Summary
Promyelocytic leukemia protein (PML), the main constituent of PML nuclear bodies, regulates various physiological processes in different cell types. However, little is known about its functions in embryonic stem cells (ESC). Here, we report that PML contributes to ESC self-renewal maintenance by controlling cell-cycle progression and sustaining the expression of crucial pluripotency factors. Transcriptomic analysis and gain- or loss-of-function approaches showed that PML-deficient ESC exhibit morphological, metabolic, and growth properties distinct to naive and closer to the primed pluripotent state. During differentiation of embryoid bodies, PML influences cell-fate decisions between mesoderm and endoderm by controlling the expression of Tbx3. PML loss compromises the reprogramming ability of embryonic fibroblasts to induced pluripotent stem cells by inhibiting the transforming growth factor β pathway at the very early stages. Collectively, these results designate PML as a member of the regulatory network for ESC naive pluripotency and somatic cell reprogramming.
Keywords: embryonic stem cells, promyelocytic leukemia protein, pluripotency, differentiation, induced pluripotent stem cells
Graphical Abstract
Highlights
-
•
PML is essential for the maintenance of naive pluripotent cells
-
•
PML prevents the naive to primed pluripotency transition
-
•
PML influences cell-fate commitment through Tbx3 regulation
-
•
PML is required for iPSCs formation via regulation of TGF signaling pathway
Kretsovali and colleagues unravel an unknown function of PML in mESC pluripotency. PML maintains ESC self-renewal by controlling the cell cycle, sustaining pluripotency factor expression levels, and preventing the switch from naive to EpiLSC pluripotent state. It also influences cell-fate decision and reprogramming efficiency through Tbx3 and TGF-β regulation, respectively.
Introduction
Pluripotent stem cells (PSC) are characterized by the ability to self-renew indefinitely and to differentiate into all of the three germ lineages—ectoderm, mesoderm, and endoderm—of the developing embryo. During the early developmental stages, mouse embryonic stem cells (mESC) are derived from pre-implantation embryos (embryonic day 3.5 [E3.5]) and possess the “naive” or “ground” pluripotent state, while mouse epiblast stem cells (EpiSC) are isolated from the post-implantation epiblast [E6.5] and represent a more differentiated state, termed “primed.” Interestingly, mESC and EpiSC can be reciprocally converted into one another, using genetic and/or epigenetic modifiers (Chenoweth et al., 2010).
The maintenance of pluripotency is principally regulated by three transcription factors, OCT4, SOX2, and NANOG, which constitute the core pluripotency network. In addition, a growing number of proteins that positively or negatively modulate the function of the core complex have been identified. The importance of the intrinsic pluripotency regulatory network genes is further highlighted by their ability to reprogram somatic cells to induced pluripotent stem cells (iPSC), a cell type resembling ESC. Notably, genes important for stem cell pluripotency are also active in cancers (Hadjimichael et al., 2015), suggesting shared regulatory mechanisms.
The promyelocytic leukemia (Pml) gene was first described in the early 1990s as being targeted by a chromosomal translocation t(15;17) in acute promyelocytic leukemia (Guan and Kao, 2015) that produces an oncogenic PML-retinoic acid receptor α (RARα) fusion protein (PML-RARα). PML protein is the key organizer of spherical subnuclear structures, named PML nuclear bodies (PML-NBs), and is expressed in a variety of tissues. Its function has been thoroughly investigated in multiple cells and it is a critical player in DNA repair, apoptosis, senescence, oncogenesis, and cancer progression (Guan and Kao, 2015). However, its role in stem cells had been neglected until recent studies unveiled intriguing findings regarding PML involvement in the hematopoietic stem cell (HSC) asymmetric divisions and maintenance through activating fatty acid oxidation (FAO) (Nakahara et al., 2014). At the same time, recent reports using embryonal carcinoma cells and ESC showed that PML is involved in the activation of Oct4 gene expression (Chuang et al., 2011) and belongs to a transcriptional repressive complex that is associated with OCT4 and NANOG (Liang et al., 2008). Although these findings link PML with important pluripotency regulators, the exact role of PML in ESC functions has yet to be clarified.
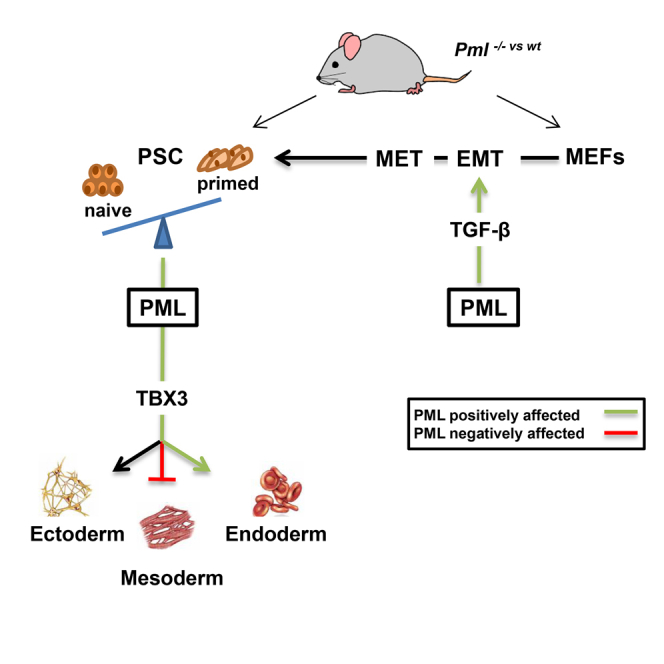
Here, we addressed the role of PML by investigating the phenotypes of ESC in which the expression of PML was experimentally up- or downregulated. We show that PML ablation impairs ESC self-renewal and pluripotency and promotes the transition from naive to primed-like pluripotent cell state. Moreover, PML depletion upregulates the expression of mesodermal markers and decreases the differentiation toward definitive endoderm. The effect of PML ablation on ESC differentiation can be rescued by TBX3 (a T-box transcription factor) overexpression. Finally, Pml−/− mouse embryonic fibroblasts (MEF) yield significantly fewer iPSC colonies compared with wild-type (WT) MEF, identifying PML as a pivotal mediator of somatic cell reprogramming.
Results
PML Is Essential for Self-Renewal and Pluripotency of ESC
Previous studies showed that ESC treatment with the histone deacetylase inhibitor trichostatin A (TSA) disrupts pluripotency and facilitates early differentiation events (Karantzali et al., 2008). Pml was included in the category of genes with reduced expression when cells started differentiating. To examine whether PML is functionally relevant in ESC, we monitored its expression levels upon induction of differentiation utilizing different approaches (embryoid bodies (EB) formation, leukemia inhibitory factor [LIF] withdrawal, retinoic acid, and TSA treatment), and in all cases we observed significantly reduced expression levels (data not shown).
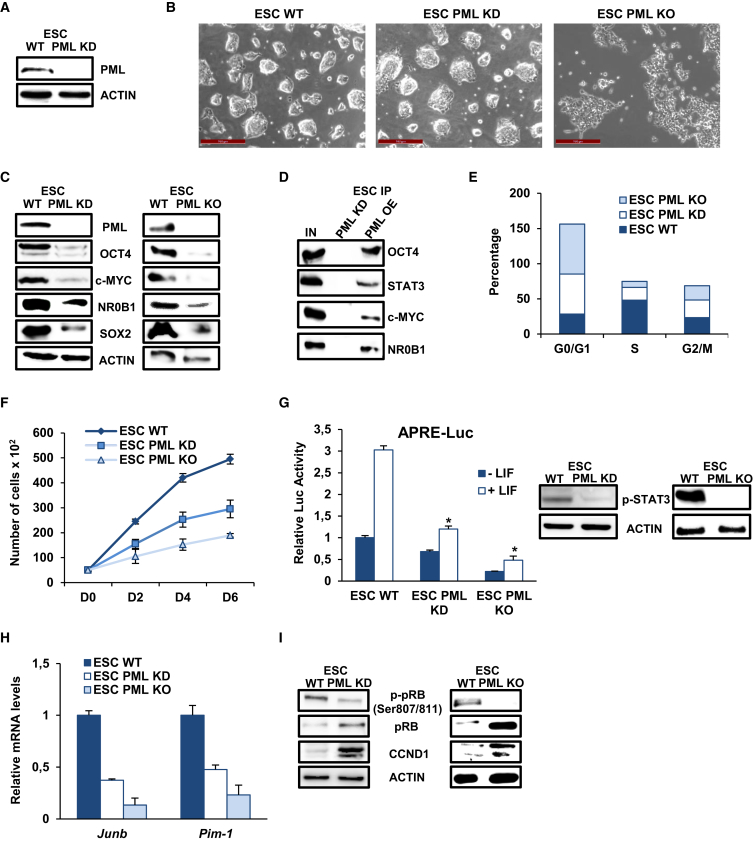
To elucidate the function of PML in the maintenance of ESC pluripotency, we first performed knockdown (KD) of PML using a short hairpin RNA (shRNA) lentiviral vector. The efficiency of PML KD was examined in three independent PML KD ESC lines, the most representative of which is shown in Figure 1A. PML KD cells propagated in both fetal bovine serum (FBS)/LIF and 2i/LIF culture conditions and sustained their ability to form dome-shaped colonies (Figures 1B and S1A). Although no morphological changes were noticed, the expression of several pluripotency factors was significantly diminished in PML KD ESC (OCT4, SOX2, c-MYC, and NR0B1) (Figure 1C). These findings were consistently observed in another ESC cell line (E14) following PML deletion (Figure S1B). To better delve into the effects of PML ablation, we isolated PML knockout (KO) ESC from Pml−/− mice. These cells displayed distinguishable flattened morphology (Figures 1B and S1A), much slower propagation in naive culture conditions, and greater reduction of pluripotency factor expression levels (Figures 1C and S1C). To further delineate the effect of PML, we generated a stable PML-overexpressing (OE) ESC line using Pml expressing vector and detected an important induction of pluripotency marker expression (Figure S1D).
Figure 1.
PML Depletion Impairs Self-Renewal and Affects the Cell Cycle
(A) PML protein level in one representative PML KD ESC clone and wild type (WT) ESC.
(B) Morphology of control (WT), PML KD, and KO ESC. Scale bar, 100 μm.
(C) Pluripotency factors protein levels upon depletion or deletion of PML.
(D) Co-immunoprecipitation of endogenous pluripotency factors (OCT4, c-MYC, STAT3, NR0B1) with endogenous PML from PML OE ESC compared with PML KD ESC.
(E) Cell-cycle analysis of WT, PML KD, and KO ESC.
(F) Growth curve of WT, PML KD, and KO ESC. Error bars indicate ±SD of four independent experiments (n = 4).
(G) The activity of APRE-Luc reporter and p-STAT3 protein expression levels in ESC WT, PML KD, and KO. Data represent the mean + SD of four independent experiments (n = 4). ∗p < 0.05.
(H) Relative mRNA levels of cell-cycle regulators Junb and Pim-1. Data are shown as mean + SD of three independent experiments (n = 3).
(I) Protein levels of cell-cycle regulators p-pRB, pRB, and CCND1 were detected.
An earlier study demonstrated that PML physically associates with NANOG and OCT4 (Liang et al., 2008). Our co-immunoprecipitation experiments using PML OE ESC verified the OCT4 interaction and further showed that PML associates with STAT3, c-MYC, and NR0B1, three essential regulators of the naive pluripotent state (Figure 1D). Considering the association with and the regulation of the expression of essential factors, we deduce that PML is involved in the maintenance of ESC stemness.
Recently it was reported that Pml-deficient MEF proliferate faster than WT MEF (Tang et al., 2013), which is in agreement with previous data showing that PML inhibits cell proliferation in cancer cells (Guan and Kao, 2015). In contrast to these observations, we noted that PML KD and KO ESC were passaged every 3 and 5 days, respectively, compared with WT (passaged every 2 days); thus we proceeded to analyze the cell-cycle phase structure in the respective cells. Interestingly, an obvious increase of the cell fraction in the G1 phase accompanied by a reduction of the S phase, but no significant alteration in G2/M phase, was detected in PML KD and KO ESC (Figures 1E and S1E). Corroborating these results, the cell proliferation rate showed a significant delay in PML KD and KO ESC (Figure 1F), while PML OE cells exhibited faster proliferation (Figure S1F).
LIF/STAT3 signaling is important for ESC-specific cell-cycle progression (rapid G1-S transition) through activation of cell-cycle regulators (e.g., Jun-b, c-myc, Pim-1) (Aksoy et al., 2007, Coronado et al., 2013, White and Dalton, 2005). We therefore examined the effect of PML on LIF/STAT3 signaling pathway using a STAT-dependent reporter (APRE-Luc), and noticed reduced activity in the absence of PML (Figure 1G) and increased activity in PML OE cells (Figure S1F). Further confirmation of this influence is provided by decreased p-STAT3 protein levels in PML KD and KO ESC (Figure 1G), whereas PML OE cells display increased levels (Figure S1G). c-Myc, among other LIF/STAT3 targets, is required for ESC self-renewal and drives cell-cycle progression (Singh et al., 2015). In addition to c-Myc (Figure 1A), Jun-b and Pim-1 mRNA levels were analyzed in PML KD and KO ESC and were notably lower in comparison with WT ESC (Figure 1H). Thus, both LIF/STAT3 and Myc-mediated mechanisms may account for the decreased cell-cycle progression upon PML ablation.
To follow up this hypothesis, we examined the impact of PML on the retinoblastoma (pRB) tumor suppressor and cell-cycle regulator. In mESC, pRB is inactivated by hyperphosphorylation and allows high E2F activity. Upon differentiation, pRB is activated and the G1-S checkpoint is established (Sage, 2012). We found that PML KD and KO ESC are characterized by reduced p-pRB protein levels as well as increased levels of total pRB (Figure 1I). In addition, the expression of CYCLIN D1 (CCND1) that is normally induced in differentiating cells (Savatier and Malashicheva, 2004) is also elevated in PML KD and KO cells (Figure 1I). Therefore, we conclude that PML influences ESC self-renewal and pluripotency as well as the G1-S transition of cell cycle.
PML Maintains the Naive Pluripotent State
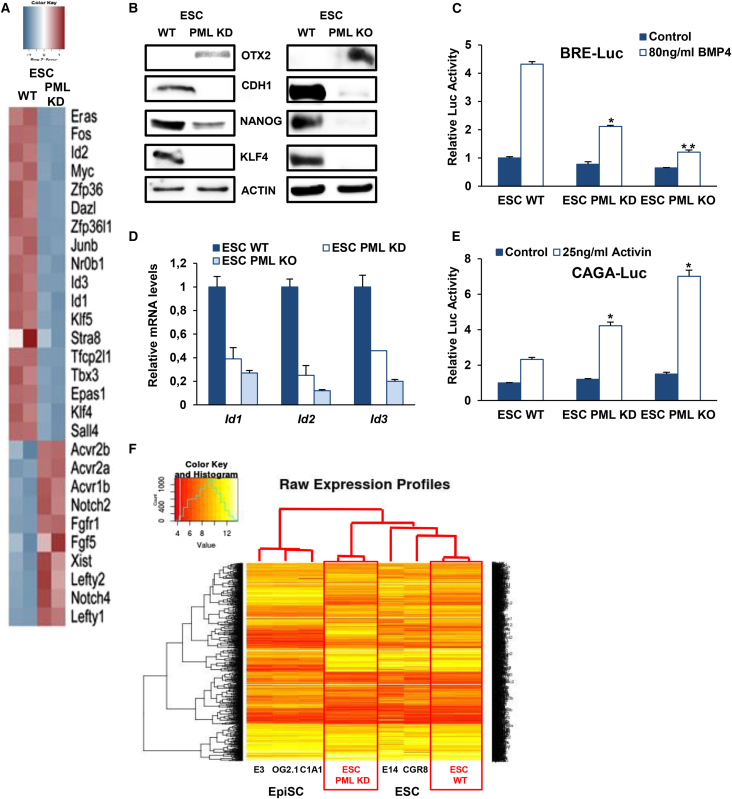
To better understand how PML depletion affects ESC identity, we performed genome-wide expression analysis from PML KD and WT ESC. As shown in Figure S2A, 3,088 genes are differentially expressed: 1,009 are upregulated (fold change > 1.5, p < 0.05) and 2,079 are downregulated (fold change < −1.5, p < 0.05) upon PML depletion. Importantly, the functional analysis using the Regulatory Network Enrichment Analysis (RNEA) software (Chouvardas et al., 2016) revealed that several signaling pathways implicated in ESC pluripotency are affected. Namely, in the absence of PML, naive pluripotency-associated bone morphogenetic protein 4 (BMP4), LIF/STAT3, and phosphatidylinositol 3-kinase (PI3K) signaling (Figures S2A and S2B) pathways are reduced, whereas the pro-differentiation ACTIVIN and fibroblast growth factor (FGF) pathways (Figure S2A), which are required for the maintenance of primed pluripotent state, are enriched. Moreover, many genes related to cell cycle (Figure S2B), chromatin organization, and DNA repair are also affected by PML depletion, suggesting that PML contributes to maintenance of ESC properties. Most importantly, several naive characteristic genes (Nr0b1, Tbx3, Klf4, Klf5) are downregulated in PML KD ESC, whereas representative primed state markers are elevated (Fgf5, Lefty1, Lefty2) (Figure 2A). These data were confirmed using RT-PCR in both PML KD and KO ESC (Figure S2C). In addition, the protein levels of OTX2, an important brain development transcription factor that is critical for ESC conversion into EpiSC (Acampora et al., 2013), are increased (Figure 2B). On the contrary, the protein levels of the essential naive state regulators NANOG, KLF4, and cadherin 1 (CDH1), recently reported to drive the transition from primed to naive pluripotency (Murayama et al., 2015), are sharply diminished (Figure 2B). Next, we verified that PML ablation negatively affects BMP4 signaling by reducing the activity of BRE-Luc reporter and the expression levels of its transcriptional targets Ids 1-3 (Figures 2C and 2D). Contrary to the effect on BMP4 and LIF/STAT3 signaling pathways, PML loss enhances the activity of Smad2/3-responsive element CAGA-Luc reporter upon stimulation with 25 ng/μL ACTIVIN A (Figure 2E). On the other hand, upon PML forced expression, naive and primed markers were raised and lessened, respectively (Figure S2D), in agreement with assays monitoring the activity of corresponding signaling pathways (Figure S2D). Our aforementioned data concerning the cell-cycle distribution are in full agreement with a recent study presenting that the switch from naive to primed state is related to increased duration of G1 phase (Coronado et al., 2013).
Figure 2.
PML Reduction Promotes an Epi-like Stem Cell State
(A) Heatmap with the top representative naive and primed genes.
(B) OTX2, CDH1, NANOG, and KLF4 protein levels prior to and after PML loss in ESC.
(C) Luciferase activity of BRE-Luc reporter upon PML depletion. Data represent the mean + SD of four independent experiments (n = 4). ∗p < 0.05, ∗∗p < 0.01.
(D) mRNA levels of BMP signaling target genes (Id1, Id2, Id3) in WT, PML KD, and KO ESC. Data are shown as mean + SD of three independent experiments (n = 3).
(E) CAGA-Luc reporter activity upon PML ablation. Error bars indicate +SD of four independent experiments (n = 4). ∗p < 0.05.
(F) Heatmap of the gene expression profiles of our WT and PML KD ESC replicates along with three EpiSC and two ESC lines.
Considering the different metabolic profiles of the two pluripotent states (Zhou et al., 2012) and the function of PML as an activator of FAO in HSC (Ito et al., 2012), we analyzed the oxygen consumption rate and the mitochondrial membrane potential (using tetramethylrhodamine methyl ester dye, TMRE) upon PML deletion. Surprisingly, PML loss in ESC leads to lower mitochondrial respiration and membrane potential compared with WT ESC (Figures S3A–S3C). This reduction is also correlated by the decrease of several FAO and oxidative phosphorylation (OXPHOS) enzyme genes in PML KD ESC (Figure S3D), in line with the metabolic changes upon ESC differentiation.
We next assessed whether PML KD cells show characteristics similar to those of other well-described EpiSC lines, including E3, OG2.1, and C1A1 (GEO: GSM699680, GSM699678, GSM699683). As shown in Figure 2F, PML KD cells were found to localize between the EpiSC and the ESC clusters (GEO: GSM1053554, GSM921484), indicating that these cells exit from naive pluripotency.
Our data illustrate that PML ensures naive pluripotency as manifested by changes in characteristic markers, signaling pathway activity, and cell metabolism following PML perturbations.
PML Impedes the Conversion from Naive ESC to EpiSC
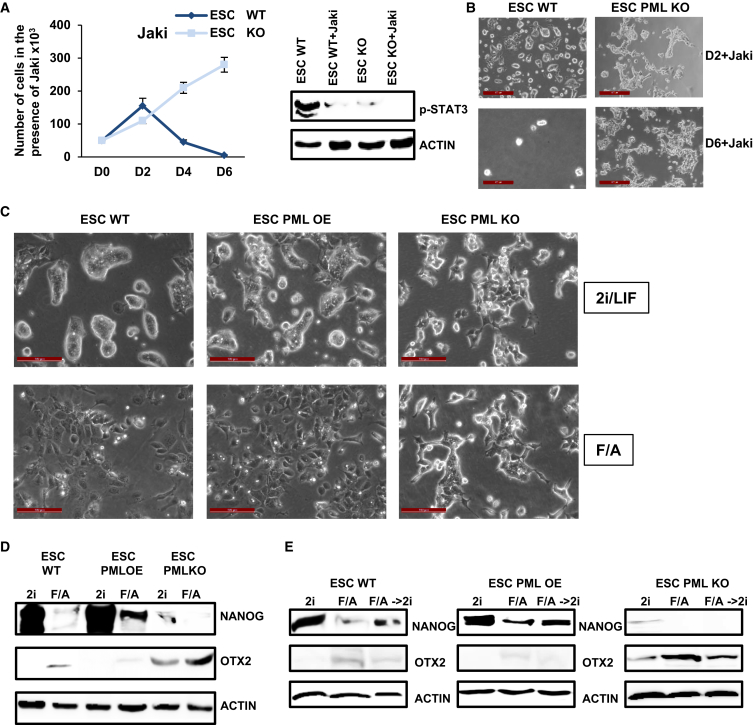
To gain more insight into the nature of Pml−/− ESC, we cultured WT and PML KO ESC in the presence of JAK inhibitor for 6 days (JAK inhibitor prevents the activation of the LIF/STAT3 pathway). PML KO, but not WT ESC, proliferated normally in this condition as shown by growth curves and cell morphology (Figures 3A and 3B), demonstrating that they behave similarly to the EpiSC and do not depend on the LIF/STAT3 pathway.
Figure 3.
PML Impedes the Conversion from Naive ESC to EpiSC
(A) Proliferation of WT and PML KD ESC with or without JAKi. Western blot showing JAKi abrogation of p-STAT3. Data represent the mean ± SD of three independent experiments (n = 3). D, day.
(B) Morphology of WT and PML KO ESC after 2 and 6 days of JAKi treatment. Scale bars, 100 μm. D, day.
(C) WT, PML KO, and PML OE ESC morphology at 6 days under 2i/LIF or F/A culture conditions. Scale bars, 100 μm.
(D) NANOG and OTX2 protein levels in WT, PML OE, and PML KO ESC. Cells were cultured either in 2i/LIF or F/A for 6 days.
(E) Western blot analysis of NANOG and OTX2 in WT, PML OE, and PML KO ESC. Cells were cultured in 2i/LIF (6 days), F/A (6 days), and F/A→ 2i/LIF (6 days F/A plus 6 days 2i/LIF).
To further evaluate whether PML has a functional role in the narrow window of ESC to EpiSC transition, we treated PML KO, OE, and WT ESC with FGF-β/ACTIVIN (F/A), previously shown to permit in vitro EpiSC derivation and maintenance. At day 6 of this conversion, WT and PML OE cells changed from dome-shaped to EpiSC-like flat-shaped colonies (Figure 3C), whereas KO cell morphology remained unchanged (Figure 3C). In control cells, NANOG and Klf4 were decreased, while OTX2 and Fgf5 were raised (Figures 3D and S3E). The silencing of PML led to a more pronounced increase of OTX2 and Fgf5, while NANOG and Klf4 were lower (Figures 3D and S3E). Conversely, PML OE ESC exit from the ground pluripotent state is significantly delayed according to the above markers (Figures 3D and S3E). These results indicate that PML KO cells are closer to EpiSC than to ESC and that forced expression of PML partially prevents the conversion to EpiSC.
We then examined whether these ESC-derived EpiLSC following propagation (at least three passages) in F/A could be reverted to naive ESC by culturing in 2i/LIF. Molecular marker analysis of PML OE cells, 6 days after this transition, showed an important induction of naive and lessening of primed markers (Figures 3E and S3F). PML KO cells exhibited no significant alterations during this transition (Figures 3E and S3F). Collectively, these data indicated that PML overexpression may favor the reversion from the EpiSC to naive pluripotent state.
Deregulated Expression Levels of PML Influence Lineage Specifications In Vitro
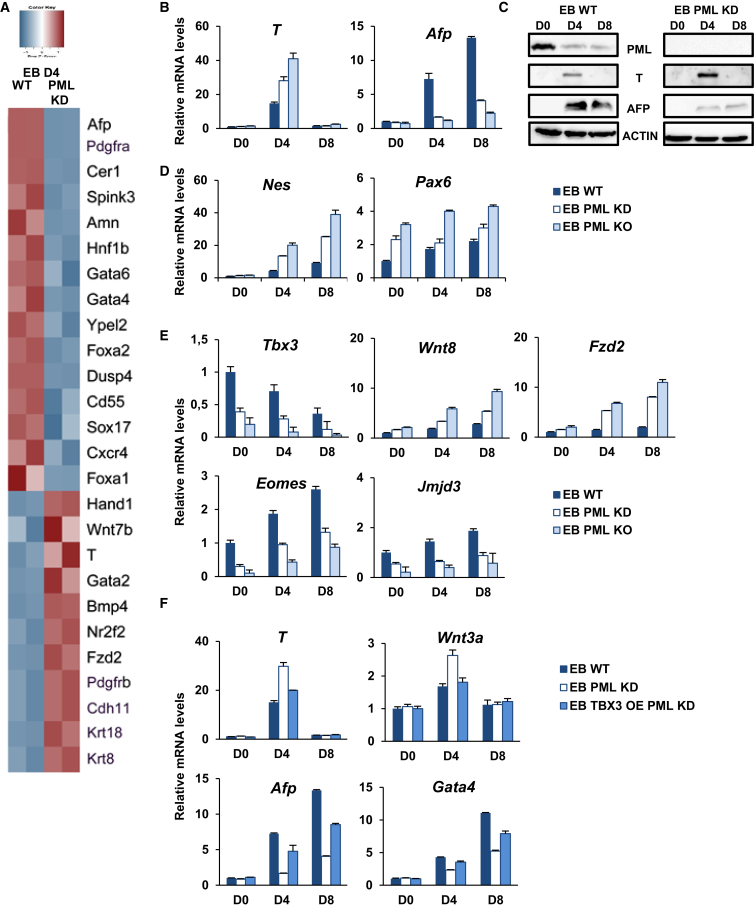
To study the role of PML in ESC fate decisions, we induced differentiation of PML KD and WT ESC through EB formation and then analyzed the gene expression profile on EB day 4, using cDNA microarray. Upon PML KD ESC differentiation, 836 genes are upregulated (fold change > 1.5, p < 0.05) and 400 genes are downregulated (fold change < −1.5, p < 0.05) (Figure S4A). Importantly, according to gene ontology (GO) (Figure S4A), PML KD preferentially induces mesodermal and represses endodermal commitment (Figure 4A). The representative differentiation marker levels were also corroborated through RT-PCR and western blot analyses in both PML KD and KO cells (Figures 4B and 4C). In agreement with the above, the reduction of c-Myc, Nr0b1, and Nanog at EB day 4 in PML KD and KO ESC—factors known to inhibit endodermal commitment—is delayed, indicating a facilitation of endodermal differentiation (Figure S4B) (Zhang et al., 2014). Concerning ectodermal differentiation only a few markers show a notable change, the most remarkable being the upregulation of Nestin (Nes) (Figure 4D). Strikingly, Pax6, a well-studied neuroectodermal marker, is, unusually, already expressed in the undifferentiated PML KD and KO ESC (Figure 3D). This derepression is in line with the study by Regad et al. (2009), in which it was demonstrated that PML is essential for the proper transition of neuroblasts to intermediate progenitors. On the contrary, PML OE favors the induction of endoderm (Afp) differentiation markers (Figure S4C) and at the same time enhances the suppression of mesodermal (T) and ectodermal (Nes) markers (Figures S4C). Hence, our data suggest that PML is essential for proper ESC differentiation toward the three germ layers. Specifically, PML promotes endodermal differentiation and limits mesodermal lineages.
Figure 4.
PML Inhibition Promotes Mesoderm and Represses Endoderm Differentiation
(A) Heatmap displaying top endodermal genes and mesodermal genes differentially expressed in PML KD and WT EB D4 (fold change > 1.5, p < 0.05).
(B) Validation of microarray results using RT-PCR in PML KD, KO, and WT EB. Error bars indicate +SD of four independent experiments (n = 4).
(C) Western blot analysis of PML, T, and α-fetoprotein (AFP) expression in PML KD and KO compared with WT EB.
(D) Nes and Pax6 mRNA levels upon differentiation of PML KD, KO, or WT ESC. Data are shown as mean + SD of four independent experiments (n = 4).
(E) Tbx3, Wnt pathway genes, Eomes, and Jmjd3 mRNA levels upon differentiation of PML KD, KO, or WT ESC. Data represent the mean + SD of three independent experiments (n = 3).
(F) Endodermal (Afp, Gata4) and mesodermal (T, Wnt3a) gene expression levels in the course of differentiation of WT, PML KD, and TBX3 OE PML KD ESC. Error bars indicate +SD of four independent experiments (n = 4).
D, day.
A potential molecular mechanism for PML effects on differentiation might be based on the already mentioned suppression of Tbx3 by PML (Figure 4E). Tbx3 is regulated by the PI3K signaling pathway and was recently identified as a negative regulator of mesodermal commitment via targeting Brachyury (T) and Wnt pathway genes (Wnt8a, Fzd2, Wnt3a) (Waghray et al., 2015). Consistent with the aforementioned work, we found that T and Wnt effectors are overexpressed and induce mesoderm differentiation (Figures 4B and 4E) in PML KD EB. In contrast to the mesodermal lineage commitment, Tbx3 was previously reported to induce the endodermal differentiation via activation of Eomes in collaboration with JMJD3 (Kartikasari et al., 2013). In PML KD ESC, the mRNA of Eomes and Jmjd3 are decreased, in agreement with Tbx3 reduction and the aforementioned studies, leading to impaired endodermal differentiation (Figure 4E). Moreover, it was also found that TBX3 physically interacts with PML in proliferating cells, reinforcing our assumption that PML mediates its role via Tbx3 (Martin et al., 2012).
To address this question, we firstly confirmed the physical association of TBX3 and PML proteins by co-immunoprecipitation (not shown) and subsequently by generating stable PML KD ESC lines carrying pcDNA3-HA-TBX3 vector (not shown). During EB differentiation, TBX3 ectopic expression can partially restore the phenotype caused by PML deficiency. In particular, induction of mesodermal lineage specification is reduced while endoderm characteristic markers are elevated (Figure 4F). Taken together, the above results propose that TBX3 is positively regulated by PML and that repression of TBX3 partially accounts for the phenotype observed in PML ablation.
PML Is Required for Efficient Reprogramming of MEF into iPSC
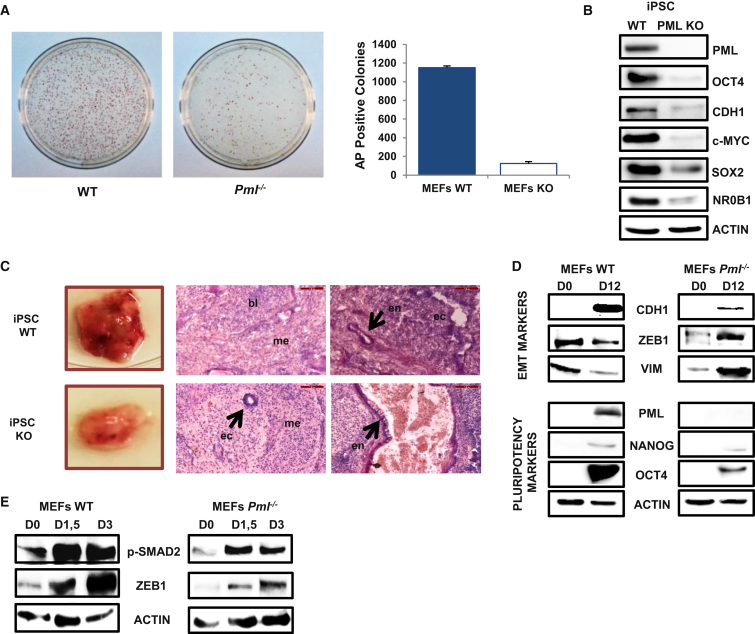
Since PML is required for ESC self-renewal and pluripotency, we investigated its contribution to the somatic cell reprogramming process. To address this issue, we isolated MEF from PML-deficient mice for iPSC generation. Pml−/− MEF and WT MEF were reprogrammed with Yamanaka factors (OSKM) and formed iPSC colonies (Takahashi and Yamanaka, 2006). PML deletion greatly decreases the efficiency of reprogramming in terms of total number of colonies (approximately 10-fold lower) and time of appearance (Figure 5A). Next, pluripotency and differentiation properties of the derived iPSC colonies were examined. Interestingly, representative undifferentiated stem cell markers are reduced in Pml−/− iPSC compared with the WT (Figure 5B) in agreement with PML KD and KO ESC. Unlike WT iPSC, Pml−/− iPSC express higher levels of developmental genes (Pax6, T) (Figure S5A). EB formation from PML KO iPSC revealed enhanced mesodermal differentiation at the expense of endodermal commitment (Figure S5B). This phenotype resembles the differentiation potential of PML KD ESC. Finally, we analyzed the pluripotency properties of PML KD iPSC with the in vivo assay of teratoma formation. WT and PML KD iPSC were injected intramuscularly in immunocompromised mice and tumors were isolated 20 days later. The germ-layer composition of the tumors was analyzed by H&E staining. Notably, PML KO iPSC form teratomas of smaller size and with more mature structures in comparison with the control (Figure 5C). This observation suggests that loss of PML impairs the pluripotency of the iPSC.
Figure 5.
PML Is Essential for Efficient Reprogramming of MEF to iPSC
(A) Alkaline phosphatase (AP) staining of iPSC colonies 28 days after OSKM lentiviral transduction. AP-positive colony numbers are shown on the right. Data are shown as mean + SD of three independent experiments (n = 3).
(B) Protein levels of pluripotency markers in WT and PML KO iPSC.
(C) Teratoma formation by intramuscular injection of WT and PML KO iPSC in immunocompromised mice. Photos show the differences between teratoma size. H&E staining analysis of the three germ layers in teratoma sections demonstrated by the respective arrows (bl, blastema; ec, ectoderm; en, endoderm; me, mesoderm). Scale bars, 100 μm.
(D) Protein expression levels of epithelial, mesenchymal (upper panel), and pluripotency markers (lower panel) at day (D) 12 of reprogramming process.
(E) Protein levels of p-SMAD2 and ZEB1 at days (D) 0, 1.5, and 3 of reprogramming process.
To specify the time window within which PML is required for the reprogramming process, we infected WT MEF with an OSKM lentiviral vector in combination with shRNA against Pml. In agreement with previous experiments, introduction of shPML reduced significantly the number of iPSC colonies compared with control (Figure S5C). PML KD efficiency was traced throughout the experiment and was sustained until day 8 (Figure S5C), supporting a defect in the early stages of reprogramming.
It is well established that reprogramming entails several molecular steps, including mesenchymal to epithelial transition (MET) in the first stage (Samavarchi-Tehrani et al., 2010). Although Pml−/− MEF express significantly lower levels of mesenchymal markers, their epithelial morphological switch at day 12 of reprogramming is defective, as shown by reduced CDH1 and increased ZEB1 and vimentin (VIM) (Figure 5D). On the contrary, WT MEF show a clear MET after OSKM transduction (Figure 5D). In addition, we analyzed the protein levels of OCT4 and NANOG at day 12 and we found that OSKM-transduced WT MEF show higher expression levels of pluripotency markers, in contrast to Pml−/− MEF (Figure 5D).
A potential mechanism for the interpretation of our data is supported by previous studies that highlight the importance of an epithelial-mesenchymal transition (EMT) that takes place in the early days of reprogramming (Liu et al., 2013, Unternaehrer et al., 2014). To investigate this hypothesis, we verified the temporary EMT in the early phase of reprogramming (days 0–3) shown by the upregulation of mesenchymal markers (ZEB1, VIM) and the presence of p-SMAD2/3. Interestingly, the EMT, as monitored by ZEB1 and p-SMAD2/3, is lower in Pml−/− MEF (Figure 5E). Thus, we concluded that PML positively contributes to the reprogramming of somatic cells by enhancing EMT at the very early stage.
Discussion
PSC offer great opportunities for human health, allowing the derivation of patient-specific tissues and providing disease models for drug screening. Thus, investigation of factors and mechanisms that regulate pluripotency constitutes a rapidly growing area of cutting-edge research. In addition to their value for regenerative medicine, pluripotency factors are promising targets to study cancer stem cells, since oncogenic transformation and cellular reprogramming share common properties (Goding et al., 2014, Hadjimichael et al., 2015).
In this report we addressed the functions of the pleiotropic regulator PML in embryonic and induced PSC. PML is highly expressed in undifferentiated ESC and its ablation induces significant changes in ESC morphology, global gene expression profile, and lineage specification decision. Specifically, PML physically interacts and regulates the expression of crucial mediators of pluripotency and contributes to the preservation of self-renewal. Furthermore, PML influences mESC cell-cycle profile, since its depletion leads to prolonged G1 phase, resembling that of differentiated cells. This effect can be attributed to the inhibition of the pro-proliferative functions of LIF/STAT3 and Myc-elicited mechanisms that we observed in lack of PML expression. In addition, GO analysis of our microarrays data highlighted PI3K pathway-related genes as being strongly downregulated in PML-deficient ESC. PI3K signaling is a well-documented mediator of proliferation, and its reduced activity in PML loss is in agreement with the effect on the ESC cell cycle.
In somatic cells PML inhibits cell proliferation through activation of pRB and p53, and this function highly contributes to its role as a tumor suppressor (Guan and Kao, 2015). These effects are mainly due to the regulation of post-translational modifications including phosphorylation and dephosphorylation by PML bodies. In mESC, pRB is constitutively inactivated by phosphorylation (Aksoy et al., 2007, Coronado et al., 2013, White and Dalton, 2005) and the p53 pathway is not active (Lee et al., 2012). Our data showed that PML is involved in maintaining pRB in an inactive state, and that PML loss impairs cell-cycle progression and favors advance to the primed-like state.
The effect of PML disruption on ESC transition from the naive to the EpiSC pluripotent state is also underpinned by transcription factor expression, specific signaling pathway deregulation, and metabolic alterations. Our transcriptomic analysis revealed a downregulation of genes encoding enzymes of FAO and OXPHOS. Thus, PML loss results in decreased respiration and mitochondrial function, another indication for exit from the naive state and progression to the EpiSC state. In addition, Pml−/− cells sustain propagation in F/A culture conditions and proliferate normally in the presence of Jaki, reinforcing the idea that they move on toward a primed-like stem cell state. Moreover, PML overexpression delays the exit from the naive state by either permitting long-term existence of a small fraction of undifferentiated naive ESC or allowing a fraction of EpiSC to regain the naive pluripotent state. These data propose that PML safeguards the naive pluripotent state.
PML is also implicated in the differentiation process. PML ablation stirred ESC toward mesodermal rather than endodermal lineage commitment through Tbx3 repression. Tbx3, well-known downstream target of the PI3K signaling pathway, is one of the most significantly repressed genes upon PML KD. Forced expression of TBX3 in PML KD ESC rescued the differentiation changes caused by PML loss. Physical interaction and reciprocal inhibition between TBX2/3 and PML was previously shown in human fibroblasts where the pro-senescence activity of PML counteracted cell proliferation driven by TBX (Martin et al., 2012). However, in ESC, TBX3 is positively regulated by PML and determines cell differentiation choices.
Finally, we demonstrate here that PML promotes iPSC generation by facilitating the early activation steps. Our proposed model for its contribution to reprogramming is that PML activates mesenchymal marker activation (EMT), which in turn enhances epithelial (MET) and pluripotent gene activation, resulting in an increase of reprogramming efficiency. In agreement with recent publications (Liu et al., 2013), we detect an initial EMT step in the reprogramming process that is compromised by PML loss. Additionally it is well established that PML is a crucial regulator of transforming growth factor β (TGF-β) signaling, since Pml−/− MEF showed a significant reduction of p-Smad2/3 and impaired expression of TGF-β target genes including mesenchymal-specific genes (Lin et al., 2004). Consequently, Pml−/− iPSC showed reduced pluripotency as manifested by the generation of smaller and more differentiated teratomas. This effect may be also attributed to telomere dysfunction that was reported in PML KD ESC (Chang et al., 2013). Functional telomeres are essential for pluripotency, and telomere shortening causes impaired teratoma formation (Huang et al., 2011).
Collectively, we present evidence that PML preserves mESC naive pluripotency and facilitates the reprogramming ability of mouse fibroblasts. This action may be related to the recently described pro-survival action of PML in breast cancer (Carracedo et al., 2012) and the involvement of PML in cancer stem cell maintenance in leukemia (Ito et al., 2008) and glioma (Zhou et al., 2015).
Our current results designate PML as a general regulator of cell fate and stemness that may have a broad impact on the exploration of PSC maintenance and differentiation, as well as on novel ways to target oncogenic processes.
Experimental Procedures
Cell Culture
The murine feeder-independent ESC line CGR8 was cultured in gelatin-coated flasks in DMEM (Gibco) supplemented with 100 U/mL LIF (ESGRO-Millipore), 2 mM L-glutamine (Gibco), 2 mM non-essential amino acids (Gibco), 0.1 mM β-mercaptoethanol (Gibco) and 15% heat-inactivated HyClone FBS (GE Healthcare Life Sciences). For EB formation, cells were trypsinized and diluted in Iscove's modified Dulbecco's medium (Gibco) supplemented with 2 mM L-glutamine (Gibco), 2 mM non-essential amino acids (Gibco), 0.1 mM β-mercaptoethanol (Gibco), and 20% heat-inactivated HyClone FBS (GE Healthcare Life Sciences), to a final concentration of 1,000 cells/20 μL. EBs were cultured without LIF as hanging drops for 2 days and subsequently collected and cultured in suspension for 6 additional days.
Generation of PML KD or OE ESC Stable Cell Lines
The generation of stably expressing PML shRNA ESC lines was achieved using PML shRNA-pLKO.1 lentiviral vector provided by Prof. Z.D. Levy of the Open University of Israel. In brief, CGR8 cells were grown to 80% confluence and infected with the lentiviruses produced either by the empty shRNA or PML shRNA vector, for 72 hr. Selection of shRNA infected cells was achieved with puromycin (2 μg/mL). A total of 60 isolated resistant colonies, evident after 2–4 weeks, were picked and further grown for screening. Puromycin-resistant clones were examined using western blot and qPCR. Respectively, PML OE ESC lines were generated using PML-IRES-GFP construct or IRES-GFP as a negative control. The isolation of the clones was performed using G418 (300 μg/mL).
Isolation of PML KO ESC
PML KO ESC were isolated from B57BL/6 Pml−/− mice. Female mice were euthanized at E3.5 stage and their uteri were immediately separated into 10 mL of DMEM on 100-mm plates. Uteri were then transferred into 2 mL of DMEM in a 35-mm dish and the blastocysts were flushed out using a 1-mL syringe. Blastocysts were collected under an inverted microscope using a 20-mL pipette and transferred to a 48-well plate containing a feeder layer of MEF mitotically inactivated 1 day earlier. After 2–3 days, blastocysts attached to the MEF feeder layer and hatched. ES medium was changed every second day while the expanded blastocysts adopted a morphology comparable with ESC (usually on day 5 or 6).
cDNA Microarray Analysis
Total RNA was extracted from PML KD and WT ESC cell lines at D0 and D4 of EB differentiation using an RNeasy Microarray Tissue Mini Kit (Qiagen). The RNA was then analyzed using Affymetrix GeneChip Mouse Gene ST 1.0 array, according the manufacturer's instructions. Microarray data were processed to extract the representative intensities from each probe set using Affymetrix Transcriptome Analysis Console software. Fold change >1.5 and p < 0.05 were used to identify differential expression between the sample groups. Prior to hierarchical clustering, log2 transformation was performed. Functional analysis was performed using RNEA (Chouvardas et al., 2016). The value p < 0.05 was employed for both significantly enriched GO terms and KEGG pathways.
iPSC Formation
Primary MEF were isolated either from C57BL/6 WT or Pml−/− mice at E13.5. For iPSC generation, MEF WT or PML KO (passage 1 or 2) were plated at 0.4–0.5 × 106 cells in a 100-mm plate and incubated overnight. The following day, cells were infected with the lentiviruses produced by TetO-FUW-OSKM (Addgene #20342) and FUW-M2rtTA vectors (Addgene #20342), for 48 hr. After 2 days, cells were reseeded on feeders and treated with 2 μg/mL doxycycline in mESC medium for induction of Yamanaka's factors. The medium was changed every other day. iPSC colonies appeared 9–11 days post infection. Alkaline phosphatase staining was performed using an Alkaline Phosphatase Detection Kit (Millipore) according to the manufacturer's instructions.
Teratoma Formation
PML KO or WT iPSC were harvested using 1× trypsin and centrifuged at 1,200 × g for 5 min. Cells were then resuspended in 1× PBS to a final concentration of 2 × 106 cells/100 μL and injected intramuscularly into 6- to 8-week-old male NOD-SCID mice (100 μL per mouse). The mice were monitored every day. Teratomas arose between 2 and 3 weeks after engraftment.
Ethical Approval for the Use of Animals
All experiments were conducted in accordance with the Laboratory Animal Care and Ethics Committee of IMBB. Animal work was approved by the IMBB Institutional Animal Care and Ethics Committee.
Statistical Analyses
Student's t test was used for all statistical analyses. Statistical significance was defined in the figures as follows: ∗p < 0.05, ∗∗p < 0.01. Values are presented as the mean ± SD.
Author Contributions
C.H. designed and performed experiments, data analysis, discussion, and writing; K.C., G.I.T., and T.M. contributed to the experimental work and discussion; A. Klonizakis and C.N. performed microarrays analysis; J.P. contributed to discussion and writing; A. Kretsovali supervised the study and contributed to writing.
Acknowledgments
We would like to thank G. Vretzos and D. Tsoukatou for technical assistance, as well as D. Vassou for her contribution in gene expression profiling (IMBB), E. Deligianni for her involvement in TMRE-stained figures analysis, T. Kosteas for help with the isolation of mouse blastocysts, and Prof. C. Spilianakis for critical suggestions. We also thank Prof. N. Tavernarakis for providing reagents for monitoring oxygen consumption and mitochondria function. This work was funded by Thalis-MIS380247-MIREG (NSRF 2007-2013), Umbistem 11SYN_10_668, Fondation Sante, “PROGRAMMATIC AGREEMENTS BETWEEN RESEARCH CENTRES – GSRT 2015-2017” (SIEMENS Biology Activity -Biophotonics) and IMBB internal funding.
Published: April 6, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.03.006.
Accession Numbers
The accession number of microarray data reported in this article is GEO: GSE93922.
Supplemental Information
References
- Acampora D., Di Giovannantonio L.G., Simeone A. Otx2 is an intrinsic determinant of the embryonic stem cell state and is required for transition to a stable epiblast stem cell condition. Development. 2013;140:43–55. doi: 10.1242/dev.085290. [DOI] [PubMed] [Google Scholar]
- Aksoy I., Sakabedoyan C., Bourillot P.Y., Malashicheva A.B., Mancip J., Knoblauch K., Afanassieff M., Savatier P. Self-renewal of murine embryonic stem cells is supported by the serine/threonine kinases Pim-1 and Pim-3. Stem Cells. 2007;25:2996–3004. doi: 10.1634/stemcells.2007-0066. [DOI] [PubMed] [Google Scholar]
- Carracedo A., Weiss D., Leliaert A.K., Bhasin M., de Boer V.C., Laurent G., Adams A.C., Sundvall M., Song S.J., Ito K. A metabolic prosurvival role for PML in breast cancer. J. Clin. Invest. 2012;122:3088–3100. doi: 10.1172/JCI62129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F.T., McGhie J.D., Chan F.L., Tang M.C., Anderson M.A., Mann J.R., Andy Choo K.H., Wong L.H. PML bodies provide an important platform for the maintenance of telomeric chromatin integrity in embryonic stem cells. Nucleic Acids Res. 2013;41:4447–4458. doi: 10.1093/nar/gkt114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenoweth J.G., McKay R.D., Tesar P.J. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev. Growth Differ. 2010;52:293–301. doi: 10.1111/j.1440-169X.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- Chouvardas P., Kollias G., Nikolaou C. Inferring active regulatory networks from gene expression data using a combination of prior knowledge and enrichment analysis. BMC Bioinformatics. 2016;17:181. doi: 10.1186/s12859-016-1040-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang Y.S., Huang W.H., Park S.W., Persaud S.D., Hung C.H., Ho P.C., Wei L.N. Promyelocytic leukemia protein in retinoic acid-induced chromatin remodeling of Oct4 gene promoter. Stem Cells. 2011;29:660–669. doi: 10.1002/stem.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado D., Godet M., Bourillot P.Y., Tapponnier Y., Bernat A., Petit M., Afanassieff M., Markossian S., Malashicheva A., Iacone R. A short G1 phase is an intrinsic determinant of naive embryonic stem cell pluripotency. Stem Cell Res. 2013;10:118–131. doi: 10.1016/j.scr.2012.10.004. [DOI] [PubMed] [Google Scholar]
- Goding C.R., Pei D., Lu X. Cancer: pathological nuclear reprogramming? Nat. Rev. Cancer. 2014;14:568–573. doi: 10.1038/nrc3781. [DOI] [PubMed] [Google Scholar]
- Guan D., Kao H.Y. The function, regulation and therapeutic implications of the tumor suppressor protein, PML. Cell Biosci. 2015;5:60. doi: 10.1186/s13578-015-0051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjimichael C., Chanoumidou K., Papadopoulou N., Arampatzi P., Papamatheakis J., Kretsovali A. Common stemness regulators of embryonic and cancer stem cells. World J. Stem Cells. 2015;7:1150–1184. doi: 10.4252/wjsc.v7.i9.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Wang F., Okuka M., Liu N., Ji G., Ye X., Zuo B., Li M., Liang P., Ge W.W. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011;21:779–792. doi: 10.1038/cr.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Bernardi R., Morotti A., Matsuoka S., Saglio G., Ikeda Y., Rosenblatt J., Avigan D.E., Teruya-Feldstein J., Pandolfi P.P. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453:1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K., Carracedo A., Weiss D., Arai F., Ala U., Avigan D.E., Schafer Z.T., Evans R.M., Suda T., Lee C.H. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat. Med. 2012;18:1350–1358. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantzali E., Schulz H., Hummel O., Hubner N., Hatzopoulos A., Kretsovali A. Histone deacetylase inhibition accelerates the early events of stem cell differentiation: transcriptomic and epigenetic analysis. Genome Biol. 2008;9:R65. doi: 10.1186/gb-2008-9-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartikasari A.E., Zhou J.X., Kanji M.S., Chan D.N., Sinha A., Grapin-Botton A., Magnuson M.A., Lowry W.E., Bhushan A. The histone demethylase Jmjd3 sequentially associates with the transcription factors Tbx3 and Eomes to drive endoderm differentiation. EMBO J. 2013;32:1393–1408. doi: 10.1038/emboj.2013.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.F., Su J., Ang Y.S., Carvajal-Vergara X., Mulero-Navarro S., Pereira C.F., Gingold J., Wang H.L., Zhao R., Sevilla A. Regulation of embryonic and induced pluripotency by aurora kinase-p53 signaling. Cell Stem Cell. 2012;11:179–194. doi: 10.1016/j.stem.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S.Y., Qin J., Wong J., Cooney A.J., Liu D. Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 2008;10:731–739. doi: 10.1038/ncb1736. [DOI] [PubMed] [Google Scholar]
- Lin H.K., Bergmann S., Pandolfi P.P. Cytoplasmic PML function in TGF-beta signalling. Nature. 2004;431:205–211. doi: 10.1038/nature02783. [DOI] [PubMed] [Google Scholar]
- Liu X., Sun H., Qi J., Wang L., He S., Liu J., Feng C., Chen C., Li W., Guo Y. Sequential introduction of reprogramming factors reveals a time-sensitive requirement for individual factors and a sequential EMT-MET mechanism for optimal reprogramming. Nat. Cell Biol. 2013;15:829–838. doi: 10.1038/ncb2765. [DOI] [PubMed] [Google Scholar]
- Martin N., Benhamed M., Nacerddine K., Demarque M.D., van Lohuizen M., Dejean A., Bischof O. Physical and functional interaction between PML and TBX2 in the establishment of cellular senescence. EMBO J. 2012;31:95–109. doi: 10.1038/emboj.2011.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama H., Masaki H., Sato H., Hayama T., Yamaguchi T., Nakauchi H. Successful reprogramming of epiblast stem cells by blocking nuclear localization of beta-catenin. Stem Cell Rep. 2015;4:103–113. doi: 10.1016/j.stemcr.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara F., Weiss C.N., Ito K. The role of PML in hematopoietic and leukemic stem cell maintenance. Int. J. Hematol. 2014;100:18–26. doi: 10.1007/s12185-014-1518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T., Bellodi C., Nicotera P., Salomoni P. The tumor suppressor Pml regulates cell fate in the developing neocortex. Nat. Neurosci. 2009;12:132–140. doi: 10.1038/nn.2251. [DOI] [PubMed] [Google Scholar]
- Sage J. The retinoblastoma tumor suppressor and stem cell biology. Genes Dev. 2012;26:1409–1420. doi: 10.1101/gad.193730.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., Sung H.K., Beyer T.A., Datti A., Woltjen K., Nagy A., Wrana J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Savatier P., Malashicheva A. Cell-cycle control in embryonic stem cells. In: Lanza R., Gearhart J., Hogan B., Melton D., Pedersen R., Thomson J., West M., editors. vol. 1. Elsevier Academic; 2004. pp. 53–62. (Handbook of Stem Cells). [Google Scholar]
- Singh A.M., Sun Y., Li L., Zhang W., Wu T., Zhao S., Qin Z., Dalton S. Cell-cycle control of bivalent epigenetic domains regulates the exit from pluripotency. Stem Cell Rep. 2015;5:323–336. doi: 10.1016/j.stemcr.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tang M.K., Liang Y.J., Chan J.Y., Wong S.W., Chen E., Yao Y., Gan J., Xiao L., Leung H.C., Kung H.F. Promyelocytic leukemia (PML) protein plays important roles in regulating cell adhesion, morphology, proliferation and migration. PLoS One. 2013;8:e59477. doi: 10.1371/journal.pone.0059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unternaehrer J.J., Zhao R., Kim K., Cesana M., Powers J.T., Ratanasirintrawoot S., Onder T., Shibue T., Weinberg R.A., Daley G.Q. The epithelial-mesenchymal transition factor SNAIL paradoxically enhances reprogramming. Stem Cell Rep. 2014;3:691–698. doi: 10.1016/j.stemcr.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghray A., Saiz N., Jayaprakash A.D., Freire A.G., Papatsenko D., Pereira C.F., Lee D.F., Brosh R., Chang B., Darr H. Tbx3 controls Dppa3 levels and exit from pluripotency toward Mesoderm. Stem Cell Rep. 2015;5:97–110. doi: 10.1016/j.stemcr.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J., Dalton S. Cell cycle control of embryonic stem cells. Stem Cell Rev. 2005;1:131–138. doi: 10.1385/SCR:1:2:131. [DOI] [PubMed] [Google Scholar]
- Zhang J., Liu G., Ruan Y., Wang J., Zhao K., Wan Y., Liu B., Zheng H., Peng T., Wu W. Dax1 and Nanog act in parallel to stabilize mouse embryonic stem cells and induced pluripotency. Nat. Commun. 2014;5:5042. doi: 10.1038/ncomms6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Choi M., Margineantu D., Margaretha L., Hesson J., Cavanaugh C., Blau C.A., Horwitz M.S., Hockenbery D., Ware C. HIF1alpha induced switch from bivalent to exclusively glycolytic metabolism during ESC-to-EpiSC/hESC transition. EMBO J. 2012;31:2103–2116. doi: 10.1038/emboj.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Cheng L., Shi Y., Ke S.Q., Huang Z., Fang X., Chu C.W., Xie Q., Bian X.W., Rich J.N. Arsenic trioxide disrupts glioma stem cells via promoting PML degradation to inhibit tumor growth. Oncotarget. 2015;6:37300–37315. doi: 10.18632/oncotarget.5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.