Cavagnero S. Chain length dependence of apomyoglobin folding: structural evolution from misfolded sheets to native helices. Biochemistry. 2003;42:7090–7099. doi: 10.1021/bi0273056. [DOI] [PubMed] [Google Scholar]
- 45.Jennings P.A., Finn B.E., Matthews C.R. A reexamination of the folding mechanism of dihydrofolate reductase from Escherichia coli: verification and refinement of a four-channel model. Biochemistry. 1993;32:3783–3789. doi: 10.1021/bi00065a034. [DOI] [PubMed] [Google Scholar]
- 46.Burns-Hamuro L.L., Dalessio P.M., Ropson I.J. Replacement of proline with valine does not remove an apparent proline isomerization-dependent folding event in CRABP I. Protein Sci. 2004;13:1670–1676. doi: 10.1110/ps.03317804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veitshans T., Klimov D., Thirumalai D. Protein folding kinetics: timescales, pathways and energy landscapes in terms of sequence-dependent properties. Fold. Des. 1997;2:1–22. doi: 10.1016/S1359-0278(97)00002-3. [DOI] [PubMed] [Google Scholar]
- 48.Sugita Y., Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 49.Kim S.J., Yoon J.S., Skach W.R. Translational tuning optimizes nascent protein folding in cells. Science. 2015;348:444–448. doi: 10.1126/science.aaa3974. [DOI] [PubMed] [Google Scholar]
- 50.Thirumalai D., Klimov D.K., Woodson S.A. Kinetic partitioning mechanism as a unifying theme in the folding of biomolecules. Theor. Chem. Accounts Theory Comput. Model. 1997;96:14–22. [Google Scholar]
- 51.Hu S., Wang M., He M. Genetic code-guided protein synthesis and folding in Escherichia coli. J. Biol. Chem. 2013;288:30855–30861. doi: 10.1074/jbc.M113.467977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.